Abstract
This report describes the first application of the novel NMR-based machine learning tool ‘Small Molecule Accurate Recognition Technology’ (SMART 2.0) for mixture analysis and subsequent accelerated discovery and characterization of new natural products. The concept was applied to the extract of a filamentous marine cyanobacterium known to be a prolific producer of cytotoxic natural products. This environmental Symploca extract was roughly fractionated, and then prioritized and guided by cancer cell cytotoxicity, NMR-based SMART 2.0, and MS2-based molecular networking. This led to the isolation and rapid identification of a new chimeric swinholide-like macrolide, symplocolide A, as well as the identification of swinholide A, samholides A-I and several new derivatives. The planar structure of symplocolide A was confirmed to be a structural hybrid between swinholide A and luminaolide B by 1D/2D NMR and LCMS2 analysis. A second example applies SMART 2.0 to the characterization of structurally novel cyclic peptides, and compares this approach to the recently appearing ‘atomic sort’ method. This study exemplifies the revolutionary potential of combined traditional and deep learning-assisted analytical approaches to overcome longstanding challenges in natural products drug discovery.
Graphical Abstract
Natural Products (NPs) of terrestrial and marine organisms have been a highly valuable source of leads for biomedical applications. About 50% of FDA approved drugs can trace their origin to NPs, and notably, marine NPs have proportionately produced a much higher success rate than other sources of drug leads (e.g. 19 approved marine derived or inspired agents are on the market today).1–3 Filamentous marine cyanobacteria are especially rich in structurally-diverse and biologically active NPs,4,5 such as those showing differential cytotoxicity against various cancer cell lines6,7. In this regard, the genus Symploca is a prolific producer of cytotoxic NPs such as dolastatin 108,9, symplocamide A10, and others11–16.
Drug discovery and development is expensive ($2 billion on average), time-consuming (13–15 years from concept to market), and risky (clinical success rate of 12%).17 Informatic tools are therefore being developed to make the discovery of new leads more efficient, to repurpose known agents, to target new metabolites on the basis of genomic analysis, to rapidly reveal mechanisms of action, and to optimize the pharmaceutical properties of drug leads. For example, the emerging research area of genome mining18 featuring bioinformatic tools such as AntiSMASH 5.019 and BiGSCAPE20 provide an orthogonal approach to NP discovery,21 although an approved drug discovered by this approach has not yet been marketed. The rapid identification of molecular structure, either known or new, is a significant challenge in NP discovery, and a variety of partial solutions are emerging using disparate analytical approaches.
For example, liquid chromatography tandem mass spectrometry-based (LCMS/MS) dereplication tools, such as the Global Natural Products Social molecular networking (GNPS; http://gnps.ucsd.edu), MS2LDA, and SIRIUS22–28 represent paradigm shifts in NP research. These new tools are facilitating the targeted isolation of new NPs as well as rapidly dereplicating known ones. Nevertheless, unambiguous identification of new NPs still requires characterization by NMR spectroscopy to comply with the minimum standards for novel metabolite annotation.30,31. Recent efforts have focused on combining various NMR techniques with mass spectrometry and in silico data bases.32,33 Previously. we reported a novel approach that involved training a deep convolutional neural network (CNN) of siamese architecture with 2,048 1H–13C HSQC spectra mined from the supporting information sections of the Journal of Natural Products.34 This trained system was used to analyze new spectra, place them within 10-dimensional SMART cluster space, and accelerate the structure elucidation of a series of cyclic lipopeptides.34,35
Encouraged, we subsequently developed automated pipelines to produce constructed HSQC spectra from data tables as well as predicted HSQC spectra from published structures. SMART 2.0 was trained on 25,434 HSQC spectra from NPs of the JEOL database (https://www.j-resonance.com/en/nmrdb/). The spectra were mapped into a 180-dimensional embedding space using a CNN (SqueezeNet).36 We mapped an additional 27,642 spectra that were computed using the ACD Labs predictor to create the HSQC spectra (see page S6 for details) from randomly chosen, mainly marine NPs (e.g. NP Atlas37 and NPASS), into the 180-D space. The resulting 53,076 NP HSQC spectra cover approximately 15% of the currently known NPs. Further experiments demonstrated the remarkable robustness of the SMART analysis towards different NMR solvents (Figure S1), and the value of calculated HSQC spectra in the SMART analysis (Figure S2).
Here, we demonstrate this unique cheminformatic tool to automatically characterize a complex NP from a cyanobacterial extract mixture for the first time. Resultantly, the extract was fractionated and the isolation of novel NPs undertaken guided by NMR-based SMART mixture analysis, MS2 molecular networking, and cytotoxicity against NCI-H460 human lung cancer cells in vitro. This led to the discovery of a new swinholide class of NP, named ‘symplocolide A’ (compound 1, Scheme 1, Scheme S1), as well as the description of several known compounds in this structural class.
Scheme 1.
