Abstract
Bloodstream infections (BSIs) are some of the most devastating preventable complications in critical care units. Of the bacterial causes of BSIs, Escherichia coli is the most common among Enterobacteriaceae. Bacteria resistant to therapeutic antibiotics represent a significant global health challenge. In this study, we present whole genome sequence data of 22 E. coli isolates that were obtained from bloodstream infection patients admitted to Cipto Mangunkusumo National Hospital, Jakarta, Indonesia. These data will be useful for analysing the serotypes, virulence genes, and antimicrobial resistance genes of E. coli. DNA sequences of E. coli were obtained using the Illumina MiSeq platform. The FASTQ raw files of these sequences are available under BioProject accession number PRJNA596854 and Sequence Read Archive accession numbers SRR10761126–SRR10761147.
Keywords: E. coli, Bloodstream infection, Whole genome sequencing, Cipto Mangunkusumo National Hospital, Jakarta, Indonesia
Specifications table
| Subject | Bacterial Sequencing |
| Specific subject area | Genomics |
| Type of data | Genome sequences (DNA-Seq raw reads) |
| How data were acquired | Illumina MiSeq sequencing platform (Illumina, San Diego, CA, USA) |
| Data format | Raw sequences (FASTQ) |
| Parameters for data collection | Genomic DNA was extracted from purified cultures of Escherichia coli and quantified, following which libraries were prepared and quality checked for sequencing. |
| Description of data collection | DNA extraction was performed using the Geneaid Presto™ Mini gDNA Bacteria Kit (with lysozyme) (Geneaid, New Taipei City, Taiwan). DNA isolate purity was quantified with a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) using a target-specific Qubit assay (dsDNA BR Assay Kit, Thermo Fisher Scientific). Libraries were prepared using the Nextera™ DNA Flex Library Prep Kit (Illumina®) and library quality was examined using The Agilent 4200 TapeStation system (G2991AA) (Agilent, Santa Clara, CA, USA). Sequencing was performed using the Illumina MiSeq system. |
| Data source location | Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia |
| Data accessibility | Raw data (FASTQ) files of Escherichia coli have been deposited in the National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/, under BioProject database: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA596854, BioSample database: (https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=596854), and Sequence Read Archive (SRA) database: https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=596854 with accession number: SRR10761126–SRR10761147. |
Value of the data
-
•
These data shed light on the molecular biology of E. coli found in bloodstream infections.
-
•
These data provide insights into antibiotic resistance in E. coli, which will be beneficial to clinicians and patients.
-
•
The data will help us understand the genomic mechanisms underlying the severity of E. coli-caused bloodstream infections.
1. Data Description
E. coli (Gram-negative bacterial commensals) naturally exists in the human gastrointestinal tract. Pathogenicity, virulence, and multidrug resistance features of pathogenic E. coli are routinely obtained by commensal E. coli through horizontal transfer and other mechanisms. Virulent E. coli share pathogenic factors, virulence, and resistance with less virulent strains, causing overlapping pathogenesis beyond their natural capability [1].
