Abstract
Introduction
Aerobic training has a beneficial effect on enhancing liver functions. Autophagy might potentially play a role in preventing excessive lipid accumulation, regulating oxidative stress, and inflammation in the liver.
Objective
To investigate the potential linking role of autophagy-related gene expressions and protein levels with histopathology changes in Wistar rat livers after treadmill training under different intensities.
Methods
20 rats were divided into 4 groups (control, low intensity, moderate intensity, and high intensity). 8 weeks of treadmill training was conducted with a frequency of 5 days per week, for a duration of 30 min per day. Liver histopathology was studied using hematoxylin-eosin, and oil red O staining. RNA and protein from the liver tissues were extracted to examine the autophagy-related gene (LC3, p62) and protein levels (Beclin, ATG5, LC3, p62). The gene expressions of CPT1a, CD36, FATP 2,3,5, GLUT2, and FGF21 were also studied.
Results
Different intensities of training might potentially modulate autophagy-related gene expressions in rat livers. LC3 and p62 mRNA expressions in moderate and high intensities decreased compared to control. Beclin, ATG5, and LC3 protein level increased compared to control, while p62 protein level decreased compared to control. Whereas for the other genes, we found an increase in CPT1a, but we did not observed any changes in the expression of the other genes. Interestingly, autophagy-related gene expressions might be correlated with the changes of sinusoidal dilatation, cloudy swelling, inflammation, and lipid droplets of the liver tissues.
Conclusion
Moderate and high intensities of training induce autophagy activity, combined with a shift in metabolic zonation in liver that might be potentially correlated with lipophagy. Our results showed the potential interplay role between autophagy and liver histopathology appearances as a part of the adaptation process to training.
Keywords: Biological sciences, Exercise, Animal physiology, Health sciences, Physiology, Treadmill training, Lipid droplets, Autophagy, Lipophagy, Histopathology
Biological sciences; Exercise; Animal physiology; Health sciences; Physiology; Treadmill training; Lipid droplets; Autophagy; Lipophagy; Histopathology
1. Introduction
The liver is the primary site for metabolism of macronutrients, which makes it susceptible to fatty liver, injury, and inflammation [1]. Lipids from dietary intake are emulsified and hydrolyzed in the intestinal tract by the bile acids. Enterocytes absorb the hydrolyzed lipids and pack them into nascent chylomicrons. They enter the bloodstream, where they receive apoC-II in order to activate lipoprotein lipase for digesting chylomicron triacylglycerols into fatty acids and glycerol [2]. Fatty acids are partially stored in adipose tissues, where lipolysis may occur to produce free fatty acids in certain conditions through the control of insulin and molecular signals. Free fatty acids are metabolized through three major pathways: (1) esterification of free fatty acids into triglycerides, as well as sequestration into LD (lipid droplets); (2) recycled into very low density lipoprotein (VLDL) [3]; and β-oxidation in hepatocyte mitochondria [4, 5]. These free fatty acids are transported to hepatocyte via fatty acid transporter proteins (FATP), with FATP2 and FATP5 acting as the major facilitators for the transport [6]. The CD36/fatty acid translocase is also known as a facilitator for the fatty acid uptake [7, 8].
Under normal conditions, β-oxidation of the short-, medium-, and long-chain fatty acids occurs in the mitochondria. In the cytosol, the long-chain fatty acids are activated and transformed into acetyl-coA, which is then shuttled across the membrane via CPT-1 (carnitine palmitoyltransferase-1) [4]. Regulation of the fatty acids also involves FGF21, especially when an individual is fasting or on a high-fat or ketogenic diet [9]. Under these conditions, the insulin resistance and fatty acids increase. The long-chain and very long-chain fatty acids are oxidized into peroxisomes and endoplasmic reticulum. Acetyl-coA is then further processed via the TCA (tricarboxylic acid cycle) into ketone bodies [10]. As an important product involved in the TCA cycle, the acetyl-coA is linking the lipid and glucose metabolism in the liver [4].
Glucose in the bloodstream is taken up via GLUT2 (glucose transporter 2) into hepatocyte, via the portal vein. GLUT2 is a membrane glucose transporter and insulin independent, with a high capacity and low affinity to glucose [11]. Phosphorylation of glucose into glucose 6 phosphate is induced by the liver GCK (glucokinase) [12]. Depending on the metabolic state, glucose 6 phosphate will be degraded during glycolysis to form 2 ATP and 2 NADH molecules per glucose molecule or be stored in the form of glycogen. Pyruvate is the glycolysis product which will further decarboxylate into acetyl-coA, which enters the TCA cycle or to be used as a substrate for de novo lipogenesis (DNL) [4].
Autophagy has been reported as an important regulator of lipid metabolism and storage, including regulation of LD degradation. Singh et al. revealed the importance of autophagy in the LD degradation [13, 14, 15]. LDs are deposits of the lipid ester intracellular, surrounded by phospholipid monolayers and separated from the hydrophilic cytosolic environment by perilipins. If the liver fails to control lipid accumulation in the hepatocytes, pathogenic conditions such as fatty liver can occur. Autophagy is involved in the mechanism that controls the growth of LDs in the liver [16]. Pi et al. proved that chronic training has an important role in regulating lipid homeostasis via autophagy [17].
It has been well known that chronic aerobic training is an effective method to improve health through various cellular and molecular pathways, including molecular pathways which are involved in lipid and glucose metabolisms [18, 19]. The benefit of this training is correlated with the intensity of the training itself. Lipid is preferred as the main fuel for lower intensity, while at higher intensity, the glucose becomes more prominent, according to the crossover concept [20]. Skeletal muscles use circulatory substrates in low intensity. But, in higher intensity, the source of the substrate shifts to triglycerides and glycogen in the muscles [21].
Taken together, different intensities of chronic treadmill training might be associated with histopathology changes and involved in regulating autophagy processes, specifically for lipid metabolism (hepatic lipophagy). This process is very important for preventing excessive accumulation of lipids. However, the molecular mechanism behind the effects of different intensities of chronic treadmill training on hepatic autophagy remains unclear. In this study, we postulate that different intensities of training induce differences in histopathology, which might be correlated with autophagy in the liver.
2. Material and methods
2.1. Animals
Male wistar rats (n = 20) at 8 weeks of age were ordered from PT. Biofarma, Bandung, Indonesia, and divided into four groups. The rats were group-housed at room temperature around ±22–24 °C, under a 12:12 h light-dark cycle, and allowed access to food and water ad libitum. The rats were fed with the standard chow diet that provides 47.3% carbohydrate, 20% protein, 4% fat, 4% fiber, 12% water, 12% calcium, and 0.7% phosphorus. This standard chow diet was given to maximize the effect of training-induced liver metabolism, and to minimize the confounding factors that occur due to dietary fat and carbohydrate compositions. All procedures were conducted according to the stipulated guidelines for the use and care of laboratory animals [22], and were approved by the Research Ethics Committee of the Faculty of Medicine, Universitas Kristen Maranatha-Rumah Sakit Immanuel Bandung No 093/KEP/V/2019.
2.2. Treadmill training protocol
All rats were habituated to the environment for two weeks, and continued with habituation on the treadmill for two weeks using standardized treadmill. Following adopted protocol from Lesmana et al. 2016, we used treadmill speed of 20 m/min for 30 min as baseline since it stimulated lactate threshold and considered as moderate intensity. Then based on lactate threshold, we defined the training protocol: speed at 10 m/min for 30 min as low intensity (sub-lactate); and speed at 30 m/min for 30 min as high intensity (supra-lactate) [23]. To test the training-induced adaptation at different intensities (n = 5), the rats ran on the treadmill for 8 weeks, 5 days/week, 30 min/day. All training sessions were performed during the light cycle between 8 and 12 AM. Non-training rats served as the sedentary control (n = 5). At the end of the study, the rats were anesthetized with 5% inhaled isoflurane until one minute after the breathing stopped. The serum was isolated from the heart's blood flow, whereas the livers were collected, weighed, and either: 1) snap frozen in liquid nitrogen and stored at -80 °C for further use, or 2) fixed in formalin for hematoxylin eosin and oil red O staining. Harvest time was done immediately after the last training sessions in each group.
