Circadian rhythms play an important role in plant water use efficiency.
Abstract
In plants, water use efficiency (WUE) is a complex trait arising from numerous physiological and developmental characteristics. Here, we investigated the involvement of circadian regulation in long-term WUE in Arabidopsis (Arabidopsis thaliana) under light and dark conditions. Circadian rhythms are generated by the circadian oscillator, which provides a cellular measure of the time of day. In plants, the circadian oscillator contributes to the regulation of many aspects of physiology, including stomatal opening, rate of photosynthesis, carbohydrate metabolism, and developmental processes such as the initiation of flowering. We investigated the impact of the misregulation of numerous genes encoding various components of the circadian oscillator on whole plant, long-term WUE. From this analysis, we identified a role for the circadian oscillator in WUE. It appears that the circadian clock contributes to the control of transpiration and biomass accumulation. We also established that the circadian oscillator within guard cells can contribute to long-term WUE. Our experiments indicate that knowledge of circadian regulation will be important for developing crops with improved WUE.
World population growth is increasing the demand for fresh water for agriculture, with climate change predicted to exacerbate this competition for water resources (Ruggiero et al., 2017). One strategy to sustainably increase agricultural production involves the improvement of crop water use (Condon et al., 2004; Xoconostle-Cazares et al., 2010; Hu and Xiong, 2014; Ruggiero et al., 2017). Because up to 97% of water taken up from the soil by plants is lost through stomatal transpiration (Yoo et al., 2009; Na and Metzger, 2014), the manipulation of transpiration represents an excellent candidate for designing crops with increased water use efficiency (WUE; Bertolino et al., 2019).
Plant water loss can be manipulated through changes in the regulation of stomatal opening and by altering stomatal density and patterning (Pei et al., 1998; Hugouvieux et al., 2001; Schroeder et al., 2001; Hetherington and Woodward, 2003; Yoo et al., 2010; Lawson and Blatt, 2014; Franks et al., 2015; Caine et al., 2019). In addition to stomatal responses to environmental cues such as light, temperature, and phytohormones, there are circadian rhythms of stomatal opening (Gorton et al., 1989; Hennessey and Field, 1991). Circadian rhythms are self-sustaining biological cycles with a period of ∼24 h. These rhythms are thought to adapt plants to daily cycles of light and dark, by anticipating daily changes in the environment, and coordinating cellular processes. In land plants, circadian rhythms are generated by several interlocked transcription-translation feedback loops known as the circadian oscillator (Hsu and Harmer, 2014). The phase of the circadian oscillator is adjusted continuously to match the phase of the environment through the process of entrainment, in response to light, temperature, and metabolic cues (Somers et al., 1998a; Millar, 2004; Salomé and McClung, 2005; Haydon et al., 2013; Webb et al., 2019). Additionally, the circadian oscillator communicates an estimate of the time of day to circadian-regulated features of the cell, initially through transcriptional regulation (Harmer et al., 2000). The known circadian oscillator controls circadian rhythms of stomatal opening because mutations that alter the circadian period or cause circadian arrhythmia lead to equivalent alterations in the circadian rhythm of stomatal opening (Somers et al., 1998b; Dodd et al., 2004, 2005). The circadian oscillator is also involved in the responses of guard cells to environmental cues such as drought and low temperature (Dodd et al., 2006; Legnaioli et al., 2009).
Circadian rhythms are often studied under conditions of constant light. However, the circadian oscillator is also important for the regulation of stomatal opening under cycles of light and dark. For example, constitutive overexpression of the circadian oscillator component CIRCADIAN CLOCK ASSOCIATED1 (CCA1; CCA1-ox) alters the daily regulation of stomatal opening such that stomatal conductance increases steadily throughout the photoperiod (Dodd et al., 2005). In comparison, stomatal conductance in wild-type plants remains relatively uniform during the photoperiod and is lower than in CCA1-ox (Dodd et al., 2005). Similarly, guard cell-specific overexpression of CCA1 generally causes greater stomatal opening during the light period, and alters drought response phenotypes (Hassidim et al., 2017). Modeling suggests that under light/dark cycles, the circadian oscillator contributes at the canopy scale to daily rhythms in stomatal aperture and carbon assimilation in bean (Phaseolus vulgaris) and cotton (Gossypium hirsutum; Resco de Dios et al., 2016).
The contribution of the circadian oscillator to both stomatal opening and biomass accumulation (Dodd et al., 2005; Graf et al., 2010) suggests that the circadian oscillator might make an important contribution to long-term WUE. WUE is the ratio of carbon incorporated through photosynthesis into biomass to the amount of water lost through transpiration. At the single leaf level, instantaneous, intrinsic WUE is often measured with gas exchange techniques and expressed as net CO2 assimilation per unit of water transpired (Vialet-Chabrand et al., 2016; Ruggiero et al., 2017; Ferguson et al., 2018). However, such measurements do not provide an accurate representation of WUE over the plant lifetime, which is influenced by features such as leaf position, dark respiration, and time-of-day changes in instantaneous WUE (Condon et al., 2004; Tomás et al., 2014; Medrano et al., 2015; Ferguson et al., 2018). It is important to note that high WUE under well-watered conditions is not the same as drought resistance, because drought resistance relates to the capacity to maintain transpirational water supply under water-limited conditions through strategies such as expanded root systems (Blum, 2009). This means that WUE does not correlate reliably with drought resistance (Kobata et al., 1996).
Given that the circadian oscillator affects stomatal opening and biomass accumulation (Gorton et al., 1989; Hennessey and Field, 1991; Dodd et al., 2005; Graf et al., 2010; Edwards and Weinig, 2011; Edwards et al., 2012), we hypothesized that specific components of the circadian oscillator might make an important contribution to the long-term WUE of plants. Although the circadian oscillator influences stomatal opening and biomass accumulation, the influence of the circadian oscillator upon long-term WUE of plants remains unknown. This is an important question for understanding roles for circadian regulation in crops, because the long-term WUE ultimately determines the amount of water that is required for a given yield of the crop. Therefore, we investigated the impact of the misregulation of parts of the circadian oscillator upon the long-term WUE of Arabidopsis (Arabidopsis thaliana). We identified that the circadian oscillator has profound effects upon the long-term WUE of plants. Importantly, some alterations in oscillator function increase long-term WUE, suggesting potential targets for future improvements of crop WUE.
RESULTS
Circadian Oscillator Components Contribute to Long-Term WUE
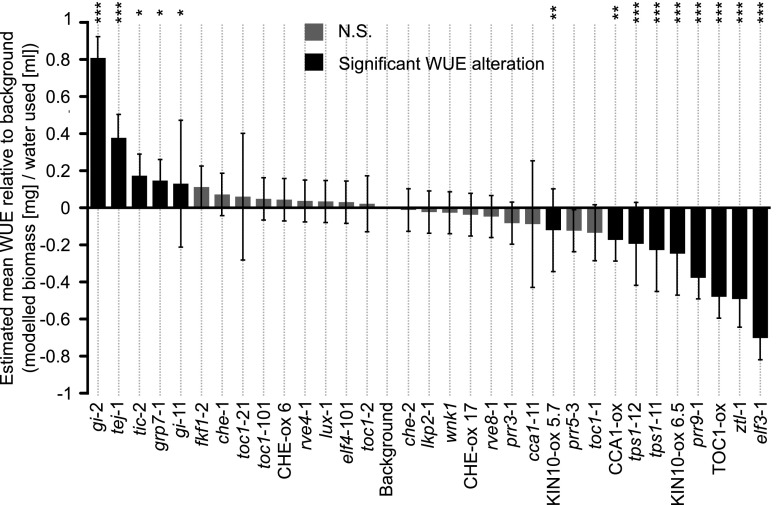
We identified that correct regulation of genes encoding circadian oscillator components makes an important contribution to WUE. Thirty-two single mutants or overexpressors of genes associated with circadian regulation, representing 21 circadian oscillator-associated components, were surveyed for WUE alterations (Fig. 1) using a method described in Wituszyńska et al. (2013). Of the mutants or overexpressors examined, 44% had a significantly different WUE from the wild type (Fig. 1; Table 1; Supplemental Fig. S1). Mutants of EARLY FLOWERING3 (elf3-1), PSEUDO-RESPONSE REGULATOR9 (prr9-1), TREHALOSE-6-PHOSPHATE SYNTHASE1 (tps1-11), tps1-12, and ZEITLUPE (ztl-1), as well as CCA1, TIMING OF CAB EXPRESSION1 (TOC1), and SNF1-RELATED PROTEIN KINASE1.1 (KIN10) overexpressors, had significantly lower WUE than the wild type (Fig. 1; Table 1; Supplemental Table S1). Mutants of GIGANTEA (gi-2), gi-11, GLY RICH PROTEIN7 (grp7-1), POLY(ADP-RIBOSE)GLYCOHYDROLASE1 (tej-1), and TIME FOR COFFEE (tic-2) had significantly greater WUE than the wild type (Fig. 1; Table 1; Supplemental Table S1). This suggests that misregulating the expression of circadian clock components CCA1, ELF3, GI, GRP7, PRR9, TEJ, TIC, TOC1, and ZTL can change whole plant long-term WUE (Fig. 1; Table 1). WUE was also altered by changing the expression of the energy signaling pathway components TPS1 and KIN10 that also provide inputs to the circadian oscillator (Fig. 1; Table 1; Supplemental Fig. S1; Shin et al., 2017; Frank et al., 2018). Overall, these data show that correct expression of a variety of circadian-clock–associated genes contributes to long-term WUE of Arabidopsis.
Figure 1.
