Sulfated salicinoids in poplar are produced by sulfotransferase1 and may function in sulfur homeostasis and plant defense.
Abstract
Salicinoids form a specific class of phenolic glycosides characteristic of the Salicaceae. Although salicinoids accumulate in large amounts and have been shown to be involved in plant defense, their biosynthesis is unclear. We identified two sulfated salicinoids, salicin-7-sulfate and salirepin-7-sulfate, in black cottonwood (Populus trichocarpa). Both compounds accumulated in high amounts in above-ground tissues including leaves, petioles, and stems, but were also found at lower concentrations in roots. A survey of salicin-7-sulfate and salirepin-7-sulfate in a subset of poplar (Populus sp.) and willow (Salix sp.) species revealed a broader distribution within the Salicaceae. To elucidate the formation of these compounds, we studied the sulfotransferase (SOT) gene family in P. trichocarpa (PtSOT). One of the identified genes, PtSOT1, was shown to encode an enzyme able to convert salicin and salirepin into salicin-7-sulfate and salirepin-7-sulfate, respectively. The expression of PtSOT1 in different organs of P. trichocarpa matched the accumulation of sulfated salicinoids in planta. Moreover, RNA interference-mediated knockdown of SOT1 in gray poplar (Populus × canescens) resulted in decreased levels of sulfated salicinoids in comparison to wild-type plants, indicating that SOT1 is responsible for their formation in planta. The presence of a nonfunctional SOT1 allele in black poplar (Populus nigra) was shown to correlate with the absence of salicin-7-sulfate and salirepin-7-sulfate in this species. Food choice experiments with leaves from wild-type and SOT1 knockdown trees suggest that sulfated salicinoids do not affect the feeding preference of the generalist caterpillar Lymantria dispar. A potential role of the sulfated salicinoids in sulfur storage and homeostasis is discussed.
In response to biotic and abiotic stresses, plants produce a multitude of specialized metabolites, with more than 200,000 structures known to date (Pichersky and Lewinsohn, 2011). The high number and the structural diversity of specialized metabolites are mainly achieved by modifications of a limited number of core structures. Such modifications include glycosylation, methylation, acetylation, or sulfation (e.g. Varin et al., 1992; Saito et al., 2013). Many specialized metabolites such as terpenes, flavonoids, or cyanogenic glycosides are widely distributed through the plant kingdom; others, however, are restricted to single plant orders or families. Benzoxazinoids, for example, are mainly restricted to the Poaceae, while glucosinolates are exclusively formed in the Brassicales (Halkier and Gershenzon, 2006; Niemeyer, 2009). Salicinoids represent another example of family-specific plant specialized metabolites, being restricted to the Salicaceae (Philippe and Bohlmann, 2007; Chen et al., 2009; Boeckler et al., 2011).
Like other specialized metabolites, salicinoids are defined by their chemical core structure (Boeckler et al., 2011). The basic structural element is 2-(hydroxymethyl)phenyl-β-d-glucopyranoside (salicin), and derivatives of this core structure are referred to as complex salicinoids (Boeckler et al., 2011). Common modifications of the core structure are mostly esterifications with organic acids at the primary alcohol function of the 2-hydroxybenzyl alcohol, or at the 2′ or 6′ position of the glucopyranose (Boeckler et al., 2011). Thus far, more than 20 different salicinoids have been described in various poplars (Populus sp.), willows (Salix sp.), and a few other related species (Boeckler et al., 2011). Some salicinoids, for instance salicin and salicortin, are widely distributed in the Salicaceae, whereas others show a more species-specific accumulation, for example idescarpin in the Japanese orange cherry (Idesia polycarpa; Picard et al., 1994; Chou et al., 1997; Feistel et al., 2017).
Salicinoids have mainly been investigated because of their function as defense compounds against various herbivores (Lindroth and St. Clair, 2013). Several mammalian herbivores, e.g. mountain hares (Lepus timidus), field voles (Microtus agrestis), and elk (Cervus elaphus), show a selective feeding behavior and prefer to feed on low salicinoid-containing plants over salicinoid-rich plants (Tahvanainen et al., 1985a; Bailey et al., 2006; Heiska et al., 2007). Salicinoids also function as efficient feeding deterrents and fitness reducers for generalist insects, including lepidopteran herbivores (Lindroth and Peterson, 1988; Lindroth et al., 1988; Feistel et al., 2017) and coleopteran herbivores (Tahvanainen et al., 1985b; Kelly and Curry, 1991). In contrast to induced defenses that are exclusively expressed rapidly upon herbivory, salicinoids are mainly considered as constitutive defense (Boeckler et al., 2011; Rubert-Nason et al., 2015), although some studies reported moderately increased accumulation levels after herbivory (Clausen et al., 1989; Stevens and Lindroth, 2005; Young et al., 2010; Rubert-Nason et al., 2015; Fabisch et al., 2019). Whether such increased accumulation is caused by de novo biosynthesis or by translocation of preformed salicinoids, is still unclear (Boeckler et al., 2011; Massad et al., 2014).
Although salicinoids have been known and investigated for a long time, their biosynthesis remains elusive. Inhibition of the Phe ammonia lyase in bay willow (Salix pentandra) shoot tips led to reduced accumulation of salicinoids (Ruuhola and Julkunen-Titto, 2003). Feeding experiments with isotopically labeled precursors in purple willow (Salix purpurea; Zenk, 1967) and black poplar (Populus nigra; Babst et al., 2010) revealed that salicinoids, as other C6-C1 benzenoids, are derived from cinnamic acid. Chain-shortening, ortho-hydroxylation, reduction, glucosylation, and esterifications have been proposed as reaction steps in the salicinoid pathway, however, their exact sequence and the involved enzymes are still not clear (Tsai et al., 2006; Babst et al., 2010). Recently, a poplar acyltransferase has been reported that catalyzes the formation of benzyl benzoate (Chedgy et al., 2015), a compound hypothesized as a key intermediate in the formation of complex salicinoids such as salicortin or tremulacin (Babst et al., 2010).
The recent identification of two sulfated salicinoids, salicin-7-sulfate in willows (Noleto-Dias et al., 2018) and idesin hydrogen sulfate in I. polycarpa (Chou et al., 1997), indicates that salicinoids can act as substrates for sulfotransferases (SOTs; EC 2.8.2). In general, SOTs catalyze the transfer of a sulfuryl group from the cofactor 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to hydroxyl groups of a variety of different substrates (Klein and Papenbrock, 2004; Hirschmann et al., 2014). In plants, SOTs form midsized gene families with e.g. 35 members in rice (Oryza sativa; Chen et al., 2012) and 21 members in Arabidopsis (Arabidopsis thaliana; Klein and Papenbrock, 2004; Hirschmann et al., 2014). Sequence comparisons of SOTs revealed common sequence features at the primary sequence level, determining four distinct regions, I–IV (Marsolais and Varin, 1995; Klein and Papenbrock, 2004; Hirschmann et al., 2014). The regions I, II, and IV are proposed to be involved in the binding of the cofactor PAPS, while the function of region III has not been completely elucidated (Kakuta et al., 1998; Hirschmann et al., 2014). However, the prediction of SOT substrate specificity based on the primary sequence is still limited (Klein and Papenbrock, 2004; Luczak et al., 2013; Hirschmann et al., 2014). So far, only a few plant SOTs have been characterized in detail. Arabidopsis AtSOT10, for example, catalyzes the sulfation of brassinosteroids, AtSOT15 accepts 11- and 12-hydroxyjasmonate as substrate, AtSOT12 produces sulfated salicylic acid, and AtSOT16, AtSOT17, and AtSOT18 produce sulfated glucosinolates (Gidda et al., 2003; Piotrowski et al., 2004; Hirai et al., 2005; Marsolais et al., 2007; Baek et al., 2010). Sulfated metabolites are known to have various functions, being involved in plant defense and developmental processes, or acting as sulfur storage compounds (e.g. Faulkner and Rubery, 1992; Falk et al., 2007; Wang et al., 2014b). However, whether and how sulfation alters the impact of salicinoids as defense compounds is unclear. Moreover, SOTs that produce sulfated salicinoids in the Salicaceae have not been described.
The aim of our study was (1) to investigate the occurrence of sulfated salicinoids in the Salicaceae family, and (2) to elucidate the biochemical and genetic mechanisms underlying the formation of sulfated salicinoids in black cottonwood (Populus trichocarpa). Untargeted and targeted liquid chromatography/mass spectrometry (LC-MS) enabled us to identify and quantify sulfated salicinoids in different Salicaceae species and plant tissues. Using phylogenetic analysis and RNA sequencing (RNA-seq) data, we identified SOT genes in P. trichocarpa. Heterologous expression, biochemical characterization, and RNA interference (RNAi)-mediated knockdown of PtSOT1 revealed that the encoded enzyme catalyzes the sulfation of salicinoids in vitro and in vivo. To elucidate a potential function of sulfated salicinoids in plant defense, we performed feeding preference assays with gypsy moth (Lymantria dispar) caterpillars, a generalist insect herbivore known to feed on poplar trees. Moreover, we measured sulfur and sulfate in P. trichocarpa leaves and calculated the amounts of total sulfur and sulfate tied in to assess the role of salicinoids as sulfur-storage compounds.
RESULTS
Identification of Salicin-7-Sulfate and Salirepin-7-Sulfate in Poplar
In our attempt to study the formation of salicinoids in the Salicaceae, we searched for possible intermediates in methanol (MeOH) extracts made from leaves of P. trichocarpa, gray poplar (Populus × canescens), and the basket willow (Salix viminalis) using LC-MS (Supplemental Fig. S1). Both poplar species produce a wide array of salicinoids when compared with S. viminalis, which was reported to be poor in salicinoids (Palo, 1984). One major peak in the chromatograms of P. trichocarpa and P. × canescens that was absent in S. viminalis, represented a compound with a molecular weight of 366 (Supplemental Fig. S1). The accurate mass of the unknown compound (measured as 365.0543, [M-H]− fragment) suggested two possible molecular formulae: C13H20O8P2 (calculated [M-H]−, 365.0561, Δ = 4.7 ppm) and C13H18O10S (calculated [M-H]−, 365.0548, Δ = 1.2 ppm). However, the abundant [M-H+2]− isotopic mass peak of [M-H]− (6.7%; Supplemental Fig. S2) indicated a sulfur-containing structure. The compound was purified from P. x canescens leaves and identified by nuclear magnetic resonance (NMR) spectroscopy as salicin-7-sulfate (Supplemental Fig. S3), which was recently reported in willows (Noleto-Dias et al., 2018).
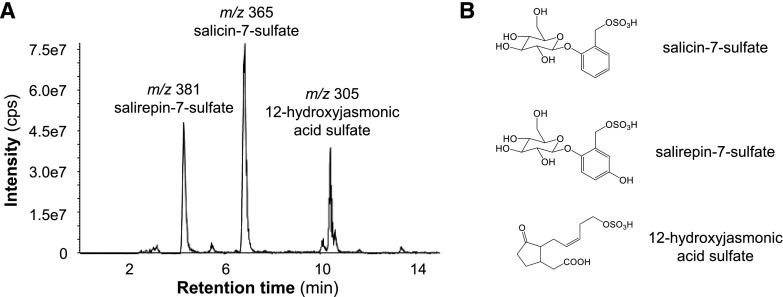
To screen for further sulfated salicinoids, we performed a precursor ion scan experiment with the sulfate (m/z 97) as the target fragment using a MeOH extract made from P. trichocarpa leaves. Besides salicin-7-sulfate, two intense peaks appeared in the chromatogram (Fig. 1). One of the compounds had a molecular weight of 306 and was identified as 12-hydroxyjasmonic acid sulfate by comparison with an authentic standard kindly provided by Claus Wasternack (Leibnitz Institute of Plant Biochemistry). The other compound had a molecular weight of 382 and was thus hypothesized to be a hydroxylated derivative of salicin-7-sulfate. The accurate mass of this compound was m/z 381.0497 [M-H]− with a proposed sum formula of C13H18O11S (calculated [M-H]−, m/z 381.0497, Δ = 0.01 ppm). After purification of the compound from P. × canescens leaves, the structure was determined by NMR spectroscopy to be salirepin-7-sulfate, a previously undescribed compound (Supplemental Fig. S3).
