Abstract
Heme catabolism is an important biochemical process that many bacterial pathogens utilize to acquire iron. However, tetrapyrrole catabolites can be reactive and often require further processing for transport out of the cell or conversion to another useful cofactor. In previous work, we presented in vitro evidence of an anaerobic heme degradation pathway in Escherichia coli O157:H7. Consistent with reactions that have been reported for other radical S-adenosyl-L-methionine methyltransferases, ChuW transfers a methyl group to heme by a radical-mediated mechanism and catalyzes the β-scission of the porphyrin macrocycle. This facilitates iron release and the production of a new linear tetrapyrrole termed “anaerobilin”. In this work, we describe the structure and function of ChuY, an enzyme expressed downstream from chuW within the same heme utilization operon. ChuY is structurally similar to biliverdin reductase and forms a dimeric complex in solution that reduces anaerobilin to the product we have termed anaerorubin. Steady state analysis of ChuY exhibits kinetic cooperativity that is best explained by a random addition mechanism with a kinetically preferred path for initial reduced nicotinamide adenine dinucleotide phosphate binding.
Graphical Abstract
Iron is an essential nutrient that pathogenic bacteria must acquire during infection to ensure their survival in a mammalian host. The ability to acquire iron, either directly or from iron-protoporphyrin IX, contributes to the virulence and pathogenesis of the organism.1,2 Iron is a reactive element, and the transport and storage mechanisms for iron are tightly regulated in eukaryotes. Therefore, the ability to scavenge heme from the host organism and further degrade it to acquire additional iron for cellular processes represents a significant competitive advantage for the pathogen.3,4 The canonical pathway for enzymatic heme degradation is initiated by the conversion of heme to biliverdin IXα by heme oxygenase.5,6 In eukaryotic systems, biliverdin is further reduced at the meso carbon by a biliverdin reductase prior to glucuronidation and removal from the cell.7 Although the entire canonical pathway is not found in proteobacteria, bacterial heme oxygenases have been identified.8–10 In many microorganisms, the degradation products are used as precursors for the chromophore in phytochromes that act as photosensors for light-dependent gene regulation.11–13 Moreover, mechanisms must be in place to avoid accumulation of reactive catabolites to prevent cellular damage.
Previous work from our group has provided evidence that ChuW from the enterohemorrhagic Escherichia coli O157:H7 is a radical S-adenosyl-L-methionine (SAM) methyltransferase that functions in anaerobic heme degradation.14 The product of ChuW-catalyzed heme degradation is a photoreactive tetrapyrrole that we termed “anaerobilin”. Given that the sequence of ChuY is somewhat homologous to that of biliverdin reductase (BVR) but is void of BVR activity, it stood to reason that the physiological function of ChuY could be to catalyze the reduction of anaerobilin to a less reactive metabolite. In this work, we provide evidence consistent with this hypothesis. Steady state analyses of ChuY suggest the enzyme exhibits .0kinetic cooperativity that is best explained by a random association model with a kinetically preferred path for reduced nicotinamide adenine dinucleotide phosphate (NADPH) binding prior to the tetrapyrrole. The X-ray crystal structure of ChuY, determined to 2.1 Å resolution, reveals several features consistent with the proposed biochemical function and other biochemical properties of the enzyme.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Enzymatic Assay of ChuY.
ChuY was expressed by growing the E. coli BL21 DE3 strain containing an isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible expression plasmid that carried the gene encoding ChuY with a six-His tag (pET15a). In short, a 20 mL starter culture was grown for 6 h before six 1 L flasks containing TB media were inoculated. Cultures were grown at 37 °C while being shaken at 200 rpm until the OD at 600 nm reached 0.8. IPTG was added to a final concentration of 500 μM. Cultures were incubated at 18 °C overnight while being shaken at 200 rpm and harvested by centrifugation before being stored at −80 °C. The frozen cell pellets were solubilized in buffer containing 20 mM Tris (pH 8.1), 300 mM KCl, and 10% glycerol with 0.05 mM PMSF, 0.05 mg/mL lysozyme, and 50 μg/mL DNase. Cells were lysed with a French pressure cell, and the lysate was centrifuged at 65000g for 1.5 h. The supernatant was applied to Talon resin, washed with 2 column equivalents of water, and 8 column equivalents of solubilization buffer. After the column had been washed with 3 column equivalents of 10 mM imidazole, ChuY was eluted with buffer containing 250 mM imidazole. Fractions containing ChuY were pooled, buffer exchanged to remove imidazole, and then concentrated. The protein was analyzed by sodium dodecyl sulfate–polyacryla-mide gel electrophoresis, Biuret analyses, and absorption at 280 nm before aliquots were frozen at −80 °C.
Crystallization, Data Collection, Phasing, Model Building, and Refinement of ChuY.
Diffraction quality crystals of ChuY were obtained when purified enzyme (1 μL at 15 mg/mL) was mixed in a 1:1 ratio with a precipitating solution containing 10% 1,4-butanediol, 100 mM Bis-Tris buffer (pH 5.5), 19% PEG 3350, and 150 mM lithium sulfate (80 μL reservoir) in a hanging drop experiment. Crystals appeared within 48 h after incubation at 18 °C. Monochromatic data collection with 0.98 Å X-rays was performed at SER-CAT on the 22-BM beamline at the Advanced Photon Source (Argonne, IL). Data were processed using SGXPRO, and 5% of the reflections were set aside for cross-validation (Table 1).15 A BLAST search against the Protein Data Bank (PDB) structures revealed that the structure of a Rossmann-fold NAD(P)-binding family protein from Shigella flexneri was 89% similar to that of ChuY. A polyalanine model of this structure (PDB entry 3QVO) was used as a successful molecular replacement model for initial phasing using PHENIX.16 Successive rounds of model building and refinement were performed using COOT17,18 and PHENIX.16
Table 1.
Data Collection and Refinement Statistics for ChuY
| PDB entry | 5FFQ |
| space group | P21 |
| wavelength | 0.98 |
| resolution range (Å) | 50.0–2.0 |
| outer shell | 2.07–2.0 |
| no. of unique observations | 30647 |
| completeness (%) | 98.9(90.3)a |
| Rsym (%)b | 0.08(0.36) |
| redundancy | 3.8(2.3) |
| I/σ | 14.3(2.8) |
| unit cell dimensions | |
| a (Å) | 50 |
| b (Å) | 63 |
| c (Å) | 66 |
| α (deg) | 90 |
| β (deg) | 106 |
| γ (deg) | 90 |
| no. of protein atoms | 3087 |
| no. of solvent atoms | 306 |
| resolution limits (Å) | 50.0–2.0 |
| Rcryst (%) | 17.8 |
| Rfree (%) | 23.9 |
| rmsd for bonds (Å) | 0.008 |
| rmsd for angles (deg) | 1.07 |
| average B factor (Å2) | 27.3 |
Numbers in parentheses denote values for the outermost resolution shell.
Rsym = Σhkl[ΣI(|Ihkl,I − ⟨Ihkl⟩|)]/Σhkl,I⟨Ihkl⟩, where Ihkl is the intensity of an individual measurement of the reflection with indices hkl and 〈Ihkl〉 is the mean intensity of that reflection.
Sedimentation Velocity.
ChuY (1 mM) was dialyzed into 25 mM HEPES (pH 7.5) and 200 mM KCl. After dialysis, 400 μL of a diluted sample (24 μM) or reference buffer solution was loaded into 12 mm double-sector Epon centerpieces equipped with quartz windows and equilibrated at 20 °C in an AN60 Ti rotor for 1 h. Sedimentation velocity studies were performed in an Optima XLA analytical ultracentrifuge using a rotor speed of 50000 rpm at 20 °C. Data were recorded at a wavelength of 280 nm using a radial step size of 0.003 cm. The partial specific volume, density, and viscosity were calculated to be 0.73478 mL/g, 1.00961 g/mL, and 0.01017 P, respectively, using SEDNTERP.19 SEDFIT20 was used to model and analyze the sedimentation data. Modeled data were fit as a continuous sedimentation coefficient c(s) distribution using the baseline, meniscus, frictional coefficient, systematic time-invariant noise, and radial-invariant noise. The weight-average sedimentation coefficient (sw) for each species was determined by integrating each peak in the c(s) distribution. The theoretical sedimentation coefficient (S) values were calculated with HYDROPRO21 using the ChuY atomic coordinates. The root-mean-square deviation (rmsd) value for the experiment was <0.002 OD.
Mass Spectrometry of Anaerorubin.
Deuteroanaerobilin and anaerobilin were prepared as previously described.14 Isolated substrates (50 μM) were mixed with 200 μM NADPH and 5 μM ChuY. After 1 h, the reaction mixture was applied to a C1-sep pack (Silicycle) before being washed with 1 mL of methanol, water, and then reaction buffer. The column was washed with 5 mL of water before being eluted in methanol. Products were analyzed using nanospray ionization mass spectrometry (NSI-MS) by direct infusion into a linear ion trap mass spectrometer (LTQ-Orbitrap Discovery, Thermo Fisher Scientific) using a nanoelectrospray source at a syringe flow rate of 0.40 μL min−1 and a capillary temperature set to 210 °C. Fragmentation by collision-induced dissociation (CID) in MS/MS and MSn was used with a normalized collision energy of 35–40%.