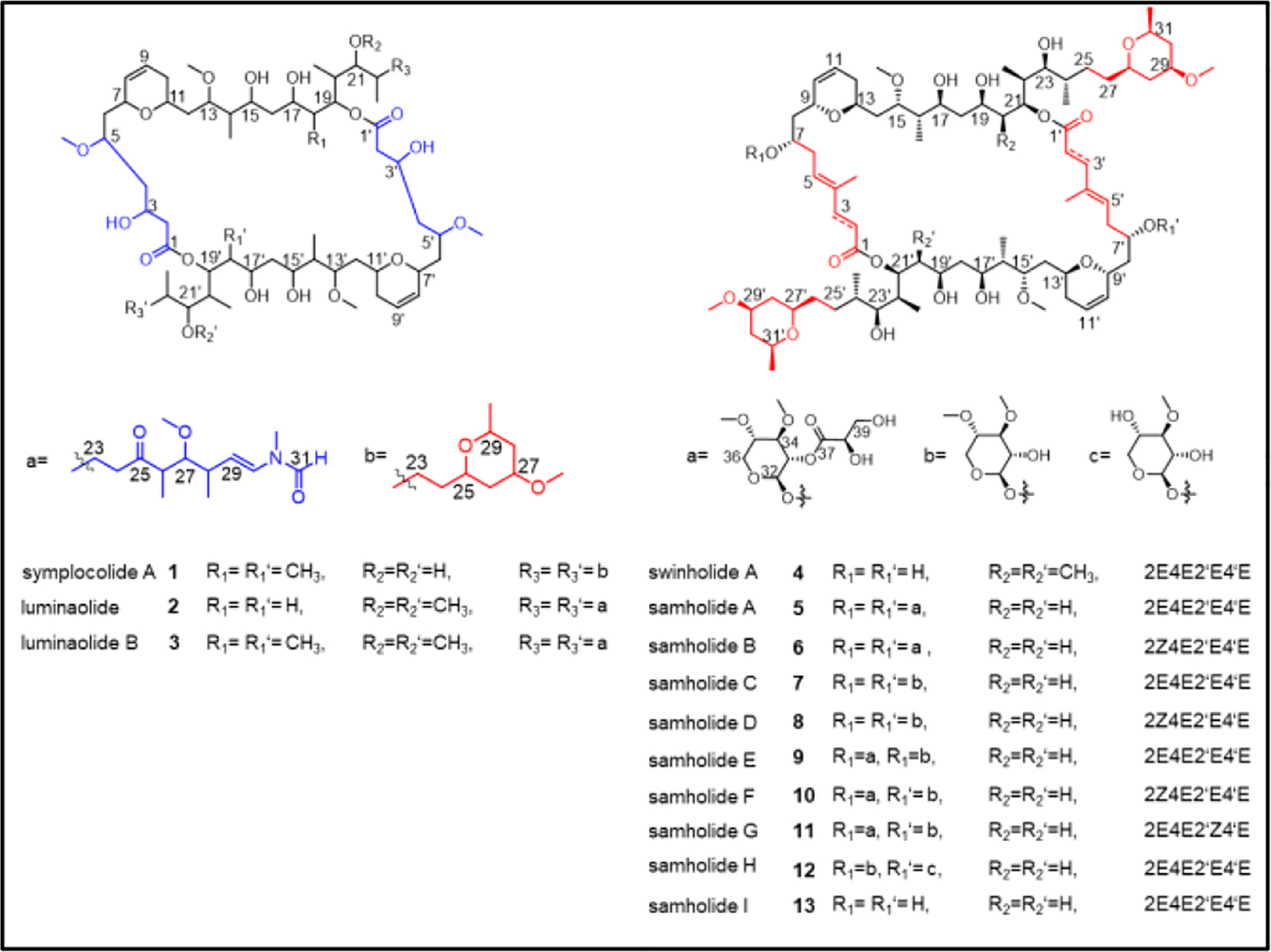
Structures of symplocolide A (1), luminaolide (2)42, luminaolide B (3)43, swinholide A (4), and samholides A-I (5–13).44 All compounds except 2 and 3 were detected in this study.
Tufts of a filamentous marine cyanobacterium, morphologically identified as Symploca sp., were collected near American Samoa. The preserved collection was repetitively extracted (2/1 CH2Cl2/MeOH) and fractionated using vacuum liquid chromatography (VLC) to obtain nine fractions of increasing polarity (A-I). All fractions and the crude extract were screened for cytotoxicity to H-460 human lung cancer cells.38 Besides the crude extract, only fraction H showed strong cytotoxicity, while fraction I was moderately active (Figures 1a and S3). For initial SMART 2.0 analysis of the most cytotoxic crude fraction H, a NUS-ASAP-HSQC39–41 was recorded in 13 minutes on about 1 mg material in a 1.7 mm TCI MicroCryoProbe™ (599.10 MHz)s to obtain the correlations of the major components of that fraction. After auto-peak picking of the 45 most intense features, 11 out of the top 12 structures returned by SMART 2.0 were macrolides including the known cyanobacterial metabolites swinholide A and isoswinholide A (Figures 1b–d).
Figure 1.
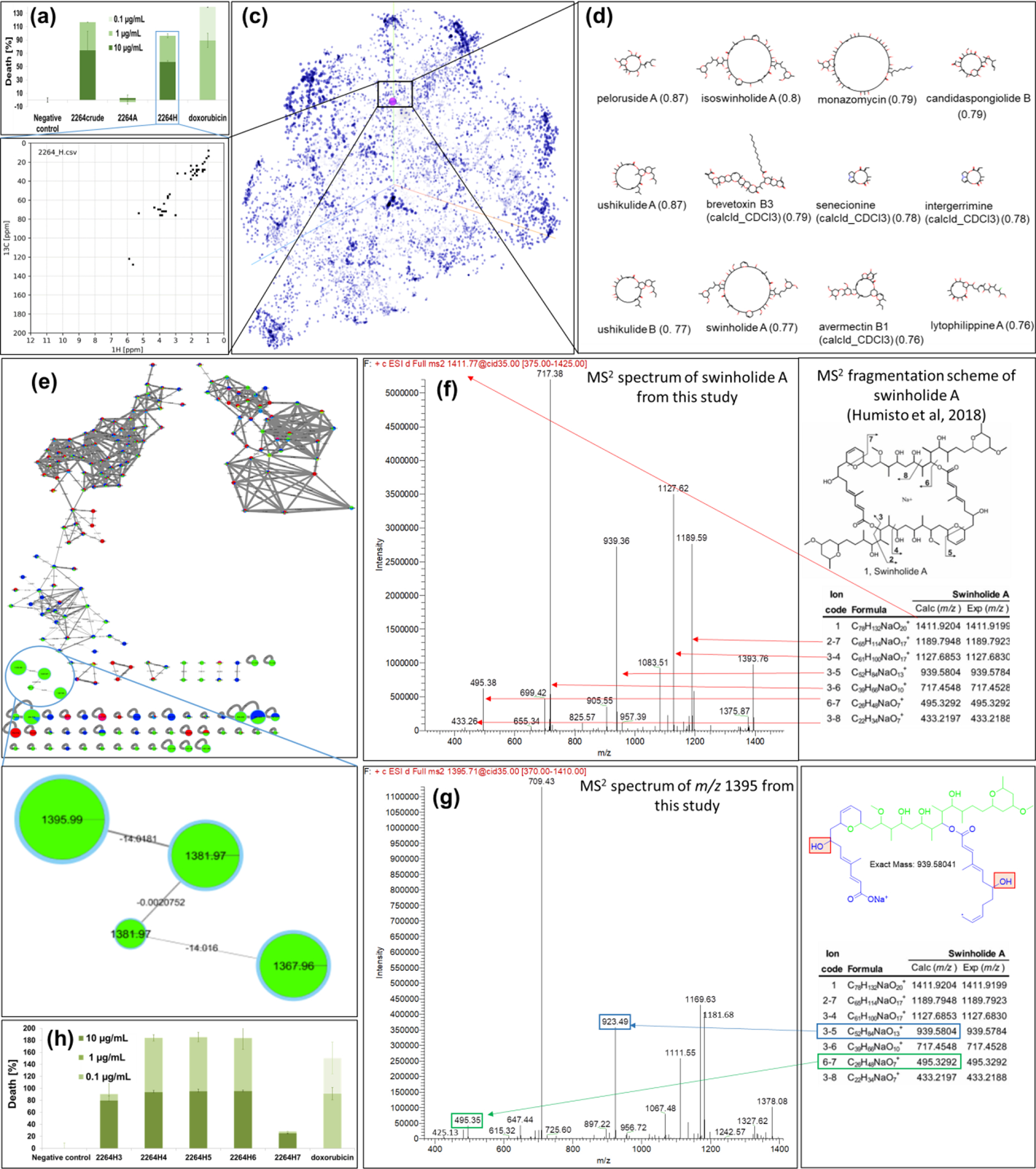
(a,h) Cytotoxicity assay against H460 lung cancer cells reveals VLC fraction H (a) and subfractions H4–H6 (h), respectively, as the most potent at 1 and 10 μg/mL (see also Figure S3). (b) Digitized HSQC spectrum of most cytotoxic fraction H. (c) t-SNE embedding of the HSQC spectrum into the 180 dimensional cluster space of 53,076 nodes (reduced here to 10,000 for clarity) representing natural products trained on the basis of their 1H-13C-HSQC spectra. Swinholide A and its closest neighbors are highlighted (purple nodes, black rectangle). (d) SMART 2.0 results (top12 structures based on cosine similarity score) of fraction H suggests that it contains macrolides from the swinholide class. (e) Molecular networking analysis of cyanobacterial subfractions H3–H7. Highlighted cluster with putatively new m/z feature 1395.9 (H4, green nodes). (f) MS2 analysis of swinholide A29 and comparison to (g) the feature detected as new m/z 1395.9 (symplocolide A) show fragmentation patterns that reveal structural insights (same fragment highlighted in green, distinctive fragment highlighted in blue) for both compounds.
