Abstract
Here we describe methods for synthesizing a cationic, multi-arm Avidin (mAv) nano-construct that has a wide range of applications in drug delivery and imaging of negatively charged tissues. We use Avidin-biotin technology that gives the flexibility for conjugating biotinylated Dexamethasone to mAv by simple mixing at room temperature. We also describe methods to control hydrolysis rates of ester linkers to enable sustained (and tunable) drug release rates in therapeutic doses.
-
•
Multi-arm structure provides multiple sites for covalent conjugation of drugs
-
•
Use of Avidin-biotin reaction gives multi-arm nano-construct a modular design enabling conjugation and delivery of similar sized biotinylated drugs.
Keywords: Avidin, Biotin, PEGs, Ester linkers, Negatively charged tissues, Drug delivery
Graphical abstract
Specification table
| Subject Area: | Pharmacology, Toxicology and Pharmaceutical Science |
| More specific subject area: | Charge-based drug delivery platform for targeting negatively charged tissues |
| Method name: | Avidin-biotin technology to synthesize multi-arm nano-construct for drug delivery |
| Name and reference of original method: | • T. He, C. Zhang, A. Vedadghavami, S. Mehta, H.A. Clark, R.M. Porter, A.G. Bajpayee, Multi-arm Avidin nano-construct for intra-cartilage delivery of small molecule drugs, JCR, (2020). 10.1016/j.jconrel.2019.12.020 |
| • A.G. Bajpayee, M.A. Quadir, P.T. Hammond, A.J. Grodzinsky, Charge based intra-cartilage delivery of single dose dexamethasone using Avidin nano-carriers suppresses cytokine-induced catabolism long term, Osteoarthritis Cartilage, 24 (2016) 71–81. 10.1016/j.joca.2015.07.010 | |
| Resource availability: | •10.1016/j.jconrel.2019.12.020 |
| •10.1016/j.joca.2015.07.010 |
Method details
Overview
Targeted drug delivery to joint tissues like cartilage remains a challenge that has prevented clinical translation of promising osteoarthritis (OA) drugs [1]. Local intra-articular (IA) injection of drugs suffers from rapid clearance from the joint space and slow diffusive transport through the dense, avascular cartilage matrix comprising of negatively charged aggrecan-glycosaminoglycans (GAGs). The high negative fixed charge density (FCD) of cartilage provides a unique opportunity to use electrostatic interactions for enhancing transport, uptake, and retention of cationic drug carriers [2]. We recently showed that there exists an optimal net positive charge to deliver a drug of given size to a tissue of known FCD that will result in rapid penetration through the full thickness of tissue, highest intra-tissue uptake and long-term retention [3]. Optimal net positive charge on the carrier is chosen to enable weak and reversible binding with the intra-tissue negatively charged groups such that the drug and its carrier can penetrate through the full tissue thickness and not get stuck in the tissue's superficial zones. Despite weak binding, the high negative FCD of aggrecan-GAGs inside cartilage greatly increases the residence time of optimally charged cationic drug carriers. Similarly, cationic glycoprotein, Avidin, due to its optimal net size (< 10 nm hydrodynamic diameter) and charge (between +6 and +20) was shown to penetrate through full thickness of rabbit cartilage following IA injection resulting in a high intra-cartilage uptake ratio of 180 (implying 180x higher concentration of Avidin inside cartilage than surrounding fluid at equilibration) and was still found to be present through the full-thickness of cartilage two weeks following its IA administration in a rabbit anterior cruciate ligament transection model of post-traumatic OA [4], [5], [6].
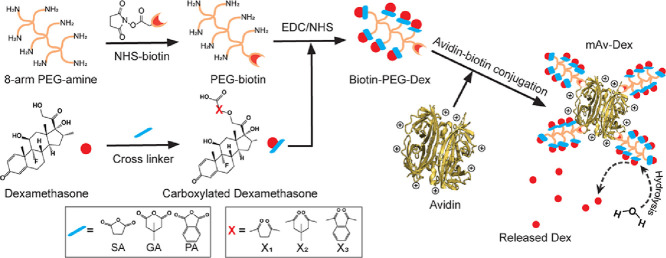
Here we synthesize a multi-arm nano-construct of Avidin (mAv) by conjugating it with four biotinylated 8-arm PEGs using Avidin-biotin binding that provided 28 sites for covalent conjugation of small molecule drugs. We conjugate mAv to Dexamethasone (mAv-Dex), a broad spectrum glucocorticoid, using a combination of hydrolysable ester linkers derived from succinic anhydride (SA), 3,3-dimethylglutaric anhydride (GA) and phthalic anhydride (PA) in a 2:1:1 molar ratio that enabled 50% drug release within 38.5 ± 1.5 h followed by sustained release in therapeutic doses over two weeks. We show that mAv-Dex can rapidly penetrate through the full thickness of cartilage in high concentration, have long intra-cartilage residence time in both healthy and arthritic cartilage via weak-reversible binding with negatively charged aggrecan-GAGs and effectively reverse cytokine induced catabolic activity significantly greater than free (unmodified) Dex; these results are discussed in detail in our recent paper published in the Journal of Controlled Release [7]. In the current article, we present a detailed method for synthesizing mAv nano-construct and ester linkers with different hydrolysis rates for conjugating a small molecule OA drug, Dex.
mAv can be conjugated with a broad array of small molecule OA drugs and their combinations for sustained delivery to chondrocytes enabling OA treatment with a single injection, thereby eliminating toxicity issues associated with multiple high dose injections [8]. Drug release rates can be tuned by using a combination of linkers presented here, depending on the type of drug, tissue and the desired release rates and dosing. This charge-based platform can also be used for delivering a wide range of drugs to other tissues with similar properties, such as meniscus, intervertebral disk, and fracture callus.