Steady State Kinetics Determined by ChuY.
Typical UV–vis assays were performed using a 3 mL quartz cuvette prepared anaerobically and sealed with a rubber septum. Deuteroanaerobilin, prepared as previously described,14 and NADPH were added to a buffer containing 20 mM Tris (pH 8.1), 300 mM KCl, and 10% glycerol for a 2 mL reaction. Assays were initiated by injecting isolated ChuY using a gastight syringe. Spectra were recorded using an Agilent 8453 diode array spectrophotometer with a Peltier temperature controller set to 25 °C and a glass slide to reduce the level of exposure of DAB to ultraviolet light. Spectra were recorded in 1 s intervals for 15 min. Activity was determined by monitoring initial changes at 439 and 780 nm using extinction coefficients previously reported. Data were fit using nonlinear regression in PRISM (GraphPad Software Inc., San Diego, CA). The substrate saturation curves were fit to either a hyperbolic or a sigmoidal model based on residual analysis (for hyperbolic kinetics, h = 1):
| (1) |
The deuteroanaerobilin substrate curves of ChuY revealed substrate inhibition and were fit to
| (2) |
Equilibrium Binding Assays.
Intrinsic tryptophan fluorescence quenching of ChuY was monitored to assess the binding of NADPH. Fluorescence binding titrations were performed at 22 °C in reaction buffer [20 mM Tris (pH 8.1), 150 mM KCl, and 5% glycerol] containing 400 nM ChuY. Fluorescence was measured using a Shimadzu RF-5301 spectrofluorometer with excitation and emission wavelengths of 280 and 354 nm, respectively, and slit widths set to 5 mm, and a glass slide was used in the emission path. Raw data were corrected for the inner filter effect as previously described.22 Because of the moderate affinity for NADPH (<4 μM), we used the following relationship to calculate unbound NADPH (NADPHfree):
| (3) |
| (4) |
| (5) |
The corrected ΔF was plotted versus the free NADPH concentration. The dissociation constant (Kd) and the cooperativity were determined by fitting the data to a sigmoidal model based on residual analysis:
| (6) |
RESULTS
Crystal Structure of Apo-ChuY.
ChuY is associated with the heme utilization operon and is downstream from chuW (Figure 1). Bioinformatic analysis of the ChuY sequence suggests that the enzyme is a NAD(P)H oxidoreductase. The native crystal structure was determined to 2.1 Å resolution by molecular replacement using an NAD(P)H oxidoreductase from Shigella flexneri (PDB entry 3QVO) as the starting model (Table 1). The asymmetric unit consists of two monomers related by a 2-fold symmetry axis (Figure 2A). The core structure contains a Rossman fold with seven β sheets sandwiched by three α helices on each side. The surface representation of the fold reveals a large pocket that, presumably, binds the substrates. The majority of the electron density for ChuY crystals is well-defined with the exception of a glycine-rich loop near the predicted location of NADPH binding. The B factors are also consistent with a flexible loop that may stabilize once NADPH is bound. After all the protein molecules had been placed, the difference (Fo − Fc) map indicated that a molecule significantly larger than water was interacting with the side chain of R27. The density also suggested a tetrahedral structure, and therefore, phosphate was modeled in the density. After further refinement, B factors throughout the molecule averaged 40 Å2 with subtle differences between the oxygen and phosphorus atom of the phosphate anion. However, considering that lithium sulfate was used as an additive in the crystallization condition, the density could also be modeled as sulfate with similar refinement statistics.
Figure 1.
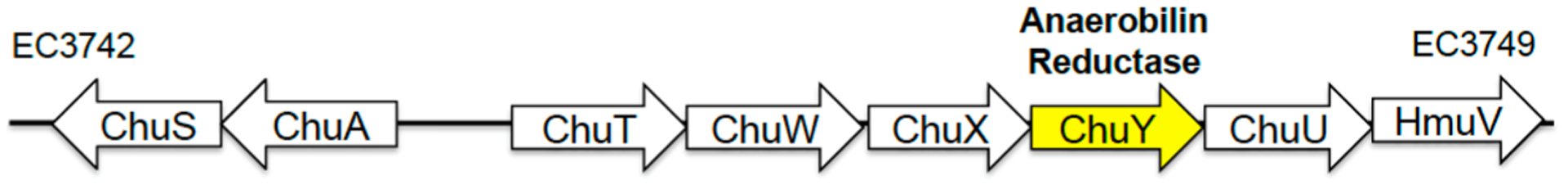
Organization of the heme uptake and utilization operon in E. coli O157:H7. The enzyme of interest to this work is the product of the chuY gene that we show to be an anaerobilin reductase.
Figure 2.
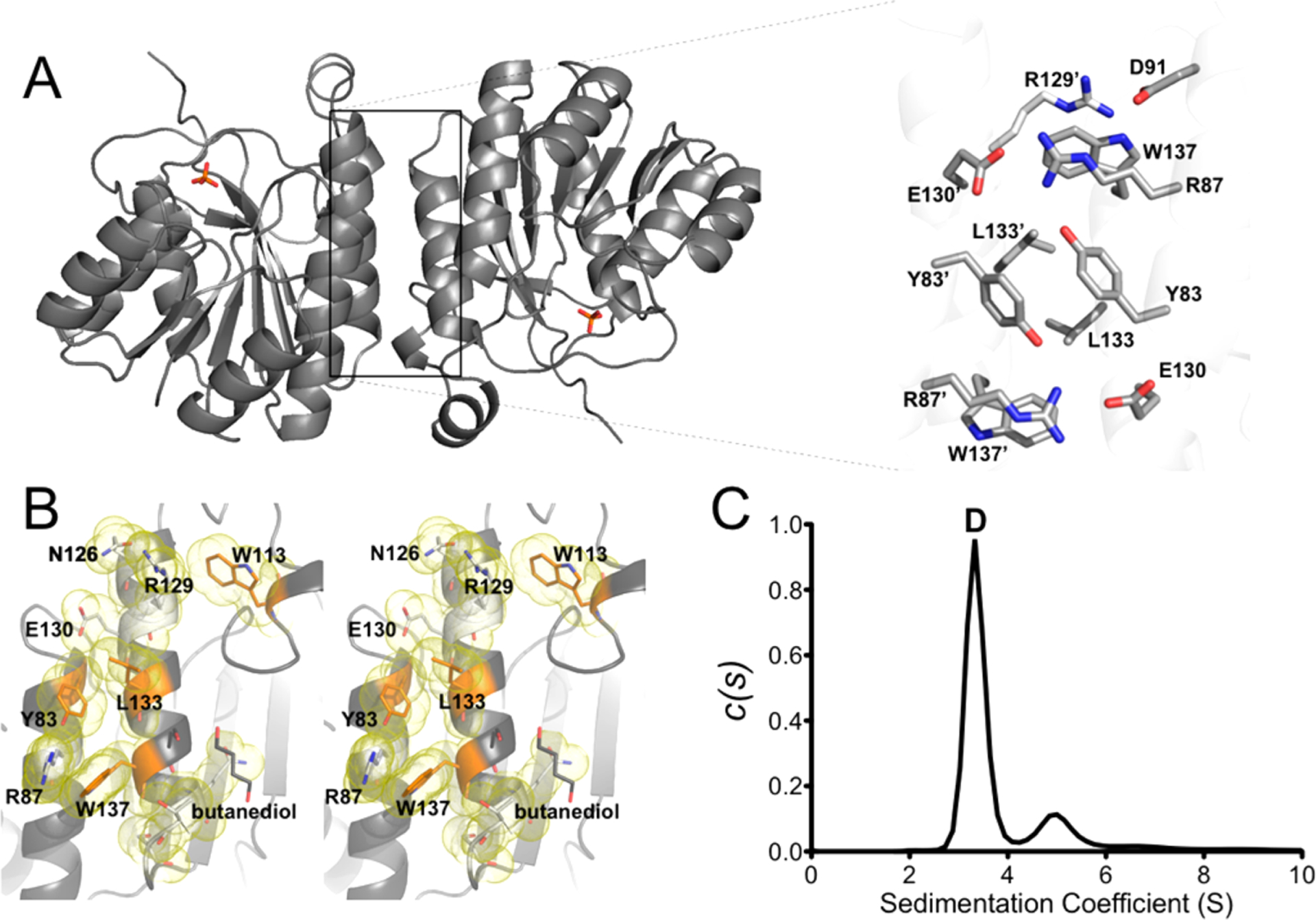
(A) Cartoon representation of the ChuY model with an expanded view of the dimer interface residues and a number of interface side chains shown as sticks and labeled. (B) Wall-eyed stereoview for one interface with the interface residues colored orange and their respective van der Waals surface shown as a yellow cloud. Amino acids that are significant for the dimer interface (see Table 2) are labeled, and side chains originating from different subunits are identified with an apostrophe. (C) c(s) sedimentation distribution of ChuY revealing a dominant species at 3.4 S (90%) and a species at 5.0 S (10%) consistent with a dimer and a tetramer, respectively.