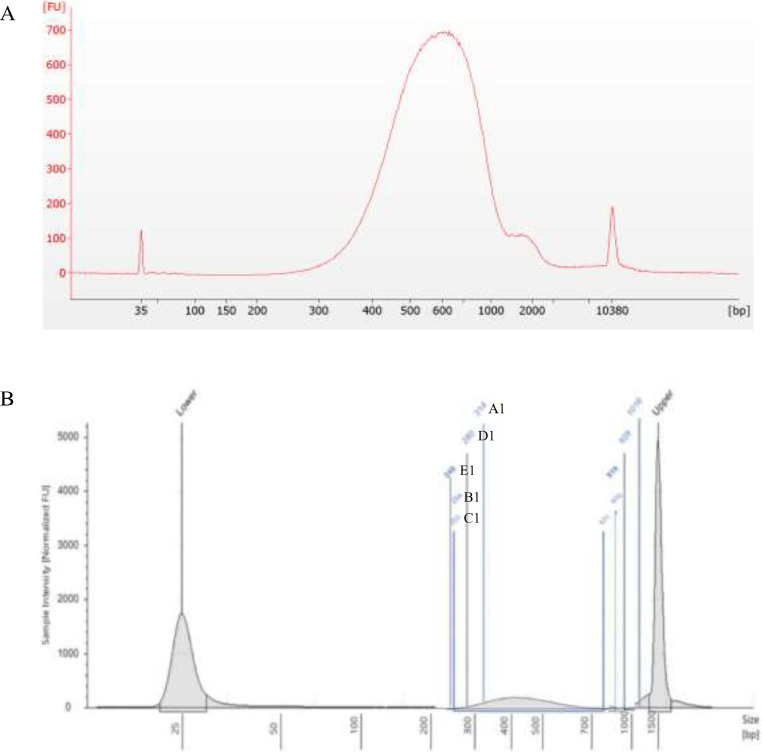
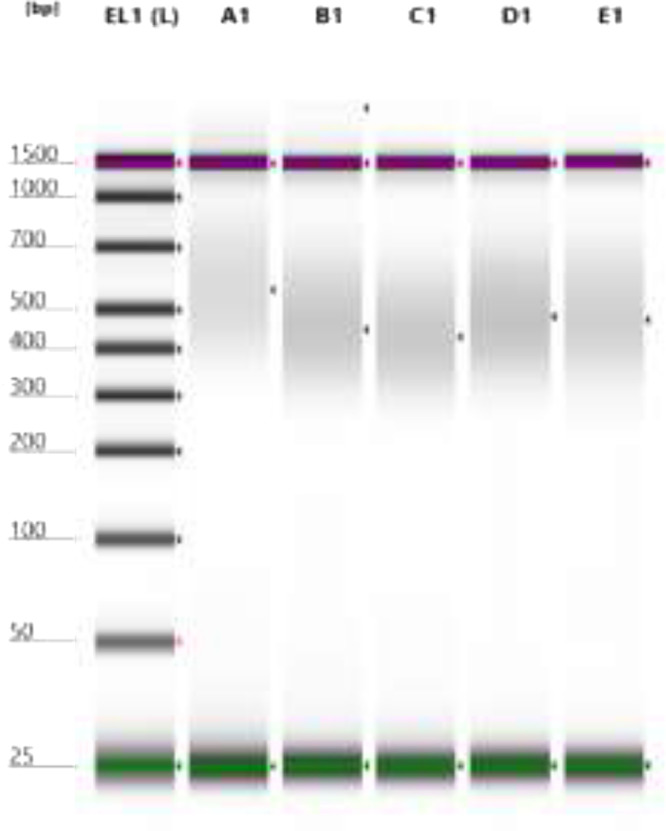
We present whole genome sequence data of 22 E. coli isolates obtained from bloodstream infection patients admitted to Cipto Mangunkusumo National Hospital, Jakarta, Indonesia. Purified E. coli DNA was quantified using Qubit 3.0 (Table 1). The quality of library preparations was checked using the Agilent 4200 TapeStation system and found to be equal to that of the reference (Fig. 1 and Table 2). Electrophoresis strengthened these results (Fig. 2). Sequencing was performed using the Illumina MiSeq system.
Table 1.
DNA library purification and concentrations using Qubit 3.0.
| Sample | Purity 260/280 | Qubit 3.0 (C) (μg/ml) |
|---|---|---|
| RSCM_EC_0102 | 2 | 25.9 |
| RSCM_EC_0203 | 2.028 | 42.5 |
| RSCM_EC_0305 | 1.808 | 19.7 |
| RSCM_EC_0406 | 1.939 | 28.1 |
| RSCM_EC_0507 | 1.914 | 40.7 |
| RSCM_EC_0608 | 1.9 | 30.2 |
| RSCM_EC_0709 | 2.038 | 30.5 |
| RSCM_EC_0911 | 2 | 19.6 |
| RSCM_EC_1013 | 2.043 | 33 |
| RSCM_EC_1114 | 2.05 | 49.3 |
| RSCM_EC_1316 | 1.943 | 57 |
| RSCM_EC_1418 | 1.955 | 21.7 |
| RSCM_EC_1526 | 2.059 | 29.7 |
| RSCM_EC_1628 | 1.95 | 16.3 |
| RSCM_EC_1732 | 2.053 | 51 |
| RSCM_EC_1833 | 2 | 42.6 |
| RSCM_EC_1935 | 1.944 | 39.9 |
| RSCM_EC_2036 | 1.864 | 49.3 |
| RSCM_EC_2137 | 2 | 30 |
| RSCM_EC_2240 | 2 | 36.9 |
| RSCM_EC_2341 | 2 | 36 |
| RSCM_EC_2442 | 2 | 43.2 |
Fig. 1.