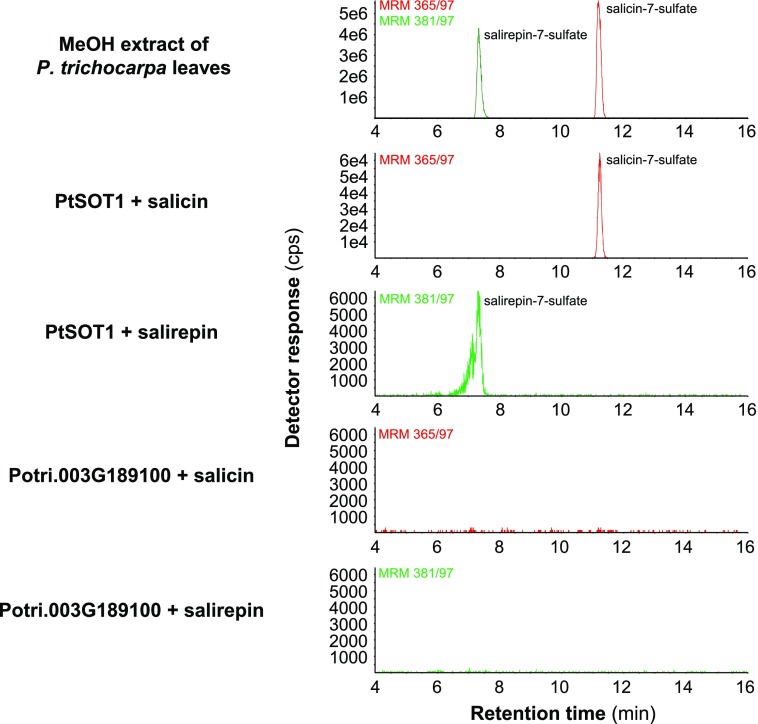
Figure 1.
Sulfated organic compounds accumulate in leaves of P. trichocarpa. A precursor ion scan experiment with the target mass m/z 97 [M-H]− (sulfate fragment) was performed to screen for sulfated organic compounds in P. trichocarpa leaves (A). Structural formulas of the three identified sulfated compounds (B). The 12-hydroxyjasmonic acid sulfate was identified by comparisons with an authentic standard. Structures of salicin-7-sulfate and salirepin-7-sulfate were verified by NMR. cps, counts per second.
Salicin-7-Sulfate and Salirepin-7-Sulfate Are Broadly Distributed within the Salicaceae and Mainly Accumulate in Above-Ground Organs
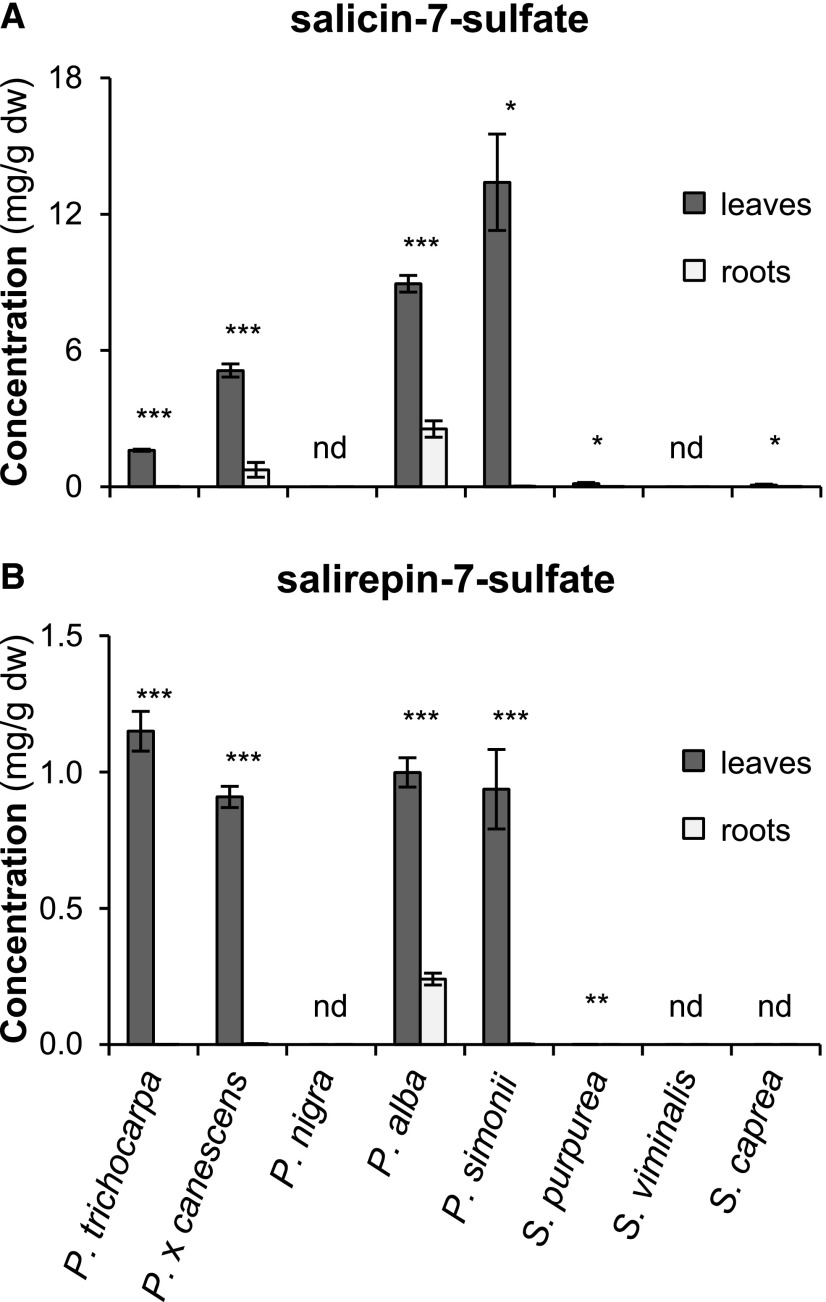
To study the distribution of sulfated salicinoids within the Salicaceae, we measured and quantified salicin-7-sulfate and salirepin-7-sulfate in leaves and roots of five poplar and three willow species. Both compounds were detected in MeOH extracts made from leaves of P. trichocarpa, P. × canescens, silver poplar (Populus alba), Simon’s poplar (Populus simonii), and S. purpurea (Fig. 2; Supplemental Table S1). Leaves of the goat willow (Salix caprea) contained only salicin-7-sulfate and no salirepin-7-sulfate (Fig. 2; Supplemental Table S1). Common to all species able to produce sulfated salicinoids was a significantly higher concentration of these compounds in leaves in comparison to roots (Fig. 2; Supplemental Table S1). In S. purpurea, the accumulation of salirepin-7-sulfate was detectable in leaves but not in roots (Fig. 2; Supplemental Table S1). P. nigra and S. viminalis, however, showed no accumulation of sulfated salicinoids, either in leaves or in roots (Fig. 2). Further analysis of four additional P. trichocarpa genotypes confirmed the consistent occurrence of sulfated salicinoids in this species (Supplemental Table S2). Moreover, salicin-7-sulfate could be detected in leaves of 50 trembling aspen (P. tremuloides) genotypes (Supplemental Fig. S4) and in the bark of I. polycarpa (Supplemental Table S2), a species from the Flacourtiaceae (or Salicaceae sensu lato) that, together with the genus Bennettiodendron, forms the sister group to Populus-Salix (Liu et al., 2016). The presence of salicin-7-sulfate and salirepin-7-sulfate in I. polycarpa suggests that the formation of these compounds evolved early in the evolution of the Salicaceae or even in a common ancestor of the Salicaceae and Flacourtiaceae.
Figure 2.
Salicin-7-sulfate and salirepin-7-sulfate concentrations in leaves (dark gray bars) and roots (light gray bars) of different poplar and willow species. The compounds were extracted with MeOH from freeze-dried plant material and analyzed using LC-MS/MS. Means and se (n = 5–7) are shown. Asterisks indicate statistical significance between the two parts of the same species as assessed by paired t-test or Wilcoxon signed rank test (* P ≤ 0.05, ** P ≤ 0.01, and *** P ≤ 0.001). A, Salicin-7-sulfate: P. trichocarpa (P < 0.001, t = 34.372); P. × canescens (P < 0.001, t = 14.388); P. alba (P < 0.001, t = 18.303); P. simonii (P = 0.031, W = −21.00); S. purpurea (P = 0.029, t = 2.843); S. caprea (P = 0.047, W = −24.00). B, Salirepin-7-sulfate: P. trichocarpa (P < 0.001, t = 17.009); P. x canescens (P < 0.001, t = 25.130); P. alba (P < 0.001, t = 13.306); P. simonii (P < 0.001, t = 7.045); S. purpurea (P = 0.001, t = 5.531). dw, dry weight; nd, not detected.
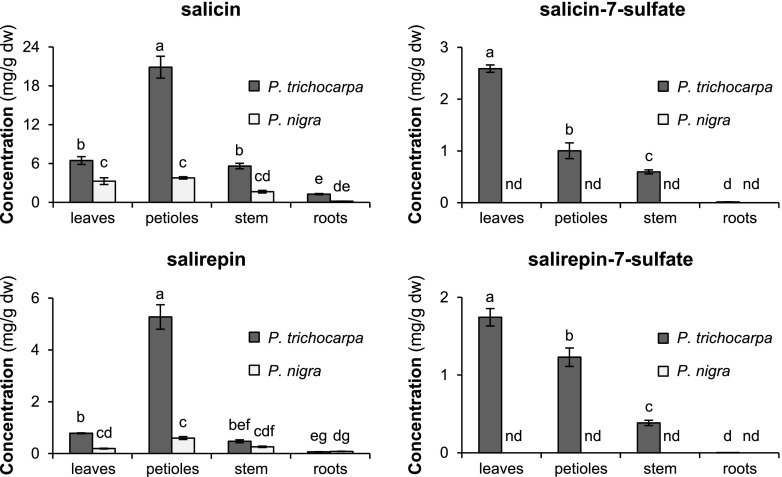
A more detailed survey including leaf blades, petioles, stems, and roots was performed to determine the organ-specific distribution of salicin-7-sulfate and salirepin-7-sulfate in the two poplar species P. trichocarpa and P. nigra. In P. trichocarpa, highest accumulation of the two sulfated salicinoids was found in leaves, followed by petioles and stems, while roots accumulated only low amounts of these compounds (Fig. 3; Supplemental Table S3). The potential biosynthetic precursors of salicin-7-sulfate and salirepin-7-sulfate, salicin, and salirepin, respectively, showed a similar pattern with highest accumulation in petioles, less abundant accumulation in leaves and stems, and lowest accumulation in roots of P. trichocarpa (Fig. 3; Supplemental Table S3). Although salicin and salirepin were found in all organs of P. nigra investigated, sulfated salicinoids could not be detected in any of them (Fig. 3; Supplemental Table S3).
Figure 3.
Salicin, salicin-7-sulfate, salirepin, and salirepin-7-sulfate concentrations in different organs of P. trichocarpa (dark gray bars) and P. nigra (light gray bars) trees. Leaf blades (leaves) and petioles originated from LPI3 to LPI10 leaves. Stem material was harvested between leaf LPI3 and LPI10. Compounds were extracted with MeOH from freeze-dried plant material and analyzed using liquid chromatography/tandem mass spectrometry (LC-MS/MS) and high-performance liquid chromatography with ultraviolet detector (HPLC-UV). Means and se (n = 6–8) are shown. Differences between organs and species were analyzed by a two-way repeated measures ANOVA and a posthoc test (Tukey’s test). Different letters indicate significant differences (P < 0.05). dw, dry weight; nd, not detected.
