A transient RNAi-based genetic screen helps elucidate the mechanisms of plant cell death control.
Abstract
A wide variety of intrinsic and extrinsic cues lead to cell death with unclear mechanisms. The infertility of some death mutants often hurdles the classical suppressor screens for death regulators. We have developed a transient RNA interference (RNAi)-based screen using a virus-induced gene silencing approach to understand diverse cell death pathways in Arabidopsis (Arabidopsis thaliana). One death pathway is due to the depletion of a MAP kinase (MAPK) cascade, consisting of MAPK kinase kinase 1 (MEKK1), MKK1/2, and MPK4, which depends on a nucleotide-binding site Leu-rich repeat (NLR) protein SUMM2. Silencing of MEKK1 by virus-induced gene silencing resembles the mekk1 mutant with autoimmunity and defense activation. The RNAi-based screen toward Arabidopsis T-DNA insertion lines identified SUMM2, MEKK2, and Calmodulin-binding receptor-like cytoplasmic kinase 3 (CRCK3) to be vital regulators of RNAi MEKK1-induced cell death, consistent with the reports of their requirement in the mekk1-mkk1/2-mpk4 death pathway. Similar with MEKK2, overexpression of CRCK3 caused dosage- and SUMM2-dependent cell death, and the transcripts of CRCK3 were up-regulated in mekk1, mkk1/2, and mpk4. MEKK2-induced cell death depends on CRCK3. Interestingly, CRCK3-induced cell death also depends on MEKK2, consistent with the biochemical data that MEKK2 complexes with CRCK3. Furthermore, the kinase activity of CRCK3 is essential, whereas the kinase activity of MEKK2 is dispensable, for triggering cell death. Our studies suggest that MEKK2 and CRCK3 exert concerted functions in the control of NLR SUMM2 activation and MEKK2 may play a structural role, rather than function as a kinase, in regulating CRCK3 protein stability.
Plants and microbes are engaged in a continuous battle for survival. Plants have evolved sophisticated defense mechanisms to detect the telltale molecules produced by pathogens and to initiate defense responses to ward off potential infections (Peng et al., 2018). In the first layer, plasma membrane-localized pattern recognition receptors detect conserved pathogen- or microbe-associated molecular patterns, and induce pattern-triggered immunity (Yu et al., 2017; Saijo et al., 2018). Adapted pathogens deliver an arsenal of effectors into host cells to damp host immune responses and promote parasitism (Dou and Zhou, 2012; Macho and Zipfel, 2015). Facing these challenges, plants have evolved a second layer of defenses whereby a plethora of intracellular nucleotide-binding site Leu-rich repeat (NBS-LRR) receptors (NLRs) detect those effectors directly or indirectly and initiate effector-triggered immunity (Cui et al., 2015), which sometimes results in localized programmed cell death called hypersensitive response (Coll et al., 2011).
Activation of MAPK cascades is one of the earliest responses in plant immune signaling (Meng and Zhang, 2013). A conventional MAPK cascade is assembled by three interlinked kinases: MAPKs (MPKs) are activated by MAPK kinases (MAPKKs, or MKKs), which are further phosphorylated and activated by MAPK kinase kinases (MAPKKKs, or MEKKs; Zhang et al., 2018). Generally, activated MPKs phosphorylate downstream targets, such as transcriptional factors and enzymes, to regulate gene expression and other cellular activities. In Arabidopsis (Arabidopsis thaliana), one MAPK cascade consisting of MAPKKK3/5, MKK4/5, and MPK3/6 plays diverse roles in plant immune responses, such as ethylene and camalexin synthesis, and stomatal immunity (Tena et al., 2011; Thulasi Devendrakumar et al., 2018). MPK3/6 were reported to regulate ethylene production at both transcriptional and posttranscriptional levels. Activated MPK6 phosphorylates and stabilizes 1-AMINOCYCLOPROPANE-1-AARBOXYLIC ACID SYNTHASE 2 (ACS2) and ACS6, two rate-limiting enzymes in ethylene biosynthesis, leading to enhanced ACS proteins and increased ethylene production (Liu and Zhang, 2004). Besides, MPK3/6 also phosphorylate WRKY33, a transcription factor that binds to the promoters of ACS2 and ACS6, to regulate the expression of ACS2 and ACS6 (Li et al., 2012). Another MAPK cascade comprising MEKK1, MKK1/2, and MPK4 is also activated during plant defense responses (Gao et al., 2008; Qiu et al., 2008). This signaling pathway was considered to negatively regulate plant immunity. Plants carrying the active form of MPK4 showed compromised disease resistance to bacterial pathogens, and MPK4 activity negatively regulates certain responses of plant pattern-triggered immunity and effector-triggered immunity (Berriri et al., 2012). In addition, MPK4 phosphorylates a trihelix transcription factor ARABIDOPSIS SH4-RELATED3 to suppress the expression of a subset of flg22-induced genes (Li et al., 2015a).
The Arabidopsis mekk1, mkk1/2, and mpk4 mutants exhibit autoimmune phenotypes, described as dwarfism, spontaneous cell death, constitutively activated defense responses, and accumulation of reactive oxygen species (Petersen et al., 2000; Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008). The cell death of mekk1, mkk1/2, and mpk4 mutants could be suppressed by loss-of-function mutations in coiled-coil (CC)–type NLR SUPPRESSOR OF MKK1 MKK2 2 (SUMM2; Zhang et al., 2012). Recently, it was reported that the mekk1 cell death could be conditionally suppressed by a mutation in the Toll/IL-receptor–type NLR RPS6 (Takagi et al., 2019), suggesting that the MEKK1-MKK1/2-MPK4 pathway regulates the activation of both CC-NLR and Toll/IL-receptor–NLR. MEKK1 belongs to a tandemly duplicated gene family with MEKK2 and MEKK3, and the mutations in MEKK2, also called SUMM1, suppressed the autoimmunity and defense responses of mekk1, mkk1/2, and mpk4 (Kong et al., 2012; Su et al., 2013). MEKK2 is also a substrate of MPK4 (Kong et al., 2012). Furthermore, the abundance of MEKK2 transcripts is tightly associated with the autoimmunity observed in the mekk1, mkk1/2, and mpk4 mutants (Su et al., 2013). MPK4 also associates and phosphorylates CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 3 (CRCK3), which is required for the autoimmune phenotype of mekk1, mkk1/2, and mpk4 (Zhang et al., 2017). Both MEKK2 and CRCK3 function upstream of SUMM2, and CRCK3 is proposed to be guarded by SUMM2 for defense activation (Kong et al., 2012; Zhang et al., 2017).
We previously reported a RNA interference (RNAi)-based genetic screen by virus-induced gene silencing (VIGS) to identify suppressors of cell death mediated by two closely related receptor-like kinases: BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), also called SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (SERK3) and SERK4 (de Oliveira et al., 2016; Yu et al., 2019). In this study, we explored the possibility to apply this screen to understand the mechanism in mekk1 cell death. We report here that silencing of MEKK1 by VIGS resulted in autoimmune phenotypes resembling the mekk1 mutant. With this RNAi-based genetic screen of Arabidopsis T-DNA insertion lines, we identified several mutants as suppressors of MEKK1-mediated cell death and the corresponding genes for these mutants were named LETHALITY SUPPRESSORS OF MEKK1 (LETUM, or LET). Genetic analysis revealed that LET4 is MEKK2, LET5 is SUMM2, and LET6 encodes CRCK3. Biochemical and genetic analysis indicates that the kinase activity of MEKK2 is dispensable, but the kinase activity of CRCK3 is important to regulate MEKK1-mediated cell death. Similar with MEKK2, CRCK3 transcription is up-regulated in the mekk1, mkk1/2, and mpk4 mutants, and overexpression of CRCK3 triggered dosage-dependent cell death. Furthermore, MEKK2 associates with CRCK3 and positively regulates the protein homeostasis of CRCK3, which is essential to elicit NLR SUMM2-dependent cell death. In addition, neither MEKK2, CRCK3, nor SUMM2 is required for RNAi BAK1/SERK4- or BAK1-INTERACTING RECEPTOR-LIKE KINASE 1 (BIR1)-mediated cell death, suggesting diverse signaling pathways involved in cell death regulation. Thus, our RNAi-based genetic screen is a useful tool to understand plant cell death regulation, and our studies reveal the concerted action of MEKK2 and CRCK3 in the control of NLR SUMM2 activation.
