Graphical abstract
Keywords: CML, RNA-seq, AS, hnRNPA1
Abstract
Chronic myeloid leukemia (CML) is a malignancy that evolves through a multi-step process. Alternative splicing of several genes has been linked to the progression of the disease, but involvement of alternations in splicing profiles has not been reported. RNA-seq of peripheral blood mononuclear cell (PBMC) samples characterized the differentially expressed and spliced transcripts in five CML chronic phase (CP) and five blast phase (BP) patients, and five healthy controls. Global splicing alteration analysis detected 6474 altered splicing events altered between CML and healthy samples, including many of the previously reported splicing variants and showing a more profound altered splicing deregulation in BP samples. Functional clustering of differentially spliced genes in CP revealed a preferred enrichment relating to cell signaling, while the spliceosome pathway was most overrepresented in BP samples. One differentially spliced spliceosome gene hnRNPA1 showed two splice isoforms; the longer isoform contained exon 8 was preferentially expressed in the BP patients, and the short one excluding exon 8 was specific to healthy controls. Our findings suggested that alternative splicing deregulation played a central role during the progression of CML from CP to BP, and the longer isoform of hnRNPA1 might represent a diagnostic marker and therapeutic target for CML.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder arising from a pathognomonic reciprocal translocation between of the Abelson (Abl) tyrosine kinase gene on ch9 and the breakpoint cluster region (Bcr) of ch22 [1], [2], resulting in a BCR-ABL fusion gene that codes for a constitutively active tyrosine kinase which is necessary and sufficient for cell transformation. CML is triphasic: the chronic phase (CP) is a relatively long-lasting phase in which symptoms can be fairly easily controlled with tyrosine kinase inhibitor (TKI) therapies [3]. As the disease progresses, patients enter a period of increasing instability known as accelerated phase (AP), which is followed by a terminal transformation to the so-called blast phase (BP) [3], [4]. A number of genetic and epigenetic alterations and a subset of mRNA metabolism aberrations are known to be intimately associated with therapeutic resistance and leukemic progression [5].
Alternative splicing events (ASEs) are a major source of proteome diversity and are highly relevant to disease and therapy in humans, due to ASEs of pre-mRNA yield multiple gene products with distinct functions from a single coding sequence [6]. Recently, ASEs involved in multiple biological processes have emerged as important drivers that fuel leukemia progression and therapy resistance [5]. A novel in-frame splice deletion of the wnt/β-catenin pathway regulator GSK3β was identified in granulocyte-macrophage progenitors of BP samples, which enhanced β-catenin expression as well as serial engraftment potential that contributed to leukemia stem cell generation [7]. Further, the expression levels of the RNA editase ADAR1 p150 isoform, which promotes expression of the myeloid transcription factor PU.1 and induces malignant reprogramming of myeloid progenitors, was found to be increased approximately eightfold in CML-BP progenitors in comparison with normal cord blood [8]. The alternative splicing of multiple prosurvival BCL2 family genes promotes malignant transformation of myeloid progenitors into BP leukemia stem cells that are quiescent in the marrow niche and that contribute to therapeutic resistance [9]. Furthermore, the MBNL3 down-regulation-related overexpression of CD44 transcript variant 3 was reported to enhance CML-CP progenitor replating capacity [10]. It is thus clear that AS analysis can be an effective approach for identifying splice-isoform regulators of leukemic maintenance in CML progression.
High-throughput RNA sequencing (RNA-seq) approaches have been extensively applied for gene expression profiling and for splicing landscape characterization in leukemia research [10]. In CML, data obtained from sequencing have been widely used for novel fusion gene and transcript identification, transcription level evaluation, and alternative splicing variant detection [8], [9]. In the present study, we performed RNA-seq to screen for genes whose ASEs in peripheral blood mononuclear cells (PBMCs) were altered between normal and CML samples in the CP and BP. A total of 6474 CML-related ASEs were identified, and we found that inclusion of exon 8 of hnRNPA1, which encodes an RNA-binding protein and auxiliary splicing factor, was markedly increased in most of the CML samples. Consistently, western blot analysis showed that a protein isoform containing exon 8 was preferentially expressed in CML samples rather than the healthy controls (Ctrls). This finding represents the first example of a potentially association between alternative hnRNPA1 splicing with CML, which may represent a novel prognostic marker and therapeutic target.
