Redundant transcription factors directly modulate expression of a pectin metabolism gene to regulate mucilage composition during seed coat development.
Abstract
The polysaccharide pectin is a major component of the plant cell wall. The pectic glycan homogalacturonan (HG) is a proportionally small but important component of a specialized seed cell wall called mucilage. HG is synthesized in a highly methylesterified form, and, following secretion, is de-methylesterified by pectin methylesterases (PMEs). The degree of methylesterification of HG determines the structural and functional properties of pectin, but how methylesterification is regulated remains largely unknown. Here, we identified two BEL1-Like homeodomain (BLH) transcription factors, BLH2 and BLH4, as positive regulators of HG de-methylesterification in Arabidopsis (Arabidopsis thaliana) seed coat mucilage. BLH2 and BLH4 were significantly expressed in mucilage secretory cells during seed mucilage production. BLH2 and BLH4 single mutants exhibited no obvious mucilage phenotype, but the blh2 blh4 double mutant displayed significantly reduced mucilage adherence to the seed. Reduced mucilage adherence in blh2 blh4 was caused by decreased PME activity in the seed coat, which increased the degree of methylesterification of HG in mucilage. The expression of several PME metabolism-related genes, including PME58, PECTIN METHYLESTERASE INHIBITOR6, SEEDSTICK, and MYB52 was significantly altered in blh2 blh4 seeds. BLH2 and BLH4 directly activated PME58 expression by binding to its TGACAGGT cis-element. Moreover, pme58 mutants exhibited reduced mucilage adherence similar to that of blh2 blh4, and the blh2 blh4 pme58 triple mutant exhibited no additional mucilage adherence defects. Furthermore, overexpression of PME58 in blh2 blh4 rescued the mucilage adherence defect. Together, these results demonstrate that BLH2 and BLH4 redundantly regulate de-methylesterification of HG in seed mucilage by directly activating PME58.
The complex polysaccharide pectin is one of the main components of the plant cell wall. It plays important roles in plant morphogenesis, development, and defense, and is used in food processing and biomedicine (Palin and Geitmann, 2012; Peaucelle et al., 2012; Xiao and Anderson, 2013). Pectin consists of three major types of polysaccharides, namely homogalacturonan (HG), rhamnogalacturonan I (RG I), and RG II. HG is a linear homopolymer of α-1,4-linked GalUA (GalA). RG I has a backbone of alternating GalA and rhamnose (Rha). RG II has a backbone of HG linked with complex side chains (Mohnen, 2008). HG is synthesized in the Golgi apparatus. It is commonly methylesterified at the C-6 carboxyl groups, with up to 80% of the GalA residues being methylesterified (Mohnen, 2008; Harholt et al., 2010). The methyl esters can be removed by pectin methylesterases (PMEs) upon secretion to the apoplast (Micheli, 2001; Pelloux et al., 2007; Jolie et al., 2010). PME activity is modulated by the proteinaceous PME inhibitors (PMEIs), which can form a reversible complex with the PME enzymes (Micheli, 2001; Jolie et al., 2010). De-methylesterified HGs can be cross-linked by calcium ions (Ca2+) to form compact egg-box structures or become susceptible to degradation by polygalacturonases and pectin lyases, depending on the pattern of de-methylesterification (Micheli, 2001; Pelloux et al., 2007; Wolf et al., 2009). Therefore, the degree and pattern of de-methylesterification of HG critically determines the structural and functional properties of pectin (Willats et al., 2001; Wolf et al., 2009). However, little is known about the regulatory mechanisms of the de-methylesterification of HG.
During seed development of myxospermous species (e.g. Arabidopsis [Arabidopsis thaliana] and Plantago ovata), a large quantity of mucilage is synthesized and deposited in the outermost epidermal cells of the seed coat, which are also called the mucilage secretory cells (MSCs; Western et al., 2000; Western, 2012; Phan et al., 2016). In Arabidopsis, the production of mucilage begins at 6 to 7 d post anthesis (DPA). Large amounts of mucilage polysaccharides are secreted to the apoplast at the junction of the outer tangential and radial primary walls (Western et al., 2000; Windsor et al., 2000; Haughn and Chaudhury, 2005; North et al., 2014). The mucilage continues to accumulate and leads to the formation of a volcano-shaped cytoplasmic column in the center of the MSCs (Beeckman et al., 2000; Western et al., 2000; Windsor et al., 2000; Young et al., 2008). At 10–13 DPA, a secondary cell wall called columella is formed, filling the cytoplasmic column, and is connected to the reinforced radial cell wall (Western et al., 2000; Windsor et al., 2000). As the seed desiccates, the mucilage is dehydrated and stored between the outer cell wall and columella (Western et al., 2000; Windsor et al., 2000). When the dry seed is imbibed, the rehydrated mucilage expands and ruptures the outer cell wall, forming a gelatinous capsule surrounding the seed. After release, the Arabidopsis seed mucilage comprises two layers, namely the outer water-soluble layer, which is diffuse and easily detached from the seed, and the inner adherent layer that is denser and tightly attached to the seed (Western et al., 2000). Both layers are composed primarily of RG I, as well as small quantities of HG, cellulose, and hemicelluloses (Macquet et al., 2007; Harpaz-Saad et al., 2011; Mendu et al., 2011; Sullivan et al., 2011; Yu et al., 2014; Voiniciuc et al., 2015a, 2015b; Hu et al., 2016a; Ralet et al., 2016). Therefore, the Arabidopsis seed mucilage is considered to be a specialized type of cell wall, and it represents a powerful system to study the synthesis, modifications, and interactions of cell wall polysaccharides (Arsovski et al., 2010; Haughn and Western, 2012; North et al., 2014; Francoz et al., 2015; Voiniciuc et al., 2015c).
Although HG is present as a minor component in Arabidopsis seed mucilage, there is increasing evidence that the appropriate degree of methylesterification (DM) of HG is required for maintenance of mucilage architecture (Rautengarten et al., 2008; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013; Ezquer et al., 2016; Shi et al., 2018). A number of studies have demonstrated that perturbations of the DM of HG in seed mucilage by modifying PME activity lead to various defects in mucilage architecture. For example, mutations in PECTIN METHYLESTERASE INHIBITOR6 (PMEI6) lead to a decreased DM of HG in mucilage and result in delayed mucilage release (Saez-Aguayo et al., 2013). Similarly, mutations of SUBTILISIN-LIKE SER PROTEASE1.7 (SBT1.7) lead to increased PME activity in seeds and a reduced DM of HG in seed mucilage, resulting in a mucilage release defect (Rautengarten et al., 2008). SBT1.7 is hypothesized to function in the degradation of PME or the activation of certain PMEI (Rautengarten et al., 2008). There are at least seven PME genes dominantly expressed in the seed coat (Louvet et al., 2006; Wolf et al., 2009; Levesque-Tremblay et al., 2015; Turbant et al., 2016). However, thus far, only PME58 has been demonstrated to function in HG de-methylesterification of seed mucilage. Disruptions of PME58 result in decreased PME activity in seeds and an increased DM of HG in seed mucilage (Turbant et al., 2016). In addition, a modified distribution of sugars between the adherent and water-soluble layers is detected in pme58 mucilage upon EDTA extraction (Turbant et al., 2016).
Recently, several transcription factors have been shown to modulate seed mucilage structure through regulating the DM of HG in mucilage (North et al., 2014; Francoz et al., 2015; Golz et al., 2018). For example, the MADS-box transcription factor SEEDSTICK (STK) negatively regulates the de-methylesterification of HG in seed mucilage through direct regulation of the expression of PMEI6 (Ezquer et al., 2016). The stk mutants have significantly increased PME activity in seeds and dramatically decreased the DM of HG in seed mucilage, leading to defects in mucilage extrusion (Ezquer et al., 2016). Similarly, MYB52 negatively regulates the de-methylesterification of HG in seed mucilage by directly activating the expression of PMEI14, PMEI6, and SBT1.7 (Shi et al., 2018). Disruption of MYB52 also results in increased PME activity in seeds and a decreased DM of HG in seed mucilage (Shi et al., 2018). The transcription factors identified thus far are negative regulators controlling the de-methylesterification of HG in mucilage. However, other transcription factors regulating the de-methylesterification of HG in mucilage, especially those directly modulating the expression of PME genes in this process, remain to be identified.
The BEL1-Like homeodomain (BLH) and KNOTTED-like homeobox (KNOX) transcription factors are collectively called three amino acid loop extension (TALE) proteins, and they play crucial regulatory roles in many important processes including embryogenesis, cell differentiation, and organ morphogenesis (Hamant and Pautot, 2010). Various studies indicate that BLH and KNOX proteins interact to form heterodimers, which enables them to be localized in the nucleus and modulate gene expression (Bellaoui et al., 2001; Bhatt et al., 2004; Cole et al., 2006). In Arabidopsis, the BLH family consists of 13 members. BEL1 is required for the morphogenesis of the ovule (Reiser et al., 1995). ARABIDOPSIS THALIANA HOMEOBOX 1 is involved in the regulation of photomorphogenesis of seedlings (Quaedvlieg et al., 1995). BLH6 is involved in the regulation of secondary cell wall development (Liu et al., 2014). BLH2/SAWTOOTH1 (SAW1) and BLH4/SAW2 redundantly regulate the morphogenesis of leaf margins (Kumar et al., 2007). However, the functions of these BLH proteins in other organs or tissues (i.e. seed coat) remain to be determined.
In this study, we report that BLH2 and BLH4 act redundantly to positively regulate the de-methylesterification of HG in seed mucilage. The blh2 blh4 double mutant exhibited significantly reduced mucilage adherence on vigorous shaking due to the increased DM of HG in mucilage. We provided several lines of biochemical and genetic evidence to demonstrate that BLH2 and BLH4 positively regulated PME activity primarily through directly activating the expression of PME58.
RESULTS
Expression of BLH2 and BLH4 in Seed Coat Coincides with Mucilage Production
We previously identified a subset of genes that are differentially expressed during seed mucilage production through reanalyzing the microarray datasets of laser-capture microdissected Arabidopsis seed samples (GSE12404; Le et al., 2010; Hu et al., 2016a). Among these genes, BLH2 and BLH4 were dramatically up-regulated during the seed coat differentiation process, indicative of a potential role in seed mucilage production or structure maintenance.
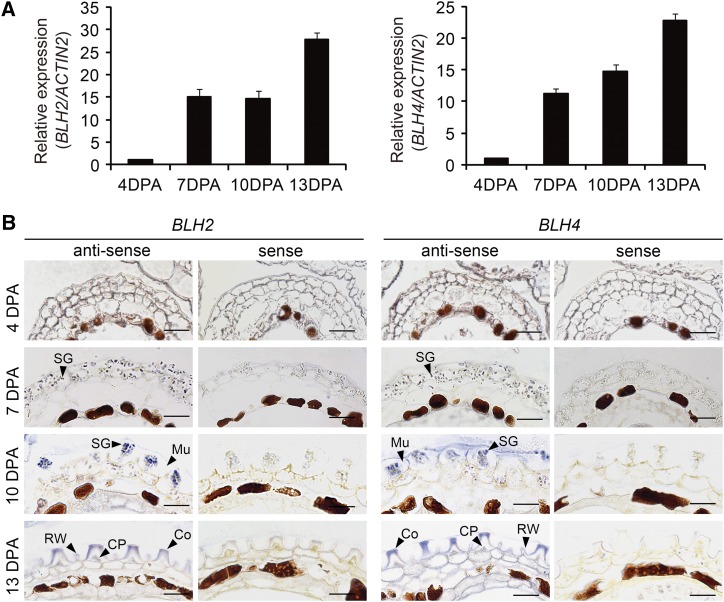
We first examined the expression of BLH2 and BLH4 in siliques at different developmental stages ranging from 4 to 13 DPA by reverse-transcriptase quantitative PCR (RT-qPCR) analysis. The transcript levels of BLH2 and BLH4 were relatively low at 4 DPA, but dramatically increased at 7 DPA when mucilage biosynthesis was initiated (Western et al., 2000; Windsor et al., 2000). Thereafter, the transcripts of BLH2 and BLH4 continued to increase at 10 DPA and a >20-fold level was reached at 13 DPA (Fig. 1A). These results suggest that the expression of BLH2 and BLH4 coincides with the process of seed mucilage production.