Experimental
Materials
10 kDa 8-arm polyethylene glycol (PEG) amine hydrochloride salt was purchased from Advanced Biochemicals (Lawrenceville, GA). N—Hydroxysuccinimido (NHS)-biotin, 1-ethyl-3-(3 dimethylaminopropyl) carbodiimide hydrochloride (EDC), N-hydroxysulfosuccinimide (NHSS), Avidin and Avidin-Texas Red conjugated, 4′-hydroxybenzene-2-carboxylic acid (HABA), 3.5 kDa molecular weight cut-off (MWCO) and 7.0 kDa MWCO SnakeSkin dialysis tubing, 2x Tris-Glycine Native sample buffer, acetonitrile, pyridine, trifluoroacetic acid, acetic acid, sodium hydroxide and 1 N hydrochloric acid solution was purchased from Thermo Fisher Scientific (Waltham, MA). Dexamethasone (Dex), succinic anhydride (SA), 3,3-dimethylglutaric anhydride (GA), phthalic anhydride (PA), dimethyl sulfoxide-d6 (DMSO-d6) containing 0.03% (v/v) tetramethylsilane, fluorescein isothiocyanate isomer I (FITC), dimethylaminopyridine (DMAP), potassium iodide, iodine, barium chloride, β-mercaptoethanol and other salts were purchased from Sigma-Aldrich (St. Louis, MO). 2x Laemmli Sample Buffer, 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels (12-well), Coomassie Brilliant Blue R-250 were purchased from Bio-Rad (Hercules, CA).
Biotinylation of 8-arm PEG
The first step is to add one biotin to 8-arm PEG. PEG was biotinylated by reaction with NHS-biotin following scheme (1). Briefly, 10 mg (0.001 mmol, 1.0 equiv.) of 8-arm PEG was dissolved in 500 µL of nanopure water and 1.7 mg (0.005 mmol, 5.0 equiv.) of NHS-biotin was dissolved in 500 µL of DMSO. NHS-biotin solution was then added dropwise to the PEG solution (5:1 molar ratio) and reacted for 2 h under gentle rotation (10 rpm, HulaMixer Sample Mixer, Thermo Fisher Scientific, Waltham, MA) at room temperature. Excess NHS-biotin was removed from the PEG-biotin conjugate solution using dialysis (7.0 kDa MWCO) for 24 h against phosphate buffer saline (PBS). A 5:1 molar ratio of NHS-biotin and 8-arm PEG was chosen as it enabled conjugation of one biotin per PEG. The extent of biotinylation was confirmed using the HABA dye assay [9] as described below.
| (1) |
Characterization of biotinylated PEG
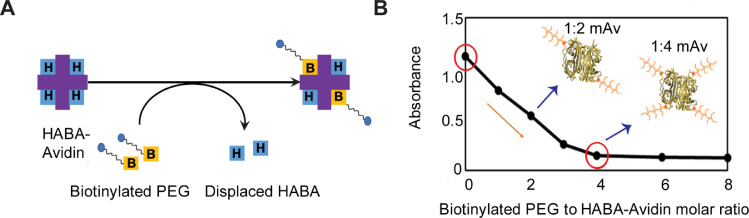
The extent of biotinylation of PEG (PEG-biotin) and the loading of PEG-biotin on Avidin were determined by using the HABA colorimetric assay [9]. Changes in absorbance of HABA-Avidin complex at 500 nm due to competitive displacement by the biotinylated PEG was used to estimate the degree of biotinylation (Fig. 1A). HABA dye was dissolved in 10 mL of nanopure water (2.42 mg/mL) and filtered using 0.2 µm filter. Excessive HABA dye was added to Avidin solution to a final concentration of 0.82 mg/mL (initial absorbance of 1.2 detected by Synergy H1 Microplate Reader, BioTek, Winooski, VT). 20 µL of graded concentrations of PEG-biotin were added to 180 µL of HABA-Avidin complex (1:1 through 8:1 molar ratio of PEG-biotin to HABA-Avidin) that competitively displaced HABA from biotin binding sites of Avidin thereby reducing the absorbance value. 100% PEGylation of Avidin was achieved when the change in absorbance achieved a plateau. As shown in Fig. 1B, the reduction in absorbance value with increasing molar ratio of biotinylated PEG to HABA-Avidin from 1:1 to 4:1, following which a plateau is achieved meaning that a majority of biotin sites on Avidin were occupied by PEG-biotin indicates the formation of 1:4 mAv. Using the Beer-Lambert Law [9], an average of 1.28 ± 0.02 biotin per PEG molecule was estimated. In addition, matrix-assisted laser-desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) was used to confirm the molecular weight of biotinylated PEG. 10 µL of 8-arm PEG, or 10 µL of 8-arm PEG-biotin (1 mg/mL) was mixed with 10 µL sinapinic acid matrix, and the change in molecular weight before and after biotinylation of 8-arm PEG was confirmed using MALDI-TOF spectrometry (Bruker Microflex II).
Fig. 1A.
Schematic of HABA dye assay: HABA binds with Avidin resulting in high absorbance value but is competitively displaced by biotin or biotinylated PEG reducing the absorbance value. B. Titration curve of biotinylated PEG with HABA-Avidin mixture. Absorbance value dropped with increasing biotinylated PEG: HABA-Avidin molar ratio, and a plateau was achieved after 4:1 molar ratio confirming that all four binding sites of Avidin (Av) were occupied by PEGs to form 1:4 mAv configuration. 1:2 mAv has 2 PEGs conjugated to Avidin.
Synthesis of mAv nano-construct
1:4 mAv containing four 8-arm PEGs was synthesized by mixing 4 mol of biotinylated PEG with 1 mol of Avidin in nanopure water for 30 min under gentle rotation (10 rpm, HulaMixer Sample Mixer) at room temperature. The HABA dye assay (Fig. 1B) was used to confirm that all four biotin binding sites on Avidin were occupied with PEG-biotin as subsequent addition of PEG-Biotin to the Avidin-HABA mixture did not change the absorbance value. This confirmed the successful synthesis of 1:4 mAv.
Characterization of mAv
Conjugation of PEG-biotin to Avidin was also confirmed by using native polyacrylamide gel electrophoresis (PAGE) in 7.5% separating gel. Since temperature-dependent dissociation of Avidin tetramer was witnessed when temperature was higher than 25 °C [10], it was common to find an Avidin monomer band (~17 kDa) using SDS-PAGE even if we didn't denature the protein samples before loading. Therefore, native PAGE in reverse polarity was used to analyze Avidin conjugates. In brief, 12 µL of protein samples (~7.5 µg protein) were mixed with 4 µL of 2x Tris-Glycine Native sample buffer (Novex) without heating. Since the isoelectric point of Avidin is 10.5 and the protein mobility depends on both its own charge and molecular weight in the native PAGE gel, the electrode polarity had to be reversed (anode was inserted at the top of gel and cathode was inserted at the bottom of gel). Electrophoresis was performed for approximately 4 h in 1x solution of non-SDS tris-base running buffer at 200 V, 40 mA and 4 °C [11].
