Abstract
Introduction
Alport syndrome is a hereditary glomerulonephritis that results from the disruption of collagen α345(IV) heterotrimerization caused by mutation in COL4A3, COL4A4 or COL4A5 genes. Many clinical studies have elucidated the correlation between genotype and phenotype, but there is still much ambiguity and insufficiency. Here, we focused on the α345(IV) heterotrimerization of α5(IV) missense mutant as a novel factor to further understand the pathophysiology of Alport syndrome.
Methods
We selected 9 α5(IV) missense mutants with typical glycine substitutions that clinically differed in disease progression. To quantify the trimerization of each mutant, split nanoluciferase-fused α3/α5 mutants and α4 were transfected into the cells, and intracellular and secreted heterotrimer were detected by luminescence using an assay that we developed previously.
Results
Trimer formation and secretion patterns tended to be similar to the wild type in most of the mutations that did not show proteinuria at a young age. On the other hand, trimer secretion was significantly reduced in all the mutations that showed proteinuria and early onset of renal failure. One of these mutants has low ability of intracellular trimer formation, and the others had the defect of low-level secretion. In addition, the mutant that is assumed to be nonpathogenic has similar trimer formation and secretion pattern as wild-type α5(IV).
Conclusion
The result of cell-based α345(IV) heterotrimer formation assay was largely correlated with clinical genotype–phenotype. These trimerization assessments provide additional phenotypic considerations and may help to distinguish between pathogenic and nonpathogenic mutations.
Keywords: Alport syndrome, COL4A5 mutation, collagen trimerization, genotype–phenotype, split nanoluciferase
Alport syndrome is a hereditary kidney disease caused by a mutation in the COL4A3, COL4A4, or COL4A5 gene that encodes type IV collagen alpha 3, 4, or 5 (α3/α4/α5[IV]), respectively, which are the major components of the glomerular basement membrane (GBM).1, 2, 3, 4 Type IV collagen α3/α4/α5(IV) chains form α345(IV) heterotrimer intracellularly. The folded heterotrimer is secreted and becomes a part of the GBM. Any of the posttranslational α3/α4/α5-chain mutants in Alport syndrome cannot form α345(IV) heterotrimer and all α chains (α3/α4/α5[IV]) are absent in the GBM.5, 6, 7
Approximately 80% to 85% of patients with Alport syndrome have X-linked inheritance (XLAS) of a mutation in the COL4A5 gene on chromosome Xq26–48.8,9 There are several large clinical studies about the correlation between genotype and phenotype in male patients with XLAS. These studies have shown that mutations with large deletion (or truncated) mutations, such as nonsense and frameshift mutations, are more severe than the missense mutation of α5(IV) chain.10, 11, 12 Glycine-substituted type of missense mutation in collagenous domain is most common in XLAS; however there are many phenotypic variations even in only glycine-substituted missense mutations. Moreover, some male patients with XLAS with missense mutation have positive α5(IV) expression in GBM and show a markedly mild clinical picture.13,14
Immunostaining of renal or skin biopsy specimens of type IV collagen α345 have been used for predicting the prognosis of patients with Alport syndrome. However, pathological diagnosis can be invasive and inconclusive, and it is not indicated for all patients with mutations. Although the phenotype variability in missense mutation is presumed to be due to the ability of α345(IV) heterotrimer formation, noninvasive methods for determining the state of α345(IV) trimer has not yet been used. Therefore, the details of trimer formation of α5(IV) missense mutant underlying the patient’s phenotype are still unknown.
We previously established a system to detect α345(IV) heterotrimer using split nanoluciferase binary (or complementation) technology (NanoLuc BiT).15 Large BiT (LgBiT) and small (SmBiT) subunits are fused to α5(IV) and α3(IV), respectively. In the presence of α3/α4/α5(IV) chains, α3/4/5(IV) forms a heterotrimer that is reflected as quantifiable luminescence due to the proximity of α3(IV)-SmBiT and α5(IV)-LgBiT. Moreover, α3(IV)-SmBiT, α5(IV)-LgBiT alone, or the combination of these in the absence of α4(IV) do not produce luminescence15; therefore, this system essentially measures the full-length α345(IV) heterotrimers in cell-based analysis. Here, we used this system to examine the relationship between the clinical phenotype of patients and the heterotrimer formation of α345(IV) in α5(IV) missense mutations. Each mutant showed a characteristic trimerization pattern and most of which tended to correlate with the clinical phenotype. These results indicate that the evaluation of α345(IV) trimerization could become one of the key indicators of genotype–phenotype correlation in XLAS.
Methods
Compliance With Ethical Standards
All procedures were reviewed and approved by the Institutional Review Board of Kobe University School of Medicine. Informed consent was obtained from patients or their parents.
Patients
Patients enrolled in this study were referred to Kobe University Hospital for clinical evaluation or genetic analysis. Most of them were followed in various local hospitals in Japan. They were suspected as having Alport syndrome from their pathological findings or family histories. All clinical and laboratory findings were obtained from the patients’ medical records at the point when genetic analysis was performed.