RESULTS
Silencing of MEKK1 Activates Spontaneous Cell Death and Defense Responses
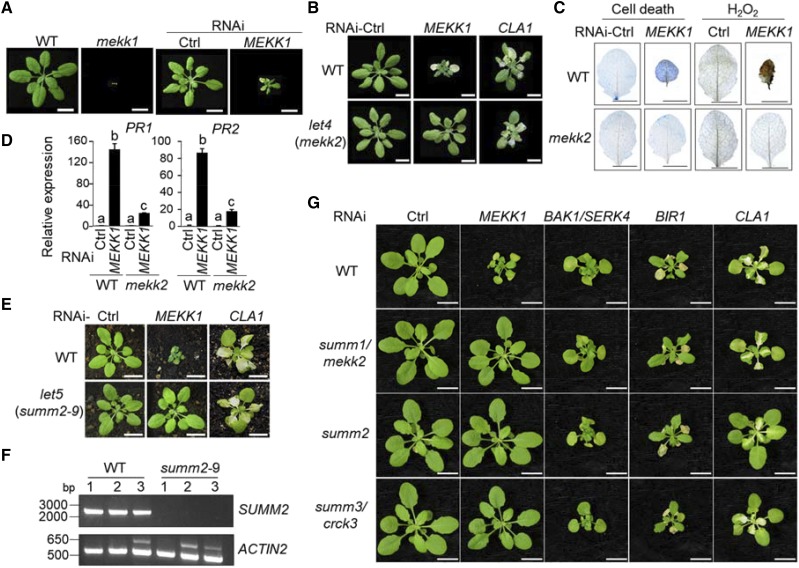
MEKK1 was previously reported to play an important role in plant defense responses (Asai et al., 2002). The mekk1 mutant exhibited a seedling-lethal phenotype, accompanied with constitutive defense responses, such as elevated accumulation of hydrogen peroxide (H2O2) and defense-related genes (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008). We tested whether silencing of MEKK1 by Agrobacterium-mediated VIGS could phenocopy the mekk1 mutants. As shown in Figure 1, A and B, at 2 weeks after inoculation, Arabidopsis Col-0 wild-type plants inoculated with Agrobacteria carrying a tobacco rattle virus –based VIGS vector targeting MEKK1 exhibited a severe cell death phenotype with a reduced plant stature, and curly and collapsed leaves, similar to the mekk1 mutant (Fig. 1, A and B). The plants inoculated with control vector carrying GFP behaved similarly with noninoculated plants (Fig. 1A). VIGS of a chloroplast development gene, CLOROPLASTOS ALTERADOS 1 (CLA1; Gao et al., 2011), induced the leaf albino phenotype, which was included as a visible marker for silencing efficiency (Fig. 1B). We further carried out the trypan blue staining to examine the cell death and 3,3′-diaminobenzidine (DAB) staining to determine the accumulation of H2O2 in MEKK1-silenced and control plants. When compared with the leaves from control plants, the leaves from MEKK1-silenced plants displayed extensive cell death and accumulation of H2O2 (Fig. 1C). Defense marker genes, including pathogenesis-related 1 (PR1) and PR2 were constitutively expressed in the mekk1 mutant (Ichimura et al., 2006). Analysis of the expression of PR1 and PR2 by reverse transcription quantitative PCR (RT-qPCR) revealed that their expression was drastically increased in the MEKK1-silenced plants (Fig. 1D). Taken together, these data indicate that silencing of MEKK1 via VIGS resulted in constitutively activated cell death and spontaneous defense responses, similar to the mekk1 mutants.
Figure 1.
The mekk2 and summ2 mutants suppress the cell death mediated by silencing of MEKK1. A, Silencing of MEKK1 by VIGS triggers cell death phenotype resembling the mekk1 mutant. Col-0 wild-type (WT) and mekk1 were grown on soil for 3 weeks (left two). Plant phenotypes of Col-0 wild-type plants are shown 2 weeks after VIGS of a vector control (Ctrl) or MEKK1. Bars = 1 cm. B, The let4/mekk2 mutant suppresses growth defects triggered by RNAi-MEKK1. Plant phenotypes of wild-type and mekk2 after VIGS of MEKK1. CLA1 is a visible marker for VIGS efficiency. Plant pictures were digitally extracted and placed on a black background in (A) and (B). Bars = 1 cm. C, The mekk2 mutant suppresses cell death and H2O2 accumulation triggered by RNAi-MEKK1. Plant true leaves after VIGS of MEKK1 were stained with trypan blue for cell death (left) and 3,3′-diaminobenzidine (DAB) for H2O2 accumulation (right). Bars = 0.5 cm. D, The mekk2 mutant suppresses PR1 and PR2 expression triggered by RNAi-MEKK1. The expression of PR1 and PR2 was normalized to the expression of UBQ10. The data are shown as mean ± se from three independent repeats. The different letters denote statistically significant difference according to one-way ANOVA followed by Tukey test (P < 0.05). E, The let5/summ2-9 mutant suppresses growth defects triggered by RNAi-MEKK1. Plant phenotypes of wild-type and let5/summ2-9 after VIGS of MEKK1 or CLA1. Bars = 1 cm. F, PCR analysis of SUMM2 in let5. At top, SUMM2 was not amplified in the let5 mutant with primers amplifying full length genomic DNA. At bottom, ACTIN2 control is shown. G, The mekk2, summ2, and crck3 mutants specifically suppressed RNAi MEKK1, but not BAK1/SERK4 or BIR1 cell death. Plant phenotypes of Col-0 plants and mutants are shown 2 weeks after VIGS of a vector control, MEKK1, BAK1/SERK4, BIR1, or CLA1. Bars = 1 cm.
The mekk2 and summ2 Mutants Suppress the Cell Death Triggered by Silencing of MEKK1, But Not BAK1/SERK4 or BIR1
The availability of the sequence-indexed T-DNA insertion library in Arabidopsis has greatly advanced plant biology research (Alonso et al., 2003). The fast and efficient VIGS to transiently silence endogenous genes enabled us to carry out multiple suppressor screens of Arabidopsis T-DNA insertion lines, and to compare the divergence and convergence of cell death signaling pathways mediated by key immune regulators, such as BAK1/SERK4 and MEKK1. Multiple candidates that showed suppression of the cell death caused by MEKK1-silencing were isolated from screening of ∼10,000 Arabidopsis T-DNA insertion lines. The corresponding genes for these mutants were named as LETUM (LET). Of the eight mutants we identified, the let4 mutant (SALK_150039C) potently suppressed the dwarfism and lethality induced by silencing of MEKK1 (Fig. 1B). The cell death suppression was not due to the impaired RNA silencing machinery in the let4 mutant because silencing of CLA1 induced leaf albino phenotype in both wild type and let4 (Fig. 1B). The let4 mutant, which was renamed as mekk2, harbors a T-DNA fragment inserted at 385 bp upstream of the start codon of At4g08480, encoding MEKK2. Several ethyl methanesulfonate mutagenized mutant alleles of MEKK2 have been identified as suppressors of mkk1/2 cell death, thus MEKK2 was also named as SUMM1 (Kong et al., 2012). The mekk2 T-DNA insertion mutant (SALK_150039C) was also able to suppress mpk4-mediated cell death (Su et al., 2013). Because MEKK1 and MEKK2 are closely linked (∼12 kb apart), it is not feasible or very tedious to test whether mekk2 could suppress mekk1 cell death by generating the mekk1mekk2 double mutant with genetic crosses (Kong et al., 2012). Our VIGS approach could silence genes independent of their genetic distance, and we convincingly show here that the mutation in mekk2 suppressed mekk1 cell death.
Staining results indicated that overaccumulation of H2O2 and dead cells induced by silencing of MEKK1 were abolished in the mekk2 mutant compared with wild-type plants (Fig. 1C). Additionally, constitutive expression of PR1 and PR2 was also markedly suppressed in the mekk2 mutant when MEKK1 was silenced (Fig. 1D). Consistent with the previous reports (Kong et al., 2012; Su et al., 2013), the cell death and defense activation induced by silencing of MEKK1 also depends on MEKK2, which further supports the feasibility to use VIGS approach to understand mekk1-mediated cell death.
The let5 mutant identified from this screen is SALK_062374, which suppressed the growth defect and cell death caused by silencing of MEKK1 (Fig. 1E). The let5 mutant (SALK_062374) was originally annotated to bear a T-DNA insertion in At3G15720, which encodes a pectin lyase-like superfamily protein. However, recent TDNA-Seq with next generation sequencing identified additional T-DNA insertions in the SALK_062374 genome (https://www.arabidopsis.org/servlets/TairObject?id=4664962&type=germplasm). One T-DNA insertion is located in the CC region of SUMM2 (At1G12280; Supplemental Fig. S1, A and B). To confirm whether there is a T-DNA insertion in the SUMM2 gene in let5, we PCR amplified SUMM2 full length genomic DNA in the wild type and let5 mutant. The SUMM2 gene could be amplified from wild type, but not let5 (Fig. 1F), suggesting that let5 carries a T-DNA insertion in SUMM2. We thus named let5 as summ2-9 (Supplemental Fig. S1A).
We have shown before that components required for BAK1/SERK4 cell death may not be involved in MEKK1 or BIR1 cell death (de Oliveira et al., 2016). We tested here whether MEKK2 and SUMM2 are involved in BAK1/SERK4- or BIR1-mediated cell death by VIGS assays. Although the mekk2 or summ2 mutants suppressed RNAi MEKK1 cell death, they did not affect the cell death caused by RNAi BAK1/SERK4 or BIR1 (Fig. 1G). The data strengthen the notion that diverse mechanisms underlie cell death regulation mediated by MEKK1, BAK1/SERK4, and BIR1.
