N-terminal acetylation regulates SIB1 protein stability to ensure its function in salicylic acid-primed cell death.
Abstract
N-terminal (Nt) acetylation (NTA) is an ample and irreversible cotranslational protein modification catalyzed by ribosome-associated Nt-acetyltransferases. NTA on specific proteins can act as a degradation signal (called an Ac/N-degron) for proteolysis in yeast and mammals. However, in plants, the biological relevance of NTA remains largely unexplored. In this study, we reveal that Arabidopsis (Arabidopsis thaliana) SIGMA FACTOR-BINDING PROTEIN1 (SIB1), a transcription coregulator and a positive regulator of salicylic acid-primed cell death, undergoes an absolute NTA on the initiator Met; Nt-acetyltransferase B (NatB) partly contributes to this modification. While NTA results in destabilization of certain target proteins, our genetic and biochemical analyses revealed that plant NatB-involved NTA instead renders SIB1 more stable. Given that the ubiquitin/proteasome system stimulates SIB1 degradation, it seems that the NTA-conferred stability ensures the timely expression of SIB1-dependent genes, mostly related to immune responses. Taking our findings together, here we report a noncanonical NTA-driven protein stabilization in land plants.
The N termini of proteins often undergo a variety of cotranslational and posttranslational modifications (Gibbs et al., 2014; Giglione et al., 2015). In eukaryotes, N-terminal (Nt) acetylation (NTA), catalyzed by Nt-acetyltransferases (NATs), is one of the most abundant and irreversible cotranslational protein modifications (Aksnes et al., 2016; Ree et al., 2018). NATs enzymatically transfer the acetyl moieties (CH3CO) from acetyl-CoA to the free α-amino group (NH3+) at the Nt end of nascent polypeptides, thereby changing the chemical properties of the proteins (Aksnes et al., 2015). NATs are generally composed of one specific catalytic subunit and in some cases also possess at least one auxiliary subunit contributing a ribosome anchor, substrate specificity, and interaction with nascent polypeptides (Aksnes et al., 2016; Ree et al., 2018). The substrate specificity of NATs is mainly determined by the first two amino acids at the protein N terminus (Aksnes et al., 2016).
To date, eight NAT complexes (NatA to NatH) have been identified in eukaryotes (Drazic et al., 2018; Aksnes et al., 2019). Three NATs (NatA/B/C) are responsible for the vast majority of NTA. NatA specifically acetylates the N terminus carrying an exposed small amino acid residue (Ala, Ser, Thr, Cys, and Val), which appears after the cleavage of the initiator Met (iMet) by Met aminopeptidases (MetAPs; Mullen et al., 1989; Arnesen et al., 2009). NatB acetylates the iMet with acidic/hydrophilic residues (MD, MN, ME, and MQ), whereas NatC acetylates the iMet followed by hydrophobic/amphipathic residues (ML, MI, MF, MW, MV, MM, MH, and MK; Polevoda et al., 1999; Polevoda and Sherman, 2001; Starheim et al., 2009; Van Damme et al., 2012; Ree et al., 2018). Interestingly, NatG (AtNaa70) is localized in the plastids, and it posttranslationally acetylates proteins with Met-, Ala-, Ser-, and Thr-starting N termini (Dinh et al., 2015). Furthermore, based on the recent Nt acetylome analyses, the first two amino acid residues also largely determine the likelihood of a particular protein being Nt acetylated.
NTA widely influences protein dynamics, such as protein turnover (Hwang et al., 2010; Shemorry et al., 2013; Myklebust et al., 2015; Xu et al., 2015; Sheikh et al., 2017), protein folding (Bartels et al., 2011; Trexler and Rhoades, 2012; Holmes et al., 2014), protein-protein interactions (Singer and Shaw, 2003; Coulton et al., 2010; Scott et al., 2011; Arnaudo et al., 2013; Monda et al., 2013; Yang et al., 2013), and subcellular localization of target proteins (Behnia et al., 2004, 2007; Setty et al., 2004; Forte et al., 2011). Recent studies also pointed out that NTA modulates protein half-life (Hwang et al., 2010; Shemorry et al., 2013; Park et al., 2015), a phenomenon dubbed the N-end rule pathway (Varshavsky, 2011; Tasaki et al., 2012; Gibbs et al., 2014; Dissmeyer et al., 2018; Nguyen et al., 2018). The protein N termini (including iMet and exposed Ala, Ser, Thr, Cys, or Val residues) can be Nt acetylated via NATs, wherein the acetyl moiety subsequently acts as an Ac/N-degron. The Ac/N-recognins (mostly E3 ligases) explicitly recognize the acetyl moiety for the ubiquitin/proteasome system (UPS), promoting protein degradation. (Hwang et al., 2010; Shemorry et al., 2013; Aksnes et al., 2016; Nguyen et al., 2018; Ree et al., 2018).
The biological relevance of NTA in plant proteins relative to yeast and mammals is not yet established. Individual NAT knockdown and knockout mutants in Arabidopsis (Arabidopsis thaliana) exhibit severe developmental and physiological defects. For instance, the loss of NatA subunits, such as Naa10 (catalytic subunit) and Naa15 (auxiliary subunit), causes embryo lethality (Linster et al., 2015). In addition, NatA knockdown plants show abscisic acid-mediated physiological responses including stomata closure, priming their resistance to drought stress (Linster et al., 2015). The mutants of NatB subunits, naa20 and naa25, exhibit several developmental defects, such as leaf reticulation, early flowering, aborted or unfertilized ovules in the siliques, and increased susceptibility to osmotic and salt stress (Ferrández-Ayela et al., 2013; Huber et al., 2020). Similarly, the loss of Naa30 (NatC catalytic subunit) leads to a decreased PSII quantum yield (Pesaresi et al., 2003). The aberrant phenotypes observed in various NAT mutants are indicative of the significance of NTA in plants. Previously, the acetylated iMet was thought to serve as an Ac/N-degron for successive proteolysis in yeast and mammals. However, a recent study demonstrated a noncanonical function of NTA in plants. The NTA determines the turnover rate of a Nod-like receptor protein, SUPPRESSOR OF NPR1 CONSTITUTIVE1 (SNC1), a well-known immune receptor (Zhang et al., 2003). SNC1 is translated into two protein variants via alternative translational initiation sites, resulting in either iMMD or iMD at the N terminus. NatA acetylates the iMet of the iMMD variant (generally considered as a substrate of NatC), whereas NatB acetylates the iMet of the iMD variant. Based on the canonical Ac/N-end rule pathway revealed in yeast and mammals, both acetylated iMD and iMMD theoretically would serve as an Ac/N-degron (Aksnes et al., 2016). This may consequently result in the degradation of both variants through the Ac/N-end rule pathway. However, NTA of the two SNC1 variants results in opposing effects on protein stability: the acetylated iMMD, as usual, serves as an Ac/N-degron, whereas the acetylated iMD virtually works as a stabilizing signal (Xu et al., 2015).
In this study, we reveal that the SIGMA FACTOR-BINDING PROTEIN1 (SIB1), a transcription coregulator targeted to both the nucleus and chloroplasts via a combined Nt nuclear localization signal peptide and plastid transit peptide, undergoes an absolute NTA on iMet. SIB1 has been implicated in both reactive oxygen species- and salicylic acid (SA)-mediated stress responses (Lai et al., 2011; Kim et al., 2012; Lv et al., 2019). While the steady-state level of SIB1 (both the transcript and protein) is nearly negligible under nonstress conditions, an increase of cellular SA and/or reactive oxygen species leads to a rapid and transient accumulation of SIB1 in both the nucleus and chloroplasts. The nucleus-localized SIB1 interacts with the transcription factors (TFs) WRKY33 and WRKY57 to induce subsets of defense-related genes, while the chloroplast-localized SIB1 represses the expression of photosynthesis-associated genes (Xie et al., 2010). Previously, we also illustrated that SIB1 contributes to the initiation of the SA-dependent uncontrolled cell death in the Arabidopsis lesion simulating disease1 (lsd1) mutant (Lv et al., 2019). Here, we now demonstrate that the NTA on SIB1, partly catalyzed by NatB, stabilizes the protein, enabling its function as a transcription coregulator. On the contrary, the UPS retains SIB1 abundance at a minimum level in order to avoid an excessive cellular stress response. This impact of NTA on SIB1 stability is in contrast to the typical Ac/N-end rule pathway in yeast and mammals, which is known to promote proteolysis of target proteins. However, this noncanonical NTA pathway, rendering the substrate more stable, largely resembles the fate of the acetylated iMD variant of SNC1 (stable form), which also undergoes NatB-mediated NTA and proteolysis via the UPS. Thus, we propose that the NTA, catalyzed partly by NatB, may serve as a stabilizing signal at least for the stress-associated proteins SIB1 and SNC1.
