DNA replication initiation sites in plants associate most strongly with AT-rich and highly accessible chromatin, and not with genes or a particular epigenetic signature.
Abstract
The selection and firing of DNA replication origins play key roles in ensuring that eukaryotes accurately replicate their genomes. This process is not well documented in plants due in large measure to difficulties in working with plant systems. We developed a new functional assay to label and map very early replicating loci that must, by definition, include at least a subset of replication origins. Arabidopsis (Arabidopsis thaliana) cells were briefly labeled with 5-ethynyl-2′-deoxy-uridine, and nuclei were subjected to two-parameter flow sorting. We identified more than 5500 loci as initiation regions (IRs), the first regions to replicate in very early S phase. These were classified as strong or weak IRs based on the strength of their replication signals. Strong initiation regions were evenly spaced along chromosomal arms and depleted in centromeres, while weak initiation regions were enriched in centromeric regions. IRs are AT-rich sequences flanked by more GC-rich regions and located predominantly in intergenic regions. Nuclease sensitivity assays indicated that IRs are associated with accessible chromatin. Based on these observations, initiation of plant DNA replication shows some similarity to, but is also distinct from, initiation in other well-studied eukaryotic systems.
DNA replication starts at multiple locations along eukaryotic chromosomes to ensure timely genome duplication, but the exact locations where replication starts are not generally known. In Saccharomyces cerevisiae, replication initiates adjacent to a conserved motif known as the autonomously replicating sequence (Stinchcomb et al., 1979). However, no simple consensus motif associated with origins has been found in metazoans. Origins in fly (Drosophila melanogaster), mouse (Mus msculus), and human (Homo sapiens) are often correlated with DNase I hypersensitive (DHS) regions, GC-rich DNA, G-quadruplex (G4) structures and genes (Cadoret et al., 2008; Hansen et al., 2010; Cayrou et al., 2011; Besnard et al., 2012; Gindin et al., 2014; Valton et al., 2014; Smith et al., 2016). In contrast, much less is known about initiation of DNA replication in plants. The DNA replication machinery and chromatin structure are largely conserved between plants and animals, but there are important differences between the two kingdoms. For example, the spatiotemporal distribution of replicating DNA differs between plant and metazoan cells (Bass et al., 2015). Moreover, plants lack homologs of lamin, CCCTC-binding factor, and geminin, which play key roles in chromatin organization and initiation of DNA replication (Shultz et al., 2007; Liu et al., 2016; Thorpe and Charpentier, 2017). Hence, we cannot assume that DNA replication origins that have been characterized in metazoans are reflective of initiation regions in plants (Savadel and Bass, 2017).
Short nascent strand (SNS) analysis, which characterizes intermediates generated during leading-strand DNA synthesis, is a common protocol for genome-wide identification of DNA replication origins. However, these experiments have proved challenging, even in favorable animal model systems (Gilbert, 2010; Hyrien, 2015; Miotto, 2017). The technique depends critically on the quality of the lambda exonuclease digestion (Foulk et al., 2015), and it is difficult to map small amounts of nascent DNA without contamination from broken DNA and Okazaki fragments. The difficulty in obtaining reproducible results is illustrated by reports for a number of human cell lines that show little overlap between studies and sometimes between biological replicates within a study (for review, see Gilbert, 2010; Hyrien, 2015; Miotto, 2017). In plant systems, shearing forces necessary to break the cell wall and high levels of endogenous nucleases (Obara et al., 2001) increase the difficulty of isolating intact DNA necessary for SNS assays. Because nascent strands are present at very low levels relative to total DNA, even small amounts of contamination with other DNA fragments complicate interpretation.
Previous efforts to map origins in Arabidopsis (Arabidopsis thaliana), using SNS analysis as a validation method or as the primary assay (Costas et al., 2011; Vergara et al., 2017; Sequeira-Mendes et al., 2019), suffer the same low reproducibility common to SNS experiments in other systems. In addition, the first origin study in Arabidopsis subjected cells to Suc starvation and hydroxyurea treatment, both of which can alter origin usage (Taylor, 1977; Ge et al., 2007; Gilbert, 2007). As a consequence, it is not known if the conclusions from these experiments accurately describe DNA replication under normal conditions. To address this concern, we developed a new functional assay by combining brief labeling with 5-ethynyl-2’-deoxy-uridine (EdU; Bass et al., 2015; Ramachandran and Henikoff, 2016; Dvořáčková et al., 2018; Macheret and Halazonetis, 2018; Tubbs et al., 2018; Macheret and Halazonetis, 2019) with flow sorting (Wear et al., 2016; Wear et al., 2017; Concia et al., 2018) to isolate nuclei in which replication has just begun, so that label is incorporated close to the points of initiation. This was followed by immunoprecipitation and characterization of the labeled DNA. Our assay is minimally invasive, involving only the supply of an exogenous nucleoside precursor and constitutes a direct assay for early replicating loci. We call these loci initiation regions (IRs) rather than origins because our approach does not map initiation events at the level of individual nucleotides. However, because IRs are the first regions of the genome to replicate during S phase, they must, by definition, contain at least an early firing subset of replication origins.
RESULTS
Identification of Arabidopsis Initiation Regions
Our approach is based on flow sorting of nuclei from Arabidopsis suspension cells pulse labeled with EdU to obtain a population in which label has been incorporated only during very early S phase. Labeled DNA fragments from these nuclei define regions in which replication begins and include early firing origins. They will also include flanking DNA that may or may not play a role in the initiation of replication. We therefore designated them as initiation regions (IRs) rather than replication origins. An outline of the protocol is shown in Figure 1A. Arabidopsis cells were labeled with EdU for 30 min followed by fixation and nuclei isolation. The nuclei were subjected to Click chemistry to covalently link incorporated EdU to an AF488 fluorophore and stained with 4′,6-diamino-phenylindole (DAPI; Salic and Mitchison, 2008). Nuclei at different stages of the cell cycle were resolved by flow cytometry based on DNA content (DAPI) and EdU incorporation (AF488; Fig. 1B; Supplemental Fig. S1). The G1 and G2 nuclei populations were separated by their DNA content. Nuclei labeled during a 30-min pulse but with DNA content similar to G1 were sorted into two populations, early (E) and very early (VE) S phase. Nuclei with high AF488 fluorescence, which indicates that they incorporated EdU during a large portion of the labeling period, represent E S phase (Fig. 1B, blue box). In contrast, nuclei with a G1 DNA content but low AF488 fluorescence must have entered S-phase near the end of the EdU labeling period and are thus in VE S phase (Fig. 1B, orange box). Hence, labeled DNA in VE nuclei will be much closer to initiation sites, and peaks of VE replication activity are likely to include early firing origins of DNA replication.
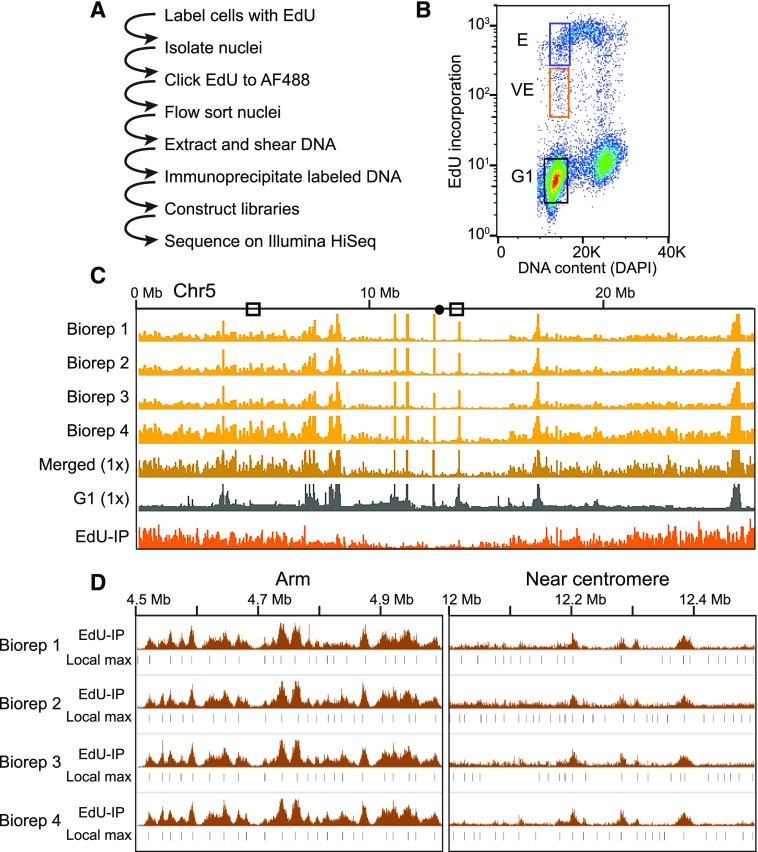
Figure 1.
Experimental strategy and flow sorting. A, Outline of EdU-seq protocol. B, Bivariate flow cytometry profile of nuclei sorted by EdU incorporation (measured by AF488 fluorescence, y axis) and DNA content (measured by DAPI fluorescence, x axis), including sorting gates used to define the G1 (black box), VE (orange box), and E (blue box) populations. C, Browser profiles of VE signal from four independent biological replicates (Biorep) across Chromosome 5 (gold tracks, scale 0–100 read density signal) illustrating the reproducibility between replicates. The merged read data normalized to 1× genome coverage (dark gold), the G1 control (gray), and the final “sequenceability” normalized file (orange) are also shown (scales 0–5 normalized signal ratio). D, Reproducibility of local maxima (Local max) across replicates. The 1× “sequenceability” normalized data, which we designate as EdU-IP (scale = 0–5 normalized signal ratio), and the local maxima are shown for each replicate.