The MEKK2 Kinase Mutant Is Able to Activate Cell Death Responses
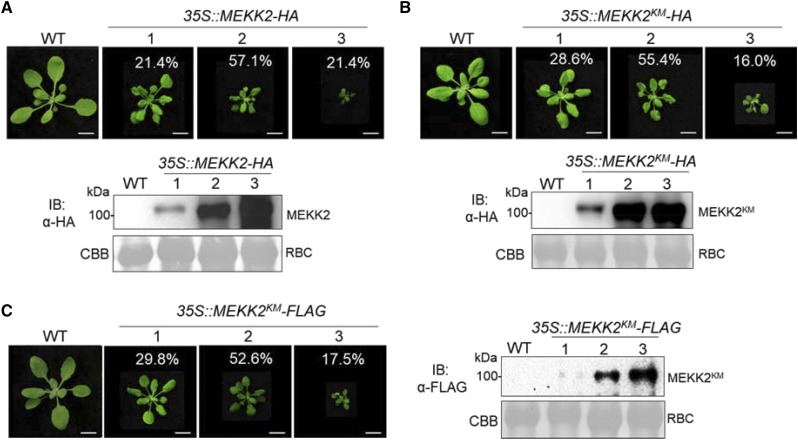
MEKK2 shows 64% identity with MEKK1 at the amino acid level (Supplemental Fig. S2). The conservation of the kinase domain of MEKK1 and MEKK2 is particularly high (Supplemental Fig. S2). When expressed in Arabidopsis protoplasts, MEKK1 strongly activated MAPKs as shown by α-pERK antibody that detects phosphorylated MAPKs (Supplemental Fig. S3). However, we did not observe the activation of MAPKs by MEKK2, although MEKK2 proteins were expressed well and even stronger than MEKK1 (Supplemental Fig. S3). Those observations prompted us to test whether the kinase activity of MEKK2 is required for its function in cell death control. Overexpression of MEKK2 led to constitutive cell death and defense responses (Kong et al., 2012; Su et al., 2013). We generated a binary vector harboring the full-length coding sequence of MEKK2 under the control of a double cauliflower mosaic virus 35S promoter with a double HA epitope at its C terminus (2x35S::MEKK2-HA). We further generated a MEKK2 kinase mutant (KM) variant with the conserved Lys (K) residue in the kinase ATP-binding loop mutated to Met (K529M; 2x35S::MEKK2KM-HA). The 2x35S::MEKK2-HA and 2x35S::MEKK2KM-HA constructs were introduced into the wild-type Col-0 background. As previously reported (Kong et al., 2012; Su et al., 2013), we observed that overexpression of MEKK2 transgenic plants were small and dwarfed with reduced plant architecture and cell death apparent in leaves (Fig. 2A). Interestingly, MEKK2KM transgenic plants were also small and dwarfed, morphologically indistinguishable from MEKK2 transgenic plants (Fig. 2B). The severity of the dwarfism and cell death was positively associated with an increasing level of MEKK2 or MEKK2KM protein expression (Fig. 2, A and B). As the phenotypes varied among the individual transgenic plants, we screened and carefully characterized a large number of transgenic plants. We obtained 42 transgenic plants carrying 2x35S::MEKK2-HA, which showed positive signals by α-HA immunoblots. We further classified them into three categories according to the phenotypic severity: 21.4% (9 out of 42) plants exhibited severe dwarf and cell death phenotypes, 57.1% (24 out of 42) showed moderate dwarf phenotypes, and 21.4% (9 out of 42) exhibited weak dwarfism (Fig. 2A). Similarly, for 2x35S::MEKK2KM-HA transgenic plants, among 56 transgenic plants with positive signals by α-HA immunoblots, 16.0% (9 out of 56) were severe dwarf with cell death, 55.4% (31 out of 56) were moderate dwarf, and 28.6% (16 out of 56) were weak dwarf (Fig. 2B). Similarly, overexpression of MEKK2KM under the control of a double cauliflower mosaic virus 35S promoter with a double FLAG epitope at its C terminus (2x35S::MEKK2KM-FLAG) also induced plant growth defects and cell death (Fig. 2C). Among 57 transgenic plants with positive signals by α-FLAG immunoblots, 17.5% (10 out of 57) were severe dwarf with cell death, 52.6% (30 out of 57) were moderate dwarf, and 29.8% (17 out of 57) were weak dwarf (Fig. 2C). These results indicate that both wild-type MEKK2 and the kinase mutant MEKK2KM could trigger cell death when overexpressed in plants. It is likely that the kinase activity of MEKK2 is not required for its function in regulating plant cell death.
Figure 2.
Overexpression of both wild-type (WT) and kinase mutant of MEKK2 induces cell death. A, Overexpression of MEKK2 induces dwarf phenotype. Four-week-old soil-grown plants were photographed. Transgenic plants of 35S::MEKK2-HA in wild type show severe dwarfism, and were grouped into three categories. Each category was calculated as a percentage of the total dwarf plants. Bottom shows the protein expression of MEKK2-HA from the above plants detected by immunoblot (IB) with an α-HA antibody. Coomassie Brilliant Blue (CBB) staining was used as the loading control. Bars = 1 cm. B, Overexpression of MEKK2KM, a kinase-dead mutant (Lys-529 to Met), triggers plant dwarfism. Five-week-old soil-grown plants were photographed. Transgenic plants of 35S::MEKK2KM-HA in wild type show severe dwarfism, and were grouped into three categories. Each category was calculated as a percentage of the total dwarf plants. Immunoblots with an α-HA antibody show MEKK2KM-HA protein expression and CBB staining was used as the loading control (bottom). Bars = 1 cm. C, Overexpression of MEKK2KM-FLAG triggers plant dwarfism. Five-week-old soil-grown plants were photographed. Transgenic plants of 35S::MEKK2KM-FLAG in wild type show severe dwarfism, and were grouped into three categories. Each category was calculated as a percentage of the total dwarf plants. Immunoblots with an α-FLAG antibody show MEKK2KM-FLAG protein expression and CBB staining was used as the loading control (right). Plant pictures were digitally extracted and placed on a black background. Bars = 1 cm.
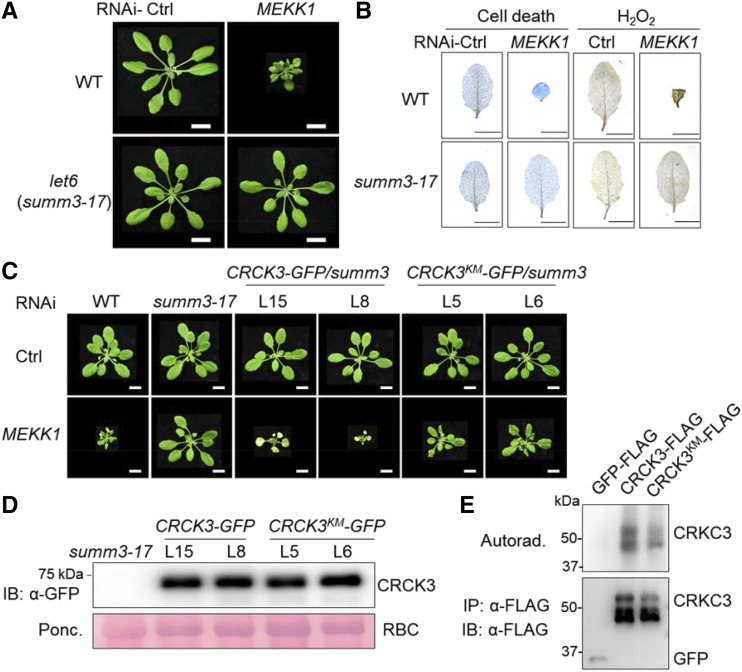
The Genomic DNA of CRCK3 Complements let6 in Regulating MEKK1-Mediated Cell Death
The let6 mutant (SALK_039370) isolated during this VIGS-based suppressor screen also suppressed the cell death phenotypes induced by silencing of MEKK1 (Fig. 3, A and B). The T-DNA fragment is located in the third intron of At2G11520 (Supplemental Fig. S4A), which was recently identified as SUMM3 (CRCK3), and let6 was named as summ3-17 (Zhang et al., 2017). Similar with mekk2 and summ2, let6/summ3-17 did not affect the cell death induced by RNAi BAK1/SERK4 or BIR1 (Fig. 1G). To confirm that CRCK3 is LET6, we transformed the full-length complementary DNA (cDNA) of CRCK3 (referred as cCRCK3) under the control of 35S promoter tagged with a double FLAG epitope at its C terminus (p35S::cCRCK3-FLAG) into the let6/summ3-17 mutant. More than 70 transformants were obtained from the hygromycin resistant screens. The presence of cCRCK3 construct was confirmed by PCR analysis in the transgenic lines (Supplemental Fig. S4B). To our surprise, cCRCK3-FLAG proteins were undetectable by immunoblots in all the transgenic lines. In addition, none of transgenic lines could complement the cell death phenotype induced by RNAi MEKK1 in summ3-17 (Supplemental Fig. S4C).
Figure 3.
Kinase activity of CRCK3 is important for its function in MEKK1-mediated cell death. A, The let6/summ3-17 mutant suppresses MEKK1-silencing induced growth defect. Bars = 1 cm. B, The summ3-17 mutant suppresses cell death (left) and H2O2 accumulation (right) triggered by RNAi-MEKK1. Plant true leaves after VIGS of MEKK1 were stained with trypan blue for cell death and DAB for H2O2 accumulation. Bars = 0.5 cm. C, Phenotype of 5-week-old Arabidopsis wild type (WT), summ3-17, and indicated transgenic lines after silencing of MEKK1. L15 and L8 are lines expressing GFP-tagged wild-type CRCK3 genomic DNA in the summ3-17 mutant. L5 and L6 are lines expressing the kinase dead CRCK3KM-GFP (Lys-253 to Glu) in the summ3-17 mutant. Bars = 1 cm. Plant pictures were digitally extracted and placed on a black background in A and C. D, Immunoblot analysis of the protein levels of CRCK3-GFP and CRCK3KM-GFP in the transgenic lines in (C). Ponceau (Ponc.) staining of Rubisco (RBC) was used as the loading control (Ctrl). E, CRCK3 has kinase activity. CRCK3-FLAG or CRCK3KM-FLAG was expressed in Arabidopsis protoplasts, and immunoprecipitated with α-FLAG agarose beads for an in vitro kinase assay. The autoradiograph (Autorad.; top) shows kinase activity, and the immunoblot (bottom) shows protein expression. GFP-FLAG was included as a control.