ChuY Is a Dimer in Solution.
The dimeric interaction is strongest in the noncrystallographic symmetry contact within the asymmetric unit. PISA analysis calculated this dimer interface to be 672.9 Å2 with a free energy of −8.8 kJ mol−1. The interface is composed of hydrogen bonds and salt bridges with only three nonpolar residues from each monomer contributing to the interaction. Specific residues involved in the electrostatic interactions and residues buried within the dimer interface are described in Table 2. Despite the calculated surface area and free energy of dimerization, the free energy P value associated with the interface specificity is 0.554 (values of <0.5 represent a likely specific interaction) and suggested a nonspecific interface interaction. To directly assess the oligomeric structure of ChuY, we performed sedimentation velocity analytical ultracentrifugation to determine the dominant species in solution (Figure 2C). After comparison of the predicted sedimentation coefficient calculated by HYDROPRO21 (3.39 S), the observed coefficient value of 3.36 S confirms ChuY is a stable dimer in solution despite the appearance of a weak interface in the crystal structure. We do observe some tetrameric species at approximately 4.9 S, but upon further inspection of the packing observed in the crystal lattice, no additional evidence was seen to support such an oligomeric structure. While this work was under review, the structure for a homologous ChuY enzyme from another pathogenic E. coli was reported.23 Although the authors indicated that their enzyme existed as a monomer in solution, we found our dimer within the unit cell of their crystallographic model (PDB entry 5GUY).
Table 2.
| Electrostatic Interactions between Chains | |||
|---|---|---|---|
| Arg87 (NH1 and NH2) ↔ Glu130 (Oε1) | |||
| Arg87 (NH2) ↔ Glu130 (Oε2) | |||
| Buried Residues | |||
| Tyr83 | Leu84 | Ile90 | Asp91 |
| Glu94 | Gly110 | Trp113 | Gln126 |
| Thr132 | Leu133 | Ser136 | Trp137 |
| Gln139 | Thr140 | Ser141 | Gln194 |
ChuY Is Structurally Homologous to Biliverdin IXβ Reductases.
ChuY belongs to the atypical short chain dehydrogenase (aSDR) protein family. This family of enzymes lacks some of the conserved residues that would otherwise compose the catalytic tetrad responsible for catalysis in the aldehyde NADPH oxidoreductases.24 This classification makes it difficult to determine function based on the primary sequence or fold. However, the sequence of ChuY suggests homology to biliverdin IX β-reductase (BVβR). Thus, we aligned the backbone α-carbon atoms of the BVβR–NADP model (PDB entry 1HE3) with our native ChuY structure (PDB entry 5FFQ) (Figure 3A). After performing the structural alignment and displaying only the NADP+ molecule from the BVβR structure as well as the electrostatic representation of the ChuY model, we see positively charged surface residues that could interact with the negative charges of the nicotinamide cofactor (Figure 3B). This suggests a similar binding mode for NADPH in ChuY. In fact, consistent with what has been observed for BVβR, there is a lysine residue following two glycines that likely coordinate the 2′-phosphate of NADPH when bound. This may suggest that ChuY is in an open conformation in our crystallographic model.
Figure 3.
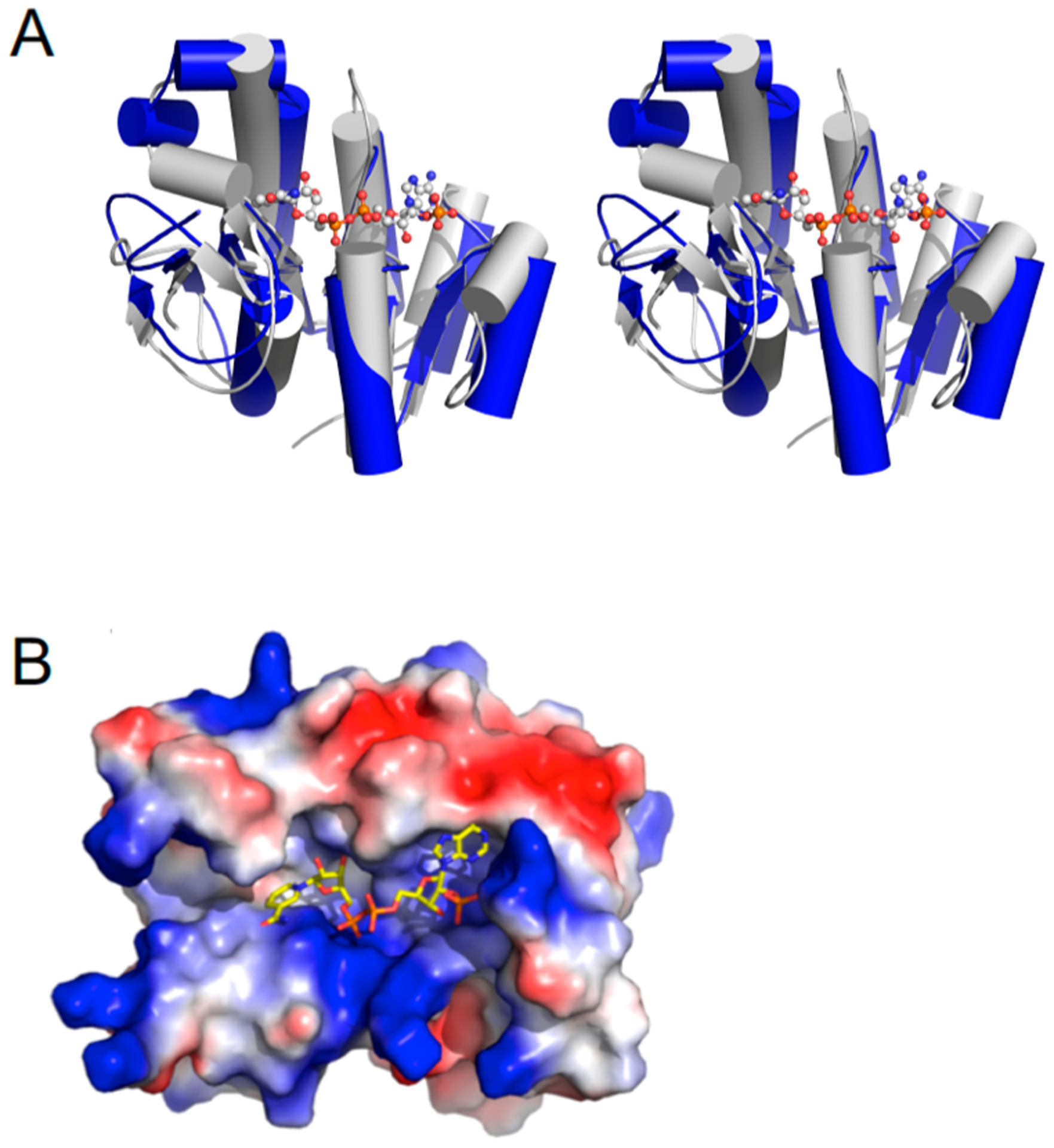
Structural alignment of ChuY with biliverdin β reductase (PDB entry 1HE3). (A) Wall-eyed stereoview showing a cartoon representation of ChuY (blue) aligned with biliverdin β-reductase (gray) by the Cα atoms that resulted in a rmsd of 3.06 Å2. (B) Same overlay showing only the cofactor from the biliverdin reductase model and the electrostatic surface representation of the ChuY model.
Characterization of Anaerorubin and Deuteroanaerorubin.
As we have previously reported,14 both of the substrates for ChuY, specifically anaerobilin (product of heme turnover by ChuW) and deuteroanaerobilin (DAB, product of deuteroheme turnover by ChuW), are light sensitive compounds. Therefore, as a precaution, the products of ChuY turnover with either substrate were also handled anaerobically and in the dark to prevent any potential light- or oxygen-driven modification of the tetrapyrrole. In both cases, turnover of anaerobilin or DAB by ChuY resulted in a significant decrease in absorption in the 400–800 nm visible range.14 To provide additional insight into the reaction being catalyzed by ChuY, we analyzed both reaction products, anaerorubin or deuteroanaerorubin (DAR), using electrospray ionization mass spectrometry. DAR resulted in a dominant mass peak at m/z 529 with an additional signal at m/z 531 (Figure 4A). Both masses are in agreement with a reduction reaction; however, it is unclear if two subsequent reductions are occurring for the larger mass. ChuY also reduces anaerobilin, as demonstrated by the MS spectra of isolated anaerorubin, which indicates a four-electron reduction (Figure 5). This product is likely from two subsequent reductions.