2.3. Biochemistry assay
Serum AST/aspartate aminotransferase, ALT/alanine aminotransferase, CHOL/cholesterol, TG/triglyceride, and HDL/high density lipoproteins were measured via spectrophotometry using a commercially available kit (Randox, United Kingdom), according to the manufacturer's instructions. Samples were exposed to the mixture from the kit, and were incubated at 37 °C for 5 min. The absorbance was measured at 340 nm for AST and ALT, and 500 nm for CHOL, TG, and HDL using a microplate reader (M200 Pro, Tecan, Morrisville, NC).
2.4. Histopathological analysis
2.4.1. Hematoxylin and eosin (H&E staining)
Parts of the liver were excised, and fixed in a buffered paraformaldehyde solution (4%). After being dehydrated in a grade alcohol series and embedded in paraffin wax, 2-μm thick sections were prepared and stained with H&E for histopathology visualization using a LEICA ICC50 HD microscope at 400x magnification. An expert pathologist conducted a blind histopathological analysis of the liver samples. The samples were evaluated for the level of congestion/sinusoidal dilatation, cloudy swelling/injury, and inflammation. The level of congestion or sinusoidal dilatation was graded as follows: 0 = No congestion/sinusoidal dilatation, 1 = Mild congestion or centrilobular (zone III) sinusoidal dilatation, 2 = Moderate congestion or centrilobular (zone II) sinusoidal dilatation, 3 = Severe congestion or centrilobular (zone I) sinusoidal dilatation. The level of cloudy swelling/hepatic injury was graded as follows: 0 = No cloudy swelling/hepatic injury, 1 = Mild cloudy swelling/hepatic injury (zone III), 2 = Moderate cloudy swelling/hepatic injury (zone II), 3 = Severe cloudy swelling/hepatic injury (zone I). The severity of the inflammation was graded as follows: 0 = No hepatic inflammation, 1 = Mild hepatic inflammation or periportal inflammation, 2 = Moderate hepatic inflammation or periportal and intraparenchym inflammation, 3 = Severe hepatic inflammation or periportal and intraparenchym inflammation with bridging necrosis. These characteristics were adapted from another study with some modification applied [24]. Geometric representation of a hepatic lobule (Zone I, II, III) and a schematic representation of a sinusoid, as well as the corresponding zonation associated with lipid metabolism in the liver is shown in the Supplemental Figure 3 [1].
2.4.2. Oil red O staining
0.5 g oil red O (OA, Sigma-Aldrich) was dissolved in 100 ml isopropanol to make the oil red O stock stain. 30 ml of the stock stain was diluted with 20 ml of distilled water to make the oil red O working solution. The liver tissues were cut at 2 mm, fixed in formalin, paraffinized and hydrated. After deparaffinized and rehydrated the slides with xylene and ethanol, we stained them with freshly prepared oil red O working solutions for 15 min. They were then rinsed with 60% isopropanol, and then lightly stained with hematoxylin for 4 min. After rinsing with distilled water, Entellan (Merck, Germany) was applied to mount the slides. The lipid droplets (LDs) were stained in red and were assessed using a LEICA ICC50 HD microscope. The number of lipid droplets per field was obtained from the average number of lipid droplets per 10 fields of each Wistar rat liver slide. The number of lipid droplets calculation was performed using the multipoint function of ImageJ.
2.5. RNA extractions and semi-quantitative PCR
The total RNA was extracted from frozen liver tissues with the use TRIsure reagent (Bioline, United Kingdom) according to the manufacturer's instructions. The concentration and quality of the RNA extractions were analyzed by measurements at 268/280 nm absorbance spectrophotometry (M200 Pro, Tecan, Morrisville, NC). The One Step RT PCR Kit (Bioline, United Kingdom) was used to conduct semiquantitative PCR. The GAPDH was used as a housekeeping gene. Electrophoresis Gels were visualized using BluePad Detection systems and Image J was used for visualization and quantification of the PCR band. Table 1 provides a list of primer sequences used in this study.
Table 1.
Primers used for Semi quantitative-PCR analysis.
| Gene Symbol | Primer Sequence (5′ to 3′) Upper strand: sense Lower strand: antisense |
Product Size (bp) |
Annealing (oC) | Cycle | References |
|---|---|---|---|---|---|
| LC3 | GGTCCAGTTGTGCCTTTATTGA | 153 | 59,5 | 35 | [43] |
| GTGTGTGGGTTGTGTACGTCG | |||||
| p62 | CTAGGCATCGAGGTTGACATT | 116 | 56 | 35 | [44] |
| CTTGGCTGAGTACCACTCTTATC | |||||
| CPT1a | GTCCCAGCTGTCAAAGATACCG | 245 | 58 | 35 | [45] |
| ATGGCGTAGTAGTTGCTGTTAACC | |||||
| CD36 | CCTCGGATGGCTAGCTGATT | 189 | 57.5 | 35 | [46] |
| AGAGCACTTGCTTCTTGCCA | |||||
| FATP2 | AGTACATCGGTGAACTGCTTCGGT | 174 | 61 | 35 | [47] |
| TGCCTTCAGTGGAAGCGTAGAACT | |||||
| FATP3 | CTGGGACGAGCTAGAGGAAG | 104 | 58 | 35 | [48] |
| GCTGAGGCCAGAGGTCTAAC | |||||
| FATP5 | TTCAGGGACCACTGGACTTCCAAA | 105 | 61 | 35 | [49] |
| ACCACATCATCAGCTGTTCTCCCA | |||||
| GLUT2 | GGCTAATTTCAGGACTGGTT | 278 | 55 | 35 | [50] |
| TTTCTTTGCCCTGACTTCCT | |||||
| FGF21 | CACACCGCAGTCCAGAAAG | 77 | 58 | 35 | [51] |
| GGCTTTGACACCCAGGATT | |||||
| GAPDH | GTTACCAGGGCTGCCTTCTC | 177 | 61 | 35 | [52] |
| GATGGTGATGGGTTTCCCGT |
2.6. Western blotting
We dissected liver tissues from the rats, weighed the tissue, and lysed them in lysis buffers mixed with protease inhibitors. The protein samples were centrifuged, and then combined with sample buffers (containing beta-mercaptoetanol) followed by heat denaturation processes (95 °C) for five minutes. Equal amounts of samples underwent electrophoresis in a SDS-PAGE Gel for 60 min at 120 V, and then transferred to a nitrocellulose membrane for one hour at 200 miliAmpere at room temperature. These samples were then blocked overnight using a 1% blocking reagent (Skim Milk) in a Tris-buffered with 0.1% Tween 20. Immunoblotting was performed using a rabbit monoclonal LC3 (#12741) and rabbit polyclonal p62 (#5114) which was purchased from Cell Signaling; mouse monoclonal Beclin (MAB5295-SP) and ATG5 (MAB5294-SP) from R & D Systems; as well as a mouse monoclonal Glyceraldehyde-3-Phosphate Dehydrogenase/GAPDH (AM4300) from ThermoFisher Scientific, with dilution ratio of 1:1000. The secondary antibody was used accordingly with constant dilution ratio of 1:15000. The anti-mouse (31432) and anti-rabbit (31460) antibodies were purchased from ThermoFisher Scientific. The protein expressions were detected using a chemiluminescence reagent (GE Healthcare, USA), imaged (LI-COR C-DiGit Chemiluminescence; and LI-COR LICOR ODYSSEY®+ CLx Infrared Imaging System, USA). The band intensities were determined using the LiCOR Quantification Software. Every blot was stripped using the stripping buffer from Thermo Scientific following the recommended procedures, and re-probed using an internal control primary antibody, GAPDH, as an internal control to monitor the integrity of the protein [25].