Targeting these macrolides for isolation, fraction H was purified by C-18 solid phase extraction into sub-fractions H1–H7; these were analyzed using SMART 2.0 mixture analysis as well as MS2-based molecular networking. The latter revealed a highly abundant and distinct cluster of four nodes with the major m/z 1395.9 in active fraction H4 (Figure 1e). Isolation and structure elucidation of this compound led to the discovery of a new cytotoxic macrolide (Scheme 1), named symplocolide A (1), representing a structural chimera between swinholide A (4) and luminaolide B (3).43 Analysis of the MS2 spectra of metabolites from these fractions using GNPS led to identification of swinholide A (4), the glycosylated samholides A-I44 (5–13, Scheme 1 Figures S12–22), as well as several putatively new analogs swinholides L and M, samholides J-M, and symplocolides B-D (Figures S23–30).
Similarities in the MS2 fragmentation patterns of 1 and 4 (Figures 1f and g)29 suggested that they differed in their carbon backbones (see blue part of the feature m/z 939.49, Figures 1g, S4). This hypothesis was confirmed following isolation and unambiguous structure determination by 2D NMR analysis and HRMS (Figures S5–S11, Table S1). These analyses disclosed 1 to possess the backbone skeleton of 3 and the side chain structure of 4, thus constituting a chimera of these two macrolides.
All known swinholide-type compounds, irrespective of origin (sponge, cyanobacteria, algae, nudibranch), possess a highly analogous monomeric carbon skeleton and identical configurations at comparable chiral centers (Scheme 1); this has been confirmed in two cases by X-ray crystallography45,46 as well as total synthesis.47 These swinholide-type metabolites are produced by highly similar trans AT polyketide synthase biosynthetic gene clusters.29,43 Thus, the stereochemistry of 1 is assigned by analogy given the close similarities between the 1H and 13C NMR chemical shifts and coupling constants of compound 1 with those of previously reported swinholide-like macrolides (Tables S2, S3, Scheme S1).42–44,48
Recently, Duggan et al. developed an ‘atomic sort’ method wherein HSQC spectra from two different NP extracts were compared with a database of 1,207, mostly human primary metabolites using Euclidian distance scores, and the method was able to detect novel structural features of marine NPs.49 This approach yielded a new analog of a known antiprotozoal polyketide (gracilioether L, see SMART results in Figure S32).50 They also demonstrated the method with re-isolation of known antibacterial cyclic heptapeptide, cyclomarin A.51–53 However, three of four “novel features” detected by this method in cyclomarin A were false positives in terms of structural novelty (e.g. regular methyl group, methylene next to primary alcohol and alpha-carbon methine of N-methylleucine).