The circadian clock regulates long-term WUE of Arabidopsis under light/dark cycles. Estimated WUE of each genotype relative to the estimated WUE of its background, derived from linear mixed effects models that combine all 18 separate batches of experimentation, each having five to 15 replicate plants per genotype. Statistical analysis was derived from pairwise post hoc comparisons between mutants and corresponding backgrounds, using the Kenward–Roger method for determining degrees of freedom and Tukey method for P-value adjustment (*P < 0.05; **P < 0.01; and ***P < 0.001; not significant [N.S.], P ≥ 0.05). Error bars, mean ± se, reflect modeled data, rather than the range of values sampled from the plants. Black bars, P < 0.05; gray bars, P ≥ 0.05.
Table 1. Genotypes with misregulated circadian clock-associated genes that have altered long-term WUE.
Findings are summarized from Figure 1 and derived from statistical significance within linear mixed effects models.
| Significantly Greater WUE than Background | Significantly Lower WUE than Background | No Significant WUE Alteration Compared to Background |
|---|---|---|
| gi-2 | CCA1-ox | cca1-11 |
| gi-11 | elf3-1 | che-1 |
| grp7-1 | KIN10-ox 5.7 | che-2 |
| tej-1 | KIN10-ox 6.5 | CHE-ox 6 |
| tic-2 | prr9-1 | CHE-ox 17 |
| TOC1-ox | elf4-101 | |
| tps1-11 | fkf1-2 | |
| tps1-12 | lkp2-1 | |
| ztl-1 | lux-1 | |
| prr3-1 | ||
| prr5-3 | ||
| rve4-1 | ||
| rve8-1 | ||
| toc1-1 | ||
| toc1-2 | ||
| toc1-21 | ||
| toc1-101 | ||
| wnk1 |
Within this experiment, each background accession had a distinct WUE (C24: 3.01 ± 0.07 mg mL−1; Col-0: 2.22 ± 0.02 mg mL−1; L. er.: 1.60 ± 0.04 mg mL−1; and Ws: 1.91 ± 0.06 mg mL−1; Supplemental Fig. S2). These differences between WUE of different Arabidopsis accessions are consistent with previous studies of WUE, stomatal function, and stomatal density in Arabidopsis (Nienhuis et al., 1994; Woodward et al., 2002; Dodd et al., 2004; Masle et al., 2005; Karaba et al., 2007; Ruggiero et al., 2017; Ferguson et al., 2018).
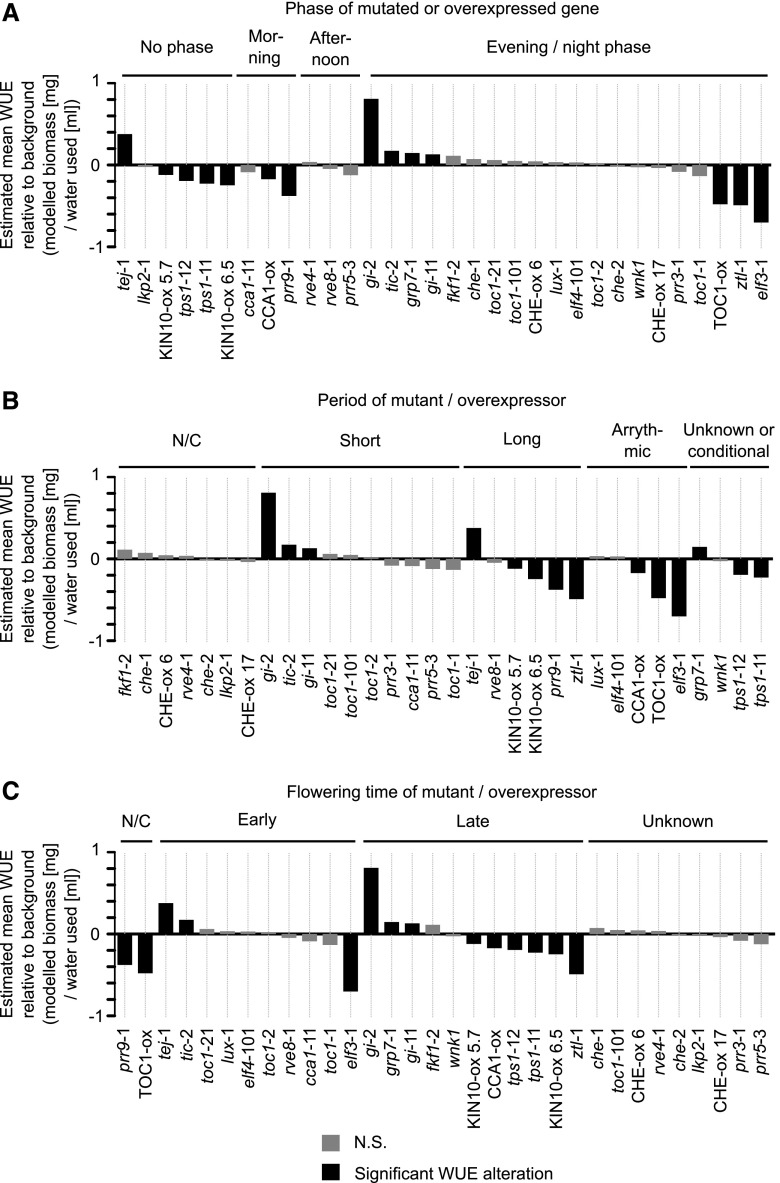
We hypothesized that variations in WUE might be associated with specific circadian phenotypes in the mutants and overexpressors that we tested. For example, mutations in circadian clock genes expressed with a particular phase (e.g. morning-expressed or evening-expressed genes) might have a more pronounced effect on WUE. Likewise, the nature of the circadian period change or flowering time change resulting from misexpression of each oscillator component might be associated with certain changes in WUE. To test this, we compared the data from our WUE screen with the circadian phase of expression of each mutated or overexpressed gene. We also compared the direction of change of WUE to the period and flowering time phenotypes that arise from each mutant or overexpressor (Hicks et al., 1996; Fowler et al., 1999; Schultz et al., 2001; Doyle et al., 2002; Nakamichi et al., 2002; Yanovsky and Kay, 2002; Imaizumi et al., 2003; Más et al., 2003; Murakami et al., 2004; Farré et al., 2005; Hazen et al., 2005; Baena-González et al., 2007; Ding et al., 2007a, 2007b; Niwa et al., 2007; Streitner et al., 2008; Wang et al., 2008; Baudry et al., 2010; Nakamichi et al., 2010; Rawat et al., 2011; Wahl et al., 2013; Hsu and Harmer, 2014). We note that the phenotypes reported by these studies were often identified under constant conditions, with flowering time experiments performed under short or long photoperiods, whereas our experiments occurred under cycles of 8-h light/16-h darkness.
There was no obvious relationship between the circadian phenotypes reported to arise from each mutant or overexpressor investigated and the WUE of each of these lines (Fig. 2). For example, mutating night-phased oscillator components can either decrease or increase WUE (Fig. 2A). Mutants that cause long circadian periods and short circadian periods can both increase and decrease WUE, although WUE was unaltered in the mutants that are reported to not alter the period (Fig. 2B). Furthermore, mutants and overexpressors that cause both early and delayed flowering can each increase and decrease WUE (Fig. 2C).
Figure 2.
Relationship between circadian clock-associated characteristics and WUE. A to C, Estimated WUE grouped according to phase of expression of each mutated or overexpressed gene (A), and the period (B) or flowering time (C) alteration caused by mutation or overexpression of each gene indicated. Shading of bars on graphs indicates statistical significance. Studies describing the phase of expression, period, and flowering time of the genotypes tested are identified in the main text. We note that the phase of expression and period data used for this analysis were often identified in previous studies under constant conditions, in contrast to our experiments under light/dark cycles. Estimated WUE relative to its corresponding background is derived from linear mixed effects models combining all 18 separate batches of experimentation, each having five to 15 replicate plants per genotype. Statistical analysis was derived from pairwise post hoc comparisons between mutants and corresponding backgrounds, using the Kenward–Roger method for determining degrees of freedom and Tukey method for P-value adjustment. Genotypes having P < 0.05 from this test are identified as significantly different from the background (black bars). N/C, no change; N.S., not significant.