Figure 4.
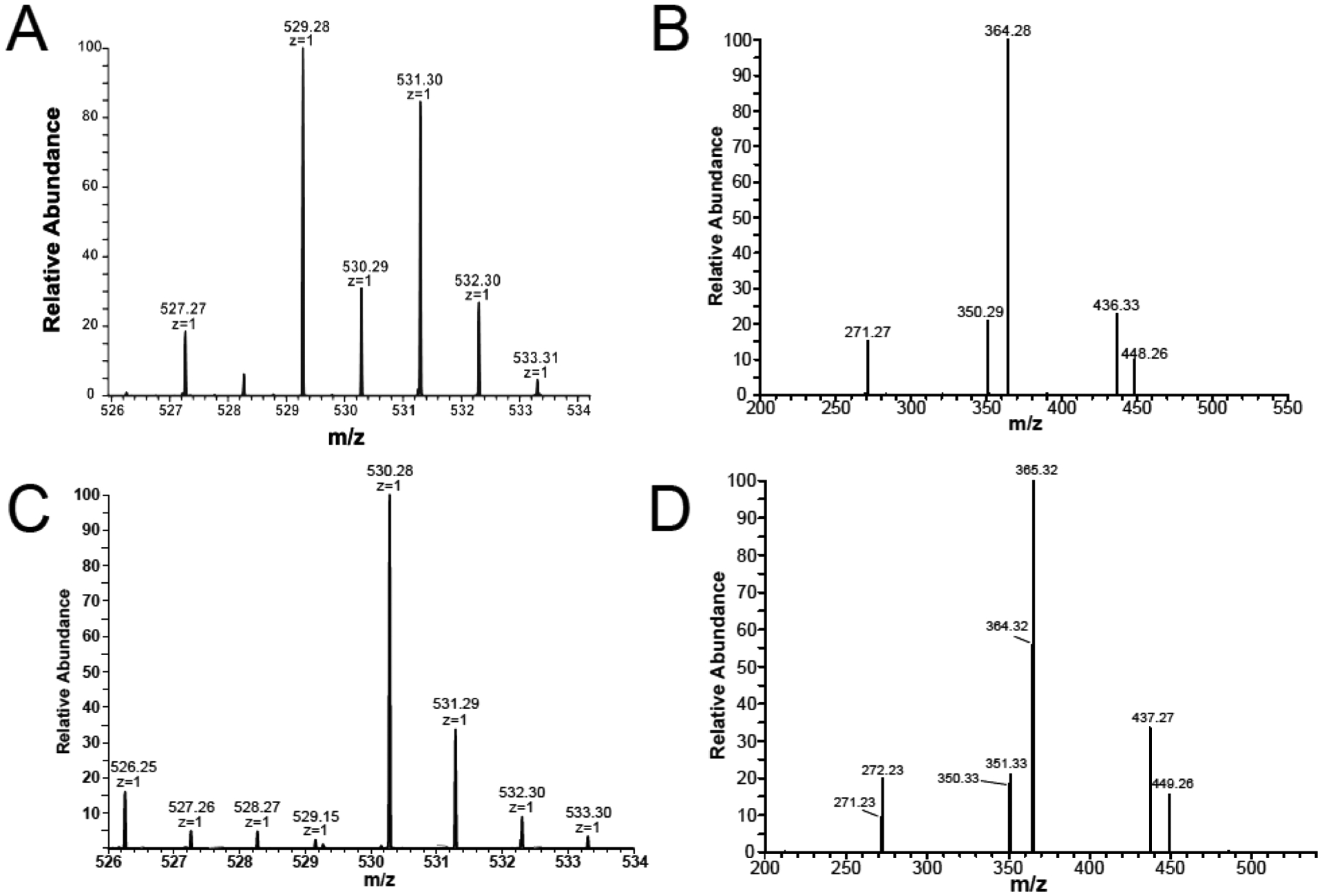
Mass spectrometry of deuteroanaerorubin (DAR). (A) MS analysis of deuteroanaerorubin (DAR) resulted in a [M + H]+ mixture of m/z 529 and 531. (B) MS/MS fragmentation of DAR revealed a dominant fragment at m/z 364 corresponding to the loss of a pyrrole ring. (C) MS of [methyl-13C]DAR attained by reacting ChuY with DAB prepared using [methyl-13C]SAM indicated retention of the isotopic label with a dominant [M + H]+ of m/z 530. (D) Fragmentation of the isotopically labeled DAR resulting in a dominant species at m/z 365 and 364 indicating that the label could be lost upon fragmenting DAR.
Figure 5.
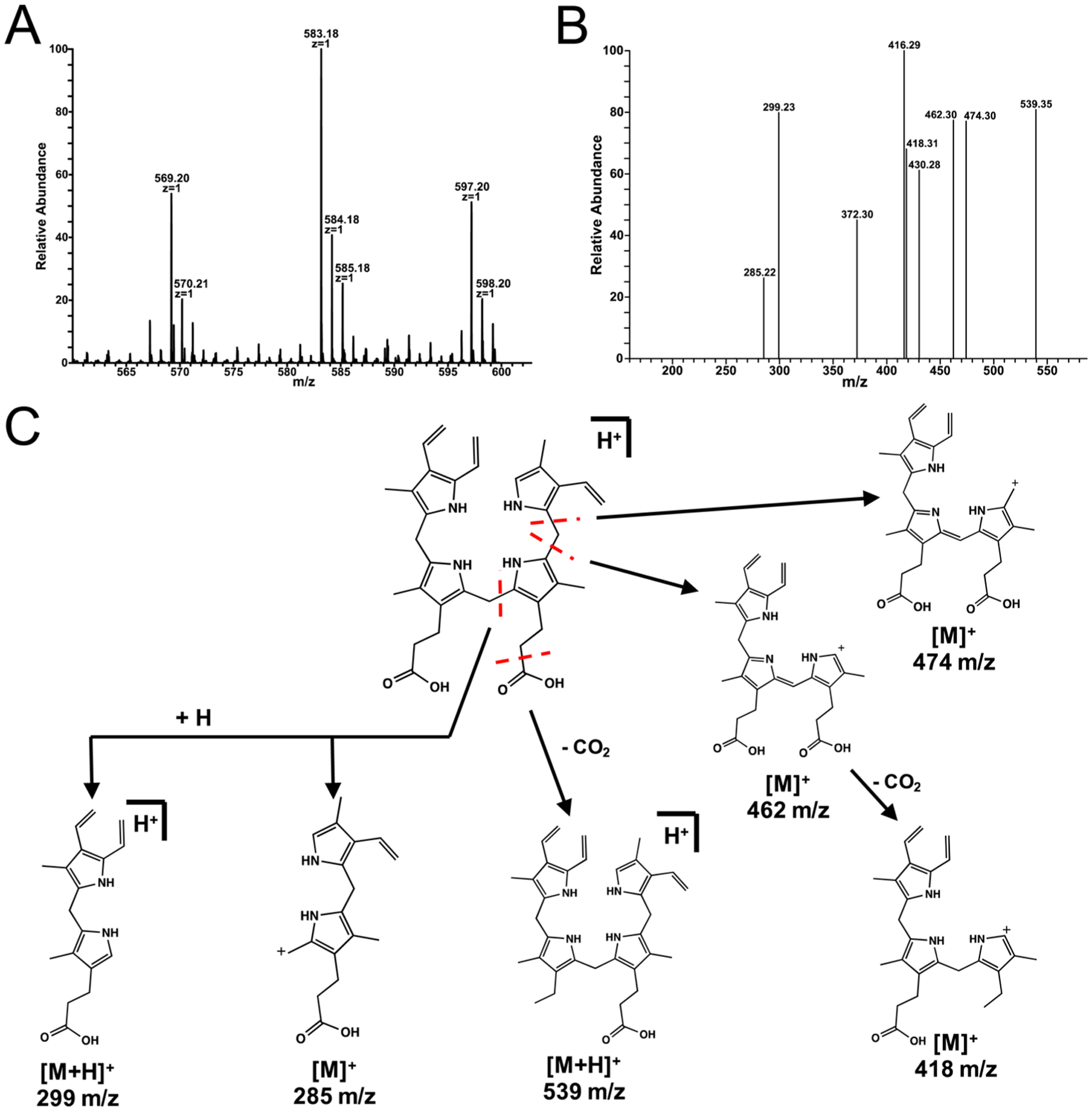
Mass spectrometry of anaerorubin was performed by isolation of the product of ChuY (2 μM) turnover with 30 μM anaerobilin as the substrate in the presence of excess NADPH. (A) Isolated products were analyzed by NSI-MS by direct infusion and revealed an [M + H]+ at m/z 583. (B) MS/MS of anaerorubin revealed multiple fragments corresponding to the reduction of the tetrapyrrole. (C) Proposed fragmentation products for anaerorubin.