To demonstrate SMART 2.0 for mixture analysis and early prioritization for rare structural motifs, we used the NMR table from the SI in Duggan et al.49 to evaluate the HSQC data of the crude extract as well as pure cyclomarin A (not in the SMART database). Intriguingly, the top two hits for cyclomarin A and top twenty hits for the crude extract, consisting of 289 C-H correlations in the HSQC spectrum, revealed the closely related ilamycins C1 and C2 as candidate structures.54 Ilamycins harbor a very similar and rare N-(1,1-dimethyl-2,3-epoxypropyl)-tryptophan moiety as cyclomarin A [substructure search of NP Atlas database, (https://www.npatlas.org/) returned no additional NPs with this amino acid moiety].37 Furthermore, ilamycins contain a similar 4-hexenoic acid residue as cyclomarin A (Figures 2, S33). This example underscores that while features such as heteroatoms and quaternary carbons are not directly measured in the 1H-13C HSQC experiment, CNNs can detect these features indirectly via analyzing chemical shift patterns as well as the relative positions of correlations. Thus, CNNs can extract information from non-peak areas whereas spectroscopists mainly rely on visible peaks for interpretation. This aspect likely explains the extraordinary robustness of SMART 2.0, as it very accurately predicts structures with chemical moieties that are not directly visible in the HSQC spectrum. Finally, each analysis performed by SMART, from NMR data acquisition to table construction and structure prediction, took less than 30 minutes (the SMART structure prediction takes approximately 8 seconds). This compares favorably with the time-scale of LCMS measurements and is a great improvement over the weeks to months typically required for the structural analysis of highly modified cyclic peptides such as cyclomarin A.
Figure 2.
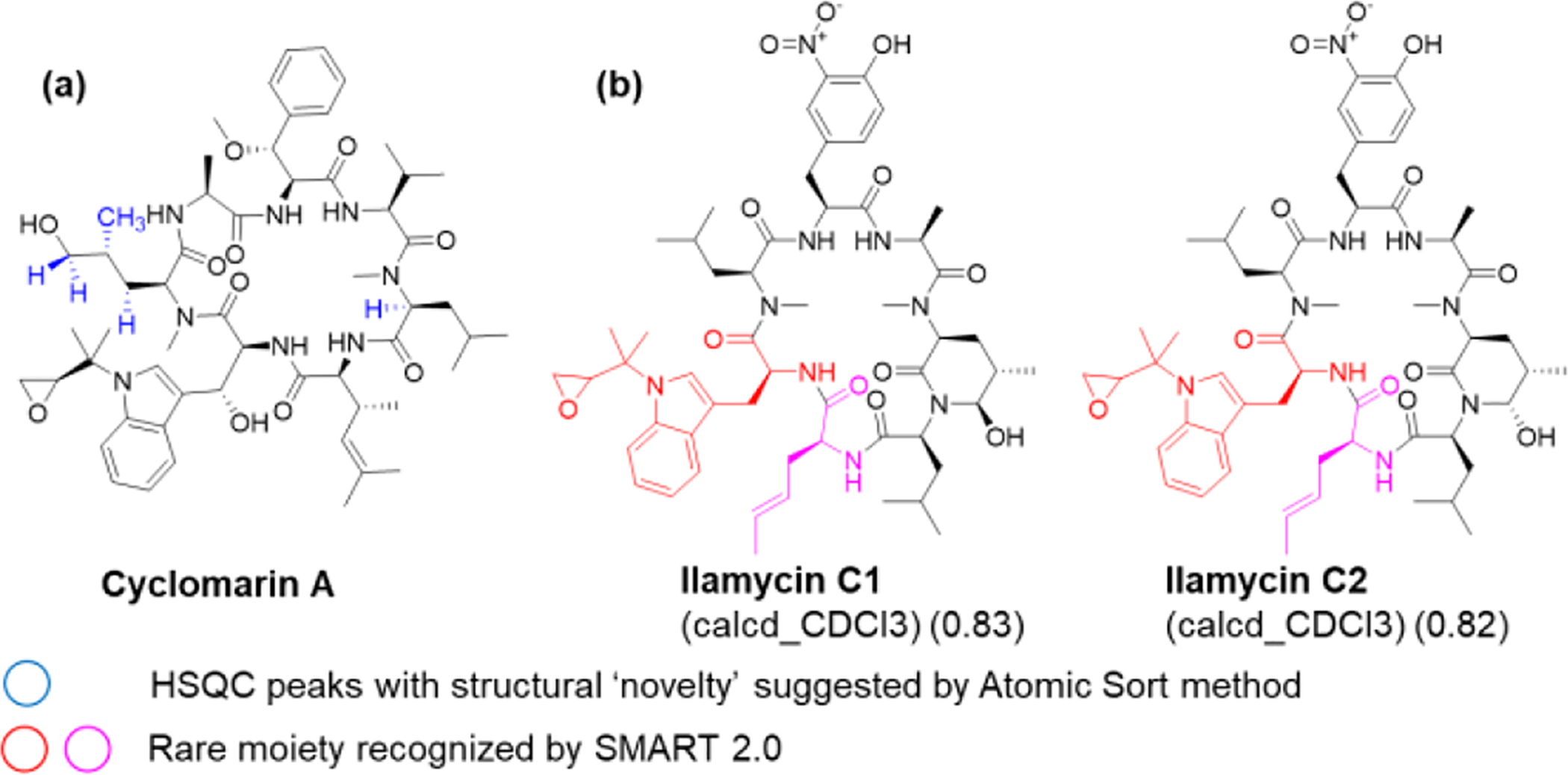
Comparison of the ‘atomic sort’ method and SMART 2.0 to detect novel/rare structural elements. (a) HSQC correlations suggesting structural novelty by ‘atomic sort’ method (highlighted in blue). (b) Top two results of SMART 2.0 analysis querying cyclomarin A (experimental HSQC data from reference49) to detect related compounds that include rare structural moieties (highlighted in red, purple).
In this report we introduce a highly improved user friendly version of the SMART tool and demonstrate its remarkable ability to rapidly recognize natural product structure types. SMART 2.0 is now available to the academic community in Python (https://smart.ucsd.edu/classic).