After sorting, DNA from each nuclei population was purified, and EdU-AF488 labeled DNA from E and VE nuclei was immunoprecipitated before sequencing. Biological replicates were highly reproducible (Spearman correlation coefficients: VE = 0.97, E = 0.97, and G1 = 0.96; Fig. 1C, Chromosome 5, four gold tracks; Supplemental Fig. S2A; Supplemental Table S1) and were merged for further analysis. Read counts were adjusted to 1× coverage (Fig. 1C, dark gold track). The merged track was then normalized to the G1 reference (Fig. 1C, gray track) to control for collapsed repeat artifacts and variation in sequenceability (Fig. 1C, bottom orange track). Separately visualizing the EdU-IP signals from the VE and E gates shows broad regions of enrichment in E, but sharper, more discrete peaks in VE, indicating that the VE gate captured nuclei as they entered S phase (Fig. 2, A–C).
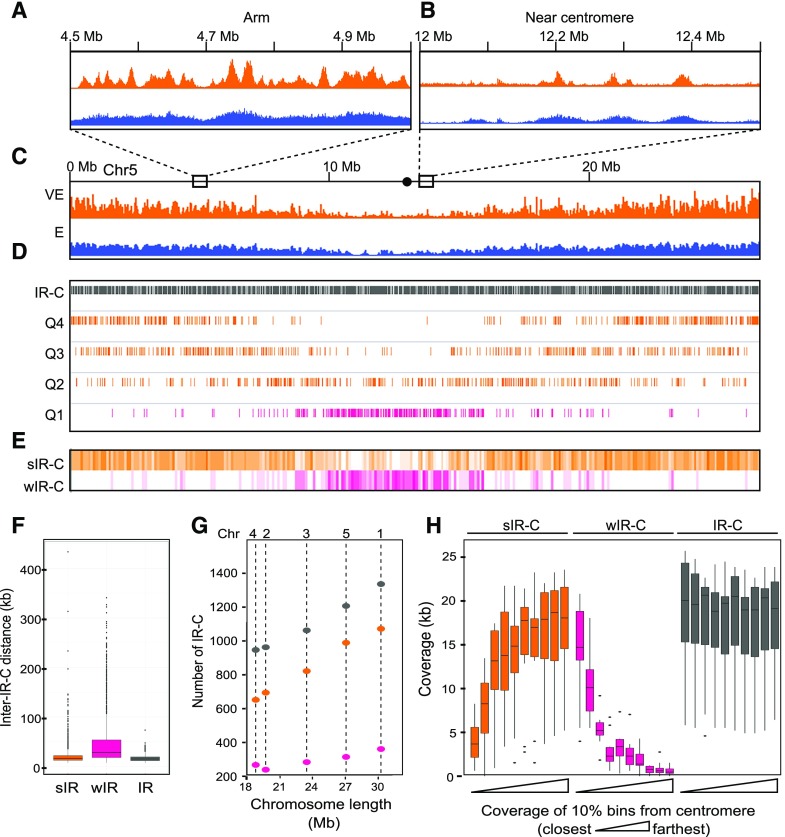
Figure 2.
Genomic distribution of replication signal and the distribution of IR-Cs. A to C, EdU signal from VE (orange) and E ( blue) S phase in 500-kb regions from an arm (A) or the centromere region (B) of Chromosome 5 (scale 0–5 normalized signal ratio; C). The dot in the schematic is the centromere. D, IR-Cs on chromosome 5. Positions of all IRs (gray) and the EdU intensity quartiles are shown. The top three quartiles of normalized EdU signal are orange, and the bottom quartile is pink. E, Coverage heat maps of strong (orange) and weak (pink) IR-Cs. F, The distribution of distances between IR-Cs displayed as a boxplot, the centerline represents the median, the box the interquartile range, the whiskers the range of distances, and the points represent outliers. G, The number of IR-Cs per chromosome (Chr) as a function of chromosome length. H, Coverage of IR-Cs as a function of distance from the centromere for all chromosomes. The data are combined for both chromosome arms and plotted in bins representing 10% of the distance from centromere to telomere. In each case, the leftmost boxplot represents the bin closest to the centromere.
Even though the biological replicates were merged, identifying local maxima on each individually normalized biological replicate further shows the reproducibility of the method. Reads from all four biological replicates were independently adjusted to 1× coverage and normalized for sequenceability relative to the G1 control (Fig. 1D, brown signal tracks) and local maxima identified as 300-bp bins (Fig. 1D, black bars).
Bins representing local maxima in the merged VE profile were designated as initiation region centers (IR-Cs). Because DNA replication likely initiates at or near these peaks of VE replication activity, we used the IR-C bins as focal points for further analysis, while recognizing that actual origins may be located elsewhere within the VE replication peak.
IR-Cs were then divided into quartiles based on the strength of the VE EdU-IP signal (Fig. 2D). The top three quartiles, Q2–Q4, which all showed differential Micrococcal nuclease sensitive (DNS) peaks well above that of random genomic controls (see results in "IRs Are Associated with Open Chromatin" and Supplemental Fig. S3), were combined and designated as strong IR-Cs (sIR-Cs; Fig. 2D, orange tracks). In contrast, IRs from the lowest quartile, Q1, had DNS peaks below the genomic mean (Supplemental Fig. S3) and, thus, were analyzed separately and designated as weak IR-Cs (wIR-Cs; Fig. 2D, pink track).
In addition, Q1 of IRs are predominantly located in the centromere and pericentromere, regions known to be heterochromatic, while Q3 and Q4 IRs are found predominantly in euchromatin. Some Q2 IRs are found in the pericentromeric region, but they are also scattered throughout the chromosome arms (Supplemental Fig. S4), and are characterized by the higher mean DNS sensitivity typical of euchromatin. In this respect, they are more similar to Q3 and Q4 than they are to Q1. We therefore decided that grouping Q2 with Q 3 and Q4 was a sensible way to compare what seems to be two major populations of IRs.
sIR-Cs and wIR-Cs Are Distributed Differently across Arabidopsis Chromosomes
Maps of sIR-C and wIR-C coverage (Fig. 2E; Supplemental Fig. S4A) show that the sIRs are primarily in the arms and the wIRs are mainly in the centromeric regions of the five Arabidopsis chromosomes. When all IR-Cs are analyzed together, they are fairly evenly distributed with a median inter-IR-C distance of 15.3 kb with an interquartile range (IQR) of 12 to 16.3 kb (Fig. 2F, gray box). When sIR-Cs and wIR-Cs are analyzed separately, the median distance between sIR-Cs is 16.5 kb (IQR 12.6 to 19.3 kb; Fig. 2F, orange box), while the median distance between wIR-Cs is 28.5 kb (IQR 18.3 to 53.7 kb; Fig. 2F, pink box). Consistent with the even distribution of sIR-Cs, there is a strong positive relationship between chromosome length and the number of sIR-Cs (r2 = 0.98; Fig. 2G). The number of wIRs is less strongly correlated with chromosome length, as might be expected from their association with (peri)centromeric regions (r2 = 0.83; compare with Chr4 and Chr2 in Fig. 2G).
To further explore the relationship between IR-Cs and the centromere, each of the 10 chromosome arms was divided into 10 bins and the coverage by IR-Cs was calculated for each bin relative to its distance from the centromere. Figure 2H shows that the distribution of IR-C coverage mirrors the pattern seen in the density heat maps, with the wIR-Cs clustered in the first two windows nearest the centromere, while the sIR-Cs are concentrated in the distal arms and less abundant near the centromere.
IRs Are AT-Rich Sequences Flanked by Relatively AT-Poor Regions
The mean AT content of the sIR-Cs and wIR-Cs is 70.0% and 66.8%, respectively, both of which are statistically higher than the overall AT content of the five chromosomes, which ranges between 63.7% and 64.1% (one sample t test and Wilcoxon rank-sum test, P-values < 0.001). An elevated AT content could be a feature of IRs or result from the use of a thymidine analog to label replicating DNA. To address this, the 300-bp IR-Cs and their flanking regions were compared with multiple datasets representing randomly selected bins with comparable AT content (CAT regions) and their flanking sequences. The IR-Cs were also compared with separate datasets derived from bins randomly selected without considering AT content (random genomic regions). Flanking regions provide important context to the IR-Cs and show how regions with similar AT content at the 300-bp scale can exhibit different nucleotide distribution patterns at larger scales.
Figure 3, A and B, show plots of the mean AT content at and around the sIR-Cs and wIR-Cs, respectively, as well as mean plots for 10 random genomic datasets and 10 CAT datasets. Both the wIR-Cs and sIR-Cs are AT rich and flanked at ±6 kb with relatively AT-poor sequences, while CAT regions are not flanked by such regions. Moreover, the CAT regions have much less EdU-IP signal than sIR-Cs of similar AT content (Supplemental Fig. S2, B–E). The differences in EdU-IP signal between sIR-Cs and their CAT datasets strongly suggest that the sIR-Cs do not merely reflect random incorporation of EdU into AT-rich DNA and, instead, represent a subset of AT-rich regions with unique properties that replicate during VE S phase. Additionally, a comparison of Supplemental Figure S2, B and D, shows that the EdU IP signal increases by more than 100% between flanking regions and the sIR-C peak, while the AT content of the same two regions differs by only 11%, again indicating that AT content is not the sole driver of EdU-IP signal in IRs. The pattern of AT-rich DNA surrounded by regions with elevated GC content suggests that AT content at an IR-C site does not by itself define an initiation region and that sequences beyond the 300-bp IR-C are also important.