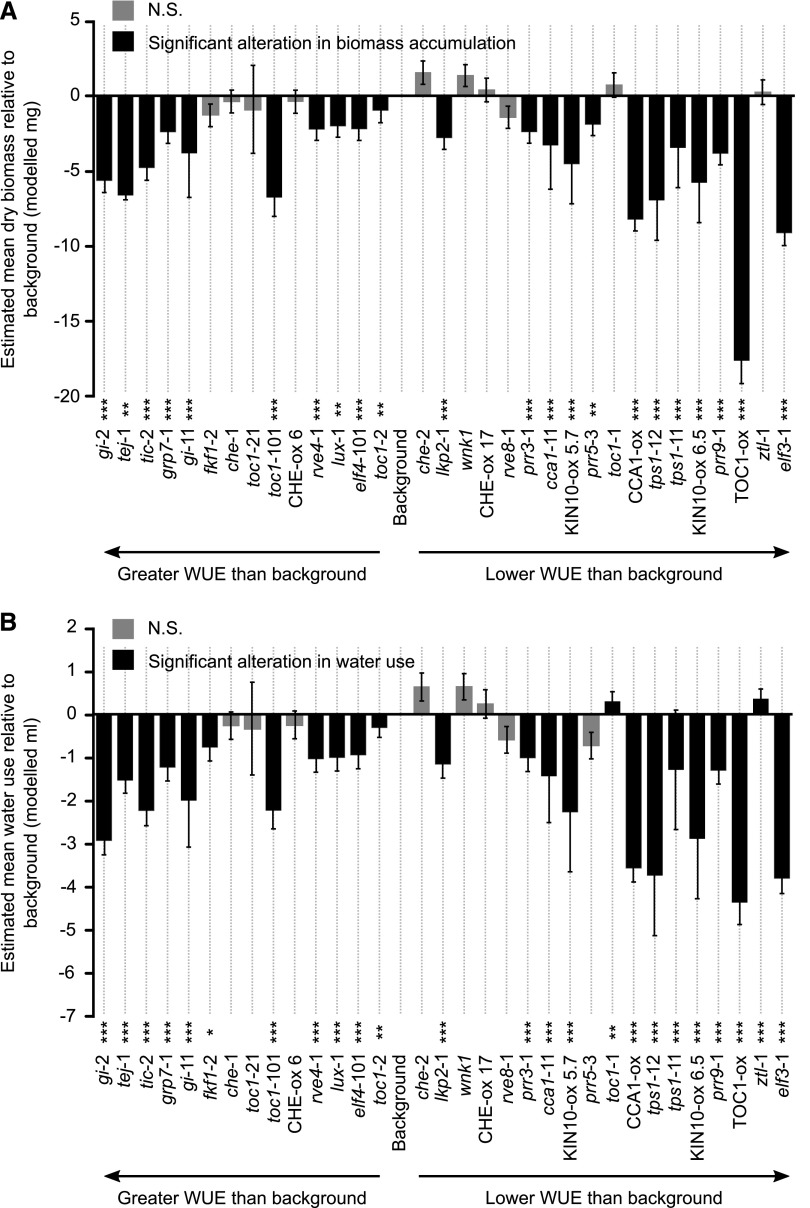
We were interested to determine whether the WUE alterations caused by misregulation of circadian oscillator gene expression arose from changes in either biomass accumulation or transpiration. Genotypes with low biomass accumulation generally had lower water use (Fig. 3). The results for tej-1 (Fig. 3) should be treated with caution because the model fits were very poor (Supplemental Table S1). No mutants or overexpressors tested increased the biomass relative to the corresponding background genotype (Fig. 3A). Together, this indicates that the altered WUE phenotypes in some genotypes with misregulated circadian clocks was not due to an alteration in just one of either water use or biomass accumulation (Fig. 3). Instead, the altered WUE of lines with misregulated circadian clock genes appears to be due to the net effect of altered biomass accumulation and altered transpiration in these genotypes (Fig. 3).
Figure 3.
Manipulating the expression of genes associated with circadian regulation alters estimated WUE by changing both water use and biomass accumulation. A and B, Biomass accumulation loss (A) and water loss (B) for each genotype relative to its respective background over the course of the experiments. Genotypes are ordered according to their WUE calculated in Figure 1. Data are derived from linear mixed effects models that combine all 18 separate batches of experimentation, each having five to 15 replicate plants per genotype. Statistical analysis was derived from pairwise post hoc comparisons between mutants and corresponding backgrounds, using the Kenward–Roger method for determining degrees of freedom and Tukey method for P-value adjustment (*P < 0.05; **P < 0.01; and ***P < 0.001; not significant [N.S.], P > 0.05). Note the model fit for the tej-1 mutant in POLY(ADP-RIBOSE)GLYCOHYDROLASE1 was poor (Supplemental Table S1). Genotypes are ordered from greatest (left) to lowest (right) estimated WUE. Error bars, mean ± se, reflect modeled data, rather than the range of values sampled from the plants.
Circadian Regulation of WUE Combines Multiple Traits
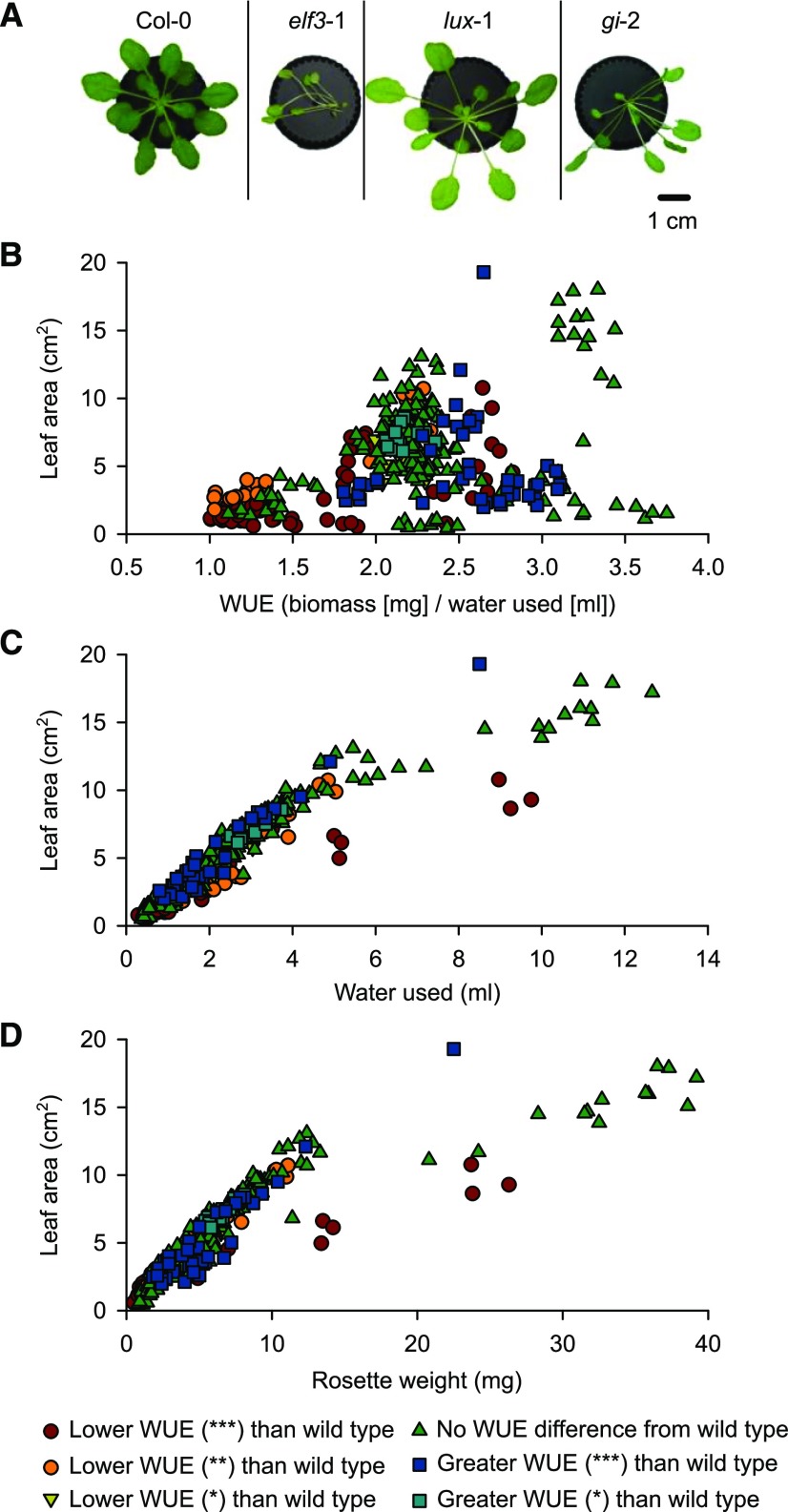
Mutation or overexpression of components of the circadian oscillator can cause changes in the development of Arabidopsis, such as alterations in rosette size, leaf shape, and petiole length (Fig. 4A; Zagotta et al., 1992; Schaffer et al., 1998; Wang and Tobin, 1998; Dodd et al., 2005; Ruts et al., 2012; Rubin et al., 2018). These changes are likely to have implications for gas exchange because, for example, spatially separated leaves are predicted to transpire more water (Bridge et al., 2013). We investigated whether the changes in WUE that were identified by our screen might arise from differences in rosette architecture between the circadian-clock–associated mutants and overexpressors and the corresponding backgrounds. There was a weak positive correlation between rosette leaf surface area and WUE (r = 0.400; r2 = 0.160; P < 0.001; Fig. 4B). Therefore, this suggests that ∼16% of variability in WUE can be explained by the variations in rosette leaf surface area that arise from misregulation of the circadian oscillator.
Figure 4.
The circadian oscillator alters WUE partially by changing rosette architecture. A, Altering circadian-associated gene expression can affect rosette architecture and size, as illustrated for mutants of elf3-1, lux-1, and gi-2 in the Col-0 background. Image backgrounds removed electronically for clarity. Vertical lines separate individual photographs that are organized into a single figure section. B to D, Variation in rosette leaf surface area across the genotypes investigated explained: 16% of variation in WUE (P < 0.001, r = 0.400, r2 = 0.160; B), 83% of variation in transpiration (P < 0.001, r = 0.912, r2 = 0.832; C), and 73% of variation in rosette dry biomass (P < 0.001, r = 0.857, r2 = 0.734; D). Data derive from 18 separate experimental batches, having five to 15 replicate plants per genotype per batch. For each genotype relative to its background, *P < 0.05; **P < 0.01; and ***P < 0.001. Correlation data (r2) were analyzed using Pearson correlation tests.
In comparison, rosette leaf surface area was strongly correlated with each of the individual parameters of water used and dry biomass accumulated. The variation in rosette surface area accounted for 83% of the variability in water transpired across the genotypes (Fig. 4C). Furthermore, the variation in rosette surface area accounted for 73% of the variability in biomass accumulation across the genotypes (Fig. 4D), which is unsurprising given that larger leaves are likely to contain more biomass.
This demonstrates that one way that circadian regulation affects WUE is through the influence of the circadian oscillator upon plant development and rosette architecture, but this variation in leaf area does not account for the majority of the influence of circadian regulation upon WUE. It also further supports the notion that the influence of the circadian oscillator upon WUE is complex, and cannot be explained by variation in one of water use or biomass accumulation alone.