The MS/MS spectrum of the m/z 529 peak associated with DAR indicates the reduction occurs at the β- or δ-meso position, according to the conventional naming scheme for a porphyrin (Figure 4C). Moreover, the observation of an even mass fragment at m/z 364 indicates an odd number of nitrogens are present. This can be explained by the reduction of either the β- or δ-meso carbon atoms and fragmentation at one of the bridging bonds, resulting in a molecule that contains only three pyrrole rings. To probe the structure of DAR further, we used [methyl-13C]-S-adenosylmethionine ([methyl-13C]SAM) to prepare the deuteroanaerorubin substrate with the isotopic label incorporated (Figure 4B). ChuY turnover of the labeled substrate results in an [M + 1] m/z 530 peak confirming incorporation of the isotopic label. Upon fragmentation of [methyl-13C]anaerorubin, we observed retention of approximately 66% of the isotopic label and 33% without, relative to the fragmented peak (m/z 365 and 364, respectively) (Figure 4D). The inescapable conclusion is that ChuY, much like BVR, is catalyzing a NADPH-dependent reduction of a tetrapyrrole. Moreover, in the absence of the additional vinyl groups (i.e., heme vs deuteroheme), deuteroanaerobilin may adopt two different orientations when binding to the active site of ChuY.
ChuY Displays Kinetic Cooperativity with the DAB Substrate.
Although we have previously reported the spectrophotometric assay monitoring the NADPH-dependent reduction of anaerobilin or DAB by ChuY, we did not explore the kinetics of either reaction further. Absorption changes in the UV–visible region are observed only when both substrates are present, and a typical assay for the NADPH-dependent reduction of DAB is shown in Figure 6. Specifically, an enzyme-dependent decrease in the 439 and 780 nm features was observed (Figure 6A,B). The loss of the Soret and longer wavelength features is consistent with a reduction of DAB and disruption of the conjugation within the tetrapyrrole. We recorded substrate saturation curves for NADPH in the presence of 50 μM DAB by monitoring the two wavelength maxima for the tetrapyrrole (439 and 780 nm). The rates of DAB reduction were plotted using extinction coefficients of 95 and 31.5 mM−1 cm−1, respectively. In addition, all rates were obtained within the first 100 s of the assay as a Selwyn’s test25 indicated enzyme death at longer time intervals. The available kinetic data show that E. coli ChuY exhibits strongly sigmoidal kinetics for NADPH and were fit to the Hill equation (eq 1), with Kmapp values of 3.5 and 3.7 μM and a Hill coefficient of 2.5 for the respective wavelengths (Figure 6A,B). These data suggested positive cooperativity with respect to the substrate NADPH. However, cooperativity observed in steady state kinetics is not definitive evidence of a thermodynamic process and must be confirmed by equilibrium binding studies.26–28
Figure 6.
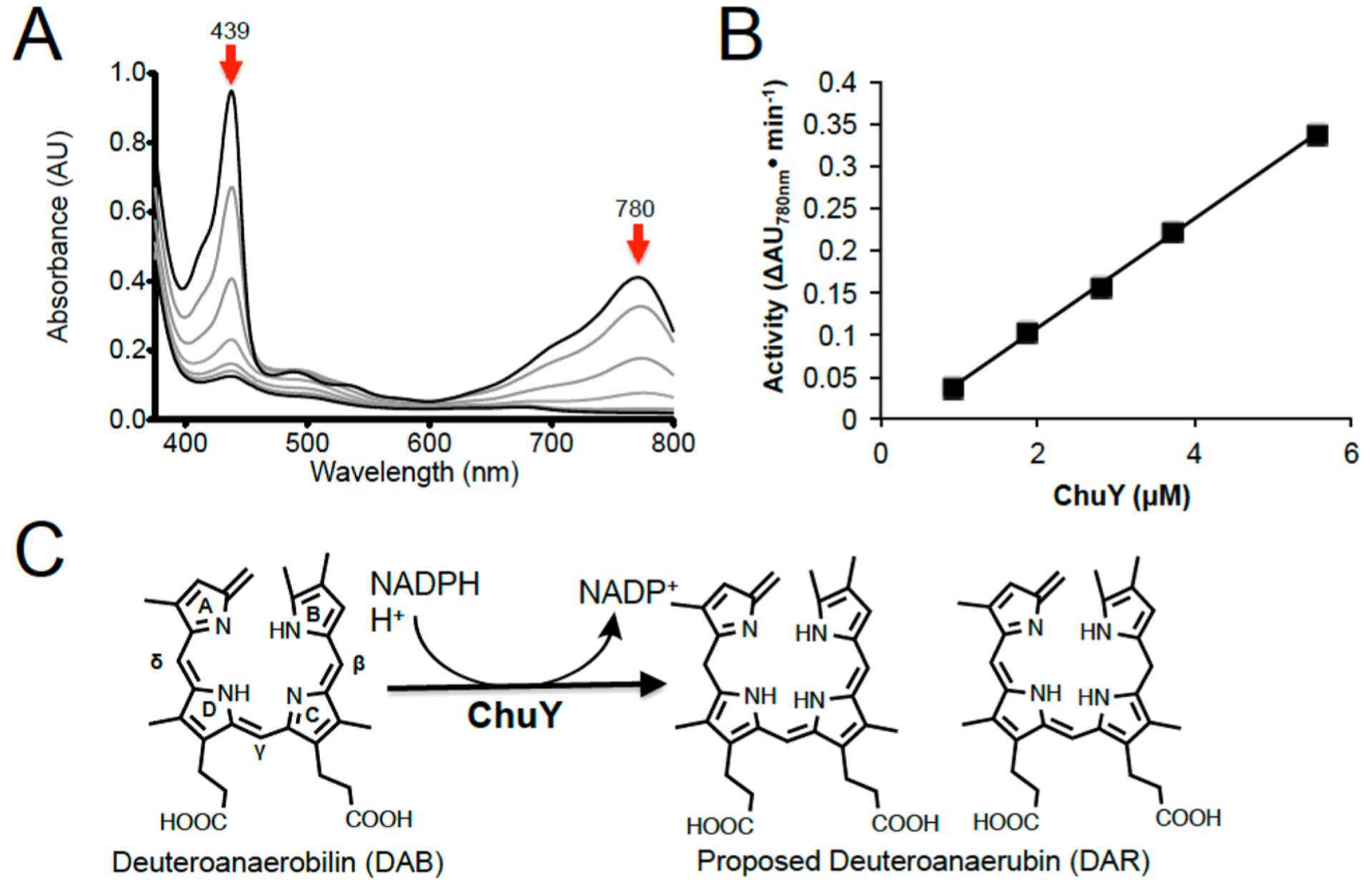
ChuY activity assays using deuteroanaerobilin (DAB) as a substrate and the proposed reaction scheme. (A) UV–visible spectrum of the ChuY (1 μM) assay with deuteroanaerobilin (15 μM) and NADPH (100 μM). Upon addition of NADPH, the magnitudes of the features at 440 and 780 nm decrease with time. (B) Initial rate of ChuY activity with respect to enzyme concentration. (C) Proposed reaction scheme based on the mass spectrometry data shown in Figure 4.
Hence, we applied intrinsic protein fluorescence to study the binding of the cofactor to ChuY. Upon excitation at 280 nm, we observe a tryptophan emission signal at 354 nm. Addition of NADPH caused quenching of the tryptophan emission peak in a concentration-dependent manner. The change in fluorescence intensity was plotted with respect to the concentration of free NADPH, and the data fit to a hyperbola (Figure 7C). All of the thermodynamic and kinetic parameters are listed in Table 3. Because we observe cooperativity in steady state kinetics and not in equilibrium binding experiments, we acquired a saturation curve for DAB to provide further insight into a kinetic model for ChuY function. The initial concentration of DAB was calculated by the extinction coefficient at 780 nm at initial time points of the assay and plotted against the respective rates. NADPH was supplied to ten times the Kd (30 μM) in assays, which exhibited a Vmax that decreased with increasing concentrations of DAB (Figure 6E). The data were modeled with an equation describing substrate inhibition (eq 2) and displayed a Kmapp of 1.6 ± 0.3 μM and a Kiapp of 14 ± 2 μM (Table 3). Considering the sigmoidal substrate saturation curve for NADPH, the observation of substrate inhibition for deuteroanaerobilin suggested kinetic cooperativity. To test the possibility of a kinetic preference for binding NADPH, we performed the same DAB substrate saturation curve with a concentration more than 200 times the Kd of NADPH (500 μM) in the assay to probe the substrate inhibition effect (Figure 7B). Providing excess NADPH should result in a shift in the equilibrium favoring NADPH binding prior to DAB. This would result in a change in the apparent Ki (Kiapp) if the substrate inhibition is not due to a thermodynamic event or substrate solubility. The data attained were modeled with the substrate inhibition equation (eq 2). Although the Vmax and Kmapp were similar to those of the previous DAB saturation, the Kiapp was estimated to be 107 μM, albeit with a significant error due to the concentration range of DAB that was tested.