2.7. Statistics
The data was expressed as the mean ± standard of error mean (SEM) and analyzed using the SPSS 20.0 statistical software (SPSS Inc., USA). The comparisons between groups were analyzed using the One Way ANOVA or Kruskal Wallis, followed by post hoc analysis LSD, or the Mann Whitney test (for data without normal distributions), with a p < 0.05 in order to be considered statistically significant.
3. Results
3.1. Effects of training on percentage increase in body weight, liver weight, and liver weight/body weight ratio
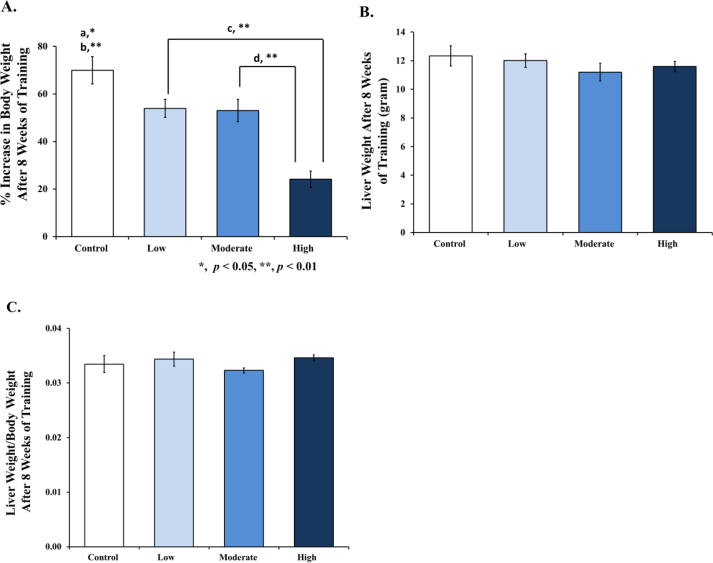
All groups have a similar body weight at the beginning of the research study (200 ± 50 g). After termination, the body and liver weights were recorded, and the percentage increase in the body weight and ratio of the liver weight/body weight was calculated. At the end of the research, it was seen across all training groups (low, moderate, and high) that a significant decrease in body weight (53.95% ± 3.80; 53.05% ± 4.68; 24.11% ± 3.47) was found compared to the control (69.97% ± 5.74), as shown in Figure 1A. The liver weight (Figure 1B) and the liver weight/body weight ratio (Figure 1C) showed no difference compared to the control, respectively.
Figure 1.
Evaluation of the percentage increase in body weight, liver weight, and liver weight/body weight ratio in the rats after 8 weeks' training with different intensities. [A] % Increase in body weight was significantly lower in the low and moderate intensities compared to control (a), high intensity compared to control (b), between low intensity and high intensity, (c) and between moderate intensity and high intensity (d). [B] Liver weight after 8 weeks of treadmill training showed no significant differences between all training groups compared to control. [C] Liver weight/body weight ratio after 8 weeks of treadmill training also showed no significant differences between all training groups, compared to control. Data was presented as an average mean ± standard error of mean (SEM) with p < 0.05 being considered as significant (∗) and p < 0.01 considered as very significant (∗∗).
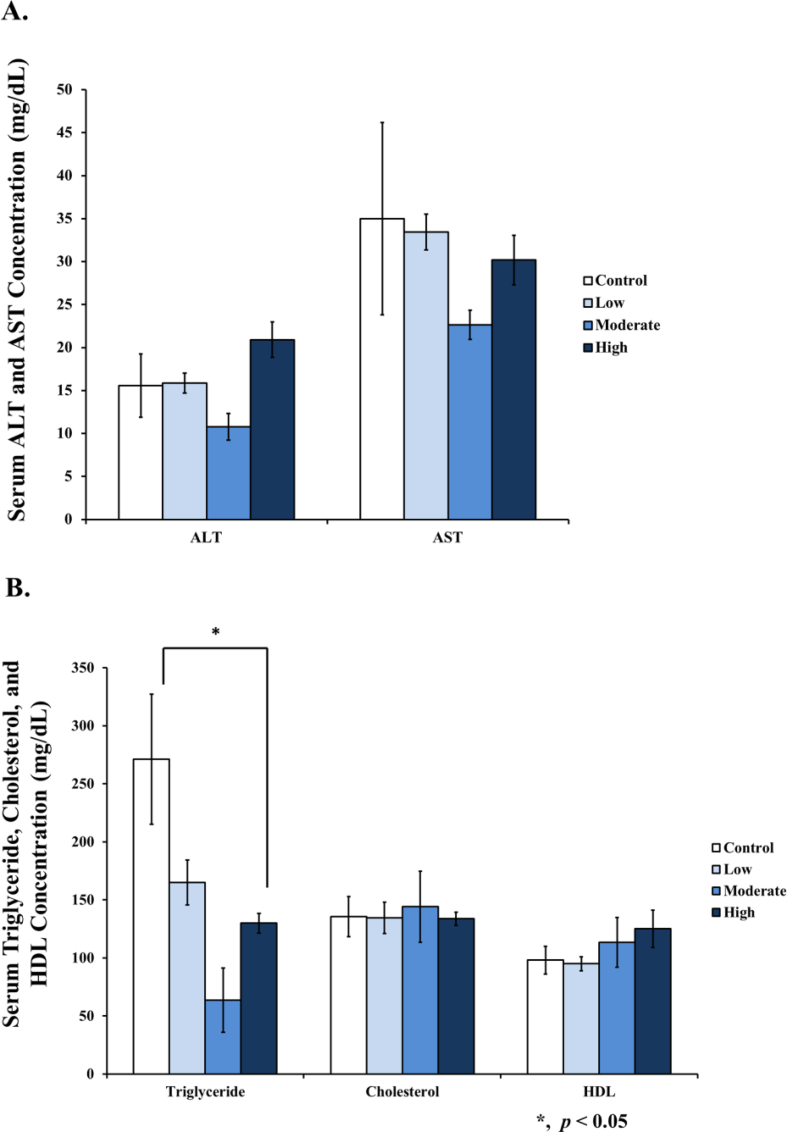
3.2. Training decreased triglyceride serums, but No change in cholesterol, HDL, AST, and ALT
We found that triglycerides decreased in moderate and high intensities of training compared to the control, but no differences were noticed among the total cholesterol, HDL, AST, and ALT samples in the serums of the Wistar rats (Figure 2).
Figure 2.
Levels of serum AST, ALT, triglyceride, cholesterol, and HDL after 8 weeks of treadmill training with different intensities. [A] No change of serum AST and ALT levels in all groups. [B] The high intensity group significantly decreased in terms of the serum triglyceride, but no change in the serum cholesterol and HDL was seen. No change of serum triglyceride in the low intensity, and no significant change of the cholesterol and HDL levels across all training groups was found, compared to the control. Data was presented as an average mean ± standard error of mean (SEM), with p < 0.05 being considered as significant (∗).
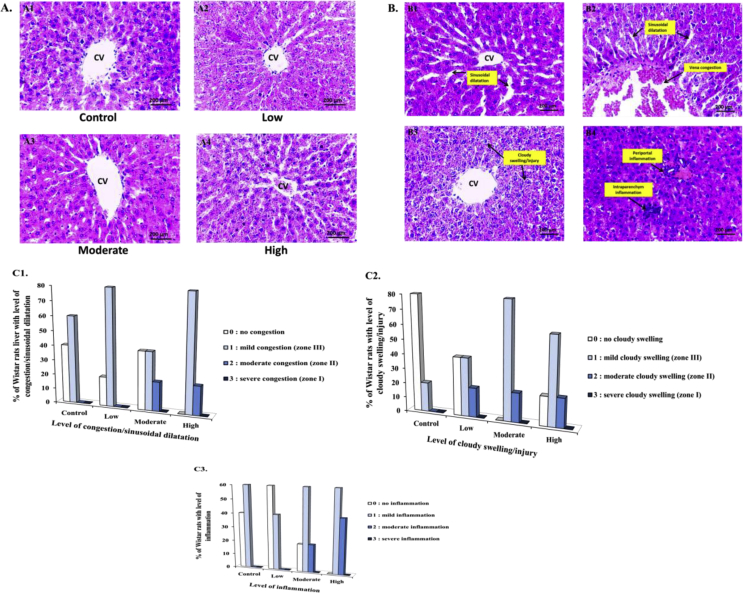
3.3. Effects of training on liver histopathology
Liver histopathology appearances in all groups are shown in Figure 3A, and the characteristics of the congestion/sinusoidal dilatation, cloudy swelling/injuries, and inflammation in all groups are presented in Figure 3B.