Contribution of Circadian Regulation in Guard Cells to WUE
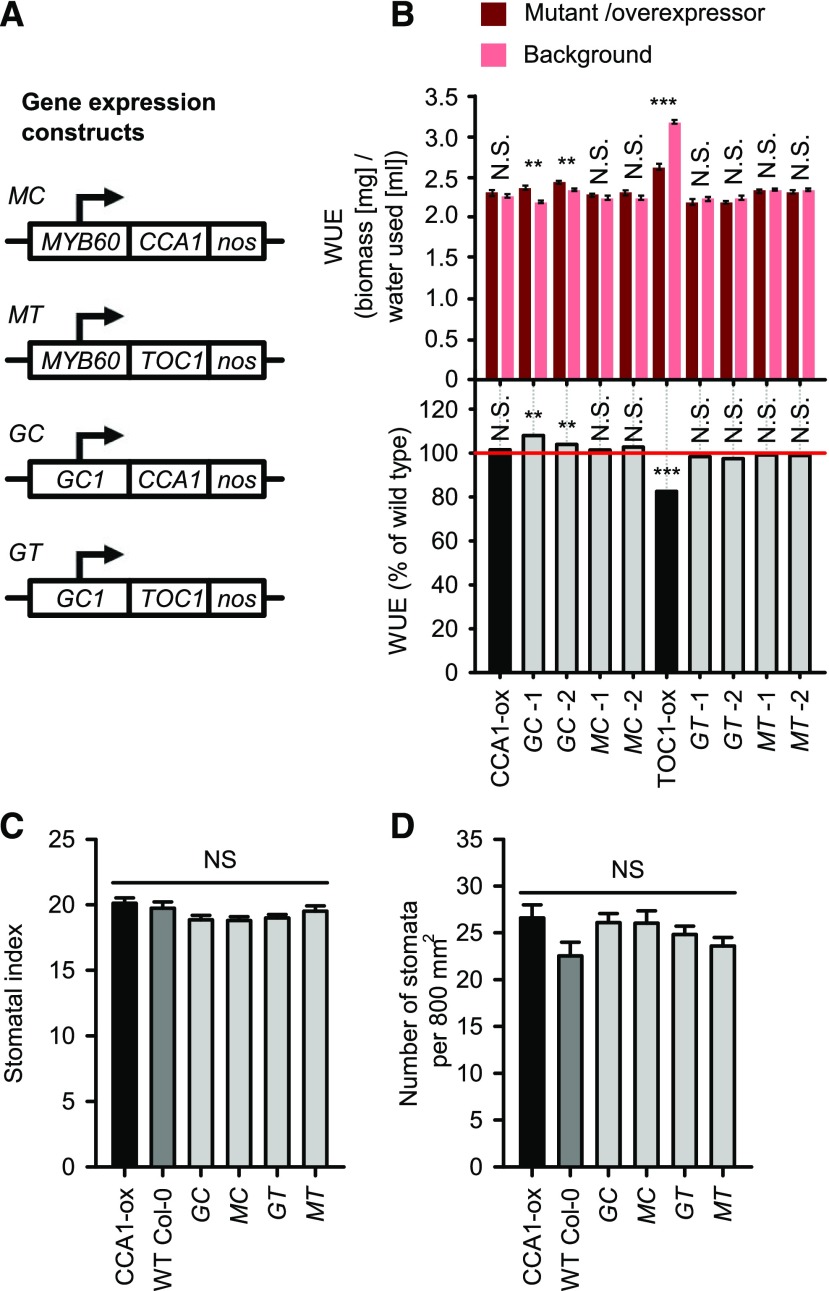
Next, we investigated whether the circadian oscillator within guard cells contributes to long-term WUE. There is evidence that guard cells contain a circadian oscillator that regulates stomatal opening (Gorton et al., 1989; Hassidim et al., 2017). To investigate the contribution of the guard cell circadian oscillator to WUE, we overexpressed two circadian oscillator components (CCA1 and TOC1) in guard cells, using two guard cell-specific promoters (the promoters of At1g22690 [GC1] and MYB DOMAIN PROTEIN60 [MYB60]) for each of CCA1 and TOC1 (Fig. 5A; Cominelli et al., 2005, 2011; Galbiati et al., 2008; Yang et al., 2008; Nagy et al., 2009; Meyer et al., 2010; Bauer et al., 2013; Rusconi et al., 2013). GC1 is a guard cell-specific promoter that is relatively unresponsive to a variety of environmental cues (cold, light, abscisic acid [ABA], gibberellin; Yang et al., 2008). We used the full-length MYB60 promoter sequence, because truncated and chimeric versions of this promoter appear to have weaker activity and/or become rapidly downregulated by dehydration and ABA (Francia et al., 2008; Cominelli et al., 2011; Rusconi et al., 2013). This produced four sets of transgenic lines: GC1::CCA1:nos (GC), GC1::TOC1:nos (GT), MYB60::CCA1:nos (MC), and MYB60::TOC1:nos (MT; Fig. 5A). We termed these guard cell-specific plants. We confirmed the guard cell specificity of the GC1 and MYB60 promoters in our hands, by driving GFP under the control of these promoters. GFP accumulation was restricted to the guard cells (Supplemental Fig. S3, A and B). There was not a circadian oscillation in the activity of either the GC1 or MYB60 promoter under our experimental conditions (Supplemental Fig. S3C), demonstrating that these promoters were appropriate for constitutive overexpression of circadian oscillator components within guard cells in our experiments.
Figure 5.
Long-term WUE can be altered by misexpression of circadian oscillator genes within stomatal guard cells. A, Constructs used to overexpress CCA1 or TOC1 coding sequences under the control of the At1g22690 (GC1) or MYB60 promoters. B, Guard cell CCA1 overexpression can increase WUE. WUE expressed as absolute WUE and percentage of the wild type (normalized to wild type as 100%, red reference line). Data were analyzed by one-way ANOVA (P < 0.001) followed by pairwise post hoc Tukey comparisons between transgenic lines and their corresponding backgrounds (**P < 0.01 and ***P < 0.001; not significant [N.S.], P > 0.05; n = 5–15; mean ± se). C and D, Guard cell CCA1 or TOC1 overexpression does not affect (C) stomatal index nor (D) stomatal density. Data were analyzed with ANOVA and Tukey’s post hoc tests (N.S. = P > 0.05; eight replicate plants per genotype with two images analyzed from each of two fully expanded leaves, giving a total of n = 19–32 images for analysis; mean ± se). Bar colors identify the whole plant overexpressor control (black), wild-type control (dark gray), and guard cell-specific overexpressor genotypes (light gray).
To further verify the guard cell-specific overexpression of CCA1 and TOC1 in the guard cell-specific plants, we examined CCA1 and TOC1 transcript accumulation within guard cells. Under constant light conditions, we measured CCA1 transcript accumulation in epidermal peels at dusk (when CCA1 transcript abundance is normally low in the wild type) and TOC1 transcript accumulation at dawn (when TOC1 transcript abundance is normally low in the wild type). Guard cell CCA1 overexpressors had greater CCA1 transcript abundance in epidermal peels at dusk than the wild type (Supplemental Fig. S3D), and guard cell TOC1 overexpressors had greater TOC1 transcript abundance at dawn than the wild type (Supplemental Fig. S3D). These data indicate that CCA1 and TOC1 were overexpressed within the guard cells of the guard cell-specific CCA1 or TOC1 overexpressor plants that we generated, respectively.
We investigated the effect on WUE of overexpression of CCA1 and TOC1 within guard cells. Two independent GC1::CCA1 lines (GC-1 and GC-2) were significantly more water-use efficient than the wild type (GC-1: P < 0.001; GC-2: P = 0.002; Fig. 5B). GC-1 and GC-2 were 8% and 4% more water-use efficient than the wild type, respectively (Fig. 5B). In comparison, two independent MYB60::CCA1 lines did not have greater WUE than the wild type (P > 0.05; Fig. 5B). This suggests that overexpressing CCA1 in guard cells can increase whole plant long-term WUE in a promoter-specific manner. Overexpression of TOC1 in guard cells with both the GC1 and MYB60 promoters did not alter WUE (P > 0.05; Fig. 5B). This suggests that decreased WUE in constitutive TOC1-ox plants (Figs. 1 and 5B) might not be explained by overexpression of TOC1 within the guard cells, and that this decreased WUE might instead be due to TOC1 overexpression in other cell types. In this particular experiment we did not see an alteration in CCA1-ox WUE relative to its background, contrasting Figure 1. This could suggest that the WUE alteration in CCA1-ox is fairly small or variable. Because the stomatal density was unaltered relative to the wild type in the guard cell overexpressors of CCA1 and TOC1 (Fig. 5, C and D), the WUE phenotypes that we identified from these lines might be caused by alterations in processes within guard cells, such as those regulating stomatal aperture, rather than altered stomatal density.
DISCUSSION
Pervasive Influence of the Circadian Oscillator upon WUE
Our data indicate that the circadian oscillator is important for regulating the long-term WUE of Arabidopsis. Misregulation of several functional subsections of the circadian oscillator altered the WUE. Misexpression of morning (PRR9 and CCA1), late day (GI), and evening (TOC1, ZTL, and ELF3) components of the circadian oscillator all perturb WUE under our experimental conditions (Fig. 1). Additionally, mutation of TEJ and GRP7 alters WUE (Fig. 1). Therefore, oscillator components that impact WUE are not confined to a specific expression phase or architectural feature (e.g. morning loop) within the multiloop circadian oscillator. Misexpression of genes encoding some proteins that provide environmental inputs to the circadian oscillator (ELF3, TPS1, ZTL, and KIN10; Covington et al., 2001; Kim et al., 2007; Shin et al., 2017; Frank et al., 2018) also alters WUE (Fig. 1). Together, this suggests that the entire circadian oscillator can influence WUE, and that alterations in water use that are caused by mutations to the circadian oscillator are not confined to a specific subloop of the circadian oscillator, or restricted to its input or output pathways. One explanation for these circadian-system–wide alterations in WUE relates to the nature of feedback within the circadian oscillator. The complex feedback and interconnectivity of the circadian oscillator means that individual components of the circadian oscillator that directly influence stomatal function or water use are likely to be altered by mutations that are distal to that component. Therefore, if correct circadian timing is required for optimum WUE, multiple components of the circadian oscillator are likely to influence WUE. Alternatively, because mutation of a number of components of the circadian oscillator had no effect upon WUE, it is possible that the oscillator components that influence WUE do so through roles in directly regulating outputs of the circadian oscillator, such as by regulating genes involved in stomatal function.