RESULTS
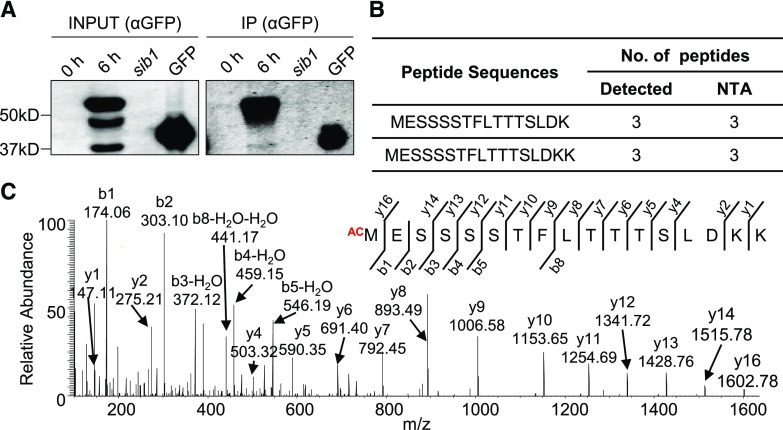
The SIB1 Protein Undergoes Nt Acetylation
Nucleus-targeted SIB1 facilitates the expression of both photosynthesis-associated and immune-related genes, while the chloroplast-localized SIB1 represses a subset of plastid-encoded photosynthesis-associated genes (Morikawa et al., 2002; Xie et al., 2010; Lai et al., 2011; Jiang and Yu, 2016; Lv et al., 2019). Increased intracellular SA content and exogenous SA result in a rapid accumulation of SIB1 at both mRNA and protein levels (Supplemental Fig. S1, A and B). Treatment of transgenic sib1 plants expressing GFP-tagged SIB1 (SIB1-GFP), driven by the native SIB1 promoter (pSIB1), with 1 mm SA resulted in the accumulation of the SIB1 protein, which reached a maximum at ∼6 h followed by a gradual decline (Supplemental Fig. S1B), coinciding with the previous report (Lv et al., 2019). This exogenous SA also induced PATHOGENESIS-RELATED1 (PR1), a marker protein for SA response, which was detectable after 9 h (Supplemental Fig. S1B). Interestingly, the immunoblot assay identified multiple forms of the SIB1 protein: the two lower bands correspond with the predicted nuclear (∼44 kD) and plastidic (∼37 kD) forms, while the uppermost (and most abundant) form was almost 10 kD bigger than the nuclear form of SIB1 (Fig. 1A; Supplemental Fig. S1B; Lv et al., 2019). Subsequent subcellular fractionation assays confirmed that the uppermost form is exclusively localized in the nucleus while the predicted nuclear form of SIB1 mostly appeared in the cytosol (Supplemental Fig. S2). This result suggests that SIB1 may undergo further modification upon nuclear localization or that the nuclear targeting of SIB1 may rely on further modification. The higher abundance of the potentially modified SIB1 relative to the protein form in the cytosol prompted us to hypothesize that, if modified, the change may stabilize SIB1. As a stress-responsive transcriptional coregulator, its modification(s) may confer a proper functionality in the regulation of stress-related TFs within a short period. Such stress-related proteins involved in gene regulation tend to degrade rapidly to avoid the undesirable activation of stress responses (Huang et al., 2013). Nonetheless, the detection of the unpredicted large-sized SIB1 stimulated us to examine any possible modifications of SIB1.
Figure 1.
iMet of SIB1 undergoes NTA. Transgenic sib1 plants harboring pSIB1::SIB1-GFP were grown on Murashige and Skoog medium under continuous light conditions. Five-day-old plants initially grown on normal medium were transferred to SA-containing Murashige and Skoog medium (1 mm SA). The samples were harvested before (0 h) and after a 6-h SA treatment. A, IP-immunoblot assay shows multiple bands of SIB1-GFP proteins. SIB1-GFP proteins were identified by anti-GFP (αGFP) antibody. Seedlings of the sib1 mutant and a transgenic line expressing a GFP-tagged small subunit of Rubisco (GFP) were used as controls for the IP assay. B, iMet of SIB1 was acetylated in all detected Nt peptides by IP-MS analysis. C, MS spectrum of Nt peptide 1MESSSSTFLTTTSLDKK17 showing acetylation on iMet.
To reveal possible modifications in SIB1, sib1 transgenic seedlings expressing pSIB1::SIB1-GFP were first grown on SA-free medium and then transferred to fresh medium containing 1 mm SA. After a 6-h treatment, these seedlings were collected and total protein was extracted to concentrate SIB1-GFP proteins using a GFP-Trap coupled to magnetic agarose beads. The enriched proteins were digested and subjected to mass spectrometry (MS) analysis to find modification(s) in SIB1; a suite of parameters such as NTA, phosphorylation, and ubiquitination was included. The MS analysis revealed a substantial coverage of SIB1 (∼59%; Supplemental Fig. S3A) and an NTA on iMet in all detected Nt peptides of SIB1 (Fig. 1, B and C; Supplemental Fig. S3B). Considering that NTA is implicated in protein turnover, subcellular localization, and protein-protein interactions, we assumed that iMet acetylation of SIB1 might be necessary for either of these aspects.
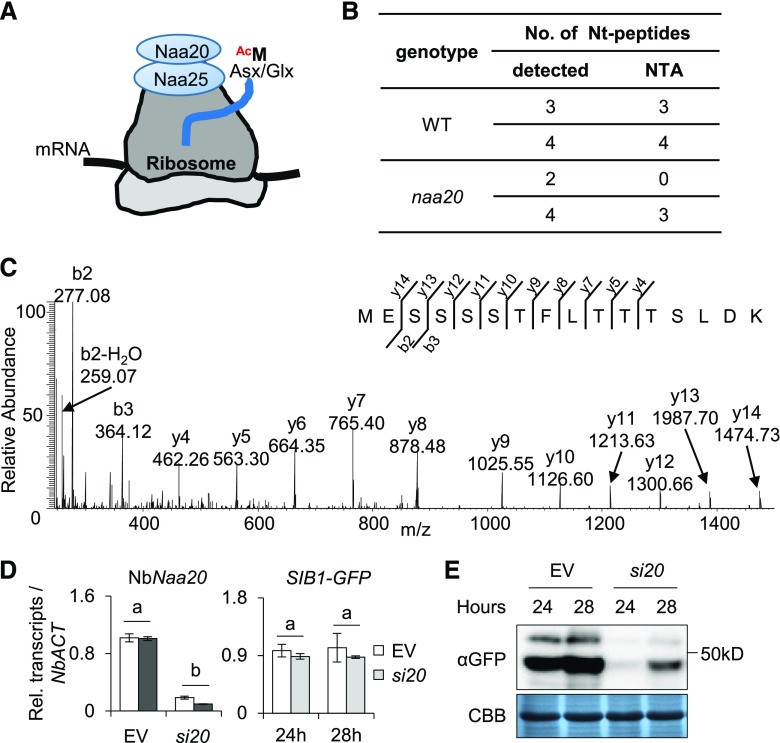
NTA Stabilizes SIB1
NATs catalyze NTAs and their substrate specificity is mostly determined by the characteristics of the first two Nt residues (Aksnes et al., 2016; Ree et al., 2018). As aforementioned, NatB is known to catalyze the NTA of those proteins starting with MD, ME, MN, and MQ, and ME-starting proteins are thought to undergo NTA with nearly 100% efficiency in yeast and mammals (Aksnes et al., 2016; Ree et al., 2018). Because the iMet of SIB1 protein is followed by E, we assumed that NatB might catalyze the NTA of SIB1, which would consequently affect the fate of SIB1. NatB is composed of two subunits, the catalytic subunit Naa20 and the auxiliary subunit Naa25, which cotranslationally acetylates MD/E/N/Q-starting N termini directly at the ribosome (Fig. 2A). As Naa20 serves as a catalytic subunit of NatB, we examined NTA of SIB1 in an Arabidopsis naa20 null mutant (Supplemental Fig. S4). The GFP-tagged Arabidopsis SIB1 was transiently overexpressed under the control of the cauliflower mosaic virus 35S promoter (35S::SIB1-GFP) in the mesophyll protoplasts isolated from 3-week-old naa20 and wild-type plants. SIB1-GFP proteins were then enriched using the GFP-trap coupled with magnetic agarose beads and subjected to MS analysis for NTA identification. The resulting MS analysis revealed an absolute NTA in all Nt peptides of SIB1 detected in the wild type (Fig. 2B; Supplemental Table S1). In naa20 protoplasts, both acetylated and nonacetylated Nt peptides of SIB1 were detected (Fig. 2, B and C; Supplemental Table S1), indicating that NatB only partly contributes to the NTA of SIB1.
Figure 2.
The SIB1 protein is stabilized via NTA, which is partly catalyzed by NatB. A, The NatB complex (Naa20 and Naa25) catalyzes the NTA on nascent peptides starting with Met-acidic/hydrophilic (Asx/Glx) N termini (MD, MN, ME, and MQ) at the ribosome. B, The NTA status of the SIB1 protein after transient expression in protoplasts. Proteins isolated from wild-type (WT) and naa20 knockout mutant mesophyll protoplasts were enriched by IP and subjected to MS analysis. C, MS spectrum of nonacetylated Nt peptide 1MESSSSTFLTTTSLDK16 detected in the naa20 mutant. D, The transcript levels of NbNaa20 and SIB1-GFP in si20 or empty vector. EV, N. benthamiana plants infiltrated with the VIGS empty vector, which serves as a negative control; si20, NbNaa20-silenced N. benthamiana plants. The SIB1-GFP was amplified by using SIB1-specific forward primer and GFP-specific reverse primer. NbACT was used as an internal standard. The data represent means of three independent biological replicates, and error bars indicate sd. Lowercase letters indicate statistically significant differences between the mean values (P < 0.05, one-way ANOVA with posthoc Tukey’s honestly significant difference [HSD] test). E, NbNaa20 was silenced in N. benthamiana by the VIGS approach. The expression levels of the SIB1-GFP fusion protein were detected using anti-GFP (αGFP) antibody. Coomassie Brilliant Blue staining (CBB) was used as a loading control.