Native gel was stained using iodine solution and Coomassie Brilliant Blue R-250. Gel was fixed by fixing solution (10% acetic acid, 30% water and 50% ethanol) and then washed with deionized (DI) water for 20 min. Gel was then incubated in 5% barium chloride solution for 15 min followed by 3 washes in DI water. Subsequently, the gel was stained with potassium iodide and iodine solution (dissolve 2.0 g potassium iodide (KI) in 50 mL DI water and then add 1.3 g of iodine to KI solution when the solution has cooled to room temperature) for 5 min to identify free or conjugated PEG. Following destaining iodine solution by DI water, the gel was then stained with Coomassie Brilliant Blue R-250 for Avidin, and de-stained three times in 100 mL of 10% acetic acid solution for 1 h.
PEGylation of Avidin in 1:2 and 1:4 mAv was further confirmed by using H—Class Acquity ultra performance liquid chromatography (UPLC) (Waters Corp, Milford, MA) equipped with an Acquity UPLC BEH200 Size Exclusion Column (200 Å, 1.7 µm column, 4.6 × 300 mm) with 20 mM ammonium bicarbonate buffer as the mobile phase at 0.2 mL/min. Avidin was detected at 280 nm. Zeta potential of Avidin and mAv was measured in nanopure water at 0.45 mg/mL concentration using a Zetasizer Nano-ZS90.
Synthesis of controlled release ester linkers to conjugate Dexamethasone (Dex) to biotinylated PEG
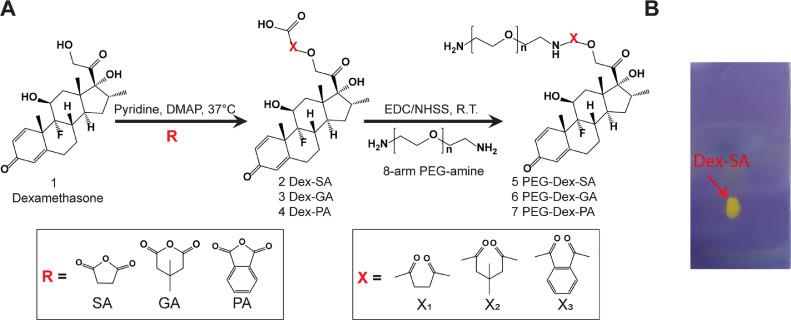
Dex was covalently conjugated to the PEGs of 1:4 mAV nano-construct by using a combination of ester linkers that enabled sustained (and tunable) Dex release over two weeks. Three carboxylated derivatives of Dex were prepared by reacting 36.0 mg Dex (0.092 mmol, 1.0 equiv.) with 46 mg of SA, 52.0 mg of GA or 67.0 mg of PA (0.458 mmol, 5.0 equiv.) in presence of 2 mg DMAP (0.015 mmol, 0.2 equiv.) as a catalyzer in 1 mL of pyridine (Fig. 2A). The reaction for Dex-SA was conducted in a round bottom flask purged with nitrogen gas for 24 h at room temperature [12]. For Dex-GA and Dex-PA, the reaction time was 48 h at 37 °C. Following completion of the reaction, pyridine was evaporated with constant purging of nitrogen gas, and 4 mL of the cold solution containing 25 mL water and 10 mL concentrated HCl was added to the flask to precipitate Dex-SA, Dex-GA and Dex-PA out. A white precipitate was observed, which was stirred for 10 min and then centrifuged at 10,000 g for 5 min for 5 cycles. In each cycle, the supernatant was replaced with fresh cold solution. The final products of Dex-SA, Dex-GA, and Dex-PA (Compounds 2, 3, 4 respectively) were lyophilized, weighed and stored at −20 °C for future use. Their structures were confirmed using Proton Nuclear Magnetic Resonance (1H NMR). Modification of three kinds of Dex were verified using 500 MHz 1H NMR (Varian Inova. Agilent Technologies). 1–2 mg of mixture of PEG or carboxylated derivatives of Dex (Compounds 2, 3, 4) was dissolved in 700 µL DMSO‑d6. The obtained NMR data was analyzed using MestRe Nova software. The carboxyl groups incorporated in Dex were verified using thin layer chromatography (TLC). First, the baseline was marked about 1.0 cm from the bottom of TLC plate (Silica gel on TLC Al foils, 5 cm × 10 cm, Sigma-Aldrich, St. Louis, MO). Then, a capillary was used to draw in a small quantity of 10 mg/mL product solution, which was placed at least 1.0 cm from the edge of the TLC plate. The plate was then placed in a chamber containing developing solvent (80% methanol and 20% chloroform). The chamber was covered with a lid to allow for the solvent front to reach top of the plate. The TLC plate was removed and immersed into Bromocresol Blue solution to stain carboxyl groups on Dex. Fig. 2B shows a yellow spot on the TLC plate demonstrating that the carboxyl group was successfully incorporated into Dex.
Fig. 2A.
Schematic of Dex conjugation with 8-arm PEG-amine using crosslinkers (R: SA, GA and PA) to form different ester linker spacers (X: X1, X2 and X3). B. Yellow point stained by Bromocresol Blue in TLC plate demonstrating that carboxyl group was successfully incorporated into Dex.