It is known that introns can affect gene expression in various ways (Le Hir et al., 2003; Rose, 2008). There are six introns in the gDNA region of CRCK3. We generated a construct harboring the full-length genomic DNA of CRCK3 containing all six introns with a C-terminal GFP tag under the control of 35S promoter, and transformed 35S::CRCK3-GFP into summ3-17 plants. The CRCK3-GFP proteins could be detected by immunoblots in these transgenic lines, and the cell death phenotype induced by MEKK1 silencing was restored to a level similar to or even more severe than that of wild-type Col-0 (Fig. 3, C and D), suggesting that the introns of CRCK3 might be important for its expression in planta.
The Kinase Activity of CRCK3 Is Important for its Function in Regulating Cell Death
To determine whether the kinase activity of CRCK3 is necessary for its function in the MEKK1-mediated cell death pathway, we mutated a conserved Lys residue in the ATP-binding loop of CRCK3 to Glu (K253E, CRCK3 kinase mutant CRCK3KM) and generated transgenic lines expressing CRCK3KM in the summ3-17 mutant background. Two representative lines with CRCK3KM-GFP protein expression level similar to CRCK3-GFP were used for the subsequent complementation analysis (Fig. 3D). Three weeks after silencing of MEKK1, the CRCK3KM-GFP transgenic lines only showed a weak cell death phenotype, whereas CRCK3-GFP transgenic plants showed a much more severe cell death phenotype (Fig. 3C), suggesting that unlike the wild-type CRCK3, CRCK3KM was not able to complement the summ3-17 mutant for RNAi MEKK1-triggered cell death. We examined whether CRCK3KM bears reduced kinase activity in plants. We first expressed CRCK3-FLAG and CRCK3KM-FLAG in protoplasts, and immunoprecipitated for an in vitro kinase assay. As shown in Figure 3E, CRCK3KM-FLAG exhibited a substantially reduced phosphorylation level compared with CRCK3-FLAG (Fig. 3E). Taken together, the data indicate that CRCK3KM could not fully complement the phenotype of summ3-17, and the kinase activity of CRCK3 is required for its full function in the MEKK1-mediated cell death pathway.
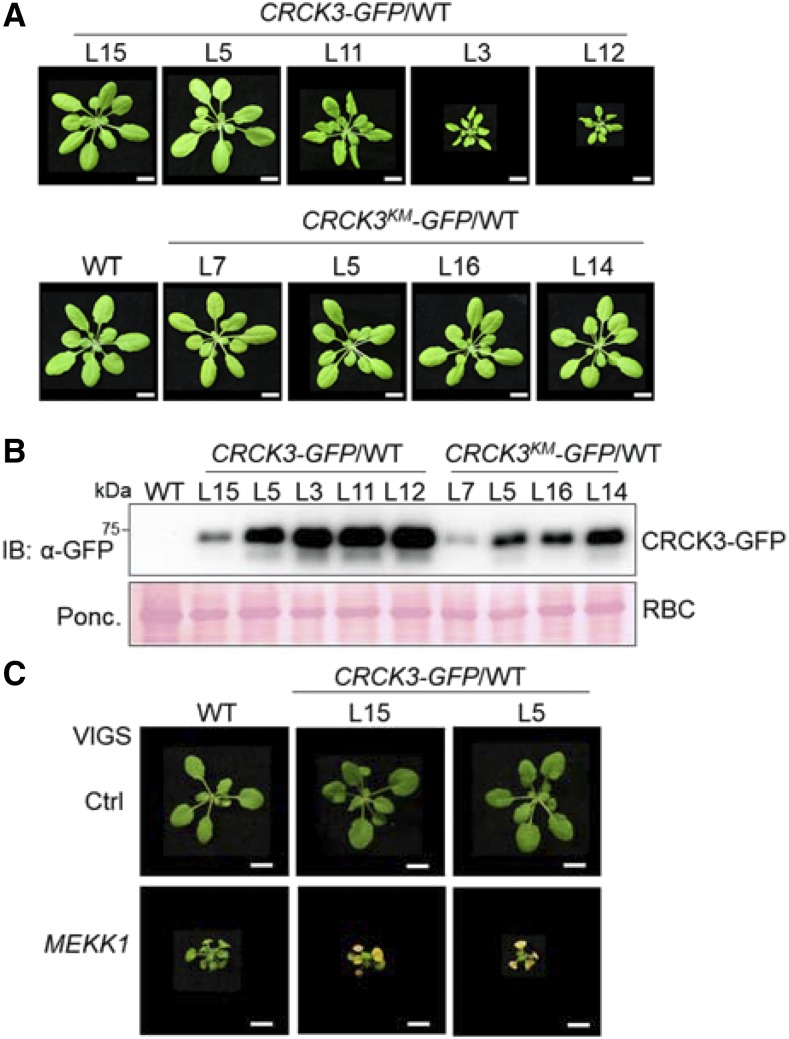
Overexpression of CRCK3 Induces a Kinase Activity-Dependent Cell Death in a Protein Dosage-Dependent Manner
When we introduced the 35S::CRCK3-GFP construct into the wild-type Col-0 background, we observed that transgenic plants with elevated CRCK3-GFP protein expressions showed dwarfism and cell death. Of the 73 transgenic lines obtained in the T1 generation, seven displayed phenotypes that resembled wild type (like L15 and L5 in Fig. 4A), 40 exhibited stunted growth and twisted leaves (like L11 in Fig. 4A), whereas 26 plants were small (like L3 and L12 in Fig. 4A). Trypan blue and DAB staining indicated the cell death and H2O2 accumulation in the leaves of plants exhibiting dwarf phenotypes (Fig. 5, A and B). Immunoblot analysis indicated that the level of plant dwarfism and cell death was positively associated with the protein level of CRCK3-GFP in transgenic plants: lines expressing low or moderate level of CRCK3 proteins resembled wild-type phenotype and those with high level of CRCK3 proteins exhibited severe dwarf phenotype (Fig. 4B). These results indicate that overexpression of CRCK3 leads to constitutively activated cell death in a protein dosage-dependent manner.
Figure 4.
Kinase activity of CRCK3 is required for its cell death inducibility. A, Phenotypes of 4-week-old wild-type (WT), CRCK3-GFP, and CRCK3KM-GFP transgenic lines in wild-type background. Bars = 1 cm. B, Immunoblot analysis of CRCK3-GFP and CRCK3KM-GFP proteins in the indicated transgenic lines in (A) with an α-GFP antibody. Ponceau (Ponc.) staining of Rubisco (RBC) was used as the protein loading control (Ctrl). C, Silencing of MEKK1 causes more severe cell death in CRCK3-overexpressing plants. Phenotypes of Col-0 wild type, and CRCK3-GFP/wild-type L15 and L5 (two independent lines) transgenic plants are shown 2 weeks after VIGS of a vector control or MEKK1. Bars = 1 cm. Plant pictures were digitally extracted and placed on a black background in A and C.
Figure 5.
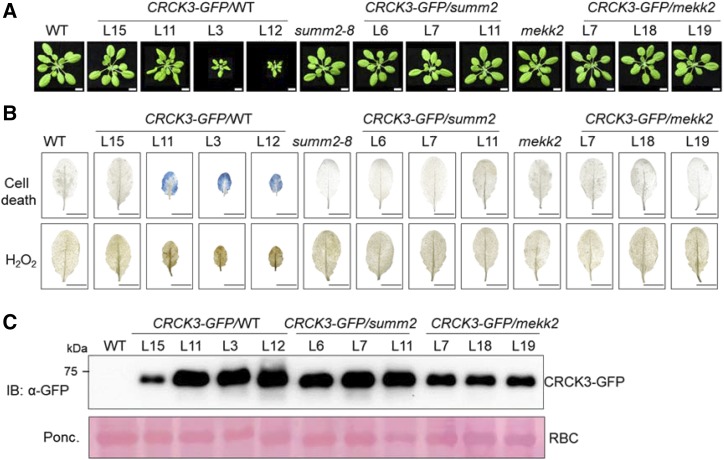
Overexpression of CRCK3 induces a SUMM2 and MEKK2-dependent cell death. A, Phenotypes of representative transgenic lines with overexpression of CRCK3-GFP in wild-type (WT), summ2-8, and mekk2 backgrounds. Pictures were taken from 4-week-old soil-grown plants. Bars = 1 cm. Plant pictures were digitally extracted and placed on a black background. B, Cell death (top) and H2O2 production (bottom) of leaves from transgenic lines in (A). Leaves from 5-week-old plants were used for trypan blue staining for cell death and DAB staining for H2O2 production. Bars = 0.5 cm. C, Immunoblot analysis of the expression of CRCK3-GFP proteins in the representative transgenic lines in A. CRCK3 protein levels were examined via an α-GFP antibody. Ponceau (Ponc.) staining of Rubisco (RBC) serves as the protein loading control (Ctrl).