To further corroborate the role of NatB on SIB1 NTA, we generated virus-induced gene silencing (VIGS) lines in Nicotiana benthamiana, specifically targeting NbNaa20 (an Arabidopsis Naa20 homolog). The 35S::SIB1-GFP construct was expressed via Agrobacterium tumefaciens infiltration in different lines of N. benthamiana, including the VIGS-empty vector control and the VIGS-NbNaa20-silenced (si20) lines. Reverse transcription-quantitative PCR (RT-qPCR) showed the substantial repression of NbNaa20 in cognate VIGS lines, whereas the transcript level of SIB1-GFP was nearly comparable (Fig. 2D). Despite the similar transcript levels, the immunoblot assay showed significantly reduced amounts of SIB1-GFP protein in the si20 background as compared with the empty vector line (Fig. 2E). In general, the NatB-acetylated N termini are potential substrates of the Ac/N-end rule pathway, facilitating protein degradation. However, our result suggests that the NTA of SIB1 likely acts as a stabilizing signal.
Since SA induces SIB1 and its cotranslational NTA, one may suggest SA-driven NatB activation or its stabilization, which would consequently alter the stress-related transcriptome. To investigate the impact of SA on NatB, N. benthamiana leaves transiently overexpressing Myc-tagged Naa20 protein were treated with SA. The subsequent immunoblot assay found a slightly increased level of Naa20-Myc protein with SA treatment (Supplemental Fig. S5A). The presence of PR1 in the negative control is attributable to the basal plant stress response to the A. tumefaciens-mediated transient gene expression assay. It was difficult to say, without further examination, whether such an SA-mediated subtle increase of Naa20-Myc impacts SIB1 NTA. Alternatively, then, we examined the impact of SA on the relative abundance of non-SA-inducible SIB1-GFP protein expressed under the control of the 35S promoter. The transgenic wild-type plants harboring 35S-driven SIB1-GFP were treated with SA, and the subsequent immunoblot assay found comparable SIB1 protein levels, irrespective of the presence or absence of SA (Supplemental Fig. S5B). This result, therefore, suggests that SA seems to affect SIB1 transcription primarily and that SA may not alter NatB (as well as other NTAs) activity.
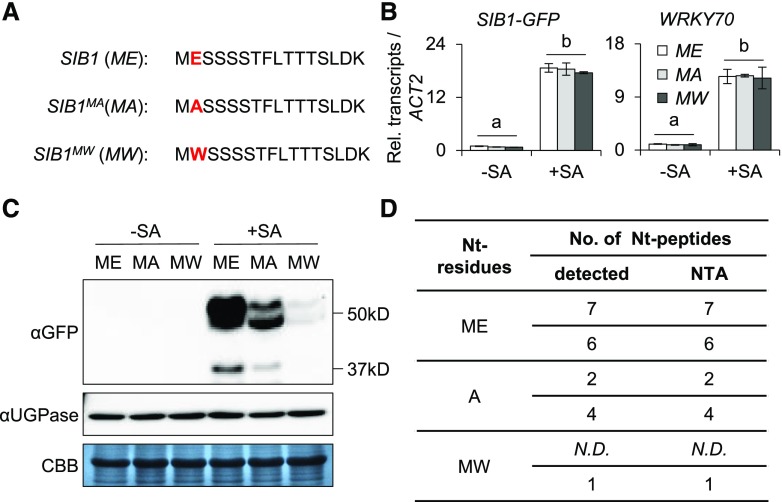
The Second Amino Acid of SIB1 Is Decisive for NTA
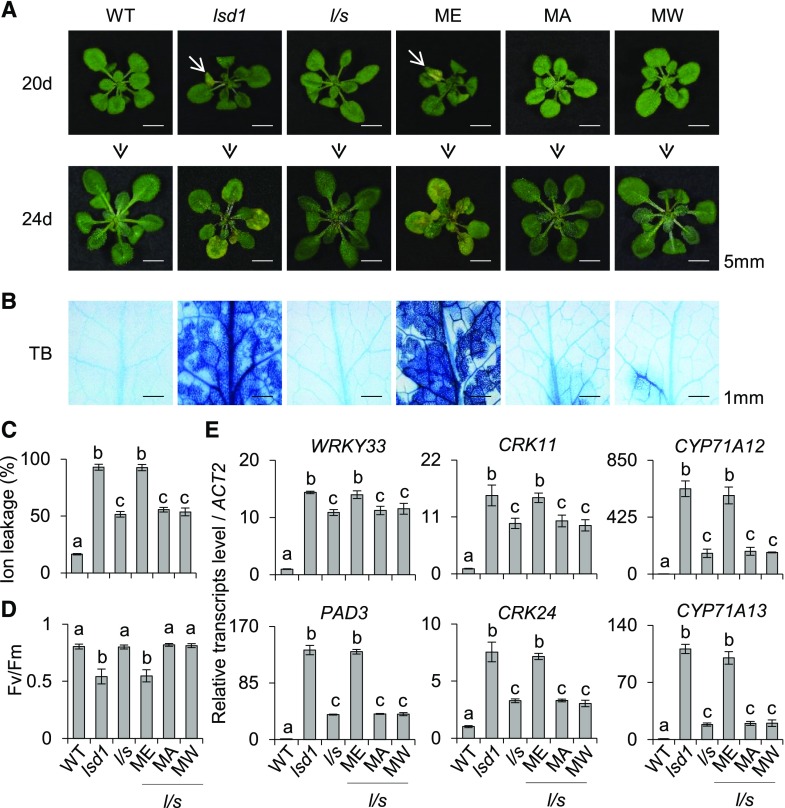
The substrate specificities of NATs are determined by the first two amino acids of the target protein (Ree et al., 2018). Consequently, the substitution of the second residue would principally modulate the NTA, thus influencing the stability and/or biological function of the target protein. By utilizing this approach, the NTA status of a given protein can be modulated to understand its biological relevance. To verify the critical role of Nt residues and the biological relevance of SIB1 NTA, we generated SIB1 variants by substituting the second residue (E) with Ala (A; a small aliphatic residue with 95% NTA frequency) or Trp (W; a bulky hydrophobic residue with 60% NTA frequency). This approach resulted in the generation of the two SIB1 variants SIB1MA and SIB1MW (Fig. 3A). By substituting the E to A, we attempted to shift the NTA from iMet to a penultimate site, which would promote SIB1 degradation via the Ac/N-end rule pathway. In contrast, the substitution of E to W would reduce the probability of NTA of iMet. Acetylated and nonacetylated MW proteins are supposed to be degraded via the Ac/N-end and Arg/N-end rule pathways (another well-known branch of the N-end rule pathway that targets the nonacetylated MW-starting proteins), respectively (Kendall and Bradshaw, 1992; Hwang et al., 2010; Xiao et al., 2010; Bonissone et al., 2013; Shemorry et al., 2013; Kim et al., 2014; Xu et al., 2015; Nguyen et al., 2018). Therefore, we anticipated a comparatively faster decline of SIB1MW. To examine the fates of these SIB1 variants, we utilized the lsd1 mutant wherein SIB1 largely contributes to cell death upon its rapid accumulation in response to the increased cellular SA content (Lv et al., 2019). Two independent transgenic lsd1 sib1 lines harboring pSIB1-driven GFP-tagged SIB1MA or SIB1MW were generated. First, the 5-d-old seedlings of these transgenic lines, including SIB1MA-GFP lsd1 sib1, SIB1MW-GFP lsd1 sib1, and SIB1-GFP lsd1 sib1, were treated with 1 mm SA. Along with WRKY70, a renowned SA-responsive TF, the SIB1-GFP transcripts were comparably induced in all examined lines (Fig. 3B). However, the content of SIB1MA and SIB1MW proteins showed a significant reduction relative to SIB1 (Fig. 3C). In particular, the SIB1MW variant was barely detectable in the immunoblot assay.
Figure 3.
The substitution of the second amino acid alters the stability of SIB1. A, Nt sequences of intact and modified SIB1. B, The transcript level of SIB1-GFP after SA treatment. Five-day-old transgenic lsd1 sib1 plants expressing different forms of GFP-tagged SIB1 driven by the SIB1 native promoter were treated with 1 mm SA. The transcript levels of SIB1-GFP from the indicated genotypes were examined 2 h after SA treatment using RT-qPCR. SIB1-GFP was amplified using a SIB1-specific forward primer and a GFP-specific reverse primer. ACTIN2 (ACT2) was used as an internal standard. The SA-responsive gene WRKY70 was used as a positive control. The data represent means of three independent biological replicates, and error bars indicate sd. Lowercase letters indicate statistically significant differences between the mean values (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test). C, The expression levels of the GFP-tagged native and modified SIB1 proteins. Fusion proteins were detected by immunoblot assay using anti-GFP (αGFP) antibody after a 6-h SA treatment. Coomassie Brilliant Blue staining (CBB) and UDP-Glc-pyrophosphorylase (UGPase) were used as loading controls. D, The NTA status of intact and modified SIB1 protein. Proteins isolated from transgenic plants were enriched by IP and subjected to MS analysis. The data represent two independent biological replicates. N.D., Not detectable.