Figure 7.
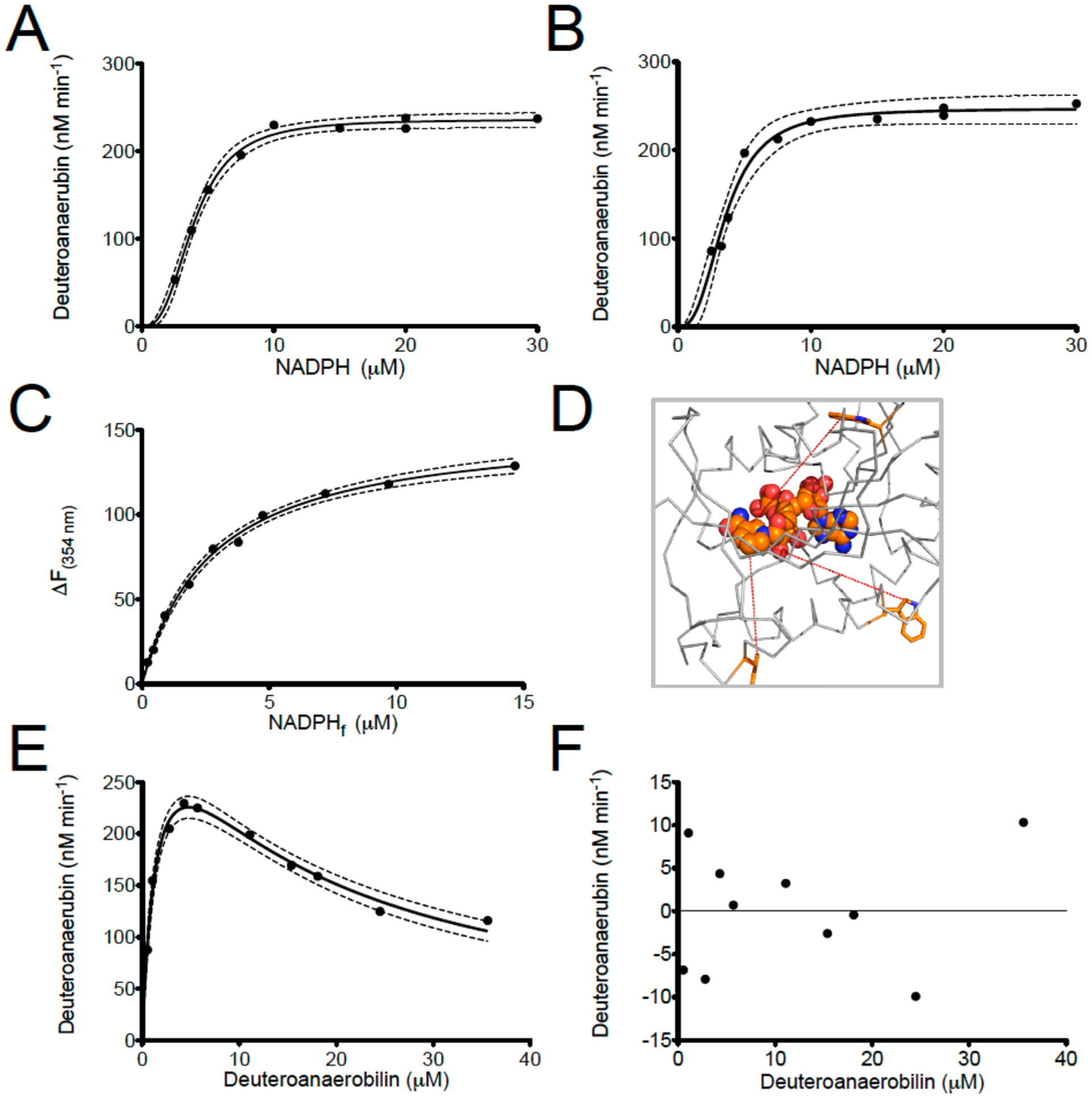
Steady state analysis and equilibrium binding studies for ChuY. (A and B) NADPH saturation kinetics were performed by monitoring the decrease in the 440 nm (A) or 780 nm (B) absorption maximum using 5 μM DAB and 50 nM ChuY. The relative rates were plotted with respect to the concentration of NADPH. The saturation plots were fit to the Hill equation (eq 1) and analyzed by residual analysis. (C) Equilibrium binding assays were performed by intrinsic tryptophan fluorescence quenching and monitoring the change in emission maximum for ChuY tryptophan fluorescence at 354 nm with respect to NADPH concentration. (D) Spatial arrangement of the tryptophan residues in ChuY (orange) in relation to the predicted NADPH-binding site. (E) DAB saturation kinetics in the presence of 30 μM NADPH. The plot was fit to an equation for substrate inhibition (eq 2). (F) Residual analysis of the substrate inhibition curve from panel E.
Table 3.
| ligand | steady state parametersa | equilibrium bindingb | ||||
|---|---|---|---|---|---|---|
| Km app (μM) | Hill coefficient | kcat (s−1) | Ki app (μM) | Hill coefficient | Kd (μM) | |
| NADPH | 3.5 ± 0.2 | 2.5 ± 0.5 | 0.082 ± 0.002c | – | 1.0d | 2.7 ± 0.2d |
| NADPH | 3.7 ± 0.2 | 2.6 ± 0.3 | 0.082 ± 0.002c | – | ||
| DAB | 1.6 ± 0.3e | 1.0e | 0.0127 ± 0.009e | 14 ± 2e | ||
| DAB | 1.2 ± 0.2f | 1.0f | 0.014 ± 0.01f | 108 ± 67f | ||
Fit to eq 1 or 2 in Experimental Procedures.
NADPH binding fit to eq 4 in Experimental Procedures.
Based on a NADPH saturation curve with DAB not fully saturated.
Based on intrinsic tryptophan fluorescence quenching and fit to equation 6 (see Figure 3C).
Based on DAB saturation kinetics with 10Kd of NADPH.
Based on DAB saturation kinetics with 200Kd of NADPH.
DISCUSSION
The metabolic products of heme degradation have important roles in pathogenic microorganisms as well as higher organisms.29–31 The accumulation of biliverdin during oxygenic heme degradation is largely controlled through the activity of biliverdin reductases (BVRs). BVRs are responsible for the conversion of biliverdin to bilirubin, which provides antioxidant benefits and allows for the removal of tetrapyrroles from the cell.32,33 BVRs are prevalent in eukaryotic heme degradation pathways and play a role in regulating the levels of bilirubin in the cell, which can act as an antioxidant. Recently, the mechanistic diversity of heme oxygenases that have been characterized in bacterial systems has increased significantly with notably different degradation products.34 All of these systems require molecular oxygen, and unlike the canonical heme oxygenase, not all of the corresponding biliverdin reductases have been characterized. In fact, with the exception of those of cyanobacteria, ChuY is the first example of a verdin reductase characterized from a bacterial heme degradation system that is similar to BVR. Finally, it is worth noting that when ChuY was deleted from pathogenic E. coli CFT073, a significant decrease in infection potential was observed.23 Specifically, on the basis of cell counts, the ability of E. coli CFT073 to infect human embryonic kidney cells was significantly weakened in the absence of ChuY. A rational explanation for this observation is that the accumulation of heme or anaerobilin is toxic to the pathogen.
Our data suggest that ChuY uses NADPH as a cosubstrate to reduce anaerobilin and deuteroanaerobilin (DAB). We have further shown that ChuY is structurally homologous to fetal BVβR (Figure 3A,B). Binding studies also support the proposal that ChuY uses NADPH rather than NADH as the preferred nicotinamide cofactor. This was also reported to be the case for ChuY from pathogenic E. coli CFT073.23 Specifically, we did not observe any significant quenching of the tryptophan fluorescence for ChuY when up to 80 μM NADH was used in the binding experiment, whereas the NADPH binding isotherm indicated a dissociation constant of 2.7 μM. Kim et al. reported a dissociation constant of 0.2 μM for NADPH binding to ChuY from E. coli CFT073 using isothermal titration calorimetry.
While E. coli CFT073 infects the human urinary tract, E. coli O157:H7 is found in the distal human intestine. In this environment, the ability to adapt quickly to changes in the oxygen tension and nutrient availability is essential to successful colonization and infection. In fact, the nutrient availability can vary depending on the precise area inhabited within the intestine. For pathogenic E. coli, or other Gram-negative enteric pathogens like Vibrio cholerae, they will preferentially take up residence in the tight mucosal layer. This is an environment populated by numerous strict anaerobes. Therefore, the ability to degrade heme under both aerobic and anaerobic conditions would provide a pathogen with a significant selective advantage.