The kinase activity of CRCK3 is important for its function in regulating MEKK1-mediated cell death (Fig. 3). To investigate whether the kinase activity of CRCK3 is also important in CRCK3 overexpression-induced cell death, we introduced 35S::CRCK3KM-GFP into the wild-type background and found that none of the transgenic lines displayed abnormal growth phenotypes (Fig. 4A). Interestingly, immunoblot analysis indicated that the proteins of CRCK3KM-GFP accumulated to a relatively lower level than those of wild-type CRCK3-GFP (Fig. 4B). We could not identify 35S::CRCK3KM-GFP plants with protein levels similar to 35S::CRCK3-GFP plants with strong cell death. The data suggest that the kinase activity of CRCK3 is required for its function in the activation of cell death.
To further determine whether overexpression of CRCK3 could promote cell death, we silenced MEKK1 by VIGS in 35S::CRCK3-GFP/wild-type transgenic plants L15 and L5, which phenotypically resembled wild-type Col-0. Substantially, L15 and L5 transgenic plants displayed more severe cell death than wild-type plants when MEKK1 was silenced (Fig. 4C), further supporting the important role of CRCK3 level in mekk1 cell death.
Both SUMM2 and MEKK2 Are Required for Cell Death Caused by Overexpression of CRCK3
SUMM2 is a NLR functioning genetically downstream of MEKK2 and CRCK3 (Zhang et al., 2012; Zhang et al., 2017). We next investigated whether cell death caused by overexpression of CRCK3 requires SUMM2 by transforming 35S::CRCK3-GFP into the summ2-8 mutant, which is morphologically similar to wild-type plants. Unlike the transgenic lines in the wild-type background, the 35S::CRCK3-GFP transgenic lines in the summ2-8 mutant background exhibited normal plant growth phenotypes similar to wild-type or summ2-8 plants (Fig. 5A). In addition, the extensive cell death and overaccumulation of H2O2 were not observed in the 35S::CRCK3-GFP transgenic lines in the summ2-8 background (Fig. 5B). The protein levels of CRCK3-GFP in the summ2-8 mutant were comparable with those in wild-type plants (Fig. 5C). These data indicate that CRCK3 overexpression-induced cell death depends on SUMM2.
It has been reported that CRCK3 is required for the autoimmune phenotypes induced by overexpression of MEKK2 (Zhang et al., 2017), suggesting that CRCK3 functions genetically downstream of MEKK2 in the mekk1 cell death pathway. Thus, it is plausible to speculate that the autoimmune phenotypes triggered by overexpression of CRCK3 are independent of MEKK2. Surprisingly, when we transformed 35S::CRCK3-GFP into mekk2 plants, all of 35S::CRCK3-GFP/mekk2 transgenic plants (more than 60 independent transgenic lines were isolated and characterized) showed similar growth phenotypes as wild-type plants (Fig. 5A), and no cell death or overaccumulation of H2O2 was detected in the transgenic lines (Fig. 5B). The data indicate that CRCK3-induced autoimmune phenotypes also genetically depend on MEKK2. Together, CRCK3 and MEKK2 are intricately linked with each other for the cell death inducibility, pinpointing the possibility of a protein complex containing both CRCK3 and MEKK2. In addition, the protein levels of CRCK3-GFP in the mekk2 mutant were slightly lower than those in the wild type or the summ2-8 mutant (Fig. 5C).
CRCK3 Transcription Is Up-Regulated in the mekk1, mkk1/2, and mpk4 Mutants
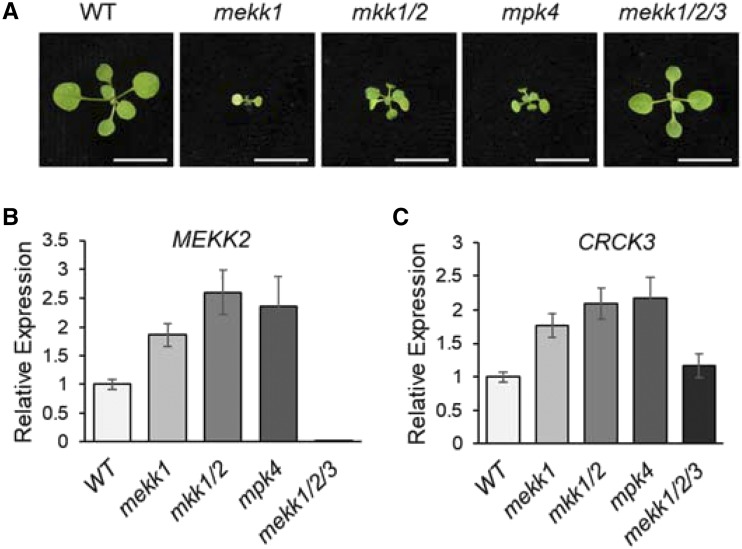
Apparently, similar with MEKK2, the expression level of CRCK3 is critical for cell death inducibility. It has been shown that the cell death and autoimmune phenotypes in the mekk1, mkk1/2, and mpk4 mutants are associated with the up-regulation of MEKK2 transcripts (Su et al., 2013). A modest increase in MEKK2 transcription could induce defense responses (Su et al., 2013). We therefore investigated whether CRCK3 transcripts are also mis-regulated in the mekk1, mkk1/2, and mpk4 mutants (Fig. 6A). Consistent with the previous report (Su et al., 2013), the transcripts of MEKK2 were up-regulated about 2-fold higher in the mekk1, mkk1/2, and mpk4 mutants compared with wild-type plants (Fig. 6B). Similarly, the transcripts of CRCK3 were also about 2-fold higher in the mekk1, mkk1/2, and mpk4 mutants compared with those in wild-type plants (Fig. 6C). The mekk1/2/3 mutant, which bears a deletion of MEKK1, MEKK2, and MEKK3, and suppresses the mekk1 cell death phenotype (Su et al., 2013), showed a similar expression level of CRCK3 as wild-type plants (Fig. 6C). Thus, the increased CRCK3 transcripts in the mekk1 mutant are also likely suppressed by the mekk2 mutation. The data suggested that the transcript levels of both CRCK3 and MEKK2 are associated with mekk1, mkk1/2, and mpk4 cell death, and the expression of CRCK3 and MEKK2 might be coregulated. Indeed, the expression pattern of CRCK3 and MEKK2 is very similar as observed in the Arabidopsis eFP Brower (Supplemental Fig. S5).
Figure 6.
The transcript levels of CRCK3 are up-regulated in the mekk1, mkk1/2, and mpk4 mutants. A, Phenotypes of 16-d-old wild type (WT), mekk1, mkk1/2, mpk4, and mekk1/2/3. Bars = 1 cm. B and C, Expression levels of MEKK2 and CRCK3 in the indicated seedlings as determined by RT-quantitative PCR. Values were normalized to the expression levels of UBQ10. The data are shown as mean ± se from three independent repeats.
MEKK2 Associates with and Stabilizes CRCK3
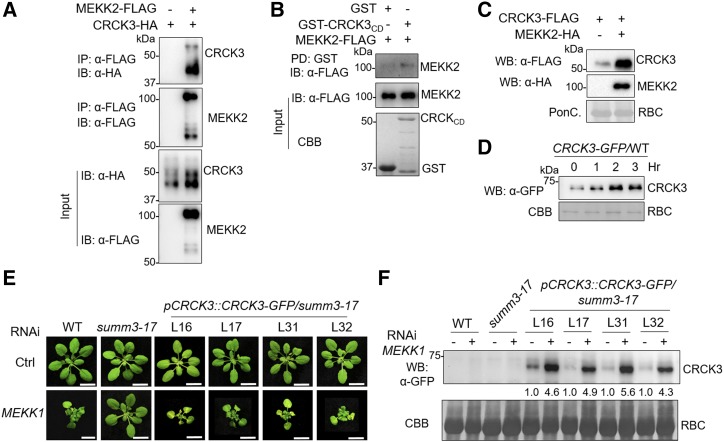
The genetic interaction between CRCK3 and MEKK2 led us to hypothesize that MEKK2 forms a complex with CRCK3. To test whether MEKK2 associates with CRCK3, a coimmunoprecipitation (Co-IP) assay was performed. MEKK2-FLAG and CRCK3-HA were transiently expressed in Arabidopsis protoplasts, and the proteins were immunoprecipitated with α-FLAG affinity beads. We then examined whether CRCK3 was in the precipitates by immunoblotting with an α-HA antibody. As shown in Figure 7A, the CRCK3-HA proteins were detected only in the sample containing MEKK2-FLAG, not in the negative control. Furthermore, an in vitro pull-down assay was performed with MEKK2-FLAG transiently expressed in Arabidopsis protoplasts as the bait against the cytoplasmic domain of CRCK3 fused with GST at its N terminus (GST-CRCK3CD). As shown in Figure 7B, GST-CRCK3CD, but not GST protein itself, was able to pull down MEKK2-FLAG. Taken together, the data suggest that CRCK3 and MEKK2 form a complex.
Figure 7.