The reduced accumulation of the modified SIB1 proteins as compared with the native form seems attributable to their altered stability. To further analyze a direct link between NTA and their protein stability, we checked the NTA status of the modified SIB1-GFP proteins by MS analysis. Label-free quantitation indeed confirmed the reduction of SIB1MA and SIB1MW proteins (Supplemental Fig. S6). The MS analysis revealed that all the Nt peptides of the iMet detected in SIB1 were Nt acetylated (Fig. 3D; Supplemental Fig. S7A). For SIB1MA, the Nt peptides with the exposed A showed an unequivocal NTA (Fig. 3D; Supplemental Fig. S7B), indicating that the iMet of SIB1MA was first cleaved by MetAPs followed by a NatA-mediated acetylation of the exposed A. In the case of SIB1MW, we could only detect acetylated iMet with a substantially reduced protein abundance (Fig. 3D; Supplemental Fig. S7C), which was actually anticipated given their susceptibility for degradation by both Ac/N-end and Arg/N-end rule pathways (Aksnes et al., 2016; Nguyen et al., 2018; Ree et al., 2018).
We also generated an NTA-avoiding SIB1 variant by substituting the second residue with Pro (P; a hydrophobic residue conferring 0% NTA frequency). MetAPs remove the iMet of MP-starting N termini, and the exposed P evades NTA (Aksnes et al., 2016; Nguyen et al., 2018; Ree et al., 2018). Accordingly, we attempted to generate an NTA-free N terminus for SIB1 in N. benthamiana leaves. We then examined the stability of the NTA-free SIB1MP form relative to intact SIB1. Immunoprecipitation (IP) coupled with the immunoblot assay showed clearly lower amounts of SIB1MP-GFP relative to SIB1-GFP protein, but with a nearly comparable transcript level between intact and modified SIB1-GFP (Supplemental Fig. S8, A–C). Furthermore, the MS result found that while all the Nt peptides of SIB1-GFP (more than 60 peptides detected in two replicates) are Nt acetylated, all the Nt peptides of SIB1MP-GFP (20 peptides detected in two replicates) present exposed P without NTA. This result further suggested that the acetylated iMet of SIB1 is essential for its protein stability. Taken together, the NTA status and protein stability comparison revealed that SIB1MA, SIB1MW, and SIB1MP are seemingly less stable relative to SIB1, ensuring that the NTA of SIB1 serves as a stabilizing signal instead of serving as a degron promoting SIB1 proteolysis via the canonical Ac/N-end rule pathway (Supplemental Fig. S8D).
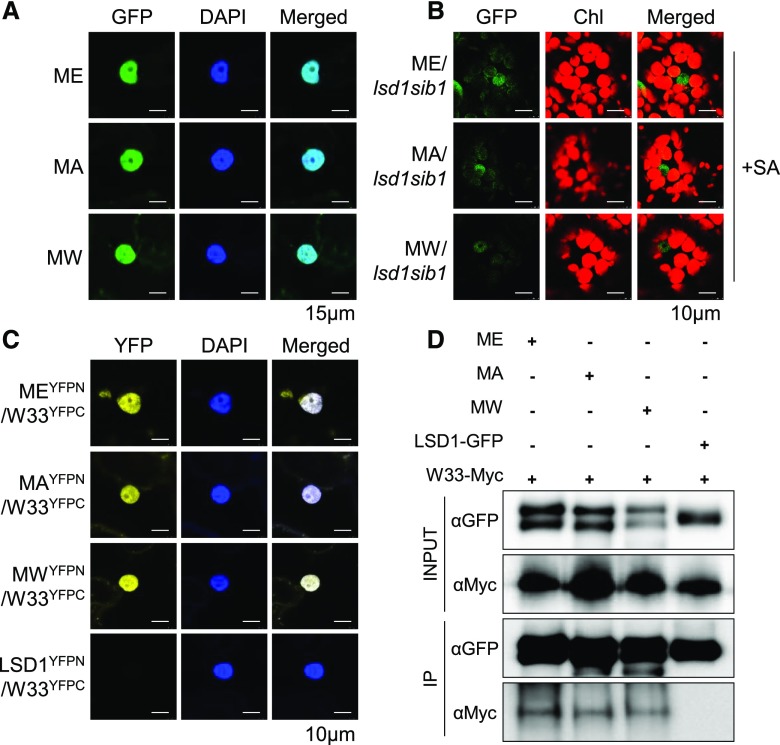
NTA Only Alters SIB1 Stability
To examine whether substitutions of the second residue impact any other aspects of the SIB1 dynamics, which may indirectly change its stability, we first analyzed the nuclear localization of the transiently overexpressed GFP-tagged SIB1MA and SIB1MW along with SIB1-GFP in N. benthamiana leaves (Fig. 4A). In addition, stable transgenic plants expressing pSIB1-driven GFP-tagged SIB1MA or SIB1MW or SIB1 were analyzed to monitor their nuclear localization upon induction by exogenous SA (Fig. 4B). Confocal imaging of the GFP signal found the native as well as the modified SIB1 proteins in the nucleus in both experimental conditions (Fig. 4, A and B). We then carried out a bimolecular fluorescence complementation (BiFC) assay and a coimmunoprecipitation (Co-IP) analysis in N. benthamiana leaves to confirm whether these substitutions affect the known physical interaction between SIB1 and WRKY33. The results show that the modified SIB1 variants also interact with WRKY33 in the nucleus (Fig. 4, C and D). The nuclear localization and interaction of all three SIB1 variants with WRKY33 suggest that the NTA status of SIB1 only affects protein stability.
Figure 4.
Substitution of the second residue does not affect the nuclear localization and protein-protein interactions of SIB1. A and B, Native and modified SIB1-GFP proteins are all targeted to the nucleus. The nuclear GFP signal was detected in N. benthamiana leaves (A) as well as in SA-treated transgenic Arabidopsis plants (B). Chl, Chlorophyll; DAPI, 4′,6-diamidino-2-phenylindole, a fluorescent stain for nuclear localization. C and D, In vivo interaction between SIB1 (ME), SIB1MA (MA), SIB1MW (MW), and WRKY33 (W33) by BiFC (C) and Co-IP analyses (D) upon transient coexpression in N. benthamiana leaves. LSD1 was used as a negative control, as it does not interact with WRKY33.
Given that SIB1 is a dually targeted protein, NTA may influence its import into the plastid and may alter the ratio of the nuclear and cytosolic fractions. However, both native SIB1 (SIB1ME) and its NTA variant (SIB1MA and SIB1MW) SIB1 proteins show a correlated abundance of nuclear, cytosolic, and plastid forms (Fig. 3C), which indicates that the altered NTA does not change the plastid import competence. Collectively, these results suggest that SIB1 NTA does not affect the subcellular targeting and protein-protein interaction but rather determines the protein stability.
NTA-Stabilized SIB1 Fully Complements the lsd1 sib1 Double Mutant
Inactivation of SIB1 significantly attenuates the cell death in the lsd1 mutant, which is fully complemented by expressing pSIB1::SIB1-GFP (Lv et al., 2019). Because NTA renders SIB1 more stable, we anticipated that unstable SIB1 variants might fail to complement the lsd1 sib1 double mutant. Thus, we analyzed the macroscopic and molecular phenotypes of lsd1 sib1 transgenic plants expressing GFP-tagged SIB1, SIB1MA, or SIB1MW. Before the onset of cell death, the transcript levels of SIB1-GFP in all these lines were induced to almost comparable levels (Supplemental Fig. S9A). By contrast, the immunoblot assay showed a noticeable difference in protein abundance (Supplemental Fig. S9B). The variance between the protein abundance in the nucleus, despite comparable transcript levels of intact and modified SIB1, indicates that the modified SIB1 proteins are likely undergoing degradation via the Ac/N-end rule pathway. Consistent with the protein abundance, the expression of intact SIB1 fully restored the lsd1-conferred lesions in the lsd1 sib1 double mutant (Fig. 5A). By contrast, the compromised foliar chlorosis and cell death phenotypes in lsd1 sib1 plants remained unchanged in the SIB1MA lsd1 sib1 and SIB1MW lsd1 sib1 lines (Fig. 5A). The determination of cell viability via Trypan Blue (TB) staining and electrolyte leakage analysis, and the determination of maximum photochemical efficiency of PSII (Fv/Fm) for chlorosis, further confirmed the differential cell death phenotypes (Fig. 5, B–D). The SIB1 protein potentiates the DNA-binding activity of WRKY33, which subsequently induces a suite of immune-related genes (Lai et al., 2011; Mao et al., 2011; Birkenbihl et al., 2017; Lv et al., 2019). Thus, we also checked the expression of six immune-related genes, including WRKY33 and its downstream genes PHYTOALEXIN DEFICIENT3 (PAD3), CYSTEINE-RICH RECEPTOR-LIKE PROTEIN KINASE11 (CRK11) and CRK24, and CYTOCHROME P450 FAMILY71 SUBFAMILY A POLYPEPTIDE12 (CYP71A12) and CYP71A13, which are all known to be up-regulated prior to the onset of cell death in the lsd1 mutant. As expected, both SIB1MA and SIB1MW failed to induce these genes (Fig. 5E), suggesting a dosage effect of SIB1 for its functionality.
Figure 5.