Library size profiles using the Agilent 4200 TapeStation system. A) Reference Guide [2] and B) Experimental E. coli libraries.
Table 2.
E. coli library regions quality-checked using the Agilent 4200 TapeStation system.
| Sample | From (bp) | To (bp) | Average Size (bp) | Conc. (μg/μl) | Region Molarity (nmol/l) | % of Total |
|---|---|---|---|---|---|---|
| RSCM_EC_0203 (A1) | 314 | 1016 | 593 | 3.15 | 8.69 | 67.5 |
| RSCM_EC_0305 (B1) | 254 | 910 | 489 | 4.95 | 16.5 | 79.2 |
| RSCM_EC_0911 (C1) | 253 | 877 | 459 | 4.57 | 16.2 | 79.7 |
| RSCM_EC_1114 (D1) | 280 | 928 | 509 | 5.53 | 17.7 | 84.9 |
| RSCM_EC_1316 (E1) | 248 | 919 | 504 | 4.41 | 14.3 | 82.4 |
Fig. 2.
Electrophoresis of five E. coli DNA library samples. EL1: Genomic DNA ladder (as reference); A1: RSCM_EC_0203; B1: RSCM_EC_0305; C1: RSCM_EC_0911; D1: RSCM_EC_1114; E1: RSCM_EC_1316).
Paired-end libraries were obtained from sequencing runs (Table 3). FASTQ raw data files have been deposited in the NCBI database under BioProject accession number PRJNA596854 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA596854), BioSample database: (https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=596854) and Sequence Read Archive (SRA) accession numbers: SRR10761126–SRR10761147 (https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=596854). These data will be useful for analysing the serotypes, virulence genes, and antimicrobial resistance genes of E. coli.
Table 3.
Descriptive information for whole genome sequencing of raw E. coli data.
| Sample | Total Raw Reads (Mb) | Total bases (Mbp) | GC Content (%) | BioSample Accession Number | SRA Accession Number |
|---|---|---|---|---|---|
| RSCM_EC_0102 | 0.56 | 311.8 | 50.4 | SAMN13640292 | SRS5880528 |
| RSCM_EC_0203 | 0.97 | 503.1 | 50.2 | SAMN13640311 | SRS5880529 |
| RSCM_EC_0305 | 1.7 | 883.5 | 50.2 | SAMN13640332 | SRS5880540 |
| RSCM_EC_0406 | 1.6 | 875.3 | 50.3 | SAMN13640527 | SRS5880543 |
| RSCM_EC_0507 | 2.2 | 1200 | 50.3 | SAMN13640528 | SRS5880544 |
| RSCM_EC_0608 | 1.3 | 655.2 | 49.6 | SAMN13640529 | SRS5880545 |
| RSCM_EC_0709 | 1.6 | 879.5 | 50.8 | SAMN13640533 | SRS5880546 |
| RSCM_EC_0911 | 1.6 | 835 | 50.5 | SAMN13640535 | SRS5880547 |
| RSCM_EC_1013 | 0.9 | 465.6 | 50.5 | SAMN13640581 | SRS5880548 |
| RSCM_EC_1114 | 1.8 | 971.4 | 50.2 | SAMN13640826 | SRS5880549 |
| RSCM_EC_1316 | 1.1 | 592.9 | 50.3 | SAMN13640832 | SRS5880530 |
| RSCM_EC_1418 | 0.90 | 512.7 | 50.0 | SAMN13640834 | SRS5880531 |
| RSCM_EC_1526 | 1.3 | 661.5 | 50.8 | SAMN13640837 | SRS5880532 |
| RSCM_EC_1628 | 0.93 | 492.0 | 49.9 | SAMN13640838 | SRS5880533 |
| RSCM_EC_1732 | 2.7 | 1500 | 50.0 | SAMN13640846 | SRS5880534 |
| RSCM_EC_1833 | 1.0 | 562.0 | 50.0 | SAMN13640847 | SRS5880535 |
| RSCM_EC_1935 | 0.92 | 466.6 | 50.7 | SAMN13640849 | SRS5880536 |
| RSCM_EC_2036 | 1.27 | 665.5 | 50.4 | SAMN13640850 | SRS5880537 |
| RSCM_EC_2137 | 0.46 | 254.2 | 50.4 | SAMN13640852 | SRS5880538 |
| RSCM_EC_2240 | 0.84 | 476.6 | 50.4 | SAMN13640877 | SRS5880539 |
| RSCM_EC_2341 | 1.4 | 749.6 | 50.6 | SAMN13640878 | SRS5880541 |
| RSCM_EC_2442 | 2.3 | 1100 | 49.9 | SAMN13640880 | SRS5880542 |
2. Experimental Design, Materials, and Methods
2.1. Sample collection and bacteria culturing