Mutational Analysis
Mutational analysis of COL4A5 was performed by targeted next-generation sequencing using a custom disease panel or conventional direct sequencing using the Sanger method for all exons and exon-intron boundaries. Regarding targeted sequencing, we designed a custom panel for targeted sequences. The gene list is shown in Supplementary Table S1. Next-generation sequencing samples were prepared using a HaloPlex Target Enrichment System Kit (Agilent Technologies, Santa Clara, CA) in accordance with the manufacturer’s instructions. Amplified target libraries were sequenced using MiSeq (Illumina, San Diego, CA) and analyzed with SureCall (v.3.0; Agilent Technologies). Detected variants were confirmed by Sanger sequencing. Sanger sequencing for COL4A5 was performed by polymerase chain reaction and direct sequencing of genomic DNA for all exons and exon-intron boundaries. Blood samples were collected from patients and/or family members, and genomic DNA was isolated from peripheral blood leukocytes using the Quick Gene Mini 80 System (Kurabo, Osaka, Japan), in accordance with the manufacturer’s instructions. For genomic DNA analysis, 51 exons of COL4A5 were amplified by polymerase chain reaction, as described previously.13 The polymerase chain reaction–amplified products were then purified and subjected to direct sequencing using a Dye Terminator Sequencing Kit (Amersham Biosciences, Piscataway, NJ) with an automatic DNA sequencer (model ABI Prism 3130; PerkinElmer, Waltham, MA).
Plasmids
The plasmids for human COL4A3, COL4A4 and COL4A5 coding sequences prepared for the nanoluciferase complementation system (Promega, Madison, WI) have been previously described.15 Briefly, a small fragment (SmBiT) and a large fragment (LgBiT) are fused to COL4A3 and COL4A5, respectively. For the C-terminal fusion nanoluciferase assay, COL4A3 was inserted in pFC36K SmBiT TK-neo Flexi vector and COLA5 was inserted in pFC34K LgBiT TK-Neo Flexi vector. For the N-terminal fusion nanoluciferase assay, COL4A3 and COLA5 were subcloned into pFN35K SmBiT TK-neo Flexi and pFN33K LgBiT TK-Neo Flexi vectors, respectively. For both C- and N-terminal fusion nanoluciferase, COL4A4 was subcloned into pEB multi-Hyg vector and fused with 3FLAG tag. Mutants of COL4A5 in pFC34K LgBiT and pFN33K LgBiT were generated by site-directed mutagenesis as previously described.16 Primer sequences are shown in Supplementary Table S2.
Cell Culture and Transfection, Luciferase Assay
HEK293T cells were obtained from the American Type Culture Collection (Manassas, VA), and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 100-U penicillin and streptomycin in culture plates coated with Cellmatrix type I-C. For nanoluciferase assay, cells were maintained in phenol red-free, low glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100-U penicillin and streptomycin, 2 mM glutamine, and 200 μM L-ascorbic acid 2-phosphate trisodium salt.
Transient transfection of plasmid DNAs was performed using TransIT LT-1 reagent (Mirus Corp., Madison, WI) when cells were sub-confluent using the protocol recommended by the manufacturer. Twenty-four hours after transfection, cells were re-plated on Falcon 96-well white/clear-bottom plates (Corning, Corning, NY) for nanoluciferase assay, and cultured for an additional 24 hours in the presence of 200 μM ascorbic acid. On the day of assay, culture media were transferred to new wells, and fresh media were added to the cells. Nano-Glo Live Cell Assay reagent (Promega) was added to the media and cells. Luminescence was measured using the GloMax Navigator system (Promega).
Statistical Analysis
Statistical parameters are indicated in the figure legends. Nanoluciferase assays were performed in quadruplicate starting from 4 separate cell cultures. All data are presented as mean ± SD. The significance of differences among groups was assessed using analysis of variance with Tukey-Kramer post hoc tests. Differences with P values < 0.05 were considered statistically significant.
Results
Missense Mutations in COL4A5 and Clinical Features
We selected 9 typical missense mutations of COL4A5 in 12 families. These mutations are substitutions of glycine residues in Gly-X-Y repeats, including those previously reported as pathogenic mutation.9,17, 18, 19, 20, 21 The information on mutation status, family history, and clinical characteristics are shown in Table 1. Most of these patients had hematuria or a family history of end-stage renal disease (ESRD) and diagnosed by genetic analysis. G230C, G869R, G1140V, and G1149V mutations were severe cases with the onset of proteinuria at the average age of 5.3 years (n = 3) and reached to ESRD at the average age of 23.3 years (n = 6) based on the available data. On the other hand, mutations in G509R, G805R, G1000V, G1030S, and G1143S mutations were cases without proteinuria at the age of diagnosis. Based on the available data, the average age at ESRD with these mutations is 41.2 years (n = 5). These less severe cases were not the consequence of treatment. We also focused on G953V mutation, which is probably a single-nucleotide polymorphism, according to the results of our numerous gene analyses and recent study.22
Table 1.