The oxygen-independent heme degradation pathway of E. coli O157:H7 consists of ChuW, a radical SAM methyltransferase, which methylates heme to form the linear tetrapyrrole anaerobilin and liberate iron. ChuY uses anaerobilin as a substrate, reducing it to the tetrapyrrole termed “anaerorubin”. Mass spectrometry analysis of the isolated anaerorubin is consistent with a four-electron reduction of anaerobilin, presumably from two subsequent turnovers (Figure 5). Interestingly when [methyl-13C]DAB, the product of [methyl-13C]deuteroheme turnover by ChuW, is utilized as the substrate for ChuY, formation of [methyl-13C]-deuteroanaerorubin resulted in only a two-electron reduction as indicated by MS analysis (Figure 4). Comparing the fragmentation patterns of DAR prepared with either [methyl-12C]- or [methyl-13C]DAB is useful for elucidating a structure. The even mass fragment observed for the dominant m/z 364 species indicates a loss of one nitrogen atom (Figure 4B). This fragment would be consistent with a reduction that occurred at the β- or δ-methine bridging carbon. The precise location of the methyl-13C group that was added in the ChuW reaction is still under investigation and could be located at position 1 or 21 of the linear tetrapyrrole based on the predicted mechanism by ChuW. However, fragmentation of [methyl-13C]DAR allows for the identification of the site of reduction relative to the location of the isotopic label. For instance, a reduction occurring on the distal side of the tetrapyrrole would result in retention of the label, whereas reduction to the adjacent methine bridging carbon would result in a loss of the label upon fragmentation. The fragmentation pattern we observe shows both species in a 3:1 ratio. Given the lack of vinyl groups on deteroheme and DAB, this may simply suggest that the substrates can enter their respective active sites in ChuW and ChuY in two different orientations as it appears that the reduction can occur at either position and/or the isotopic label may be on either side of the bilin as demonstrated by the fragmentation pattern we report here and have observed previously.14
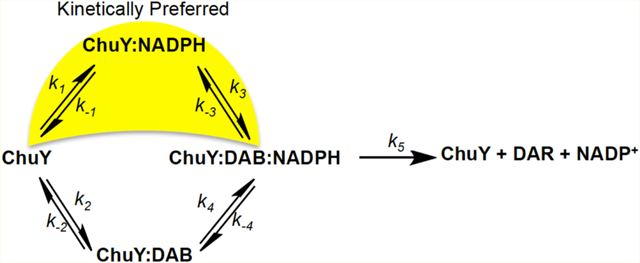
When performing steady state kinetics of E. coli ChuY using deuteroanaerobilin (DAB) as a substrate, we observe a distinct positive cooperativity with respect to NADPH saturation, but an apparent substrate inhibition with respect to DAB (Figure 7A,B,E). Although cooperativity is common for many oligomeric enzymes, sigmoidal kinetics do not unequivocally establish thermodynamic cooperativity.26–28 Moreover, such kinetic behavior is inconsistent with ChuY functioning as a monomer.23 In fact, equilibrium binding studies are the only way to provide evidence of site-to-site communication described by thermodynamic cooperativity. For example, the observation of a hyperbolic binding isotherm for binding of NADPH to ChuY would indicate a kinetic cooperative mechanism. However, kinetic cooperativity can be described by a hysteretic lag in progress curves,28,35 or it can be due to a kinetic preference for substrate binding during turnover.22 On the basis of the observation of sigmoidal kinetics with NADPH saturation and substrate inhibition with DAB, the kinetic cooperativity by ChuY is best explained by random association of substrates with a kinetically preferred path for NADPH binding (Figure 8A). This mechanism is possible if k1k3 > k2k4 and if k5 is not rate-limiting. The kinetic differences for substrate binding in the random association model provide an explanation for the apparent inhibition observed for the DAB saturation kinetics. At lower concentrations of DAB, turnover will proceed through the kinetically preferred path that involves NADPH binding prior to DAB. However, at higher concentrations of DAB, the kinetic path will gradually switch to the kinetically unfavored path of initial DAB binding. As more of the ChuY population switches to this path, the rate decreases, causing the appearance of substrate inhibition. To overcome the apparent inhibition effect and test the kinetic model, NADPH must be provided in excess to maintain the thermodynamically favorable binding pathway. To this end, by providing a concentration 200 times the Kd of NADPH, we observed a 10-fold increase in the apparent Ki app (Figure 8B) due to the majority of ChuY binding NADPH initially before proceeding through the kinetically preferred path. Because NADPH concentrations will likely be higher than those of anaerobilin in vivo, this kinetic model may provide a physiological benefit resulting in quicker turnover when anaerobilin is produced.
Figure 8.
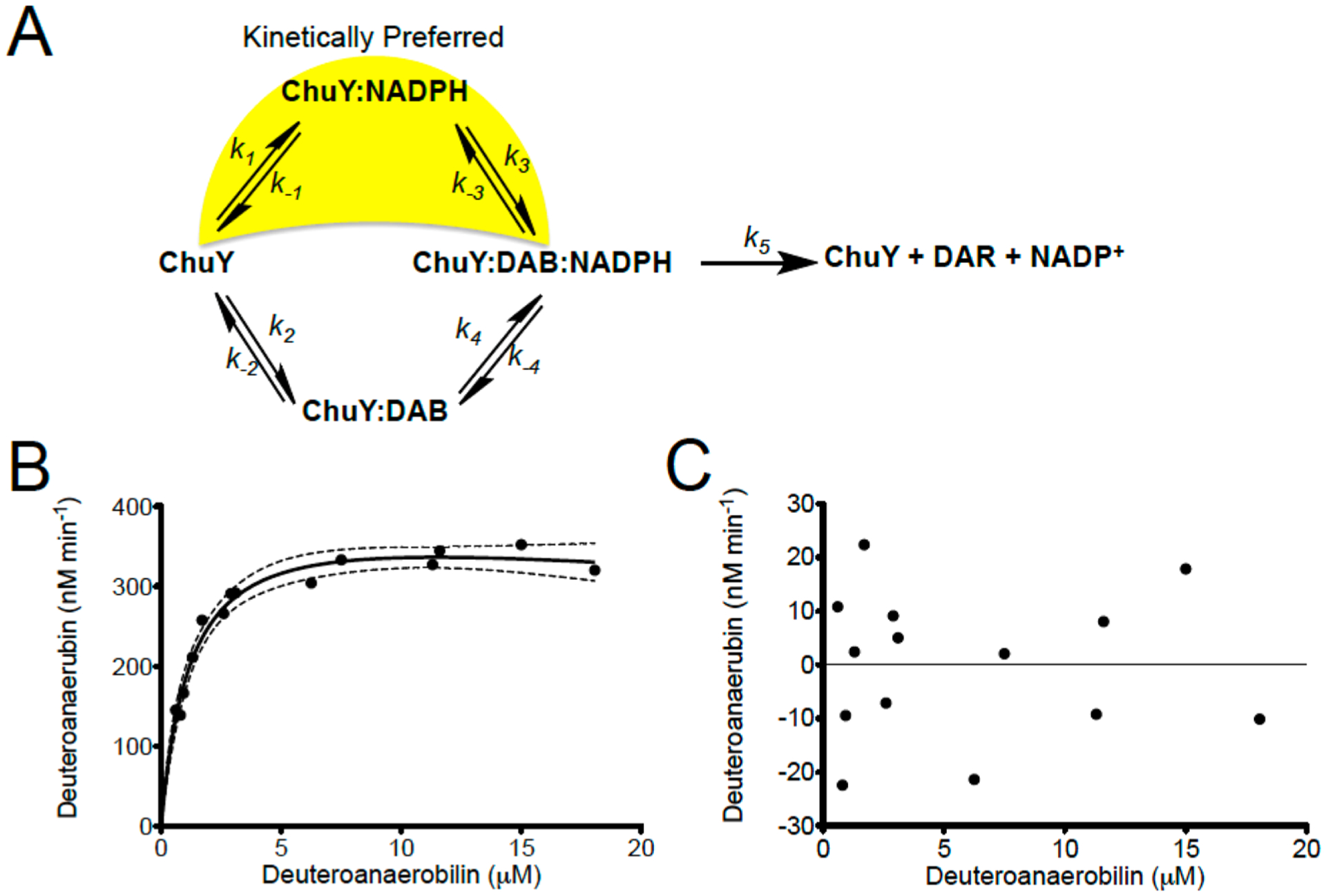
Proposed kinetic model for ChuY and experimental validation. (A) ChuY follows a random association kinetic model with a kinetic preference for NADPH binding. (B) DAB saturation kinetics were performed as described in the legend of Figure 6E using approximately 200 μM NADPH. The rates were plotted with respect to DAB concentration and fit to the equation for substrate inhibition (eq 2). (C) Residual analysis was used to assess the fit of the data.