Figure 1.
Expression patterns of BLH2 and BLH4 in developing seeds. A, RT-qPCR analysis of BLH2 and BLH4 expression in siliques at different developmental stages from 4 to 13 DPA. The ACTIN2 gene was used as the internal control. The expression level at 4 DPA was set as 1. Data are means ± sd of three biological replicates. B, In situ hybridization showing the transcripts of BLH2 and BLH4 in seed coats at different developmental stages. Mu, mucilage; SG, starch granule; Co, Columella; RW, radial cell wall; CP, cytoplasm. Scale bars = 50 μm. The experiments were independently performed twice and similar results were obtained.
To obtain more detailed information about the spatiotemporal expression patterns of BLH2 and BLH4 within the seed, in situ hybridization assays were performed with seeds staged from 4 to 13 DPA. The transcripts of BLH2 and BLH4 were substantially detected in the MSCs by their specific antisense probes with almost identical expression patterns (Fig. 1B). Relatively weak hybridization signals were detected in the MSCs at 4 DPA and 7 DPA, while more intense hybridization signals were detected in the MSCs at 10 DPA. At 13 DPA, strong hybridization signals were detected in the cytoplasm and columella of the MSCs. By contrast, the signals of sense probes were almost undetectable (Fig. 1B). These results indicate that BLH2 and BLH4 are expressed in the MSCs during the process of mucilage production.
Mucilage Adherence Is Reduced in blh2 blh4 upon Vigorous Shaking
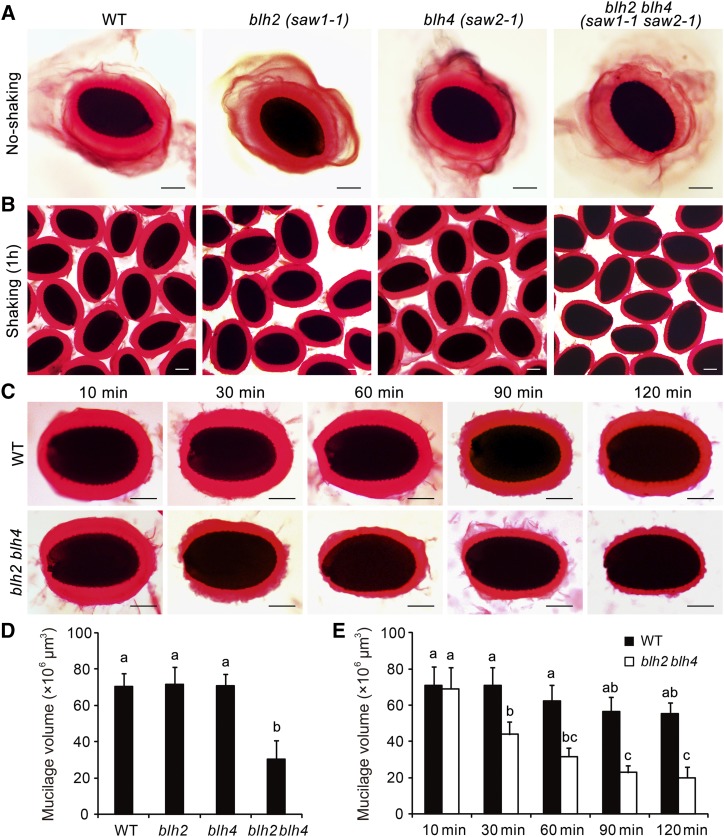
To investigate whether BLH2 and BLH4 play functional roles in mucilage production or structure maintenance, we isolated the knockout mutants of BLH2 (saw1-1) and BLH4 (saw2-1), as well as the double mutant saw1-1 saw2-1 (blh2 blh4 hereafter; Kumar et al., 2007), and examined the mucilage phenotypes by ruthenium red (RR) staining. These mutants exhibited normal mucilage release compared to the wild-type upon imbibition (Fig. 2A). However, when subjected to vigorous shaking at 200 rpm for 1 h, the blh2 blh4 double mutant displayed a significantly thinner mucilage layer compared to the wild type, while no obvious differences were observed for the single mutants in comparison to the wild type (Fig. 2, B and D). To obtain a more detailed examination of the mucilage phenotype of blh2 blh4, we further performed a continuous observation of mucilage at various time points under shaking at 200 rpm. The thickness of the adherent mucilage of blh2 blh4 was comparable to that of the wild type after 10 min of shaking, when the water-soluble mucilage layer had mostly detached (Fig. 2, C and E). As the shaking time was prolonged, the adherent mucilage of the wild type remained tightly attached to the seed, and no obvious changes in the thickness were observed even after 2 h of shaking. By contrast, the adherent mucilage of blh2 blh4 displayed a coarse surface and the thickness was significantly decreased after shaking for more than 30 min. This led to a significant reduction in the thickness of the adherent mucilage in blh2 blh4 compared to the wild type. When the shaking was extended to 90 min or more, the reduction observed in the thickness of the adherent mucilage in blh2 blh4 was even more conspicuous (Fig. 2, C and E). These observations suggest that a proportion of the adherent mucilage of blh2 blh4 was detached from the seed and dispersed into the solution during vigorous shaking. We also checked the mucilage phenotypes of the other two saw1 saw2 double mutants carrying different mutation alleles (Kumar et al., 2007). These mutants exhibited mucilage phenotypes similar to that of blh2 blh4 (Supplemental Fig. S1). These results suggest that BLH2 and BLH4 act redundantly in maintaining mucilage adherence.
Figure 2.
Mucilage phenotypes of blh2, blh4, and blh2 blh4 seeds. A, Release of mucilage in water containing 0.01% RR. B, Staining of the adherent mucilage with 0.01% RR after shaking at 200 rpm for 1 h. C, Staining of the adherent mucilage of blh2 blh4 and the wild type (WT) with 0.01% RR after shaking at 200 rpm for 10, 30, 60, 90, and 120 min. Scale bars = 150 μm. D, Mucilage volumes of wild-type, blh2, blh4, and blh2 blh4 seeds after shaking at 200 rpm for 1 h. E, Mucilage volumes of wild-type and blh2 blh4 seeds after shaking at 200 rpm for 10, 30, 60, 90, and 120 min. Data represent means ± sd of 20 seeds. Means not sharing the same letter are significantly different according to the one-way ANOVA analysis followed by Tukey’s multiple comparison test (P < 0.05). The experiments were independently performed at least three times (each with >20 seeds) and similar results were obtained.
Seed Coat Differentiation and Mucilage Production Are Unaffected in blh2 blh4
To determine whether the mucilage adherence defect of blh2 blh4 is associated with abnormalities in seed coat differentiation, we sectioned and observed the morphology of MSCs in blh2 blh4 staged from 4 DPA to 13 DPA. The results revealed that the blh2 blh4 mutant exhibited no abnormality in the differentiation of MSCs or the accumulation of mucilage in comparison to the wild type (Supplemental Fig. S2). This suggests that simultaneous disruptions of BLH2 and BLH4 do not affect MSC differentiation and mucilage production.
Sugar Distribution Is Altered in the blh2 blh4 Mucilage
The reduced adherence of the blh2 blh4 mucilage indicates possible alterations in sugar components or architecture. To test this hypothesis, we sequentially extracted the water-soluble and adherent mucilage and determined the monosaccharide compositions. Each type of monosaccharide was increased in the water-soluble mucilage of blh2 blh4 compared to the wild type, resulting in a significant increase in the total amount of sugars in the water-soluble mucilage of blh2 blh4 (Table 1). Conversely, Rha and GalA were significantly decreased in the adherent mucilage of blh2 blh4 compared to the wild type, resulting in a dramatic reduction of total sugar amounts in the adherent mucilage of blh2 blh4 (Table 1). However, there was no significant difference in the amount of each monosaccharide and total sugars for the whole mucilage between the blh2 blh4 and the wild type (Table 1). These results indicate that it is the partitioning of mucilage sugars, rather than the level of synthesis or composition, that is altered in the blh2 blh4 mucilage. The sugars were redistributed from the adherent layer to the water-soluble layer. This suggests that BLH2 and BLH4 are involved in maintaining mucilage architecture.
Table 1. Monosaccharide compositions of wild-type and blh2 blh4 mucilage.
Data represent means ± sd of three biological replicates. Asterisks indicate significant differences from the wild type as determined by one-way ANOVA (**P < 0.01).
| Sugar Contents (mg g−1 seeds) | Water-Soluble Mucilage | Adherent Mucilage | Total Mucilage | |||
|---|---|---|---|---|---|---|
| Wild Type | blh2 blh4 | Wild Type | blh2 blh4 | Wild Type | blh2 blh4 | |
| Rha | 8.31 ± 0.47 | 9.50 ± 0.01 | 4.81 ± 0.47 | 2.77 ± 0.16** | 13.12 ± 0.89 | 12.27 ± 0.16 |
| GalA | 12.00 ± 0.71 | 14.44 ± 0.80** | 4.27 ± 0.25 | 2.78 ± 0.32** | 16.27 ± 0.51 | 17.22 ± 1.13 |
| Man | 0.32 ± 0.02 | 0.38 ± 0.02 | 0.31 ± 0.08 | 0.23 ± 0.01 | 0.64 ± 0.10 | 0.61 ± 0.02 |
| Glc | 0.19 ± 0.04 | 0.24 ± 0.01 | 0.35 ± 0.06 | 0.28 ± 0.11 | 0.50 ± 0.08 | 0.52 ± 0.10 |
| Gal | 0.15 ± 0.01 | 0.23 ± 0.03 | 0.27 ± 0.02 | 0.23 ± 0.08 | 0.42 ± 0.04 | 0.50 ± 0.09 |
| Xyl | 0.43 ± 0.03 | 0.51 ± 0.01 | 0.29 ± 0.01 | 0.19 ± 0.07 | 0.72 ± 0.05 | 0.70 ± 0.09 |
| Ara | 0.03 ± 0.01 | 0.05 ± 0.01 | 0.04 ± 0.00 | 0.06 ± 0.01* | 0.07 ± 0.00 | 0.11 ± 0.01 |
| Total | 21.37 ± 1.47 | 25.39 ± 0.65** | 10.13 ± 0.27 | 6.54 ± 0.62** | 31.50 ± 1.59 | 31.93 ± 1.27 |
The DM of HG Is Increased in the blh2 blh4 Mucilage
To further explore potential structural alterations in the blh2 blh4 mucilage, we performed in situ immunolabeling assays of mucilage using different monoclonal antibodies that recognize specific polysaccharides. The labeling of RG I was performed with the CCRC-M36 antibody (Young et al., 2008; Pattathil et al., 2010). No obvious difference was observed between blh2 blh4 and wild-type mucilage when seeds were not subjected to shaking (Supplemental Fig. S3A). Considering that blh2 blh4 exhibited reduced mucilage adherence on vigorous shaking, we further performed the immunolabeling assays with seeds after shaking at 200 rpm for 1 h in water. After shaking, the halo of mucilage labeled with CCRC-M36 was much smaller and the peripheral edges were more irregular in the blh2 blh4 mucilage compared to the wild type (Supplemental Fig. S3B). These results are consistent with the mucilage phenotype of blh2 blh4 on vigorous shaking, as observed by RR staining (Fig. 2C).