Methods
Patient population, healthy controls and samples
All samples were collected and informed consent was obtained from each patient and healthy volunteer in accordance with the Declaration of Helsinki and institutional guidelines. Ethical approval was given by the medical ethics committee of the Second Affiliated Hospital of Nanchang University, China, and all experimental protocols and methods were performed in accordance with relevant guidelines and regulations. In accordance with the World Health Organization classification of neoplastic diseases of haemopoietic and lymphoid tissues, five CML-CP patients and five CML-BP patients from the Second Affiliated Hospital of Nanchang University were diagnosed CML disease in chronic and blastic phase, respectively. Healthy volunteers were confirmed after physical examination. Circulating peripheral blood and bone marrow samples were collected from 7:30 to 9:30 am for detection of routine blood, morphological classification of bone marrow cells, and fusion genes, respectively. The clinical and biological characteristic of the 10 patients and 5 Ctrls were shown in Table S1, and 17 patients and 8 Ctrls were enrolled in this study for training for validation. Two milliliters of peripheral blood were collected and anticoagulated with EDTA-K2, and then PBMCs were isolated by Ficoll density gradient. Finally, deposition of PBMCs were mixed by one-milliliter Trizol and stored at −80 °C until extraction of total RNA.
Nucleic acid extraction, library construction and sequencing
Total RNA was extracted from PBMCs by using TRIzol Reagent (Ambion, Texas, USA) following the manufacturer’s instructions. Total RNA was treated with RQ1 DNase (Promega, Wisconsin, USA) to remove DNA. The quality and quantity of the purified RNA were determined by measuring the absorbance at 260 nm/280 nm (A260/A280) using smartspec plus (BioRad, California, USA). RNA integrity was further verified by 1.5% agarose gel electrophoresis.
For each sample, 1 μg total RNA was used for RNA-seq library preparation. Polyadenylated mRNAs were purified and concentrated with oligo(dT)-conjugated magnetic beads (Invitrogen, California, USA). Purified mRNAs were fragmented at 95 °C followed by end repair and 5′ adaptor ligation. Then mRNA reverse transcription was performed with RT primer harboring 3′ adaptor sequence and randomized hexamers. The cDNAs were purified and amplified with RNA-Seq Library Preparation Kit (Gnomegen, California, USA). Products corresponding to 200–500 bps were purified, quantified, and stored at −80 °C before sequencing.
For high-throughput sequencing, the libraries were prepared following the manufacturer’s instructions and applied to Illumina Nextseq 500 system for 150 nt paired-end sequencing by ABlife. Inc (Wuhan, China).
RNA-seq raw data clean and alignment statistics
Raw reads were first discarded if containing>2-N bases, then reads were processed by clipping adaptor and removing low quality bases, reads less than 16nt were also removed. FASTX-Toolkit (Version 0.0.13) was used to get the clean reads. Quality control checks were performed by FastQC to evaluate base quality, GC content, sequence length distribution and duplication level. After that, clean reads were aligned to UCSC Homo sapiens reference genome(GRCh37/hg19) by TopHat2 [11]. Based on gene annotation of the genome, multiple mapped reads were discarded due to their ambiguous location. Uniquely localized reads were used to calculate reads per kilobase of exon model per million mapped reads (RPKM).
Differentially expressed genes (DEGs)
Differentially expressed genes between the paired groups were analyzed by using edgeR [12] package embedded in R software. For each gene, significance P-value and false discovery rate (FDR) were obtained based on the model of negative binomial distribution. Fold changes of gene expression were also estimated by the edgeR package. The criteria for DEGs have been set as |log2 fold change | >1 and FDR < 0.05.
Alternative splicing events
In this study, the known/model splice junctions (SJs) was defined by joining splice sites at the exon-intron boundaries of all annotated introns in the human GRCh37/hg19 genome, generating a total of 264,339 model/known SJs. All other SJs involving only one or none of the known splice site were considered as novel junction. SJs in the transcriptome supported by cDNA reads were identified using the TopHat2 tool [13]. AS events were detected and analyzed using ABLas pipeline for significantly regulated AS events selection, we set P-value < 0.05 and |ratio| > 0.15 as the criteria.
Identification of disease stage-regulated alternative transcript events
To assess possible stage-specific expression for each event, Student’s t-test was performed to evaluate the significance of the ratio variation of AS events. Stage-specific expression was assessed by comparing read data from each phase. The stages in this analysis were Ctrl, CP and BP. For this analysis, alternative ratio of non-intron-retention (IR) events was defined as the ratio of the number of alternative splicing reads to the sum of alternative reads plus “common” reads to increase the power to detect stage regulation. For IR events, the ratio was calculated by base depth. Those events which were significant at P-value cutoff corresponding to a false discovery rate cutoff of 5% between stages were considered stage-regulated.
Functional enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was utilized to define the enrichment of differentially expressed or spliced genes in each KEGG pathway. Hypergeometric test was performed with robust FDR correction to obtain an adjusted P-value between certain tested gene groups and genes annotated by KEGG database in the reference genome.