The sugar signaling proteins TPS1 and KIN10 influence a broad range of phenotypes, in addition to participating in circadian entrainment (Baena-González et al., 2007; Gómez et al., 2010; Paul et al., 2010; Delatte et al., 2011; Shin et al., 2017; Frank et al., 2018; Nietzsche et al., 2018; Simon et al., 2018a, 2018b). The tps1-12 TILLING mutant of TPS1 decreases stomatal aperture and increases the ABA sensitivity of guard cells (Gómez et al., 2010), whereas we found that tps1-11 and tps1-12 had lower long-term WUE than the wild type (Fig. 1). Lower biomass accumulation in tps1-11 and tps1-12 (Fig. 3A) was consistent with slow growth of these alleles (Gómez et al., 2010). Overall, this suggests that the decreased stomatal aperture of tps1-12 mutants (Gómez et al., 2010) does not translate into an overall increase in WUE, perhaps due to slower growth or misregulated ABA signaling in the tps mutants (Gómez et al., 2010). The broad range of phenotypes that are altered in tps1-11, tps1-12, and KIN10-ox indicates that these genotypes might alter WUE through mechanisms other than circadian regulation.
Potential Roles for the Evening Complex in WUE
Our finding that ELF3 can influence WUE (Fig. 1) is supported by previous evidence. Under constant light conditions, wild-type Arabidopsis has circadian rhythms of stomatal aperture, whereas elf3 stomata are constantly open and unresponsive to light and dark (Kinoshita et al., 2011). Furthermore, ELF3 negatively regulates blue-light–mediated stomatal opening (Kinoshita and Hayashi, 2011). Therefore, perturbation of the anticipation of day/night transitions or responses to environmental cues in elf3 stomata might cause long-term alterations in WUE.
ELF3 binds to the PRR9 promoter and elf3-1 has elevated PRR9 transcript abundance (Thines and Harmon, 2010; Dixon et al., 2011; Herrero et al., 2012). The low WUE of elf3-1 might potentially be caused by altered PRR9 expression, because misregulation of PRR9 also affected WUE (Fig. 1). In a similar fashion, ELF3/ELF4 signaling represses PRR7, and elf3-1 has elevated PRR7 transcript abundance (Herrero et al., 2012). Under light-dark cycles, elf3-1 also has high and constitutive GI expression (Fowler et al., 1999), and elf3-1 and gi mutants have opposite WUE phenotypes (Fig. 1). Therefore, the WUE phenotype of elf3-1 (Fig. 1) might be caused by disruption of ELF3 itself, or specific perturbations of PRR7, PRR9, and/or GI expression.
Mutating further components of the evening complex (EC; ELF4 and LUX ARRHYTHMO [LUX]) did not affect WUE (Fig. 1). This is despite these genes influencing circadian oscillator function and plant physiology (Hsu and Harmer, 2014; Huang and Nusinow, 2016), and nocturnal regulation of stomatal aperture altering WUE (Costa et al., 2015; Coupel-Ledru et al., 2016). One possibility is that the impact of elf3-1 on WUE may be greater than that of elf4 or lux because ELF3 is key to EC scaffolding, with ELF3 operating genetically downstream from ELF4 and LUX (Herrero et al., 2012; Huang and Nusinow, 2016).
The EC binds upstream of and regulates a variety of other genes that might also underlie the WUE alterations in elf3-1 mutants (Ezer et al., 2017). This includes regulators of growth, components of the photosynthetic apparatus, and genes associated with phytohormone signaling. This means that potential roles for the EC in WUE might occur through several physiological mechanisms. There also appears to be a negative relationship between temperature and EC promoter binding (Ezer et al., 2017), so it is possible that any influence of the EC upon WUE might be temperature-sensitive.
ELF4 appears to play a greater role in circadian regulation in the vascular tissue than stomatal guard cells, with vasculature expression up to 10 times higher than other tissues (Endo et al., 2014). Processes within the vasculature can affect WUE; for example, mutations in CELLULOSE SYNTHASE CATALYTIC SUBUNIT7 might impact water use through effects of the collapse of the vasculature upon guard cell size (Liang et al., 2010). Because elf3-1 affects WUE differently from elf4-101 and lux-1 (Fig. 1), ELF3 might regulate WUE independently from ELF4 and LUX.
Multiple Physiological Causes of Altered WUE in Circadian Oscillator Mutants
Our data suggest that changes in WUE caused by misexpression of circadian clock components might be due to a combination of physiological factors. Many mutants or overexpressors tested alter both biomass accumulation and water loss, often in the same direction (Fig. 3), so mutations to the circadian oscillator did not alter water use by specifically altering either carbon assimilation or transpiration. This is consistent with previous work demonstrating that both stomatal opening and CO2 fixation is perturbed in circadian arrhythmic plants under light/dark cycles (Dodd et al., 2005), and also with the finding that daily carbohydrate management is dependent upon correct circadian regulation (Graf et al., 2010). We speculate that delayed or advanced stomatal and photosynthetic responses to the day–night cycle might occur in circadian period mutants, because period mutants inaccurately anticipate the onset of dawn (Dodd et al., 2014). Circadian clock mutants might also affect WUE by changing the sensitivity of stomatal movements and photosynthesis to environmental transitions, because there is circadian gating of the responses of both stomata and photosynthesis to environmental cues (Dodd et al., 2006; Kinoshita et al., 2011; Litthauer et al., 2015; Joo et al., 2017; Cano-Ramirez et al., 2018). Some effects of the circadian oscillator upon WUE arise from alterations in leaf size that occur in some circadian oscillator mutants (Fig. 4, A and B). This suggests that developmental alterations arising from lesions in the circadian oscillator can lead to changes in WUE. Such developmental alterations might alter WUE by changing airflow around the rosette, boundary layer conductance, or internal leaf structure.
It has been reported previously that during the light period of light/dark cycles, CCA1-ox has greater stomatal conductance than the wild type and decreased CO2 assimilation and biomass accumulation (Dodd et al., 2005; Graf et al., 2010). If these alterations in growth, CO2 fixation, and transpiration persist throughout the vegetative growth phase, it might be predicted that CCA1-ox would have lower long-term WUE than the wild type. We found that both biomass accumulation and water loss were reduced significantly in CCA1-ox relative to its background (Fig. 3), with the ratio between the two parameters indicating also a significant decrease in WUE of CCA1-ox (Fig. 1). This might be due to alterations in gas exchange reported in Dodd et al. (2005), and also other developmental changes caused by CCA1 overexpression.
Contribution of Circadian Regulation in Guard Cells to WUE
We also investigated whether the circadian oscillator within guard cells contributes to long-term WUE. This involved overexpressing two circadian clock genes in guard cells using two different guard cell-specific promoters. Comparable approaches have been adopted to investigate roles of specific cell types in the functioning of the circadian system and their relationships with physiology and development (Endo et al., 2014; Shimizu et al., 2015; Hassidim et al., 2017). Under our experimental conditions, we did not identify consistent alterations in the long-term WUE of seedlings overexpressing CCA1 or TOC1 in stomatal guard cells (Fig. 5B). This could indicate that decreased long-term WUE of TOC1-ox plants (Fig. 1) arises from altered circadian regulation in cell types other than guard cells. While two lines harboring a GC1::CCA1 construct had greater WUE than the wild type, WUE was unaltered in comparable lines harboring MYB60::CCA1 (Fig. 5B). The differing WUE phenotype of GC1::CCA1 and MYB60::CCA1 might be explained by differences in promoter strength, because the GC1 promoter appears to have somewhat greater activity than the MYB60 promoter (Supplemental Fig. S3, D and E). Although both promoters are guard cell-specific in our hands (Supplemental Fig. S3), we cannot exclude the possibility of ectopic promoter activity.
Interestingly, GC1::CCA1 is reported to have greater drought sensitivity of long-term biomass accumulation than the wild type (Hassidim et al., 2017), whereas we found that GC1::CCA1 had greater WUE than the wild type (Fig. 5B). This might reflect the integration of circadian regulation into ABA signaling (Legnaioli et al., 2009; Robertson et al., 2009), or occur because guard cell circadian regulation is required for correct guard cell metabolism and/or stomatal movements under conditions of abiotic stress. For example, circadian regulation is proposed to participate in daily cycles of triacylglycerol mobilization that are important for stomatal opening (McLachlan et al., 2016). Together, these findings suggest that guard cell circadian regulation is important under both well-watered conditions and conditions of environmental stress (Fig. 5C; Robertson et al., 2009; Hassidim et al., 2017), with circadian regulation in other tissues also contributing to overall WUE. It would be informative in future to perform reverse genetic screening of the dehydration tolerance or long-term drought tolerance of sets of circadian clock mutants. However, because well-watered WUE is not a drought-tolerance trait (Blum, 2009), it possible that different circadian clock mutant alleles might confer dehydration or drought tolerance compared with those alleles that alter WUE (Fig. 1).