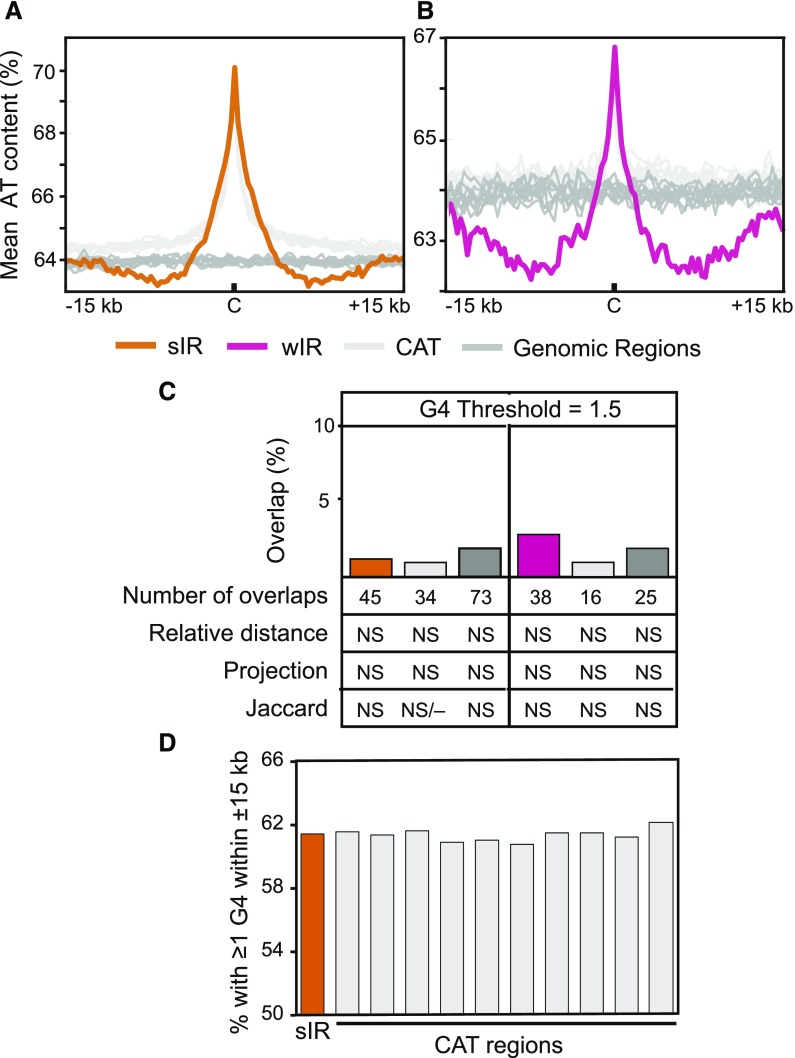
Figure 3.
IR-Cs are AT rich and not associated with G4s. A and B, Mean AT content plot for sIR-Cs (orange) and wIR-Cs (pink; B) compared with 10 random genomic datasets (dark gray), and 10 CAT datasets (light gray) regions. The plots cover ±15 kb flanking the IRs with the “C”, marking the center. The mean was calculated in 300-bp bins. C, Percent of sIRs and wIRs overlapped by at least one potential G4 (generated using G4Hunter; Bedrat et al., 2016) and the number of overlaps using a threshold of 1.5 (at least one base pair overlap was required to be called “overlapped”) along with counts of overlaps and statistical results in the rows below. The statistical results were not significant (NS). Detailed statistics of each G4 threshold dataset for sIR-Cs and wIR-Cs are in Supplemental Tables S2 and S3. The statistical analyses of the random genomic and CAT controls represent the means of 10 datasets for each, with the individual datasets shown in Supplemental Table S5. D, The percent of sIR-Cs and their CAT datasets that have at least one potential G4 within ±15 kb.
Given that G-quadruplex (G4) forming sequences have been associated with origins in animals (Besnard et al., 2012; Valton et al., 2014; Cayrou et al., 2015; Foulk et al., 2015), it was of interest to examine the relatively GC-rich areas flanking the IR-Cs for potential G4 elements. Potential G4s were identified in the Arabidopsis genome sequence with G4Hunter (Bedrat et al., 2016), using the score thresholds of 1, 1.25, 1.5, 1.75, and 2. Results obtained with a threshold of 1.5 are illustrated in Figure 3, C and D, and Supplemental Tables S2 and S3. Supplemental Table S4 shows no significant relationship (using a P-value cutoff of 0.001) between G4s and IR-Cs or their corresponding random genomic or CAT datasets. Results using other thresholds are shown in Supplemental Figure S5. No statistically significant relationship was found between the IR-Cs and G4s at any threshold. Although ∼61% of IR-Cs had a potential G4 within ±15 kb, the same was true of all 10 CAT datasets (Fig. 3D). Taken together, the data do not support an essential role for G4 forming sequences in Arabidopsis initiation regions.
IRs Are Associated with Nongenic Regions and Helitrons
The plots in Figure 4A show the density of different genomic features defined by the Araport 11 annotation (Krishnakumar et al., 2015; Cheng et al., 2017), plotted as a function of distance from the IR-C. The “Not Gene or TE” (NGT) annotation, which was also derived from Araport 11, is defined as any region of the genome not covered by a gene or a transposable element (TE; Huala et al., 2001; Buisine et al., 2008; Lamesch et al., 2012).
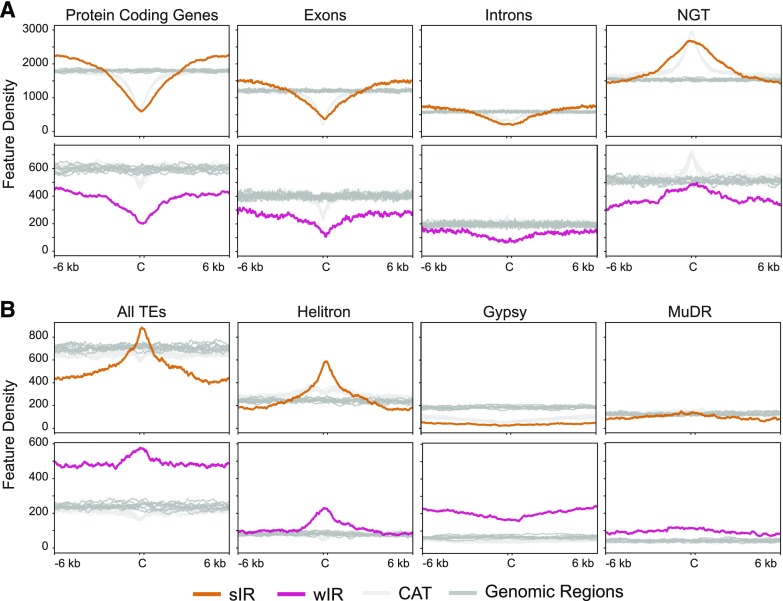
Figure 4.
Density plots of genomic features around sIR-Cs (orange) and wIR-Cs (pink) compared with their random genomic (dark gray) and CAT (light gray) regions. Density plots in ±6-kb areas surrounding the IR-Cs, in 20-bp bins. A, Density plots for genic and NGT annotations derived from Araport 11 (Krishnakumar et al., 2015; Cheng et al., 2017) for sIR-Cs and wIR-Cs. B, Density plots of all TEs and various TE families for sIR-Cs and wIR-Cs.
Density plots of features around IR-Cs and their random genomic and CAT datasets show that sIR-Cs overlap genes less than the random genomic datasets, although the depletion is similar for sIR-Cs and the CAT datasets (Fig. 4A). However, genic density increases and exceeds random genomic levels at approximately ±3 kb around sIR-Cs, while genic density around CAT regions increases but does not exceed random genomic levels. This result suggests that sIR-Cs are more likely than CAT regions to be flanked by genes beyond ±3 kb. In contrast, the NGT density plots show a strong enrichment for intergenic sequences over sIR-Cs and CAT regions, with a broader peak around the sIR-Cs indicating that the immediate genomic environment surrounding sIR-Cs is more likely to lack annotated features within ±3 kb.
The wIR-Cs also show a strong depletion for genic features that remains below the CAT and random genomic levels across the entire 12.3-kb window (Fig. 4A). The NGT profile for wIR-Cs is also below both types of control datasets. This result is consistent with the preferential location of wIR-Cs in the TE-rich and gene-poor pericentromeric and centromeric regions.
Both sIR-Cs and wIR-Cs are associated with a greater density of TEs than either the random genomic or the CAT datasets (Fig. 4B). Total TE density decreases quite rapidly on either side of the sIR-Cs, a decrease which is not seen in the vicinity of CAT or random genomic regions. The Helitron family is positively associated with sIR-Cs (P-value < 0.001, Supplemental Table S2), while the LINE, Copia, and Gypsy families show a negative association with sIR-Cs (P-value < 0.001, Supplemental Table S2). Density plots of selected families confirm these relationships (Fig. 4B). The wIR-Cs also have a positive association with Helitrons, in addition to En-Spm, MuDr, and Gypsy families (P-value < 0.001, Supplemental Table S3).
We also examined the relationships between IR-Cs and various genome annotations using GenometriCorr statistical R package (Favorov et al., 2012; Supplemental Fig. S6; Supplemental Tables S2 and S3). In addition to the features shown in Figure 4, we tested the associations of IR-Cs with 5′ UTRs, 3′ UTRs, transcription start sites, transcription termination sites and the HAT, EnSpm, LINE, and Copia TE families. The annotations were taken or derived from Araport 11. Consistent with the density plots, both projection and Jaccard test results showed that IR-Cs overlap most genic annotations (genes, exons, introns, 5′ UTRs, and 3′UTRs) less than expected (using a P-value cutoff of < 0.001 for both tests; Supplemental Fig. S6; Supplemental Tables S2 and S3). Similarly, the relative distance test indicated that sIR-Cs are farther away from genes, exons, introns, and UTRs, and wIR-Cs are farther away from genes, than would be expected (P-values < 0.001 and negative empirical distribution cumulative function area from the relative distance test; Supplemental Fig. S6; Supplemental Tables S2 and S3). Projection and Jaccard tests showed that both classes of IR-Cs overlap TEs more than expected (P-values < 0.001 for both tests, Supplemental Fig. S6; Supplemental Tables S2 and S3), while the relative distance test showed that the sIR-Cs, but not wIRs, are closer to TEs than would be expected (P-value < 0.001 and negative empirical distribution cumulative function area correlation, Supplemental Fig. S6; Supplemental Tables S2 and S3). All three tests showed that sIR-Cs overlap NGT regions more than would be expected and that both sIR-Cs and wIR-Cs are closer to NGT regions than would be expected. To explore potential asymmetry in the various correlations, the IR-Cs and annotations were interchanged as query and reference for each test (Supplemental Tables S2 and S3), with no change in the overall patterns.
