MtSSPdb hosts a compendium of small secreted peptide sequences with annotations and an RNA-seq-based gene expression atlas for Medicago truncatula, a plant small secreted peptide prediction tool, and phenotyping data from synthetic peptide screens in planta.
Abstract
A growing number of small secreted peptides (SSPs) in plants are recognized as important regulatory molecules with roles in processes such as growth, development, reproduction, stress tolerance, and pathogen defense. Recent discoveries further implicate SSPs in regulating root nodule development, which is of particular significance for legumes. SSP-coding genes are frequently overlooked, because genome annotation pipelines generally ignore small open reading frames, which are those most likely to encode SSPs. Also, SSP-coding small open reading frames are often expressed at low levels or only under specific conditions, and thus are underrepresented in non-tissue-targeted or non-condition-optimized RNA-sequencing projects. We previously identified 4,439 SSP-encoding genes in the model legume Medicago truncatula. To support systematic characterization and annotation of these putative SSP-encoding genes, we developed the M. truncatula Small Secreted Peptide Database (MtSSPdb; https://mtsspdb.noble.org/). MtSSPdb currently hosts (1) a compendium of M. truncatula SSP candidates with putative function and family annotations; (2) a large-scale M. truncatula RNA-sequencing-based gene expression atlas integrated with various analytical tools, including differential expression, coexpression, and pathway enrichment analyses; (3) an online plant SSP prediction tool capable of analyzing protein sequences at the genome scale using the same protocol as for the identification of SSP genes; and (4) information about a library of synthetic peptides and root and nodule phenotyping data from synthetic peptide screens in planta. These datasets and analytical tools make MtSSPdb a unique and valuable resource for the plant research community. MtSSPdb also has the potential to become the most complete database of SSPs in plants.
Plant small secreted peptides (SSPs) are crucial intercellular messenger molecules that regulate a multitude of processes (Matsubayashi, 2014). SSPs are typically encoded within preproteins of 100 to 250 amino acids, that are subsequently processed into shorter bioactive peptides of ∼5 to 50 residues (Lease and Walker, 2006; Breiden and Simon, 2016; de Bang et al., 2017) that act at very low, often nanomolar physiological concentrations (Murphy et al., 2012).
SSPs have emerged as an important class of regulatory molecules involved in plant growth, development, plant-microbe interactions, and stress tolerance (Czyzewicz et al., 2013; Nakaminami et al., 2018; Takahashi et al., 2018). This is of particular significance for legumes, since recent discoveries show that SSPs regulate symbiotic root nodulation (Djordjevic et al., 2015; Nishida et al., 2018; Kereszt et al., 2018) and root development (Araya et al., 2016; Patel et al., 2018). SSPs are also involved in reproductive development, embryogenesis, and pathogen interaction, among many other plant processes (Matsubayashi, 2014; Breiden and Simon, 2016). Due to their various effects in plants, SSPs are of interest as potential tools to improve plant performance, including as supplements to improve fertilizer-use efficiency for instance.
Legumes are key components of sustainable agricultural systems, since they form symbioses with soil bacteria that fix atmospheric nitrogen, reducing dependency on synthetic nitrogen fertilizers, with clear benefits to agricultural producers and the environment (Graham and Vance, 2003; Valentine et al., 2017). Medicago truncatula has been chosen as a premier model legume because it is closely related to economically important forage species such as alfalfa (Medicago sativa; Young and Udvardi, 2009), and it is invaluable for cross-legume genomic comparison studies (Tang et al., 2014). The M. truncatula sequencing project began in 2003; its bacterial artificial chromosome (BAC)-based genomic assembly was released in 2011 (Mt3.5; Young et al., 2011), and an optical map-based assembly using Illumina and 454 sequences was released in 2014 (Mt4.0; Tang et al., 2014). The Mt4.0 assembly has 50,894 genes (31,661 with high confidence and 19,233 with low confidence), with an ∼82% overlap with the previous genome annotation (Mt3.5), but there are still gaps, unanchored scaffolds, and 13,367 genes annotated as encoding hypothetical proteins (Tang et al., 2014). More recently, a M. truncatula genome assembly (MtrunA17r5.0-ANR) based on high-depth PacBio sequencing, comprising a total of 51,316 gene models that also includes a significant number of noncoding genes, was published (Pecrix et al., 2018). However, that assembly was not focused on SSP discovery, and many SSP-coding genes are not included.