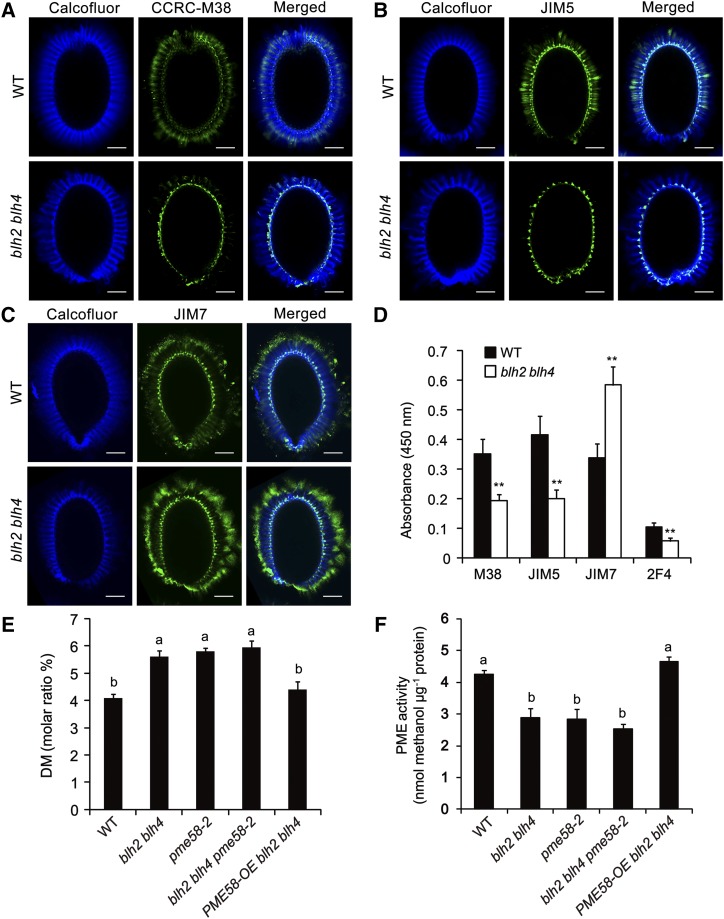
We next analyzed the pattern of HG methylesterification in the blh2 blh4 mucilage by in situ immunolabeling assays. The monoclonal antibodies CCRC-M38, JIM5, and JIM7, which recognize the sparsely methylesterified, moderately methylesterified, and highly methylesterified HGs, respectively (Willats et al., 2000; Pattathil et al., 2010), were employed in the immnolabeling assays. The signals of CCRC-M38 and JIM5 were distributed at the tips of columella and alongside the ray structure in the inner regions of the adherent mucilage in the wild type (Fig. 3, A and B). The signal patterns are comparable to previous observations (Macquet et al., 2007; Rautengarten et al., 2008; Shi et al., 2018). By contrast, the signals of CCRC-M38 and JIM5 were only detectable in the columella in blh2 blh4 (Fig. 3, A and B), suggesting a substantial reduction of the sparsely and moderately methylesterified HGs in the blh2 blh4 mucilage. The JIM7 signals were mainly detected in the columella and the outer periphery of the adherent layer in both blh2 blh4 and wild-type mucilage (Fig. 3C), as previously reported (Macquet et al., 2007; Rautengarten et al., 2008; Shi et al., 2018). However, the blh2 blh4 mucilage exhibited much stronger labeling intensity of JIM7 compared to the wild type (Fig. 3C), indicating an increase in highly methylesterified HG in the blh2 blh4 mucilage. Moreover, we quantitatively determined the signal intensities of CCRC-M38, JIM5, and JIM7 antibodies by enzyme-linked immunosorbent assays (ELISAs) using adherent mucilage extracts. The results confirmed that the blh2 blh4 mucilage exhibited significantly decreased binding of CCRC-M38 and JIM5, and increased binding of JIM7 compared with the wild type (Fig. 3D). These results of in situ immunolabeling and ELISA assays suggest that the DM of HG is increased in the blh2 blh4 mucilage. To verify this hypothesis, we quantitatively determined the DM of HG in the blh2 blh4 and wild-type mucilage. The results showed that the DM of HG in the wild-type mucilage was ∼4%, which is comparable to previous reports (Rautengarten et al., 2008; Saez-Aguayo et al., 2013; Shi et al., 2018). However, the DM of HG in the blh2 blh4 mucilage was ∼5.5%, an increase of 37.5% compared to the wild type (Fig. 3E). These results indicate that BLH2 and BLH4 are involved in the modulation of the DM of HG in seed mucilage.
Figure 3.
blh2 blh4 increased the DM of HG in mucilage and decreased PME activity in the seed coat. A to C, Immunolabeling signals (green fluorescence) of CCRC-M38 for sparsely methylesterified HG (A), JIM5 for moderately methylesterified HG (B), and JIM7 for highly methylesterified HG (C) in the adherent mucilage of the wild type (WT) and blh2 blh4. The cellulosic ray-structure was counterstained with Calcofluor White (cyan fluorescence). Scale bars = 50 μm. Three independent experiments (each with >20 seeds) were performed for each antibody and similar results were obtained. D, ELISA analysis of the adherent mucilage from the wild-type and blh2 blh4 seeds using the CCRC-M38, JIM5, JIM7, and 2F4 antibodies. Data represent means ± sd of three biological replicates. Asterisks denote significant differences from the wild type based on one-way ANOVA analysis (**P < 0.01). E and F, Determination of the DM of HG in mucilage (E) and PME activity in the seed coat (F). Seed coat samples (containing some attached endosperm) were used to determine PME activity. Data represent means ± sd of three biological replicates. Means with different letters are significantly different according to the one-way ANOVA analysis followed by Tukey’s multiple comparison test (P < 0.05).
Since the status of HG methylesterification influences the binding of HG by Ca2+, we further performed ELISA assay using the 2F4 antibody that specifically binds to HG cross-linked Ca2+ (Liners et al., 1989). The blh2 blh4 mucilage exhibited significantly decreased 2F4 binding capacity compared to the wild type (Fig. 3D). These results suggest that the increased DM of HG in the blh2 blh4 mucilage leads to decreased binding of HG by Ca2+.
blh2 blh4 Mucilage Exhibited Altered Cellulose Labeling
In addition to the alterations in HG and RG I labeling, the blh2 blh4 mucilage also exhibited altered cellulose labeling. The signals of Calcofluor white, a fluorescent probe that binds to β-glycans, were regularly distributed in the feathery ray structures emanating from the top of columella, and diffuse labeling was commonly observed between the rays in the wild-type mucilage (Fig. 3; Supplemental Figs. S3 and S4). However, the diffuse labeling was absent in the blh2 blh4 mucilage, while cellulose rays were retained, with the terminal end of the rays curled inward (Fig. 3; Supplemental Figs. S3 and S4). This phenotype was more conspicuous especially on vigorous shaking at 200 rpm for 1 h (Supplemental Figs. S3 and S4). These results suggest that the labeling pattern of cellulose was affected in the blh2 blh4 mucilage.
Furthermore, we performed immunolabeling of crystalline cellulose with the carbohydrate binding module family 3a (CBM3a; Dagel et al., 2011). The CBM3a signals were located closer to the seed surface in the blh2 blh4 mucilage than in wild-type mucilage, filling the vacant regions of the diffuse labeling of Calcofluor in the blh2 blh4 mucilage (Supplemental Fig. S4, A1–L1). In particular, this phenomenon became more noticeable when seeds were subjected to vigorous shaking at 200 rpm for 1 h (Supplemental Fig. S4, A2–L2). Therefore, the altered pattern of CBM3a signals in the blh2 blh4 mucilage is apparently associated with the absence of diffuse cellulose-labeling. Nevertheless, quantification of the crystalline cellulose content revealed no significant difference between the blh2 blh4 and wild-type mucilage (Supplemental Fig. S5), suggesting that BLH2 and BLH4 might not interfere with crystalline cellulose synthesis.
PME Activity Is Decreased in the blh2 blh4 Seed Coat
To explore whether the increased DM of HG in the blh2 blh4 mucilage is associated with potential alterations in PME activity in the seed coat during mucilage production, we quantitatively determined PME activity in the blh2 blh4 and wild-type seed coats at 7 to 10 DPA. The results showed that PME activity was decreased by 33% in the blh2 blh4 seed coats compared to the wild type (Fig. 3F), indicating that BLH2 and BLH4 might positively modulate PME activity in the seed coat during mucilage production.
BLH2 and BLH4 Regulate the Expression of PME-Related Genes
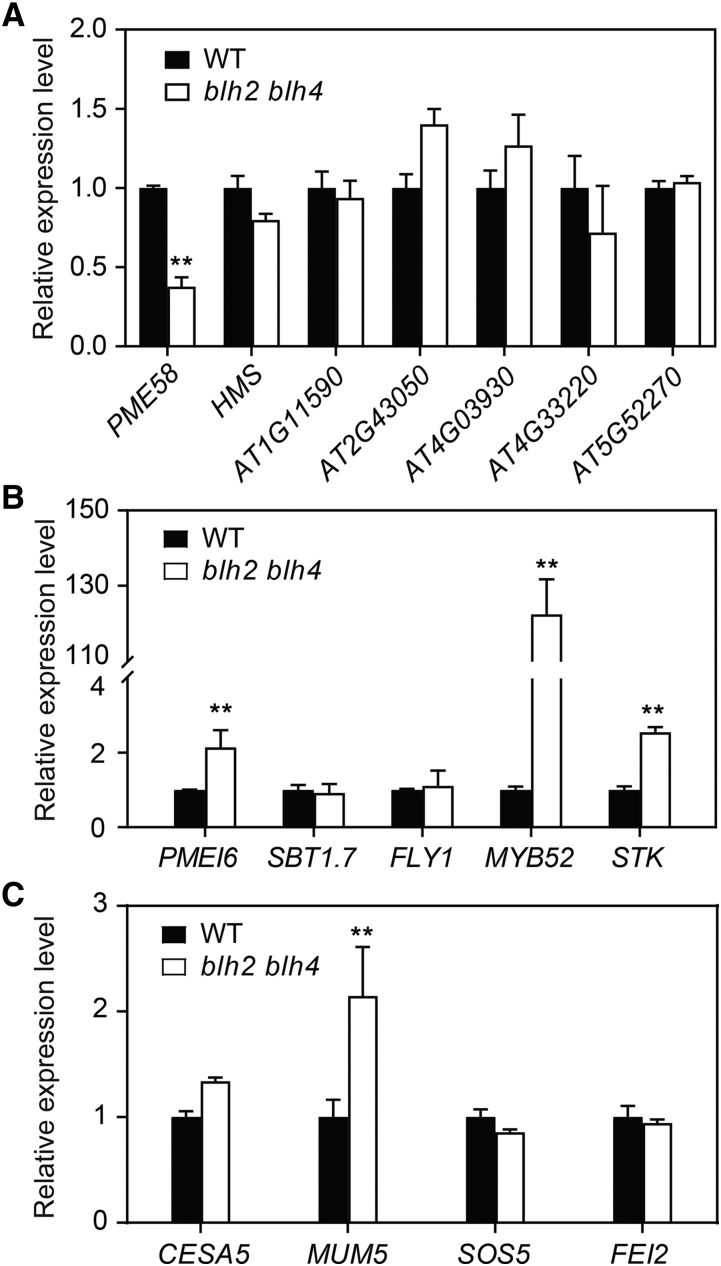
To discover the molecular mechanisms underlying the decreased PME activity in blh2 blh4 seed coats, we first examined the expression levels of PME-encoding genes in blh2 blh4 seeds at 7 to 10 DPA by RT-qPCR analysis. It has been reported that at least seven PME genes are expressed in seeds (Louvet et al., 2006; Wolf et al., 2009; Levesque-Tremblay et al., 2015; Turbant et al., 2016). Among them, PME58 is involved in the de-methylesterification of HG in seed mucilage (Turbant et al., 2016). Our results showed that the expression level of PME58 was significantly reduced in blh2 blh4 seeds, while the expression of the other six PME genes were not obviously affected (Fig. 4A), suggesting that BLH2 and BLH4 might positively modulate the expression of PME58 in seeds. We also examined the expression of several genes encoding enzymes that modulate PME activity during seed mucilage production, including PMEI6, SBT1.7, and FLYING SAUCER1 (FLY1; Rautengarten et al., 2008; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013). The results showed that the expression level of PMEI6 was significantly increased, while SBT1.7 and FLY1 were not significantly affected in blh2 blh4 seeds (Fig. 4B). In addition, the expression levels of two transcription factor genes, STK and MYB52, were dramatically increased in blh2 blh4 seeds (Fig. 4B). Taken together, these results indicate that BLH2 and BLH4 probably regulate PME activity by modulating the expression of genes associated with PME metabolism .