Rapid structure prediction of major constituents from crude extracts and fractions greatly assists with prioritization of structurally novel or otherwise interesting NPs for further study. Because knowledge of the structural class of an unknown NP can greatly accelerate its structure elucidation, this tool can overcome several of the bottlenecks currently present in NP research. We applied SMART 2.0 on crude fractions from a cyanobacterial extract that helped target the isolation of a new chimeric macrolide, symplocolide A, as well as dereplicate swinholide A and related compounds. The hybridic nature of symplocolide A suggests a novel biosynthetic pathway, an aspect that is under continuing study as the genetic basis for molecular diversification in a compound class is of great interest.43,55 Further, we demonstrated here that SMART 2.0 can detect rare structural features from crude extracts and fractions. We envision that the continued development of SMART 2.0 can revolutionize aspects of NP drug discovery, especially when combined with orthogonal information derived from genome mining of the biosynthetic gene clusters, accurate mass and fragmentation data, and fingerprint analyses of molecular substructures via the GNPS platform, MS2LDA and Sirius.22,27,28
Supplementary Material
ACKNOWLEDGMENT
The authors are grateful for funding from NIH grant R01 GM107550 to G.W.C., P.C.D., and W.H.G. and Gordon and Betty Moore Foundation grant GBMF7622 to G.W.C, P.C.D., and W.H.G. We thank N. Moss and B. Miller for assistance with collection of the Symploca sp, and G. Arevalo for extraction and VLC fractionation. We also thank P. Landon for purchasing and assembling our Graphic Processing Unit (GPU) machine.
Footnotes
The authors declare no conflict of interest. Chen Zhang, Garrison W. Cottrell, and William H. Gerwick are the cofounders of NMR Finder LLC. Mingxun Wang is the founder of Ometa Labs LLC.
REFERENCES
- 1.Gerwick WH; Moore BS Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chemistry & Biology 2012, 19 (1), 85–98. 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman DJ; Cragg GM Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat.l Prod 2016, 79 (3), 629–661. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 3.Pereira F Have Marine Natural Product Drug Discovery Efforts Been Productive and How Can We Improve Their Efficiency? Expert Opin. Drug Discov 2019, 14 (8), 717–722. 10.1080/17460441.2019.1604675. [DOI] [PubMed] [Google Scholar]
- 4.Carroll AR; Copp BR; Davis RA; Keyzers RA; Prinsep MR Marine Natural Products. Nat. Prod. Rep 2019, 36 (1), 122–173. 10.1039/C8NP00092A. [DOI] [PubMed] [Google Scholar]
- 5.Vijayakumar S; Menakha M Pharmaceutical Applications of Cyanobacteria—A Review. J. Acute Med 2015, 5 (1), 15–23. 10.1016/j.jacme.2015.02.004. [DOI] [Google Scholar]
- 6.Khalifa SAM; Elias N; Farag MA; Chen L; Saeed A; Hegazy MEF; Moustafa MS; Abd El-Wahed A; Al-Mousawi SM; Musharraf SG; Chang FR; Iwasaki A; Suenaga K; Alajlani M; Goransson U; El-Seedi HR, Marine Natural Products: A Source of Novel Anticancer Drugs. Mar Drugs 2019, 17 (9), 491 10.3390/md17090491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang I-S; Zimba PV Cyanobacterial Bioactive Metabolites—A Review of Their Chemistry and Biology. Harmful Algae 2019, 86, 139–209 10.1016/j.hal.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Luesch H; Moore RE; Paul VJ; Mooberry SL; Corbett TH Isolation of Dolastatin 10 from the Marine Cyanobacterium Symploca Species VP642 and Total Stereochemistry and Biological Evaluation of Its Analogue Symplostatin 1. J. Nat. Prod 2001, 64 (7), 907–910. 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 9.Pettit GR Kamano Y; Herald CL; Fujii Y; Kizu H; Boyd MR; Boettner FE; Doubek DL; Schmidt JM; Chapuis JC; Michel C Isolation of Dolastatins 10–15 from the Marine Mollusk Dolabella-Auricularia. Tetrahedron 1993, 49 (41), 9151–9170. 10.1016/0040-4020(93)80003-C. [DOI] [Google Scholar]
- 10.Linington RG; Edwards DJ; Shuman CF; McPhail KL; Matainaho T; Gerwick WH Symplocamide A, a Potent Cytotoxin and Chymotrypsin Inhibitor from the Marine Cyanobacterium Symploca Sp. J. Nat. Prod 2008, 71 (1), 22–27. 10.1021/np070280x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrigan GG; Luesch H; Yoshida WY; Moore RE; Nagle DG; Paul VJ; Mooberry SL; Corbett TH; Valeriote FA Symplostatin 1: A Dolastatin 10 Analogue from the Marine Cyanobacterium Symploca Hydnoides. J. Nat. Prod 1998, 61 (9), 1075–1077. 10.1021/np980321c. [DOI] [PubMed] [Google Scholar]
- 12.Salvador LA; Biggs JS; Paul VJ; Luesch H Veraguamides A-G, Cyclic Hexadepsipeptides from a Dolastatin 16-Producing Cyanobacterium Symploca Cf. Hydnoides from Guam. J. Nat. Prod 2011, 74 (5), 917–927. 10.1021/np200076t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmons TL; McPhail KL; Ortega-Barría E; Mooberry SL; Gerwick WH Belamide A, a New Antimitotic Tetrapeptide from a Panamanian Marine Cyanobacterium. Tetrahedron Lett. 2006, 47 (20), 3387–3390. 10.1016/j.tetlet.2006.03.082. [DOI] [Google Scholar]