COL4A5 missense mutations and clinical details
| Amino acid change | Nucleotide | Exon | Patient ID; familial history | Gender | Gene analysis | Age at diagnosis (yr) | Age at ESRD (yr) | Hematuria | Proteinuria age of onset (yr) | Received treatment | α5(IV) expression |
|---|---|---|---|---|---|---|---|---|---|---|---|
| p.Gly230Cys | c.688G>T | 13 | 1 | M | ○ | 17 | 17 | + | + (3) | No | |
| 1 Grandfather | M | NA | 26 | NA | NA | NA | |||||
| p.Gly509Arg | c.1525G>C | 23 | 2 | M | ○ | 14 | + | − | No | ||
| 2 Brother | M | NA | 13 | + | − | No | |||||
| 2 Maternal grandfather | M | NA | 60s | NA | NA | NA | |||||
| p.Gly805Arg | c.2413G>A | 30 | 3 | M | ○ | 33 | + | − | No | +a | |
| 3 Relative | M | NA | 25 | + | − | No | |||||
| p.Gly869RArg | c.2605G>A | 31 | 4 | M | ○ | 12 | + | + (12) | Yes | −a | |
| p.Gly1000Val | c.2999G>T | 34 | 5 Father | M | ○ | 37 | + | − | No | +b | |
| p.Gly1030Ser | c.3088G>A | 35 | 6 Brother | M | ○ | 48 | + | − | NA | ||
| 6 Nephew | M | ○ | 20 | + | − | No | |||||
| 7 Father | M | NA | 20s | NA | NA | NA | |||||
| p.Gly1140Val | c.3419G>T | 38 | 8 | M | ○ | 2 | + | + (1) | No | ||
| 8 Mother's cousin | M | NA | 40 | 24 | NA | NA | NA | ||||
| 8 Mother's cousin | M | NA | 30 | 24 | NA | NA | NA | ||||
| p.Gly1143Ser | c.3427G>A | 38 | 9 | M | ○ | 9 | + | − | No | ||
| 9 Cousin | M | NA | 15 | + | − | No | |||||
| 9 Cousin | M | NA | 12 | + | − | No | |||||
| 9 Maternal grandfather | M | NA | 45 | NA | NA | NA | |||||
| 10 | M | ○ | 5 | + | − | No | |||||
| 10 Grandfather | M | NA | 66 | 49 | NA | NA | NA | ||||
| 10 Mother's cousin | M | NA | 46 | 32 | NA | NA | NA | ||||
| 10 Mother's cousin | M | NA | 43 | NA | NA | NA | |||||
| 11 Son | M | NA | 7 | NA | NA | No | |||||
| 11 Father | M | NA | 73 | NA | NA | No | |||||
| p.Gly1149Val | c.3446G>T | 38 | 12 Father | M | ○ | 23 | + | + (NA) | NA | ||
| 12 Uncle | M | NA | 26 | + | + (NA) | NA | |||||
| p.Gly953Val | c.2858G>T | 33 | Possibility of SNPs |
ESRD, end-stage renal disease; M, male; NA, not available; SNP, single-nucleotide polymorphism.
In the bottom row is a possible SNP found in multiple people who harbor this mutation.
Open circles indicate that gene analysis has been performed. +, α5(IV) expression observed; –, α5(IV) expression not observed.
Staining result of renal biopsy.
Staining result of skin biopsy.
α345(IV) Trimer Formation of C-Terminal-Tagged α5(IV) Mutant
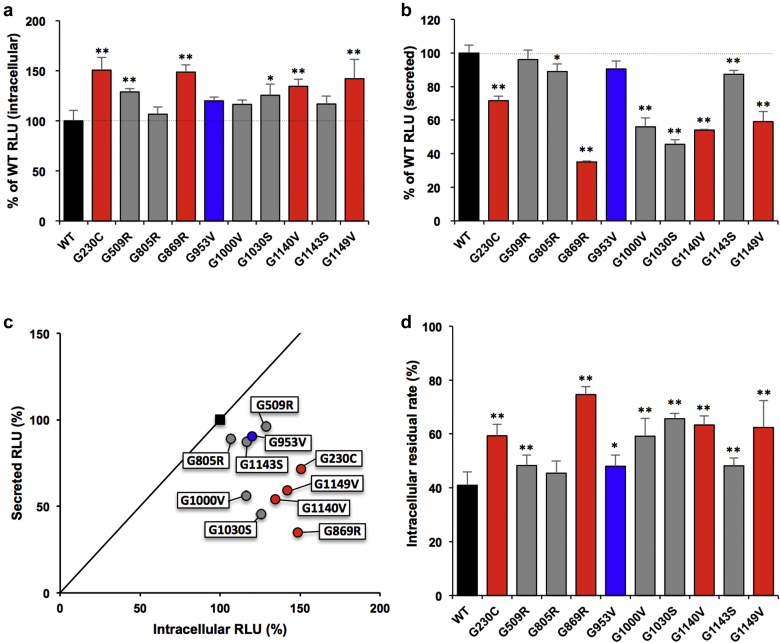
To correlate the mutation in COL4A5 with its heterotrimer formation, we evaluated α345(IV) trimerization of LgBiT C-terminal-tagged α5(IV) mutants in Table 1 using our established split nanoluciferase (NanoLuc) complementation system. C-terminal of type IV collagen has the noncollagenous 1 domain, which is important for initiation of trimerization. C-terminal LgBiT tag was attached next to the noncollagenous 1 domain. Intracellular analysis showed that all mutants can form more or equal complexes than the wild type. G230C, G509R, G869R, G1140V, and G1149V showed a significant increase in intracellular complexes compared with wild type (>130% relative light units [RLU]) (Figure 1a). Nevertheless, their extracellular secretion was reduced except in G509R, G805R, and G953V, whereas in G1143S, secretion was slightly reduced (>80% RLU). The extracellular secretion of G869R, G1000V, G1030S, G1140V, and G1149V showed a significant decrease (<60% RLU) compared with wild type (Figure 1b). To visualize the patterns of each mutant, scatterplot analysis of the intracellular/secreted RLU ratio was performed. The single-nucleotide polymorphism (G953V) and mutants with less severe phenotype (G509R, G805R, G1143S), except G1000V and G1030S, showed similar pattern as the wild type. In contrast, most mutants with severe phenotypes (G230C, G869R, G1140V, G1149V) were in the lower right (Figure 1c). Furthermore, complexes of all these mutants with severe phenotypes and exceptional mild phenotypes (G1000V, G1030S) tended to remain in the cells (>50% of the total complexes) (Figure 1d). These data suggest that the extracellular secretion of complexes was suppressed.