CONCLUSION
We show that circadian regulation contributes to whole plant long-term WUE under cycles of day and night. This control occurs partly through the influence of components of the circadian oscillator upon rosette architecture. Mutation or overexpression of CCA1, TOC1, ELF3, GI, GRP7, PRR9, TEJ, TIC, and ZTL altered WUE under our experimental conditions. The roles of these genes in WUE may be independent or overlapping, and their WUE phenotypes might be due to direct effects of these genes, or indirect effects on transcript and/or protein abundance of other circadian clock gene(s). Misregulation of the expression of CCA1 HIKING EXPEDITION (CHE), FLAVIN-BINDING KELCH REPEAT F-BOX1 (FKF1), LOV KELCH PROTEIN2 (LKP2), REVEILLE4 (RVE4), RVE8, PRR3, PRR5, ELF4, LUX, and WITH NO LYS KINASE1 (WNK1) did not appear to alter WUE under our experimental conditions.
Our results have broad implications. First, our data suggest that alterations in circadian function that arise during crop breeding could have the potential to alter WUE; therefore, manipulation of the functioning of the circadian oscillator might represent a pathway to tune the WUE of crops. Second, our results indicate that circadian regulation in a single cell type can have implications for whole-plant physiology. Third, our experiments with guard-cell misregulation of circadian oscillator genes suggest that the circadian oscillator within guard cells and other cell types influences WUE. Fourth, our findings suggest that circadian regulation potentially alters a single trait (WUE) by affecting many aspects of physiology, along with leaf area. Overall, our study demonstrates that the circadian oscillator is important for the WUE of Arabidopsis plants during their entire vegetative growth period. In future, it will be informative to distinguish the contribution to overall WUE of circadian regulation within additional cell types, such as the mesophyll, vascular tissue, and root cell types. It will also be important to identify specific mechanisms underlying the WUE phenotypes, and determine the extent to which these findings scale to crop species.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface-sterilized as described in Noordally et al. (2013). For experiments investigating stomatal density and index, seeds were stratified for 3 d at 4°C, then sown on compost mix comprising a 3:1 ratio of coarsely sieved Levington Advance F2 seed compost (Everris) and horticultural silver sand (Melcourt), supplemented with 0.4 g L−1 thiacloprid insecticide granules (Exemptor; Everris). Seedlings were germinated in controlled environment chambers (Reftech, Netherlands) under an 8-h photoperiod at 70% humidity, 20°C, and photon flux density of 100 μmol m−2 s−1 of overhead lighting supplied by cool-white fluorescent tubes (Reftech, Netherlands). For experiments investigating long-term WUE, seeds were sown within a custom Falcon tube system, then stratified. The genotypes that were screened for WUE alterations are identified in Supplemental Table S2 (Zagotta et al., 1992; Millar et al., 1995; Wang and Tobin, 1998; Fowler et al., 1999; Somers et al., 2000; Strayer et al., 2000; Panda et al., 2002; Alonso et al., 2003; Eriksson et al., 2003; Hall et al., 2003; Imaizumi et al., 2003, 2005; Khanna et al., 2003; Más et al., 2003; Michael et al., 2003; Hazen et al., 2005; Kikis et al., 2005; Baena-González et al., 2007; Ding et al., 2007a, 2007b; Pruneda-Paz et al., 2009; Gómez et al., 2010; Rawat et al., 2011). For all experiments, at least two completely independent experimental repeats were performed per genotype and per treatment, with multiple replicate plants within each of the experimental repeats.
Generation of Transgenic Lines
To create the GC1::CCA1:nos (GC), GC1::TOC1:nos (GT), MYB60::CCA1:nos (MC), and MYB60::TOC1:nos (MT) constructs, the CaMV nos terminator sequence was ligated between the SpeI and NotI restriction sites in the pGREENII0229 binary vector (Hellens et al., 2000). The GC1 upstream sequence (−1,894 to −190) or MYB60 upstream sequence (−1,724 to −429) was then ligated between the KpnI and ApaI restriction sites of pGREENII0229. Finally, the CCA1 coding sequence or TOC1 coding sequence, obtained using reverse transcription PCR (RT-PCR), was ligated between the restriction XhoI and XmaI sites. Primers used are identified in Supplemental Table S3. Constructs were transformed into Col-0 wild-type Arabidopsis using transformation with Agrobacterium tumefaciens strain GV3101. Transformants were identified by screening for phosphinothricin resistance, and then further validated using genomic DNA PCR. Homozygous lines were identified via phosphinothricin (BASTA; Bayer Crop Science) resistance, and two independently transformed homozygous lines were investigated in detail per genotype.
Guard cell specificity of promoter activity was investigated using GC1::GFP:nos and MYB60::GFP:nos promoter-reporter lines (Supplemental Fig. S3, A–C), which were created as above with the GFP coding sequence ligated between the XhoI and XmaI restriction sites. Leaf discs (5 mm diameter) from seedlings or mature plants were mounted on microscope slides with distilled water, and examined for GFP fluorescence using confocal microscopy (DMI6000; Leica). The following settings were used: argon laser at 20% capacity; 488-nm laser at 48% capacity with a bandwidth of 505 to 515 nm; gain of 1,250; offset at 0.2%; 20× or 40× objective; zoom ×1 to ×4.
Measurement of WUE
The WUE assay was adapted from Wituszyńska et al. (2013). Plants were grown for 6 weeks in modified 50-mL Falcon tubes, under an 8-h photoperiod at 70% humidity, 20°C, and photon flux density of 100 μmol m−2 s−1 of overhead lighting supplied by cool-white fluorescent tubes (Reftech). The Falcon tube systems consisted of a 50-mL Falcon tube filled with 37.5 mL of a 1:1 ratio of compost, consisting of perlite and 35 mL of Milli-Q water (Merck), with the remaining volume filled with a 1:1 ratio of compost and Milli-Q water (Supplemental Fig. S4). Each Falcon tube lid had a 2-mm–diameter hole drilled in its center to allow plant growth. The lid was spray-painted black (Hycote) because we found that the orange color of the Falcon tube lid caused leaf curling (Supplemental Fig. S4). The system was wrapped in aluminum foil to exclude light (Supplemental Fig. S4). Ten to 15 seeds were sown through the Falcon tube lid using a pipette. After stratification, Falcon tube systems were placed under growth conditions using a randomized experimental design. Seven d after germination, seedlings were thinned to one per Falcon tube system, and initial Falcon tube weight was recorded. The seedling-thinning step was sensitive to seedling damage for genotypes with substantially altered morphologies (e.g. tps1 mutants), reducing the number of replicates available for some genotypes. After 6 weeks of growth, rosette leaf surface area was measured by photography (D50; Nikon) and Fiji software (imagej.net/Fiji), rosette dry weight was measured (4 d at 60°C), and final Falcon tube weight was recorded. All experiments were stopped before flowering occurred, with the 8-h photoperiod being used to delay flowering as much as possible. Plants were not obviously stressed during the experiment (e.g. leaves did not become purple due to strong anthocyanin accumulation, and plants did not wilt or become contaminated with mildew; Supplemental Fig. S4). Negative controls (Falcon tube systems without plants) were used to assess soil water evaporation over 18 experimental batches, with an overall mean weight loss of 0.513 g ± 0.004 g over 6 weeks for plant-free Falcon tubes.
Plant WUE was calculated as follows:
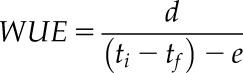 |
where d is the rosette dry weight at the end of the experiment (mg); ti and tf are the Falcon tube weight at the start and end of the experiment, respectively (g); and e is the amount of water evaporation directly from the compost (g). WUE is derived as mg biomass per mL−1 water lost. These calculations assumed that 1 g of weight change was equivalent to a change of 1 mL of water. For examination of individual batches of data (Fig. 1), the WUE of each circadian oscillator genotype was normalized to its respective background to control for variation in the WUE of each background accession and expressed as a percentage of that background. Statistical comparisons with the background lines occurred before normalization. For quantitative investigation of the entire dataset, linear mixed effects models were used (below).
Measurement of Stomatal Density
Plants were grown for 7 to 8 weeks on compost mix. Dental paste (Coltene) was applied to the abaxial surface of fully expanded leaves. Transparent nail varnish (Rimmel) was applied to these leaf molds once they had set, and then peeled away from the mold using clear adhesive tape (Scotch Crystal). Stomatal and pavement cells were counted within an 800 × 800 μm square at the center of each leaf half, using an epifluorescence microscope (HAL100; Zeiss) and Volocity (Perkin Elmer) and Fiji software. For each experimental repeat, two leaves were sampled per plant and eight plants sampled per genotype. Stomatal index was calculated as follows:
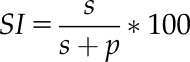 |
where SI is the stomatal index; s is the number of stomata in the field of view (800 × 800 µm); and p is the number of pavement cells in the field of view.
RNA Extraction and RT Quantitative PCR
RNA extractions, complementary DNA synthesis, and RT quantitative (q) PCR were performed according to Simon et al. (2018a), except that ∼10 seedlings were used per RNA sample and analysis was performed using an MXPro 3005 real-time PCR system (Agilent Technologies) with 5× HOT FIREPol EvaGreen qPCR Mastermix (Solis Biodyne). RT-qPCR primers are provided in Supplemental Table S4. Rhythmic features within qPCR data were identified with the BioDare2 platform (Zielinski et al., 2014), using the Fast Fourier Transform Non-Linear Least Squares method. One independently-transformed line of each guard cell-specific circadian clock gene overexpressor was also investigated using RT-qPCR conducted on RNA isolated from epidermal peels. Abaxial leaf epidermis was detached, then washed in 10 mm MES (pH 6.15, adjusted using 10 m KOH) to remove RNA derived from ruptured epidermal cells. Each RNA sample was derived from 20 epidermal peels (five plants, four leaves per plant) that were collated and flash-frozen in liquid nitrogen. Guard cell RNA was extracted using the RNeasy UCP Micro Kit (Qiagen) according to manufacturer’s instructions, with the following modification: guard cell lysis was performed by adding glass beads (425–600-μm diameter, acid-washed, from Sigma-Aldrich) and 350 μL of RULT buffer to the sample, then vortexed for 5 min.