Synthesized carboxylated derivatives of Dex were then conjugated to PEG-biotin using EDC/NHS chemistry (Fig. 2A) to form compounds 5, 6 and 7. Briefly, 5.0 mg of Dex-SA, Dex-GA or Dex-PA (0.010 mmol, 100.0 equiv.) was dissolved initially in 120 µL of DMSO and added 600 µL of 2-morpholinoethanesulfonic acid (MES) dropwise. Then, 19.2 mg of EDC (0.104 mmol, 104.0 equiv.) and 21.7 mg NHSS (0.092 mmol, 92.0 equiv.) were added to Dex-SA, Dex-GA and Dex-PA solution, and all of them were purged with nitrogen to activate the reaction for 30 min. Subsequently, 1.0 mg of PEG-biotin (0.100 µmol, 1.0 equiv.) was added to each of the solutions and reacted for 2 h at room temperature, purged with nitrogen gas. Upon completion of the reaction, the final product was dialyzed using 7.0 kDa MWCO membrane to remove the excessive reagents under 4 °C for 24 h. The pure product was then lyophilized and stored at −20 °C for future purposes. The formation of these three chemical compounds 5, 6 and 7 was then confirmed using 1H-NMR following the same procedures of 1H-NMR mentioned above.
Dex loading content
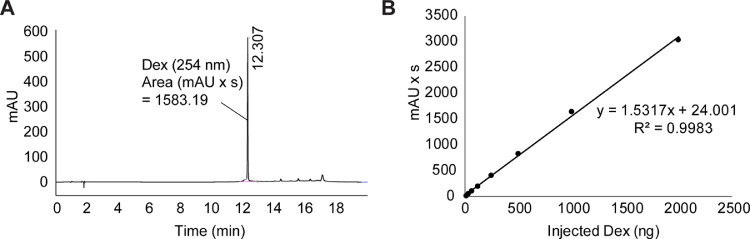
To estimate Dex loading, compounds 5, 6, and 7 were hydrolyzed using 0.1 N hydrochloric acid overnight and neutralized against 0.1 N sodium hydroxide. The amount of Dex released was quantified by HPLC (Agilent Technologies 1260 infinity II) equipped with a Variable Wavelength Detector using a Poroshell 120 EC—C18 4.6 × 150 mm column. A gradient of solvent A (0.1% trifluoroacetic acid (TFA) in water) and solvent B (0.1% TFA in acetonitrile) was used. The concentration of solvent B was increased linearly from 5% to 65% over 15 min. Column temperature of 30 °C and a flow rate of 1.0 mL/min were used. Dex was eluted at 12.3 min (254 nm) and peak area of UV absorbance (mAU * s) was used for calculation (Fig. 3A). Standard curve plotting absorbance vs injected standard Dex is shown in Fig. 3B. Compounds 5, 6, and 7 were conjugated to Avidin to form 1:4 mAv and the drug loading content (DLC) of the conjugate was calculated as:
Fig. 3A.
Reverse Phase-HPLC analysis of Dex. B. Standard curve of injected standard Dex (ng) vs absorbance (mAU x s).
Drug release study
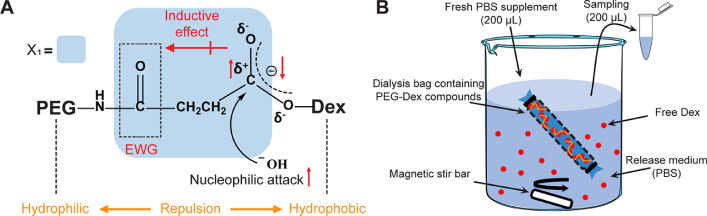
First, we conjugated Dex to PEG using hydrolysable ester linkers derived from SA (Fig. 2A). However, the fast-release ester linker in PEG-Dex-SA had a half-life of 6.8 ± 0.2 h. The hydrolysis of an ester bond begins when hydroxide ions of water attack the electrophilic carbon in the ester bond (Fig. 4A), breaking the p-π conjugation of ester bond creating a tetrahedral intermediate [13]. Since the adjacent carbonyl group from the amide bond tends to compete with and withdraw electrons from the ester bond (inductive effect) resulting in a decrease in the ester bond's electron density [14], the carbon in ester bond becomes more electrophilic and reactive to nucleophilic attack from hydroxide ion causing a faster release. Furthermore, hydrophobic Dex conjugated to hydrophilic polymer PEG can produce repulsive forces, thereby accelerating the separation of Dex from PEG. As for PEG-Dex-GA, the GA cross linker will increase the carbon spacer length between the ester and amide bonds thus weakening the inductive effect of the carbonyl group. The two methyl groups in the GA branched chain can also donate electrons, thereby stabilizing the ester bond. Similarly, the phenyl group in PA can donate numerous electrons to the ester bond (PEG-Dex-PA). We, therefore, used ester linkers derived from SA, GA and PA by physically mixing them in 2:1:1 molar ratio of Dex to formulate controlled release PEG-Dex (2:1:1) or abbreviated as mAv-Dex (2:1:1) after conjugation with Avidin, which enabled 50% release of Dex in 38.5 h followed by a sustained release of the remaining drug over the next two weeks. Dex release rates from Dex-PEG-biotin were estimated in PBS at pH 7.4, 37 °C using dialysis tubing (7.0 kDa MWCO) with continuous shaking under sink conditions: Dex concentration was kept 10x lower than the saturation solubility of Dex in PBS (Fig. 4B). At different time intervals (0 h, 0.5 h, 1 h, 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 24 h, 48 h, 72 h, 96 h, 120 h and 144 h), 200 µL of release media was used to estimate the Dex concentration by HPLC, which was replaced by equal amount of fresh release media.
Fig. 4A.
Mechanism of fast hydrolysis of PEG-Dex-SA in PBS (pH 7.4). Carbonyl in amide bond, an electron withdrawing group (EWG), withdraws electrons from methylene and ester bonds (inductive effect) thereby decreasing ester bond's electron density and making it more electrophilic (δ+) and reactive to nucleophilic attack from hydroxide ion (−OH) causing faster hydrolysis of the ester bond. Repulsion between hydrophilic PEG and hydrophobic Dex further strains the ester linker making it unstable. When X1 is replaced by X2 or X3, the carbon spacer length is increased that weakens the inductive effect of carbonyl and donates more electrons to stabilize the ester bond. This also reduces the repulsive effects between PEG and Dex. B. In-vitro setup to study drug release rates.