- 14.Naman CB; Rattan R; Nikoulina SE; Lee J; Miller BW; Moss NA; Armstrong L; Boudreau PD; Debonsi HM; Valeriote FA; Dorrestein PC; Gerwick WH Integrating Molecular Networking and Biological Assays To Target the Isolation of a Cytotoxic Cyclic Octapeptide, Samoamide A, from an American Samoan Marine Cyanobacterium. Journal of natural products 2017, 80 (3), 625–633. 10.1021/acs.jnatprod.6b00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams PG; Yoshida WY; Moore RE; Paul VJ Tasipeptins A and B: New Cytotoxic Depsipeptides from the Marine Cyanobacterium Symploca Sp. J. Nat. Prod 2003, 66 (5), 620–624. 10.1021/np020582t. [DOI] [PubMed] [Google Scholar]
- 16.Williams PG; Yoshida WY; Moore RE; Paul VJ Micromide and Guamamide: Cytotoxic Alkaloids from a Species of the Marine Cyanobacterium Symploca. J. Nat. Prod 2004, 67 (1), 49–53. 10.1021/np030215x. [DOI] [PubMed] [Google Scholar]
- 17.Pereira F; Aires-de-Sousa J Computational Methodologies in the Exploration of Marine Natural Product Leads. Mar. Drugs 2018, 16 (7), 236 10.3390/md16070236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medema MH; Fischbach MA Computational Approaches to Natural Product Discovery. Nat. Chem. Biol 2015, 11 (9), 639–648. 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blin K; Shaw S; Steinke K; Villebro R; Ziemert N; Lee SY; Medema MH; Weber T antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47 (1), 81–87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro-Muñoz JC; Selem-Mojica N; Mullowney MW; Kautsar SA; Tryon JH; Parkinson EI; De Los Santos ELC; Yeong M; Cruz-Morales P; Abubucker S; Roeters A; Lokhorst W; Fernandez-Guerra A; Cappelini LTD; Goering AW; Thomson RJ; Metcalf WW; Kelleher NL; Barona-Gomez F; Medema MH A Computational Framework to Explore Large-Scale Biosynthetic Diversity. Nat. Chem. Biol 2019, 1–9. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min CK; Machado H; Jang KH; Trzoss L; Jensen PR; Fenical W Integration of Genomic Data with NMR Analysis Enables Assignment of the Full Stereostructure of Neaumycin B, a Potent Inhibitor of Glioblastoma from a Marine-Derived Micromonospora J. Am. Chem. Soc 2018, 140 (34), 10775–10784. 10.1021/jacs.8b04848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M; Carver JJ; Phelan VV; Sanchez LM; Garg N; Peng Y; Nguyen DD; Watrous J; Kapono CA; Luzzatto-Knaan T; Porto C; Bouslimani A; Melnik AV; Meehan MJ; Liu WT; Criisemann M; Boudreau PD; Esquenazi E; Sandoval-Calderon M; Kersten RD; Pace LA; Quinn RA; Duncan KR; Hsu CC; Floros DJ; Gavilan RG; Kleigrewe K; Northen T; Dutton RJ; Parrot D; Carlson EE; Aigle B; Michelsen CF; Jelsbak L; Sohlenkamp C; Pevzner P; Edlund A; McLean J; Piel J; Murphy BT; Gerwick L; Liaw CC; Yang YL; Humpf HU; Maansson M; Keyzers RA; Sims AC; Johnson AR; Sidebottom AM; Sedio BE; Klitgaard A; Larson CB; Boya CA; Torres-Mendoza D; Gonzalez DJ; Silva DB; Marques LM; Demarque DP; Pociute E; O’Neill EC; Briand E; Helfrich EJN; Granatosky EA; Glukhov E; Ryffel F; Houson H; Mohimani H; Kharbush JJ; Zeng Y; Vorholt JA; Kurita KL; Charusanti P; McPhail KL; Nielsen KF; Vuong L; Elfeki M; Traxler MF; Engene N; Koyama N; Vining OB; Baric R; Silva RR; Mascuch SJ; Tomasi S; Jenkins S; Macherla V; Hoffman T; Agarwal V; Williams PG; Dai JQ; Neupane R; Gurr J; Rodriguez AMC; Lamsa A; Zhang C; Dorrestein K; Duggan BM; Almaliti J; Allard PM; Phapale P; Nothias LF; Alexandrovr T; Litaudon M; Wolfender JL; Kyle JE; Metz TO; Peryea T; Nguyen DT; VanLeer D; Shinn P; Jadhav A; Muller R; Waters KM; Shi WY; Liu XT; Zhang LX; Knight R; Jensen PR; Palsson BO; Pogliano K; Linington RG; Gutierrez M; Lopes NP; Gerwick WH; Moore BS; Dorrestein PC; Bandeira N Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 2016, 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst M; Kang KB; Caraballo-Rodríguez AM; Nothias L-F; Wandy J; Chen C; Wang M; Rogers S; Medema MH; Dorrestein PC; van der Hooft JJJ MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 2019, 9 (7), 144 10.3390/metabo9070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva RR; Wang M; Nothias L-F; van der Hooft JJJ; Caraballo-Rodríguez AM; Fox E; Balunas MJ; Klassen JL; Lopes NP; Dorrestein PC Propagating Annotations of Molecular Networks Using in Silico Fragmentation. PLOS Comput. Biol 2018, 14 (4), e1006089 10.1371/journal.pcbi.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohimani H; Gurevich A; Mikheenko A; Garg N; Nothias L-F; Ninomiya A; Takada K; Dorrestein PC; Pevzner PA Dereplication of Peptidic Natural Products through Database Search of Mass Spectra. Nat. Chem. Biol 2017, 13 (1), 30–37. 10.1038/nchembio.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M; Jarmusch AK; Vargas F; Aksenov AA; Gauglitz JM; Weldon K; Petras D; da Silva R; Quinn R; Melnik AV; van der Hooft JJJ; Caraballo Rodríguez AM; Nothias LF; Aceves CM; Panitchpakdi M; Brown E; Di Ottavio F; Sikora N; Elijah EO; Labarta-Bajo L; Gentry EC; Shalapour S; Kyle KE; Puckett SP; Watrous JD; Carpenter CS; Bouslimani A; Ernst M; Swafford AD; Zúñiga EI; Balunas MJ; Klassen JL; Loomba R; Knight R; Bandeira N; Dorrestein PC MASST: A Web-Based Basic Mass Spectrometry Search Tool for Molecules to Search Public Data. Nat. Biotechnol 2020, doi: 10.1038/s41587-019-0375-9. [DOI] [Google Scholar]