Figure 3.
Photomicrographs of the liver section in the control and training groups after 8 weeks of treadmill training with different intensities. [A1-4] Representative photomicrographs of the general appearances from the liver parenchyma after H&E staining (400x) in the control (A1), low intensity (A2), moderate intensity (A3), and high intensity samples (A4). [B1-4] Representative photomicrographs after H&E staining (400x) showing sinusoidal dilatation (B1), vena congestion and sinusoidal dilatation (B2), cloudy swelling/injury (B3), and periportal and intraparenchym inflammation (B4). [C1-3] Percentage of Wistar rat liver with level of congestion/sinusoidal dilatation [C1], cloudy swelling/injury [C2], and inflammation [C3].
In Figure 3C, percentage of Wistar rat liver with the histopathology scoring of congestion/sinusoidal dilatation (Figure C1), cloudy swelling/injury (Figure C2), and inflammation (Figure C3) after chronic training with different intensities is depicted. As for the level of congestion/sinusoidal dilatation, we found that 40% had no congestion, and 60% of the control samples had mild congestions. For the low intensity samples, 20% had no congestion, and 80% had mild congestion. For the moderate intensity group, 40% had no congestion, 40% had mild congestion, and 20% had moderate congestion. Finally, for the high intensity group, 80% had mild congestion, and 20% had moderate congestion. As for cloudy swelling/injury, we found that 80% had no cloudy swelling, and 20% had mild cloudy swelling in the control. On the other hand, in the low intensity group, 40% had no cloudy swelling, 40% had mild cloudy swelling, and 20% had moderate cloudy swelling. In the moderate intensity group, 80% had mild cloudy swelling, and 20% had moderate cloudy swelling. In the high intensity group, 20% had no cloudy swelling, 60% had mild cloudy swelling, and 20% had moderate cloudy swelling. As for the inflammation, we found that 40% had no inflammation, and 60% had mild inflammation in the control. In the low intensity group, 60% had no inflammation, and 40% had mild inflammation. In the moderate intensity group, 20% had no inflammation, 60% had mild inflammation, and 20% had moderate inflammation. Finally, in the high intensity group, 60% had mild inflammation, and 40% had moderate inflammation.
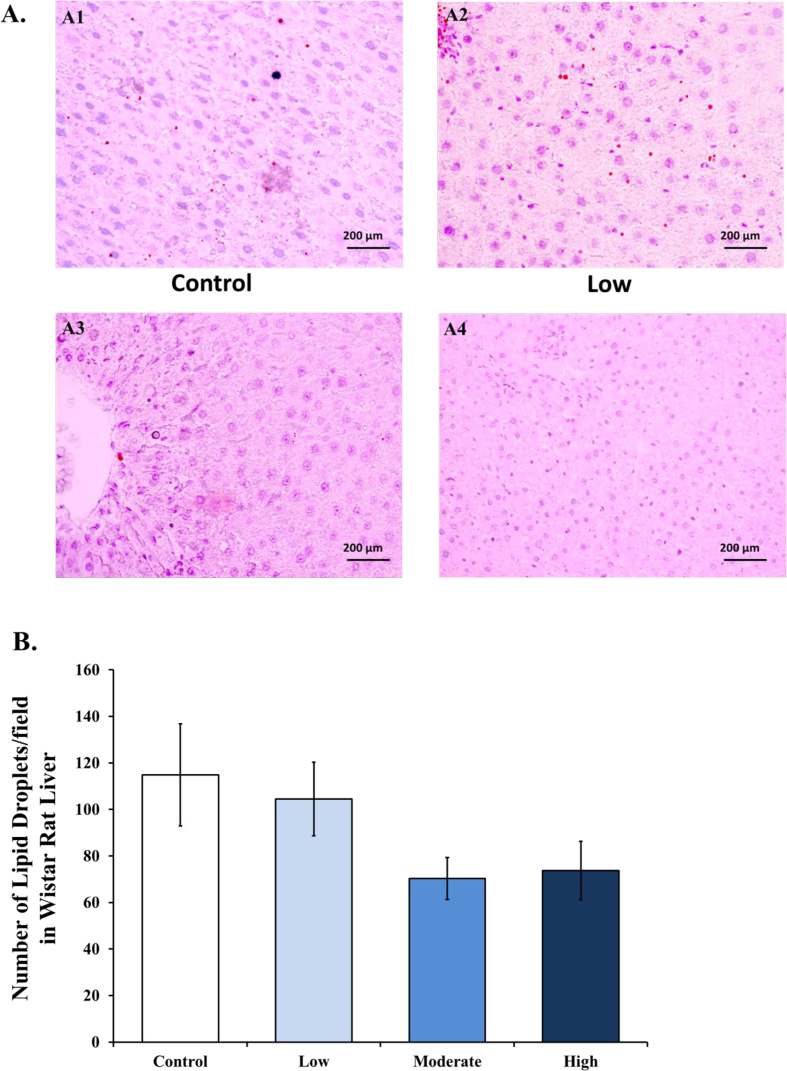
Chronic training with moderate and high intensities decreased lipid droplets compared to control and low intensities groups, as depicted by oil red O staining of the liver section (Figure 4A). Number of lipid droplets were calculated in each slide of liver section presented as graph in Figure 4B.
Figure 4.
Lipid droplets in Wistar rat liver decreases in moderate and high intensities groups compared to control after 8 weeks of treadmill training. [A1-4] Representative photomicrographs from the liver parenchyma after oil red O staining (400x) in the control (A1), low intensity (A2), moderate intensity (A3), and high intensity samples (A4). [B] The average number of lipid droplets calculated from each slide of Wistar rat liver after oil red O staining (400x) in the control, low intensity, moderate intensity, and high intensity groups.