Results and discussion
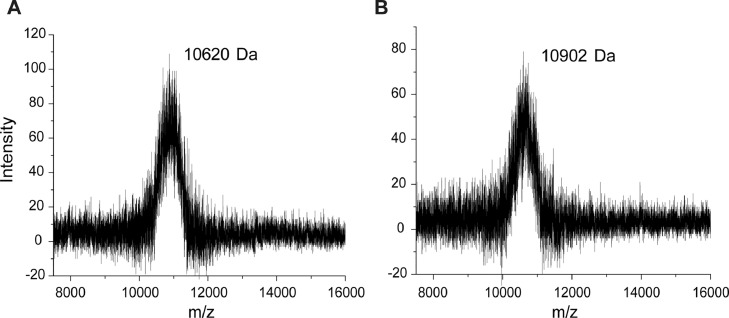
MALDI-TOF MS for biotinylated PEG
In Fig. 5A, the molecular weight of PEG was 10,620 Da; this increased to 10,902 Da following biotinylation (Fig. 5B) showing that an average 1.15 biotins per PEG were present. Using the Beer-Lambert Law [9], an average of 1.28 ± 0.02 biotins per PEG molecule was estimated (Fig. 1B), which is consistent with the mass spectrometry data.
Fig. 5.
Confirmation of A. PEG and B. biotinylated PEG using MALDI-TOF MS. The calculated mass of PEG is 10,620 Da and biotinylated PEG is 10,902 Da.
Characterization of mAv-Dex
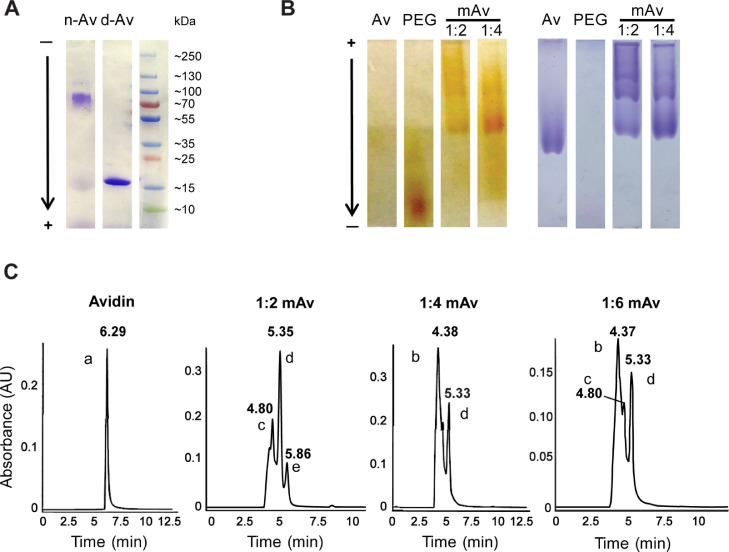
We first tried using SDS-PAGE (4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 12-well) to confirm the Avidin tetramer. However, Fig. 6A shows that Avidin had denatured, (d-Av, treated by β-mercaptoethanol and boiling) resulting in an Avidin monomer band (~17 kDa) in the gel. The Avidin tetramer still dissociated despite efforts to prevent denaturing by mixing it with only 2x Laemmli Sample Buffer (n-Av),. Therefore, native PAGE and UPLC were used instead to confirm PEGylation of mAv. Fig. 6B shows native PAGE gel in reverse polarity used to confirm PEGylation in 1:2 and 1:4 mAv containing two or four 8-arm PEGs, respectively. PEG was stained yellow with iodine and protein was stained blue with Coomassie Brilliant Blue R-250. In PEG-staining (left), bands only appear in the PEG and mAv channels. However, in protein-staining (right), bands only appear in the Avidin and mAv channels. Therefore, bands at the same position in the mAv channels with both PEG-staining and protein-staining verified the formation of mAv. Furthermore, UPLC confirmed that a majority of the population in 1:4 mAv or 1:6 mAv had 4 PEGs conjugated to Avidin (peak ‘b’ at 4.38 min or 4.37 min) followed by a secondary population of mAv with 2 PEGs (peak ‘d’ at 5.33 min) (Fig. 6C). UPLC of 1:2 mAv also confirmed that the majority of Avidin conjugated with two PEGs (peak ‘d’ at 5.35 min) followed by configurations containing three PEGs (peak ‘c’ at 4.80 min) and one PEG (peak ‘e’ at 5.86 min). No peak for native Avidin (6.29 min) was found in 1:4 mAv and 1:2 mAv confirming that Avidin was successfully PEGylated. The similar peak shape between 1:4 mAv and 1:6 mAv confirmed that increased molar ratio of biotinylated PEG to Avidin from 1:4 to 1:6 didn't increase the percentage of Avidin with 4 PEGs conjugated (peak ‘b’) in this mAv system. Therefore, 1:4 molar ratio of biotinylated PEG to Avidin was finally chosen to synthesize mAv nano-construct. Due to this heterogeneity, the structural properties determined represent the collective behavior of different populations in each formulation.
Fig. 6A.
SDS-PAGE gel (4–20%) of non-denatured Avidin (n-Av) and denatured Avidin (d-Av) stained with Coomassie Brilliant Blue R-250. B. Native PAGE gel (7.5%) of Avidin (Av), PEG, 1:2 mAv and 1:4 mAv under reverse polarity stained with (left) iodine for PEGs and with (right) Coomassie Brilliant Blue R-250 for protein. C. UPLC analysis of standard Avidin shows peak ‘a’ at 6.29 min. 1:6 mAv and 1:4 mAv formulation have similar peak shape showing a majority of mAv with 4 PEGs (peak ‘b’ at 4.38 min or 4.37 min) and a secondary population of mAv with 2 PEGs (peak ‘d’ at 5.33 min). In addition, 1:2 mAv has three peaks, a majority of mAv with 2 PEGs (peak ‘d’ at 5.35 min), a secondary population of mAv with 3 PEGs (peak ‘c’ at 4.80 min) and a minority of mAv with 1 PEGs (peak ‘e’ at 5.86 min).
PEGylation did not reduce mAv's zeta potential (ζ) suggesting minimum shielding of cationic charge (Table 1). Its net size was within the 10 nm size cut-off determined for nanoparticles to penetrate through the full-thickness of cartilage [15]. The hydrodynamic diameter of Avidin and mAv was estimated from their molecular weights using the Stokes-Einstein equation [15]. The net size of mAv was within the 10 nm limit enabling it to penetrate through the full thickness of cartilage similar to unmodified Avidin.