- 27.Wandy J; Zhu Y; van der Hooft JJJ; Daly R; Barrett MP; Rogers S Ms2lda.Org: Web-Based Topic Modelling for Substructure Discovery in Mass Spectrometry. Bioinformatics 2018, 34 (2), 317–318. 10.1093/bioinformatics/btx582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dührkop K; Fleischauer M; Ludwig M; Aksenov AA; Melnik AV; Meusel M; Dorrestein PC; Rousu J; Böcker S SIRIUS 4: A Rapid Tool for Turning Tandem Mass Spectra into Metabolite Structure Information. Nat. Methods 2019, 16 (4), 299–302. 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 29.Humisto A; Jokela J; Liu L; Wahlsten M; Wang H; Permi P; Machado JP; Antunes A; Fewer DP; Sivonen K The Swinholide Biosynthesis Gene Cluster from a Terrestrial Cyanobacterium, Nostoc Sp. Strain UHCC 0450. Appl. Environ. Microbiol 2017, 84 (3). 10.1128/AEM.02321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J Identifying Small Molecules via High Resolution Mass Spectrometry: Communicating Confidence. Environ. Sci. Technol 2014, 48 (4), 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 31.Sumner LW; Amberg A; Barrett D; Beale MH; Beger R; Daykin CA; Fan TW-M; Fiehn O; Goodacre R; Griffin JL; J. L; Hankemeier T; Hardy N; Harnly J; Higashi R; Kopka J; Lane AN; Lindon JC; Marriott P; Nicholls AW; Reily MD; Thaden JJ; Viant MR Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3 (3), 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C; Zhang B; Timári I; Somogyi Á; Li DW; Adcox HE; Gunn JS; Bruschweiler-Li L; Brüschweiler R Accurate and Efficient Determination of Unknown Metabolites in Metabolomics by NMR-Based Molecular Motif Identification Anal Chem. 2019, 91 (24), 15686–15693. 10.1021/acs.analchem.9b03849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfender J-L; Nuzillard J-M; van der Hooft JJJ; Renault J-H; Bertrand S Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography–High-Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem 2019, 91 (1), 704–742. 10.1021/acs.analchem.8b05112. [DOI] [PubMed] [Google Scholar]
- 34.Zhang C; Idelbayev Y; Roberts N; Tao Y; Nannapaneni Y; Duggan BM; Min J; Lin EC; Gerwick EC; Cottrell GW; Gerwick WH Small Molecule Accurate Recognition Technology (SMART) to Enhance Natural Products Research. Sci. Rep 2017, 7 (1). 10.1038/s41598-017-13923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlpine JB; Chen S-N; Kutateladze A; MacMillan JB; Appendino G; Barison A; Beniddir MA; Biavatti MW; Bluml S; Boufridi A; Butler MS; Capon RJ; Choi YH; Coppage D; Crews P; Crimmins MT; Csete M; Dewapriya P; Egan JM; Garson MJ; Genta-Jouve G; Gerwick WH; Gross H; Harper MK; Hermanto P; Hook JM; Hunter L; Jeannerat D; Ji NY; Johnson TA; Kingston DGI; Koshino H; Lee HW; Lewin G; Li J; Linington RG; Liu MM; McPhail KL; Molinski TF; Moore BS; Nam JW; Neupane RP; Niemitz M; Nuzillard JM; Oberlies NH; Ocampos FMM; Pan GH; Quinn RJ; Reddy DS; Renault JH; Rivera-Chavez J; Robien W; Saunders CM; Schmidt TJ; Seger C; Ben S. h. e. n.; Steinbeck C; Stuppner H; Sturm S; Taglialatela-Scafati O; Tantillo DJ; Verpoorte R; Wang BG; Williams CM; Williams PG; Wist J; Yue JM; Zhang C; Xu ZR; Simmler C; Lankin DC; Bisson J; Pauli GF The Value of Universally Available Raw NMR Data for Transparency, Reproducibility, and Integrity in Natural Product Research. Nat Prod Rep. 2019, 36 (1), 35–107. 10.1039/c7np00064b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iandola FN; Han S; Moskewicz MW; Ashraf K; Dally WJ; Keutzer K SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and <0.5MB Model Size. arXiv:1602.07360v4 2016. [Google Scholar]
- 37.van Santen JA; Jacob G; Singh AL; Aniebok V; Balunas MJ; Bunsko D; Neto FC; Castaño-Espriu L; Chang C; Clark TN; Little JLC; Delgadillo DA; Dorrestein PC; Duncan KR; Egan JM; Galey MM; Haeckl FPJ; Hua A; Hughes AH; Iskakova D; Khadilkar A; Lee JH; Lee S; LeGrow N; Liu DY; Macho JM; McCaughey CS; Medema MH; Neupane RP; O’Donnell TJ; Paula JS; Sanchez LM; Shaikh AF; Soldatou S; Terlouw BR; Tran TA; Valentine M; van der Hooft JJJ; Vo DA; Wang MX; Wilson D; Zink KE; Linington RG The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci 2019, 5 (11), 1824–1833. 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alley MC; Scudiero DA; Monks A; Hursey ML; Czerwinski MJ; Fine DL; Abbott BJ; Mayo JG; Shoemaker RH; Boyd MR Feasibility of Drug Screening with Panels of Human Tumor Cell Lines Using a Microculture Tetrazolium Assay. Cancer Res. 1988, 48 (3), 589–601. [PubMed] [Google Scholar]