The Accumulation of Salicin-7-Sulfate and Salirepin-7-Sulfate Is Not Influenced by Leaf Herbivory of the Generalist Feeding Caterpillar Lymantria dispar
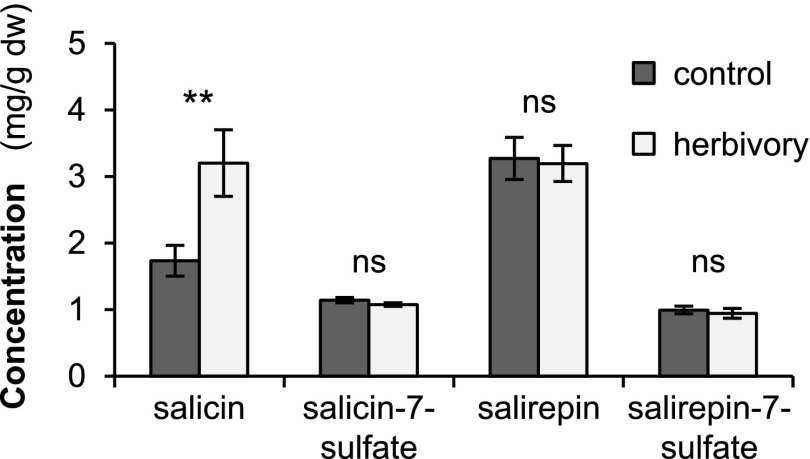
Similar to nonsulfated salicinoids, salicin-7-sulfate and salirepin-7-sulfate might act as defense compounds against insect herbivores. To test whether their formation is influenced by insect herbivory, we analyzed the amounts of sulfated and nonsulfated salicinoids in undamaged P. trichocarpa leaves and leaves that had been fed upon by L. dispar caterpillars for nearly 24 h. LC-MS/MS analysis revealed that the herbivore treatment did not affect the accumulation of salicin-7-sulfate and salirepin-7-sulfate (Fig. 4). Furthermore, salirepin and the complex salicinoids salicortin, tremulacin, 6′-o-benzoylsalicortin, and homaloside D were not influenced by the herbivore treatment (Fig. 4; Supplemental Table S4). However, the accumulation of salicin was significantly increased in response to herbivory (Fig. 4).
Figure 4.
Salicin, salicin-7-sulfate, salirepin, and salirepin-7-sulfate concentrations in undamaged (control) and L. dispar caterpillar-damaged (herbivory) leaves of P. trichocarpa. Compounds were extracted with MeOH from freeze-dried plant material and analyzed using LC-MS/MS or HPLC-UV. Means and se (n = 7–8) are shown. Asterisks indicate statistical significance as assessed by Student’s t-test (**P < 0.01). Salicin (P = 0.009, t = −3.065); salicin-7-sulfate (P = 0.152, t = 1.523); salirepin (P = 0.846, t = 0.198); salirepin-7-sulfate (P = 0.588, t = 0.555). dw, dry weight; ns, not significant.
PtSOT1 Catalyzes the Formation of Salicin-7-Sulfate and Salirepin-7-Sulfate In Vitro
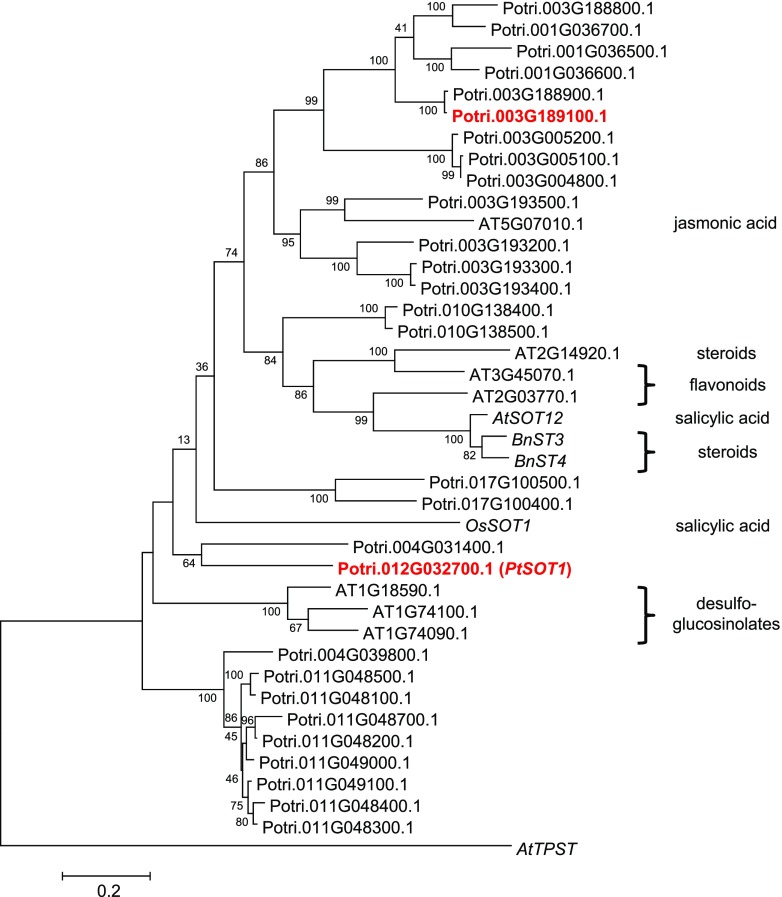
To identify SOT genes responsible for the formation of salicin-7-sulfate and salirepin-7-sulfate, a BLAST software search was performed with the previously characterized salicylic acid sulfotransferase AtSOT12 from Arabidopsis (Baek et al., 2010) as query and the P. trichocarpa genome as template. This analysis revealed 28 putative poplar SOT gene candidates that grouped together with SOT genes from other plants in a phylogenetic tree (Fig. 5). Because the sulfated salicinoids accumulated mostly in above-ground tissues of P. trichocarpa (Figs. 2 and 3), we hypothesized that the responsible biosynthetic genes are highly expressed in leaves. The analysis of the identified SOT genes in a transcriptome made from P. trichocarpa leaves (Günther et al., 2019) revealed two candidates (Potri.012G032700 and Potri.003G189100) with high expression in leaves (Supplemental Fig. S5). An amino acid alignment of Potri.012G032700, Potri.003G189100, and characterized SOTs from rice and Arabidopsis confirmed that the poplar candidate enzymes contained the conserved regions important for the catalytic activity of SOTs (Marsolais and Varin, 1995; Klein and Papenbrock, 2004; Hirschmann et al., 2014; Supplemental Fig. S6). For biochemical characterization of Potri.012G032700 and Potri.003G189100, we heterologously expressed the full-length genes in Escherichia coli and performed enzyme assays with affinity-purified recombinant N-terminal His-tag fusion proteins, the potential substrates salicin or salirepin, and the cosubstrate PAPS. LC-MS/MS analysis of enzyme assays revealed that Potri.012G032700 converted salicin and salirepin into salicin-7-sulfate and salirepin-7-sulfate, respectively, in vitro (Fig. 6). In contrast, recombinant Potri.003G189100 showed no enzymatic activity with the tested substrates (Fig. 6). Interestingly, salicyl alcohol, the aglycone of salicin, was not accepted as substrate, neither by Potri.012G032700 nor by Potri.003G189100 (Supplemental Fig. S7). Based on its in vitro activity, Potri.012G032700 was designated as PtSOT1. Because Potri.003G189100 showed no activity with the tested substrates (Fig. 6; Supplemental Fig. S7), the gene was not further considered in this study.
Figure 5.
Cladogram analysis of putative poplar SOT genes and characterized SOT genes from other plants. The tree was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model implemented in the software MEGA6 (Tamura et al., 2013). Bootstrap values (n = 1,000) are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were first+second+third+Noncoding. All positions with <80% site coverage were eliminated. Arabidopsis AtTPST (NM_100677) was used as an outgroup. Poplar SOTs investigated in this study are highlighted in red. Substrates of characterized SOTs are shown next to the tree on the right side. AtSOT12, Arabidopsis NM_126423; BnST3, B. napus AF000307; BnST4, B. napus AY442306; OsSOT1, O. sativa LOC_Os11g30910.
Figure 6.
Catalytic activity of PtSOT1 (Potri.012G032700) and Potri.003G189100. PtSOT1 and Potri.003G189100 were heterologously expressed in E. coli as His-tag fusion proteins and recombinant proteins were purified using affinity chromatography. Purified proteins were incubated with the potential substrates salicin or salirepin, and the cosubstrate PAPS as sulfate group donor. Reaction products were analyzed using LC-MS/MS. cps, counts per second.
The Expression of PtSOT1 Regulates the Formation of Sulfated Salicinoids in P. trichocarpa
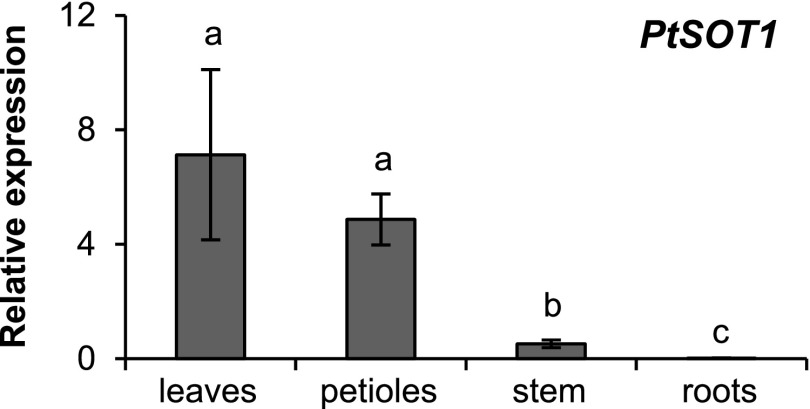
Reverse transcription quantitative PCR (RT-qPCR) was used to measure the transcript accumulation of PtSOT1 in different tissues of P. trichocarpa. PtSOT1 was highly expressed in leaf blades and petioles, but showed less expression in stems, and only trace expression in roots (Fig. 7). This expression pattern matched the accumulation of salicin-7-sulfate and salirepin-7-sulfate in the respective tissues of P. trichocarpa (Fig. 3), suggesting that the formation of the two compounds is mainly regulated by PtSOT1 transcript levels.
Figure 7.
Gene expression analysis of PtSOT1. Transcript accumulation of PtSOT1 in different organs of P. trichocarpa was measured using RT-qPCR. Means and se are shown (n = 6). Leaf blades (leaves) and petioles originated from LPI3 to LPI10 leaves. Stem material was harvested between LPI3 and LPI10 leaves. HIS was used as a housekeeping gene (Wang et al., 2014a). Differences between organs were analyzed by a one-way repeated measures ANOVA and a posthoc test (Tukey’s test). Different letters indicate significant differences (P < 0.05).
The Absence of Sulfated Salicinoids in P. nigra Is Caused by a Loss-of-Function Allele of SOT1
In contrast to P. trichocarpa, P. nigra was found to contain no sulfated salicinoids, although the respective precursors salicin and salirepin could be readily detected in this species (Figs. 2 and 3). To confirm this observation, we tested 19 additional P. nigra genotypes and all showed no detectable accumulation of salicin-7-sulfate or salirepin-7-sulfate (Supplemental Fig. S8; Supplemental Table S5). Amplification of PnSOT1 from genomic DNA (gDNA) isolated from two different P. nigra genotypes revealed a frame shift mutation in both alleles, leading to a nonfunctional PnSOT1 in P. nigra (Supplemental Fig. S9). Moreover, we could not detect any PnSOT1 transcript accumulation in leaves, petioles, stems, and roots of P. nigra plants (Supplemental Table S6), suggesting that the mutated gene is not expressed in this species.