Figure 1.
α345(IV) trimer assay of clinical α5(IV) mutants with C-terminal tag. (a,b) Luminescence was measured in the cells (a) or media (b) in HEK293T cells transiently expressing α3, α4, and C-terminal-tagged α5(IV) wild type (WT) or the indicated mutant. Error bars are the mean ± SD (n = 4). ∗P < 0.01, ∗∗P < 0.001 versus WT (Tukey-Kramer test). (c) Scatterplot of the % relative light units (RLU) of intracellular (a)/secreted (b) from cells expressing WT or mutant α5 chain. Solid line: Y = X. (d) Intracellular residual rate calculated from the measured value of RLU of intracellular/(intracellular + secreted) from cells expressing WT or mutant α5(IV) chain. Error bars indicate the mean ± SD (n = 4). ∗P < 0.01, ∗∗P < 0.001 versus WT (Tukey-Kramer test). (a–d) Black, WT; blue, possibly of single-nucleotide polymorphisms; gray, α5(IV) mutants with mild pathology; red, α5(IV) mutants with severe pathology.
α345(IV) Trimer Formation of N-Terminal-Tagged α5(IV) Mutant
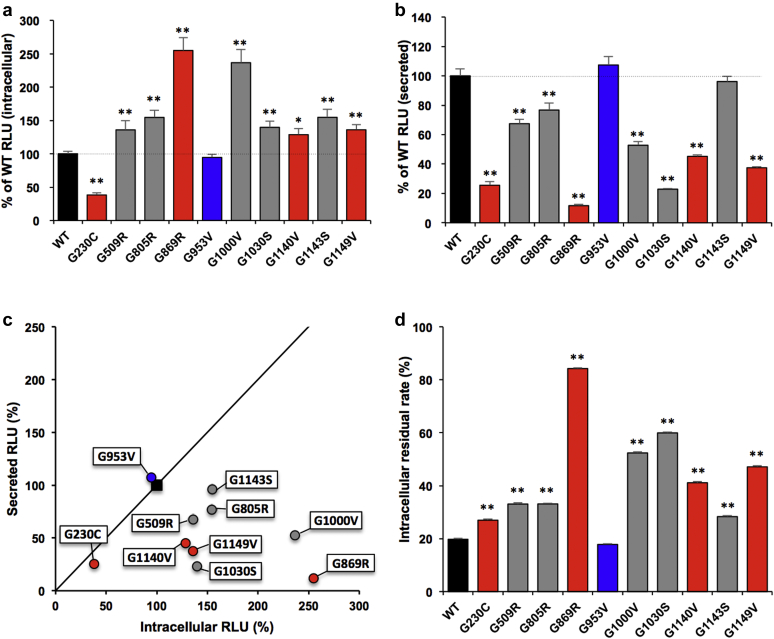
Besides C-terminal trimerization assay, we evaluated the trimerization of mutants tagged with LgBiT at the N-terminal side. The N-terminal region of type IV collagen is called the 7S domain, which is the end of trimerization. Therefore, it is assumed that N-terminal analysis can detect the final form of the complexes. Intracellular complex formation was significantly decreased in G230C mutant and increased in most mutants except G953V compared with wild type (Figure 2a). Secreted complexes were significantly decreased in most mutants except G953V and G1143S. In particular, the secretion of G230C, G869R, G1000V, G1030S, G1140V, and G1149V complexes were significantly decreased (<60% RLU) compared with wild type (Figure 2b). In the scatter plot, single-nucleotide polymorphism (G953V) was almost the same as the wild type, and mutants with mild phenotypes (G509R, G805R, G1143S) showed relatively similar trends as the wild type. On the other hand, G230C, one of the severe phenotypic mutants, was in the lower left on scatter plot. This means that complex formation was inhibited rather than secretion in G230C (Figure 2c). Most of the wild-type complexes that formed up to the N-terminus were mostly secreted (residual rate is approximately 20%). In contrast, complexes of mutants with severe phenotype (G869R, G1140V, G1149V) and some mutants with mild phenotypes (G1000V, G1030S) tended to remain in the cells (Figure 2d), which was consistent with C-terminal analysis.