It is important to note that short open reading frames (ORFs) were largely overlooked and omitted in both the Mt3.5 and Mt4.0 annotations. Newly generated RNA-sequencing (RNA-seq) data can provide expression evidence for such omitted genes. With the intent to mine for genes omitted from published versions of the M. truncatula genome assembly, we reannotated the M. truncatula genome (both Mt3.5v5 and Mt4.0v1 genome assemblies) using 64 RNA-seq libraries (de Bang et al., 2017). In addition, hidden Markov models (HMMs) of known SSP families were used to scan both genome assemblies for SSP genes. Relying on the improved procedure, 4,439 SSP-coding genes were identified in M. truncatula, including 2,455 novel SSPs not previously reported in the literature (de Bang et al., 2017). The multistep analytical procedure employed in this study (Boschiero et al., 2019) was particularly successful in the prediction of new small ORFs that were overlooked in standard genome annotation (Zhou et al., 2013).
To host the reannotated genes, SSP families, and related knowledge (de Bang et al., 2017), we developed a comprehensive database named the M. truncatula Small Secreted Peptide Database (MtSSPdb). The main highlights and features of MtSSPdb are (1) a compendium of 48 known SSP gene families and >200 putative SSP families, which were curated from 4,439 potential or confirmed SSP-coding genes from the above-mentioned reannotation procedure; (2) an online prediction tool that is able to predict SSPs for user-submitted large-scale protein sequences using a protocol similar to that described in de Bang et al. (2017); (3) a comprehensive transcriptome database for SSP genes with analytical tools; and (4) a catalog of trait information for a collection of SSPs tested on roots and nodulated plants. MtSSPdb also hosts all novel gene models in M. truncatula that were identified by the reannotation procedure (de Bang et al., 2017). MtSSPdb is an important resource for the plant scientific community and has the potential to become the most complete database of SSPs in plants.
RESULTS
Database Content
M. truncatula SSP Genes and Families
The M. truncatula genome (both Mt3.5v5 and Mt4.0v1 genome assemblies) was recently reannotated, and in total, 70,094 nonredundant genes were predicted, including 7,771 newly annotated gene loci (de Bang et al., 2017). The reannotation corrected many previously predicted gene models and helped to identify additional genes (de Bang et al., 2017). From the nonredundant genes, 4,425 gene loci were flagged as candidate SSP-coding genes based on different criteria, including protein length, signal peptide prediction, and homology with previously known SSPs or HMMs identified in previously known SSP families. A total of 1,970 of these SSP-coding genes were homologs of 46 previously established SSP gene families, while an additional 2,455 candidate SSP-coding genes were classified under the “Focal List,” which contained potential novel SSPs. Importantly, from the potentially novel SSPs, 56% were found to have a putative ortholog in at least one of 16 plant species, including many that appear to be legume specific (de Bang et al., 2017).
Among the items on this Focal List, a new gene family identified was called Peptide Suppressing Nodulation (PSN), encompassing four members (de Bang et al., 2017). Moreover, 14 new SSPs in M. truncatula were added from the recently described IRON MAN (IMA) family (Grillet et al., 2018), making a grand total of 4,439 putative SSP-coding genes (1,988 SSP homologs and 2,451 putative novel SSPs) and 48 SSP gene families (Fig. 1A).
Figure 1.
MtSSPdb content. A, Proportion of M. truncatula reannotated genes identified as known or putative SSPs. B, List of RNA-seq datasets available at Medicago SSP-GEA.
MtSSPdb provides HMMs for most SSP families. All 4,439 SSP-coding genes belong to 262 SSP gene families (48 known and 214 putative SSP families) that are well-described in the MtSSPdb and include family HMM models, profile logos for visualization, and gene family members. The 48 known gene families include 1,988 genes, and the 214 putative SSP families include 2,451 potentially novel SSPs that were previously unreported in the literature. The smallest SSP families are Plant Elicitor Peptides (PEPs), Casparian Strip Integrity Factor (CIF), and Subtilisin-embedded PEP (SUBPEP), with only one M. truncatula gene; and the largest families are Nodule-specific Cys Rich Group B (NCR-B) and NCR-A, with 428 and 361 genes, respectively, totaling 789 genes in MtSSPdb.