The modified SIB1 proteins do not complement the lsd1 sib1 mutant. A, Images were taken from continuous light-grown 20- and 24-d-old wild-type (WT), lsd1, and lsd1 sib1 (l/s) plants as well as transgenic lsd1 sib1 plants expressing the different variants of pSIB1-driven GFP-tagged SIB1. B, Cell death was visualized by TB staining. C and D, Ion leakage for the quantification of cell death (C) and Fv/Fm for chlorosis (D) were determined. First and second leaves from 24-d-old plants were harvested for measuring ion leakage and Fv/Fm. Six leaves per genotype were used for each measurement. Data represent means from three independent measurements, and error bars indicate sd. Lowercase letters indicate statistically significant differences between mean values at each of the indicated time points (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test). E, The relative transcript levels of immune-related genes in 20-d-old plants were analyzed using RT-qPCR. ACTIN2 (ACT2) was used as an internal standard. The data represent means of three independent biological replicates, and error bars indicate sd. Lowercase letters indicate statistically significant differences between mean values at each of the indicated time points (P < 0.05, one-way ANOVA with posthoc Tukey’s HSD test).
SIB1 Undergoes 26S Proteasome-Mediated Degradation
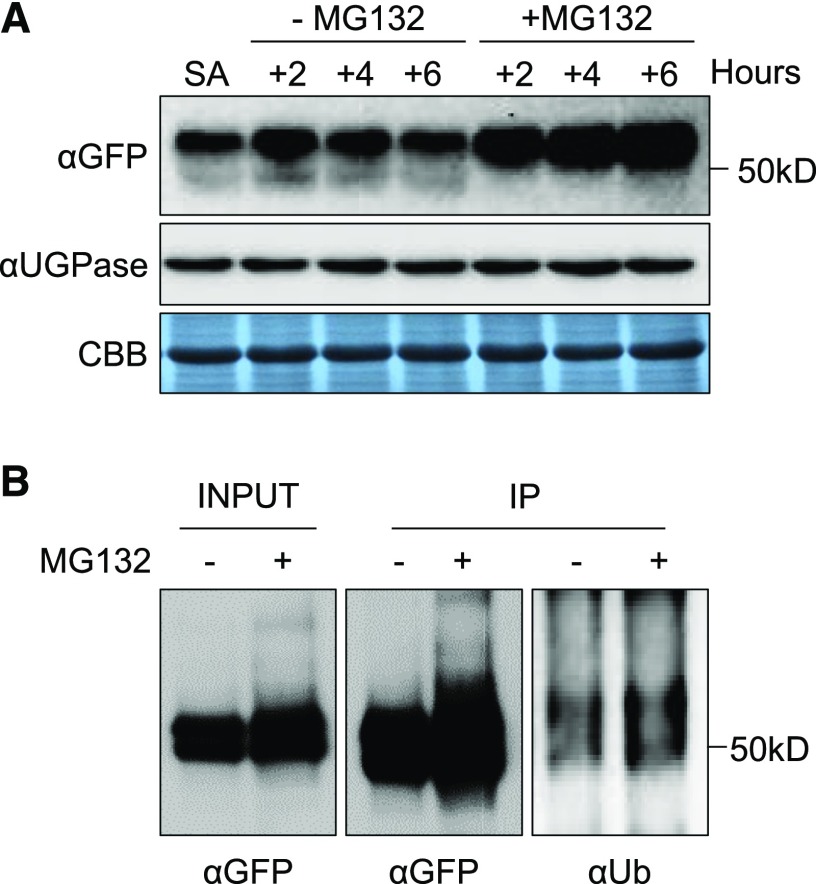
It seems that the NAT-catalyzed NTA plays a vital role in sustaining the protein level of SIB1. Our data show that the SIB1 protein undergoes degradation (Supplemental Fig. S1), probably upon induction of its downstream target genes. Considering that the molecular mass of the uppermost band of SIB1-GFP in the immunoblot assay showed a molecular shift of roughly 10 kD from its predicted mass (Fig. 1A) and that the UPS marks substrate proteins with a polyubiquitin chain for proteolysis by the 26S proteasome (Vierstra, 2009; Gibbs et al., 2014; Dissmeyer et al., 2018), this posttranslational modification may lead to the UPS-dependent SIB1 degradation. To explore this, we first looked for potential ubiquitination sites of SIB1 using UbiSite (http://csb.cse.yzu.edu.tw/UbiSite/). In agreement with our hypothesis, two Lys residues, Lys-17 (K17) and Lys-57 (K57), show high probability for ubiquitination. To investigate whether the UPS is responsible for SIB1 degradation, we supplemented 100 µm MG132, a potent and cell-permeable 26S proteasome inhibitor, to 5-d-old pSIB1::SIB1-GFP transgenic seedlings pretreated with 1 mm SA for 4 h. MG132 treatment led to a notable intensification of the immunoblot signal from the largest SIB1 form as compared with the nontreated samples (Fig. 6A). The accumulation of the uppermost band upon MG132 treatment indicates UPS-mediated SIB1 degradation. However, we failed to detect any polyubiquitinated forms of SIB1 via MS analysis, presumably because of insufficient amounts of SIB1-GFP proteins. We, therefore, enriched SIB1-GFP proteins using IP and visualized them with a longer exposure duration of the immunoblot using anti-GFP and anti-ubiquitin antibodies. Enrichment and longer exposure showed the presence of smears in input and IP-enriched samples, indicating the polyubiquitination of SIB1-GFP fusion proteins (Fig. 6B). Additionally, the MS analysis of IP-enriched proteins also revealed a potential interactome of the SIB1 protein in the nucleus that includes several UPS-related proteins (Supplemental Table S2). These observations support the hypothesis that SIB1 undergoes UPS-dependent degradation. Also, the presence of an uppermost band of SIB1 as early as its induction suggests that Nt-acetylated SIB1 is rapidly ubiquitinated either directly upon its translocation into the nucleus or after its dissociation from the TFs (Fig. 7). The two respective modifications seem to fine-tune the SIB1 turnover, where NTA ensures stability and proper function while ubiquitination marks the protein for its gradual degradation. Our results reveal a novel mechanism of how the stability of stress-related proteins, especially related to immunity, are tightly regulated in synchrony via NTA and the UPS.
Figure 6.
The SIB1 protein undergoes UPS-mediated proteolysis. Five-day-old transgenic sib1 plants expressing pSIB1-driven GFP-tagged SIB1 were first treated with 1 mm SA for 4 h. Then, these seedlings were treated with 100 μm MG132 for the indicated times. A, MG132 inhibits SIB1 degradation. Samples were harvested at the indicated time points for immunoblot analysis. SA indicates 4 h of SA treatment (first sample on the left), and the other samples (additional hours of SA treatment as indicated) were prepared in the presence or absence of MG132. B, Exogenous SA promotes the ubiquitinylation of SIB1. SIB1-GFP proteins extracted from 6-h MG132-treated and nontreated samples were first enriched with an IP assay. The SIB1-GFP and ubiquitinylated SIB1-GFP proteins were identified by immunoblot analysis using anti-GFP (αGFP) and anti-ubiquitin (αUb) antibody, respectively.
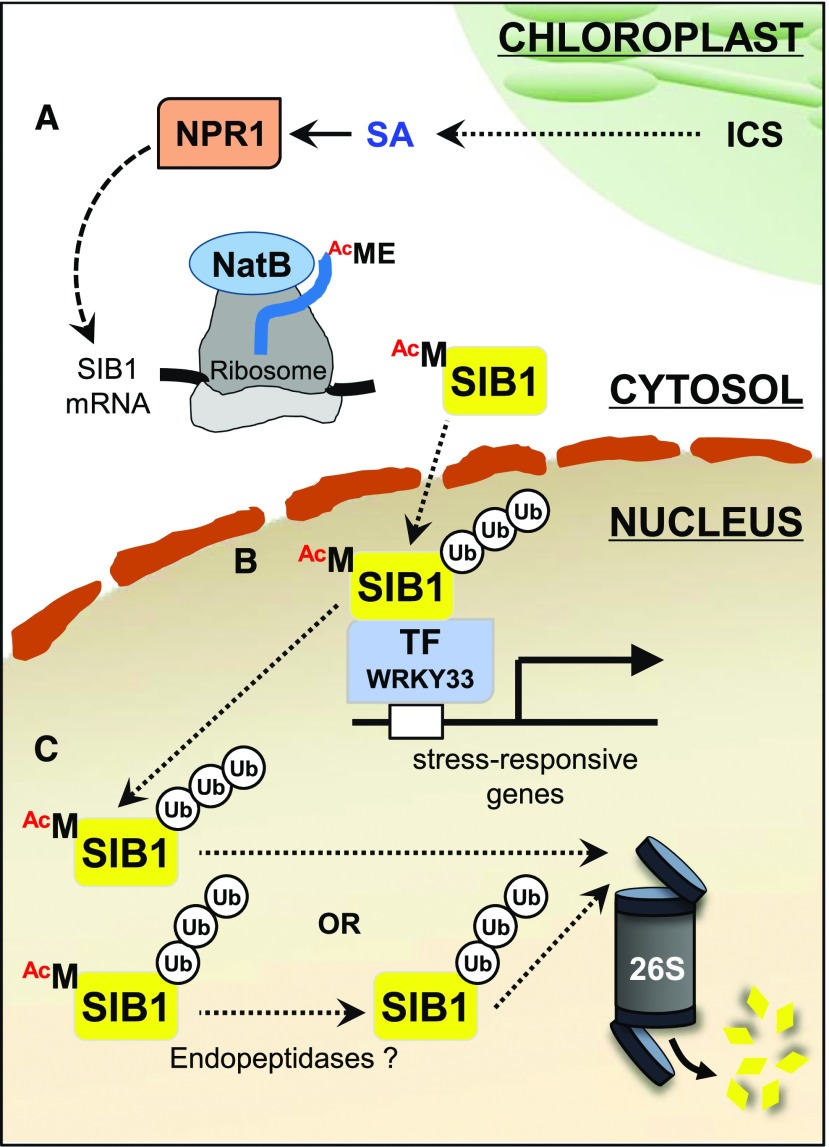
Figure 7.