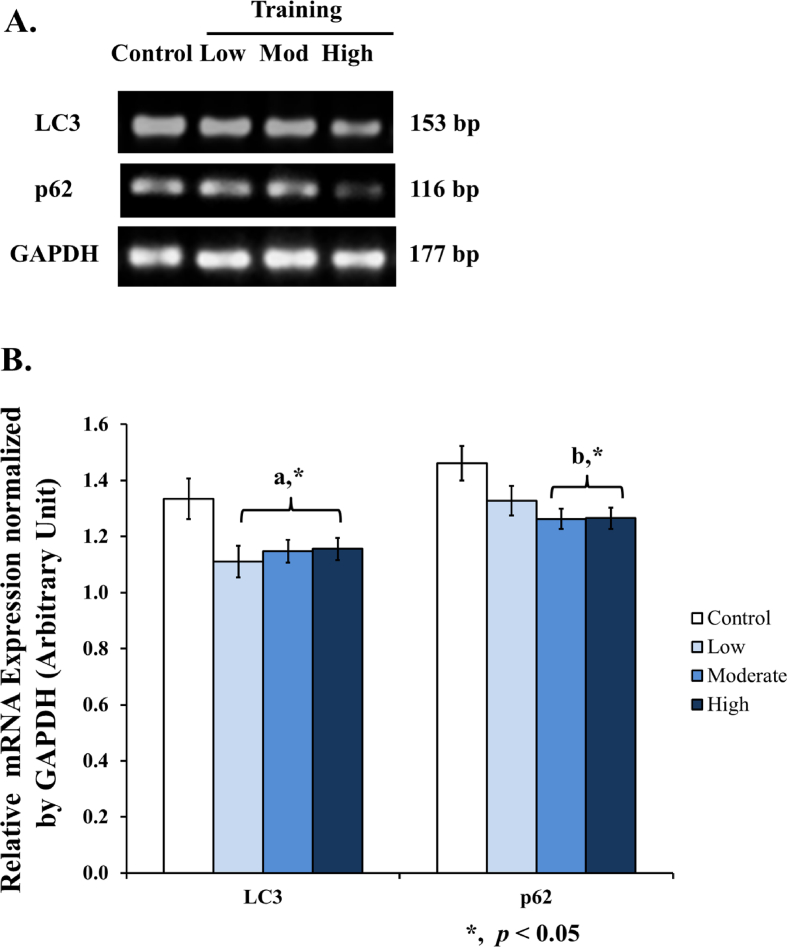
3.4. LC3 and p62 mRNA expressions in wistar rat liver
In this study, we found a significant decrease of LC3 mRNA expression (Figures 5A and 5B) in low (0.83-fold), moderate (0.86-fold), and high intensities (0.87-fold). There was also a decrease of p62 mRNA expression (Figures 5A and 5B) in moderate (0.86-fold) and high intensities (0.87-fold).
Figure 5.
Autophagy-related mRNA expression in the liver after 8 weeks of treadmill training with different intensities. [A] Eight weeks of treadmill training with different intensities modulates LC3 and p62 mRNA expression in rat liver. [B] A significant decrease of LC3 mRNA expression was found in all training groups compared to control (a). A significant decrease of p62 mRNA expression was found in moderate and high intensity compared to control (b). Data was presented as average mean ± standard error minimum (SEM) with p < 0.05 considered as significant (∗) and p < 0.01 considered as very significant (∗∗). (∗∗). Representative full, non-adjusted gel images are shown as Supplementary Figure 1.
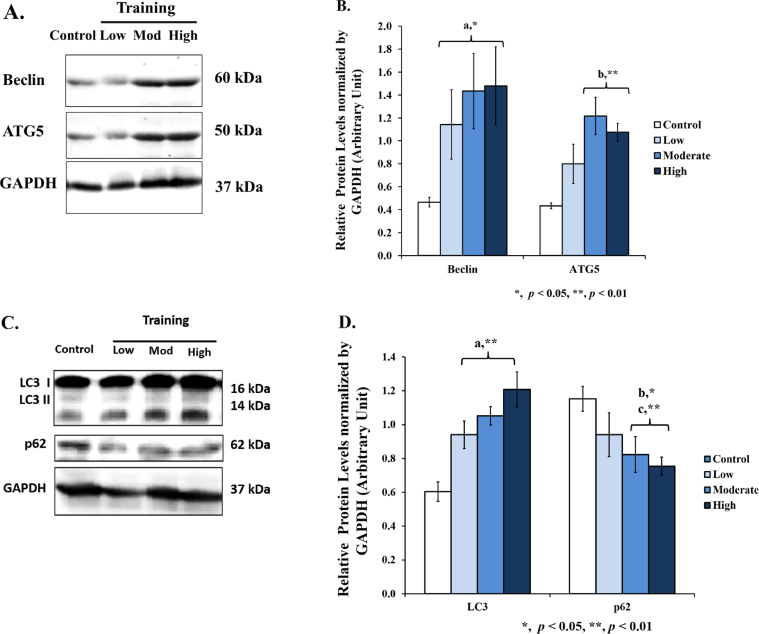
3.5. Beclin, ATG5, LC3 and p62 protein levels in wistar rat liver
We also performed a Western blot analysis, and the results showed a significant increase of Beclin protein levels in low (2.45–fold), moderate (3.08 fold), and high intensities (3.17-fold); ATG5 protein levels in moderate (2.81-fold) and high intensities (2.49-fold) (Figure 6A dan 6B). As for LC3 protein levels, we found a significant increase in low (1.56-fold), moderate (1.74-fold), and high intensities (twofold); and a significant decrease of p62 protein levels in moderate (0.71-fold) and high intensities (0.65-fold) (Figures 6C and 6D). Increase of Beclin, Atg5, and LC3 protein levels following by a decrease of p62 protein level are reflecting an increase of autophagy activity.
Figure 6.
Autophagy-related protein levels in the liver after 8 weeks of treadmill training with different intensities. [A] Eight weeks of treadmill training with different intensities increases Beclin and ATG5 protein levels in rat liver. [B] A significant increase of Beclin protein levels was found in all training groups compared to control (a). A significant increase of ATG5 protein levels was found in moderate and high intensities compared to control (b). [C] Eight weeks of treadmill training with different intensities modulates LC3 and p62 protein levels in rat liver. [D] A significant increase of LC3 protein levels was found in all training groups compared to control (a). A significant decrease of p62 protein levels was found in moderate (b) and high intensities compared to control (c). Data was presented as average mean ± standard error minimum (SEM) with p < 0.05 considered as significant (∗) and p < 0.01 considered as very significant (∗∗). Representative full, non-adjusted blot images are shown as Supplementary Figure 2.
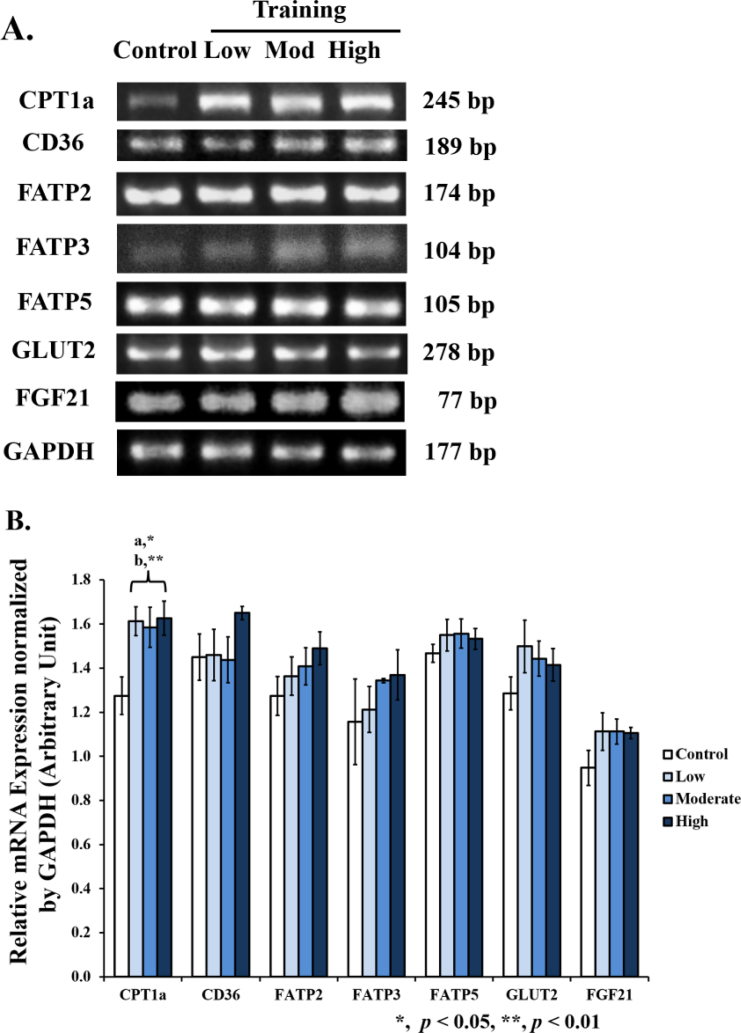
3.6. Increased CPT1a mRNA expressions, but No change in CD36, FATP2,3,5, GLUT2, and FGF21 mRNA expression in the liver of wistar rats
We found an increased CPT1a mRNA expression across all training groups (low 1.27-fold, moderate 1.24-fold, and high 1.28-fold), but the mRNA expression of the CD36, FATP2,3,5, and GLUT2 was largely unchanged in the liver of the training groups (Figure 7).