Validation of RNA-seq results
To validate the RNA-seq data, a number of DEGs and splice types between paired groups were selected for expression level comparison by quantitative real time PCR (qRT-PCR). Finally, fifteen DEGs having consistent expression level within groups and high expression level were performed with PBMCs from both CML patients and controls. For qRT-PCR, total RNA remaining from RNA-seq library preparation was treated with RQ1 DNase (Promega, Wisconsin, USA) to remove DNA, and cDNA was synthesized with M-MLV Reverse Transcriptase (Vazyme, Jiangsu, China) using 1 μg of total RNA and oligo dT primer. RT-qPCR was performed with the gene-specific primers listed in Table S4, using a QuantStudio 6 Flex System (ABI). The levels of mRNA expression of all the genes were normalized against the expression levels of GAPDH mRNA.
Western blot analysis
Whole-cell lysate was subjected to western blot analysis following a previously described procedure [14]. For the preparation of total cell lysates, the PBMCs were lysed in RIPA buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1.0% deoxycholate, 1% Triton X-100, 1 mM EDTA and 0.1% SDS. The samples were centrifuged (12,000×rpm, 5 min) and the supernatants were further analyzed on a 12% SDS-PAGE gel and subsequently transferred to a PVDF membrane (Millipore, Massachusetts, USA). The following antibodies were used for western blotting: Tubulin (1:1000, AC015), SUMO1 (1:500, A2130) and UBE2Q2 (1:500, A9992) and were purchased from ABclonal Biotechnology. HNRNPA1 (1:500, A12446) and TRA2B (1:500, 23832–1-AP) were purchased from Proteintech group. All the antibodies were originated from rabbit. The immunoreactive proteins were detected by ECL chemiluminescence system (Clinx, Shanghai, China) with default settings and the Tubulin was set as the normalized control.
Statistical analysis
To describe the biological functional significance of selected gene sets, we used Hypergeometric Test to calculate the enrichment of Gene Ontology (GO) terms and KEGG pathways. Other statistical analysis was performed using the R software (https://www.r-project.org/) unless otherwise stated. Significance of differences was evaluated with either Student’s t-test, when only two groups were compared, or hypergeometric test for venn diagram. No statistical methods were used to predetermine sample size. P-value < 0.05 was considered as statistical significance, and the star number represents the significant P-value: *P < 0.05, **P < 0.01 and ***P < 0.001. Hierarchical clustering was performed by Cluster3.0 or heatmap function in R.
Availability of supporting data
The datasets supporting the results of this article are available in NCBI’s gene Expression Omnibus and are accessible through GEO series accession number GSE100026.
Results
Transcriptome profiling reveals distinctly different patterns of gene expression between healthy and CML patient groups
After removing adaptors and discarding low quality reads, 555.1 million clean reads were retained (Table S2) and about 60 percent of reads were successfully mapped to exonic regions (Fig. 1A), suggesting that the RNA-seq data was highly reliable. To compare reads distribution in different genomic locations, the proportion of reads was plotted, showing that exonic and intronic reads were distinctly higher in CML-BP specimens and healthy PBMCs, respectively when compared with each other (Fig. 1A, P-value < 0.05, t-test). In addition, principal component analysis (PCA) revealed that genome-wide gene expression profiles of the CML-CP and -BP patients were separated by the second component and they both were clearly separated from the Ctrls by the first component (Fig. 1B). The clear distinction in the gene expression profiles between CML-BP patients and Ctrls, and between two phases of patient samples, suggested that the experimental replicates were strongly correlated and were appropriate for the subsequent analyses.
Fig. 1.
The distinct difference of CML sample and healthy control. (A) The box plot represents distribution of uniquely mapped reads to the exonic and intronic regions. Dots in the figure depicts the percentage of reads from individual sample. P < 0.05; asterisk (*) indicates significant difference (non-paired two tail t-test). (B) Principal component analysis (PCA) of 15 distinct samples based on normalized mRNAs expression level. The samples were grouped by CML stages, and the ellipse for each group represents the confidence ellipse.
Distinct stage-specific gene expression profiles during CML progression
To explore the difference in gene expression patterns between CML patients and Ctrls, 7866 and 5448 DEGs were identified in the CML-CP and -BP samples when compared to controls, respectively (complete gene list in Tables S3, S4). Overlap of the two sets of DEGs showed that 3682 of them were shared by both the CML-BP and -CP patients, indicating that a large common subset of genes is differentially expressed in CML samples regardless of the stages (Fig. 2A, P-value < 0.001, hypergeometric test). Meanwhile, by hierarchical clustering analysis, we found gene clusters more expressed in either CPH22U or BP samples were also observed (Fig. 2B). To validate gene expression level, the results generally agreed with the estimated gene abundance (RPKM) values, thus reinforcing the reliability of the sequencing data (Fig. 2C; Table S5).
Fig. 2.