The biological significance of NTA toward SIB1 function. A, Upon the increase in cellular SA content, mostly synthesized via the chloroplast-established isochorismate (ICS) pathway, NPR1 induces the expression of SIB1. The cotranslational NTA modification via NatB renders SIB1 proteins more stable. B, The nucleus-localized Nt-acetylated SIB1 interacts with stress-related TFs to modulate the expression of their target genes. C, UPS-mediated SIB1 degradation through the 26S proteasome controls the steady-state level of SIB1. UB, Ubiquitin.
DISCUSSION
NTA is a widespread cotranslational protein modification contributing to a broad range of cellular processes, including development, photosynthesis, stress responses, and immune regulation (Pesaresi et al., 2003; Ferrández-Ayela et al., 2013; Linster et al., 2015; Xu et al., 2015; Huber et al., 2020). The interpretation of NTA-primed functionality of different proteins has just started to emerge in plants. A recent report sheds light on the importance of NTA in stabilizing the immune receptor protein SNC1 (Xu et al., 2015). Likewise, in our study, we found that NTA stabilizes the stress/immune-related protein SIB1, enabling its prompt function and the related stress response (Fig. 7, A and B). Based on the substrate specificity of NATs illustrated in yeast and mammals, NatB is supposed to be the NAT catalyzing the NTA of SIB1. However, the MS analysis detected both nonacetylated and acetylated Nt peptides of SIB1 in the naa20 mutant (Fig. 2, B and C; Supplemental Table S1). Although this result indicates that NatB is involved in the NTA of SIB1, it also suggests that functional redundancy may exist among the Arabidopsis NATs, which partially compensates the loss of NatB function. This assumption is in line with the fact that the Arabidopsis genome comprises 25 putative NATs (Dinh et al., 2015). Characterization of the putative NATs in Arabidopsis would certainly provide insights toward this notion (redundancy versus specificity).
In the canonical Ac/N-end rule pathway, the specific Nt-acetylated proteins are recognized by Ac/N-recognin (normally an E3 ligase), which targets these proteins for UPS-mediated proteolysis (Hwang et al., 2010; Shemorry et al., 2013; Nguyen et al., 2018). However, the plant Ac/N-end rule pathway is poorly understood to date. Through the substitution of the penultimate amino acid (Figs. 3 and 4; Supplemental Figs. S6–S8), we noticed that, in SIB1, NTA is able to act as either a stabilizing or a destabilizing signal to regulate protein half-life in plants. Modified SIB1 variants in which the iMet is followed by small residues (A) or bulky hydrophobic residues (W) follow the canonical N-end rule pathway and undergo protein degradation. Besides, the NTA-free SIB1MP variant appeared to be very unstable. On the contrary, if the iMet comes with acidic residues (D and E), as in native SIB1 and the iMD variant of SNC1, the NTA of iMet instead stabilizes these proteins. Thus, together with the previous findings on SNC1, our findings suggest that the impact of NTA in plants might be more complicated than the widely accepted roles of NTA in yeast and mammals (Hwang et al., 2010; Shemorry et al., 2013; Xu et al., 2015).
Furthermore, considering the irreversibility of NTA on cellular proteins (Aksnes et al., 2016; Ree et al., 2018), the NTA probably accompanies the majority of cellular proteins throughout their lifespan. Despite this, most of the Nt-acetylated proteins remain stable and long-lived, which also suggests a potential conditionality of the Ac/N-end rule pathway. Apparently, some evidence in yeast and mammals demonstrates that the steric shielding or sequestration of NTA through a rapid intramolecular protein folding, protein-protein interaction, or subcellular compartmentation protects the Nt-acetylated proteins from the Ac/N-end rule pathway-mediated degradation (Hwang et al., 2010; Varshavsky, 2011; Shemorry et al., 2013; Kim et al., 2014; Park et al., 2015). Using the PredictProtein server (https://www.predictprotein.org/), we analyzed the structure annotation of SIB1, and the resulting prediction shows that the N terminus of SIB1 is exposed, hydrophilic, and disordered without any potential protein-protein interaction domains, indicating that it might be difficult for SIB1 to shield its N terminus via intramolecular protein folding or protein-protein interaction. Taken together, our research delivers a new possibility between the NTA and the protein stability in plants: the NAT-catalyzed Nt-acetylated iMet followed by acidic residues serves as a stabilizing signal rather than an Ac/N-degron to protect the target protein from degradation.
In eukaryotes, the UPS largely contributes to cellular protein homeostasis. Proteins targeted to UPS-mediated degradation undergo ubiquitination in response to internal and external cues (Vierstra, 2009; Gibbs et al., 2014; Dissmeyer et al., 2018). Generally, the proteins targeted by the Ac/N-end rule pathway undergo UPS-mediated degradation. The endoplasmic reticulum-inserted DEGRADATION OF ALPHA2 10 (yeast) and MEMBRANE ASSOCATED RING-CH-TYPE FINGER6 (also called TEB4; mammals) E3 ubiquitin ligases (Ac/N-recognins) specifically recognize the Ac/N-degrons and enable polyubiquitylation and the subsequent degradation via the 26S proteasome (Hwang et al., 2010; Shemorry et al., 2013; Nguyen et al., 2018). In this study, we found that the UPS counteracts the NAT-mediated SIB1 stabilization (Fig. 6), suggesting that NTA and the UPS may finely coordinate the steady-state level of SIB1. Perhaps, once stress responses reach certain physiological levels, the UPS facilitates SIB1 degradation upon its disassociation from the TFs. Alternatively, the removal of acetylated iMet via an endopeptidase may stimulate SIB1 degradation via the UPS (Fig. 7C). These intertwined regulations may play an essential role in sustaining cellular homeostasis under fluctuating environmental conditions. In this regard, and since both SIB1 and SNC1 are involved in plant stress response, it would be interesting to extend such study (NTA and the UPS) to a larger group of immune/stress-related proteins that only appear to emerge upon stress treatment and then undergo degradation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutant seeds of lsd1 (SALK_042687), sib1 (SM_3.30596), and naa20 (SAIL_323_B05) were obtained from the Nottingham Arabidopsis Stock Centre. Arabidopsis seeds were sterilized by soaking in 1.6% (v/v) hypochlorite solution for 3 min and washed five times with sterile water. Seeds were then plated on Murashige and Skoog medium (Duchefa Biochemie) containing 0.7% (w/v) agar (Duchefa Biochemie). After 4 d of stratification at 4°C in darkness, plates were placed in a growth chamber (CU-41L4; Percival Scientific) programmed for continuous light conditions (100 μmol m−2 s−1 at 22°C ± 2°C).
Chemical Treatments
For SA treatment, 5-d-old seedlings grown under continuous light were transferred to Murashige and Skoog medium containing 1 mm SA (Sigma-Aldrich). For MG132 treatment, the seedlings were immersed in water containing 100 μm MG132 (MedChemExpress) to block the 26S proteasome-mediated protein degradation, and the set of seedlings treated with water (0.1% [v/v] dimethyl sulfoxide) was used as a negative control.
Plasmid Construction and Generation of Transgenic Plants
The stop codon-less genomic SIB1 DNA covering the 1.8-kb promoter region and the coding sequences of SIB1 and Naa20 were amplified by PCR (primers are listed in Supplemental Table S3) and cloned into a pDONR221 Gateway vector (Thermo Scientific) through the Gateway BP reaction (Thermo Scientific). Subsequently, the cloned genes were inserted into the Gateway-compatible plant binary vector pGWB504 (no promoter, C-sGFP; –R1-CmR-ccdB-R2-sGFP–), pGWB505 (35S, C-sGFP; –R1-CmR-ccdB-R2-sGFP–), and pGWB620 (35S, C-10xMyc; –R1-CmR-ccdB-R2-10xMyc–) through the Gateway LR reaction (Thermo Scientific; Nakagawa et al., 2007). The same cloning process was applied for constructing the different versions of SIB1 (SIB1MA, SIB1MW, and SIB1MP variants) through PCR-based substitution of the coding sequence corresponding to the second amino acid residue. Each vector was transformed by electroporation into Agrobacterium tumefaciens strain GV3101. Transgenic plants in the lsd1 sib1 background were generated using A. tumefaciens-mediated transformation via the floral dip method (Clough and Bent, 1998), and homozygous transgenic plants were selected on Murashige and Skoog medium containing 25 mg L−1 hygromycin (Merck Millipore).
Protein Extraction and Immunoblot Analysis
Plant tissue was ground in liquid nitrogen to a fine powder. The protein extraction buffer (50 mm Tris-HCl [pH 7.5], 150 mm NaCl, 5 mm EDTA, 10% [v/v] glycerol, 1% [v/v] Nonidet P-40, 1% [w/v] deoxycholate, 0.1% [w/v] SDS, 100 μm MG132, and 1× Complete protease inhibitor cocktail [Roche]) was added at a 1:3 ratio (tissue:buffer) to the powder and mixed well. The supernatant was harvested after two centrifugations at 13,000 rpm for 20 min in a table-top centrifuge. Protein content in all the samples was then quantified using the Pierce BCA protein assay kit (Thermo Scientific). The resulting proteins were resuspended in 4× SDS sample buffer and denatured for 10 min at 95°C. Equal amounts of proteins were separated by 10% SDS-PAGE gels and blotted onto Immun-Blot polyvinylidene difluoride membrane (Bio-Rad). The SIB1-GFP fusion protein was immunochemically detected using a mouse anti-GFP monoclonal antibody (1:5,000; Roche, catalog no. 11814460001). PR1 and UGPase proteins were detected using rabbit anti-PR1 (1:5,000; Agrisera, catalog no. AS10 687) and rabbit anti-UGPase (1:10,000; Agrisera, catalog no. AS05 086) antibodies, respectively.