Figure 4.
BLH2 and BLH4 modulate the expression of PME-related genes in seeds. A and B, Expression analysis of PME genes (A) and genes encoding modulators of PME (B) in wild-type (WT) and blh2 blh4 seeds at 7 to 10 DPA. C, Expression of CESA5, MUM5, SOS5, and FEI2 in wild-type and blh2 blh4 seeds at 7 to 10 DPA. Data represent means ± sd of three biological replicates. Asterisks denote significant differences from the wild type based on one-way ANOVA analysis (**P < 0.01).
The altered labeling pattern of cellulose in the blh2 blh4 mucilage prompted us to examine the expression of CELLULOSE SYNTHASE 5 (CESA5), MUCILAGE MODIFIED5 (MUM5), SALT OVERLY SENSITIVE5 (SOS5), and FEI2, mutations of which lead to phenotypes in cellulose labeling that are similar or opposite to blh2 blh4 (Griffiths et al., 2014, 2016; Ralet et al., 2016). The results showed that MUM5 expression was increased in blh2blh4 seeds, while the expression of CESA5, SOS5, and FEI2 was not significantly affected (Fig. 4C).
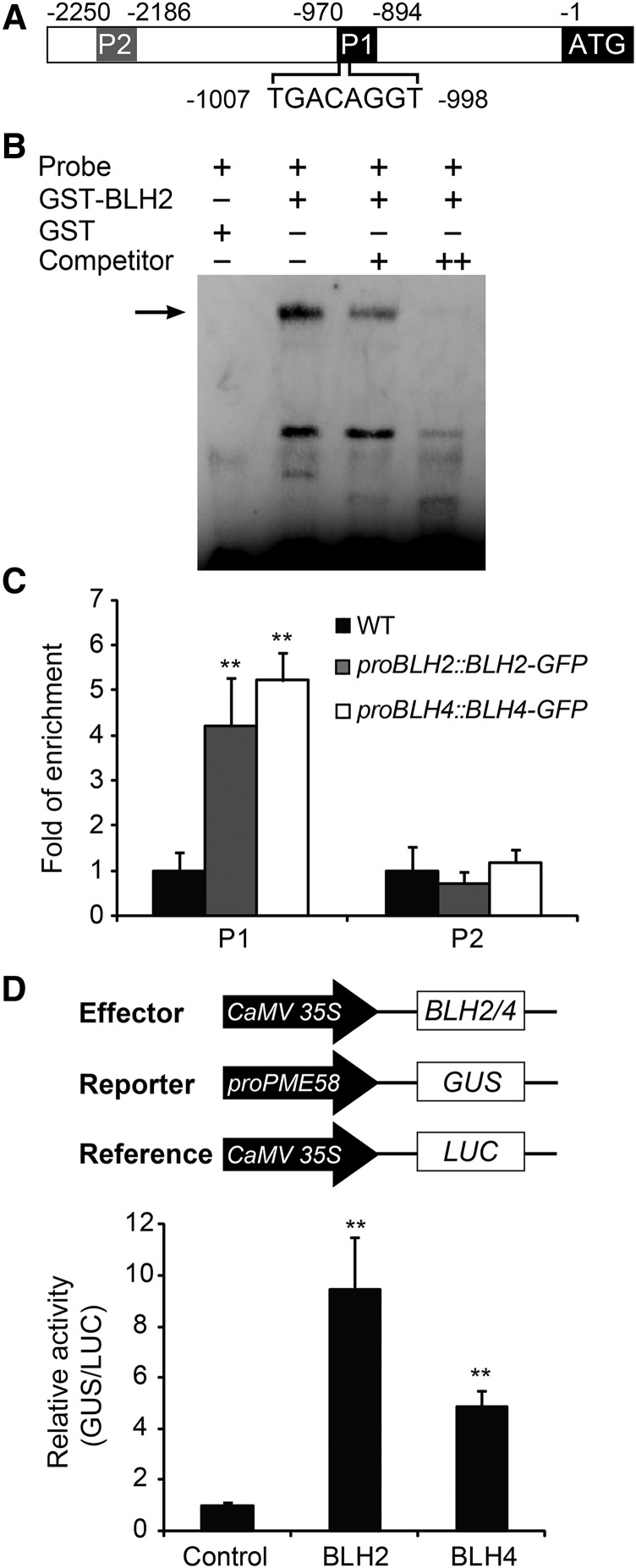
BLH2 and BLH4 Directly Target PME58
BLH transcription factors are reported to bind to DNA sequences containing a TGAC core motif (Smith et al., 2002; Tioni et al., 2005; Viola and Gonzalez, 2009). For instance, ATH1, a homolog of BLH2 and BLH4 in Arabidopsis, preferentially binds to the sequence 5′-TGACAGGT-3′ (Viola and Gonzalez, 2006). We examined the promoter sequences of the differentially expressed PME-related genes in blh2 blh4, and found that the PME58 promoter contains a TGACAGGT cis-element at 998 bp upstream of the translation initiation codon (Fig. 5A). To test whether BLH2 and BLH4 bind to the TGACAGGT cis-element in the PME58 promoter, we first performed electrophoretic mobility shift assays (EMSAs). Since BLH2 and BLH4 are functionally redundant and share high sequence identity (58%) in amino acids, a recombinant glutathione S transferase (GST) and BLH2 fusion protein (GST-BLH2) was employed in the EMSA. The results showed that BLH2 could bind to the PME58 promoter probe containing the TGACAGGT cis-element (Fig. 5B). In addition, the binding affinity was efficiently competed out by the unlabeled probe, suggesting that the binding is highly specific. Subsequently we examined the binding of BLH2 and BLH4 to the PME58 promoter in vivo through chromatin immunoprecipitation (ChIP) assays. We generated transgenic lines of BLH2-GFP and BLH4-GFP driven by their native promoters in the blh2 blh4 background (proBLH2::BLH2-GFP and proBLH4::BLH4-GFP; Supplemental Fig. S6, A and B), and siliques of the transgenic plants at 7 to 10 DPA were used in the ChIP assays. Staining of seed mucilage with RR showed that seeds from the transgenic plants were indistinguishable from wild-type seeds after vigorous shaking for 60 min (Supplemental Fig. S6C). This suggests that the BLH2-GFP and BLH4-GFP fusion proteins were biologically active, and that each can complement the absence of endogenous BLH2 and BLH4 proteins. The results of ChIP-qPCR showed that the PME58 promoter region containing the TGACAGGT cis-element was significantly enriched in the precipitated DNA from the proBLH2::BLH2-GFP and proBLH4::BLH4-GFP siliques compared to the wild type. By contrast, no significant enrichment was detected for the promoter region without the binding site (Fig. 5C). These results indicate that BLH2 and BLH4 directly bind to the PME58 promoter through specifically recognizing the TGACAGGT cis-element.
Figure 5.
BLH2 and BLH4 directly bind to the promoter of PME58 and activate its expression. A, Schematic representation of PME58 promoter and the position of the TGACAGGT cis-element. P1 and P2 indicate the regions tested in ChIP-qPCR assays. B, EMSA assays showing that the GST-BLH2 fusion protein binds to the PME58 promoter probe containing the TGACAGGT cis-element. Arrow indicates the shifted bands. The experiments were independently performed three times and similar results were obtained. C, ChIP-qPCR assays showing the binding of BLH2 and BLH4 to the PME58 promoter in vivo. Data represent enrichment values normalized to ACTIN8. D, Transient transcriptional regulation assays showing that BLH2 and BLH4 activate the expression of the PME58 promoter. Data are given as the ratio of GUS activity to luciferase (LUC) activity calculated relative to the empty vector control. Error bars indicate the sd of three biological replicates. Asterisks denote significant differences from the wild type (WT) as determined by one-way ANOVA analysis (**P < 0.01).
To further confirm the regulatory role of BLH2 and BLH4 on PME58 expression, a transient transcriptional regulation assay was performed using Arabidopsis mesophyll protoplasts. BLH2 and BLH4 individually driven by the CaMV 35S promoter were employed as effectors. The GUS gene driven by the PME58 promoter was used as the reporter, and the Renilla luciferase (LUC) gene driven by the CaMV 35S promoter was used as the internal control for normalization (Fig. 5D). The data showed that the GUS activity produced by the PME58 promoter was strongly activated 10- and 5-fold by BLH2 and BLH4, respectively, relative to the negative control (Fig. 5D). Together, these results demonstrate that BLH2 and BLH4 could directly bind to the promoter of PME58 and activate its expression.