Figure 2.
α345 (IV) trimer assay of clinical α5(IV) mutants with N-terminal tag. (a,b) Luminescence was measured in the cells (a) or media (b) in HEK293T cells transiently expressing α3, α4, and N-terminal-tagged α5(IV) WT or the indicated mutant. Error bars are the mean ± SD (n = 4). ∗P < 0.01, ∗∗P < 0.001 versus WT (Tukey-Kramer test). (c) Scatterplot of the % relative light units (RLU) of intracellular (a)/secreted (b) from cells expressing WT or mutant α5 chain. Solid line: Y = X. (d) Intracellular residual rate calculated from the measured value of RLU of intracellular/(intracellular + secreted) from cells expressing WT or mutant α5(IV) chain. Error bars indicate the mean ± SD (n = 4). ∗P < 0.01, ∗∗P < 0.001 versus WT (Tukey-Kramer test). Black, WT; blue, possibly of single-nucleotide polymorphisms; gray, α5(IV) mutants with mild pathology; red, α5(IV) mutants with severe pathology.
Discussion
In the present study, we evaluated the trimer formation and secretion of typical α5(IV) glycine-substituted missense mutants. Glycine substitutions occur in glycine-X-Y, which is a basic repeat sequence of the collagenous domain and accounts for 85% of patients with missense-type Alport syndrome.23 Because glycine is the smallest amino acid that can be buried inside the triple helix, its substitution can cause structural abnormalities.24,25 The structure of α5(IV) and α345(IV) triple-helix may be changed depending on the amino acid type and position of substitution. Although these changes probably contribute to the phenotype variabilities of missense-type Alport syndrome, it is difficult to prove the degree of structural differences with only genetic information. There are studies that tried to analyze the α5(IV) chain protein state and structure. One study using the co-immunoprecipitation method detected the reduction of α5(IV) chain with glycine substitution in α345(IV) heterotrimer complex.26, 27, 28 Another study using circular dichroism spectroscopy showed that 2 different types of glycine substitution within the same region have different secondary structure of α5(IV) partial recombinant proteins.29 Although these studies hinted at the genotype–phenotype correlation, these studies were relatively qualitative analysis and the number of mutations was limited. Therefore, they are still insufficient to represent the complexity of actual α345(IV) trimer formation. To address these limitations of previous studies, we established the α345(IV) trimer evaluation system using split nanoluciferase, and our approach can assess the state of trimerization of many mutants.15
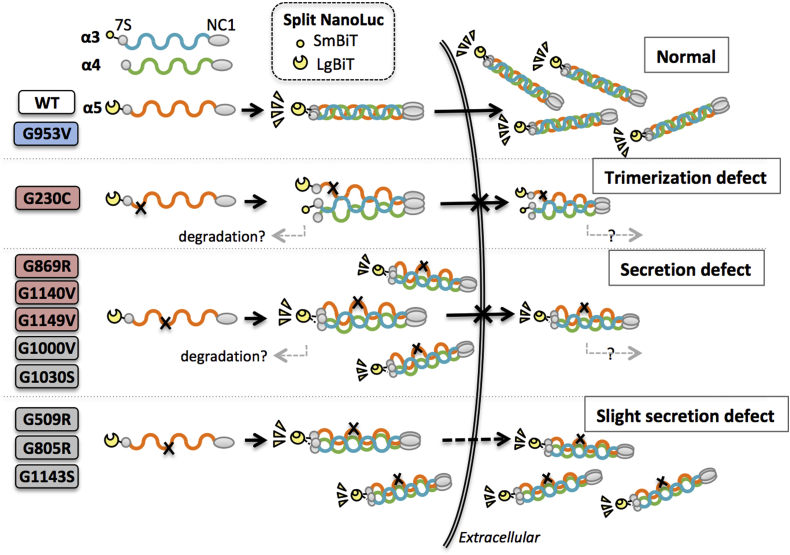
In the present study, we focused on missense mutations in the broad area of collagenous domain of α5(IV) chain that have different clinical severity. These mutations also include those that cause different severity despite substitutions with the same amino acid (e.g., G805R vs. G869R) or mutations at close positions (e.g., G1140V vs. G1143S). Figure 3 summarizes the results of the trimer assays of 10 mutants and shows the defect of these mutants. Overall, there was no association between the same amino acid substitutions or mutations at close positions. The following is a discussion of each mutant.
Figure 3.