Table 1.
Zeta potential (ζ) and net size (diameter) of Avidin, 1:2 mAv and 1:4 mAv. Data shown as Mean ± SD.
| Formulation | Avidin | 1:2 mAv | 1:4 mAv |
|---|---|---|---|
| ζ (mV) | 18.3 ± 0.5 | 20.3 ± 0.3 | 25.3 ± 0.7 |
| Diameter (nm) | ~7.0 | ~7.6 | ~8.1 |
. 1H NMR spectra of carboxylated derivatives of Dex and PEG-Dex compounds
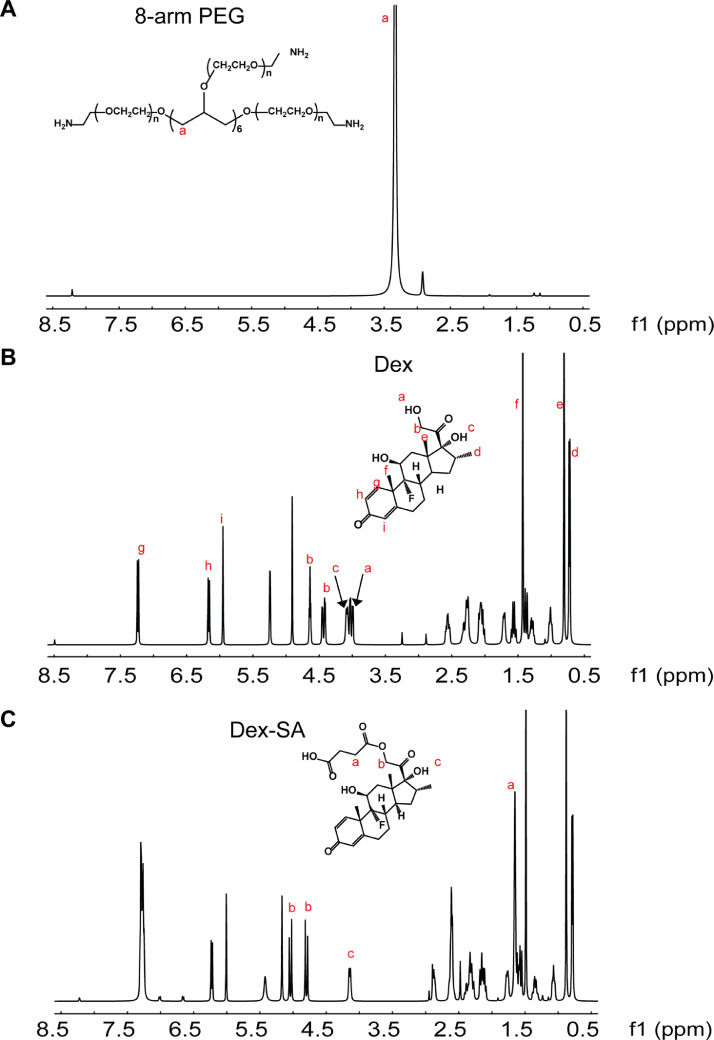
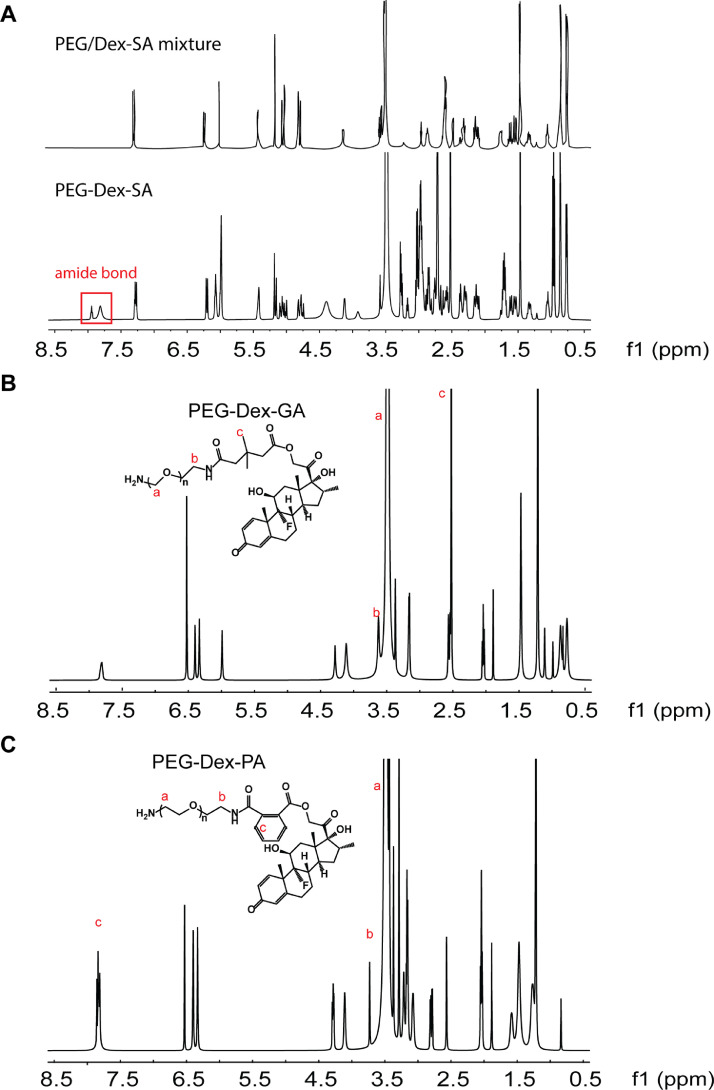
The chemical structures of synthesized Dex-SA, PEG-Dex-SA, PEG-Dex-GA and PEG-Dex-PA were confirmed by 1H NMR. The 1H NMR data of standard 8-arm PEG and Dex is presented and main proton peaks were identified in Fig. 7A and 7B. Based on the NMR spectrum of standard sample Dex, the disappearance of hydroxyl signal (4.0 ppm), shift of protons ‘b’ from 4.4 ppm, 4.6 ppm to 4.8 ppm, 5.0 ppm, and additional signal of protons ‘a’ at 1.6 ppm suggests the formation of ester in Dex-SA (Fig. 7C). The 1H NMR spectrum of the newly PEG chain introduced to Dex-SA was identified at 3.3 ppm (‘a’). At the same time, formation of amide bond was confirmed at ~7.8 ppm in PEG-Dex-SA group whereas PEG/Dex-SA physical mixture didn't display this peak (Fig. 8A). The 1H NMR spectra of PEG-Dex-GA and PEG-Dex-PA were identical to that of PEG-Dex-SA. The appearance of protons ‘c’ in Fig. 8B and 8C indicates the presence of side chains (-CH3) and benzene ring in PEG-Dex-GA and PEG-Dex-PA, respectively.