Together, the results show that the sIR-Cs and wIR-Cs are negatively associated with genes and enriched in NGT regions. Somewhat similar results were also seen for the CAT control datasets, showing that they are also negatively associated with genes and exons and positively associated with NGT regions (P-values < 0.001, Supplemental Tables S5 and S6). This result suggests that these associations are characteristic of AT-rich regions, of which IRs are a subset. However, the density plots in Figure 4 demonstrate that the genome environments around sIRs, in particular, are distinct from those surrounding other AT-rich regions.
IR-Cs Are Depleted for Selected Acetylated Histones
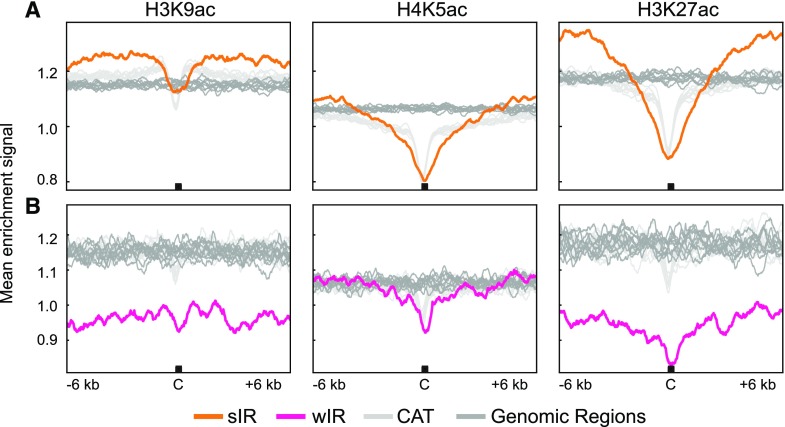
Initiation regions have been associated with acetylated histones in yeast and metazoan systems. We examined the relationships between IRs and H3K9ac, H4K5ac, and H3K27ac because of their association with origins in other systems (Costas et al., 2011; Eaton et al., 2011; Picard et al., 2014). Genome-wide maps of the histone acetylation marks were used to create mean enrichment profiles centered around sIR-Cs (Fig. 5A) and wIR-Cs (Fig. 5B) with their random genomic and CAT datasets. The mean signal for all three acetylated histones at and around random genomic regions was fairly uniform. However, localized depletion was observed at sIR-Cs and their CAT regions. The sIR profiles showed broader valleys of depletion for H4K5ac and H3K27ac relative to those for CAT regions, which could reflect reduced nucleosome occupancy. However, the signals for all 3 acetylated histones are higher in flanking regions of sIR-Cs than they are for CAT or random genomic control regions. This is particularly striking for the H3K27ac signal, which is strongly elevated starting at ±3 kb compared with both control datasets. The relative enrichment of these modified histones in flanking regions differentiates sIRs from both control datasets and suggests that these flanking regions are functionally significant. The low levels of H3K9ac and H3K27ac in wIRs are consistent with their predominant localization in centromeric regions, where acetylated histones are generally depleted (Sequeira-Mendes et al., 2014; Wang et al., 2015).
Figure 5.
Mean analysis of selected acetylated histones at IRs. A, Mean enrichment signal for H3K9ac (left), H4K5ac (center), and H3K27ac (right) at and around sIR-Cs (orange), 10 random genomic datasets (dark gray), and 10 CAT datasets (light gray) regions. B, Mean enrichment signal for each acetylated histone at and around the wIR-Cs (pink), 10 random genomic datasets (dark gray), and 10 CAT datasets (light gray) regions. The plots are ±6-kb windows around the IR-Cs with the C marking the center. The mean was calculated in 20-bp bins.
We further assessed the epigenetic environment of IR-Cs by examining their distribution across genome-wide Arabidopsis chromatin state maps created by Wang et al. and Sequeira-Mendes et al. (2014; Supplemental Fig. S7, B and C, respectively; Supplemental Tables S2 and S3). The coverage by each chromatin state was calculated for the genome as a whole and for the IR-Cs. The sIR-Cs and wIR-Cs are both generally depleted in genic euchromatin in both chromatin state maps (P-values < 0.001, see Supplemental Tables S2 and S3). In contrast, the sIR-Cs are enriched in nongenic, AT-rich chromatin with H3K27 methylation in both maps (CS4 in Supplemental Fig. S7B and CS2, CS4, and CS5 in Supplemental Fig. S7C; P-value < 0.001, Supplemental Table S2). Surprisingly, none of the chromatin states associated with sIR-Cs are enriched for the acetylated H3 marks shown in Figure 5, although H4K5ac was widespread across all chromatin states in the Wang map (Wang et al., 2015). The wIR-Cs are associated with the two heterochromatin states in both maps (P-value < 0.001, Supplemental Table S2), a result that is expected given their localization in centromeric and pericentromeric regions. The sIR-Cs are most strongly associated with the Wang CS4 (Wang et al., 2015) chromatin state (P-value < 0.001, Supplemental Table S2), which is not defined by a unique mark or set of marks, suggesting that histone modifications or patterns thus far cataloged may not, in themselves, play a large role in defining initiation regions.
IRs Are Associated with Open Chromatin
To examine chromatin accessibility of IRs, we first used published DHS maps for Arabidopsis seedlings (Sullivan et al., 2014). DHS sites have been associated with replication origins in other eukaryotes (Urnov et al., 2002; Cadoret et al., 2008; Gindin et al., 2014). Approximately 20% of sIR-Cs and 5% of wIR-Cs overlapped one of the seedling DHS sites. For the sIR-Cs, this is significantly higher than expected (P-values < 0.001, Supplemental Fig. S8; Supplemental Tables S2 and S3). However, the overlap is less than expected if initiation occurred primarily at these sites. Moreover, the CAT datasets also show significant association with DHS sites (P-value < 0.001), suggesting that AT content per se, as well as differences between seedlings and cultured cells, could be confounding this analysis (Supplemental Table S7).
Micrococcal nuclease (MNase) digestion permits a holistic, quantitative analysis of chromatin accessibility on a genome-wide basis (Vera et al., 2014; Pass et al., 2017), and references cited therein). To apply this analysis to Arabidopsis suspension cells, unsorted nuclei (which are primarily in G1 phase) were subjected to heavy, moderate, or light MNase digestion, and protected DNA fragments were isolated and sequenced. DNA protected during heavy digestion is considered to be occupied by stable and persistent nucleosomes, while DNA protected only during light digestion is associated with proteins that are bound transiently or with lower affinity. Additionally, size fractionation can differentiate between nucleosome-sized protected fragments (140–200 bp) and smaller fragments protected by other proteins. Work in other systems has indicated that these smaller fragments often represent binding sites for transcription factors or prereplication complexes (Vera et al., 2014; Belsky et al., 2015).
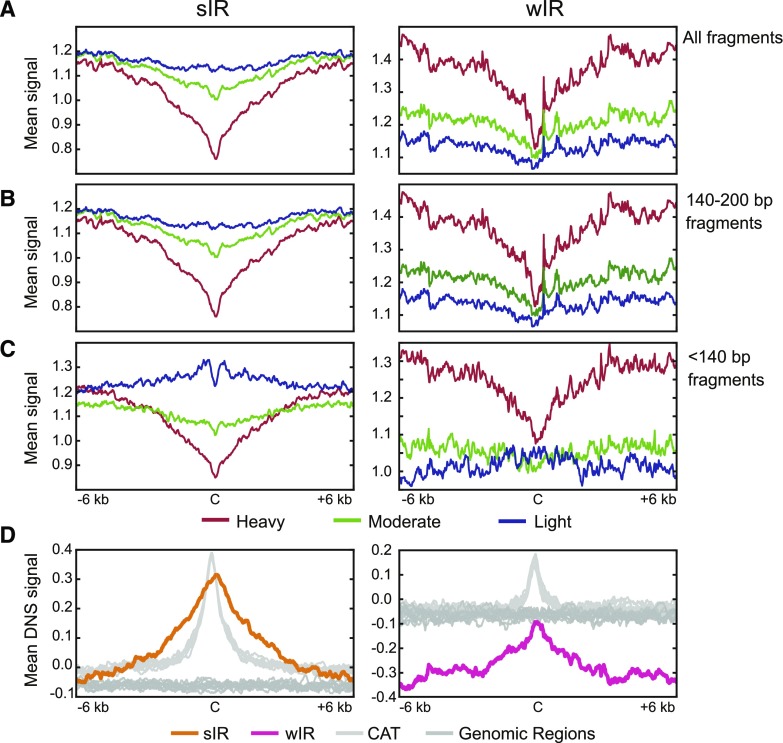
The mean average signal for all protected fragments at IR-Cs and in their surrounding regions was plotted for the three levels of digestion (Fig. 6A). In addition, signals from protected fragments longer and shorter than 140 bp were plotted separately (Fig. 6, B and C). The patterns of the 140- to 200-bp signal (Fig. 6B) are very similar to the combined signals for all size classes (Fig. 6A; for a more critical comparison between the two profiles see Supplemental Fig. S9). Signals from 0- to 140-bp fragments in the heavy and moderate digests (Fig. 6C) are also similar to their total chromatin pattern, except that they are lower around wIRs. However, for sIRs, the light digestion profiles for short fragments are strikingly different. In sIRs, the mean abundance of these short, protected fragments (light digestion signal) increases as it nears the sIR-C, but then dips sharply at the sIR-C itself (Fig. 6C, sIR-C is the black bar labeled “C”). The overall pattern for all digestions and fragment sizes is consistent with a model in which IR-Cs and their flanking sequences are depleted of nucleosomes, with depletion progressively greater toward the IR-C. The pattern is also consistent with the occurrence of smaller, less stably bound proteins on either side of the IR-C. The MNase digestion profiles of the sIRs and wIRs are readily distinguishable from the profiles for the CAT and random genomic datasets (Supplemental Fig. S10). Strikingly, the double peak pattern for <140-bp fragments seen in the light digest of sIRs is not observed for the CAT controls, which instead show a single peak centered over the control sequences (Supplemental Fig. S10C).
Figure 6.