Downregulation of SOT1 by RNAi in P. x canescens Led to Reduced Formation of Sulfated Salicinoids
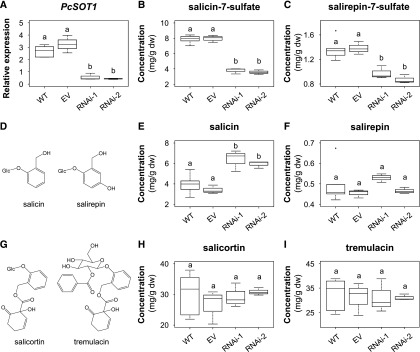
To study the role of SOT1 in the formation of sulfated salicinoids in more detail, we used RNAi to downregulate its expression in P. × canescens, a species usually used for Agrobacterium-mediated poplar transformation (Leple et al., 1992). Two independent transgenic SOT1 knockdown lines (RNAi-1 and RNAi-2) were generated and compared to wild-type trees and trees carrying an empty vector (EV). Transcript expression analysis of SOT1 by RT-qPCR showed that the gene was significantly less expressed in leaves of the knockdown lines in comparison to wild-type and EV trees (Fig. 8). LC-MS/MS analysis of leaf MeOH extracts made from 6-week–old trees revealed a significantly reduced concentration of salicin-7-sulfate and salirepin-7-sulfate in both transgenic SOT1 lines in comparison to wild-type and EV plants (Fig. 8). Along with the sulfated salicinoids, we measured the accumulation of their potential biosynthetic precursors salicin and salirepin. Whereas the salicin concentration was significantly increased in the SOT1 knockdown lines compared to wild-type and EV leaves, the concentration of salirepin was not influenced. Moreover, the accumulation of the complex salicinoids salicortin and tremulacin was not affected by the downregulation of SOT1 (Fig. 8). The analysis of salicin-7-sulfate and salirepin-7-sulfate in leaves of 9- and 12-week–old trees showed a pattern comparable to 6-week–old trees (Supplemental Fig. S10), indicating that the observed phenotype was stable for at least six weeks.
Figure 8.
RNAi-mediated knock-down of SOT1 in P. × canescens. Transcript accumulation of P. × c. sulfotransferase1 (A), and salicin-7-sulfate (B), salirepin-7-sulfate (C), salicin (E), salirepin (F), salicortin (H), and tremulacin (I) concentration in leaves of wild-type (WT) and transgenic P. × canescens trees are shown. The chemical structures are displayed in (D) and (G). Data are displayed for 6-week–old trees. Gene expression was measured using RT-qPCR, with UBQ (Ramírez-Carvajal et al., 2008) as a housekeeping gene. Compounds were extracted with MeOH from freeze-dried leaf material and analyzed using LC-MS/MS and HPLC-UV. Medians ± quartiles, and outliers are shown. dw, dry weight; Differences among wild type, EV, and RNAi knockdown lines were analyzed by a one-way ANOVA and a posthoc test (Tukey’s test) or by Kruskal–Wallis one way ANOVA on ranks (n = 3–6). Different letters above each box indicate significant differences (P < 0.05).
The Reduced Accumulation of Salicin-7-Sulfate and Salirepin-7-Sulfate in SOT1 RNAi Plants Did Not Influence the Feeding Preference of the Generalist Herbivore L. dispar
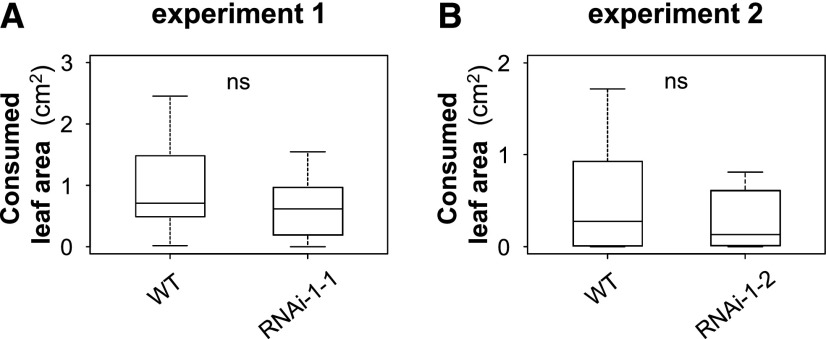
Salicinoids are effective feeding deterrents against various herbivores (e.g. Lindroth and Peterson, 1988; Lindroth et al., 1988). Because sulfation is known to alter the physical and biological properties of plant metabolites (e.g. Gidda et al., 2003; Wang et al., 2014b), the question arose whether sulfated salicinoids also act as feeding deterrents. To address this question, we performed feeding preference assays, offering L. dispar caterpillars leaf discs from P. × canescens wild-type and SOT1 knockdown lines. By analyzing the consumed leaf area, we could show that the caterpillars had no feeding preference for either the wild-type or the SOT1 knockdown line (Fig. 9).
Figure 9.
A and B, Feeding preference of L. dispar caterpillars (L2) for leaf discs of P. × canescens wild type (WT) versus sulfotransferase1 (RNAi) knockdown lines. Consumed leaf area was measured after 21 h of feeding. Medians ± quartiles are shown. ns, not significant. Statistical significance was assessed by paired t-test or Wilcoxon signed rank test (n = 11–14). Experiment 1 (P = 0.172, t = −1.470); experiment 2 (P = 0.380, W = 24.00).
As shown above, a side effect of the RNAi-mediated knockdown of SOT1 was a significantly increased accumulation of the salicin-7-sulfate precursor salicin (Fig. 8). To exclude the possibility that the increased salicin concentration compensated for a possible decrease in deterrence caused by reduced amounts of salicin-7-sulfate, we performed choice assays with artificially applied salicin. By applying increasing amounts of salicin (from 0.1 mg g−1 fresh weight [FW] to 1.5 mg g−1 FW) to P. × canescens wild-type leaf discs, we could show that an application of 1.5 mg g−1 FW is required to shift the feeding preference of the caterpillars to control leaf discs (Supplemental Fig. S11). This amount corresponds to an increase of ∼5 mg g−1 dry weight of salicin. Because the difference in salicin accumulation between wild-type and SOT1 knockdown lines was smaller (∼2.5 mg g−1 dry weight, Fig. 8), we assume that the altered accumulation of salicin did not influence the behavior of the caterpillars in our feeding preference assays.
Salicin-7-Sulfate and Salirepin-7-Sulfate Represent a Substantially Sulfur Pool in P. trichocarpa Leaves
Sulfur is an essential macronutrient, which is required for the synthesis of amino acids such as Cys and Met, but also for the formation of essential prosthetic groups (e.g. Takahashi et al., 2011). To gain insights into a potential role of sulfated salicinoids in sulfur homeostasis in poplar, we analyzed the amounts of total sulfur and free and hydrolysable sulfate in P. trichocarpa leaves. An elemental analysis of freeze-dried poplar leaf material revealed a concentration of 0.68% total sulfur, and sulfate measurements showed that 73.51% of the total sulfur was in the form of sulfate (0.50% of leaf dry weight). Considering the concentrations of salicin-7-sulfate (2.59 mg g−1 dry weight) and salirepin-7-sulfate (1.74 mg g−1 dry weight; Fig. 3) analyzed in the same tissue, we calculated that 5.47% of the total sulfur and 7.47% of free and hydrolysable sulfate are tied up in sulfated salicinoids.
DISCUSSION
Salicinoids are a group of highly abundant specialized metabolites in the Salicaceae (Boeckler et al., 2011), investigated mainly for their function as defense compounds. However, little is known about their biosynthesis. As part of a search for intermediates of salicinoid biosynthesis in poplar, we encountered two sulfated salicinoids, the previously reported salicin-7-sulfate (Noleto-Dias et al., 2018) and salirepin-7-sulfate, described here. Sulfation is mediated by SOTs, a group of enzymes catalyzing the transfer of a sulfuryl group from PAPS to an acceptor molecule (Klein and Papenbrock, 2004; Hirschmann et al., 2014, 2017). Thus, we hypothesized that SOT enzymes might contribute to the biosynthesis of sulfated salicinoids and aimed to identify and biochemically characterize the responsible proteins in poplar.
Salicin-7-Sulfate and Salirepin-7-Sulfate Are Two Widespread Salicinoids in Poplar
The structural diversity of specialized metabolites is mainly driven by modifications of common core structures by processes such as glycosylation, methylation, acetylation, or sulfation (Varin et al., 1992; Saito et al., 2013). Sulfated specialized metabolites are known to be involved in plant defense and plant development (Faulkner and Rubery, 1992; Wang et al., 2014b). A sulfated metabolite, salicin-7-sulfate, was recently described to occur in different willow species (Noleto-Dias et al., 2018) and was here isolated from various poplars (Fig. 2). Moreover, by performing LC-MS precursor ion scan analyses, we identified two further sulfated compounds in poplar, salirepin-7-sulfate (Fig. 1; Supplemental Fig. S3) and the jasmonic acid derivative 12-hydroxy jasmonic acid sulfate, a compound known from many other plant species (Gidda et al., 2003; Miersch et al., 2008). The sulfated salicinoids were found to occur in a range of poplar organs and species, and to be present in substantial amounts, sometimes at concentrations similar to those of their nonsulfated counterparts (Figs. 2 and 3). Because precursor ion scan analyses rely on a specific fragmentation pattern, we cannot rule out the presence of other sulfated compounds in poplar. Sulfated flavonoids, for instance, are commonly distributed within the plant kingdom (Varin and Ibrahim, 1989; Varin et al., 1992; Falcone Ferreyra et al., 2012) and thus may also occur in poplar and willow species. Very recently, a novel method that uses neutral loss scan mass spectrometry for the identification of sulfated flavonoids in plant extracts has been described by Kleinenkuhnen et al. (2019). Applying this method to poplar and willow extracts might be a worthwhile approach for further discovery of sulfated compounds in the Salicaceae.
A Functional SOT1 Allele Mediates the Formation of Salicin-7-Sulfate and Salirepin-7-Sulfate in Poplar
Sulfotransferases have been described in a number of plant species including Arabidopsis, rice, and clasping yellowtops (Flaveria chloraefolia), where they are involved in the sulfation of desulfoglucosinolates, salicylic acid, and flavonoids, respectively (Varin et al., 1992; Piotrowski et al., 2004; Wang et al., 2014b). The genome of P. trichocarpa possesses 28 putative SOT genes (Fig. 5) that form a typical midsized gene family as described for other species (Klein and Papenbrock, 2004; Chen et al., 2012; Hirschmann et al., 2014). Phylogenetic and transcriptome analysis led to the identification of two putative poplar SOT candidate genes mainly expressed in leaves, the organs with the highest amounts of sulfated salicinoids (Figs. 2 and 5; Supplemental Fig. S5). Biochemical characterization revealed that one of the candidates, PtSOT1, catalyzes the sulfation of salicin and salirepin in vitro using PAPS as the cofactor (Fig. 6). Notably, PtSOT1 did not accept the aglycone salicyl alcohol as substrate, indicating that sulfation occurs after glucosylation as the last step in the biosynthesis of sulfated salicinoids (Supplemental Fig. S7). The RNAi-mediated knockdown of SOT1 in P. × canescens revealed that SOT1 is responsible for the formation of salicin-7-sulfate and salirepin-7-sulfate in planta (Fig. 8; Supplemental Fig. S10). Moreover, the presence of a nonfunctional SOT1 allele in P. nigra, a species not able to produce sulfated salicinoids (Figs. 2 and 3; Supplemental Figs. S8 and S9; Supplemental Table S5), suggests that the SOT1 locus is likely responsible for the formation of salicin-7-sulfate and salirepin-7-sulfate in other poplar and willow species.
In addition to salicin-7-sulfate and salirepin-7-sulfate, P. trichocarpa accumulates significant amounts of 12-hydroxy jasmonic acid sulfate in leaves (Fig. 1). Potri.003G189100, the enzyme inactive with salicin and salirepin (Fig. 6), was found to be highly expressed in leaves (Supplemental Fig. S5); however, it is only distantly related to the Arabidopsis 12-hydroxy jasmonic acid SOT At2G14920.1 (Fig. 5) and thus most likely not involved in the formation of 12-hydroxy jasmonic acid sulfate. Phylogenetic analysis revealed a small subgroup of four putative poplar SOT genes that cluster together with At2G14920.1 (Fig. 5). Thus, these genes might encode 12-hydroxy jasmonic acid SOTs in poplar. Future research is needed to elucidate the involvement of specific SOTs in the formation of 12-hydroxy jasmonic acid sulfate.