The discrete gene expression pattern. (A) Venn diagram illustrates the common DEGs between three groups (***P < 0.001, hypergeometric test). (B) A hierarchical clustering dendrogram of DEGs from three groups. Red: relatively high expression; Blue: relatively low expression. The RPKM values was normalized by row. (C) RT-qPCR experimental validation of differentially expressed genes between CML (n = 5) and healthy controls (Ctrl) (n = 5). *P < 0.05; **P < 0.01; ***P < 0.001; t-test. (D) Top 10 enriched KEGG pathways of the significantly up-regulated and down-regulated genes in CML-BP compared with CML-CP (Corrected P < 0.05).
The enriched functions of the DEG sets were analyzed using GO and KEGG tools, and the up-regulated genes from CML-BP vs. CML-CP cases were enriched in primary immunodeficiency and hematopoietic cell lineage; and the down-regulated genes were enriched in osteoclast differentiation, leukocyte transendothelial migration patheway and a number of signaling pathways(Fig. 2D). Additionally, the enriched pathways were quite similar in both CML progression stages. (Figure S1).
Patient-specific alternative splicing profiles for previously reported candidate CML-related ASEs
We analyzed exons and SJs, and found that, overall, 81% and 68.2% of the annotated exons and SJs were detected (Fig. 3A). By applying ABLas algorithm, we identified 14,483 known AS events, accounting for 35.60% of the total annotated events (Fig. 3A, Table S6). In addition, 53,183 novel AS events occurred in 12,855 genes were detected (Table S6). Classification of AS events revealed that IR events were the most enriched type of splicing events, showing slightly decrease with disease progression (Fig. 3B). This observation is consistent with the decreased distribution of intron reads in CML samples (Fig. 1A), suggesting that introns were more frequently retained in PBMCs of the Ctrls rather than that of CML patients.
Fig. 3.
Survey of alternative splicing in chronic myeloid leukemia. (A) Detection of exon, SJ, and alternative splicing events by RNA-seq. Annotated exon, SJ, and alternative splicing events were identified by aligning RNA-seq reads as described in the materials and methods. Pie charts depict the proportions of the exon, SJ, and alternative splicing events in all the samples. (B) Classification of different types of alternative splicing events. The histogram depicts the percentage of each alternative splice type in five individuals of each group. 3pMXE, 3 prime mutual exclusion exon; 5pMXE, 5 prime mutual exclusion exon; A3SS, alternative 3 prime splicing site; A5SS, alternative 5 prime splicing site; ES: exon skipping; intronR, intron retention; MXE, mutual exclusion exon. (C) Known alternative splicing events in the FES proto-oncogene (FES) transcript. Alterations in reads coverage are indicated by line and number. Right panel showed the inclusion ratio of exon3 and exon13 in three groups.
Analysis of ASEs in PBMCs from CML patients and healthy control subjects revealed numerous, previously reported CML progression-related ASEs. Alternative splicing isoforms expression correlates with cell differentiation, leukemia stem cell generation [2], [7], [15], such as the c-FES (deletion of both exon 3 and exon1, Fig. 3C), GSK3β, ABL1, WT1, c-myc transcriptional repressor FIR (PUF60) genes and so on, were detected in most of samples (listed in the Tables 1 and S7). The detection of known AS events suggests that our method for AS analysis was feasible and the results were reliable.
Table 1.
The known ASEs with the malignant phenotype of leukemia.
| Gene symbol | Type | Funcation |
|---|---|---|
| FES | ES(exon 3 deletion) | cell differentiation in myeloid leukemia [2] |
| FES | ES(exon 11 deletion) | |
| ABL1 | CE(partial insertion of 35nt from intron 8) | imatinib resistance [37] |
| GSK3β | ES(exon 9 deletion) | leukemia stem cell generation [7] |
| GSK3β | ES(exon 11 deletion) | |
| WT1 | ES(exon 5 deletion) | the maintenance of a malignant phenotype in leukemias [15] |
| PUF60 | ES(exon 2 deletion) | colorectal carcinogenesis and leukemogenesis [38] |
Over a thousand CML-related ASEs occur commonly in the BP and CP stages
Phase-specific splice isoforms are of great importance, as they can be potent drivers of disease evolution. To identify CML progression-related AS events, we compared the splicing pattern of CML samples with Ctrl samples by Student's t-test (corrected P < 0.05) and found that 4,222 and 3,456 AS events exhibited a difference in the isoform ratio for CML-BP and CML-CP, respectively (Fig. 4A). It is noteworthy that 1,204 AS events from 1,047 genes were differentially spliced regardless of CML phase (Fig. 4A Tables S7, S8). By examining the ratio between the alternative and model isoforms of these differentially spliced events across all 15 samples, a heatmap was generated to display the distribution of splicing alterations (Fig. 4B). We observed a cluster of AS events showing a decreased alternative isoform ratio in both CML-BP and -CP samples (CML-loss), as well as a cluster specifically lost in BP samples (BP-loss). We also identified CP-gained and BP-gained AS clusters (Fig. 4B). In summary, BP samples showed a larger alternative splicing deregulation than CP samples, when compared with normal controls.