Mean analysis comparing heavy, moderate, and light MNase digestions by fragment size. A, Mean heavy (red), moderate (green), and light (blue) digestion profiles at and around sIR-Cs (left) and wIR-Cs (right). B, Mean digestion plots comprising only 140- to 200-bp fragments. Plots in (A) and (B) look similar; however, Supplemental Figure S9 shows they are not identical. C, Mean digestion plots comprising only fragments less than 140 bp. D, Mean differential nuclease sensitivity (DNS) signal at and around sIR-Cs (left, orange line) and wIR-Cs (right, pink line) and their random genomic regions (dark gray) and CAT regions (light gray). DNS signal is the difference between the light digestion signal and the heavy digestion signal (scale −2 to 2), where positive values represent more open chromatin than negative values. The mean was calculated in 20-bp bins, and the plots are of ±6-kb windows around the IR-Cs with the C marking the center.
A simple way to combine the data from the MNase digestions into a single metric to assess the “openness” of the chromatin is to subtract the heavy digestion signal from the light digestion signal, which allows visualization of fragments protected only during light digestion, separate from fragments that remain protected during stronger digestions. This procedure creates a genome-wide DNS map (Supplemental Fig. S4B), in which positive signals represent more open chromatin, and negative signals represent more compact chromatin (Vera et al., 2014). The heterochromatic centromere regions have predominately negative DNS signal while chromosome arms contain many more regions of positive DNS signal (Supplemental Fig. S4, B and C).
A composite plot of DNS signal strength at and around IR-Cs shows that the sIR-Cs have approximately 6-fold higher DNS signal than their random genomic datasets, while the wIR-Cs have similar DNS signal compared with their random genomic datasets (Fig. 6D). However, both sIR-Cs and wIR-Cs are found at DNS signal peaks, suggesting they are local regions of open chromatin, flanked by regions with gradually increasing compaction. It is interesting to note that the DNS profile of sIRs has a wider peak than the profile for their control CAT datasets, suggesting that sIRs form broader regions of accessible chromatin than randomly selected AT-rich regions. Hence, although AT rich regions have a role in creating open chromatin, as previously reported (Iyer and Struhl, 1995; Thåström et al., 1999; Field et al., 2008; Hughes et al., 2012; Pascuzzi et al., 2014), sIRs clearly represent a distinct subset of such regions.
The wIR-Cs are also located within a region of locally increased DNS. Even though the wIR-Cs (Fig. 6D) have a DNS signal similar to that of random genomic regions, they are flanked by regions of decreased DNS. Thus, although wIRs are mostly located in compact, heterochromatic regions, wIR-Cs represent local regions of relatively more open chromatin, flanked by regions of gradually increasing compaction.
DISCUSSION
Two-parameter flow sorting allowed us to obtain high resolution profiles of DNA replication initiation activity in VE S phase (Fig. 2, A and B). We defined IR-Cs as local peaks of replication activity in VE S phase. We infer that replication must initiate in or near these regions. Importantly, as replication proceeds from VE to E S phase, IR-Cs become associated with less prominent, much broader peaks of replication signal, in a manner consistent with spreading of replication activity into neighboring regions. Such bidirectional spreading would be expected according to all current models of DNA replication, and thus represents a significant validation of our functional assay.
Comparison of IRs to Other Arabidopsis Studies
An earlier paper and follow-up studies from the Gutierrez lab using Arabidopsis Ler-0 suspension cells defined as origins a set of loci that are GC-rich sequences associated with the 5′ ends of genes and enriched for the acetylated histone H4K5ac (Costas et al., 2011; Xing et al., 2014; Sequeira‐Mendes and Gutierrez, 2015; Vergara et al., 2017; Sequeira-Mendes et al., 2019). To compare our IRs with the loci defined in these studies, we plotted replication signal enrichment from our data around the loci defined by Costas et al. (2011) and Vergara et al. (2017). Neither set of loci defined by the Gutierrez group showed local increases in replication signal across a ±15-kb window, despite the fact that their loci were identified using BrdU, a thymidine analog similar to EdU (Supplemental Fig. S11A). We conclude that replication profiles in the Ler-0 ecotype under the conditions used in these two studies differ from those in the Col-0 ecotype under our conditions.
The initial study by Costas et al. (2011) used 24 h of Suc starvation followed by readdition of Suc to synchronize the cell cultures, which were labeled 2.5 h later with BrdU in the presence of hydroxyurea, an inhibitor of nucleotide biosynthesis, which has been shown to cause DNA damage and induce DNA repair (Sakano et al., 2001; Banh and Hales, 2013). Hence, much of the BrdU incorporation could reflect repair activity more than DNA replication. Moreover, stress treatments such as Suc starvation and the slowing of fork elongation by hydroxyurea (Merrick et al., 2004; Alvino et al., 2007) have both been shown to alter origin usage (Taylor, 1977; Ge et al., 2007; Gilbert, 2007). As previously noted, our method avoids both growth stress and chemical inhibitors and provides a direct functional assay for genomic regions where replication is first initiated.
Reproducibility has long been an issue in studies of replication origins in both plants and animals. Our EdU-Seq datasets were highly reproducible as demonstrated by Spearman’s correlation coefficients >97% across the four biological replicates (Supplemental Table S1, far right column). Given the high correlation, IR-Cs were more robustly identified using merged biological replicate data. However, when the replicates were analyzed separately, 82% of the sIR-Cs and 46% of the wIR-Cs were independently identified in at least three of the four biological replicates (Supplemental Fig. S2A). The Gutierrez group used a single BrdU dataset, in which they identified either 1,534 or 3,230 putative origins depending on the peak calling algorithm used (MACS and MACS2, respectively; Costas et al., 2011; Vergara et al., 2017). The same group reported a high level of variability between biological replicates when using the SNS assay to identify putative origins in Arabidopsis Col-0 seedlings (Sequeira-Mendes et al., 2019). Results with SNS assays have often been difficult to reproduce, even in animal systems (Gilbert, 2010; Hyrien, 2015; Miotto, 2017). In plants, the low percentage of cells replicating DNA at any given time in seedlings, and the presence of endogenous nucleases in vacuoles may contribute to variability by decreasing the signal to noise level in SNS assays (Obara et al., 2001).
Nucleotide Content, Labeling Pattern, and Sequence Context of IR-Cs
The IRs identified make up a distinct subset of AT-rich sequences in the Arabidopsis genome. The central AT-rich sequences in sIR-Cs are flanked by more GC-rich regions starting at approximately ±3 kb from the center, while their CAT datasets maintain higher than genomic levels of AT content for ±15 kb (Fig. 3A). Central regions of the wIRs are narrower and slightly less AT-rich, and their flanking regions of elevated GC content are somewhat larger than those of the sIR-Cs (Fig. 3B). The profile of EdU-IP signals at and around the sIR-Cs is stronger and broader than for the corresponding CAT regions, while the wIR-Cs have less EdU-IP signal than their corresponding CAT regions (Supplemental Fig. S2, B–E). Taken together, the differences between the IR profiles and CAT regions provide strong support for the conclusion that EdU local maxima in the VE profile represent functionally unique regions.
Although metazoan origins are generally described as GC rich (Cadoret et al., 2008; Cayrou et al., 2011; Besnard et al., 2012; Valton et al., 2014); AT-rich replication initiation peaks embedded in GC-rich regions have been described in human cells (Karnani et al., 2010). In addition, the S. cerevisiae autonomously replicating sequence, which binds to the origin recognition complex (ORC), is an AT-rich motif (Stinchcomb et al., 1979). In Schizosaccharomyces pombe the ORC4 protein contains an AT-hook domain, and origins are AT rich (Lee et al., 2001). In addition, DNA unwinding elements, which are thought to be adjacent to ORC binding sites, are AT rich (Ishimi and Matsumoto, 1994; Chowdhury et al., 2010; Gai et al., 2010). AT-rich regions have also been shown to disfavor nucleosome occupancy, consistent with origins being located preferentially in nucleosome-free regions (Lubelsky et al., 2011; Belsky et al., 2015; Rodriguez et al., 2017).
Several reports have suggested that metazoan origins are associated with G4 quadraplexes and CpG islands (Cayrou et al., 2011; Langley et al., 2016). It has been proposed that G4 structures within 1 kb of origins are important for nucleosome phasing and impact origin selection (Foulk et al., 2015). Although the regions flanking Arabidopsis IR-Cs are more GC rich than the random genome datasets (Fig. 3, A and B), they still contain >50% AT ± 6 kb from the center, distinguishing them from the GC-rich regions described for metazoan origins. Hence, it seems unlikely that flanking regions around IR-Cs are involved in phasing nucleosomes in Arabidopsis. This idea is underscored by the lack of statistically significant overlap of IR-Cs by potential G4 quadruplex structures, and by similar rates of association for IR-Cs and CAT regions (Fig. 3, C and D; Supplemental Fig. S5, E and F; projection test P-value = 0.0797 for G4s called at 1.5 threshold and sIR-Cs; for G4 datasets called at other thresholds see Supplemental Tables S2 and S3; for associations between G4s called at 1.5 threshold and random genomic and CAT datasets see Supplemental Table S4).
Genomic and Chromatin Context of IR-Cs
IRs are predominantly located in intergenic regions associated with open chromatin. IR-Cs also tend to be flanked by genes and are moderately enriched in TEs. In most respects, these features differ from those of the loci characterized by the Gutierrez group. Approximately 80% of the IR-Cs show no overlap with genes (P-value < 0.001), but are more likely than their CAT regions to have genes in their flanking regions (Fig. 4A). The sIR-Cs are scattered throughout the euchromatic arms and associate with gene-depleted chromatin states and early replication timing (P-value < 0.001, Supplemental Fig. S7), while the wIR-Cs colocalize with heterochromatic chromatin states (P-values < 0.001) and late replication timing (P-value < 0.001) in the pericentromere and centromere regions (Concia et al., 2018).