Mucilage Adherence Is Reduced in pme58 Mutants upon Vigorous Shaking
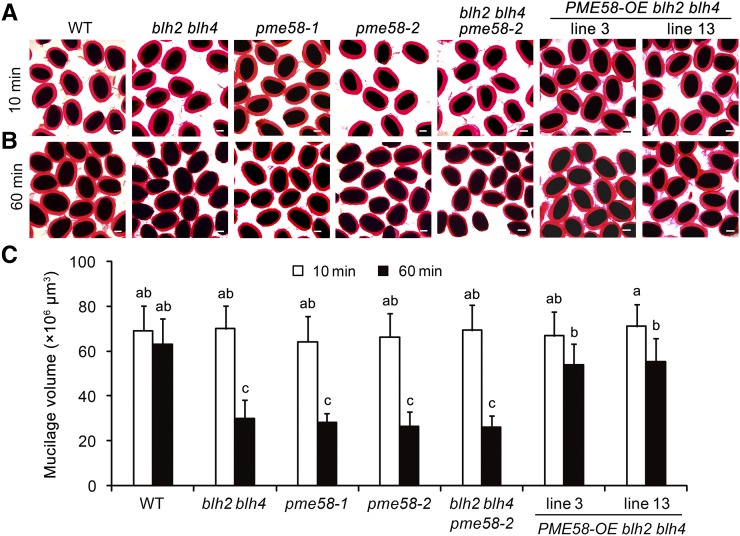
Since blh2 blh4 and pme58 mutants have similar alterations in the DM of HG in mucilage and PME activity in the seed coat (Fig. 3, E and F), we sought to explore whether the pme58 mutants exhibit a mucilage adherence defect similar to that observed for blh2 blh4 on vigorous shaking. No obvious difference in the thickness of the adherent mucilage was observed between the pme58 mutants and the wild type after 10 min of shaking at 200 rpm (Fig. 6A). However, the adherent mucilage of pme58 mutants exhibited a significant reduction in thickness compared to the wild type after shaking at 200 rpm for 60 min (Fig. 6, B and C). Moreover, the degree of reduction in mucilage thickness of pme58 mutants after shaking for 60 min was comparable to that observed in blh2 blh4 (Fig. 6C). These results indicate that the mucilage adherence of pme58 mutants is reduced on vigorous shaking, similar to that of blh2 blh4.
Figure 6.
Genetic analysis of the regulation of PME58 by BLH2/BLH4. A and B, Mucilage phenotypes of wild-type (WT), blh2 blh4, pme58-1, pme58-2, blh2 blh4 pme58-2, and PME58-OE blh2 blh4 seeds after shaking at 200 rpm for 10 min (A) and 60 min (B). Scale bars = 150 μm. C, Mucilage volumes of wild-type, blh2 blh4, pme58-1, pme58-2, blh2 blh4 pme58-2, and PME58-OE blh2 blh4 seeds after shaking at 200 rpm for 10 min and 60 min. Data represent means ± sd of 20 seeds. Means not sharing the same letter are significantly different according to one-way ANOVA analysis followed by Tukey’s multiple comparison test (P < 0.05). The experiments were independently performed three times (each with >20 seeds) and similar results were obtained.
Genetic Analysis of the Regulation of PME58 Expression by BLH2/BLH4
As shown above, the molecular data indicate that BLH2 and BLH4 directly and positively regulate the expression of PME58. To genetically verify the regulation of BLH2 and BLH4 on PME58 expression, and to clarify whether the mucilage adherence phenotype of blh2 blh4 is dependent on PME58 function, we constructed the blh2 blh4 pme58-2 triple mutant and analyzed the mucilage phenotype. The results showed that the blh2 blh4 pme58-2 triple mutant phenocopied the mucilage adherence defects of the blh2 blh4 and pme58 mutants on vigorous shaking (Fig. 6, A and B). Additionally, the triple mutant exhibited a degree of reduction in adherent mucilage thickness similar to that for the blh2 blh4 and pme58 mutants after shaking at 200 rpm for 60 min (Fig. 6C). These observations indicate that simultaneous mutation of PME58, BLH2, and BLH4 has no further impact on mucilage adherence relative to the blh2 blh4 and pme58 mutants. In other words, the impaired PME58 function is the main contributor to the reduced mucilage adherence of blh2 blh4. Moreover, determination of PME activity in the seed coat and the DM of HG in mucilage showed that the blh2 blh4 pme58-2 triple mutant had degrees of alteration similar to those of the blh2 blh4 and pme58 mutants (Fig. 3, E and F). Taken together, this genetic evidence demonstrates that the function of BLH2/BLH4 in regulating HG de-methylesterification is dependent on the activation of PME58 expression.
Overexpression of PME58 Rescues Mucilage Adherence Defects of blh2 blh4
To further confirm that the reduced mucilage adherence of blh2 blh4 was due to the impaired expression of PME58, we generated overexpression lines of PME58 in the blh2 blh4 background (PME58-OE blh2 blh4; Supplemental Fig. S7) and examined the mucilage phenotype. The thickness of the adherent mucilage of PME58-OE blh2 blh4 seeds was comparable to that of the wild type after shaking at 200 rpm for 1 h (Fig. 6, A–C). Furthermore, determination of the DM of HG in mucilage and PME activity in the seed coat showed that PME58-OE blh2 blh4 restored the DM and PME activity to the wild-type level (Fig. 3, E and F). Correspondingly, immunolabeling of HG with CCRC-M38, JIM5, and JIM7 antibodies revealed that the pattern of HG methylesterification in the adherent mucilage of PME58-OE blh2 blh4 closely resembled that of the wild type (Supplemental Fig. S8). These results indicate that overexpression of PME58 could complement the mucilage adherence defects of blh2 blh4 by restoring the HG methylesterification pattern in mucilage. However, the altered cellulose labeling in the blh2 blh4 mucilage was not restored by PME58 overexpression (Supplemental Fig. S8), suggesting that the altered cellulose labeling in the blh2 blh4 mucilage is not associated with the impaired expression of PME58 or altered mucilage adhesion.
Regulation of PME58 by BLH2/BLH4 Is Spatiotemporal Specific
It has been reported that PME58 is preferentially expressed in siliques (Louvet et al., 2006; Wolf et al., 2009; Levesque-Tremblay et al., 2015; Turbant et al., 2016). In particular, promoter activity analysis showed that PME58 is specifically expressed in seed coat cells (Turbant et al., 2016). These findings are consistent with the public transcriptome data (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), which show that the PME58 transcript is specifically higher in floral organs (Supplemental Fig. S9A), with especially high expression in the seed coat (Supplemental Fig. S9B). To confirm the expression pattern of PME58, we examined its expression levels across different tissues by RT-qPCR analysis. The PME58 transcripts could be detected in floral buds, flowers, and siliques, but were barely detectable in vegetative tissues (Supplemental Fig. S9C). Next, we investigated whether PME58 expression in floral buds and flowers is also regulated by BLH2 and BLH4. RT-qPCR analysis showed that the expression levels of PME58 in these tissues were not obviously affected in blh2 blh4 compared to the wild type (Supplemental Fig. S9C). Interestingly, although PME58 expression was reduced in blh2 blh4 siliques from 7 to 14 DPA, its expression was not obviously affected in siliques at 1 to 6 DPA (Supplemental Fig. S9C). These results suggest that BLH2/BLH4 transcriptionally regulate PME58 expression in a spatiotemporal-specific manner.
Moreover, BLH2 and BLH4 have been previously reported to regulate leaf margin morphogenesis, and the blh2 blh4 mutant has increased leaf serrations (Kumar et al., 2007). To investigate whether the function of BLH2 and BLH4 in regulating leaf margin development is associated with HG de-methylesterification, we determined PME activity in the blh2 blh4 and wild-type leaves. The results showed that PME activity in the blh2 blh4 leaves is comparable to that in the wild type leaves (Supplemental Fig. S10), suggesting that BLH2 and BLH4 might not affect the de-methylesterification of HG in leaves.
DISCUSSION
BLH2 and BLH4 Act Redundantly in Maintaining Mucilage Adherence via Regulating the DM of HG
BLH2 and BLH4 have been previously reported to function redundantly in leaf margin morphogenesis (Kumar et al., 2007). In this work, we show that BLH2 and BLH4 also act redundantly to maintain mucilage adherence. BLH2 and BLH4 were expressed in the MSCs during seed mucilage production with almost identical expression patterns (Fig. 1). Single mutants of BLH2 and BLH4 exhibited no phenotypes in seed mucilage, while the blh2 blh4 mutant displayed reduced mucilage adherence on vigorous shaking (Fig. 2). Correspondingly, a redistribution of sugars from the adherent layer to the water-soluble layer was detected in the blh2 blh4 mucilage on vigorous shaking (Table 1). Nevertheless, the blh2 blh4 mutant exhibited no abnormalities in MSC differentiation, mucilage production and release, or monosaccharide contents (Fig. 2; Table 1; Supplemental Fig. S1), indicating that the biosynthesis and monosaccharide compositions of mucilage were not affected in blh2 blh4. Therefore, the reduced adherence of the blh2 blh4 mucilage was speculated to have originated from changes in the macromolecular structure of the pectic carbohydrate complex. In support of this hypothesis, immunolabeling and biochemical assays demonstrated that the DM of HG in the blh2 blh4 mucilage was significantly increased compared to the wild type (Fig. 4). Therefore, we speculated that the reduced adherence of the blh2 blh4 mucilage is associated with the increased DM of HG.
Several studies have proven that the DM of HG plays an important role in controlling mucilage structure or extrusion. Mutations of PMEI6, SBT1.7, FLY1, and STK lead to mucilage extrusion defects due to decreased DM of HG in mucilage (Rautengarten et al., 2008; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013; Ezquer et al., 2016). De-methylesterified HGs are prone to be cross-linked by Ca2+ ions to form a compact egg-box structure (Moustacas et al., 1991; Micheli, 2001; Wolf et al., 2009). It is hypothesized that the increased amounts of de-methylesterified HGs in these mutants could promote the formation of Ca2+ bridges and strengthen the structure of the pectin gel matrix, thus reducing the mucilage expansion potential (Saez-Aguayo et al., 2013; Voiniciuc et al., 2013; Ezquer et al., 2016; Shi et al., 2018). This is supported by the fact that imbibition of these mutant seeds with EDTA, a Ca2+ chelator, facilitates mucilage extrusion (Rautengarten et al., 2008; Arsovski et al., 2009; Saez-Aguayo et al., 2013; Voiniciuc et al., 2013). Conversely, in the current study, we demonstrated that the increased DM in the blh2 blh4 mucilage was associated with decreased binding of HG by Ca2+ (Fig. 3D), which could reduce the formation of the egg-box structure and weaken the strength of the pectin gel matrix, eventually leading to decreased mucilage adherence to seeds.
The redistribution of sugars from the adherent layer to the water-soluble layer in the blh2 blh4 mucilage could be mainly attributed to the reduced adherence of the inner layer on vigorous shaking (Table 1). Since the extraction of the water-soluble mucilage was performed by vigorous shaking at 200 rpm for 1 h, it is likely that a proportion of the adherent mucilage of blh2 blh4 was detached from seeds and thus included in the water-soluble fraction. Similarly, redistribution of sugars from the adherent layer to the water-soluble layer was also observed for the pme58 mutant upon EDTA extraction (Turbant et al., 2016). Conversely, an opposite tendency in sugar distribution was detected in mutants with reduced DM of HG in mucilage (e.g. pmei6, fly1, and myb52). In these mutants, sugars were redistributed from the water-soluble layer to the adherent layer (Saez-Aguayo et al., 2013; Voiniciuc et al., 2013; Shi et al., 2018). These findings suggest that HG methylesterification influences the attachment of polysaccharides in the adherent layer.
Collectively, it can be concluded that apart from cellulose and xylan, which anchor mucilage to the seed (Sullivan et al., 2011; Voiniciuc et al., 2015a; Hu et al., 2016a, 2016b; Ralet et al., 2016), the HG methylesterification also affects the mucilage adherence. Our study demonstrated that BLH2 and BLH4 function redundantly in establishing an appropriate DM of HG in mucilage which is required for the maintenance of mucilage adherence.