Figure 7.
CPT1a, CD36, FATP, GLUT2, and FGF21 mRNA expressions in the liver after 8 weeks of treadmill training with different intensities. [A] Eight weeks of treadmill training with different intensities increased the CPT1a mRNA expressions in the rat liver, but there was no change in the CD36, FATP2, FATP3, FATP5, GLUT2, and FGF21 mRNA expressions. [B] A significant increase of the CPT1a mRNA expressions was found in moderate (a), low, and high intensity samples (b) compared to control. No significant change was found for the CD36, FATP, GLUT2, and FGF21 in the mRNA expression between all training groups and control. Data was presented as an average mean ± standard error minimum (SEM), with p < 0.05 considered as significant (∗) and p < 0.01 considered as very significant (∗∗). Representative full, non-adjusted gel images are shown as Supplementary Figure 1.
4. Discussion
The liver has the capability to store and assemble glucose, oxidize lipids, and package excess lipids to be stored in tissues [1]. Ninety percent of endogenous glucose production occurs in the liver, so the liver is crucial for maintaining glucose homeostasis [26, 27]. As for lipid metabolism, lipoprotein particles form chylomicrons to digest lipids from the intestines before sending them to other organs. Lipoprotein lipase at the liver extracts fatty acids from chylomicrons to digest the lipids from chylomicron and transport them to the hepatocytes via FATP 2, 3, 5, and CD36 [1]. The next process is esterification of fatty acids, which produces triglyceride and cholesterol ester within the endoplasmic reticulum (ER), and the ER bilayer from the outer leaflet distends, creating precipitation of unique spherical cytosolic organelles in the ER to form a LD, at the key sites of the hepatic lipid metabolism [28, 29].
Singh et al. have convincingly shown that the consolidation of LD to autophagosome would increase free fatty acids for the use of β-oxidation in the mitochondria [14]. More importantly, research also showed that autophagic machinery was able to recognize LD from other structures in the cytoplasm, using a process called hepatic lipophagy. The role of hepatic lipophagy, a form of autophagy that targets the LD, is seen to be important to prevent excessive lipid accumulation in the liver [15]. In lipophagy, an activated membrane bilayer shallows the LD, creating autophagosome which is delivered to the lysosome, where the fatty acids are separated from triglyceride by acid lipases [28].
The goal of this study is to examine the autophagy-related gene expressions and protein levels in the Wistar rat livers after training under different intensities and to know if the autophagy is correlated with the histopathology and metabolism in the liver. We observed a decrease of LC3 and p62 mRNA expressions in moderate and high intensity groups. As for protein levels, we found a significant increase of Beclin, ATG5, LC3, and a decrease of p62 protein levels in moderate and high intensities compared to the control (Figures 4 and 5). Based on protein levels, we concluded that autophagy process in the liver was increased in moderate and high intensities compared to control. Recent studies have confirmed that autophagy increased by training and was beneficial against NAFLD (nonalcoholic fatty liver disease) [17]. Targets of autophagy are LDs, which then transfer to lysosomes for degradation [13, 14, 15]. In this study we also found decreased triglyceride serum levels in moderate and high intensity samples (Figure 2), along with increased autophagy in the same groups, which means autophagy might be possibly involved in the hepatic lipid metabolism by inducing lipophagy.
Animal studies have shown an increased palmitate oxidation in the liver, through the increase of hepatic CPT1a after training. It is an enzyme to help transport fatty acids from the cytosol across the membrane of the mitochondria [30, 31]. We also found an increased hepatic CPT1a in all training groups compared to the control (Figure 7). But we found no change in CD36, FATP2, FATP3, FATP5, GLUT2, and FGF21 gene expressions (Figure 7), suggesting that the change of lipid and glucose metabolism might be involved in another pathway, for example, FABP, SREBP1, ACC1, FAS, PPARα, FINS, FBG [17, 30, 31].
The result from the histopathology examination (Figure 3C) showed a slight increase of moderate congestion/sinusoidal dilatation (20% in moderate and high intensity compared to 0% in control), moderate cloudy swelling/injury (20% in all training groups compared to 0% in control), and moderate inflammation (20% in moderate and 40% in high compared to 0% in control). Inflammation could be defined as an immune response in order to establish homeostasis in various tissues after stimulation by many kinds of inducers [32, 33]. Training as inducer could lead to local and systemic inflammation, which is mediated by cytokine and growth factor signaling [34]. It also induced muscle tissue trauma and will be repaired through the combined actions of many organs, for example, the brain, kidney, immune system, and liver [35, 36].
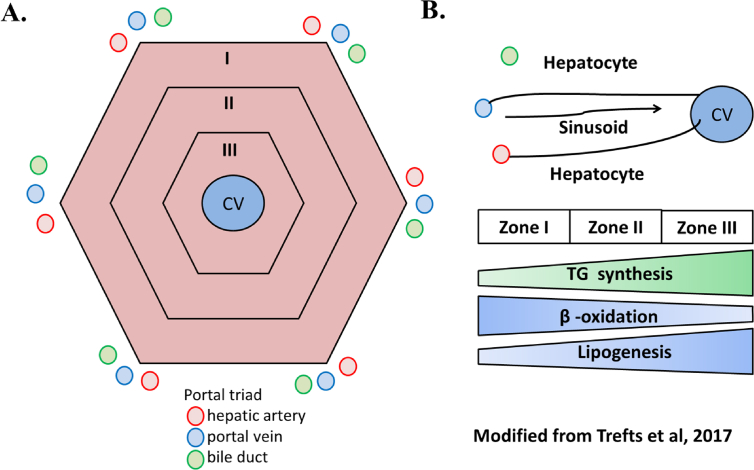
The liver has metabolic zones (zone I, II, and III), which are organized by gradient formation and relevant metabolic processes that occur in each of these zones (Figure 8) [1]. These zones can shift in size and location if altered blood flow or hepatocellular damage occurs [1]. In this study, the vena congestion/sinusoidal dilatation and cloudy swelling/injury shifted to zone 2/intermediate zone (from mild to moderate), indicating that the metabolic zone might also shift from lipogenesis and TG synthesis to β-oxidation. This shift was more prominent in moderate and high intensities compared to low and control samples. Oil red O staining from liver tissues also showed a decrease of LDs in moderate and high intensities compared to control (Figure 4).
Figure 8.
Geometric and schematic representation of a hepatic lobule and metabolic zonation. [A] Geometric representation of a hepatic lobule (zone I, II, III). [B] Schematic representation of a sinusoid and the corresponding zonation associated with lipid metabolism in the liver (modified from Trefts et al., 2017).
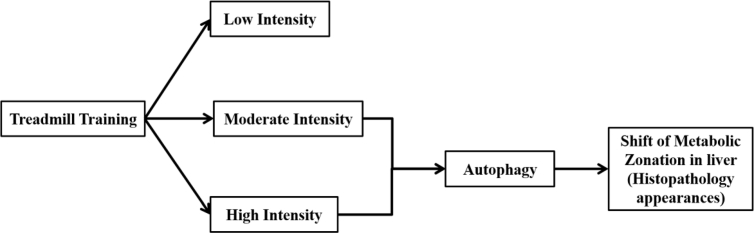
There is a limited study about the importance of training intensities in determining lipid metabolism in liver. We had found several studies that directly linking the exercise to the clinical diseases treatment like Non-Alcoholic Fatty Liver Disease/NAFLD in mice [37] and in human [5, 38, 39, 40]. Research about training intensities showed a significant effect of high intensity interval training in reducing liver fat in NAFLD [41], while other study in overweight/obese adults showed no difference between different regimens of training intensities [42]. In those studies, important perspective about physiological role of different training intensities itself were not well elucidated. Therefore in the present study, we had provided a schematic figure to simplify the potential physiological mechanism of different training intensities in regulating lipid metabolism in liver via autophagy (Figure 9).