Sulfated Salicinoids Do Not Act as Strong Feeding Deterrents against the Generalist Herbivore Lymantria dispar
Salicinoids are known as effective defense compounds acting against various folivorous insects (e.g. Lindroth et al., 1988; Hemming and Lindroth, 1995; Feistel et al., 2017). To test if sulfated salicinoids act as feeding deterrents, we performed preference assays with leaf discs of P. × canescens wild-type and SOT1 knockdown lines offered to L. dispar caterpillars. However, we could not observe a feeding preference for reduced sulfated salicinoid accumulation (Fig. 9). Because downregulation of SOT1 led to an increased salicin concentration (Fig. 8), the deterrence of this compound might obscure a feeding preference for low levels of sulfated salicinoids. However, preference assays with artificially applied salicin revealed that ∼5 mg g−1 dry weight of salicin are required to shift the feeding preference of the caterpillars toward control leaf discs. This amount is above the observed salicin increase in the SOT1 knockdown lines of ∼2.5 mg g−1 dry weight and even above the overall salicin concentration in the plant tissue itself (Figs. 3 and 8). Thus, it seems unlikely that the increased amount of salicin masked the effect of decreased sulfated salicinoids in the SOT1 knockdown plants. Altogether, our results imply that sulfated salicinoids and salicin itself are not strong feeding deterrents against L. dispar caterpillars.
In general, the available data on the effectiveness of salicin, the structurally simplest salicinoid, as a defense compound are contradictory. For example, for the tiger swallowtail (Papilio glaucus glaucus) and the southern armyworm (Spodoptera eridania) it was shown that salicin had only marginal effects on growth rate and developmental time, whereas the two complex salicinoids salicortin and tremulacin had a significant negative impact (Lindroth and Peterson, 1988; Lindroth et al., 1988). However, artificial application of salicin let to a significantly decreased weight gain in performance assays with L. dispar caterpillars (Boeckler et al., 2016). Similar to salicin, it might be possible that sulfated salicinoids have an impact on caterpillar development, although L. dispar caterpillars had no pronounced feeding preference toward decreased levels of sulfated salicinoids (Fig. 9). Besides generalist feeders, salicin-7-sulfate and salirepin-7-sulfate may also influence the behavior of specialized feeding herbivorous insects. Larvae of the red poplar leaf beetle (Chrysomela populi), for instance, are able to sequester and metabolize salicin to salicylaldehyde and use this compound as a protectant against predators and microbial infection (Pasteels et al., 1983; Gross et al., 2002; Michalski et al., 2008). Sulfation may inhibit larval sequestration by competitive inhibition of larval transporters and metabolic enzymes, and thus impact the feeding preference of the beetle larvae.
Salicin-7-Sulfate and Salirepin-7-Sulfate May Act as Sulfur Storage Pools in Poplar
Sulfur is an essential macronutrient in primary metabolism for the synthesis of Cys, Met, coenzymes, and prosthetic groups among other compounds (e.g. Takahashi et al., 2011). Because of its importance in the primary metabolism, plants tightly regulate their sulfur status and uptake (Kopriva, 2006; Davidian and Kopriva, 2010; Takahashi et al., 2011). Sulfur deprivation, for instance, usually leads to an increased sulfur uptake, while an excess amount of reduced sulfur (e.g. hydrogen sulfide) leads to a decreased sulfur uptake (Herschbach et al., 1995; Lappartient and Touraine, 1996; Westerman et al., 2001). The main sulfur source for plants is sulfate, which is taken up from the soil by the plant roots (Davidian and Kopriva, 2010). However, the release of sulfate from sulfated specialized metabolites might also be important for sulfur homeostasis. We could show that substantial amounts of total sulfur (5.47%) and sulfate (7.47%) are tied up in sulfated salicinoids in poplar leaves. Notably, a similar number (6.4% total sulfur) has been reported for glucosinolates, a group of highly abundant sulfur-containing specialized metabolites in the Brassicaceae (Blake-Kalff et al., 1998; Halkier and Gershenzon, 2006). Principally known as defense compounds, it has been long-term–speculated that glucosinolates may function as a sulfur-storage pool (Hirai et al., 2004, 2005; Falk et al., 2007). Glucosinolate accumulation is highly dependent on the sulfur availability and sulfur status of the plant (Hirai et al., 2004, 2005; Falk et al., 2007). Moreover, the expression of glucosinolate biosynthetic genes and genes involved in the primary sulfur metabolism are regulated by the same R2R3-MYB transcription factors (Yatusevich et al., 2010). Because the amounts of total sulfur tied up in glucosinolates and salicinoids in Arabidopsis and poplar, respectively, are comparable, it is tempting to speculate that salicin-7-sulfate and salirepin-7-sulfate act as sulfur storage compounds in poplar. Like glucosinolates, the formation of sulfated salicinoids might also be dependent on the sulfur status of the plant and is likely influenced by changing environmental conditions. Flooding, for example, can alter the microbiome community by increasing the abundance of sulfur-reducing bacteria, which use sulfate as electron acceptors and produce hydrogen sulfide (Muyzer and Stams, 2008; Randle-Boggis et al., 2018). Poplar and willow trees often grow in wetland or even riparian soils, where flooding can occur. Thus, storing high amounts of sulfur as sulfated salicinoids could be a strategy to overcome episodes of flooding with low sulfate availability. This might be a common pattern, because sulfation of specialized metabolites is known to occur more frequently near watery landscapes (Barron et al., 1988), and we showed that sulfated salicinoids broadly occur within the Salicaceae family (Fig. 2). Investigating the exact roles of sulfated salicinoids in sulfur metabolism and the sulfur status of poplar trees are worthwhile aims for future research.
MATERIALS AND METHODS
Plant and Insect Material
Trees of black cottonwood (Populus trichocarpa, clone ‘Muhle-Larsen’; P&P Baumschule), black poplar (Populus nigra, clones 1–19 and f169; Boeckler et al., 2013), gray poplar (Populus × canescens, clone INRA 7171-B4; Jörg-Peter Schnitzler, German Research Centre for Environmental Health), silver poplar (Populus alba; ‘Nivea’, Eggert Baumschulen), Simon’s poplar (Populus simonii; Eggert Baumschulen), basket willow (Salix viminalis; www.pflanzmich.de), purple willow (Salix purpurea; Max Planck Institute for Chemical Ecology), and goat willow (Salix caprea; Max Planck Institute for Chemical Ecology) were propagated from monoclonal stem cuttings and grown under summer conditions in the greenhouse (day, 23°C to 25°C; night, 19°C to 23°C; 50% to 60% relative humidity; 16-h/8-h light/dark cycle) in a 1:1 mixture of sand and clay granules (Klasmann-Deilmann), until they reached ∼60 to 90 cm in height. Bark from the P. trichocarpa clones ‘Muhle Larsen’, ‘625’, ‘262/63’, ‘Brühl4’, ‘606’, and from the Japanese orange cherry (Idesia polycarpa; Max Planck Institute for Chemical Ecology) were harvested from young twigs of ∼1.5-m tall trees. Trembling aspen (Populus tremuloides) leaf material was harvested in June 2017 from 50 randomly selected genotypes of the WisAsp common garden project in Wisconsin (Barker et al., 2019). Leaf material of P. tremuloides × Populus grandidentata was obtained from the same resource.
Leaves were numbered according to the leaf plastochron index (LPI; Frost et al., 2007). For analysis of different Salicaceae species, leaf blades of LPI3 to LPI10, and roots were harvested, immediately flash-frozen in liquid nitrogen, then lyophilized, pulverized, and stored at 4°C. For analysis of the tissue-specific distribution of the sulfated salicinoids in P. trichocarpa and P. nigra, leaves of LPI3 to LPI10, their petioles, the stem in between these leaves and the roots, were harvested and immediately flash-frozen in liquid nitrogen and ground with one part stored at −80°C until further processing, while the other part was lyophilized, and stored at 4°C. For analysis of the sulfated salicinoids in different P. nigra genotypes and in P. × canescens SOT1 knockdown lines, leaves (LPI5 and LPI6, P. nigra; LPI3 to LPI5, P. × canescens) were harvested and further processed as described above for P. trichocarpa and P. nigra tissues. Leaves of P. × canescens SOT1 knockdown plants, wild-type plants, and EV plants were harvested at a plant age of 6-, 9-, and 12-weeks (corresponding to a tree height of ∼30, 70, and 120 cm, respectively).
Gypsy moth (Lymantria dispar) egg batches were kindly provided by Hannah Nadel (APHIS), and hatched caterpillars were reared on an artificial diet (Gypsy moth diet; MP Biomedicals).
Herbivore Treatment
L. dispar caterpillars were fed with P. trichocarpa leaves for one week and starved for 24 h before the onset of the experiment. For the herbivore treatment, each caterpillar (fourth to fifth instar) was encaged on the LPI5, LPI7, LPI9, and LPI11 leaf of a single P. trichocarpa tree. Caterpillars were allowed to feed for 21 h (5 pm to 2 pm). For the control treatment, empty cages were placed on the LPI5, LPI7, LPI9, and LPI11 leaves. Plant material was harvested as described above for different Salicaceae species.
MeOH Extraction of Plant Material
Metabolites were extracted from 10 mg of freeze-dried plant material by adding 1 mL of 100% MeOH containing 0.8 mg mL−1 of phenyl-β-d-glucopyranoside (Sigma-Aldrich) and 40 ng mL−1 of D6-abscisic acid (D6-ABA; Santa Cruz Biotechnology) as internal standards. Samples were shaken for 30 s in a paint shaker (Scandex) and afterward for 30 min at 200 rpm on a horizontal shaker (IKA Labortechnik). After centrifugation, the supernatants were split for HPLC-UV and LC-MS/MS measurements and were subsequently analyzed.
LC-MS Analysis of Plant MeOH Extracts and Enzyme Products
Full Scan Analysis
Chromatographic separation was achieved on an Agilent 1100 Series LC system, using an EC 250/4.6 Nucleodur Sphinx column (RP 5 μm; Macherey-Nagel), with aqueous formic acid (0.2% [v/v]) and acetonitrile as mobile phases A and B, respectively. The mobile phase flow rate was 1 mL/min. The elution profile was: 0 to 22 min, 14% to 58% B; 22.1 to 25 min, 100% B; 25.1 to 30 min, 14% B. The column temperature was maintained at 25°C. The LC system was coupled with an Esquire 6000 ESI-Ion Trap Mass Spectrometer (Bruker Daltonics). Compounds were analyzed in alternating ionization (positive/negative) mode with a skimmer voltage of 40 V/−40 V, a capillary exit voltage of 113.5 V/−121 V, and a capillary voltage of −3,000/3,000 V. Nitrogen was used as drying gas (11 L/min, 330°C) and nebulizer gas (pressure 35 psi).
Full Scan Analysis For Purification
Chromatography and mass spectrometry analysis was done as described above for “Full Scan Analysis,” except the mobile phases consisted of 0.05% (v/v) formic acid in water (A), and acetonitrile (B), and the elution profile was: 0 to 8 min, 14% to 30% B; 8.1 to 11 min, 100% B; and 11.1 to 15 min, 14% B.
Precursor Ion Scan Analysis
For the precursor ion scan analysis, an Agilent 1200 HPLC system (Agilent Technologies) coupled to an API 3200 tandem mass spectrometer (Applied Biosystems) was used. Chromatographic separation was achieved as described above for “Full Scan Analysis,” with the exception that the column temperature was maintained at 20°C. The mass spectrometer was equipped with a turbospray ion source, operated in negative ionization mode. The ion spray voltage was maintained at −4,500 eV and the turbo gas temperature was set at 700°C. Nebulizing gas was set at 70 psi, curtain gas at 30 psi, heating gas at 60 psi, and collision gas at 10 psi. A precursor ion scan (precursor of m/z 97) was performed to identify further sulfated compounds. A mass range from 80 to 800 D was scanned, the declustering potential (DP) was set at −35 V, and the collision energy (CE) ranged between −55 V to −60 V. The software Analyst 1.5 (Applied Biosystems) was used for data acquisition and processing.
Accurate Mass Determination
Analytes were separated using a Dionex Ultimate 3000 RS Pump system (Thermo Fisher Scientific) equipped with an EC 100/2 Nucleodur C18 Isis column (1.8 μm; Macherey-Nagel). The mobile phases consisted of 0.1% (v/v) formic acid in water (A) and acetonitrile (B), with a flow rate of 0.3 mL/min. The column temperature was maintained at 25°C. The elution profile was: 0 to 0.5 min, 5% B; 0.5 to 11 min, 5% to 60% B; 11.1 to 12 min, 100% B; 12.1 to 15 min, 5% B. The LC system was coupled to a timsTOF mass spectrometer (Bruker Daltonics) equipped with a turbospray ion source (capillary voltage, 3,500 V). Nitrogen was used as drying gas (10 L/min, 230°C) and nebulizer gas (1.8 bar). Analysis was done in negative ionization mode, scanning a mass range from m/z 50 to 1,500. Sodium formate adducts were used as internal calibrators.