Trimerization and secretion behavior of α5(IV) WT and mutants. Monomers assemble at the noncollagenous 1 (NC1) domain to form heterotrimers. Proximity of the NanoBiT (SmBiT and LgBiT) tags on trimer formation produces quantifiable luminescence. The scheme shows classification of each mutant trimerization: single-nucleotide polymorphism (blue), mutant with severe phenotype (red), and mutant with mild phenotype (gray), by N-terminal-tag assays. WT and G953V can trimerize and are secreted normally. Mutant α5(IV) monomers that cannot be trimerized (trimerization defect), such as G230C, are not secreted. Other variants can partially trimerize intracellularly but are not secreted normally (secretion defect). The level of secretion may depend on the state and control mechanism of each mutant trimerization, which is likely to affect the severity of the disease. Unsecreted monomers and complexes may be degraded by undetermined mechanisms, whereas trimerization and secretion are assumed to be important rate-limiting processes.
G230C
Intracellular complex formation was decreased in the N-terminal trimer assay. α345(IV) trimerization is initiated by the assembly of C-terminal noncollagenous 1 domain. G230C was partially trimerized on the C-terminal side, but not on the N-terminal side. As a result, α345(IV) secretion was greatly reduced, and this result correlated with the severe phenotype of the patients. The mean age at ESRD is 21.5 years (n = 2), with 1 patient presenting onset of proteinuria at the age of 3 years (Table 1).
G509R, G805R, and G1143S
More than 60% of complexes were secreted extracellularly compared with wild type in both tag assays. These mutant complexes probably retained similar properties of the completely formed α345(IV) trimer and had only slight inhibition of secretion. The partial reduction correlated with clinically mild phenotypes, that is, no early onset of proteinuria was observed (Table 1). One of the patients with G805R missense mutant was α5(IV) chain-positive at GBM in renal biopsy. Previous studies have confirmed the expression of α5(IV) in 29% of male patients with XLAS.13,14 In these cases, proteinuria and end-stage renal failure occurred significantly in older ages. Thus, if α345(IV) secretion is maintained to some extent, as in G509R, G805R, and G1143S, α5(IV) expression might be positive and the phenotype is likely to be less severe.
G869R, G1000V, G1030S, G1140V, and G1149V
Intracellular complexes were increased but extracellular complexes were decreased in both N- and C-terminal trimerization assays. These results suggest that G869R, G1000V, G1030S, G1140V, and G1149V have structural abnormalities within the collagenous domain, resulting in significant inhibition of secretion as α345(IV) trimers. Among them, G869R, G1140V, and G1149V mutants present cases of severe phenotype with proteinuria onset at 12 years old or younger based on the available information. The secretion of G869R complexes were markedly inhibited, and more than 80% of the complexes remained in the cells (Figures 1d and 2d). These data from our cell-based assay revealed obvious functional defect in G869R missense mutant and probably show typical clinical characteristics of the Alport syndrome phenotype. This might be a reason that G869R is one of the highly reported mutations of Alport syndrome, and in the LOVD database all 21 cases are classified as pathogenic (or likely pathogenic).30
G1000V and G1030S
Contrary to our expectation, these were mutations that inhibited α345(IV) trimer secretion in our cell-based trimerization assay even though the patients with G1000V and G1030S mutation did not exhibit proteinuria at a young age. There are reports showing that the median age of ESRD with G1030S mutation is 38 years.9,31 On the other hand, 1 patient with this mutation had ESRD onset in his 20s (Table 1). The differences in the observed severity among these patients point to a more stratified and complex clinical picture that our assay system may not be able to predict with 100% accuracy, which is a limitation of this assay. Although this in vitro assay system may help to determine the pathogenicity/nonpathogenicity of the variant, it may not be able to accurately predict the degree of severity of the phenotype due to mitigating or exacerbating factors existing in vivo. For G1000V, there is a report of α5(IV) expression in skin biopsy of benign familial hematuria with G1000V mutation. In this study, α5(IV) was distributed diffusely but weakly in the father and mosaically in the daughter.19 This partial decrease in α5(IV) expression may be correlated to our study showing that G1000V mutant complex secretion was 56% (C-terminal tag) and 53% (N-terminal tag) compared with wild type. In cases when α345(IV) trimer secretion is maintained at approximately 50%, further consideration is required to understand the degree of phenotype. As shown in our study using the N-terminal tag assay, G1000V can accumulate more complexes in the cell (Figure 2a). Also, in our unpublished data, the secreted trimers complexed at the N-terminus are relatively stable even of mutants. Therefore, when small amounts of complexes continue to be secreted, it may be expressed as basement membrane. However, in this study, we have not clarified whether mutant α345(IV) trimers can form a basement membrane after secretion.
G953V
This mutation has been considered as a pathogenic variant of Alport syndrome for a long time because it is a typical glycine substitution.21,32, 33, 34 However, our recent gene analysis and reports from other groups have indicated that G953V is a single-nucleotide polymorphism variant and is nonpathogenic.22 Consistent with those views, G953V exhibited the same trimer formation and secretion abilities as the wild type in our cell-based trimerization assay. We note that G953V is the only mutant without a significant difference in both N- and C-terminal trimer assays. This result indicated that our cell-based assay may be able to predict nonpathogenic mutation without extensive genomic polymorphism analysis. Moreover, the recent development of genetic analysis technology has enabled comprehensive screening. Interestingly, type IV collagen gene mutations have been frequently found in patients with familial glomerulonephritis of unknown cause.35 Our cell-based trimer formation assay may become crucial in determining pathogenicity and nonpathogenicity for novel mutations as genetic diagnosis is advanced.