To examine chromatin accessibility of IRs, we used MNase-derived DNS profiles (Supplemental Fig. S4, B and C). In contrast with DHS sites, which occur as sharp, discrete, and very local peaks, DNS profiles provide a continuous, quantitative readout of relative accessibility across the entire genome (Vera et al., 2014). Because the DNS signal represents the difference between and light and heavy digests, it allows us to focus on regions that, while nucleosome free, may contain smaller, less tightly bound proteins. Mean DNS profiles show that IR-Cs are centered under a peak of positive signal, which gradually decreases in flanking regions, suggesting that both the peak and, to a lesser extent, the flanking regions have an open chromatin structure. CAT regions for the sIR-Cs also show a positive DNS signal, but the CAT peak is higher and narrower than the sIR peak (Fig. 6D). The differences in the two profiles indicate that sIRs occupy broader regions of open chromatin than the CAT regions, which could reflect a role for accessible chromatin around the sIR-Cs in the formation of prereplication and preinitiation complexes, as well as in the recruitment of replisome proteins. In the case of wIRs, the mean DNS signal at the wIR-C is similar to that of the random genomic datasets and becomes gradually more negative in flanking regions. This result is consistent with locally more open regions in an environment of tightly packed heterochromatin (Fig. 6D, right column). Thus, a general feature of both types of IR-Cs is that they occur in regions of increased “openness” relative to surrounding chromatin.
The Arabidopsis genome is densely packed with genes. It is therefore interesting that IR-Cs are negatively associated with genes (exons and introns, P-values < 0.001; Supplemental Tables S2 and S3), while the loci described as origins by the Gutierrez group (Costas et al., 2011; Vergara et al., 2017) are positively associated with genes and exons (P-values < 0.001, Supplemental Fig. S11B; Supplemental Tables S8–S10). Consistent with these observations, NGT (Not Gene or TE) regions are enriched around IR-Cs and depleted around the Gutierrez loci (Supplemental Fig. S11C; P-values < 0.001, Supplemental Tables S2, S3, and S8–S10). TEs as a whole are moderately enriched at IR-Cs and somewhat depleted at the Gutierrez loci (Supplemental Fig. S11C; P-values < 0.001, Supplemental Tables S2, S3, and S8–S10). The TE-associated loci described by Vergara et al. (2017) show the expected high association with TEs (Supplemental Fig. S11C; P-values < 0.001, Supplemental Table S9), but their TE family enrichment pattern is distinct from that for IRs (Supplemental Fig. S11D; Supplemental Tables S2 and S3). There is also a difference between the local chromatin accessibility around IR-Cs and that around the Gutierrez group loci (Supplemental Fig. S11E), with IRs characterized by broad positive peaks indicating relatively large regions of accessible chromatin while the Gutierrez group loci are characterized by as relatively sharp negative peaks indicative of locally compact chromatin. This difference is particularly striking because of the strong association of origins with accessible chromatin in animals (Field et al., 2008; Hansen et al., 2010; Eaton et al., 2011; Smith et al., 2016).
Are IRs Origins of Replication?
Several lines of evidence support the idea that the peaks of VE EdU-IP signal include origins of replication. The sIRs are consistently different from other AT-rich regions in the Arabidopsis genome, indicating that they are not random EdU-labeled sequences. In addition, sIRs are most highly represented in zones of early replication (Supplemental Fig. S7A; P-value < 0.001, Supplemental Table S2), consistent with a function as early firing origins. Comparing the VE and E signal tracks along chromosome length displays bidirectional spreading of replication signal, consistent with replication initiation in VE and subsequent elongation in E (Fig. 2, A and B). Moreover, sIRs show many similarities to previously described eukaryotic origins, including their euchromatic environment and association with local regions of open chromatin. However, Arabidopsis sIRs also have some unique characteristics, such as their AT richness, intergenic location and association with Helitrons. In contrast, wIRs are mostly located in the centromere and pericentromere, where most replication occurs during late S phase (Concia et al., 2018). Given their location in regions where replication activity at the beginning of S phase is unexpected, the significance of the wIRs remains uncertain. However, wIRs do not represent a collection of random locations, suggesting they are not merely noise. The wIRs might contain inefficient early firing origins, or origins that fire in early S phase but whose forks move slowly or become stalled in heterochromatin and are reactivated only during late S phase. The greater average distance between wIR-Cs compared with sIR-Cs is consistent with the theory that replication time is a function of origin density (Gindin et al., 2014). Our results do not exclude the possibility that later firing origins may exist that could not be detected in very early S phase. However, if one includes the wIRs, the number and spacing of IRs along the chromosomes is sufficient to account for replication of the entire genome. Taken together, our results suggest that IRs represent locations involved in DNA replication initiation, and that broad chromatin accessibility is an important characteristic of these regions.
MATERIALS AND METHODS
Arabidopsis Cell Culture
Arabidopsis (Arabidopsis thaliana) Col-0 suspension cells were grown in 250-mL baffled flasks containing 50 mL of growth medium (3.2 g/L Gamborgs B-5 medium, 3 mm MES, 3% [v/v] Suc, 1.1 mg L−1 2,4-dichlorophenoxyacetic acid). The cultures were maintained at 23°C under continuous light on a rotary shaker (160 rpm) and subcultured by 1:10 dilution into fresh medium at 7-d intervals (Lee et al., 2010).
EdU Labeling and Cell Collection
At 7 d after subculture, 25 mL of Arabidopsis suspension culture was diluted 1:1 into fresh medium and grown under standard conditions for 16 h. Each diluted culture was then labeled with 10 µm 5-ethynyl-2’-deoxy-uridine (EdU) for 30 min under the same conditions. Cells from 18 to 24 flasks were collected by pouring the cultures over two layers of miracloth and washed with cold 1× phosphate-buffered saline. The cells were scraped off the miracloth and suspended in a 1% (v/v) solution of paraformaldehyde for 10 min, swirling occasionally to keep them from settling. Fixation was stopped by the addition of Gly to a final concentration of 125 mm with a 5-min incubation. The fixed cells were collected on two layers of miracloth and washed with 1× phosphate-buffered saline. The cells were then divided into 50-mL tubes, with two flasks of cells in each tube, flash frozen in liquid nitrogen, and stored at −80°C. For each biological replicate 24 flasks were collected.
Nuclei Isolation via Flow Cytometry and DNA Purification
Incorporated EdU was conjugated to AF488 using the Click-iT EdU Alexa Fluor 488 Imaging Kit by ThermoFisher Scientific (C10337) and using the prescribed protocol, and total DNA was stained with 2 mg L−1 DAPI before flow sorting of nuclei on an InFlux sorter (BD Biosciences) as previously described (Wear et al., 2016; Concia et al., 2018). Crude nuclei preparations were purified using Percoll gradients (Folta and Kaufman, 2006; Wear et al., 2016). Four biological replicates were prepared and sorted on different days (see Supplemental Fig. S1 for sort gating details, sort purity, and the number of nuclei sorted from each gate). Sorted nuclei were stored at −20°C. DNA was extracted from sorted nuclei using two phenol:chloroform extractions and then sheared to ∼250 bp using a Covaris S220 Ultrasonicator as described previously (Concia et al., 2018).
Immunoprecipitation and Sequencing of EdU-labeled DNA
AF488-containing (replicating) DNA fragments were immunoprecipitated as described (Lee et al., 2010; Concia et al., 2018) using a rabbit antibody to Alexa Fluor 488 (Life Technologies, catalog no. A11094). Unlike BrdU, EdU can be immunoprecipitated without denaturing the DNA, allowing for cleaner DNA preparation and for the DNA to be directly transformed into sequencing libraries. Libraries were constructed with the NEXTflex Illumina ChIP-Seq Library Prep Kit (Bio Scientific), using the ultra-low input protocol. After adapter ligation, the libraries were amplified with 18 cycles of PCR. For each experiment, individual samples and biological replicates were barcoded and pooled. The libraries were sequenced on an Illumina HiSeq 2000 platform.
Chromatin Extraction and Immunoprecipitation
Suspension cells were grown and fixed as described above but without EdU labeling. For each biological replicate, one-half flask of cells was used. Chromatin was extracted and immunoprecipitation performed as previously described (Lee et al., 2010) except that chromatin was sheared using a Covaris S220 Ultrasonicator with the SonoLab7 software and the following parameters: peak incident power 200, duty cycle 10, cycles per burst 200, time per treatment 60 s, number of treatments 3. The antibodies were antihistone H3 acetyl K27 (Abcam, cat no. ab4729), antihistone H3 acetyl K5 (Abcam, cat no. ab51997), and antihistone H3 acetyl K9 (Abcam, cat no. ab 12179).
Differential Nuclease Sensitivity Assay
Suspension cells were grown and fixed as described for the chomatin immunoprecipitation sequencing (ChIP-seq) experiments. Four biological replicates were collected, and nuclei were isolated using the blending and Percoll method as described above for flow sorting. After the second Percoll gradient, the pelleted nuclei were placed in a 15-mL tube and brought to 15-mL volume with MNase Digestion buffer (50 mm HEPES, pH 7.6; 12.5% [v/v] glycerol; 25 mm KCl; 4 mm MgCl2; 1 mm CaCl2). Nuclei were washed twice by centrifuging at 2000g for 15 min at 4°C, and the supernatant was discarded. Final nuclear pellets were resuspended in 500 µL of MNase Digestion buffer per original flask of cells and flash frozen for −80°C storage. The rest of the differential nuclease sensitivity assay was performed as described (Vera et al., 2014).
Data Analysis
All data, including processed files, were hosted and can be found on iPlant, now CyVerse (Goff et al., 2011; https://de.cyverse.org/de/?type=data&folder=/iplant/home/shared/ncsu-plantreplication/public/Arabidopsis/Arabidopsis_initiation_of_DNA_replication). The raw sequencing files can be found at National Center for Biotechnology Information Sequence Read Archive under the accessions: PRJNA421176 (EdU-seq); PRJNA470543 (H4K5ac ChIP-seq); PRJNA609130 (H3K27ac ChIP-seq); PRJNA609131 (H3K9ac ChIP-seq); PRJNA472593 (MNase-seq). The data were loaded and analyzed on the Stampede and Stampede2 supercomputers at the TX Advanced Computing Center and locally on a Mac OSX 10.12.3 machine.