Fig. 7.
1H NMR spectra of (A) 8-arm PEG, (B) Dex, (C) Dex-SA in Dimethyl sulfoxide-d6. The lower-case labels “a~i” indicate the resonance peaks corresponding to each proton in the structure.
Fig. 8.
1H NMR spectra of (A) PEG/Dex mixture and PEG-Dex-SA, (B) PEG-Dex-GA and (C) PEG-Dex-PA in Dimethyl sulfoxide-d6. The lower-case labels “a~c” indicate the resonance peaks corresponding to each proton in the structure.
Drug loading content of PEG-Dex compounds and drug release rates
The amount of Dex conjugated to PEG was determined by analytical reverse-phase HPLC. As shown in Table 2, PEG-Dex-SA, PEG-Dex-GA and PEG-Dex-PA had 6.6 ± 0.5, 1.6 ± 0.4 and 3.3 ± 0.5 molecules of Dex on one molecule of 8-arm PEG, respectively. After conjugating to Avidin, the DLC for mAv-Dex-SA was calculated as 15.7 ± 1.0%, and 3.8 ± 0.9% and 7.8 ± 0.1%, for mAv-Dex-GA and mAv-Dex-PA, respectively.
Table 2.
Hydrolysis half-lives of ester linkers between 8-arm PEG and carboxylated derivatives of Dex. Molar ratio of Dex conjugated with PEG for each configuration and the corresponding drug loading content (DLC). Data shown as Mean ± SD.
| Ester linker | PEG-Dex-SA | PEG-Dex-GA | PEG-Dex-PA | PEG-Dex (2:1:1) |
|---|---|---|---|---|
| Half-life (h) | 6.8 ± 0.2 | 79 ± 1.8 | 86 ± 2.3 | 38.5 ± 1.5 |
| Dex: PEG (molar ratio) | 6.6 ± 0.5:1 | 1.6 ± 0.4:1 | 3.3 ± 0.5:1 | 4.6 ± 0.5:1 |
| DLC (%) | 15.70 ± 1.0 | 3.81 ± 0.9 | 7.85 ± 0.1 | 11.1 ± 0.8 |
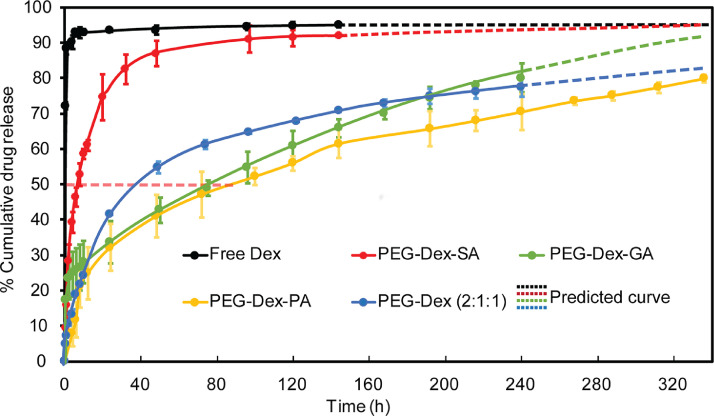
The drug release profiles of PEG-Dex compounds were shown in Fig. 9. About 70% of Dex was released from PEG-Dex-SA in PBS within the first 24 h resulting in a short release half-life (t1/2) of about 6.8 h. The adjacent carbonyl group in amide bond and hydrophobic-hydrophilic intermolecular reaction resulted in the fast hydrolysis of ester bond in PEG-Dex-SA. We addressed this issue by increasing the carbon spacer length between the ester and adjacent amide bond by replacing SA with GA or PA to form carboxylic acid derivatives of Dex. In PEG-Dex-GA, increased carbon spacer length (from -C-C- to -C-C—C-) and the two methyl groups in the GA branched chain weakened the inductive effect of carbonyl group by donating more electrons to stabilize the ester bond. The resulting half-life of ester hydrolysis increased to 79 ± 1.8 h. Similarly, the phenyl group in PEG-Dex-PA can donate electrons to the ester bond thereby increasing the release half-life to 86 ± 2.3 h. Controlled release PEG-Dex (2:1:1) had a release half-life of 38.5 ± 1.5 h. The products were then lyophilized and stored at −20 °C until further use.
Fig. 9.
Dex release rates from PEG-Dex compounds at 37 °C, pH 7.4 in PBS. Controlled release PEG-Dex (2:1:1) represents combination of ester linkers synthesized from SA, GA and PA in 2:1:1 molar ratio.
A single 10 µM dose of controlled release mAv-Dex (2:1:1) was chosen to achieve a therapeutic minimum concentration of 10 nM Dex in culture media based on intra-cartilage uptake of mAv-Dex and Dex release rates from mAv-Dex. Detailed calculations are provided in He et al., Journal of Controlled Release [7]. mAv-Dex effectively suppressed the cytokine induced catabolic activity significantly greater than Dex alone in a cartilage explant culture model of OA [7]. Methods to quantify intra-cartilage transport of mAv or in-vitro cartilage culture setup are not described here. Readers are directed to the following references for methods on tissue transport [3,6,16] and in-vitro cartilage OA model [12,17].
Conclusion
Here we present a protocol for synthesizing cartilage penetrating cationic multi-arm Avidin (mAv) nano-construct that provides multiple sites for covalent loading of Dex using hydrolysable ester linkers. We present a method for designing more stable ester linkers by increasing carbon spacer length between the ester and adjacent amide bond by replacing SA with GA or PA. The controlled release mAv-Dex formulation containing ester derivatives from SA, GA and PA in 2:1:1 molar ratio showed a release half-life of 38.5 ± 1.5 h providing sustained drug release over at least 10 days.