Data Analysis
Experiments were conducted in a series of 18 separate experimental batches, each including a set of mutants and their corresponding backgrounds. This subdivision ensured high quality experimental attention to each replicate within a large-scale study, and the management of plant growth space. To investigate the nature of any differences between the mutants and their backgrounds in a manner that accounted for experimental batch variation in the backgrounds, we used a linear mixed effects modeling approach. Because there are differences between the WUE of Arabidopsis background accessions (Nienhuis et al., 1994; Woodward et al., 2002; Dodd et al., 2004; Masle et al., 2005; Karaba et al., 2007; Ruggiero et al., 2017; Ferguson et al., 2018; Supplemental Fig. S2), separate models were generated for each background to avoid comparing accessions with unequal underlying WUE. WUE data were analyzed by fitting a linear mixed models using the software package lme4 (R package lme4 v1.1-21; Bates et al., 2015) within the R statistical computing platform v3.6.2 (R Core Team, 2019). Using the lmer function, “Mutant” was assigned as a fixed effect (xf) and the “Batch” as a random effect (xr) to test for the effect of the mutations on the physiological parameters, while controlling for differences between batches. Separate models for each background accession were created with R code:
For the dataset from each Arabidopsis background, diagnostic residual plots suggested that the model fits were appropriate for larger datasets (Columbia-0, C24, Landsberg erecta, and Ws), and for analytical consistency the same model was applied across the entire dataset. Genotypes within two experimental batches (including toc1-101 and TOC1-ox) were analyzed within a separate model because the backgrounds had rather greater water loss and biomass accumulation than the other experimental batches, so were incorporated into a separate model to obtain the best possible fit. Conditional R2 was obtained using the r.squaredGLMM function in the software package MuMln v1.43.15 (https://CRAN.R-project.org/package=MuMIn; Nakagawa and Schielzeth, 2013). The emmeans function (previously “lsmeans”; Searle et al., 1980; https://CRAN.R-project.org/package=emmeans; v1.4.3.01) was used to subsequently obtain an estimated marginal mean for each mutant, and conduct post hoc pairwise comparisons between mutant and its corresponding background, using the Kenward–Roger method for determining degrees of freedom and Tukey method for P-value adjustment:
With this analysis, this output from group comparisons indicate the statistical significance of any differences of the modeled mutants from the modeled means. This is indicated where relevant on the figures.
Accession Numbers
Arabidopsis Genome Initiative identifiers for the genes mentioned in this study are: CCA1 (At2g46830), CHE (At5g08330), ELF3 (At2g25930), ELF4 (At2g40080), FKF1 (At1g68050), GC1 (At1g22690), GI (At1g22770), GRP7 (At2g21660), KIN10 (At3g01090), LKP2 (At2g18915), LUX (At3g46640), MYB60 (At1g08810), PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2AA3, At1g13320), PRR3 (At5g60100), PRR5 (At5g24470), PRR9 (At2g46790), RVE4 (At5g02840), TEJ (At2g31870), TIC (At3gt22380), TOC1, At5g61380), TPS1 (At1g78580), WNK1 (At3g04910), ZTL (At5g57360).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The circadian clock regulates long-term WUE of Arabidopsis under light/dark cycles.
Supplemental Figure S2. Differences in the WUE of background accessions.
Supplemental Figure S3. Genotyping of plants with guard cell-specific overexpression of CCA1 or TOC1.
Supplemental Figure S4. Assay used to measure long-term whole plant WUE in Arabidopsis.
Supplemental Table S1. Quality of fit of linear mixed effects models to experimental data.
Supplemental Table S2. Arabidopsis genotypes screened for WUE.
Supplemental Table S3. Primers used for cloning.
Supplemental Table S4. Primers used for RT-qPCR.
Acknowledgments
We thank Kester Cragg-Barber, James Chen, Ioanna Kostaki, Jean-Charles Isner, Deirdre McLachlan, Peng Sun, Ashutosh Sharma, and Dora Cano-Ramirez for technical advice during experimentation. We thank Marc Knight, Tracy Lawson, and Steven Penfield for discussions and advice concerning data interpretation and analysis. We thank Keara Franklin, Alex Webb, Paloma Mas, Steve Kay, Isabelle Carre, Takato Imaizumi, Filip Rolland, Ian Graham, Stacey Harmer, and Steven Penfield for donating seed lines.
Footnotes
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (grant nos. BB/J014400/1, BB/M009122/1, and GEN BB/P013511/1) and the Wolfson Foundation.
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J(2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S(2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67: 48 [Google Scholar]
- Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua N-H, Tobin EM, et al. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Bertolino LT, Caine RS, Gray JE(2019) Impact of stomatal density and morphology on water-use efficiency in a changing world. Front Plant Sci 10: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A.(2009) Effective use of water (EUW) and not water-use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Res 112: 119–123 [Google Scholar]
- Bridge LJ, Franklin KA, Homer ME(2013) Impact of plant shoot architecture on leaf cooling: A coupled heat and mass transfer model. J R Soc Interface 10: 20130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caine RS, Yin X, Sloan J, Harrison EL, Mohammed U, Fulton T, Biswal AK, Dionora J, Chater CC, Coe RA, et al. (2019) Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol 221: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Ramirez DL, Saskia de Fraine T, Griffiths OG, Dodd AN(2018) Photosynthesis and circadian rhythms regulate the buoyancy of Marimo Lake balls. Curr Biol 28: R869–R870 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G, Tonelli C(2011) DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol 11: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C(2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Condon AG, Richards RA, Rebetzke GJ, Farquhar GD(2004) Breeding for high water-use efficiency. J Exp Bot 55: 2447–2460 [DOI] [PubMed] [Google Scholar]
- Costa JM, Monnet F, Jannaud D, Leonhardt N, Ksas B, Reiter IM, Pantin F, Genty B(2015) Open All Night Long: The dark side of stomatal control. Plant Physiol 167: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupel-Ledru A, Lebon E, Christophe A, Gallo A, Gago P, Pantin F, Doligez A, Simonneau T(2016) Reduced nighttime transpiration is a relevant breeding target for high water-use efficiency in grapevine. Proc Natl Acad Sci USA 113: 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA(2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese-Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H(2011) Growth arrest by trehalose-6-phosphate: An astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiol 157: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Doyle MR, Amasino RM, Davis SJ(2007a) A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Millar AJ, Davis AM, Davis SJ(2007b) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19: 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ(2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Dalchau N, Gardner MJ, Baek S-J, Webb AAR(2014) The circadian clock has transient plasticity of period and is required for timing of nocturnal processes in Arabidopsis. New Phytol 201: 168–179 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Jakobsen MK, Baker AJ, Telzerow A, Hou S-W, Laplaze L, Barrot L, Poethig RS, Haseloff J, Webb AAR(2006) Time of day modulates low-temperature Ca signals in Arabidopsis. Plant J 48: 962–973 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Parkinson K, Webb AAR(2004) Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol 162: 63–70 [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR(2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM(2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Edwards CE, Ewers BE, McClung CR, Lou P, Weinig C(2012) Quantitative variation in water-use efficiency across water regimes and its relationship with circadian, vegetative, reproductive, and leaf gas-exchange traits. Mol Plant 5: 653–668 [DOI] [PubMed] [Google Scholar]
- Edwards CE, Weinig C(2011) The quantitative-genetic and QTL architecture of trait integration and modularity in Brassica rapa across simulated seasonal settings. Heredity 106: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA(2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ(2003) Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218: 159–162 [DOI] [PubMed] [Google Scholar]
- Ezer D, Jung J-H, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017) The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA(2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Ferguson JN, Humphry M, Lawson T, Brendel O, Bechtold U(2018) Natural variation of life-history traits, water use, and drought responses in Arabidopsis. Plant Direct 2: e00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J(1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia P, Simoni L, Cominelli E, Tonelli C, Galbiati M(2008) Gene trap-based identification of a guard cell promoter in Arabidopsis. Plant Signal Behav 3: 684–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano-Ramirez DL, Chembath A, et al. (2018) Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr Biol 28: 2597–2606.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks PJW, W Doheny-Adams T, Britton-Harper ZJ, Gray JE(2015) Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol 207: 188–195 [DOI] [PubMed] [Google Scholar]
- Galbiati M, Simoni L, Pavesi G, Cominelli E, Francia P, Vavasseur A, Nelson T, Bevan M, Tonelli C(2008) Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. Plant J 53: 750–762 [DOI] [PubMed] [Google Scholar]
- Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA(2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64: 1–13 [DOI] [PubMed] [Google Scholar]
- Gorton HL, Williams WE, Binns ME, Gemmell CN, Leheny EA, Shepherd AC(1989) Circadian stomatal rhythms in epidermal peels from Vicia faba. Plant Physiol 90: 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM(2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al. (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA(2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hassidim M, Dakhiya Y, Turjeman A, Hussien D, Shor E, Anidjar A, Goldberg K, Green RM(2017) CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the circadian control of stomatal aperture. Plant Physiol 175: 1864–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AAR(2013) Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502: 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA(2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM(2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hennessey TL, Field CB(1991) Circadian rhythms in photosynthesis. Plant Physiol 96: 831–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM, Woodward FI(2003) The role of stomata in sensing and driving environmental change. Nature 424: 901–908 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA(1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL(2014) Wheels within wheels: The plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L(2014) Genetic engineering and breeding of drought-resistant crops. Annu Rev Plant Biol 65: 715–741 [DOI] [PubMed] [Google Scholar]