Nuclear/Cytoplasm Subcellular Fractionation Assay
The subcellular fractionation was conducted according to the method described before (Xu and Copeland, 2012) with some modifications. Two-week-old transgenic plants overexpressing SIB1-GFP were used to extract nuclei and cytoplasm fractions. Around 1 g of leaf tissue was ground to a fine powder in liquid nitrogen, which was further homogenized using 2 mL of cold lysis buffer (20 mm Tris-HCl [pH 7.5], 25% [v/v] glycerol, 20 mm KCl, 2 mm EDTA, 2.5 mm MgCl2, 250 mm Suc, and 1× Complete protease inhibitor cocktail). The mixture was sequentially filtered through a 100-μm and a 40-μm nylon mesh. Then nuclei were pelleted at 1,500g at 4°C for 10 min. The supernatant was collected as the cytosolic fraction after one more centrifugation step at 10,000g at 4°C for 15 min. The pellet containing nuclei was washed three times with 3 mL of NRBT buffer (20 mm Tris-HCl [pH 7.5], 25% [v/v] glycerol, 2.5 mm MgCl2, 0.2% [v/v] Triton X-100, and 1× Complete protease inhibitor cocktail) and then three times with NRB buffer (NRBT buffer without Triton X-100) at 1,500g at 4°C for 10 min. The cytosolic fraction and nuclei pellet were then resuspended in 2× SDS loading buffer and boiled at 95°C for immunoblot assay.
IP Coupled with MS Analysis
Total proteins were isolated using the protein extraction buffer described above. The protein extract (25 mg of the total protein extract) was incubated with 25 µL of GFP-Trap magnetic agarose beads (GFP-TrapMA, Chromotek) for 2.5 h at 4°C with constant rotation. The beads were then washed five times with the washing buffer (10 mm Tris-HCl [pH 7.5], 150 mm NaCl, 0.5 mm EDTA, and 1× Complete protease inhibitor cocktail). After the last wash, 5 µL of the beads was eluted in 100 μL of 2× SDS protein sample buffer by incubating at 95°C for 10 min. The eluates were separated by 10% SDS-PAGE, and SIB1-GFP was detected by immunoblot analyses using a mouse anti-GFP monoclonal antibody (1:5,000; Roche, catalog no. 11814460001). Ubiquitylation was identified using a mouse anti-ubiquitin monoclonal antibody (1:5,000; Abcam, catalog no. ab7254).
The remaining 20 µL of beads was used for the MS analysis as described previously (Dogra et al., 2019). The mass spectra were submitted to the Mascot Server (version 2.5.1, Matrix Science) for peptide identification and scanned against the Arabidopsis protein sequences downloaded from the TAIR Web site (http://www.arabidopsis.org/). Peptide mass tolerance was 20 ppm, fragment mass tolerance was 0.02 D, and a maximum of two missed cleavages was allowed. Carbamidomethylation of Cys was set as a fixed modification, while Nt acetylation, oxidation of Met, and phosphorylation of Ser, Trp, and Tyr were defined as variable modifications. The significance threshold for search results was set at P < 0.05 and ions score cutoff of 15.
Transient Expression in Protoplasts
The transient expression was performed as described before (Yoo et al., 2007). The SIB1-GFP coding sequence was amplified by PCR and inserted into a transient expression vector (pSAT6 plasmid) using the ClonExpress II One Step Cloning Kit (Vazyme) based on the manufacturer’s recommendations. The sequences of all primers used in the transient expression assay are listed in Supplemental Table S3. For transient expression assay, Arabidopsis mesophyll protoplasts from the wild type and the naa20 mutant (6 × 105) were transfected with 200 μg of plasmid and incubated overnight. Protoplast samples were used to perform the Co-IP coupled with MS analysis.
VIGS
VIGS was performed as described before (Bachan and Dinesh-Kumar, 2012). Briefly, 300-bp fragments of Nicotiana benthamiana Naa20 were amplified using PCR and cloned into the VIGS vector pTRV2 through restriction digestion and ligation steps. The construct (pTRV2::NbNaa20) was then transformed into A. tumefaciens GV3101. The A. tumefaciens strain containing pTRV2::NbNaa20 was mixed with the A. tumefaciens strain containing the VIGS vector pTRV1 at a 1:1 ratio and coinfiltrated into 2-week-old N. benthamiana leaves. After 3 weeks, the expression of NbNaa20 was examined using RT-qPCR. Once the silencing of the target gene was confirmed, SIB1-GFP was transiently expressed, and the leaves were harvested at the indicated time points. Leaves coinfiltrated with the pTRV2 empty vector and VIGS vector pTRV1 were used as a negative control. The primers for pTRV2 constructs and RT-qPCR are listed in Supplemental Table S3.
Cell Death Measurement
Cell death was measured using TB staining and electrolyte leakage methods as described previously (Lv et al., 2019). For TB staining, plant leaves were immersed into TB staining solution and boiled for 2 min. After 12 h of incubation at room temperature with gentle horizontal agitation, the nonspecific staining was removed with the TB destaining solution. The destained plant tissues were then kept in 50% (v/v) glycerol and documented. For the determination of electrolyte leakage, first or second leaves of plants were harvested at the indicated time points and immersed into 5 mL of deionized water. After 12 h of incubation at room temperature, the conductivity of the solution was measured with an Orion Star A212 conductivity meter (Thermo Scientific). For each measurement, six leaves per genotype were used, and the experiment was repeated three times.
Measurement of Fv/Fm
The Fv/Fm was measured with the FluorCam system (FC800-C/1010GFP, Photon Systems Instruments) containing a CCD camera and an irradiation system according to the instrument manufacturer’s instructions.
Subcellular Localization and Confocal Laser-Scanning Microscopy
The subcellular localizations of SIB1, SIB1MA, and SIB1MW proteins were visualized by transient overexpression in N. benthamiana as well as by SA-induced expression in stable transgenic Arabidopsis lines. For transient overexpression, native and modified GFP-tagged SIB1 were expressed in N. benthamiana leaves. The GFP and 4′,6-diamidino-2-phenylindole fluorescence signals were detected by confocal laser-scanning microscopy using a Leica TCS SP8 (Leica Microsystems) 30 h after infiltration. For detecting expression in transgenic lsd1 sib1 plants expressing intact and modified SIB1 under the control of the native SIB1 promoter, 5-d-old seedlings treated with 1 mm SA and 16-d-old plants prior to the onset of cell death were used for detecting fluorescence signals. All the images were acquired and processed using Leica LAS AF Lite software version 2.6.3.
RNA Extraction and RT-qPCR
Total RNA was extracted with the Universal Plant Total RNA Extraction Kit (Spin-column; BioTeke) and spectrophotometrically quantified at 260 nm with the NanoDrop 2000 (Thermo Fisher Scientific). Total RNA (1 μg) was reverse transcribed using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper; Vazyme) based on the manufacturer’s recommendations. The qPCR was carried out with ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). Relative transcript level was calculated by the ddCt method and normalized to the ACTIN2 (At3g18780) transcript level. The primer sequences used in this study are listed in Supplemental Table S3.
BiFC Assay
BiFC assays were carried out using a split-YFP system in N. benthamiana leaves as described previously (Lv et al., 2019). In brief, the pDONR/Zeo entry vectors carrying the coding sequences of SIB1, SIB1MA, SIB1MW, lsd1, and WRKY33 were recombined into the split-YFP vectors (pGTQL1221 or pGTQL1211) through the Gateway LR reaction (Thermo Scientific). Then, A. tumefaciens cultures containing the appropriate BiFC constructs were mixed and infiltrated into 4-week-old N. benthamiana leaves. After 24 h of infiltration, the absence or presence of the YFP signal was monitored using a confocal microscope (Leica TCS SP8, Leica Microsystems).
Co-IP Assay to Detect Protein-Protein Interaction
All the constructs we used in this assay have already been described above or before (Lv et al., 2019). The different combinations of selected vectors were coexpressed in 4-week-old N. benthamiana leaves by A. tumefaciens-mediated infection. Protein extraction and Co-IP methods were described above. The eluates were separated by 10% SDS-PAGE, and the interaction between coexpressed proteins was determined by immunoblot analyses using a mouse anti-GFP monoclonal antibody (1:5,000; Roche, catalog no. 11814460001) and a mouse anti-Myc monoclonal antibody (1:10,000; Cell Signaling Technology, catalog no. 2276).