Fig. 4.
Identification of CML-associated alternative splicing events. (A) Venn diagram illustrates the differentially spliced AS events between two paired groups. ***P < 0.001, hypergeometric test. (B) Distinct splicing pattern of different samples. A hierarchical clustering dendrogram of differently spliced events. Red: relatively high ratio; Blue: relatively low ratio. The ratio values were normalized by row. (C) RT-qPCR experimental validation of differentially spliced events among CML-BP (n = 5), CML-CP(n = 5) and normal controls (n = 5). Same statistical method in Fig. 2D was used. (D) Bubble plot showing the top ten enriched KEGG pathways of genes with different AS events in three paired groups.
Among 28 significantly regulated AS events, we enabled the identification of the expected transcripts in the corresponding samples. Especially, their ratio of alternative splicing in these three groups was consistent with the expression trend of our sequencing analysis results, and 14 selected transcripts were confirmed be significantly regulated AS events(P < 0.05) (Fig. 4C).
Genes harboring CML-related ASEs are enriched in signaling and spliceosome pathways
To explore the biological functions of the genes whose AS events were associated with CML phases, KEGG annotations were applied. This analysis revealed that genes differentially spliced between CML-CP and Ctrls were enriched in 23 pathways (corrected P < 0.05). Four of the most enriched 10 pathways were signaling related, including phosphatidylinositol signaling system, mTOR signaling pathway, B cell receptor signaling pathway, TNF signaling pathway, and T cell receptor signaling pathway (Fig. 4D), therefore implying that genes involved in signal transduction were frequently and differentially spliced in CML-CP. As one of the genes involved in phosphatidylinositol signaling, PIP4K2A was required for acute myeloid leukemia cell proliferation and survival [16], while the constitutively active of mTOR signaling gene AKT depletes hematopoietic stem cells and induces leukemia in mice [17] (Fig. S2). Given that genes in these pathways are important mediators of cytokine signaling implicated in the regulation of hematopoiesis, their mis-splicing may give rise to signal transduction abnormalities in CML.
As for CML BP, 29 pathways were enriched with spliceosome as the most abundant representative (Fig. 4D). Well-known splice complex genes U2AF1, U2AF2, SF3B2, SF3B3 and SR family member SRSF1[18], as well as heterogeneous nuclear ribonucleoparticle protein-encoding genes HNRNPA1, HNRNPA3, HNRNPC, and HNRNPK were included in this pathway, implying that mis-splicing of genes implicated in spliceosome assembly and alternative splicing regulation may to some extent account for the disease progression. Epstein-Barr virus (EBV) infection emerged as the second-most enriched pathway. Given that several malignancies are induced by unknown interactions between this virus and the host B lymphocytes [19], and especially that detection of EBV is relatively common in leukemic children and is significantly associated with a decline in the overall survival [20], this finding may provide a clue for elucidating the vague relationship of EBV infection and leukemic progression. Besides, the T and B cell receptor signaling pathway, and proteasome were also significantly enriched. In addition, the functional clusters most highly correlated with disease phase (BP relative to CP) included spliceosome, nucleotide excision repair, proteasome, ubiquitin mediated proteolysis, RNA transport and cell cycle (Fig. 4D).
The longer splice isoform of hnRNPA1 is accumulated at higher levels in PBMCs from CML patients vs controls
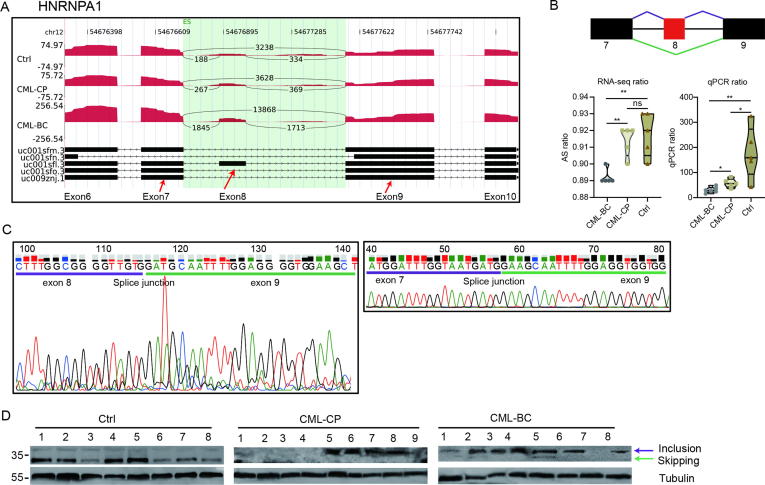
Alternative splicing deregulation of spliceosome pathways in BP samples indicates a key role of spliceosome deregulation in CML progression. We then chose hnRNPA1 to further study the deregulated alternative splicing of spliceosome pathway components in BP samples. Interestingly, our RNA-seq analysis revealed the skipped ratio of exon 8 (ES) was significantly decreased in BP samples (Fig. 5A). The ES ratio from RT-qPCR also showed that the ratio of exon 8 skipping decreased from control to CML-CP, from CML-CP to CML-BP (P < 0.05, Fig. 5B).