Targeted Analysis For Quantification
Chromatographic separation was achieved with an Agilent 1260 Infinity II LC system (Agilent) equipped with a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 μm; Agilent), using aqueous formic acid (0.05% [v/v]) and acetonitrile as mobile phases A and B, respectively. The mobile phase flow rate was 1.1 mL per min. The elution profile was: 0 to 0.5 min, 5% B; 0.5 to 6.0 min, 5% to 37.4% B; 6.02 to 7.5 min, 80% to 100% B; 7.5 to 9.5 min, 100% B; 9.52 to 12 min, 5% B. The column temperature was maintained at 20°C. The LC system was coupled to a QTRAP 6500 Tandem Mass Spectrometer (Sciex) equipped with a turbospray ion source, operated in negative ionization mode. The ion spray voltage was maintained at −4,500 eV and the turbo gas temperature was set at 700°C. Nebulizing gas was set at 60 psi, curtain gas at 40 psi, heating gas at 60 psi, and collision gas at medium level. Multiple reaction monitoring was used to monitor analyte parent ion → product ion formation for salicin-7-sulfate: m/z 365 → 97.0 (DP −30 V; CE −25 V); salirepin-7-sulfate: m/z 381 → 97.0 (DP −30 V; CE −55 V); salirepin: m/z 301 → 139 (DP −30 V; CE −18 V), and D6-ABA: 269 → 159.2 (DP −30 V; CE −22 V). Sulfated salicinoids and salirepin were quantified relative to the signal of the internal standard D6-ABA, by applying experimentally determined response factors (RF; salicin-7-sulfate RF: 1.9; salirepin-7-sulfate RF: 3.0; salirepin RF: 111.47). The software Analyst 1.6.3 (Applied Biosystems) was used for data acquisition and processing.
Analysis of Enzyme Products
Products of SOT enzymes were measured using an Agilent 1200 HPLC system coupled to an API 3200 tandem mass spectrometer (Applied Biosystems). For analysis of enzyme assays with salicin and salirepin as substrates, the chromatographic separation was achieved with 0.2% (v/v) formic acid in water (A) and acetonitrile (B) as mobile phases and an EC 250/4.6 Nucleodur Sphinx column (RP, 5 μm; Macherey-Nagel). The column temperature was maintained at 20°C. The elution profile was: 0 to 0.5 min, 5% B; 0.5 to 20.5 min, 5% to 45% B; 20.52 to 22 min, 100% B, 22.1 to 26 min, 5% B, with a flow rate of 1 mL/min. The mass spectrometer was operated in negative ionization mode, with the same settings as described for the “Precursor Ion Scan Analysis.” For analysis of enzyme assays with salicyl alcohol as substrate, the chromatographic separation was conducted using a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 μm; Agilent), and aqueous formic acid (0.05% [v/v], A) and acetonitrile (B) as mobile phases. The elution profile was: 0 to 0.5 min, 5% B; 0.5 to 6 min, 5% to 37.4% B; 6.02 to 7.5 min, 100% B, 7.6 to 10 min, 5% B. The flow rate was 1.1 mL/min, and the column temperature was maintained at 25°C. The mass spectrometer was operated in negative ionization mode. The ion spray voltage was maintained at −4,500 eV and the turbo gas temperature was set at 650°C. Nebulizing gas was set at 60 psi, curtain gas at 25 psi, heating gas at 60 psi, and collision gas at 7 psi. For both chromatographic separations, multiple reaction monitoring mode was used to monitor precursor ion → product ion reactions for each analyte as follows: salicin-7-sulfate: m/z 365 → 97.0 (DP −35 V; CE −55 V); salirepin-7-sulfate: m/z 381 → 97.0 (DP −35 V; CE −55 V); salicyl alcohol-sulfate m/z 203 → 97.0 (DP −35 V; CE −25 V). The software Analyst 1.5 (Applied Biosystems) was used for data acquisition and processing.
HPLC-UV Analysis for Salicinoid Quantification and Purification of Sulfated Salicinoids
Salicinoid Quantification
Salicinoids were analyzed and quantified by HPLC-UV (200 nm) as described in Boeckler et al. (2013) for the compounds salicin, salicortin, tremulacin, and homaloside D. The 6′-o-benzoylsalicortin was analyzed and quantified by HPLC-UV (200 nm) as described in Lackner et al. (2019).
Purification of Sulfated Salicinoids
The purification of sulfated salicinoids via preparative HPLC was conducted on a model no. 1200 HPLC system (Agilent Technologies), with a UV/VIS-detector, connected to a model no. SF-2120 Super Fraction Collector (Advantec/MSF). Chromatographic separation was achieved as described above in “Full Scan Analysis for Purification” using a model no. EC 250/4.6 Nucleodur 100-5 C18 EC column (Macherey-Nagel). The UV absorption was detected at 200 nm and used for determination and collection of the respective peaks.
Purification of Salicin-7-Sulfate and Salirepin-7-Sulfate from P. × canescens Leaves
Ground and freeze-dried P. × canescens leaf material (12 g) was extracted twice with 100% MeOH (2 × 120 mL). The extract was evaporated using nitrogen gas to a volume of 10 mL and then diluted with water to a final concentration of 2% (v/v) MeOH. The extract was first purified by solid phase extraction using a CHROMABOND HR-XA mixed-mode anion-exchange column (85 μm, 6 mL/500 mg; Macherey-Nagel) and, after washing, eluted with 75% (v/v) formic acid in MeOH. The eluate was dried with nitrogen gas. After reconstitution with water, the mixture was further purified using a CHROMABOND C18 EC solid phase extraction column (45 mL/5,000 mg; Macherey-Nagel). Sulfated salicinoids were sequentially eluted with 2% (v/v) and 10% (v/v) MeOH in water, respectively, and dried with nitrogen gas. Fractions containing the sulfated salicinoids were reconstituted with 100% MeOH and separated using HPLC on an EC 250/4.6 Nucleodur 100-5 C18 EC column (Macherey-Nagel). The purified sulfated salicinoids were collected, dried with nitrogen gas, and subjected to NMR analysis. For details on HPLC separation and LC-MS analysis of the fractions, please see above for “Purification of Sulfated Salicinoids” and “Full Scan Analysis for Purification,” respectively.
NMR General Experimental Procedure
NMR spectra (1H-NMR, 1H-1H COSY, 1H-13C HSQC, and 1H-13C HMBC) were measured on an Avance III HD 500 NMR spectrometer, operating at 500.13 MHz for 1H and 125.75 MHz for 13C, and an Avance III HD 700 NMR spectrometer, operating at 700.13 MHz for 1H and 175.75 MHz for 13C (Bruker BioSpin). Both spectrometers were equipped with a TCI CryoProbe (5 mm for Avance III HD 500 and 1.7 mm for Avance III HD 700). Spectrometer control and data processing was accomplished using the software Topspin ver. 3.6.1 (Bruker BioSpin). All NMR measurements were recorded at 25°C in D2O, and chemical shifts were left uncorrected.
Determination of Total Sulfur and Sulfate in P. trichocarpa Plant Material
Total sulfur was determined in 30 mg of freeze-dried P. trichocarpa leaf material with a vario EL elemental analyzer (Elementar Analysensysteme). Sulfate was extracted from 40 mg of freeze-dried P. trichocarpa leaf material by adding 4 mL of double-distilled water. The extracts were incubated for 12 h at 40°C and 100 rpm. Afterward, the extracts were centrifuged twice (21,000g for 15 min at 4°C) and were incubated in between the centrifugation steps for 10 min at 4°C. The collected supernatants were filtered over a 0.45-μm membrane (0.45 μm of Filtropur S; Sarstedt) and stored at 4°C until further analysis. The sulfate concentration was analyzed via ion chromatography using a model no. Dionex DX-500 system (Thermo Fisher Scientific).
RNA Extraction and Reverse Transcription
Total RNA was isolated from frozen and ground plant material using the InviTrap Spin Plant RNA Kit (Stratec) according to manufacturer’s instructions. RNA concentration was assessed using a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific). RNA was treated with DNaseI (Thermo Fisher Scientific) before complementary DNA (cDNA) synthesis. Single-stranded cDNA was prepared from 1 μg of DNase-treated RNA using SuperScript III reverse transcriptase and oligo (dT12-18) primers (Invitrogen).
Identification and Cloning of Putative Poplar Sulfotransferase Genes
Putative poplar SOT genes were identified by BLAST software analysis using AtSOT12 (NM_126423) from Arabidopsis (Arabidopsis thaliana) as query and the poplar genome as reference (Tuskan et al., 2006, http://www.phytozome.net/poplar). The complete open reading frames of the two candidate genes, Potri.012G032700 and Potri.003G189100, were amplified from leaf cDNA. Because only Potri.003G189100 could be inserted into the Escherichia coli expression vector pET100/D-TOPO (Thermo Fisher Scientific), Potri.012G032700 was synthesized as a codon-optimized sequence and subsequently cloned into the same expression vector (Thermo Fisher Scientific). The cloned Potri.003G189100 gene was fully sequenced.
Phylogenetic Analysis, and Amino Acid and DNA Alignment
An alignment of all putative P. trichocarpa SOT genes and characterized SOT genes from Arabidopsis, rice (Oryza sativa), and rapeseed (Brassica napus) was constructed using the MUSCLE (codon) algorithm (gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.5; clustering method, UPGMB algorithm) implemented in the program MEGA6 (Tamura et al., 2013). Tree reconstruction was done with MEGA6 using a Maximum Likelihood algorithm (model/method, Kimura 2-parameter model; substitutions type, nucleotide; rates among sites, uniform rates; gaps/missing data treatment, partial deletion; site coverage cutoff, 80%). A bootstrap resampling analysis with 1,000 replicates was performed to evaluate the tree topology.
An amino acid alignment of Potri.012G032700 (PtSOT1) and Potri.003G189100, together with Arabidopsis SOT12 (NP_178471; Baek et al., 2010) and rice SOT1 (LOC_Os11g30910; Wang et al., 2014b) was constructed with MEGA6 and visualized with BioEdit. A DNA alignment of PtSOT1 and PnSOT1 (f41 and f169) was also constructed and visualized with the softwares MEGA6 and BioEdit, respectively.
gDNA Extraction and Cloning of PnSOT1
gDNA was extracted from ground and freeze-dried P. nigra f41 and f169 leaf material, using the Invisorb Spin Plant Mini Kit (Stratec), according to the manufacturer’s instructions. gDNA concentration was assessed using a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific). A PCR was conducted for amplification of PnSOT1 using gene-specific primers for PtSOT1 (Supplemental Table S7). The amplified PCR product was inserted into the expression vector pET100/D-TOPO (Thermo Fisher Scientific) and the cloned genes were fully sequenced.
RNA-Seq and RT-qPCR Analysis
For gene expression analysis of the 28 putative poplar SOTs, we reanalyzed the data set (Sequence Read Archive accession no. PRJNA516861; https://www.ncbi.nlm.nih.gov/sra/docs/) of an RNA-seq experiment as recently described in Günther et al. (2019).
For RT-qPCR analysis, cDNA was prepared as described above and diluted 1:10 with water. For the amplification of a SOT1 gene fragment with a length of 107 bp, primers were designed having a Tm ≥ 60°C, a GC content between 50% and 60%, and a primer length of 20 nucleotides (Supplemental Table S7). The specificity of the primers was confirmed by agarose gel electrophoresis, melting curve analysis, standard curve analysis, and by sequence verification of cloned PCR amplicons. Ubiquitin (UBQ), actin, elongation factor1 alpha, histone superfamily protein H3 (HIS), and tubulin were tested as reference genes (Ramírez-Carvajal et al., 2008; Xu et al., 2011; Wang et al., 2014a). Comparison of ∆Cq values and the corresponding sd revealed HIS as the most suitable reference gene for expression analysis in P. trichocarpa and P. nigra samples (Supplemental Tables S8 and S9). Based on previous studies from Irmisch et al. (2013) and Günther et al. (2019), UBQ was used as the reference gene for expression analysis in (non) transgenic P. × canescens leaves. The following PCR conditions were applied for analysis of SOT1 expression: Initial incubation at 95°C for 3 min followed by 40 cycles of amplification (95°C for 10 s, 57°C for 20 s, and 65°C for 10 s). Reference gene expression analysis was performed with an initial incubation at 95°C for 3 min followed by 40 cycles of amplification (95°C for 10 s and 60°C for 10 s). For all measurements, plate reads were taken at the end of the extension step of each cycle and data for the melting curves were recorded at the end of cycling from 60°C to 95°C. All samples were run on the same PCR machine (Bio-Rad CFX Connect Real-Time PCR Detection system; Bio-Rad) in an optical 96-well plate, using Brilliant III SYBR Green QPCR Master Mix (Stratagene). For analyzing gene expression in P. trichocarpa and P. nigra, six biological replicates were analyzed, each as technical triplicates. Expression analysis of the P. × canescens knockdown, wild-type, and EV trees was conducted for all biological replicates in technical triplicates.