Statistical Analyses
Statistical analyses for the significance of the differences among the mean values of different genotypes were tested by Student’s t test with unequal variances (P < 0.05) using Microsoft Excel for Office 365 MSO while comparing up to two samples and by one-way ANOVA with posthoc Tukey’s HSD test (P < 0.05) using an online tool (https://astatsa.com/OneWay_Anova_with_TukeyHSD/) when more than two samples were compared. Samples included in the analysis were arranged based on at least three biological replicates, except in MS analysis, where at least two independent biological replicates were tested.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org) under the following accession numbers: SIB1 (AT3G56710), LSD1 (AT4G20380), Naa20 (AT1G03150), Naa25 (AT5G58450), WRKY70 (AT3G56400), WRKY33 (AT2G38470), PAD3 (AT3G26830), CRK11 (AT4G23190), CRK24 (AT4G23320), CYP71A12 (AT2G30750), and CYP71A13 (AT2G30770).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SA-induced SIB1 presents multiple bands in immunoblot assay.
Supplemental Figure S2. The nuclear and cytoplasmic forms of SIB1.
Supplemental Figure S3. MS data of SIB1 NTA.
Supplemental Figure S4. SAIL_323_B05 is a knockout mutant of Arabidopsis Naa20 (At1g03150).
Supplemental Figure S5. The impact of SA on Naa20 and SIB1 proteins.
Supplemental Figure S6. The protein abundance of native and modified SIB1-GFP proteins detected by MS analysis.
Supplemental Figure S7. The mass spectra of Nt-acetylated peptides from intact and modified SIB1-GFP proteins.
Supplemental Figure S8. The distinct fates of intact and modified SIB1 proteins.
Supplemental Figure S9. The GFP-tagged intact and modified SIB1 are induced before the onset of cell death in the lsd1 mutant.
Supplemental Table S1. MS data of SIB1 NTA detected in wild-type and naa20 protoplast.
Supplemental Table S2. List of the proteins associated with SIB1.
Supplemental Table S3. List of primer sets used in this study.
Acknowledgments
We thank the Core Facility of Proteomics, Shanghai Center for Plant Stress Biology, for carrying out the MS analysis.
Footnotes
This work was supported by the Strategic Priority Research Program from the Chinese Academy of Sciences (grant no. XDB27040102), the 100-Talent Program of the Chinese Academy of Sciences, and the National Natural Science Foundation of China (grant no. 31871397) to C.K. and by the Research Fund for International Young Scientists Program of the National Natural Science Foundation of China (grant no. 31850410478) and the President’s International Fellowship Initiative postdoctoral fellowship from the Chinese Academy of Sciences (grant no. 2019PB0066) to V.D.
References
- Aksnes H, Drazic A, Marie M, Arnesen T(2016) First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci 41: 746–760 [DOI] [PubMed] [Google Scholar]
- Aksnes H, Hole K, Arnesen T(2015) Molecular, cellular, and physiological significance of N-terminal acetylation. Int Rev Cell Mol Biol 316: 267–305 [DOI] [PubMed] [Google Scholar]
- Aksnes H, Ree R, Arnesen T(2019) Co-translational, post-translational, and non-catalytic roles of N-terminal acetyltransferases. Mol Cell 73: 1097–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudo N, Fernández IS, McLaughlin SH, Peak-Chew SY, Rhodes D, Martino F(2013) The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat Struct Mol Biol 20: 1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, et al. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA 106: 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachan S, Dinesh-Kumar SP(2012) Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol Biol 894: 83–92 [DOI] [PubMed] [Google Scholar]
- Bartels T, Choi JG, Selkoe DJ(2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477: 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Barr FA, Flanagan JJ, Barlowe C, Munro S(2007) The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J Cell Biol 176: 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JRC, Munro S(2004) Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol 6: 405–413 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Roccaro M, Somssich IE(2017) Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29: 20–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA(2013) N-terminal protein processing: A comparative proteogenomic analysis. Mol Cell Proteomics 12: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP(2010) The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci 123: 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TV, Bienvenut WV, Linster E, Feldman-Salit A, Jung VA, Meinnel T, Hell R, Giglione C, Wirtz M(2015) Molecular identification and functional characterization of the first Nα-acetyltransferase in plastids by global acetylome profiling. Proteomics 15: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Rivas S, Graciet E(2018) Life and death of proteins after protease cleavage: Protein degradation by the N-end rule pathway. New Phytol 218: 929–935 [DOI] [PubMed] [Google Scholar]
- Dogra V, Li M, Singh S, Li M, Kim C(2019) Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat Commun 10: 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, Boczkowska M, Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et al. (2018) NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA 115: 4399–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrández-Ayela A, Micol-Ponce R, Sánchez-García AB, Alonso-Peral MM, Micol JL, Ponce MR(2013) Mutation of an Arabidopsis NatB N-alpha-terminal acetylation complex component causes pleiotropic developmental defects. PLoS ONE 8: e80697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte GM, Pool MR, Stirling CJ(2011) N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol 9: e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ(2014) The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol 24: 603–611 [DOI] [PubMed] [Google Scholar]
- Giglione C, Fieulaine S, Meinnel T(2015) N-terminal protein modifications: Bringing back into play the ribosome. Biochimie 114: 134–146 [DOI] [PubMed] [Google Scholar]
- Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR(2014) Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun 5: 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Miao M, Kud J, Niu X, Ouyang B, Zhang J, Ye Z, Kuhl JC, Liu Y, Xiao F(2013) SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytol 197: 1214–1224 [DOI] [PubMed] [Google Scholar]
- Huber M, Bienvenut WV, Linster E, Stephan I, Armbruster L, Sticht C, Layer DC, Lapouge K, Meinnel T, Sinning I, et al. (2020) NatB-mediated N-terminal acetylation affects growth and biotic stress responses. Plant Physiol 182: 792–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A(2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yu D(2016) The WRKY57 transcription factor affects the expression of jasmonate ZIM-domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiol 171: 2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall RL, Bradshaw RA(1992) Isolation and characterization of the methionine aminopeptidase from porcine liver responsible for the co-translational processing of proteins. J Biol Chem 267: 20667–20673 [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K(2012) Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24: 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Kim RR, Oh JH, Cho H, Varshavsky A, Hwang CS(2014) The N-terminal methionine of cellular proteins as a degradation signal. Cell 156: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z(2011) Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23: 3824–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster E, Stephan I, Bienvenut WV, Maple-Grødem J, Myklebust LM, Huber M, Reichelt M, Sticht C, Møller SG, Meinnel T, et al. (2015) Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat Commun 6: 7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv R, Li Z, Li M, Dogra V, Lv S, Liu R, Lee KP, Kim C(2019) Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31: 210–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S(2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW, Bennett EJ, Schulman BA(2013) Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure 21: 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa K, Shiina T, Murakami S, Toyoshima Y(2002) Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett 514: 300–304 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R(1989) Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J 8: 2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myklebust LM, Van Damme P, Støve SI, Dörfel MJ, Abboud A, Kalvik TV, Grauffel C, Jonckheere V, Wu Y, Swensen J, et al. (2015) Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum Mol Genet 24: 1956–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Mun SH, Lee CS, Hwang CS(2018) Control of protein degradation by N-terminal acetylation and the N-end rule pathway. Exp Mol Med 50: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SE, Kim JM, Seok OH, Cho H, Wadas B, Kim SY, Varshavsky A, Hwang CS(2015) Control of mammalian G protein signaling by N-terminal acetylation and the N-end rule pathway. Science 347: 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Gardner NA, Masiero S, Dietzmann A, Eichacker L, Wickner R, Salamini F, Leister D(2003) Cytoplasmic N-terminal protein acetylation is required for efficient photosynthesis in Arabidopsis. Plant Cell 15: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Norbeck J, Takakura H, Blomberg A, Sherman F(1999) Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J 18: 6155–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F(2001) NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J Biol Chem 276: 20154–20159 [DOI] [PubMed] [Google Scholar]
- Ree R, Varland S, Arnesen T(2018) Spotlight on protein N-terminal acetylation. Exp Mol Med 50: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA(2011) N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SRG, Strochlic TI, Tong AHY, Boone C, Burd CG(2004) Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Sheikh TI, de Paz AM, Akhtar S, Ausió J, Vincent JB(2017) MeCP2_E1 N-terminal modifications affect its degradation rate and are disrupted by the Ala2Val Rett mutation. Hum Mol Genet 26: 4132–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemorry A, Hwang CS, Varshavsky A(2013) Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell 50: 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JM, Shaw JM(2003) Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc Natl Acad Sci USA 100: 7644–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starheim KK, Gromyko D, Evjenth R, Ryningen A, Varhaug JE, Lillehaug JR, Arnesen T(2009) Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol 29: 3569–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Sriram SM, Park KS, Kwon YT(2012) The N-end rule pathway. Annu Rev Biochem 81: 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trexler AJ, Rhoades E(2012) N-Terminal acetylation is critical for forming α-helical oligomer of α-synuclein. Protein Sci 21: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, De Juan-Pardo E, Demeyer K, Hole K, Larrea E, et al. (2012) N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci USA 109: 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A.(2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20: 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD.(2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Xiao Q, Zhang F, Nacev BA, Liu JO, Pei D(2010) Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49: 5588–5599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YD, Li W, Guo D, Dong J, Zhang Q, Fu Y, Ren D, Peng M, Xia Y(2010) The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant Cell Environ 33: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Copeland C(2012) Nuclear extraction from Arabidopsis thaliana. Bio Protoc 2: e306 [Google Scholar]
- Xu F, Huang Y, Li L, Gannon P, Linster E, Huber M, Kapos P, Bienvenut W, Polevoda B, Meinnel T, et al. (2015) Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 27: 1547–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Fang Q, Wang M, Ren R, Wang H, He M, Sun Y, Yang N, Xu RM(2013) Nα-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat Struct Mol Biol 20: 1116–1118 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X(2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]