- 39.Delaglio F; Walker GS; Farley KA; Sharma R; Hoch JC; Arbogast LW; Brinson RG; Marino JP Non-Uniform Sampling for All: More NMR Spectral Quality, Less Measurement Time. Am Pharm Rev. 2017, 20 (4), 339681. [PMC free article] [PubMed] [Google Scholar]
- 40.Ndukwe IE; Shchukina A; Kazimierczuk K; Butts CP Rapid and Safe ASAP Acquisition with EXACT NMR. ChemComm 2016, 52 (86), 12769–12772. 10.1039/C6CC07140F. [DOI] [PubMed] [Google Scholar]
- 41.Schulze-Sünninghausen D; Becker J; Luy B Rapid Heteronuclear Single Quantum Correlation NMR Spectra at Natural Abundance. J. Am. Chem. Soc 2014, 136 (4), 1242–1245. 10.1021/ja411588d. [DOI] [PubMed] [Google Scholar]
- 42.Kitamura M; Schupp PJ; Nakano Y; Uemura D Luminaolide, a Novel Metamorphosis-Enhancing Macrodiolide for Scleractinian Coral Larvae from Crustose Coralline Algae. Tetrahedron Lett. 2009, 50 (47), 6606 10.1016/j.tetlet.2009.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueoka R; Uria AR; Reiter S; Mori T; Karbaum P; Peters EE; Helfrich EJN; Morinaka BI; Gugger M; Takeyama H; Matsunaga S; Piel J Metabolic and Evolutionary Origin of Actin-Binding Polyketides from Diverse Organisms. Nat. Chem. Biol 2015, 11 (9), 705–712. 10.1038/nchembio.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao Y; Li P; Zhang D; Glukhov E; Gerwick L; Zhang C; Murray TF; Gerwick WH Samholides, Swinholide-Related Metabolites from a Marine Cyanobacterium Cf. Phormidium Sp. J. Org. Chem 2018, 83 (6), 3034–3046. 10.1021/acs.joc.8b00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitagawa I; Kobayashi M; Katori T; Yamashita M; Tanaka J; Doi M; Ishida T Absolute Stereostructure of Swinholide A, a Potent Cytotoxic Macrolide from the Okinawan Marine Sponge Theonella Swinhoei. J. Am. Chem. Soc 1990, 112 (9), 3710–3712. 10.1021/ja00165a094. [DOI] [Google Scholar]
- 46.Ishibashi M; Moore RE; Patterson GML; Xu C; Clardy J Scytophycins, Cytotoxic and Antimycotic Agents from the Cyanophyte Scytonema Pseudohofmanni. J. Org. Chem 1986, 51 (26), 5300–5306. 10.1021/jo00376a047. [DOI] [Google Scholar]
- 47.Nicolaou KC; Ajito K; Patron AP; Khatuya H; Richter PK; Bertinato P Total Synthesis of Swinholide A. J. Am. Chem. Soc 1996, 118 (12), 3059–3060. 10.1021/ja954211t. [DOI] [Google Scholar]
- 48.Andrianasolo EH; Gross H; Goeger D; Musafija-Girt M; McPhail K; Leal RM; Mooberry SL; Gerwick WH Isolation of Swinholide A and Related Glycosylated Derivatives from Two Field Collections of Marine Cyanobacteria. Org. Lett 2005, 7 (7), 1375–1378. 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 49.Duggan BM; Cullum R; Fenical W; Amador LA; Rodríguez AD; La Clair JJ Searching for Small Molecules with an Atomic Sort. Angew Chem Int Ed. 2020, 59 (3), 1144–1148. 10.1002/ange.201911862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueoka R; Nakao Y; Kawatsu S; Yaegashi J; Matsumoto Y; Matsunaga S; Furihata K; van Soest RWM; Fusetani N Gracilioethers A-C, Antimalarial Metabolites from the Marine Sponge Agelas Gracilis. J. Org. Chem 2009, 74 (11), 4203–4207. 10.1021/jo900380f. [DOI] [PubMed] [Google Scholar]
- 51.Renner MK; Shen Y-C; Cheng X-C; Jensen PR; Frankmoelle W; Kauffman CA; Fenical W; Lobkovsky E; Clardy J Cyclomarins A-C, New Antiinflammatory Cyclic Peptides Produced by a Marine Bacterium (Streptomyces Sp.). J. Am. Chem. Soc 1999, 121 (49), 11273–11276. 10.1021/ja992482o [DOI] [Google Scholar]
- 52.Vasudevan D; Rao SPS; Noble CG Structural Basis of Mycobacterial Inhibition by Cyclomarin A. J. Biol. Chem 2013, 288 (43), 30883–30891. 10.1074/jbc.M113.493767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt EK; Riwanto M; Sambandamurthy V; Roggo S; Miault C; Zwingelstein C; Krastel P; Noble C; Beer D; Rao SPS; Au M; Niyomrattanakit P; Lim V; Zheng J; Jeffery D; Pethe K; Camacho LR. The Natural Product Cyclomarin Kills Mycobacterium Tuberculosis by Targeting the ClpC1 Subunit of the Caseinolytic Protease. Angew Chem Int Ed. 2011, 50 (26), 5889–5891. 10.1002/anie.201101740. [DOI] [PubMed] [Google Scholar]
- 54.Ma J; Huang H; Xie Y; Liu Z; Zhao J; Zhang C; Jia Y; Zhang Y; Zhang H; Zhang TY; Ju JH Biosynthesis of Ilamycins Featuring Unusual Building Blocks and Engineered Production of Enhanced Anti-Tuberculosis Agents. Nat. Commun 2017, 8 (1), 1–10. 10.1038/s41467-017-00419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu L; Wang B; Kulkarni A; Geders TW; Grindberg RV; Gerwick L; Håkansson K; Wipf P; Smith JL; Gerwick WH; Sherman DH Metamorphic Enzyme Assembly in Polyketide Diversification. Nature 2009, 459 (7247), 731–735. 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