MEKK2 associates with and stabilizes CRCK3. A, MEKK2 associates with CRCK3 in the Co-IP assay. MEKK2-FLAG and CRCK3-HA were transiently expressed in Arabidopsis protoplasts, immunoprecipitated with α-FLAG affinity beads (IP: α-FLAG), and immunoblotted with an α-HA (IB: α-HA) or α-FLAG (IP: α-FLAG) antibody (two at top). The protein inputs were detected before immunoprecipitation by an α-HA or α-FLAG immunoblot, respectively (two at bottom). B, MEKK2 interacts with CRCK3 in a pull-down assay. Arabidopsis protoplasts transiently expressing MEKK2-FLAG were incubated with GST or GST-CRCK3CD. The interaction between MEKK2 and CRCK3 was detected by α-FLAG immunoblot after immunoprecipitation with glutathione-agarose beads (top). The protein levels of MEKK2 and CRCK3CD were detected with an α-FLAG immunoblot or CBB staining, respectively. C, Immunoblot analysis of CRCK3-FLAG protein levels in N. benthamiana. CRCK3-FLAG was transiently coexpressed in N. benthamiana with either a control (Ctrl) vector or MEKK2-HA. Total protein extract was subjected to immunoblot analysis using an α-FLAG or α-HA antibody. The protein loading is shown by Ponceau staining of Rubisco (RBC). D, MG132 stabilizes CRCK3. The 10-d-old 35S::CRCK3-GFP/wild-type seedlings were treated with 2 μm MG132 for 0, 1, 2, or 3 h. Total protein extracts were analyzed by immunoblotting using an α-GFP antibody. CBB staining was used as a loading control. E, Phenotype of 5-week-old Arabidopsis wild type, summ3-17, and pCRCK3::CRCK3-GFP/summ3-17 complementation lines after silencing of MEKK1. L16, L17, L31, and L32 are lines expressing GFP-tagged CRCK3 genomic DNA driven by its native promoter in the summ3-17 mutant. Bars = 1 cm. Plant pictures were digitally extracted and placed on a black background. F, Silencing of MEKK1 by VIGS increased CRCK3 protein accumulation. The CRCK3 protein levels in the representative transgenic lines in D were examined via an α-GFP immunoblot. The band intensities were quantified using ImageJ software and labeled underneath the gel. The protein loading is shown by CBB staining. The above experiments were repeated three times with similar results.
The observation that the protein level of CRCK3-GFP in mekk2 was lower than that in wild-type plants (Fig. 5C) prompted us to test whether MEKK2 may positively regulate the protein accumulation of CRCK3. When we coexpressed CRCK3-HA together with MEKK2-FLAG in protoplasts for the Co-IP assay, we observed an increased CRCK3-HA protein level in the presence of MEKK2-FLAG compared with the vector control (Fig. 7A, third section). We further transiently coexpressed CRCK3-FLAG with MEKK2-HA or a vector control in Nicotiana benthamiana, and the increased CRCK3-FLAG protein level was detected in the presence of MEKK2-HA compared with the control (Fig. 7C), suggesting that MEKK2 regulates CRCK3 protein accumulation. We then examined whether CRCK3 is degraded in proteasome-dependent manner. The seedlings of the 35S::CRCK3-GFP/wild-type transgenic plants were treated with MG132, a 26S proteasome inhibitor, for different times, and subjected to immunoblot analyses. As shown in Figure 7D, MG132 treatments stabilized the accumulation of CRCK3 proteins, suggesting that CRCK3 stability is regulated by 26S proteasome-mediated degradation.
We further tested the importance of CRCK3 protein level in MEKK1-mediated cell death. We generated transgenic plants carrying CRCK3-GFP under its native promoter in the summ3-17 mutant (pCRCK3::CRCK3-GFP/summ3), and silenced MEKK1 by VIGS (Fig. 7E). Transformation of pCRCK3::CRCK3-GFP into the summ3 mutant restored the RNAi MEKK1-induced cell death as the wild-type plants (Fig. 7E). Importantly, the protein level of CRCK3-GFP was substantially increased (∼4- to 6-fold) upon silencing of MEKK1 in all transgenic lines examined (Fig. 7F). Notably, there was only about a 2-fold increase of CRCK3 transcripts in the mekk1 mutant compared with wild-type plants (Fig. 6C). Apparently, CRCK3 proteins are stabilized or translationally increased in the MEKK1-silenced plants. Thus, CRCK3 protein homeostasis plays a crucial role in the MEKK1 cell death pathway.
DISCUSSION
Modifier and suppressor screens are powerful forward genetic approaches to understand signaling pathways and uncover genetic interactions in a particular biological process. However, the beauty of this approach can be diminished when it is used to understand the functions of some essential genes due to the lethality of the null mutants. In plants, VIGS is a widely used RNAi approach for functional analysis of individual genes by knocking down the expression of endogenous genes, avoiding lethality caused by complete loss-of-functions (Burch-Smith et al., 2004; Senthil-Kumar and Mysore, 2011). In addition, this approach could silence multiple members of a gene family spontaneously to eliminate functional redundancy. We have used VIGS to silence two functionally redundant receptor-like kinases BAK1 and SERK4, which resembled the bak1/serk4 mutant plants (de Oliveira et al., 2016). VIGS could also be readily deployed in different genetic backgrounds, which could substantially expedite the tedious and time-consuming process of generating higher order mutants. For examples, we have silenced two closely related Cys-rich protein kinases CRK22 and CRK28 in the crk29 mutant to reveal their redundant functions in plant immunity (Yadeta et al., 2017). By making a random cDNA library of certain plant species, VIGS could also be used as a high throughput and fast forward genetic screen for identifying components in a biological process (Lu et al., 2003; Li et al., 2015b). In Arabidopsis, the availability of sequence index T-DNA insertion library makes it possible to identify the mutants that suppress or enhance the phenotype caused by VIGS of gene(s). The causal mutations could be easily identified based on the information of T-DNA insertions. With this approach, we have shown that protein glycosylation is important in bak1/serk4 cell death (de Oliveira et al., 2016). Thus, this unbiased and highly efficient genetic screen combines the features of both forward and reverse genetics, and provides an alternative to uncover the pathways and mechanisms regulating plant cell death, and embryonic or postembryonic seedling lethality caused by one or multiple redundant genes.
Understanding the mekk1-mkk1/2-mpk4 cell death pathway was mainly achieved through the suppressor screen of the mkk1/2 mutant, which is able to produce seeds when grown at 28°C (Gao et al., 2009; Kong et al., 2012; Zhang et al., 2017). Interestingly, certain mutants could suppress mkk1/2 cell death, but did not affect mekk1 or mpk4 cell death (Lian et al., 2018), suggesting the independent functions of individual components in the MEKK1-MKK1/2-MPK4 cascade. Unlike mkk1/2, the genetic mutants of mekk1 are unable to produce enough seeds for a suppressor screen. We show here that silencing of MEKK1 by VIGS in wild-type plants resembles the mekk1 mutant with autoimmune phenotypes. We further used this approach to screen the Arabidopsis T-DNA insertion library and identified MEKK2, SUMM2, and CRCK3 as specific regulators of RNAi MEKK1, but not BAK1/SERK4 nor BIR1-induced cell death. Previous identification of these components in the mekk1-mkk1/2-mpk4 cell death pathway supports the feasibility of our approach to understand the conserved and specific functions of MEKK1 in regulating cell death and others.
MEKK2 resides in a tandem repeat region with MEKK1 and shares 64% amino acid identity with MEKK1 (Supplemental Fig. S2). MEKK1 is indispensable for MPK4 activation, consistent with the fact that MEKK1 functions as a MAPKKK for the activation of MPK4 (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007). Consistently, MEKK1 has strong kinase activity (Asai et al., 2002; Supplemental Fig. S3). Surprisingly, the kinase activity of MEKK1 might not be required for the activation of MPK4 since the kinase-impaired mutant of MEKK1K361M could complement the mekk1 defects in terms of MPK4 activation (Suarez-Rodriguez et al., 2007). In addition, MEKK1K361M could complement the lethality of the mekk1 mutant, suggesting that the kinase activity of MEKK1 might not be essential for its function in cell death control (Suarez-Rodriguez et al., 2007). We show here that MEKK2 has little kinase activity in our assay conditions, and the kinase-impaired mutant MEKK2K529M did not affect its cell death inducibility when overexpressed in Arabidopsis wild-type plants, suggesting that kinase activity of MEKK2 may not be required for its function in cell death control (Fig. 2). Thus, both MEKK1 and MEKK2 may confer roles as structural or scaffold proteins, rather than functional kinases, in the MEKK1-MKK1/2-MPK4 cascade. Apparently, MPK4 kinase activation is associated with its function in plant immunity and cell death control (Berriri et al., 2012; Su et al., 2013). Thus, it is tantalizing to hypothesize that another MAPKKK, which might be scaffolded by MEKK1 or MEKK2, may form a conventional MAPK cascade together with MKK1/2 and MPK4 in activating the downstream signaling.
CRCKs are calcium-dependent CaM-binding receptor-like cytoplasmic kinases (RLCKs; Yang et al., 2004). RLCKs play important roles in plant immunity and development (Lin et al., 2013; Liang and Zhou, 2018). The Arabidopsis genome encodes three CRCKs (Yang et al., 2004). However, their biological functions and connections with Ca2+/CaM still remain elusive. It has been shown that Ca2+/CaM can stimulate CRCK1 kinase activity (Yang et al., 2004). Ca2+ signaling is essential in the activation of plant NLRs (Gao et al., 2013). It will be interesting to determine in the future whether Ca2+ or CaM is involved in CRCK3-mediated NLR SUMM2 activation. Several RLCKs, such as AVRPPHB SUSCEPTIBLE 1 (PBS1) and PBS1-LIKE 2 (PBL2), have been proposed to act as guardees or decoys for the activation of NLR immune receptors (Shao et al., 2003; Wang et al., 2015). CRCK3 was hypothesized as a guardee of SUMM2, and the phosphorylation level of CRCK3 by MPK4 is sensed by SUMM2 for activation (Zhang et al., 2017). We show here that overexpression of CRCK3 led to SUMM2-dependent cell death. Thus, in addition to phosphorylation, the homeostasis of CRCK3 is important for NLR SUMM2 activation.