The sequence of ChuW is homologous to that of HutW from V. cholerae; however, these enzymes form a different clade when a phylogenetic tree is constructed on the basis of sequence identity.14 Specifically, E. coli contains the anaerobilin reductase ChuY, whereas Vibrio species express the HutZ protein from the equivalent position on the heme uptake operon. There are currently two models for HutZ function; the initial characterization suggested that HutZ could functionally replace a heme oxygenase.36 However, HutZ did not appear to have such activity in enzyme assays.37 Considering the ability for HutZ to bind heme, Wyckoff et al. proposed that, while not a genuine heme oxygenase, HutZ could function as a heme chaperone. Later reports suggested HutZ did have in vitro heme oxygenase activity,36 although the products were never identified and the activity was disputed by others.38 The structure of HutZ has been determined to 2.0 Å resolution and is described as having a split barrel fold that is typical for FMN-binding proteins.38 The proposal that ChuY may play a role in iron/heme homeostasis by modulating the relative concentrations of the anaerobic heme catabolites is consistent with recent observations in other pathogenic organisms, specifically, the observation that the mechanism and rate of heme uptake in Pseudomonas aeruginosa are regulated by the relative concentrations of the different heme catabolites.29 Regardless, in this work, we have demonstrated that certain aspects of ChuY structure and function in the anaerobic heme degradation pathway parallel those of biliverdin reductase. When our data are considered in light of the fitness phenotype reported by Kim et al., further investigation of the anaerobic heme degradation pathway represents an unexplored opportunity for antibiotic development.
ACKNOWLEDGMENTS
The authors would like to thank Zac Wood and the members of the Wood lab for numerous helpful discussions. This research was supported by NSF grant MCB 0835432 to W.N.L.. Support and access to MS instrumentation was provided by an NIG NIGMS grant (P41GM103490).
ABBREVIATIONS
- Tris
tris(hydroxymethyl)aminomethane
- TCEP
tris(2-carboxyethyl)phosphine hydrochloride
- BVR
biliverdin reductase
- aSDR
atypical short chain dehydrogenase
- BVβR
biliverdin IX β-reductase
- DAB
deuteroanaerobilin
- DAR
deuteroanaerorubin
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b01099.
Proposed structures consistent with the fragmentation pattern observed for deuteroanaerorubin with and without the 13C label (PDF)
Accession Codes
Atomic coordinates for the native ChuY structure have been deposited in the Protein Data Bank as entry 5FFQ.
The authors declare no competing financial interest.
REFERENCES
- (1).Torres AG, and Payne SM (1997) Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol 23, 825–833. [DOI] [PubMed] [Google Scholar]
- (2).Torres AG, Redford P, Welch RA, and Payne SM (2001) TonB-dependent systems of uropathogenic Escherichia coli: aero-bactin and heme transport and TonB are required for virulence in the mouse. Infection and immunity 69, 6179–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Anzaldi LL, and Skaar EP (2010) Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infection and immunity 78, 4977–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wilks A (2002) Heme oxygenase: evolution, structure, and mechanism. Antioxid. Redox Signaling 4, 603–614. [DOI] [PubMed] [Google Scholar]
- (5).Tenhunen R (1972) The enzymatic degradation of heme. Semin. Hematol 9, 19–29. [PubMed] [Google Scholar]
- (6).Tenhunen R, Marver HS, and Schmid R (1968) The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc. Natl. Acad. Sci. U. S. A 61, 748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wong KP (1971) Bilirubin glucuronyltransferase. Specific assay and kinetic studies. Biochem. J 125, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ratliff M, Zhu W, Deshmukh R, Wilks A, and Stojiljkovic I (2001) Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. Journal of bacteriology 183, 6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wilks A, and Schmitt MP (1998) Expression and characterization of a heme oxygenase (Hmu O) from Corynebacte-rium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem 273, 837–841. [DOI] [PubMed] [Google Scholar]
- (10).Zhu W, Wilks A, and Stojiljkovic I (2000) Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. Journal of bacteriology 182, 6783–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wegele R, Tasler R, Zeng Y, Rivera M, and Frankenberg-Dinkel N (2004) The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem 279, 45791–45802. [DOI] [PubMed] [Google Scholar]
- (12).Davis SJ, Vener AV, and Vierstra RD (1999) Bacteriophytochromes: phytochrome-like photoreceptors from non-photosynthetic eubacteria. Science 286, 2517–2520. [DOI] [PubMed] [Google Scholar]
- (13).Bhoo SH, Davis SJ, Walker J, Karniol B, and Vierstra RD (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414, 776–779. [DOI] [PubMed] [Google Scholar]
- (14).LaMattina JW, Nix DB, and Lanzilotta WN (2016) Radical new paradigm for heme degradation in Escherichia coli O157:H7. Proc. Natl. Acad. Sci. U. S. A 113, 12138–12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fu ZQ, Rose J, and Wang BC (2005) SGXPro: a parallel workflow engine enabling optimization of program performance and automation of structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr 61, 951–959. [DOI] [PubMed] [Google Scholar]
- (16).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- (18).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Laue TM, and Pelletier SL (1986) Interactive Computer-Aided Interpretation of Sedimentation Data. Biophys. J 49, A274–A274. [Google Scholar]
- (20).Schuck P, Gillis RB, Besong TM, Almutairi F, Adams GG, Rowe AJ, and Harding SE (2014) SEDFIT-MSTAR: molecular weight and molecular weight distribution analysis of polymers by sedimentation equilibrium in the ultracentrifuge. Analyst 139, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ortega A, Amoros D, and Garcia de la Torre J (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J 101, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sanchez JE, Gross PG, Goetze RW, Walsh RM Jr., Peeples WB, and Wood ZA (2015) Evidence of Kinetic Cooperativity in Dimeric Ketopantoate Reductase from Staphylococcus aureus. Biochemistry 54, 3360–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kim H, Chaurasia AK, Kim T, Choi J, Ha SC, Kim D, and Kim KK (2016) Structural and functional study of ChuY from Escherichia coli strain CFT073. Biochem. Biophys. Res. Commun, DOI: 10.1016/j.bbrc.2016.12.008. [DOI] [PubMed] [Google Scholar]
- (24).Buysschaert G, Verstraete K, Savvides SN, and Vergauwen B (2013) Structural and biochemical characterization of an atypical short-chain dehydrogenase/reductase reveals an unusual cofactor preference. FEBS J. 280, 1358–1370. [DOI] [PubMed] [Google Scholar]
- (25).Selwyn MJ (1965) A simple test for inactivation of an enzyme during assay. Biochim. Biophys. Acta, Enzymol. Biol. Oxid 105, 193–195. [DOI] [PubMed] [Google Scholar]
- (26).Hammes GG, and Wu CW (1974) Kinetics of allosteric enzymes. Annu. Rev. Biophys. Bioeng 3, 1–33. [DOI] [PubMed] [Google Scholar]
- (27).Ricard J, and Cornish-Bowden A (1987) Co-operative and allosteric enzymes: 20 years on. Eur. J. Biochem 166, 255–272. [DOI] [PubMed] [Google Scholar]
- (28).Nest KE (1995) Cooperativity in enzyme function: equilibrium and kinetic aspects. Methods Enzymol. 249, 519–567. [DOI] [PubMed] [Google Scholar]
- (29).Mourino S, Giardina BJ, Reyes-Caballero H, and Wilks A (2016) Metabolite-driven Regulation of Heme Uptake by the Biliverdin IXbeta/delta-Selective Heme Oxygenase (HemO) of Pseudomonas aeruginosa. J. Biol. Chem 291, 20503–20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gibbs PE, Miralem T, and Maines MD (2015) Biliverdin reductase: a target for cancer therapy? Front. Pharmacol 6, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kikuchi G, Yoshida T, and Noguchi M (2005) Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun 338, 558–567. [DOI] [PubMed] [Google Scholar]
- (32).Jansen T, and Daiber A (2012) Direct Antioxidant Properties of Bilirubin and Biliverdin. Is there a Role for Biliverdin Reductase? Front. Pharmacol 3, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Greenberg DA (2002) The jaundice of the cell. Proc. Natl. Acad. Sci. U. S. A 99, 15837–15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wilks A, and Ikeda-Saito M (2014) Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Acc. Chem. Res 47, 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kadirvelraj R, Sennett NC, Custer GS, Phillips RS, and Wood ZA (2013) Hysteresis and negative cooperativity in human UDP-glucose dehydrogenase. Biochemistry 52, 1456–1465. [DOI] [PubMed] [Google Scholar]
- (36).Uchida T, Sekine Y, Matsui T, Ikeda-Saito M, and Ishimori K (2012) A heme degradation enzyme, HutZ, from Vibrio cholerae. Chem. Commun 48, 6741–6743. [DOI] [PubMed] [Google Scholar]
- (37).Wyckoff EE, Schmitt M, Wilks A, and Payne SM (2004) HutZ is required for efficient heme utilization in Vibrio cholerae. Journal of bacteriology 186, 4142–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Liu X, Gong J, Wei T, Wang Z, Du Q, Zhu D, Huang Y, Xu S, and Gu L (2012) Crystal structure of HutZ, a heme storage protein from Vibrio cholerae: A structural mismatch observed in the region of high sequence conservation. BMC Struct. Biol 12, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]