BLH2 and BLH4 Positively Modulate HG De-Methylesterification in Mucilage
We showed that BLH2 and BLH4 positively regulate PME activity in the seed coat (Fig. 3F), suggesting that BLH2 and BLH4 function in promoting HG de-methylesterification in seed coat cells. Gene expression analysis revealed that the expression of PME58 was significantly reduced in blh2 blh4 seeds. PME58 has been demonstrated to function in the de-methylesterification of HG in seed mucilage (Turbant et al., 2016). We demonstrated that BLH2 and BLH4 directly bind to the PME58 promoter and activate its expression (Fig. 5). Furthermore, we identified that pme58 mutants also exhibited reduced mucilage adherence similar to that of blh2 blh4 on vigorous shaking. Moreover, overexpression of PME58 in blh2 blh4 could rescue the mucilage adherence defects of blh2 blh4 (Fig. 6). It is noteworthy that the blh2 blh4 pme58-2 triple mutant had comparable alterations in mucilage adherence, seed coat PME activity, and DM of HG in mucilage relative to blh2 blh4 and pme58 mutants (Figs. 3, E and F, and 6). These results suggest that impaired PME58 function is the main contributor to the defects of mucilage adherence in blh2 blh4.
Transcription factors STK and MYB52 negatively regulate the de-methylesterification of HG in seed mucilage by modulating PME activity via the common target gene PMEI6 (Ezquer et al., 2016; Shi et al., 2018). Our RT-qPCR results showed that the expression levels of STK, MYB52, and PMEI6 were significantly increased in blh2 blh4 seeds (Fig. 4B), implying that BLH2 and BLH4 might negatively affect the STK-PMEI6 and MYB52-PMEI6 modules. However, it seems unlikely that STK and MYB52 are the direct targets of BLH2 and BLH4, as there was no TGACAGGT cis-element in STK and MYB52 promoters.
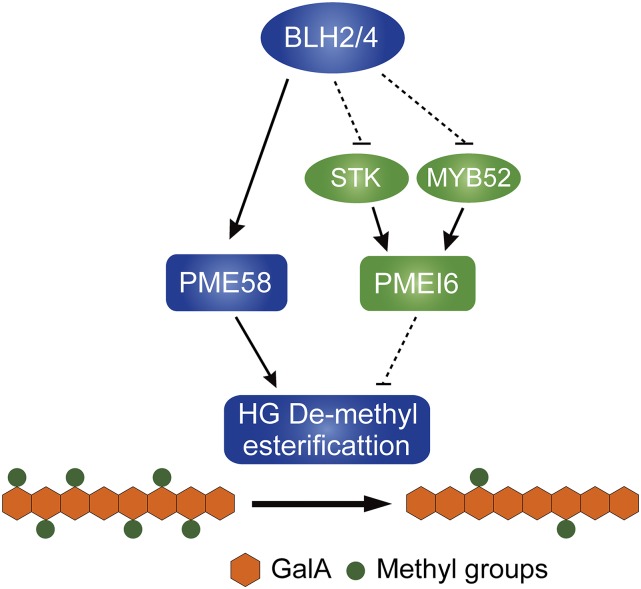
Taking all the data together, we proposed a working model of BLH2 and BLH4 in modulating the de-methylesterification of HG during seed mucilage production (Fig. 7). BLH2 and BLH4 act redundantly to directly activate the expression of PME58 that functions directly on de-methylesterification of HG in seed mucilage. Meanwhile, BLH2 and BLH4 also negatively regulate the STK-PMEI6 and MYB52-PMEI6 modules that negatively modulate the HG de-methylesterification in seed mucilage.
Figure 7.
A proposed working model of BLH2 and BLH4 in the modulation of HG de-methylesterification in seed mucilage. BLH2 and BLH4 act redundantly to directly activate the expression of PME58 that functions directly on the de-methylesterification of HG in seed mucilage. Meanwhile, BLH2 and BLH4 also negatively regulate the STK-PMEI6 and MYB52-PMEI6 modules. Solid lines indicate direct regulation or function. Dotted lines indicate indirect regulation or function.
In addition, since BLH and KNOX proteins usually form heterodimers (Bellaoui et al., 2001; Bhatt et al., 2004; Cole et al., 2006), it will be intriguing to investigate the KNOX partner(s) of BLH2/BLH4 in regulating the de-methylesterification of HG during seed mucilage production. KNOTTED-LIKE HOMEOBOX OF ARABIDOPSIS THALIANA 7 (KNAT7) seems to be one of the candidates, because the knat7 mutant has serious defects in mucilage adherence (Bhargava et al., 2013).
BLH2 and BLH4 Are Involved in Maintaining the Cellulose-Pectin Interactions in Seed Mucilage
The reduced mucilage adherence is usually accompanied by alterations in cellulose morphology, as observed in various mucilage mutants, such as cesa5, irregular xylem 7 (irx7), irx14, mum5, sos5, and cobra-like2 (cobl2; Sullivan et al., 2011; Griffiths et al., 2014; Ben-Tov et al., 2015; Voiniciuc et al., 2015a; Hu et al., 2016a, 2016b; Ralet et al., 2016). This firmly demonstrates that interactions between cellulose and pectin play an important role in maintaining the structure of seed mucilage. In the current study, the blh2 blh4 mucilage exhibited loss of diffuse cellulose-staining regions while the cellulose rays were still stained (Figs. 3 and 4; Supplemental Fig. S2), suggesting that the cellulose-pectin interactions were affected in the blh2 blh4 mucilage. The cellulose staining pattern of blh2 blh4 was similar to that of cesa5 and mum5 mutants, while contrasting with the sos5 and fei2 mutants (Griffiths et al., 2014, 2016; Ralet et al., 2016). Interestingly, the expression of MUM5 was significantly increased in blh2 blh4 seeds, while the expression of CESA5, SOS5, and FEI2 was not significantly altered (Fig. 4C). Since disruption of MUM5 results in a similar phenotype in cellulose labeling to blh2 blh4, the increased expression of MUM5 in blh2 blh4 seems unlikely to account for the blh2 blh4 phenotype. One possibility is that MUM5 is not under the direct control of BLH2 and BLH4. It might be regulated by other unidentified transcription factors that are engaged in the transcriptional network involving BLH2 and BLH4. These results suggest that BLH2/BLH4 might modulate cellulose-pectin interactions via pathways independent of MUM5, CESA5, SOS5, and FEI2. In addition, since the pme58 mutants have no phenotype in cellulose staining and overexpression of PME58 could not rescue the cellulose staining phenotype of blh2 blh4, the role of BLH2/BLH4 in maintaining cellulose structure would appear to be independent of their function in modulating the DM of HG. Further studies are needed to clarify the mechanisms of BLH2 and BLH4 in maintaining cellulose structure in seed mucilage.
CONCLUSIONS
In summary, our data unravel a regulatory mechanism of the de-methylesterification of HG in seed mucilage. BLH2 and BLH4 redundantly exert direct regulation of PME activity by activating the expression of PME58. To date, BLH2 and BLH4 represent the only two identified positive regulators of HG de-methylesterification in seed mucilage. Our findings provide insights into the regulatory mechanisms of the de-methylesterification of HG in seed mucilage and further demonstrate the importance of the de-methylesterification of HG in mucilage adherence. Furthermore, taking advantage of the phenotype of increased mucilage detachment on vigorous shaking used in this study will be helpful in identifying other regulators of the de-methylesterification of HG in seed mucilage in future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) saw1-1 (SALK_009120), saw2-1 (SALK_121117), saw1-1 saw2-1 (CS16377), saw1-2 saw2-1 (CS16378), saw1-1 saw2-2 (CS16379), pme58-1 (CS25025), and pme58-2 (SALK_055262) mutant lines were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/stocks/233532). To generate the proBLH2::BLH2-GFP blh2 blh4 and proBLH4::BLH4-GFP blh2 blh4 transgenic lines, the promoters of BLH2 (3,200 bp upstream of the ATG) and BLH4 (3,300 bp upstream of the ATG), as well as the full-length coding sequence of BLH2 and BLH4 were amplified and inserted into the KpnI site of a modified pCAMBIA-2301 vector (GFP coding sequence inserted into the BamHI site). The recombinant plasmid (Supplemental Fig. S6A) was introduced into blh2 blh4 plants by Agrobacterium tumefaciens-mediated transformation. To generate the PME58-OE blh2 blh4 transgenic lines, the full-length coding sequence of PME58 was amplified and inserted into the EcoRI site of the 35S::MYC vector (Shi et al., 2018). The resulting 35S::MYC-PME58 construct (Supplemental Fig. S7A) was introduced into blh2 blh4 plants by Agrobacterium tumefaciens-mediated transformation. Primers used for the constructs are listed in Supplemental Table S1.
Seeds were surface-sterilized and sown on one-half strength Murashige and Skoog solid medium containing 1% (w/v) Suc and 0.05% (w/v) MES monohydrate (pH 5.8). The proBLH2::BLH2-GFP blh2 blh4, proBLH4::BLH4-GFP blh2 blh4, and PME58-OE blh2 blh4 transgenic lines were screened on one-half strength Murashige and Skoog medium supplemented with kanamycin (30 mg L−1). After a 2-d treatment at 4°C for synchronization, seeds were germinated at 22°C in a chamber (16-h-light/8-h-dark, 120 µmol m−2 s−1, 65% relative humidity). Seven-day-old seedlings were transferred to soil and grown under the same conditions to set seeds. Seed lots used in individual experiments were harvested from plants grown simultaneously.
Ruthenium Red Staining and Determination of Adherent Mucilage Volumes
Mature dry seeds were imbibed in water and directly stained with 0.01% (w/v) ruthenium red (Sigma-Aldrich) for 30 min. Alternatively, the imbibed seeds were first subjected to shaking at 200 rpm for 10 min to 2 h, and then stained with 0.01% (w/v) ruthenium red for 30 min. Seeds were photographed with the BX51 light microscope (Olympus). More than 20 seeds were examined in each experiment. The length (2a) and width (2b) of each seed, as well as the length (2A) and width (2B) of the same seed plus adherent mucilage were measured using ImageJ software (V1.8). The volume of adherent mucilage was calculated by subtracting the volume of the seed (V = 4/3*π*a*b*b) from the volume of seed plus mucilage (V = 4/3*π*A*B*B).
Resin Section
Siliques at 4, 7, 10, and 13 DPA were fixed in 2.5% (w/v) glutaraldehyde in phosphate-buffered saline (PBS; pH 7.0) at 4°C overnight. After being washed with PBS, samples were postfixed in 1% (v/v) osmium tetraoxide in PBS for 1 h. The fixed samples were subsequently washed twice with PBS for 15 min, dehydrated through a gradient ethanol series (30% to 100% [v/v]), embedded in Spurr’s resin and cut into 1.0-µm sections. Sections were stained for 5 min in 0.1% (w/v) toluidine blue O dissolved in PBS and photographed with the BX51 light microscope (Olympus). More than 10 siliques at each stage were sectioned.
Mucilage Extraction and Monosaccharide Quantification
One hundred milligrams of dry seeds were imbibed with 5 mL of distilled water and shaken at 200 rpm for 1 h. The supernatant containing the water-soluble mucilage was collected. After two washes with 1 mL of distilled water, the supernatants were pooled together as the water-soluble mucilage extracts. Seeds were then resuspended in 5 mL of distilled water and subjected to ultrasonic treatment (200 W, 2 min) using an ultrasonic device with a 2 mm probe (Zhao et al., 2017). After two washes with 1 mL of distilled water, the supernatants were pooled together as the adherent mucilage extracts. The mucilage extracts were dialyzed extensively against running water for 3 d and then lyophilized. The extraction was performed in three biological replicates.