Heterologous Expression of Sulfotransferase Genes
The E. coli strain BL21 Star (DE3; Thermo Fisher Scientific) was used for heterologous expression of Potri.012G032700 (PtSOT1) and Potri.003G189100. Cultures were grown at 37°C, induced at an OD600 = 0.6 with 1 mm of IPTG, and subsequently placed at 18°C and grown for another 20 h. The cells were collected by centrifugation and disrupted by a 4 × 20 s treatment with a sonicator (model no. UW2070; Bandelin) in chilled extraction buffer (50 mm of Tris-HCl at pH 7.5, 500 mm of NaCl, 20 mm of imidazole, 1% [v/v] Tween 20, 10% [v/v] glycerol, 0.3 mg mL−1 of lysozyme, 2.5 U mL−1 of Benzonase Nuclease [Merck], and protease-inhibitor Mix HP [Serva, according to the manufacturer’s instructions]). After centrifugation (15,000g for 30 min at 4°C), the supernatant was used for further affinity-based purification with the Ni-NTA Spin Kit (Qiagen), following the manufacturer’s instructions. To determine the catalytic activity of recombinant enzymes, assays containing 50 μL of the purified protein, 42 μL of assay buffer (50 mm of Tris-HCl at pH 7.5, 500 mm of NaCl, and 10% [v/v] glycerol), 3 μL of the tested substrates (10 mm of salicin, salirepin, or salicyl alcohol), and 5 μL of PAPS (4 mm) were incubated for 2 h at 30°C under shaking at 300 rpm. The reaction was stopped by adding 200 μL of MeOH. After placing on ice for 30 min, the denatured enzymes were removed by centrifugation (15,000g for 10 min at 4°C) and the supernatant was transferred into a glass vial for LC-MS/MS analysis. Reaction products were analyzed using LC-MS/MS.
Vector Construction and Transformation of Poplar
The construction of the binary vector was described by Levée et al. (2009). The transformation of the P. × canescens clone INRA 7171-B4 followed a protocol published by Meilan and Ma (2007). To target SOT1 mRNA, two fragments between positions 114 and 389 (RNAi-1) or 327 and 471 (RNAi-2) of the coding sequence of PtSOT1 were selected. Transgenic RNAi plants were amplified by micropropagation as described by Behnke et al. (2007). Saplings of ∼10 cm high were repotted to soil (Klasmann potting substrate) and propagated in a controlled environment chamber for six weeks (day, 22°C; night, 18°C; 65% relative humidity; 16-h/8-h light/dark cycle) before they were transferred to the greenhouse. To test the level of gene silencing, RT-qPCR analysis was done on wild-type plants, EV control plants, and RNAi-plants (line RNAi-1 and line RNAi-2).
Feeding Preference Assays with Lymantria dispar Caterpillars
Feeding preference assays were performed as described in Lackner et al. (2019) with the following modifications: L. dispar caterpillars (second larval instar) were starved for 6.5 h before the assay and the caterpillars were then allowed to feed for 21 h. Two discs of P. × canescens leaves (LPI3 to LPI5) harvested from each wild-type and SOT1 knockdown lines (RNAi-1) were offered.
To test the feeding preference with artificial application of salicin (Alfa Aesar), different salicin solutions (corresponding to 0.1 mg g−1 of FW salicin, 0.5 mg g−1 of FW, 1.0 mg g−1 of FW, and 1.5 mg g−1 of FW) in ethanol were prepared. Salicin solutions or pure ethanol were applied to P. × canescens wild-type leaf discs (LPI3 to LPI5). The same setup was used as described in Lackner et al. (2019), with the modifications described above.
Statistical Analysis
Throughout the article, data are presented as means ± se. Statistical analysis was performed with the program SigmaPlot 11.0 for Windows (Systat Software) and is described in the figure and table legends for the respective experiments. Whenever necessary, the data were log transformed to meet statistical assumptions such as normality and homogeneity of variances.
Availability of Data and Materials
All supporting data are included as additional files. Constructs described in this work and datasets analyzed during this study are available from the corresponding author upon request. Sequences were deposited in the NCBI GenBank (see “Materials and Methods”).
Accession Numbers
Sequence data for PtSOT1 (MN729495), and Potri.003G189100 (MN729494) can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the corresponding identifiers.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. LC-MS analysis of MeOH extracts made from leaves of P. × canescens, P. trichocarpa, and S. viminalis.
Supplemental Figure S2. Isotopic cluster of [M-H]− peak for unknown compound (m/z 365), later identified as salicin-7-sulfate.
Supplemental Figure S3. NMR chemical shift data (in D2O) of salicin-7-sulfate (1) and salirepin-7-sulfate (2).
Supplemental Figure S4. Concentrations (μg g−1 dry weight) of salicin-7-sulfate in leaves of different P. tremuloides and P. tremuloides x P. grandidentata genotypes.
Supplemental Figure S5. Expression of putative poplar SOT genes in P. trichocarpa leaves.
Supplemental Figure S6. Amino acid sequence comparison of putative P. trichocarpa SOTs Potri.012G032700 and Potri.003G189100 with characterized SOTs from other plants.
Supplemental Figure S7. Catalytic activity of putative P. trichocarpa SOTs Potri.012G032700 (PtSOT1) and Potri.003G189100.
Supplemental Figure S8. Concentrations of sulfated and nonsulfated salicinoids in leaves of different P. nigra genotypes.
Supplemental Figure S9. DNA sequence comparison of P. trichocarpa PtSOT1 with P. nigra SOT1 cloned from two different genotypes.
Supplemental Figure S10. RNAi-mediated knockdown of SOT1 in P. × canescens.
Supplemental Figure S11. Feeding preference of L. dispar caterpillars (L2) in food choice assays with P. × canescens wild-type leaf discs and wild-type leaf discs with artificially applied salicin (salicin).
Supplemental Table S1. Concentrations (mg g−1 dry weight) of salicin, salirepin, and complex salicinoids in leaves and roots of different poplar and willow species.
Supplemental Table S2. Concentrations of sulfated salicinoids (µg g−1 dry weight), salicin, and salirepin (mg g−1 dry weight) in the bark of different P. trichocarpa genotypes and I. polycarpa.
Supplemental Table S3. Concentrations (mg g−1 dry weight) of complex salicinoids in different organs of P. trichocarpa and P. nigra trees.
Supplemental Table S4. Concentrations (mg g−1 dry weight) of complex salicinoids in undamaged (control) and L. dispar caterpillar-damaged (herbivory) P. trichocarpa leaves.
Supplemental Table S5. Concentrations (mg g−1 dry weight) of sulfated salicinoids, salicin, and salirepin in leaves of different P. nigra genotypes.
Supplemental Table S6. Expression levels of SOT1 in different organs of P. trichocarpa (Pt) and P. nigra (Pn) trees.
Supplemental Table S7. Oligonucleotides used for the amplification of full-length SOT1, Potri.003G189100, and for gene expression analysis of SOT1 by RT-qPCR.
Supplemental Table S8. Expression levels of potential housekeeping genes in different organs of P. trichocarpa trees.
Supplemental Table S9. Expression levels of potential housekeeping genes in different organs of P. nigra trees.
Acknowledgments
We thank all gardeners of the Max Planck Institute for Chemical Ecology for their help with rearing poplar and willow trees, and Marion Stäger (Max Planck Institute for Chemical Ecology) for excellent technical assistance. For rearing L. dispar caterpillars and providing different P. nigra genotypes, we thank the members of Sybille B. Unsicker’s group at the Max Planck Institute for Chemical Ecology. We thank Christopher T. Cole (University of Wisconsin–Madison) for providing P. tremuloides and P. tremuloides × P. grandidentata genotypes. Ines Hilke and Birgit Fröhlich (Max Planck Institute for Biogeochemistry) are acknowledged for help with the analysis of sulfur and sulfate.
Footnotes
This work was supported by the Max-Planck Society.
Articles can be viewed without a subscription.