Figure 9.
Schematic figure to simplify the potential physiological mechanism of different training intensities in regulating lipid metabolism in liver via autophagy.
This study provides a molecular biology approach for describing potential physiological role of training intensities in lipid metabolism. It will enrich the knowledge about the role of optimum intensity in controlling risk factor like obesity (weight management or weight loss program) or improving recovery and treatment for several illness like Non-Alcoholic Fatty Liver Disease, Fatty Liver, obesity or other metabolic diseases.
In the present study, we presented a potential link between training intensities, autophagy and lipid metabolism. Our study suggested that specific training intensities (moderate and high) might induce autophagy which play an important in lipid metabolism and potentially correlated with fat reduction in the liver. We observed that moderate and high intensities were more beneficial in the highlight of reducing fat accumulation in the liver. However, several details needs to be investigated, especially liver micrograph study using electron microscope (lipophagy); and also further study in human would be very important for clinical implementation.
The limitation of this study is we did not use female rats, so we could not determine sex-specific differences between all training groups and control. In addition, the glucose and amino acid metabolism in the liver was not investigated. This may potentially also be associated with histopathology and autophagy.
5. Conclusion
There is a very limited study about physiological role of different training intensities linked with autophagy and histopathology appearances which is potentially associated with lipid metabolism in wistar rat liver. In summary, we presented that moderate and high intensities of training induced autophagy activity and it is in line with changes in histopathology of the liver, a shift in metabolic zonation, we believe that it might be potentially correlated with lipophagy. Taken together, moderate and high intensities training are more likely beneficial for reducing fat deposition in liver. In the highlight of physiological application, we believe that moderate and high intensities training might be more suitable for weight loss and weight maintenance program. However, further study like liver micrograph examination using electron microscope is important for clarifiying lipophagy process in liver; and in addition, similar study in human would be very important for further implementation as a preventive protocol against metabolic disorders.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
Declarations
Author contribution statement
T. Lucretia: Conceived and designed the experiments; Performed the experiments.
V. Tarawan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
R. Wahyudianingsih: Analyzed and interpreted the data.
H. Ray and U. Supratman: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
J. Gunadi and F. Tanuwijaya: Performed the experiments; Wrote the paper.
I. Setiawan: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
R. Lesmana: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
I. Setiawan and R. Lesmana were supported by World Class Research Grant from Kementerian Riset Teknologi dan Pendidikan Tinggi Republik Indonesia with grant number 3854/UN6.C/LT/2019. V. Tarawan was supported by Riset Kompetensi Dosen Unpad from Universitas Padjadjaran, Indonesia with grant number 3855/UN6.C/LT/2019. H. Ray was supported by Research of Invitation from Lembaga Pengelola Dana Pendidikan (LPDP), Indonesia with grant number PRJ 50/LPDP/2019.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Susianti, Nurul Ihsani, Meita, Deni Firmansyah, and Lidya Agustina for the technical assistance during the course of the experiments. The authors also want to thank Dr. Nova Sylviana, Dr. Yuni Susanti Pratiwi, and Dr. Hanna Goenawan for their constructive comments, as well as drh Okta, dr Ardo, Azis, Risma Ully, Steven, Andi Batari, Nafisya Bella, Tiodora, Grace, Sharfina, Ririn, and Xena for their assistance in studying the Wistar rats. The authors would like to thank Enago (www.enago.com) for the English language review.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplemental Figure 1
References
- 1.Trefts E., Gannon M., Wasserman D.H. The liver. Curr. Biol. 2017;27(21):R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merkel M., Eckel R.H., Goldberg I.J. Lipoprotein lipase: genetics, lipid uptake, and regulation. J. Lipid Res. 2002;43(12):1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M. Heart disease and stroke statistics-2016 update: a report from the American heart association. Circulation. 2016;133(4):e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 4.Bechmann L.P., Hannivoort R.A., Gerken G., Hotamisligil G.S., Trauner M., Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012;56(4):952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 5.van der Windt D.J., Sud V., Zhang H., Tsung A., Huang H. The effects of physical exercise on fatty liver disease. Gene Expr. 2018;18(2):89–101. doi: 10.3727/105221617X15124844266408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge F., Zhou S., Hu C., Lobdell H.t., Berk P.D. Insulin- and leptin-regulated fatty acid uptake plays a key causal role in hepatic steatosis in mice with intact leptin signaling but not in ob/ob or db/db mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299(4):G855–G866. doi: 10.1152/ajpgi.00434.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abumrad N.A., el-Maghrabi M.R., Amri E.Z., Lopez E., Grimaldi P.A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993;268(24):17665–17668. [PubMed] [Google Scholar]
- 8.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009;2(72):re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher F.M., Maratos-Flier E. Understanding the physiology of FGF21. Annu. Rev. Physiol. 2016;78:223–241. doi: 10.1146/annurev-physiol-021115-105339. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen P., Leray V., Diez M., Serisier S., Le Bloc'h J., Siliart B. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. (Berl). 2008;92(3):272–283. doi: 10.1111/j.1439-0396.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- 11.Leturque A., Brot-Laroche E., Le Gall M. GLUT2 mutations, translocation, and receptor function in diet sugar managing. Am. J. Physiol. Endocrinol. Metab. 2009;296(5):E985–E992. doi: 10.1152/ajpendo.00004.2009. [DOI] [PubMed] [Google Scholar]
- 12.Agius L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 2008;414(1):1–18. doi: 10.1042/BJ20080595. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Lopez N., Singh R. Autophagy and lipid droplets in the liver. Annu. Rev. Nutr. 2015;35:215–237. doi: 10.1146/annurev-nutr-071813-105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R., Cuervo A.M. Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012;2012:282041. doi: 10.1155/2012/282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg A.S., Coleman R.A., Kraemer F.B., McManaman J.L., Obin M.S., Puri V. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Invest. 2011;121(6):2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pi H., Liu M., Xi Y., Chen M., Tian L., Xie J. Long-term exercise prevents hepatic steatosis: a novel role of FABP1 in regulation of autophagy-lysosomal machinery. FASEB J. 2019 doi: 10.1096/fj.201900812R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley J.A., Maughan R.J., Hargreaves M. Exercise metabolism: historical perspective. Cell Metabol. 2015;22(1):12–17. doi: 10.1016/j.cmet.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Brouwers B., Hesselink M.K., Schrauwen P., Schrauwen-Hinderling V.B. Effects of exercise training on intrahepatic lipid content in humans. Diabetologia. 2016;59(10):2068–2079. doi: 10.1007/s00125-016-4037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks G.A. Importance of the 'crossover' concept in exercise metabolism. Clin. Exp. Pharmacol. Physiol. 1997;24(11):889–895. doi: 10.1111/j.1440-1681.1997.tb02712.x. [DOI] [PubMed] [Google Scholar]