Fig. 5.
Validation of alternative splicing events of hnRNPA1. (A) Sashimi plots show AS events occurred in hnRNPA1. Reads density for alternative exon 8 were illustrated. Multiple transcripts for the gene are shown below. (B) Detection of exon 8 skipping of hnRNPA1. The schematic diagram depicts the skipping of exon 8 in hnRNPA1. The exons are denoted by black or red boxes and intron by horizontal line. RNA-seq quantification (left, exon skipping ratios) and RT-qPCR validation (right) of the AS events were shown below (**P < 0.01; *P < 0.05; t-test). (C) Validation of alternative splice junctions in hnRNPA1. Two splice junctions result in exon 8 inclusion or exclusion were shown. (D) Western blot analysis of hnRNPA1 protein isoforms. The two isoforms of hnRNPA1 were illustrated. Tubulin is a loading control.
We further used PCR method to amplify the hnRNPA1 cDNAs around the splice junction from clinical samples, and then sequenced the PCR products. This experiment confirmed that the splice sites coincided with those identified in RNA-seq results (Fig. 5C). To confirm the protein products of the two splice isoforms in this study, we performed Western blot assays using antibodies against the N terminus of hnRNPA1, which detected two protein isoforms, 38 and 34 kDa respectively (Fig. 5D). The experiments also showed that the smaller molecular weight hnRNPA1 isoform was predominant in Ctrls, which was largely diminished in CML patients. Instead, the larger hnRNPA1 isoform became predominant in CML patients, in all BP patient samples and some CP samples (Fig. 5D).
Discussion
The availability of TKIs marked a major advance in the treatment of CML by prolonging survival of patients across all stage. However, the effective diagnosis and treatment options are limted in advanced stages of the disease, which are hampered in part by factors that the complexity of genetic defects implicated in disease progression. Our RNA-seq analysis provides comprehensive perspective on alternatively spliced transcripts in CML and will be of great value in addressing the regulation of gene expression and function.
In this study, transcriptome-wide investigation of gene expression has revealed that samples were clearly distinguished by a clustering matrix based on PCA, indicating a large difference between control and two phases of CML samples. DEGs analysis identified more differentially expressed genes in the CP than BP when compared to Ctrl. The two groups shared a large common set of DEGs, revealing a dramatic and extensive change in gene expression during the chronic phase. In addition, DEGs of the two phases were enriched in highly similar functional pathways. Altogether these findings revealed that the differences in gene expression between chronic and blast phases comprise a qualitative change rather than a quantitative change, which quite contradicts to the previous report [21]. It is imperative that large series of cases should be included in further study to robustly validate the qualitative changes between CML-BP to CML-CP.
The potential mechanisms of disease transformation to AP/BC subdivided in ABL1 mutation–dependent, Bcr-abl independent molecular abnormality and changes of bone marrow microenvironment. The presence of specific isoforms resulting from the alternative splicing of genes has long been implicated in disease and therapy [22], [23], [24], [25]. In recent studies, IR was found as a pivotal type of splicing which regulated gene expression during granulopoiesis [26] and erythropoiesis [27]. Our study bolsters these findings by revealing that IR accounts for nearly a third of the ASEs across all samples and is much more common in PBMCs. Furthermore, Ctrls frequently exhibit marked increases in intronic reads and IR events relative to CML specimens. Additionally, differentially spliced variants are more prevalent in normal PBMCs of Ctrls rather than CML specimens. Similar observations have also been reported in breast cancer, which exhibits a decreased IR relative to normal controls, counter to what is found in acute myeloid leukemia [28].
Spliceosome pathways were the most enriched functional cluster of genes showing differential alternative splicing between the CML-BP and CML-CP patients. Given that spliceosomal mutations have already been found in diverse malignancies [29] and that a subset of spliceosomal components are intimately associated with cancers [30], the finding of mis-spliced genes necessary for spliceosomal machinery may be of great significance for CML research because it suggests that the splicing components orchestrating pre-mRNA splicing process may differ substantially between cancer patients and Ctrls. Mutation of SF3B1 establishes aberrant 3′splicing site selection and different branch point sequence utilization to induce aberrant splicing across multiple cancer types [31], while recurrent mutation in U2AF1 in de novo myelodysplastic syndrome patients suggests an increased risk of progression to secondary acute myeloid leukemia [32]. In our study, spliceosome genes such as SNRPA1, SF3B2, and HNRNPA1 were differentially spliced between CML-BP and CML-CP. Consequences of splicing aberration include the emergence of novel transcript variants, protein isoforms, and dysregulation at the gene and protein levels, all of which can affect global splicing through splice factors. However, the effects of aberrant splicing in splice factors themselves are largely unknown.