References
- Babst BA, Harding SA, Tsai C-J(2010) Biosynthesis of phenolic glycosides from phenylpropanoid and benzenoid precursors in Populus. J Chem Ecol 36: 286–297 [DOI] [PubMed] [Google Scholar]
- Baek D, Pathange P, Chung JS, Jiang J, Gao L, Oikawa A, Hirai MY, Saito K, Pare PW, Shi H(2010) A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ 33: 1383–1392 [DOI] [PubMed] [Google Scholar]
- Bailey JK, Schweitzer JA, Rehill BJ, Irschick DJ, Whitham TG, Lindroth RL(2006) Rapid shifts in the chemical composition of aspen forests: An introduced herbivore as an agent of natural selection. Biol Invasions 9: 715–722 [Google Scholar]
- Barker HL, Riehl JF, Bernhardsson C, Rubert-Nason KF, Holeski LM, Ingvarsson PK, Lindroth RL(2019) Linking plant genes to insect communities: Identifying the genetic bases of plant traits and community composition. Mol Ecol 28: 4404–4421 [DOI] [PubMed] [Google Scholar]
- Barron D, Varin L, Ibrahim RK, Harborne JB, Williams CA(1988) Sulphated flavonoids—an update. Phytochemistry 27: 2375–2395 [Google Scholar]
- Behnke K, Ehlting B, Teuber M, Bauerfeind M, Louis S, Hänsch R, Polle A, Bohlmann J, Schnitzler JP(2007) Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J 51: 485–499 [DOI] [PubMed] [Google Scholar]
- Blake-Kalff MM, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP(1998) Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol 118: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler GA, Gershenzon J, Unsicker SB(2011) Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 72: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Gershenzon J, Unsicker SB(2013) Gypsy moth caterpillar feeding has only a marginal impact on phenolic compounds in old-growth black poplar. J Chem Ecol 39: 1301–1312 [DOI] [PubMed] [Google Scholar]
- Boeckler GA, Paetz C, Feibicke P, Gershenzon J, Unsicker SB(2016) Metabolism of poplar salicinoids by the generalist herbivore Lymantria dispar (Lepidoptera). Insect Biochem Mol Biol 78: 39–49 [DOI] [PubMed] [Google Scholar]
- Chedgy RJ, Köllner TG, Constabel CP(2015) Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113: 149–159 [DOI] [PubMed] [Google Scholar]
- Chen F, Liu C-J, Tschaplinski TJ, Zhao N(2009) Genomics of secondary metabolism in Populus: Interactions with biotic and abiotic environments. Crit Rev Plant Sci 28: 375–392 [Google Scholar]
- Chen R, Jiang Y, Dong J, Zhang X, Xiao H, Xu Z, Gao X(2012) Genome-wide analysis and environmental response profiling of SOT family genes in rice (Oryza sativa). Genes Genomics 34: 549–560 [Google Scholar]
- Chou C-J, Lin L-C, Tsai W-J, Hsu S-Y, Ho L-K(1997) Phenyl β-d-glucopyranoside derivatives from the fruits of Idesia polycarpa. J Nat Prod 60: 375–377 [Google Scholar]
- Clausen TP, Reichardt PB, Bryant JP, Werner RA, Post K, Frisby K(1989) Chemical model for short-term induction in quaking aspen (Populus tremuloides) foliage against herbivores. J Chem Ecol 15: 2335–2346 [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S(2010) Regulation of sulfate uptake and assimilation—the same or not the same? Mol Plant 3: 314–325 [DOI] [PubMed] [Google Scholar]
- Fabisch T, Gershenzon J, Unsicker SB(2019) Specificity of herbivore defense responses in a woody plant, black poplar (Populus nigra). J Chem Ecol 45: 162–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius SP, Casati P(2012) Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk KL, Tokuhisa JG, Gershenzon J(2007) The effect of sulfur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biol (Stuttg) 9: 573–581 [DOI] [PubMed] [Google Scholar]
- Faulkner IJ, Rubery PH(1992) Flavonoids and flavonoid sulphates as probes of auxin-transport regulation in Cucurbita pepo hypocotyl segments and vesicles. Planta 186: 618–625 [DOI] [PubMed] [Google Scholar]
- Feistel F, Paetz C, Lorenz S, Beran F, Kunert G, Schneider B(2017) Idesia polycarpa (Salicaceae) leaf constituents and their toxic effect on Cerura vinula and Lymantria dispar (Lepidoptera) larvae. Phytochemistry 143: 170–179 [DOI] [PubMed] [Google Scholar]
- Frost CJ, Appel HM, Carlson JE, De Moraes CM, Mescher MC, Schultz JC(2007) Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol Lett 10: 490–498 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L(2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem 278: 17895–17900 [DOI] [PubMed] [Google Scholar]
- Gross J, Podsiadlowski L, Hilker M(2002) Antimicrobial activity of exocrine glandular secretion of Chrysomela larvae. J Chem Ecol 28: 317–331 [DOI] [PubMed] [Google Scholar]
- Günther J, Lackus ND, Schmidt A, Huber M, Stödtler HJ, Reichelt M, Gershenzon J, Köllner TG(2019) Separate pathways contribute to the herbivore-induced formation of 2-phenylethanol in poplar. Plant Physiol 180: 767–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J(2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Heiska S, Tikkanen O-P, Rousi M, Julkunen-Tiitto R(2007) Bark salicylates and condensed tannins reduce vole browsing amongst cultivated dark-leaved willows (Salix myrsinifolia). Chemoecology 17: 245–253 [Google Scholar]
- Hemming JDC, Lindroth RL(1995) Intraspecific variation in aspen phytochemistry: Effects on performance of gypsy moths and forest tent caterpillars. Oecologia 103: 79–88 [DOI] [PubMed] [Google Scholar]
- Herschbach C, De Kok LJ, Rennenberg H(1995) Net uptake of sulfate and its transport to the shoot in spinach plants fumigated with H2S or SO2: Does atmospheric sulfur affect the “inter-organ” regulation of sulfur nutrition? Bot Acta 108: 41–46 [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K(2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann F, Krause F, Baruch P, Chizhov I, Mueller JW, Manstein DJ, Papenbrock J, Fedorov R(2017) Structural and biochemical studies of sulphotransferase 18 from Arabidopsis thaliana explain its substrate specificity and reaction mechanism. Sci Rep 7: 4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann F, Krause F, Papenbrock J(2014) The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Front Plant Sci 5: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, McCormick AC, Boeckler GA, Schmidt A, Reichelt M, Schneider B, Block K, Schnitzler JP, Gershenzon J, Unsicker SB, et al. (2013) Two herbivore-induced cytochrome P450 enzymes CYP79D6 and CYP79D7 catalyze the formation of volatile aldoximes involved in poplar defense. Plant Cell 25: 4737–4754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y, Petrotchenko EV, Pedersen LC, Negishi M(1998) The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis. J Biol Chem 273: 27325–27330 [DOI] [PubMed] [Google Scholar]
- Kelly MT, Curry JP(1991) The influence of phenolic compounds on the suitability of three Salix species as hosts for the willow beetle Phratora vulgatissima. Entomol Exp Appl 61: 25–32 [Google Scholar]
- Klein M, Papenbrock J(2004) The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot 55: 1809–1820 [DOI] [PubMed] [Google Scholar]
- Kleinenkuhnen N, Büchel F, Gerlich SC, Kopriva S, Metzger S(2019) A novel method for identification and quantification of sulfated flavonoids in plants by neutral loss scan mass spectrometry. Front Plant Sci 10: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S.(2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner S, Lackus ND, Paetz C, Köllner TG, Unsicker SB(2019) Aboveground phytochemical responses to belowground herbivory in poplar trees and the consequence for leaf herbivore preference. Plant Cell Environ 42: 3293–3307 [DOI] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B(1996) Demand-driven control of root ATP sulfurylase activity and SO42− uptake in intact canola (the role of phloem-translocated glutathione). Plant Physiol 111: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leple JC, Brasileiro ACM, Michel MF, Delmotte F, Jouanin L(1992) Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A(2009) Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol 184: 48–70 [DOI] [PubMed] [Google Scholar]
- Lindroth RL, Peterson SS(1988) Effects of plant phenols of performance of southern armyworm larvae. Oecologia 75: 185–189 [DOI] [PubMed] [Google Scholar]
- Lindroth RL, Scriber JM, Hsia MTS(1988) Chemical ecology of the tiger swallowtail: Mediation of host use by phenolic glycosides. Ecology 69: 814–822 [Google Scholar]
- Lindroth RL, St. Clair SB(2013) Adaptations of quaking aspen (Populus tremuloides Michx.) for defense against herbivores. For Ecol Manage 299: 14–21 [Google Scholar]
- Liu X, Wang Z, Wang D, Zhang J(2016) Phylogeny of Populus-Salix (Salicaceae) and their relative genera using molecular datasets. Biochem Syst Ecol 68: 210–215 [Google Scholar]
- Luczak S, Forlani F, Papenbrock J(2013) Desulfo-glucosinolate sulfotransferases isolated from several Arabidopsis thaliana ecotypes differ in their sequence and enzyme kinetics. Plant Physiol Biochem 63: 15–23 [DOI] [PubMed] [Google Scholar]
- Marsolais F, Boyd J, Paredes Y, Schinas AM, Garcia M, Elzein S, Varin L(2007) Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225: 1233–1244 [DOI] [PubMed] [Google Scholar]
- Marsolais F, Varin L(1995) Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol 3-sulfotransferase. J Biol Chem 270: 30458–30463 [DOI] [PubMed] [Google Scholar]
- Massad TJ, Trumbore SE, Ganbat G, Reichelt M, Unsicker S, Boeckler A, Gleixner G, Gershenzon J, Ruehlow S(2014) An optimal defense strategy for phenolic glycoside production in Populus trichocarpa--isotope labeling demonstrates secondary metabolite production in growing leaves. New Phytol 203: 607–619 [DOI] [PubMed] [Google Scholar]
- Meilan R, Ma C(2007) Poplar (Populus spp.) In Wang K, ed, Agrobacterium Protocols, 2nd edition, Vol Vol II Humana Press, Totowa, NJ, pp 143–151 [Google Scholar]
- Michalski C, Mohagheghi H, Nimtz M, Pasteels J, Ober D(2008) Salicyl alcohol oxidase of the chemical defense secretion of two chrysomelid leaf beetles. Molecular and functional characterization of two new members of the glucose-methanol-choline oxidoreductase gene family. J Biol Chem 283: 19219–19228 [DOI] [PubMed] [Google Scholar]
- Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C(2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol 177: 114–127 [DOI] [PubMed] [Google Scholar]
- Muyzer G, Stams AJ(2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6: 441–454 [DOI] [PubMed] [Google Scholar]
- Niemeyer HM.(2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: Key defense chemicals of cereals. J Agric Food Chem 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Noleto-Dias C, Ward JL, Bellisai A, Lomax C, Beale MH(2018) Salicin-7-sulfate: A new salicinoid from willow and implications for herbal medicine. Fitoterapia 127: 166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palo RT.(1984) Distribution of birch (Betula Spp.), willow (Salix Spp.), and poplar (Populus Spp.) secondary metabolites and their potential role as chemical defense against herbivores. J Chem Ecol 10: 499–520 [DOI] [PubMed] [Google Scholar]
- Pasteels JM, Rowell-Rahier M, Braekman JC, Dupont A(1983) Salicin from host plant as precursor of salicylaldehyde in defensive secretion of Chrysomeline larvae. Physiol Entomol 8: 307–314 [Google Scholar]
- Philippe RN, Bohlmann J(2007) Poplar defense against insect herbivores. Can J Bot 85: 1111–1126 [Google Scholar]
- Picard S, Chenault J, Augustin S, Venot C(1994) Isolation of a new phenolic compound from leaves of Populus deltoides. J Nat Prod 57: 808–810 [Google Scholar]
- Pichersky E, Lewinsohn E(2011) Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol 62: 549–566 [DOI] [PubMed] [Google Scholar]
- Piotrowski M, Schemenewitz A, Lopukhina A, Müller A, Janowitz T, Weiler EW, Oecking C(2004) Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem 279: 50717–50725 [DOI] [PubMed] [Google Scholar]
- Ramírez-Carvajal GA, Morse AM, Davis JM(2008) Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol 177: 77–89 [DOI] [PubMed] [Google Scholar]
- Randle-Boggis RJ, Ashton PD, Helgason T(2018) Increasing flooding frequency alters soil microbial communities and functions under laboratory conditions. MicrobiologyOpen 7: e00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubert-Nason KF, Couture JJ, Major IT, Constabel CP, Lindroth RL(2015) Influence of genotype, environment, and gypsy moth herbivory on local and systemic chemical defenses in trembling aspen (Populus tremuloides). J Chem Ecol 41: 651–661 [DOI] [PubMed] [Google Scholar]
- Ruuhola T, Julkunen-Titto R(2003) Trade-off between synthesis of salicylates and growth of micropropagated Salix pentandra. J Chem Ecol 29: 1565–1588 [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR(2013) The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Stevens MT, Lindroth RL(2005) Induced resistance in the indeterminate growth of aspen (Populus tremuloides). Oecologia 145: 298–306 [DOI] [PubMed] [Google Scholar]
- Tahvanainen J, Helle E, Julkunen-Tiitto R, Lavola A(1985a) Phenolic compounds of willow bark as deterrents against feeding by mountain hare. Oecologia 65: 319–323 [DOI] [PubMed] [Google Scholar]
- Tahvanainen J, Julkunen-Tiitto R, Kettunen J(1985b) Phenolic glycosides govern the food selection pattern of willow feeding leaf beetles. Oecologia 67: 52–56 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R(2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S(2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y(2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Varin L, DeLuca V, Ibrahim RK, Brisson N(1992) Molecular characterization of two plant flavonol sulfotransferases. Proc Natl Acad Sci USA 89: 1286–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin L, Ibrahim RK(1989) Partial purification and characterization of three flavonol-specific sulfotransferases from Flaveria chloraefolia. Plant Physiol 90: 977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Chen J, Tian Q, Wang S, Xia X, Yin W(2014a) Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol Plant 152: 529–545 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu Y, He J, Zheng X, Hu J, Liu Y, Dai H, Zhang Y, Wang B, Wu W, et al. (2014b) STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat Commun 5: 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman S, Stulen I, Suter M, Brunold C, De Kok LJ(2001) Atmospheric H2S as sulphur source for Brassica oleracea: Consequences for the activity of the enzymes of the assimilatory sulphate reduction pathway. Plant Physiol Biochem 39: 425–432 [Google Scholar]
- Xu M, Zhang B, Su X, Zhang S, Huang M(2011) Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Anal Biochem 408: 337–339 [DOI] [PubMed] [Google Scholar]
- Yatusevich R, Mugford SG, Matthewman C, Gigolashvili T, Frerigmann H, Delaney S, Koprivova A, Flügge UI, Kopriva S(2010) Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J 62: 1–11 [DOI] [PubMed] [Google Scholar]
- Young B, Wagner D, Doak P, Clausen T(2010) Induction of phenolic glycosides by quaking aspen (Populus tremuloides) leaves in relation to extrafloral nectaries and epidermal leaf mining. J Chem Ecol 36: 369–377 [DOI] [PubMed] [Google Scholar]
- Zenk MH.(1967) Pathways of salicyl alcohol and salicin formation in Salix purpurea L. Phytochemistry 6: 245–252 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supporting data are included as additional files. Constructs described in this work and datasets analyzed during this study are available from the corresponding author upon request. Sequences were deposited in the NCBI GenBank (see “Materials and Methods”).