- 21.Romijn J.A., Coyle E.F., Sidossis L.S., Gastaldelli A., Horowitz J.F., Endert E. Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am. J. Physiol. 1993;265(3 Pt 1):E380–E391. doi: 10.1152/ajpendo.1993.265.3.E380. [DOI] [PubMed] [Google Scholar]
- 22.Council N.R. eighth ed. National Academies Press (US); Washington (DC): 2011. Guide for the Care and Use of Laboratory Animals; p. 246. [Google Scholar]
- 23.Lesmana R., Iwasaki T., Iizuka Y., Amano I., Shimokawa N., Koibuchi N. The change in thyroid hormone signaling by altered training intensity in male rat skeletal muscle. Endocr. J. 2016;63(8):727–738. doi: 10.1507/endocrj.EJ16-0126. [DOI] [PubMed] [Google Scholar]
- 24.Praphatsorn P., Thong-Ngama D., Kulaputana O., Klaikeaw N. Effects of intense exercise on biochemical and histological changes in rat liver and pancreas. Asian Biomed. 2010;4:619–625. [Google Scholar]
- 25.Pratiwi Y.S., Lesmana R., Goenawan H., Sylviana N., Setiawan I., Tarawan V.M. Nutmeg extract increases skeletal muscle mass in aging rats partly via IGF1-AKT-mTOR pathway and inhibition of autophagy. Evid. Based Complement Alternat. Med. 2018;2018:2810840. doi: 10.1155/2018/2810840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekberg K., Landau B.R., Wajngot A., Chandramouli V., Efendic S., Brunengraber H. Contributions by kidney and liver to glucose production in the postabsorptive state and after 60 h of fasting. Diabetes. 1999;48(2):292–298. doi: 10.2337/diabetes.48.2.292. [DOI] [PubMed] [Google Scholar]
- 27.Moore M.C., Coate K.C., Winnick J.J., An Z., Cherrington A.D. Regulation of hepatic glucose uptake and storage in vivo. Adv. Nutr. 2012;3(3):286–294. doi: 10.3945/an.112.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walther T.C., Farese R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilfling F., Haas J.T., Walther T.C., Farese R.V., Jr. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N., Liu Y., Ma Y., Wen D. High-intensity interval versus moderate-intensity continuous training: superior metabolic benefits in diet-induced obesity mice. Life Sci. 2017;191:122–131. doi: 10.1016/j.lfs.2017.08.023. [DOI] [PubMed] [Google Scholar]
- 31.Evangelista F.S., Muller C.R., Stefano J.T., Torres M.M., Muntanelli B.R., Simon D. Physical training improves body weight and energy balance but does not protect against hepatic steatosis in obese mice. Int. J. Clin. Exp. Med. 2015;8(7):10911–10919. [PMC free article] [PubMed] [Google Scholar]
- 32.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140(6):771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Allen J., Sun Y., Woods J.A. Exercise and the regulation of inflammatory responses. Prog. Mol. Biol. Transl. Sci. 2015;135:337–354. doi: 10.1016/bs.pmbts.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Zaldivar F., Wang-Rodriguez J., Nemet D., Schwindt C., Galassetti P., Mills P.J. Constitutive pro- and anti-inflammatory cytokine and growth factor response to exercise in leukocytes. J. Appl. Physiol. (1985) 2006;100(4):1124–1133. doi: 10.1152/japplphysiol.00562.2005. [DOI] [PubMed] [Google Scholar]
- 35.Silva F., Macedo D. Physical exercise, inflammatory process and adaptive condition: an overview. Revista Brasileira de Cineantropometria & Desempenho Humano. 2011;13:320–328. [Google Scholar]
- 36.Smith L.L. Cytokine hypothesis of overtraining: a physiological adaptation to excessive stress? Med. Sci. Sports Exerc. 2000;32(2):317–331. doi: 10.1097/00005768-200002000-00011. [DOI] [PubMed] [Google Scholar]
- 37.la Fuente F.P., Quezada L., Sepúlveda C., Monsalves-Alvarez M., Rodríguez J.M., Sacristán C. Exercise regulates lipid droplet dynamics in normal and fatty liver. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864(12):158519. doi: 10.1016/j.bbalip.2019.158519. [DOI] [PubMed] [Google Scholar]
- 38.Cuthbertson D.J., Shojaee-Moradie F., Sprung V.S., Jones H., Pugh C.J., Richardson P. Dissociation between exercise-induced reduction in liver fat and changes in hepatic and peripheral glucose homoeostasis in obese patients with non-alcoholic fatty liver disease. Clin. Sci. (London, England: 1979) 2016;130(2):93–104. doi: 10.1042/CS20150447. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan S., Kirk E.P., Mittendorfer B., Patterson B.W., Klein S. Randomized trial of exercise effect on intrahepatic triglyceride content and lipid kinetics in nonalcoholic fatty liver disease. Hepatology (Baltimore, Md) 2012;55(6):1738–1745. doi: 10.1002/hep.25548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houghton D., Thoma C., Hallsworth K., Cassidy S., Hardy T., Burt A.D. Exercise reduces liver lipids and visceral adiposity in patients with nonalcoholic steatohepatitis in a randomized controlled trial. Clin. Gastroenterol. Hepatol. – Offic. Clin. Pract. J. Am. Gastroenterol. Assoc. 2017;15(1):96–102. doi: 10.1016/j.cgh.2016.07.031. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallsworth K., Thoma C., Hollingsworth K.G., Cassidy S., Anstee Q.M., Day C.P. Modified high-intensity interval training reduces liver fat and improves cardiac function in non-alcoholic fatty liver disease: a randomized controlled trial. Clin. Sci. (London, England: 1979) 2015;129(12):1097–1105. doi: 10.1042/CS20150308. [DOI] [PubMed] [Google Scholar]
- 42.Keating S.E., Hackett D.A., Parker H.M., O'Connor H.T., Gerofi J.A., Sainsbury A. Effect of aerobic exercise training dose on liver fat and visceral adiposity. J. Hepatol. 2015;63(1):174–182. doi: 10.1016/j.jhep.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 43.Yin P., Wan C., He S., Xu X., Liu M., Song S. Transport stress causes damage in rats' liver and triggers liver autophagy. Bio Technol. Indian J. 2013;8:1561–1566. [Google Scholar]
- 44.Kowalik M.A., Perra A., Ledda-Columbano G.M., Ippolito G., Piacentini M., Columbano A. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules. Oncotarget. 2016;7(5):5788–5799. doi: 10.18632/oncotarget.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahlhoff C., Worsch S., Sailer M., Hummel B.A., Fiamoncini J., Uebel K. Methyl-donor supplementation in obese mice prevents the progression of NAFLD, activates AMPK and decreases acyl-carnitine levels. Mol. Metab. 2014;3(5):565–580. doi: 10.1016/j.molmet.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye K., Li L., Zhang D., Li Y., Wang H.Q., Lai H.L. Effect of maternal obesity on fetal growth and expression of placental fatty acid transporters. J. Clin. Res. Pediatr. Endocrinol. 2017;9(4):300–307. doi: 10.4274/jcrpe.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ono-Moore K.D., Ferguson M., Blackburn M.L., Issafras H., Adams S.H. Application of an in vivo hepatic triacylglycerol production method in the setting of a high-fat diet in mice. Nutrients. 2016;9(1) doi: 10.3390/nu9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y., Choi J., Kim E., Seok J., Lee H.-J., Yoon J.-H. Human chorionic plate-derived mesenchymal stem cells restore hepatic lipid metabolism in a rat model of bile duct ligation. Stem Cell. Int. 2017;2017:1–9. doi: 10.1155/2017/5180579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie G., Zhong W., Li H., Li Q., Qiu Y., Zheng X. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013;27(9):3583–3593. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura Y., Sugimoto M., Murayama T., Ueda Y., Kanamori H., Ono K. Inhibition of CCR2 ameliorates insulin resistance and hepatic steatosis in db/db mice. Arterioscler. Thromb. Vasc. Biol. 2008;28(12):2195–2201. doi: 10.1161/ATVBAHA.108.168633. [DOI] [PubMed] [Google Scholar]
- 51.Nygaard E.B., Vienberg S.G., Orskov C., Hansen H.S., Andersen B. Metformin stimulates FGF21 expression in primary hepatocytes. Exp. Diabetes Res. 2012;2012:465282. doi: 10.1155/2012/465282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K., Wang F., Bao J.P., Xie Z.Y., Chen L., Zhou B.Y. Tumor necrosis factor alpha modulates sodium-activated potassium channel SLICK in rat dorsal horn neurons via p38 MAPK activation pathway. J. Pain Res. 2017;10:1265–1271. doi: 10.2147/JPR.S132185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.