Read Preprocessing and Merging Biological Replicates
Reads were trimmed of adapter sequences and reads shorter than 40 bp were discarded (Cutadapt v.1.11; Martin, 2011). Reads were mapped to the Arabidopsis reference genome (TAIR10) using the bwa mem algorithm (BWA v.0.7.12; Jo and Koh, 2015) with default settings. PCR duplicates and improperly paired reads were removed (samTools v.1.2 and v.0.1.19; Li et al., 2009). Due to high Spearman’s correlation values (calculated using deepTools v.2.3.5; Ramírez et al., 2016), all biological replicates from the AF488 immunoprecipitations and MNase digestions were merged, and three replicates from the ChIP-seq experiments were merged (samTools v.1.2).
Read Depth and Sequenceability Normalization
After the biological replicates for each experiment were merged, the read coverage was normalized to 1× (deepTools v.2.3.5) and divided by the G1 reference (sorted AF488 experiment) or total genomic input (ChIP-seq and MNase-seq experiments) to control for collapsed repeats and variations in sequenceability. Before read normalization, the MNase-seq data from each level of digestion were subdivided by read length (see Supplemental Fig. S12A for read length distribution) and then divided by total genomic input to generate final normalized signal data. The total normalized signal from the heavy digestion was subtracted from the total normalized signal from the light digestion to create a genome-wide DNS map (Supplemental Fig. S4, B and C).
Identifying Local Maxima in EdU-seq Signal
Local maxima from the normalized VE EdU signal data were defined using a custom R script (Supplemental Fig. S12D). A sliding window of 51 300-bp bins was moved in 300-bp steps along the genome. When the center bin contained the highest signal of the 51 bins, that 300-bp bin was called a local maximum. Many window sizes were assessed for calling local maxima. The number of local maxima returned as a function of the number of bins in the window was plotted, and a logarithmic pattern observed (Supplemental Fig. S12B). A window size of 51 bins was chosen to balance being conservatively into the plateau without losing true local maxima. Additionally, mean plot analysis of EdU signal at local maxima showed that regardless of the number of bins used the mean peak width of EdU signal around local maxima was ∼15 kb, which is approximately the size of the 51-bin window (Supplemental Fig. S12C). After local maxima using 51 bins were identified, several filtering steps were applied, resulting in 5597 final local maxima. Nineteen local maxima that overlapped gaps in the genome assembly were excluded. Thirteen local maxima from low coverage regions or their flanking regions were excluded as technical artifacts.
Defining Initiation Regions
The local maxima were divided into quartiles based on their VE EdU-IP signal. The top three quartiles were combined into a single group and maintained separately from the bottom quartile for the remainder of the analyses (Fig. 2D). Given that the local maxima are presumed to be close to DNA replication origins, they were called IR-Cs. The IR-Cs belonging to the top three quartiles are defined as strong IR-Cs (sIR-Cs; 4197 total), while those in the bottom quartile were defined as weak IR-Cs (wIR-Cs; 1400 total).
Distribution of IRs Among and Across Chromosomes
IR-C coverage data were divided into 100-kb static bins. BedTools/2.25.0 (Quinlan and Hall, 2010) was used to calculate the number of IR-Cs per chromosome length, the distance to the next nearest IR-C, the distance from each IR-C to the centromere, and the coverage of IRs in 10% bins from the centromere to the end of the chromosome arm (Fig. 2, F–H). The centromere locations were identified by blast search of the 180-bp repeat sequence against the genome and smoothing the coverage with a 5-kb window; the resulting maximal 1-kb bin from each chromosome was used as the centromere.
AT Content Analysis
BedTools was used to calculate the AT content in 300-bp bins across the Arabidopsis genome. To compare the IR-Cs to random genomic regions, 10 independent random genomic datasets were generated using bedTools. The random datasets were designed to be comparable to IR-Cs in size (300 bp) and number (4197 for sIRs or 1400 for wIRs). Next, 10 additional independent random datasets were generated from regions in the genome that fall within the IQR of AT content for each set of IR-Cs, while the IR-Cs themselves were excluded. These datasets are referred to as CAT (Comparable AT) datasets. The deepTools suite was used to plot the mean AT content around IR-Cs (±15 kb) using each of the random datasets.
G-Quadruplex Forming Sequences
The G4Hunter (Bedrat et al., 2016) algorithm was used to identify potential G4s across the genome using a window size of 25 bp and threshold scores of 1, 1.25, 1.5, 1.7, or 2. The number of G4s called and their length distribution were plotted. The percent of IR-Cs with a G4 within ± 15 kb around the IR-C was calculated with bedTools v2.25.0.
Relationship to Genomic Features, DHS, and G4s
BedTools v2.25.0 was used to calculate the percent overlap (using a criterion of at least one base pair overlap) between IR-Cs and different genomic features from the Araport 11 annotation (Krishnakumar et al., 2015; Cheng et al., 2017), TEs (Huala et al., 2001; Buisine et al., 2008; Lamesch et al., 2012), published DHS (Sullivan et al., 2014), and predicted G4s. Relative and absolute distance, Kolmogorov-Smirnov tests, Jaccard tests, and projection tests were performed using GenometriCorr (Favorov et al., 2012) to assess relationships between IR-Cs and genomic features, DHS, and G4s.
The Kolmogorov-Smirnov test compares the relative distances between a query and a reference set of genomic regions. When combined with permutation analysis it produces a p-value to determine the level of independence between the datasets and plots an Empirical Distribution Cumulative Function, from which a correlation-like measure is calculated to show the direction of correlation. The projection test is a two-sided binomial test that looks at the probability of the query dataset intersecting the reference dataset compared with the null expectation of random intersection. The Jaccard approach is a permutation test between intervals that looks at the correlation of overlap between the query and reference datasets as a ratio of their intersection and union.
Code and Data Availability Statement
All data analysis was done using open source software with the exception of the local maxima finding script, which can be found in Supplemental Figure S12D. The datasets generated during the current study are hosted at CyVerse: https://de.cyverse.org/de/?type=data&folder=/iplant/home/shared/ncsu-plantreplication/public/Arabidopsis/Arabidopsis_initiation_of_DNA_replication.
Accession Numbers
The raw sequencing files are deposited on the National Center for Biotechnology Information Sequence Read Archive Web site under the accessions: PRJNA421176 (EdU-seq); PRJNA470543 (H4K5ac ChIP-seq); PRJNA609130 (H3K27ac ChIP-seq); PRJNA609131 (H3K9ac ChIP-seq); PRJNA472593 (MNase-seq).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nuclei sorting gating strategy and resort analysis.
Supplemental Figure S2. Mean plots of DNS signal around IRs by EdU signal quartile.
Supplemental Figure S3. Percent of IR-Cs from merged data that correspond to local maxima found in individual biological replicates and EdU-IP and AT content at IR-Cs and their comparison datasets.
Supplemental Figure S4. Coverage of IR-Cs across each chromosome and Differential Nuclease Sensitivity (DNS) signal across each chromosome.
Supplemental Figure S5. IR-Cs are not associated with G-quadurplex (G4) forming sequences.
Supplemental Figure S6. Overlap of the IR-Cs by genomic features.
Supplemental Figure S7. Percent coverage of the genome, sIR-Cs, and wIR-Cs by replication timing classes and chromatin states.
Supplemental Figure S8. Overlap of IRs by DNase I hypersensitivity (DHS) sites (Sullivan et al., 2014).
Supplemental Figure S9. Comparison of 140–200 bp subpopulation of fragments to total fragments.
Supplemental Figure S10. Mean profiles of different MNase digestions classified by fragment length at sIR-Cs and wIR-Cs compared to their random genomic and CAT regions.
Supplemental Figure S11. Comparison of genomic features and DNA replication initiation site datasets.
Supplemental Figure S12. Size distribution of DNA fragments from heavy, moderate, and light MNase digestions and effect of window size on the number of local maxima, and mean EdU-IP from VE profiles of local maxima.
Supplemental Table S1. Results from sequenced libraries and mapping.
Supplemental Table S2. Relative distance, projection test, and Jaccard test results between strong IR-Cs and genomic features.
Supplemental Table S3. Relative distance, projection test, and Jaccard test results between weak IR-Cs and genomic features.
Supplemental Table S4. Overlap and statistical relationship between G4s called using a 1.5 threshold in G4Hunter (Bedrat et al., 2016) and IR-Cs and their comparison datasets.
Supplemental Table S5. Overlap and statistical relationship between genomic features and IR-Cs and their comparison datasets.
Supplemental Table S6. Overlap and statistical relationship between TE families (Huala et al., 2001; Buisine et al., 2008; Lamesch et al., 2012) and IR-Cs and their comparison datasets.
Supplemental Table S7. Overlap and statistical relationship between DHS (Sullivan et al., 2014) and IR-Cs and their comparison datasets.
Supplemental Table S8. Relative distance, projection test, and Jaccard test results between the loci described as origins by Costas et al. (2011) and genomic features.
Supplemental Table S9. Relative distance, projection test, and Jaccard test results between the loci described as origins by Vergara et al. (2017) and genomic features.
Supplemental Table S10. Relative distance, projection test, and Jaccard test results between the TE-associated loci described as origins by Vergara et al. (2017) and genomic features.
Acknowledgments
We recognize Eric Szymanski and Patrick Mulvaney at North Carolina State University for help processing the cell cultures and isolating nuclei. We acknowledge the Texas Advanced Computing Center and Jawon Song and Greg Zynda at University of Texas at Austin for providing storage and computing resources. Research in the Martienssen laboratory is supported by the Howard Hughes Medical Institute. We recognize assistance from Cold Spring Harbor Laboratory Shared Resources, which is funded in part by the Cancer Center Support Grant (5PP30CA045508).
Footnotes
This work was supported by the NSF | BIO | Division of Integrative Organismal Systems (IOS; no. NSF IOS–1025830 to M.W.V., R.A.M., W.F.T., and L.H.-B., and no. NSF IOS–1444532 to H.W.B.).
Articles can be viewed without a subscription.