Hundreds of Alport syndrome cases and pathogenic mutations have been reported; however, there are no mutational hotspots, and the number of patients with the same mutation is very limited. Furthermore, there are different pathological interpretations, such as pathogenic, likely pathogenic, or benign, especially in missense mutations even with the same mutations. It also should be noted that these previous reports include not only individual patient differences and partial genetic analysis results, but also that their treatment history was unknown. Therefore, it is difficult to diagnose and predict patient phenotypes using only historical clinical databases. Although the split nanoluciferase assessment of mutant trimerization alone did not perfectly predict clinical severity, many trimer characteristics assessed using this system correlated with clinical pathology. Examining more mutants could provide new information for existing genotype–phenotype correlations.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant nos. JP26460098 and JP17K08309 [to MAS], JP19H03379 [to HK], JP16K19642 [to TY], JP15K09691 [to KN], and JP17H04189 [to KI]), the Japan Agency for Medical Research and Development (grant nos. 7930006 [to KN and KI]), the Alport Syndrome Research Funding Program of the Alport Syndrome Foundation and the Pedersen family, Kidney Foundation of Canada (to HK), Kumamoto University HIGO Program Research Funding Project (Fiscal Year 2015–2018 [to MK]), Grant-in-Aid for JSPS Fellows for Young Researchers (grant no. JP19J15443 [to MK]), and JSPS Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers (grant no. S2803 [to HK]).
Footnotes
Table S1. Targeted genes included in custom panel of next-generation sequencing.
Table S2. Primer sequences for mutagenesis of COL4A5.
Supplementary Material
References
- 1.Barker D.F., Hostikka S.L., Zhou J. Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science. 1990;248:1224–1227. doi: 10.1126/science.2349482. [DOI] [PubMed] [Google Scholar]
- 2.Jefferson J.A., Lemmink H.H., Hughes A.E. Autosomal dominant Alport syndrome linked to the type IV collage alpha 3 and alpha 4 genes (COL4A3 and COL4A4) Nephrol Dial Transplant. 1997;12:1595–1599. doi: 10.1093/ndt/12.8.1595. [DOI] [PubMed] [Google Scholar]
- 3.Lemmink H.H., Mochizuki T., van den Heuvel L.P. Mutations in the type IV collagen alpha 3 (COL4A3) gene in autosomal recessive Alport syndrome. Hum Mol Genet. 1994;3:1269–1273. doi: 10.1093/hmg/3.8.1269. [DOI] [PubMed] [Google Scholar]
- 4.Lemmink H.H., Nillesen W.N., Mochizuki T. Benign familial hematuria due to mutation of the type IV collagen alpha4 gene. J Clin Invest. 1996;98:1114–1118. doi: 10.1172/JCI118893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gubler M.C., Knebelmann B., Beziau A. Autosomal recessive Alport syndrome:immunohistochemical study of type IV collagen chain distribution. Kidney Int. 1995;47:1142–1147. doi: 10.1038/ki.1995.163. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi K., Yoshikawa N., Iijima K. Immunohistochemical study of alpha 1–5 chains of type IV collagen in hereditary nephritis. Kidney Int. 1994;46:1413–1421. doi: 10.1038/ki.1994.413. [DOI] [PubMed] [Google Scholar]
- 7.Hudson B.G., Tryggvason K., Sundaramoorthy M. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. New Engl J Med. 2003;348:2543–2556. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 8.Crockett D.K., Pont-Kingdon G., Gedge F. The Alport syndrome COL4A5 variant database. Hum Mutat. 2010;31:E1652–E1657. doi: 10.1002/humu.21312. [DOI] [PubMed] [Google Scholar]
- 9.Martin P., Heiskari N., Zhou J. High mutation detection rate in the COL4A5 collagen gene in suspected Alport syndrome using PCR and direct DNA sequencing. J Am Soc Nephrol. 1998;9:2291–2301. doi: 10.1681/ASN.V9122291. [DOI] [PubMed] [Google Scholar]
- 10.Jais J.P., Knebelmann B., Giatras I. X-linked Alport syndrome: natural history in. 195 families and genotype-phenotype correlations in males. J Am Soc Nephrol. 2000;11:649–657. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 11.Gross O., Netzer K.O., Lambrecht R. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant. 2002;17:1218–1227. doi: 10.1093/ndt/17.7.1218. [DOI] [PubMed] [Google Scholar]
- 12.Bekheirnia M.R., Reed B., Gregory M.C. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2010;21:876–883. doi: 10.1681/ASN.2009070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimura Y., Nozu K., Kaito H. Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV alpha5 chain. Kidney Int. 2014;85:1208–1213. doi: 10.1038/ki.2013.479. [DOI] [PubMed] [Google Scholar]