- Huang H, Nusinow DA(2016) Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet 32: 674–686 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI(2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA(2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA(2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Joo Y, Fragoso V, Yon F, Baldwin IT, Kim S-G(2017) Circadian clock component, LHY, tells a plant when to respond photosynthetically to light in nature. J Integr Plant Biol 59: 572–587 [DOI] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, Krishnan A, Nataraja KN, Udayakumar M, Pereira A(2007) Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci USA 104: 15270–15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kikis EA, Quail PH(2003) EARLY FLOWERING 4 functions in phytochrome B-regulated seedling de-etiolation. Plant Physiol 133: 1530–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH(2005) ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim W-Y, Fujiwara S, Suh S-S, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE(2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Hayashi Y(2011) New insights into the regulation of stomatal opening by blue light and plasma membrane H+-ATPase In Jeon KW, ed, International Review of Cell and Molecular Biology, Vol Vol 289 Academic Press, Cambridge, MA, pp 89–115 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Ono N, Hayashi Y, Morimoto S, Nakamura S, Soda M, Kato Y, Ohnishi M, Nakano T, Inoue S, et al. (2011) FLOWERING LOCUS T regulates stomatal opening. Curr Biol 21: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Kobata T, Okuno T, Yamamoto Y(1996) Contributions of capacity for soil water extraction and water use efficiency to maintenance of dry matter production in rice subjected to drought. Jpn J Crop Sci 65: 652–662 [Google Scholar]
- Lawson T, Blatt MR(2014) Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164: 1556–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P(2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YK, Xie X, Lindsay SE, Wang YB, Masle J, Williamson L, Leyser O, Hetherington AM(2010) Cell wall composition contributes to the control of transpiration efficiency in Arabidopsis thaliana. Plant J 64: 679–686 [DOI] [PubMed] [Google Scholar]
- Litthauer S, Battle MW, Lawson T, Jones MA(2015) Phototropins maintain robust circadian oscillation of PSII operating efficiency under blue light. Plant J 83: 1034–1045 [DOI] [PubMed] [Google Scholar]
- Más P, Kim W-Y, Somers DE, Kay SA(2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD(2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436: 866–870 [DOI] [PubMed] [Google Scholar]
- McLachlan DH, Lan J, Geilfus C-M, Dodd AN, Larson T, Baker A, Hõrak H, Kollist H, He Z, Graham I, et al. (2016) The breakdown of stored triacylglycerols is required during light-induced stomatal opening. Curr Biol 26: 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano H, Tomás M, Martorell S, Flexas J, Hernández E, Rosselló J, Pou A, Escalona J-M, Bota J(2015) From leaf to whole-plant water use efficiency (WUE) in complex canopies: Limitations of leaf WUE as a selection target. Crop J 3: 220–228 [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid KAS, Geiger D, Marten I, Martinoia E, Hedrich R(2010) AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR(2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ.(2004) Input signals to the plant circadian clock. J Exp Bot 55: 277–283 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA(1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Murakami M, Yamashino T, Mizuno T(2004) Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 45: 645–650 [DOI] [PubMed] [Google Scholar]
- Na J-K, Metzger JD(2014) Chimeric promoter mediates guard cell-specific gene expression in tobacco under water deficit. Biotechnol Lett 36: 1893–1899 [DOI] [PubMed] [Google Scholar]
- Nagy R, Grob H, Weder B, Green P, Klein M, Frelet-Barrand A, Schjoerring JK, Brearley C, Martinoia E(2009) The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J Biol Chem 284: 33614–33622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H(2013) A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4: 133–142 [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H(2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T(2002) Compilation and characterization of a novel WNK family of protein kinases in Arabidopsis thaliana with reference to circadian rhythms. Biosci Biotechnol Biochem 66: 2429–2436 [DOI] [PubMed] [Google Scholar]
- Nienhuis J, Sills GR, Martin B, King G(1994) Variance for water-use efficiency among ecotypes and recombinant inbred lines of Arabidopsis thaliana (Brassicaceae). Am J Bot 81: 943–947 [Google Scholar]
- Nietzsche M, Guerra T, Alseekh S, Wiermer M, Sonnewald S, Fernie AR, Börnke F(2018) STOREKEEPER RELATED1/G-element binding protein (STKR1) interacts with protein kinase SnRK1. Plant Physiol 176: 1773–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Ito S, Nakamichi N, Mizoguchi T, Niinuma K, Yamashino T, Mizuno T(2007) Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol 48: 925–937 [DOI] [PubMed] [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, Wetherill SJ, Kusakina J, Walton EJ, Kato M, Azuma M, Tanaka K, Hanaoka M, et al. (2013) Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339: 1316–1319 [DOI] [PubMed] [Google Scholar]
- Panda S, Poirier GG, Kay SA(2002) tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev Cell 3: 51–61 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Jhurreea D, Zhang Y, Primavesi LF, Delatte T, Schluepmann H, Wingler A(2010) Upregulation of biosynthetic processes associated with growth by trehalose 6-phosphate. Plant Signal Behav 5: 386–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Ghassemian M, Kwak CM, McCourt P, Schroeder JI(1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287–290 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA(2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL(2011) REVEILLE8 and PSEUDO-RESPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Resco de Dios V, Gessler A, Ferrio JP, Alday JG, Bahn M, Del Castillo J, Devidal S, García-Muñoz S, Kayler Z, Landais D, et al. (2016) Circadian rhythms have significant effects on leaf-to-canopy scale gas exchange under field conditions. Gigascience 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FC, Skeffington AW, Gardner MJ, Webb AAR(2009) Interactions between circadian and hormonal signalling in plants. Plant Mol Biol 69: 419–427 [DOI] [PubMed] [Google Scholar]
- Rubin MJ, Brock MT, Baker RL, Wilcox S, Anderson K, Davis SJ, Weinig C(2018) Circadian rhythms are associated with shoot architecture in natural settings. New Phytol 219: 246–258 [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Punzo P, Landi S, Costa A, van Oosten JM, Grillo S(2017) Improving plant water use efficiency through molecular genetics. Horticulturae 3: 1–22 [Google Scholar]
- Rusconi F, Simeoni F, Francia P, Cominelli E, Conti L, Riboni M, Simoni L, Martin CR, Tonelli C, Galbiati M(2013) The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J Exp Bot 64: 3361–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruts T, Matsubara S, Wiese-Klinkenberg A, Walter A(2012) Aberrant temporal growth pattern and morphology of root and shoot caused by a defective circadian clock in Arabidopsis thaliana. Plant J 72: 154–161 [DOI] [PubMed] [Google Scholar]
- Salomé PA, McClung CR(2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G(1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D(2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA(2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13: 2659–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle SR, Speed FM, Milliken GA(1980) Population marginal means in the linear model: An alternative to least squares means. J Am Stat Assoc 34: 216–221 [Google Scholar]
- Shimizu H, Katayama K, Koto T, Torii K, Araki T, Endo M(2015) Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nat Plants 1: 15163. [DOI] [PubMed] [Google Scholar]
- Shin J, Sánchez-Villarreal A, Davis AM, Du SX, Berendzen KW, Koncz C, Ding Z, Li C, Davis SJ(2017) The metabolic sensor AKIN10 modulates the Arabidopsis circadian clock in a light-dependent manner. Plant Cell Environ 40: 997–1008 [DOI] [PubMed] [Google Scholar]
- Simon NML, Kusakina J, Fernández-López Á, Chembath A, Belbin FE, Dodd AN(2018a) The energy-signaling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol 176: 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon NML, Sawkins E, Dodd AN(2018b) Involvement of the SnRK1 subunit KIN10 in sucrose-induced hypocotyl elongation. Plant Signal Behav 13: e1457913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA(1998a) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA(2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA(1998b) The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125: 485–494 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA(2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D(2008) The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J 56: 239–250 [DOI] [PubMed] [Google Scholar]
- Thines B, Harmon FG(2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci U S A 207: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás M, Medrano H, Escalona JM, Martorell S, Pou A, Ribas-Carbó M, Flexas J(2014) Variability of water use efficiency in grapevines. Environ Exp Bot 103: 148–157 [Google Scholar]
- Vialet-Chabrand S, Matthews JSA, Brendel O, Blatt MR, Wang Y, Hills A, Griffiths H, Rogers S, Lawson T(2016) Modelling water use efficiency in a dynamic environment: An example using Arabidopsis thaliana. Plant Sci 251: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M(2013) Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X(2008) The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol (Stuttg) 10: 548–562 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM(1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Webb AAR, Seki M, Satake A, Caldana C(2019) Continuous dynamic adjustment of the plant circadian oscillator. Nat Commun 10: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wituszyńska W, Ślesak I, Vanderauwera S, Szechyńska-Hebda M, Kornaś A, van der Kelen K, Mühlenbock P, Karpińska B, Maćkowski S, van Breusegem F, et al. (2013) Lesion simulating disease1, enhanced disease susceptibility1, and phytoalexin deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol 161: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward FI, Lake JA, Quick WP(2002) Stomatal development and CO2: Ecological consequences. New Phytol 153: 477–484 [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares B, Ramirez-Ortega FA, Flores-Elenes L, Ruiz-Medrano R(2010) Drought tolerance in crop plants. Am J Plant Physiol 5: 241–256 [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI(2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA(2002) Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- Yoo CY, Pence HE, Hasegawa PM, Mickelbart MV(2009) Regulation of transpiration to improve crop water use. Crit Rev Plant Sci 28: 410–431 [Google Scholar]
- Yoo CY, Pence HE, Jin JB, Miura K, Gosney MJ, Hasegawa PM, Mickelbart MV(2010) The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Shannon S, Jacobs C, Meeks-Wagner DR(1992) Early-flowering mutants of Arabidopsis thaliana. Funct Plant Biol 19: 411–418 [Google Scholar]
- Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ(2014) Strengths and limitations of period estimation methods for circadian data. PLoS One 9: e96462. [DOI] [PMC free article] [PubMed] [Google Scholar]