References
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK(2007) Replication in hydroxyurea: It’s a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banh S, Hales BF(2013) Hydroxyurea exposure triggers tissue-specific activation of p38 mitogen-activated protein kinase signaling and the DNA damage response in organogenesis-stage mouse embryos. Toxicol Sci 133: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass HW, Hoffman GG, Lee T-J, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, Thompson WF(2015) Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei. Plant Mol Biol 89: 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedrat A, Lacroix L, Mergny J-L(2016) Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res 44: 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky JA, MacAlpine HK, Lubelsky Y, Hartemink AJ, MacAlpine DM(2015) Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev 29: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, Lemaitre J-M(2012) Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19: 837–844 [DOI] [PubMed] [Google Scholar]
- Buisine N, Quesneville H, Colot V(2008) Improved detection and annotation of transposable elements in sequenced genomes using multiple reference sequence sets. Genomics 91: 467–475 [DOI] [PubMed] [Google Scholar]
- Cadoret J-C, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau M-N(2008) Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci USA 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau J-C, van Helden J, Méchali M(2015) The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res 25: 1873–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, Méchali M(2011) Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-Y, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD(2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Liu G, Kemp M, Chen X, Katrangi N, Myers S, Ghosh M, Yao J, Gao Y, Bubulya P, Leffak M(2010) The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol Cell Biol 30: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concia L, Brooks AM, Wheeler E, Zynda GJ, Wear EE, LeBlanc C, Song J, Lee T-J, Pascuzzi PE, Martienssen RA, et al. (2018) Genome-wide analysis of the Arabidopsis replication timing program. Plant Physiol 176: 2166–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas C, de la Paz Sanchez M, Stroud H, Yu Y, Oliveros JC, Feng S, Benguria A, López-Vidriero I, Zhang X, Solano R, et al. (2011) Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořáčková M, Raposo B, Matula P, Fuchs J, Schubert V, Peška V, Desvoyes B, Gutierrez C, Fajkus J(2018) Replication of ribosomal DNA in Arabidopsis occurs both inside and outside the nucleolus during S phase progression. J Cell Sci 131: jcs202416. [DOI] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM(2011) Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favorov A, Mularoni L, Cope LM, Medvedeva Y, Mironov AA, Makeev VJ, Wheelan SJ(2012) Exploring massive, genome scale datasets with the GenometriCorr package. PLOS Comput Biol 8: e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E(2008) Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLOS Comput Biol 4: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS(2006) Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat Protoc 1: 3094–3100 [DOI] [PubMed] [Google Scholar]
- Foulk MS, Urban JM, Casella C, Gerbi SA(2015) Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res 25: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai D, Chang YP, Chen XS(2010) Origin DNA melting and unwinding in DNA replication. Curr Opin Struct Biol 20: 756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ(2007) Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM.(2010) Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM.(2007) Replication origin plasticity, Taylor-made: Inhibition vs recruitment of origins under conditions of replication stress. Chromosoma 116: 341–347 [DOI] [PubMed] [Google Scholar]
- Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S(2014) A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol 10: 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, et al. (2011) The iPlant collaborative: Cyberinfrastructure for plant biology. Front Plant Sci 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA(2010) Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci USA 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. (2001) The Arabidopsis Information Resource (TAIR): A comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res 29: 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K(2012) A functional evolutionary approach to identify determinants of nucleosome positioning: A unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O.(2015) Peaks cloaked in the mist: The landscape of mammalian replication origins. J Cell Biol 208: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y, Matsumoto K(1994) Loading of a DNA helicase on the DNA unwinding element in the yeast replication origin: Mechanism of DNA replication in a model system. Biochemistry 33: 2733–2740 [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K(1995) Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H, Koh G(2015) Faster single-end alignment generation utilizing multi-thread for BWA. Biomed Mater Eng 26(Suppl 1): S1791–S1796 [DOI] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A(2010) Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng C-Y, Moreira W, Mock SA, et al. (2015) Araport: The Arabidopsis information portal. Nucleic Acids Res 43: D1003–D1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley AR, Gräf S, Smith JC, Krude T(2016) Genome-wide identification and characterisation of human DNA replication origins by initiation site sequencing (ini-seq). Nucleic Acids Res 44: 10230–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-K, Moon K-Y, Jiang Y, Hurwitz J(2001) The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc Natl Acad Sci USA 98: 13589–13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-J, Pascuzzi PE, Settlage SB, Shultz RW, Tanurdzic M, Rabinowicz PD, Menges M, Zheng P, Main D, Murray JAH, et al. (2010) Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet 6: e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D(2016) Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res 26: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Sasaki T, Kuipers MA, Lucas I, Le Beau MM, Carignon S, Debatisse M, Prinz JA, Dennis JH, Gilbert DM(2011) Pre-replication complex proteins assemble at regions of low nucleosome occupancy within the Chinese hamster dihydrofolate reductase initiation zone. Nucleic Acids Res 39: 3141–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M, Halazonetis TD(2018) Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheret M, Halazonetis TD(2019) Monitoring early S-phase origin firing and replication fork movement by sequencing nascent DNA from synchronized cells. Nat Protoc 14: 51–67 [DOI] [PubMed] [Google Scholar]
- Martin M.(2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Merrick CJ, Jackson D, Diffley JFX(2004) Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 279: 20067–20075 [DOI] [PubMed] [Google Scholar]
- Miotto B.(2017) Comment l’approche génomique aide à comprendre le processus d’initiation de la réplication. Médecine/sciences 33: 143–150 [DOI] [PubMed] [Google Scholar]
- Obara K, Kuriyama H, Fukuda H(2001) Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol 125: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascuzzi PE, Flores-Vergara MA, Lee T-J, Sosinski B, Vaughn MW, Hanley-Bowdoin L, Thompson WF, Allen GC(2014) In vivo mapping of arabidopsis scaffold/matrix attachment regions reveals link to nucleosome-disfavoring poly(dA:dT) tracts. Plant Cell 26: 102–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pass DA, Sornay E, Marchbank A, Crawford MR, Paszkiewicz K, Kent NA, Murray JAH(2017) Genome-wide chromatin mapping with size resolution reveals a dynamic sub-nucleosomal landscape in Arabidopsis. PLoS Genet 13: e1006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Cadoret J-C, Audit B, Arneodo A, Alberti A, Battail C, Duret L, Prioleau M-N(2014) The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet 10: e1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM(2010) BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Henikoff S(2016) Transcriptional regulators compete with nucleosomes post-replication. Cell 165: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T(2016) deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44(W1): W160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Lee L, Lynch B, Tsukiyama T(2017) Nucleosome occupancy as a novel chromatin parameter for replication origin functions. Genome Res 27: 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano K, Oikawa S, Hasegawa K, Kawanishi S(2001) Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide. Jpn J Cancer Res 92: 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ(2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savadel SD, Bass HW(2017) Take a look at plant DNA replication: Recent insights and new questions. Plant Signal Behav 12: e1311437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Aragüez I, Peiró R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, Gutierrez C(2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Gutierrez C(2015) Links between genome replication and chromatin landscapes. Plant J 83: 38–51 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Vergara Z, Peiro R, Morata J, Araguez I, Costas C, Giraldez RM, Casacuberta JM, Bastolla U, Gutierrez C(2019) Differences in firing efficiency, chromatin and transcription underlie the developmental plasticity of the Arabidopsis DNA replication origins. Genome Res 29: 784–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WF(2007) Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol 144: 1697–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S, et al. (2016) Distinct epigenetic features of differentiation-regulated replication origins. Epigenetics Chromatin 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW(1979) Isolation and characterisation of a yeast chromosomal replicator. Nature 282: 39–43 [DOI] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep 8: 2015–2030 [DOI] [PubMed] [Google Scholar]
- Taylor JH.(1977) Increase in DNA replication sites in cells held at the beginning of S phase. Chromosoma 62: 291–300 [DOI] [PubMed] [Google Scholar]
- Thåström A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J(1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences In J Mol Biol, Vol 288 pp 213–229 [DOI] [PubMed] [Google Scholar]
- Thorpe SD, Charpentier M(2017) Highlight on the dynamic organization of the nucleus. Nucleus 8: 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Sridharan S, van Wietmarschen N, Maman Y, Callen E, Stanlie A, Wu W, Wu X, Day A, Wong N, et al. (2018) Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell 174: 1127–1142.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Liang C, Blitzblau HG, Smith HS, Gerbi SA(2002) A DNase I hypersensitive site flanks an origin of DNA replication and amplification in Sciara. Chromosoma 111: 291–303 [DOI] [PubMed] [Google Scholar]
- Valton A-L, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou J-F, Prioleau M-N(2014) G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J 33: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera DL, Madzima TF, Labonne JD, Alam MP, Hoffman GG, Girimurugan SB, Zhang J, McGinnis KM, Dennis JH, Bass HW(2014) Differential nuclease sensitivity profiling of chromatin reveals biochemical footprints coupled to gene expression and functional DNA elements in maize. Plant Cell 26: 3883–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara Z, Sequeira-Mendes J, Morata J, Peiró R, Hénaff E, Costas C, Casacuberta JM, Gutierrez C(2017) Retrotransposons are specified as DNA replication origins in the gene-poor regions of Arabidopsis heterochromatin. Nucleic Acids Res 45: 8358–8368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D(2015) Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 25: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear EE, Concia L, Brooks AM, Markham EA, Lee T-J, Allen GC, Thompson WF, Hanley-Bowdoin L(2016) Isolation of plant nuclei at defined cell cycle stages using edu labeling and flow cytometry In Caillaud M-C, ed, Plant Cell Division Methods in Molecular Biology, Vol 1370 Springer, New York, pp 69–86 [DOI] [PubMed] [Google Scholar]
- Wear EE, Song J, Zynda GJ, LeBlanc C, Lee T-J, Mickelson-Young L, Concia L, Mulvaney P, Szymanski ES, Allen GC, et al. (2017) Genomic analysis of the DNA replication timing program during mitotic S phase in maize (Zea mays) root tips. Plant Cell 29: 2126–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YQ, Liu GQ, Zhao XJ, Zhao HY, Cai L(2014) Genome-wide characterization and prediction of Arabidopsis thaliana replication origins. Biosystems 124: 1–6 [DOI] [PubMed] [Google Scholar]