- 14.Said S.M., Fidler M.E., Valeri A.M. Negative staining for COL4A5 correlates with worse prognosis and more severe ultrastructural alterations in males with Alport syndrome. Kidney Int Rep. 2017;2:44–52. doi: 10.1016/j.ekir.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omachi K., Kamura M., Teramoto K. A split-Luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in Alport syndrome. Cell Chem Biol. 2018;25:634–643.e634. doi: 10.1016/j.chembiol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki S., Shuto T., Sato T. Inhibition of post-translational N-glycosylation by HRD1 that controls the fate of ABCG5/8 transporter. Sci Rep. 2014;4:4258. doi: 10.1038/srep04258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fallerini C., Dosa L., Tita R. Unbiased next generation sequencing analysis confirms the existence of autosomal dominant Alport syndrome in a relevant fraction of cases. Clin Genet. 2014;86:252–257. doi: 10.1111/cge.12258. [DOI] [PubMed] [Google Scholar]
- 18.Plant K.E., Green P.M., Vetrie D. Detection of mutations in COL4A5 in patients with Alport syndrome. Hum Mutat. 1999;13:124–132. doi: 10.1002/(SICI)1098-1004(1999)13:2<124::AID-HUMU4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko K., Tanaka S., Hasui M. A family with X-linked benign familial hematuria. Pediatr Nephrol. 2010;25:545–548. doi: 10.1007/s00467-009-1370-z. [DOI] [PubMed] [Google Scholar]
- 20.Renieri A., Bruttini M., Galli L. X-linked Alport syndrome: an SSCP-based mutation survey over all 51 exons of the COL4A5 gene. Am J Hum Genet. 1996;58:1192–1204. [PMC free article] [PubMed] [Google Scholar]
- 21.Knebelmann B., Breillat C., Forestier L. Spectrum of mutations in the COL4A5 collagen gene in X-linked Alport syndrome. Am J Hum Genet. 1996;59:1221–1232. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Ding J., Wang S. Reassessing the pathogenicity of c.2858G>T(p.(G953V)) in COL4A5 gene: report of 19 Chinese families. Eur J Hum Genet. 2020;28:244–252. doi: 10.1038/s41431-019-0523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertz J.M., Thomassen M., Storey H. Clinical utility gene card for: Alport syndrome - update. 2014. Eur J Hum Genet. 2015;23(9) doi: 10.1038/ejhg.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fidler A.L., Boudko S.P., Rokas A. The triple helix of collagens - an ancient protein structure that enabled animal multicellularity and tissue evolution. J Cell Sci. 2018;131(7) doi: 10.1242/jcs.203950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashtan C.E. Alport syndrome:abnormalities of type IV collagen genes and proteins. Ren Fail. 2000;22:737–749. doi: 10.1081/jdi-100101959. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi T., Kakihara T., Uchiyama M. Mutational analysis of type IV collagen alpha5 chain, with respect to heterotrimer formation. Biochem Biophys Res Commun. 2008;366:60–65. doi: 10.1016/j.bbrc.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi T., Uchiyama M. Mutant-type alpha5(IV) collagen in a mild form of Alport syndrome has residual ability to form a heterotrimer. Pediatr Nephrol. 2010;25:1169–1172. doi: 10.1007/s00467-009-1433-1. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T., Uchiyama M. Characterization of assembly of recombinant type IV collagen alpha3, alpha4, and alpha5 chains in transfected cell strains. Kidney Int. 2003;64:1986–1996. doi: 10.1046/j.1523-1755.2003.00323.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y.F., Ding J., Wang F. Effect of glycine substitutions on alpha5(IV) chain structure and structure-phenotype correlations in Alport syndrome. Biochem Biophys Res Commun. 2004;316:1143–1149. doi: 10.1016/j.bbrc.2004.02.168. [DOI] [PubMed] [Google Scholar]
- 30.Savige J., Storey H., Il Cheong H. X-Linked and autosomal recessive Alport syndrome: pathogenic variant features and further genotype-phenotype correlations. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pont-Kingdon G., Sumner K., Gedge F. Molecular testing for adult type Alport syndrome. BMC Nephrol. 2009;10:38. doi: 10.1186/1471-2369-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miao Y., Xiong J., Zhang X. Genetic diagnosis of polycystic kidney disease, Alport syndrome, and thalassemia minor in a large Chinese family. Clin Sci (Lond) 2017;131:2427–2438. doi: 10.1042/CS20170245. [DOI] [PubMed] [Google Scholar]
- 33.Wang F., Wang Y., Ding J. Detection of mutations in the COL4A5 gene by analyzing cDNA of skin fibroblasts. Kidney Int. 2005;67:1268–1274. doi: 10.1111/j.1523-1755.2005.00204.x. [DOI] [PubMed] [Google Scholar]
- 34.Mencarelli M.A., Heidet L., Storey H. Evidence of digenic inheritance in Alport syndrome. J Med Genet. 2015;52:163–174. doi: 10.1136/jmedgenet-2014-102822. [DOI] [PubMed] [Google Scholar]
- 35.Gast C., Pengelly R.J., Lyon M. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2016;31:961–970. doi: 10.1093/ndt/gfv325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.