In this study, we focused on clinical PBMCs rather than CML cell lines, which was beneficial to the mining of clinical biomarkers. Our study therefore expanded the scope of samples for CML research. The identification of two distinct hnRNPA1 protein isoforms corresponding to the exclusion and inclusion of exon 8 in PBMC samples in this study was consistent with previous reports [33], [34]. Previous studies have reported the existence of two experimentally validated protein isoforms of hnRNPA1, the full-length A1-b isoform (hnRNPA1-b) of 372 aa with a molecular mass of 38 kDa, and the shorter 320 aa 34 kDa A1-a variant (hnRNPA1-a), which is missing residues 253 to 303 [23]. These two isoforms were produced by alternative inclusion/exclusion of the exon 8 [24], [25], [26]. Exclusion of exon 8 would result in the production of a shorter transcript by 156 nucleotides, thereby potentially causing an in-frame deletion that removes 52 amino acids (5 kDa) at the protein level. We found that the accumulation of hnRNPA1-b isoform was highly specific in CML-BP patients, and also occurred in some CML-CP patients. On the contrary, the hnRNPA1-a is more specific to Ctrls, agreeing with a previous report that it is over 20-times more abundant than the full-length protein in most tissues [33]. The result reminded that the hnRNPA1 alternative splicing shown to be associated with disease progression of CML, and the expression level of hnRNPA1-b subtype also was early signs of CML-CP to CML-AP/BP in the future. Undeniably, hnRNPA1 accompanies eukaryotic pre-mRNA and mRNA metabolism, include in nascent pre-mRNA in a sequence-specific manner, promoting the annealing of cRNA strands, and regulating splice site selection, exon skipping or inclusion, nuclear export of mature mRNAs, mRNA turnover, and translation [35]. Furthermore, Danilo Perrotti’s group has overwhelmingly demonstrated a role for hnRNP A1 in CML-BP, which effected on the nucleocytoplasmic trafficking of mRNAs that encode factors essential for survival and differentiation [36]. According to the Radich leukemia Oncomine dataset, there were no differences about the hnRNPA1 expression level between CML and other hematologic malignancies, and also between relapse and remission CML patients of TKI treatment (Fig. S3A,B); However, the hnRNPA1 mRNA level in CML-BP was significantly higher than that in CML-CP/AP (P < 0.001) (Fig. S3C). The above data further confirmed the uniqueness of the hnRNPA1 in CML-BP, that is not related to treatment but only to disease progression. Although the correlation of hnRNPA1 in the progression of CML to AP/BC was early concerned, the mechanism was not deeply explored, then our research might be a new entry point. Our finding may represent the first report of the involvement of these two hnRNPA1 isoforms in disease progression, and suggests its clinical significance as biomarkers for indicating leukemic progression.
In conclusion, we revealed the transcriptome profile of PBMCs from CML patients. Our study provides new insights into gene expression and alternative splicing patterns in CML by revealing that the mis-splicing of genes involved in signaling and spliceosome pathways may have great biological relevance to disease progression. An auxiliary splicing factor hnRNPA1 was screened out as a biomarker which could distinguish the CML patients and Ctrls. The full-length splice isoform hnRNPA1b was accumulated at high levels in the PBMCs of CML-BP patients, and large sample validation is required at a later stage. Although the biological consequences of this isoform switch remain unclear, the distinct expression pattern of hnRNPA1 may provide a novel prognostic marker and therapeutic target.