Transcriptional reprogramming is one of the most important responses in plant defense (Li et al., 2016). The transcripts of both MEKK2 and CRCK3 are up-regulated in the mekk1, mkk1/2, and mpk4 mutants, and the abundance of MEKK2 and CRCK3 is tightly associated with autoimmune phenotypes (Fig. 6, B and C; Su et al., 2013). However, it is still unclear whether the up-regulation of MEKK2 and CRCK3 is responsible for or the outcome of the autoimmune phenotypes observed in the mekk1, mkk1/2, and mpk4 mutants. The transcriptional regulation of MEKK2 and CRCK3 appears to be complicated. Although MEKK2 is a substrate of MPK4 (Kong et al., 2012), the MEKK2 transcript level is regulated by MPK4 activity since overexpression of constitutively active MPK4 rescued the mekk1 cell death phenotype and restored the elevated MEKK2 expression of the mekk1 mutant to the wild-type level (Su et al., 2013). Similarly, the transcript level of CRCK3 might be regulated by MEKK2 since the increased CRCK3 expression in the mekk1 mutant was restored in the mekk1/2/3 mutant (Fig. 6C). Thus, multiple positive feedback regulations may exist in the mekk1-mkk1/2-mpk4 cell death pathway. It is also possible that the expression of MEKK2 and CRCK3 might be regulated at the posttranscriptional or translational levels. Some RNA processing-associated proteins, such as MODIFIER OF snc1 (MOS), are involved in the regulation of NLR protein SNC1-mediated autoimmune phenotypes (Palma et al., 2007). Identification and functional studies of other LETs have great potential to uncover the detailed molecular mechanisms underlying NLR SUMM2 activation and regulation at transcriptional, posttranscriptional, and posttranslational levels.
Co-IP and pull-down assays revealed that CRCK3 associates with MEKK2 in vivo and in vitro (Fig. 7, A and B), indicating that they exist in a protein complex in plants, and may require each other for functionality. Consistently, we observed that CRCK3 cell death inducibility depends on MEKK2 (Fig. 5). MEKK2 cell death inducibility also depends on CRCK3 (Zhang et al., 2017). Notably, MEKK2 positively regulates the protein stability of CRCK3 (Fig. 7C). When the dispensable role of kinase activity in MEKK2 cell death inducibility is considered, MEKK2 is likely a nonfunctional kinase, and it may function as a scaffold to stabilize CRCK3. It is possible that CRCK3 undergoes constant turnover in wild-type plants, whereas, in the mekk1 mutant, CRCK3 transcripts are increased and proteins are stabilized (Figs. 6C and 7F), partly due to the increased expression of MEKK2 scaffolding or stabilizing the complex. Notably, the pseudokinase RKS1 is required for RLCK PBL2 and NLR ZAR1-mediated immunity and to form the PBL2-RKS1-ZAR1 resistosome (Wang et al., 2019a, 2019b). The activated NLR ZAR1 may form pores in the plasma membrane, leading to cell death and oxidative stress. It is tempting to hypothesize that CRCK3-MEKK2-SUMM2 also assemble into a similar resistosome that is activated by the depletion of MEKK1-MKK1/2-MPK4 cascade, ultimately triggering cell death.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) mutant lines used here were described previously: mekk2 (SALK_150039C), summ2-8 (SAIL_1152_A06), summ2-9 (SALK_062374), summ3-17 (SALK_039370), mekk1 (SALK_052557), mpk4-2 (SALK_056245), mkk1/2, mekk1/2/3 in the Col-0 background (Zhang et al., 2012; Su et al., 2013; Zhang et al., 2017). Arabidopsis T-DNA insertion lines and library were obtained from Arabidopsis Biological Resource Center (ABRC). Arabidopsis and Nicotiana benthamiana plants were grown on soil (LP5 RSi, Sun Gro Horticulture) in a growth room at 23°C, 50% relative humidity, and 75 μE m-2s−1 light with a 12-h light/12-h dark photoperiod.
Plasmid Construction and Generation of Transgenic Plants
The VIGS constructs for MEKK1, BAK1/SERK4, BIR1, and CLA1 were reported previously (de Oliveira et al., 2016). To generate pMDC32-2x35S::cCRCK3-FLAG and pMDC32-2x35S::MEKK2-HA, the coding sequences of CRCK3 and MEKK2 were amplified by PCR from Col-0 cDNA and introduced into a plant gene expression vector pHBT containing an HA tag at the C terminus by Xba I/Sma I or BamH I/Stu I restriction enzyme digestions. The fragments were released by Bgl II/Sma I or BamH I/Stu I enzyme digestions and subcloned into modified pMDC32-FLAG or pMDC32-HA vectors. To make pCB302-35S::CRCK3-GFP and pCB302-35S::CRCK3-FLAG, the genomic fragment of CRCK3 was amplified from Col-0 genomic DNA and cloned into pHBT by Bgl II/Sma I enzyme digestions. The CRCK3 fragments were subcloned into modified pCB302-GFP or pCB302-FLAG vectors by Nhe I/Sma I digestions. The point mutations of MEKK2KM and CRCK3KM were generated using site-directed mutagenesis. To generate pCB302-pCRCK3::CRCK3-GFP, the promoter of CRCK3 was amplified from Col-0 genomic DNA and cloned into pCB302 by Sac I/BamH I enzyme digestions; then the CRCK3-GFP fragments from pHBT-p35S::CRCK3-GFP was released by Bgl II/EcoR I digestion and subcloned into this modified pCB302 vector. The Escherichia coli expression vector GST-CRCK3CD was generated by PCR amplifying the CRCK3 cytosolic domain (146–510 amino acids) from pHBT-35S::cCRCK3-HA, and subcloning it into a modified pGST vector using a one-step cloning kit (Vazyme Biotech) by BamH I and Stu I digestion. All DNA fragments cloned into vectors were confirmed via Sanger sequencing. The primers for cloning and point mutations are listed in Supplemental Table S1.
Transgenic plants were generated via the Agrobacterium-mediated floral-dip method. The transformants were screened with the herbicide BASTA (Bayer; resistance conferred by the pCB302 vector) or the antibiotic hygromycin for the pMDC32 vector.
Agrobacterium-Mediated Virus-Induced Gene Silencing Assay
The VIGS assay was performed as described previously (de Oliveira et al., 2016). In brief, the Agrobacterium strain GV3101 containing pYL156-RNA1, pYL156-MEKK1, pYL156-BAK1/SERK4, pYL156-BIR1, pYL156-CLA1, or pYL156-GFP (the vector control) was grown overnight in LB medium (50 μg mL−1 kanamycin, 50 μg mL−1 gentamycin, 10 mm MES, and 20 μm acetosyringone). The bacteria were harvested by centrifugation at room temperature and resuspended in infiltration buffer (10 mm MES, 10 mm MgCl2, and 200 μm acetosyringone). Bacterial cultures containing pYL156-MEKK1, pYL156-BAK1/SERK4, pYL156-BIR1, pYL156-CLA1, or pYL156-GFP were mixed with pYL156-RNA1 cultures at the 1:1 ratio, individually. The first pair of true leaves of 10-d-old plants were hand-infiltrated using a needleless syringe with the mixed bacterial cultures. The albino phenotype (CLA1-silencing) or cell death phenotypes showed up 2 weeks after infiltration.
Trypan Blue and DAB Staining
Staining was performed as described previously, with minor modifications (Zhou et al., 2019). For trypan blue staining, the leaves from 5-week-old plants were immersed in boiled latophenol (lactic acid/glycerol/liquid phenol/distilled water, 1:1:1:1) with 0.25 mg mL−1 trypan blue for 30 s. For 3,3′-diaminobenzidine (DAB) staining, the leaves from 5-week-old plants were immersed in 1 mg mL−1 DAB solution under vacuum pressure for 2 h, followed by an overnight incubation at room temperature in the dark. The trypan blue or DAB stained leaves were destained with destain buffer (ethanol/lactic acid/liquid phenol, 2:1:1) at 65°C for 1 h, and washed three times with 75% (v/v) ethanol. The destained leaves were photographed under the microscope.
Co-IP and transient expression assays
Arabidopsis protoplasts were transfected with different plasmids and incubated overnight. Samples were lysed in Co-IP buffer (100 mm NaCl; 1 mm EDTA; 20 mm Tris-HCl, pH 7.5; 2 mm NaF; 2 mm Na3VO4; 1 mm dithiothreitol; 0.5% [v/v] Triton X-100; 10% [v/v] glycerol; and 1× protease inhibitor). The supernatant was collected after centrifugation at 13,000 rpm at 4°C for 15 min and incubated with α-FLAG affinity beads (Sigma) at 4°C for 2 h with gentle shaking. The beads were collected and washed three times with Co-IP washing buffer (20 mm Tris-HCl, pH 7.5; 100 mm NaCl; 1 mm EDTA; 1% [v/v] Triton X-100). The immunoprecipitated and input proteins were analyzed by immunoblot with the indicated antibodies.