E. coli were isolated from blood samples of bloodstream infection patients, who varied in gender and age and were admitted to Cipto Mangunkusumo National Hospital in 2018. After isolation, the E. coli were cultured in Lactose Broth medium in the laboratory facilities at the Centre for Research and Development of Biomedical and Basic Health Technology, National Institute of Health Research and Development, Ministry of Health, Indonesia.
2.2. DNA isolation and quantification
DNA extraction was performed using a Geneaid Presto™ Mini gDNA Bacteria Kit (with lysozyme). DNA sample purity was determined from the 260/280 nm absorbance values with a ratio of 1.8–2.0 indicating a pure DNA sample [2]. Pure DNA isolates were then quantified using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific) and a target-specific Qubit assay (dsDNA BR Assay Kit, Thermo Fisher Scientific). Initially, 43 DNA samples were prepared; however, 22 samples were chosen for DNA sequencing based on their high purity and adequate concentration (Table 1).
2.3. Library preparation and quality checking
DNA libraries were prepared using the Nextera™ DNA Flex Library Prep Kit (Illumina) according to the manufacturer's protocol. Five random libraries were quality checked using the Agilent 4200 TapeStation system (G2991AA). The E. coli library profile showed similarities with that of the Reference Guide of the Nextera™ DNA Flex Library Prep Kit [2]. Fig. 1 shows typical library size profiles with an average fragment size of 600 bp when analysed with a size range of 150–1500 bp. Specific region details are shown in Table 2. The library quality was also strengthened by the electrophoresis results that showed each of the five libraries having a fragment size of approximately 300–1000 bp (Fig. 2). Thus, it was demonstrated that the quality of the E. coli libraries met the Illumina MiSeq platform requirements.
2.4. Whole genome sequencing and data
The E. coli libraries were sequenced using the Illumina MiSeq platform according to the following steps: 1) Denaturing the libraries; 2) Diluting the libraries; 3) Preparing the optional PhiX control; 4) Loading the libraries onto the reagent cartridge; and 5) Setting up the sequencing run [3]. Paired-end libraries were obtained from sequencing runs (Table 3). The data sequences were deposited in the SRA under BioProject accession number PRJNA596854.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This work was supported by a Q1Q2 Grant from Universitas Indonesia [grant number NBK-0220/UN2.R3.1/HKP.05.00/2019].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105631.
Appendix. Supplementary materials
References
- 1.Sonda T, Kumburu H, van Zwetselaar M, Alifrangis M, Mmbaga BT, Aarestrup FM. Whole genome sequencing reveals high clonal diversity of Escherichia coli isolated from patients in a tertiary care hospital in Moshi, Tanzania. Antimicrob Resist Infect Control. 2018;7(72):1–12. doi: 10.1186/s13756-018-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Illumina, Nextera DNA Flex Library Prep Reference Guide, California, 2019, pp. 2–14.
- 3.Illumina MiSeq System Denature and Dilute Libraries Guide, California, 2019, pp. 3–12.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.