This cationic multi-arm Avidin (mAv) nano-construct can enable intra-cartilage delivery of a broad array of small molecule OA drugs and their combinations to chondrocytes. Drug release rates can be modulated by using a combination of ester linkers with different rates of hydrolysis based on the type of drug, its target sites and state of disease. Avidin-biotin technology provides the flexibility for biotinylating other similar sized drugs as Dex that can then be conjugated with Avidin by simple mixing at room temperature, which can be conducted at the clinic prior to use. This is a platform technology that, for example, can also be used for delivering contrast agents for imaging of a wide range of negatively charged tissues.
Acknowledgements
This work was funded by the United States Department of Defense through the Congressionally Directed Medical Research Programs (CDMRP) [W81XWH-17-1-0085], National Institute of Health R03 EB025903-1 and the Northeastern University Tier 1 Award. We are also grateful to Dr. Jason Guo, Director of the NMR facility, Dr. Jared Auclair, Director of the Biopharmaceutical Training Lab, Prof. Heather Clark and Prof. Ke Zhang at Northeastern University for their guidance and providing access to their labs/facilities.
Declaration of Competing Interest
The Authors confirm that there are no conflicts of interest.
References
- 1.Yang M., Ding J., Zhang Y., Chang F., Wang J., Gao Z., Zhuang X., Chen X. Activated macrophage-targeted dextran–methotrexate/folate conjugate prevents deterioration of collagen-induced arthritis in mice. J. Mater. Chem. B. 2016;4:2102–2113. doi: 10.1039/c5tb02479j. [DOI] [PubMed] [Google Scholar]
- 2.Bajpayee A.G., Grodzinsky A.J. Cartilage-targeting drug delivery: can electrostatic interactions help? Nat. Rev. Rheumatol. 2017;13:183. doi: 10.1038/nrrheum.2016.210. [DOI] [PubMed] [Google Scholar]
- 3.Vedadghavami A., Wagner E.K., Mehta S., He T., Zhang C., Bajpayee A.G. Cartilage penetrating cationic peptide carriers for applications in drug delivery to avascular negatively charged tissues. Acta Biomater. 2019;93:258–269. doi: 10.1016/j.actbio.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajpayee A.G., Rodolfo E., Scheu M., Varady N.H., Yannatos I.A., Brown L.A., Krishnan Y., Fitzsimons T.J., Bhattacharya P., Frank E.H. Sustained intra-cartilage delivery of low dose dexamethasone using a cationic carrier for treatment of post traumatic osteoarthritis. Eur. Cell Mater. 2017;34:341. doi: 10.22203/eCM.v034a21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajpayee A.G., Scheu M., Grodzinsky A.J., Porter R.M. A rabbit model demonstrates the influence of cartilage thickness on intra articular drug delivery and retention within cartilage. J. Orthop. Res. 2015;33:660–667. doi: 10.1002/jor.22841. [DOI] [PubMed] [Google Scholar]
- 6.Bajpayee A.G., Wong C.R., Bawendi M.G., Frank E.H., Grodzinsky A.J. Avidin as a model for charge driven transport into cartilage and drug delivery for treating early stage post-traumatic osteoarthritis. Biomaterials. 2014;35:538–549. doi: 10.1016/j.biomaterials.2013.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T., Zhang C., Vedadghavami A., Mehta S., Clark H.A., Porter R.M., Bajpayee A.G. Multi-arm Avidin nano-construct for intra-cartilage delivery of small molecule drugs. J. Control Release. 2020;318:109–123. doi: 10.1016/j.jconrel.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang M., Feng X., Ding J., Chang F., Chen X. Nanotherapeutics relieve rheumatoid arthritis. J. Control Release. 2017;252:108–124. doi: 10.1016/j.jconrel.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 9.Green N.M. A spectrophotometric assay for Avidin and biotin based on binding of dyes by Avidin. Biochem. J. 1965;94:23C–24C. doi: 10.1042/bj0940023c. [DOI] [PubMed] [Google Scholar]
- 10.Marttila A.T., Airenne K.J., Laitinen O.H., Kulik T., Bayer E.A., Wilchek M., Kulomaa M.S. Engineering of chicken avidin: a progressive series of reduced charge mutants. FEBS Lett. 1998;441:313–317. doi: 10.1016/s0014-5793(98)01570-1. [DOI] [PubMed] [Google Scholar]
- 11.Schagger H., Cramer W., Vonjagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 12.Bajpayee A.G., Quadir M.A., Hammond P.T., Grodzinsky A.J. Charge based intra-cartilage delivery of single dose dexamethasone using Avidin nano-carriers suppresses cytokine-induced catabolism long term. Osteoarthritis Cartilage. 2016;24:71–81. doi: 10.1016/j.joca.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wade L.G., Simek J.W. Organic Chemistry. Ninth ed. Person Education; 2017. Carboxcylic acids. [Google Scholar]
- 14.Wade L.G., Simek J.W. Ninth ed. Person Education; 2017. Acids and Bases in: Organic Chemistry. [Google Scholar]
- 15.Edward J.T. Molecular volumes and the Stokes-Einstein equation. J. Chem. Educ. 1970;47:261. doi: 10.1021/ed047p261. [DOI] [Google Scholar]
- 16.Pouran B., Arbabi V., Bajpayee A.G., van Tiel J., Töyräs J., Jurvelin J.S., Malda J., Zadpoor A.A., Weinans H. Multi-scale imaging techniques to investigate solute transport across articular cartilage. J. Biomech. 2018;78:10–20. doi: 10.1016/j.jbiomech.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Mehta S., Akhtar S., Porter R.M., Onnerfjord P., Bajpayee A.G. Interleukin-1 receptor antagonist (IL-1Ra) is more effective in suppressing cytokine-induced catabolism in cartilage-synovium co-culture than in cartilage monoculture. Arthritis Res. Ther. 2019;21:238. doi: 10.1186/s13075-019-2003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]