Compliance with Ethics Requirements
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). Informed consent was obtained from all patients for being included in the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant No. 81360083, No. 81271912, No. 81560033 and No. 81860034. This work was also supported by grant from the Appreciate the Beauty of Life, Inc. (ABL2014-04012 to Y.Z.). We are very grateful to Dr. John Snyder and Isaac V. Greenhut for their professional editing on the language and discussion of the structure of this paper.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.04.016.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Goldman J.M., Melo J.V. Chronic myeloid leukemia—advances in biology and new approaches to treatment. N Engl J Med. 2003;349(15):1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- 2.Carlson A., Berkowitz J.M., Browning D., Slamon D.J., Gasson J.C., Yates K.E. Expression of c-Fes protein isoforms correlates with differentiation in myeloid leukemias. DNA Cell Biol. 2005;24(5):311–316. doi: 10.1089/dna.2005.24.311. [DOI] [PubMed] [Google Scholar]
- 3.Apperley J.F. Chronic myeloid leukaemia. The Lancet. 2015;385(9976):1447–1459. doi: 10.1016/s0140-6736(13)62120-0. [DOI] [PubMed] [Google Scholar]
- 4.Melo J.V., Barnes D.J. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- 5.Makishima H., Visconte V., Sakaguchi H., Jankowska A.M., Abu Kar S., Jerez A. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119(14):3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Blanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22(5):535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 7.Abrahamsson A.E., Geron I., Gotlib J., Dao K.H., Barroga C.F., Newton I.G. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. PNAS. 2009;106(10):3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Q., Crews L.A., Barrett C.L., Chun H.J., Court A.C., Isquith J.M. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. PNAS. 2013;110(3):1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goff D.J., Court Recart A., Sadarangani A., Chun H.J., Barrett C.L., Krajewska M. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013;12(3):316–328. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm F., Hellqvist E., Mason C.N., Ali S.A., Delos-Santos N., Barrett C.L. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. PNAS. 2015;112(50):15444–15449. doi: 10.1073/pnas.1506943112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia H., Chen D., Wu Q., Wu G., Zhou Y., Zhang Y. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim Biophys Acta (BBA)-Gene Regulat Mech. 2017 doi: 10.1016/j.bbagrm.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Renshaw J., Orr R.M., Walton M.I., Te Poele R., Williams R.D., Wancewicz E.V. Disruption of WT1 gene expression and exon 5 splicing following cytotoxic drug treatment: antisense down-regulation of exon 5 alters target gene expression and inhibits cell survival. Mol Cancer Ther. 2004;3(11):1467–1484. [PubMed] [Google Scholar]
- 16.Jude J.G., Spencer G.J., Huang X., Somerville T.D.D., Jones D.R., Divecha N. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene. 2014;34(10):1253–1262. doi: 10.1038/onc.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharas M.G., Okabe R., Ganis J.J., Gozo M., Khandan T., Paktinat M. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 2010;115(7):1406–1415. doi: 10.1182/blood-2009-06-229443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Seto E., Moosmann A., Gromminger S., Walz N., Grundhoff A., Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loutfy S.A., Abo-Shadi M.A., Fawzy M., El-Wakil M., Metwally S.A., Moneer M.M. Epstein-Barr virus and cytomegalovirus infections and their clinical relevance in Egyptian leukemic pediatric patients. Virol J. 2017;14(1):46. doi: 10.1186/s12985-017-0715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radich J.P., Dai H., Mao M., Oehler V., Schelter J., Druker B. Gene expression changes associated with progression and response in chronic myeloid leukemia. PNAS. 2006;103(8):2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garciablanco M.A., Baraniak A.P., Lasda E.L. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22(5):535. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 23.Venables J.P., Klinck R., Koh C., Gervais-Bird J., Bramard A., Inkel L. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16(6):670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 24.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Weiss W.A. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34(1):1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 26.Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154(3):583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 27.Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016;44(2):838–851. doi: 10.1093/nar/gkv1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dvinge H., Bradley R.K. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7(1):45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong Y., Krauthammer M., Halaban R. Rare SF3B1 R625 mutations in cutaneous melanoma. Melanoma Res. 2014;24(4):332–334. doi: 10.1097/CMR.0000000000000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525(7569):384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darman R.B., Seiler M., Agrawal A.A., Lim K.H., Peng S., Aird D. Cancer-associated SF3B1 hotspot mutations induce cryptic 3' splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033–1045. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 32.Graubert T.A., Shen D., Ding L., Okeyo-Owuor T., Lunn C.L., Shao J. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jean-Philippe J., Paz S., Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14(9):18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66(7):1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buvoli M., Cobianchi F., Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20(19):5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iervolino A., Santilli G., Trotta R., Guerzoni C., Cesi V., Bergamaschi A. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol. 2002;22(7):2255–2266. doi: 10.1128/mcb.22.7.2255-2266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laudadio J., Deininger M.W., Mauro M.J., Druker B.J., Press R.D. An intron-derived insertion/truncation mutation in the BCR-ABL kinase domain in chronic myeloid leukemia patients undergoing kinase inhibitor therapy. J Mol Diagnost: JMD. 2008;10(2):177–180. doi: 10.2353/jmoldx.2008.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsushita K., Kitamura K., Rahmutulla B., Tanaka N., Ishige T., Satoh M. Haploinsufficiency of the c-myc transcriptional repressor FIR, as a dominant negative-alternative splicing model, promoted p53-dependent T-cell acute lymphoblastic leukemia progression by activating Notch1. Oncotarget. 2015;6(7):5102–5117. doi: 10.18632/oncotarget.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the results of this article are available in NCBI’s gene Expression Omnibus and are accessible through GEO series accession number GSE100026.