The transient expression assays in N. benthamiana were carried out as described previously (Feng et al., 2016). Briefly, Agrobacterium strain GV3101 containing binary vectors was cultured overnight at 28°C. Bacteria were harvested and resuspended with infiltration buffer (10 mm MES, 10 mm MgCl2, and 200 μm acetosyringone) for inoculation. The leaf samples were harvested at 30 h post infiltration.
RNA Isolation and Reverse Transcription Quantitative PCR (RT-qPCR) Analysis
Total RNAs were prepared using the TRIzol reagent (Invitrogen). RNase-free DNase I (New England Biolabs) was used to remove contaminating genomic DNA. cDNAs were synthesized with M-MuLV Reverse Transcriptase (New England Biolabs) and oligo(dT) primers. RT-qPCR analysis was performed using iTaq Universal SYBR green Supermix (Bio-Rad) with an 7900HT Fast Real-Time PCR system (Applied Biosystems). The expression of each gene was normalized to the expression of UBQ10.
Protein Isolation and In Vitro Pull-Down Assay
GST and GST-CRCK3CD were purified from E. coli with a standard glutathione agarose beads (Thermo Scientific). MEKK2-FLAG was transiently expressed in protoplasts overnight and lysed with 250 μL of extraction buffer (10 mm HEPES, pH 7.5; 100 mm NaCl; 1 mm EDTA; 10% [v/v] glycerol; 0.5% [v/v] Triton X-100; and 1:200 complete protease inhibitor cocktail from Sigma). For the pull-down assay, about 10 μg GST or GST-CRCK3CD proteins were mixed with the MEKK2-FLAG cell lysate at 4°C for 1 h with gentle shaking, subsequently incubated with 20 μL of glutathione agarose beads at 4°C for another 2 h with gentle shaking. The beads were harvested by centrifugation and washed five times with the washing buffer (10 mm HEPES, pH 7.5; 100 mm NaCl; 1 mm EDTA; 10% [v/v] glycerol; 0.5% [v/v] Triton X-100). The bound proteins were released from beads by boiling in 50 μL of 2× SDS protein loading buffer for 10 min and detected by an immunoblot with an α-FLAG antibody.
In Vitro Kinase Assay
GFP-FLAG, CRCK3-FLAG, and CRCK3KM-FLAG were transiently expressed in protoplasts for overnight, and purified by α-FLAG agarose. The proteins were incubated with 20 μL of kinase assay buffer (20 mm Tris-HCl, pH 7.5; 10 mm MgCl2; 5 mm EGTA; 100 mm NaCl; 1 mm dithiothreitol; and 1 μL [γ-32P]ATP) at room temperature for 3 h with gentle shaking. The reactions were stopped by adding 4× SDS protein loading buffer. The phosphorylation of proteins was analyzed by autoradiography after separation with 10% SDS-PAGE.
Accession numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (TAIR) or GenBank/EMBL databases under the following accession numbers: CLA1(AT4G15560), CRCK3 (AT2G11520), MEKK1 (AT4G08500), MEKK2(AT4G08480), SUMM2 (AT1G12280), MKK1(AT4G26070), MKK2(AT4G29810), MPK4(AT4G01370), BIR1 (AT5G48380), BAK1 (AT4G33430), SERK4 (AT2G13790), PR1 (AT2G14610), PR2 (AT3G57260), ACTIN2 (AT3G18780), and UBQ10 (AT4G05320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SALK_062374 is summ2-9.
Supplemental Figure S2. Sequence alignment of MEKK1 and MEKK2.
Supplemental Figure S3. MEKK2 does not have detectable kinase activity.
Supplemental Figure S4. Overexpression of CRCK3 derived from cDNA cannot complement summ3-17 phenotype.
Supplemental Figure S5. The expression pattern of CRCK3 and MEKK2.
Supplemental Table S1. Primers for gene cloning, point mutation, genotyping, and RT-qPCR.
Acknowledgments
We thank Arabidopsis Biological Resource Center (ABRC), Drs. Patrick Krysan (University of Wisconsin) and Yuelin Zhang (University of British Columbia) for Arabidopsis seeds, and members in He and Shan laboratories for comments.
Footnotes
This work was supported by National Science Foundation (NSF; grant no. MCB–1906060); HHS | National Institutes of Health (NIH; grant nos. R01GM092893 to P.H. and 1R01GM097247); the Robert A. Welch foundation (grant no. A–1795 to L.S.); National Natural Science Foundation of China (NSFC; grant no. 31672142 to X.W.); and China Scholarship Council (CSC; to Y.Y., C.Y., D.G., and Y.H.).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J(2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Berriri S, Garcia AV, Frei dit Frey N, Rozhon W, Pateyron S, Leonhardt N, Montillet JL, Leung J, Hirt H, Colcombet J(2012) Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP(2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL(2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE(2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- de Oliveira MV, Xu G, Li B, de Souza Vespoli L, Meng X, Chen X, Yu X, de Souza SA, Intorne AC, de A Manhães AM, et al. (2016) Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nat Plants 2: 15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D, Zhou JM(2012) Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Feng B, Ma S, Chen S, Zhu N, Zhang S, Yu B, Yu Y, Le B, Chen X, Dinesh-Kumar SP, Shan L, He P(2016) PARylation of the forkhead-associated domain protein DAWDLE regulates plant immunity. EMBO Rep 17: 1799–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y(2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, Zhang Y(2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, Shan L, He P(2013) Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L(2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K(2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Kong Q, Qu N, Gao M, Zhang Z, Ding X, Yang F, Li Y, Dong OX, Chen S, Li X, Zhang Y(2012) The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ(2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28: 215–220 [DOI] [PubMed] [Google Scholar]
- Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, Shan L(2015a) Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27: 839–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P(2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S(2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li F, He P(2015b) Construction of a cotton VIGS library for functional genomics study. Methods Mol Biol 1287: 267–279 [DOI] [PubMed] [Google Scholar]
- Lian K, Gao F, Sun T, van Wersch R, Ao K, Kong Q, Nitta Y, Wu D, Krysan P, Zhang Y(2018) MKK6 functions in two parallel MAP kinase cascades in immune signaling. Plant Physiol 178: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zhou JM(2018) Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Lin W, Ma X, Shan L, He P(2013) Big roles of small kinases: The complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol 55: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S(2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC(2003) High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J 22: 5690–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C(2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S(2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Soukupová H, Schikora A, Zárský V, Hirt H(2006) A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J Biol Chem 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Palma K, Zhao Q, Cheng YT, Bi D, Monaghan J, Cheng W, Zhang Y, Li X(2007) Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev 21: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, van Wersch R, Zhang Y(2018) Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol Plant Microbe Interact 31: 403–409 [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu J-L, Zhou L, Yun B-W, Nielsen HB, Fiil BK, Petersen K, Mackinlay J, Loake GJ, Mundy J, Morris PC(2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. (2008) Intron-mediated regulation of gene expression. In Reddy ASN and Golovkin MV, eds, Nuclear pre-mRNA Processing in Plants. Current Topics in Microbiology and Immunology, vol 326 Springer, Berlin, pp 277–290 [Google Scholar]
- Saijo Y, Loo EP, Yasuda S(2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW(2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Su SH, Bush SM, Zaman N, Stecker K, Sussman MR, Krysan P(2013) Deletion of a tandem gene family in Arabidopsis: Increased MEKK2 abundance triggers autoimmunity when the MEKK1-MKK1/2-MPK4 signaling cascade is disrupted. Plant Cell 25: 1895–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ(2007) MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Hamano K, Takagi H, Morimoto T, Akimitsu K, Terauchi R, Shirasu K, Ichimura K(2019) Disruption of the MAMP-Induced MEKK1-MKK1/MKK2-MPK4 pathway activates the TNL immune receptor SMN1/RPS6. Plant Cell Physiol 60: 778–787 [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J(2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasi Devendrakumar K, Li X, Zhang Y(2018) MAP kinase signalling: Interplays between plant PAMP- and effector-triggered immunity. Cell Mol Life Sci 75: 2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chen S, et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J(2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: eaav5868. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, Zhou JM, Chai J(2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364: 364. [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Elmore JM, Creer AY, Feng B, Franco JY, Rufian JS, He P, Phinney B, Coaker G(2017) A cysteine-rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiol 173: 771–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Chaudhuri S, Yang L, Chen Y, Poovaiah BW(2004) Calcium/calmodulin up-regulates a cytoplasmic receptor-like kinase in plants. J Biol Chem 279: 42552–42559 [DOI] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L(2017) From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55: 109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Xu G, Li B, de Souza Vespoli L, Liu H, Moeder W, Chen S, de Oliveira MVV, Ariadina de Souza S, Shao W, et al. (2019) The receptor kinases BAK1/SERK4 regulate Ca2+ channel-mediated cellular homeostasis for cell death containment. Curr Biol 29: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Su J, Zhang Y, Xu J, Zhang S(2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol 45(Pt A): 1–10 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liu Y, Huang H, Gao M, Wu D, Kong Q, Zhang Y(2017) The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep 18: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y(2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang P, Claus LAN, Savatin DV, Xu G, Wu S, Meng X, Russinova E, He P, Shan L(2019) Proteolytic processing of SERK3/BAK1 regulates plant immunity, development, and cell death. Plant Physiol 180: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]