The determination of the monosaccharide composition in water-soluble and adherent mucilage was performed as previously described (Hu et al., 2016a). Briefly, 2 mg of mucilage was hydrolyzed with 1 mL of 2 m trifluoroacetic acid (TFA) at 121°C for 2 h. The TFA was evaporated under a stream of nitrogen. The hydrolysates were derivatized with 400 μL of 1-phenyl-3-methyl-5-pyrazolone and 400 μL of 0.3 m NaOH at 70°C for 30 min. After neutralization with 400 μL of 0.3 m HCl, the derivatives were extracted three times with chloroform and then analyzed by a Hypersil ODS-2 C18 column (4.6×250 mm; Thermo Fisher Scientific) using a Waters HPLC System. Ten μL of the derivatives were injected, eluted with 82% (v/v) PBS (0.1 m, pH 7.0) and 18% (v/v) acetonitrile at 1 mL min−1, and monitored at 245 nm. A mixture of monosaccharide containing GalA, arabinose (Ara), Rha, Gal, Glc (Glu), Man, and Xyl was included in the analysis as the standard sample. The quantification was performed in three biological replicates.
Crystalline Cellulose Content Quantification
Twenty milligrams of de-mucilaged seeds were ground to a fine powder in liquid nitrogen and de-starched using a total starch assay kit (Megazyme). After being washed sequentially with ethanol and acetone, the samples were dried under a vacuum at 60°C to obtain the alcohol insoluble residue. For the determination of the crystalline cellulose content, 5 mg of alcohol insoluble residue (or 2 mg of mucilage) was hydrolyzed in 2 mL of 2 m TFA at 121°C for 2 h. After centrifugation, the pellets were suspended in 1 mL Updegraff reagent (acetic acid:nitric acid:water, 8:1:2 [v/v/v]) and heated at 100°C for 1 h (Updegraff, 1969). After centrifugation, pellets were hydrolyzed in 3 mL of 72% (w/v) H2SO4 for 30 min. The amounts of crystalline cellulose were quantified using the phenolsulfuric acid method with a dehydration factor of 0.9 (Ge et al., 2012).
Immunolabeling Procedures
Whole seeds were blocked for 1 h with PBS containing 3% (w/v) nonfat milk powder and then incubated with 10-fold diluted primary monoclonal antibodies (JIM5, JIM7, CCRC-M36, CCRC-M38, and CBM3a) in the PBS-milk solution for 2 h at room temperature. Seeds were then gently washed three times in PBS and incubated with Alexa Fluor488-tagged donkey antirat IgG (for JIM series; Thermo Fisher Scientific) or Alexa Fluor488-tagged donkey antimouse IgG (for CCRC series; Thermo Fisher Scientific) diluted 100-fold in the PBS-milk solution for 1 h in the dark at room temperature. For CBM3a, an additional incubation with 100-fold-diluted anti-His monoclonal antibody in the PBS-milk solution for 1 h was performed before incubation with the AlexaFluor488-tagged donkey antimouse IgG. After three washes in PBS, seeds were counterstained with Calcofluor white (Sigma-Aldrich) for 5 min. More than 20 seeds were labeled in each experiment. Images were captured with a FluoView FV1000 confocal laser scanning microscope (Olympus). Calcofluor and Alexa Fluor488 were excited with a 405 nm diode laser and a 488 nm argon laser, respectively. Fluorescence emission was recorded between 410 and 500 nm for Calcofluor and between 500 and 630 nm for Alexa Fluor488. The same settings for image acquisition were used within each immunolabeling experiment.
ELISA
The CCRC and JIM antibodies were used with the conventional PBS buffer, whereas the 2F4 antibody required Tris-buffered calcium and saline (TCS) buffer (20 mm Tris-HCl [pH 8.2], 0.5 mm CaCl2, and 150 mm NaCl). One hundred microliters mucilage extractions (100 μg mL−1) were coated onto 96-well microtitre plates at 4°C overnight. Coating solutions were removed and 200 μL PBS/TCS containing 3% (w/v) nonfat milk were added to block the plates at 4°C overnight. Plates were washed and 100 μL of 10-fold diluted primary antibodies in the PBS/TCS-milk solution were added to each well. After incubation at 37°C for 2 h, plates were washed with PBS/TCS and incubated with 100-fold diluted horseradish peroxidase-conjugated antirat (JIM series) or antimouse (CCRC series) secondary antibodies in the PBS/TCS-milk solution at 37°C for 2 h. After washing with PBS/TCS, antibody binding was determined by the addition of 150 μL horseradish peroxidase -substrate (10 mL 0.1 m sodium acetate [pH 5.5], 200 μL 1% [w/v] tetramethylbenzidine, and 18 μL 6% [v/v] H2O2) to each well. The reaction was terminated after 10 min by the addition of 50 μL of 2 m H2SO4. The absorbance was measured at 450 nm.
Determination of the DM of HG in Mucilage
Whole mucilage was extracted by shaking 20 mg mature dry seeds in 400 μL of 50 mm EDTA with 25 movements min−1 using a bead mill (Tissue Lyser II, Qiagen). After the seeds were settled for 10 min, 200 μL of supernatant was transferred into a new tube and saponified with 0.25 m NaOH for 1 h at room temperature with rotation. The reaction was neutralized with 0.25 m HCl and centrifuged for 10 min at 10,000g. The amount of methanol released after the saponification reaction was measured by a colorimetric method (Klavons and Bennett, 1986). One hundred micrioliters of supernatant was transferred into a new 1.5-mL tube, oxidized with 0.5 units of alcohol oxidase (Sigma-Aldrich) for 15 min at 25°C and incubated with 100 μL of freshly prepared 0.02 m 2,4-pentanedione (dissolved in 2 m ammonium acetate and 0.05 m acetic acid) for 15 min at 60°C. After cooling on ice for 2 min, the absorbance was measured at 412 nm and quantified using a methanol standard curve. The uronic acid content was determined by the meta-hydroxydiphenyl method (Blumenkrantz and Asboe-Hansen, 1973) using GalA as the standard. Forty micrioliters of 5-fold-diluted supernatant was transferred into a new 1.5 mL microtube and hydrolyzed with 220 μL of concentrated sulfuric acid containing 0.0125M Sodium tetraborate (Sigma-Aldrich) for 5 min at 100°C. After cooling on ice, 4 μL of 0.15% (w/v) meta-hydroxydiphenyl (Sigma-Aldrich) in 0.5% (w/v) NaOH was added. Absorbance was measured at 525 nm. The DM of HG was calculated as the percentage molar ratio of methanol to uronic acid (Ralet et al., 2012).
Determination of PME Activity in the Seed Coat
Developing seeds at 7–10 DPA were harvested from about 100 siliques and scattered on glass slides. By means of pressing the developing seeds with another slide, the embryos and most of the endosperms were squeezed out of the seed coat. Then seed coats (with some endosperm attached) were carefully harvested and ground in 400 μL of extraction buffer (1 m NaCl, 12.5 mm citric acid, and 50 mm Na2HPO4, pH 6.5). The homogenate was shaken for 1 h at 4°C and the supernatant was collected by centrifugation at 20,000g for 15 min. Protein concentrations were determined according to the Bradford method (Bradford, 1976). PME activity was determined according to a previously reported method (Anthon and Barrett, 2004). Briefly, 50 μL of protein extract was added into a reaction mix containing 100 mm PBS buffer, 0.4 mg mL−1 citrus fruit pectin (≥85% esterified; Sigma-Aldrich), and 0.1 unit alcohol oxidase (Sigma-Aldrich) and incubated at 30°C for 10 min. Subsequently, 200 μL of 0.5 m NaOH solution containing 5 mg mL−1 Purpald (Sigma-Aldrich) was added. After incubation at 30°C for 30 min, 550 μL of distilled water was added to give a final volume of 1 mL, and the absorbance was measured at 550 nm. PME activity was calculated as nanomoles of methanol per micrograms of protein using a methanol standard curve.
In Situ Hybridization
Siliques at four different developing stages (4, 7, 10, and 13 DPA) were fixed, dehydrated, embedded, sectioned, and attached to adhesive slides as previously described (Hu et al., 2016a). For the preparation of Digoxigenin-labeled RNA probes, the coding regions of BLH2 and BLH4 were amplified by PCR using specific primers (Supplemental Table S1) and individually cloned into the pGEM-T vector. Digoxigenin-labeled sense and antisense RNA probes were generated in vitro from either T7 or SP6 promoter using the Digoxigenin RNA Labeling kit (Roche). The in situ hybridization procedure was performed as previously described (Mayer et al., 1998). The blocking reagent, anti-Digoxigenin antibody, and NBT/BCIP Stock Solution used in the experiments were purchased from Roche. The antisense and sense samples were operated in parallel throughout the procedure. Images were captured with the BX51 light microscope (Olympus).
Gene Expression Analysis
Total RNA was isolated from siliques and seeds using the Cetyltrimethyl Ammonium Bromide (CTAB) method. The first-strand complementary DNA was reverse transcribed using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen) according to the manufacturer’s instructions. The quantitative real-time PCR was performed using SYBR Premix Ex Taq (TaKaRa) with the LightCycler 480 detection system (Roche). The ACTIN2 gene was used as the internal control. Relative expression was calculated by the 2-∆∆Ct method (Livak and Schmittgen, 2001). Gene-specific primers are listed in Supplemental Table S1.
EMSA
To express BLH2 in E. coli, the full-length coding sequence of BLH2 was cloned into the pGEX-4T-1 vector (GE Healthcare) and introduced into E. coli BL21 cells (Novagen). The GST-BLH2 fusion protein was expressed in the transformed cells cultured at 28°C, induced with 1 mm isopropyl β-d-thiogalactoside (IPTG), and then purified with Glutathione Sepharose 4B resin (GE Healthcare). To prepare double-strand DNA probes, two complementary oligonucleotides labeled by biotin at the N terminus were mixed, denatured at 95°C for 10 min, and annealed by gradually cooling to room temperature. The EMSA reaction and detection was performed using the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
ChIP Assays
Chromatin was extracted from 7- to 10-DPA siliques of the proBLH2::BLH2-GFP and proBLH4::BLH4-GFP transgenic plants and wild-type plants. The procedures for chromatin extraction and immunoprecipitation were performed as previously described (Xu et al., 2015) with some modifications. Briefly, samples were harvested and cross-linked in 1% (w/v) paraformaldehyde under vacuum for 30 min, followed by termination of the cross-linking in 0.125 m Gly under vacuum for 5 min. After grinding in liquid nitrogen, 2 g of powder was used for chromatin extraction following a previously described method (Bowler et al., 2004). Chromatins were then sonicated to achieve an average DNA size between 0.2 and 1 kb. Immunoprecipitation was performed using the anti-GFP antibody (JL-8, Takara) conjugated with the Protein G Agarose (Millipore). Following incubation overnight, the antibody-chromatin complex was washed, eluted, and de-cross-linked. The precipitated DNA was then recovered by phenol-chloroform extraction and analyzed by qPCR using the primers listed in Supplemental Table S1.
Transcriptional Regulation Assays in Protoplasts
To assess the regulatory activity of BLH2 and BLH4 on PME58 expression in protoplasts, the full-length coding sequences of BLH2 and BLH4 were individually inserted into a modified pBI221 vector (GUS reporter gene removed) downstream of the CaMV 35S promoter to create the effector constructs. The PME58 promoter was inserted into another modified pBI221 vector (CaMV 35S promoter removed) upstream of the GUS reporter gene to create the reporter construct. The plasmid carrying the Renilla Luciferase gene driven by the CaMV 35S promoter (35S::LUC) was used as an internal control for normalization. The isolation and transformation of Arabidopsis mesophyll protoplasts were performed according to a previously reported method (Yoo et al., 2007). After transformation, the protoplasts were incubated at 22°C for 18 h in dark. GUS and LUC activities were measured and the relative GUS activity (GUS/LUC) was calculated. All experiments were performed for three biological triplicates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT4G36870 (BLH2); AT2G23760 (BLH4); AT5G49180 (PME58); AT2G47670 (MYB52); and AT4G09960 (STK).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mucilage phenotypes of the other alleles of saw1 saw2.
Supplemental Figure S2. Differentiation of seed coat epidermal cells and mucilage production in developing seeds of blh2 blh4.
Supplemental Figure S3. Immunolabeling of RG I in the blh2 blh4 mucilage.
Supplemental Figure S4. Immunolabeling of crystalline cellulose in the blh2 blh4 mucilage.
Supplemental Figure S5. Determination of crystalline cellulose content in the blh2 blh4 mucilage.
Supplemental Figure S6. Characterization of the proBLH2::BLH2-GFP and proBLH4::BLH4-GFP transgenic plants and RR staining of the transgenic seeds.
Supplemental Figure S7. Generation of the PME58-OE blh2 blh4 transgenic plants.
Supplemental Figure S8. Immunolabeling assays of HG methylesterification pattern in the adherent mucilage of PME58-OE blh2 blh4 seeds.
Supplemental Figure S9. Spatiotemporal expression pattern of PME58.
Supplemental Figure S10. Determination of PME activity in blh2 blh4 leaves.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Michael G. Hahn (Complex Carbohydrate Research Center, University of Georgia) for providing the CCRC antibody, and Paul Knox (University of Leeds) for providing the JIM5 and JIM7 antibodies.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31701069, 31770336, and 31670302), the Key Research and Development Program of Shandong (grant no. 2017CXGC0309), the Major Basic Research Project of Shandong Natural Science Foundation (grant no. ZR2018ZC0335), the China Postdoctoral Science Foundation (grant no. 2016M602207), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (grant no. 2014187), and the Taishan Scholar Program of Shandong (to G.Z.).
Articles can be viewed without a subscription.
References
- Anthon GE, Barrett DM(2004) Comparison of three colorimetric reagents in the determination of methanol with alcohol oxidase. Application to the assay of pectin methylesterase. J Agric Food Chem 52: 3749–3753 [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL(2010) Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav 5: 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL(2009) AtBXL1 encodes a bifunctional β-D-xylosidase/α-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman T, De Rycke R, Viane R, Inze D(2000) Histological study of seed coat development in Arabidopsis thaliana. J Plant Res 113: 139–148 [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW(2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13: 2455–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tov D, Abraham Y, Stav S, Thompson K, Loraine A, Elbaum R, de Souza A, Pauly M, Kieber JJ, Harpaz-Saad S(2015) COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol 167: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Ahad A, Wang S, Mansfield SD, Haughn GW, Douglas CJ, Ellis BE(2013) The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 237: 1199–1211 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H(2004) VAAMANA—a BEL1-like homeodomain protein interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328: 103–111 [DOI] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G(1973) New method for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J(2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Bradford MM.(1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W(2006) Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res 34: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagel DJ, Liu YS, Zhong L, Luo Y, Himmel ME, Xu Q, Zeng Y, Ding SY, Smith S(2011) In situ imaging of single carbohydrate-binding modules on cellulose microfibrils. J Phys Chem B 115: 635–641 [DOI] [PubMed] [Google Scholar]
- Ezquer I, Mizzotti C, Nguema-Ona E, Gotté M, Beauzamy L, Viana VE, Dubrulle N, Costa de Oliveira A, Caporali E, Koroney AS, et al. (2016) The developmental regulator SEEDSTICK controls structural and mechanical properties of the Arabidopsis seed coat. Plant Cell 28: 2478–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Burlat V, Dunand C(2015) Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Ge XM, Green VS, Zhang NN, Sivakumar G, Xu JF(2012) Eastern gamagrass as an alternative cellulosic feedstock for bioethanol production. Process Biochem 47: 335–339 [Google Scholar]
- Golz JF, Allen PJ, Li SF, Parish RW, Jayawardana NU, Bacic A, Doblin MS(2018) Layers of regulation—insights into the role of transcription factors controlling mucilage production in the Arabidopsis seed coat. Plant Sci 272: 179–192 [DOI] [PubMed] [Google Scholar]
- Griffiths JS, Crepeau MJ, Ralet MC, Seifert GJ, North HM(2016) Dissecting seed mucilage adherence mediated by FEI2 and SOS5. Front Plant Sci 7: 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JS, Tsai AY, Xue H, Voiniciuc C, Sola K, Seifert GJ, Mansfield SD, Haughn GW(2014) SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol 165: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Pautot V(2010) Plant development: A TALE story. C R Biol 333: 371–381 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Vibe Scheller H(2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, McFarlane HE, Xu S, Divi UK, Forward B, Western TL, Kieber JJ(2011) Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. Plant J 68: 941–953 [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A(2005) Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472–477 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Western TL(2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Li J, Wang X, Zhao X, Yang X, Tang Q, He G, Zhou G, Kong Y(2016a) Xylan synthesized by Irregular Xylem 14 (IRX14) maintains the structure of seed coat mucilage in Arabidopsis. J Exp Bot 67: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Li J, Yang X, Zhao X, Wang X, Tang Q, He G, Zhou G, Kong Y(2016b) Irregular xylem 7 (IRX7) is required for anchoring seed coat mucilage in Arabidopsis. Plant Mol Biol 92: 25–38 [DOI] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Van Loey AM, Hendrickx ME(2010) Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr Res 345: 2583–2595 [DOI] [PubMed] [Google Scholar]
- Klavons JA, Bennett RD(1986) Determination of methanol using alcohol oxidase and its application to methyl-ester content of pectins. J Agric Food Chem 34: 597–599 [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW(2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque-Tremblay G, Müller K, Mansfield SD, Haughn GW(2015) HIGHLY METHYL ESTERIFIED SEEDS is a pectin methyl esterase involved in embryo development. Plant Physiol 167: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liners F, Letesson JJ, Didembourg C, Van Cutsem P(1989) Monoclonal antibodies against pectin: Recognition of a conformation induced by calcium. Plant Physiol 91: 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, You S, Taylor-Teeples M, Li WL, Schuetz M, Brady SM, Douglas CJ(2014) BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 26: 4843–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Louvet R, Cavel E, Gutierrez L, Guénin S, Roger D, Gillet F, Guerineau F, Pelloux J(2006) Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 224: 782–791 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM(2007) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T(1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S(2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F.(2001) Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Mohnen D.(2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Moustacas AM, Nari J, Borel M, Noat G, Ricard J(1991) Pectin methylesterase, metal ions and plant cell-wall extension. The role of metal ions in plant cell-wall extension. Biochem J 279: 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North HM, Berger A, Saez-Aguayo S, Ralet MC(2014) Understanding polysaccharide production and properties using seed coat mutants: Future perspectives for the exploitation of natural variants. Ann Bot 114: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palin R, Geitmann A(2012) The role of pectin in plant morphogenesis. Biosystems 109: 397–402 [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook S, Höfte H(2012) Cell wall mechanics and growth control in plants: The role of pectins revisited. Front Plant Sci 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ(2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Phan JL, Tucker MR, Khor SF, Shirley N, Lahnstein J, Beahan C, Bacic A, Burton RA(2016) Differences in glycosyltransferase family 61 accompany variation in seed coat mucilage composition in Plantago spp. J Exp Bot 67: 6481–6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeek P, Smeekens S(1995) The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Crépeau MJ, Vigouroux J, Tran J, Berger A, Sallé C, Granier F, Botran L, North HM(2016) Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol 171: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralet MC, Williams MA, Tanhatan-Nasseri A, Ropartz D, Quéméner B, Bonnin E(2012) Innovative enzymatic approach to resolve homogalacturonans based on their methylesterification pattern. Biomacromolecules 13: 1615–1624 [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T(2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL(1995) The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83: 735–742 [DOI] [PubMed] [Google Scholar]
- Saez-Aguayo S, Ralet MC, Berger A, Botran L, Ropartz D, Marion-Poll A, North HM(2013) PECTIN METHYLESTERASE INHIBITOR6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell 25: 308–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Ren A, Tang X, Qi G, Xu Z, Chai G, Hu R, Zhou G, Kong Y(2018) MYB52 negatively regulates pectin demethylesterification in seed coat mucilage. Plant Physiol 176: 2737–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Boschke I, Hake S(2002) Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA 99: 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Ralet MC, Berger A, Diatloff E, Bischoff V, Gonneau M, Marion-Poll A, North HM(2011) CESA5 is required for the synthesis of cellulose with a role in structuring the adherent mucilage of Arabidopsis seeds. Plant Physiol 156: 1725–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tioni MF, Viola IL, Chan RL, Gonzalez DH(2005) Site-directed mutagenesis and footprinting analysis of the interaction of the sunflower KNOX protein HAKN1 with DNA. FEBS J 272: 190–202 [DOI] [PubMed] [Google Scholar]
- Turbant A, Fournet F, Lequart M, Zabijak L, Pageau K, Bouton S, Van Wuytswinkel O(2016) PME58 plays a role in pectin distribution during seed coat mucilage extrusion through homogalacturonan modification. J Exp Bot 67: 2177–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff DM.(1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Viola IL, Gonzalez DH(2006) Interaction of the BELL-like protein ATH1 with DNA: Role of homeodomain residue 54 in specifying the different binding properties of BELL and KNOX proteins. Biol Chem 387: 31–40 [DOI] [PubMed] [Google Scholar]
- Viola IL, Gonzalez DH(2009) Binding properties of the complex formed by the Arabidopsis TALE homeodomain proteins STM and BLH3 to DNA containing single and double target sites. Biochimie 91: 974–981 [DOI] [PubMed] [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW(2013) FLYING SAUCER1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Günl M, Schmidt MHW, Usadel B(2015a) Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol 169: 2481–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Schmidt MH, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Günl M(2015b) MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol 169: 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Yang B, Schmidt MH, Günl M, Usadel B(2015c) Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int J Mol Sci 16: 3452–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL.(2012) The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Western TL, Skinner DJ, Haughn GW(2000) Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Limberg G, Buchholt HC, van Alebeek GJ, Benen J, Christensen TMIE, Visser J, Voragen A, Mikkelsen JD, Knox JP(2000) Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides, and enzymatic degradation. Carbohydr Res 327: 309–320 [DOI] [PubMed] [Google Scholar]
- Windsor JB, Symonds VV, Mendenhall J, Lloyd AM(2000) Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J 22: 483–493 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mouille G, Pelloux J(2009) Homogalacturonan methyl-esterification and plant development. Mol Plant 2: 851–860 [DOI] [PubMed] [Google Scholar]
- Xiao C, Anderson CT(2013) Roles of pectin in biomass yield and processing for biofuels. Front Plant Sci 4: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zong W, Hou X, Yao J, Liu H, Li X, Zhao Y, Xiong L(2015) OsARID3, an AT-rich interaction domain-containing protein, is required for shoot meristem development in rice. Plant J 83: 806–817 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL(2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G(2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Qiao L, Wu AM(2017) Effective extraction of Arabidopsis adherent seed mucilage by ultrasonic treatment. Sci Rep 7: 40672. [DOI] [PMC free article] [PubMed] [Google Scholar]