It is worth mentioning that the 24 known SSP gene families that were searched for (de Bang et al., 2017) but not identified in M. truncatula, were nonetheless included in MtSSPdb. These families were discovered in several other plant species, including Arabidopsis (Arabidopsis thaliana), maize (Zea mays), or tobacco (Nicotiana tabacum), and can be useful in the study of other species.
RNA-Seq Gene Expression Data
The SSP Gene Expression Atlas (SSP-GEA) currently hosts 16 RNA-seq experiments (10 publicly available datasets and six datasets from in-house experiments) comprising 681 RNA-seq samples from 192 treatments or plant organs (Fig. 1B) covering drought, plant hormone treatments, macronutrient deficiencies, nodule/root development, symbiotic interactions, salt stress, and various plant organs for all genes (not just SSP-coding genes). SSP-GEA will be updated twice a year depending on newly available data and user suggestions. Currently, no other gene atlas is available that curates published RNA-seq data for M. truncatula. It is important to mention that the previous M. truncatula Gene Expression Atlas (MtGEA) includes only microarray data (739 arrays from 274 experiments) and the last update of that atlas was in 2015 (Benedito et al., 2008).
Synthetic Peptide Library Data
Another section in MtSSPdb is the peptide library that currently lists 155 synthetic peptides derived from 104 M. truncatula genes, including 95 SSP-coding genes from 20 known SSP families, six putative SSPs from five putative SSP families, and three non-SSPs. In this section, users can find the peptides grouped by gene family for ease of use. For each peptide, detailed information is provided about chemical composition (e.g. Mr, pI, and grand average of hydropathicity [GRAVY]) and phenotype description (Supplemental Fig. S1). The synthetic peptides were tested on three different species (M. truncatula, Arabidopsis, and Panicum virgatum) for 24 root- and nodule-related phenotypes based on five categories: descriptive traits, primary root traits, lateral root traits, total root traits, and nodule traits (Table 1). Images are available that show root phenotypes from M. truncatula plants treated with 91 different synthetic peptides compared to untreated control roots. Metadata information for each peptide are available for download, and users can request aliquots of available synthetic peptides for their research via the provided contact form. Detailed information about the synthetic peptides, SSP families, and their annotations are shown in Supplemental Table S1.
Table 1. Root and nodule-related traits evaluated in M. truncatula, Arabidopsis, and P. virgatum for 155 synthetic peptides.
At, Arabidopsis; Mt, M. truncatula; Pv, P. virgatum.
| Trait | Species Tested |
|---|---|
| Descriptive traits | |
| Root phenotype | Mt |
| Nodule phenotype | Mt |
| Ca-Spike Assay | Mt |
| Primary root traits | |
| Primary root length (cm) | Mt, At, Pv |
| Lateral root density (n/cm) | Mt, At, Pv |
| Primary root mean diameter (cm) | Mt, At, Pv |
| Primary root surface area (cm2) | Mt, At, Pv |
| Primary root volume (cm3) | Mt, At, Pv |
| Primary root straightness (vector length/total length) | Mt, At, Pv |
| Lateral root traits | |
| Total number of lateral roots | Mt |
| Total lateral root length (cm) | Mt, At |
| Number of secondary lateral roots | Mt, At, Pv |
| Total length of secondary lateral roots | Mt, At, Pv |
| Mean length of secondary lateral roots | Mt, At, Pv |
| Mean diameter of secondary lateral roots (cm) | Mt, At, Pv |
| Mean diameter of all lateral roots (cm) | Mt |
| Secondary lateral roots surface area (cm2) | Mt, At, Pv |
| Secondary lateral roots volume (cm3) | Mt, At, Pv |
| Secondary lateral roots insertion angle (°) | Mt, At, Pv |
| Total root traits | |
| Total root length (cm) | Mt, At, Pv |
| Total root surface area system (cm2) | Mt, At, Pv |
| Total root volume system (cm3) | Mt, At, Pv |
| Nodule traits | |
| Nodule number | Mt |
| Nodule density | Mt |
User Interface and Utility
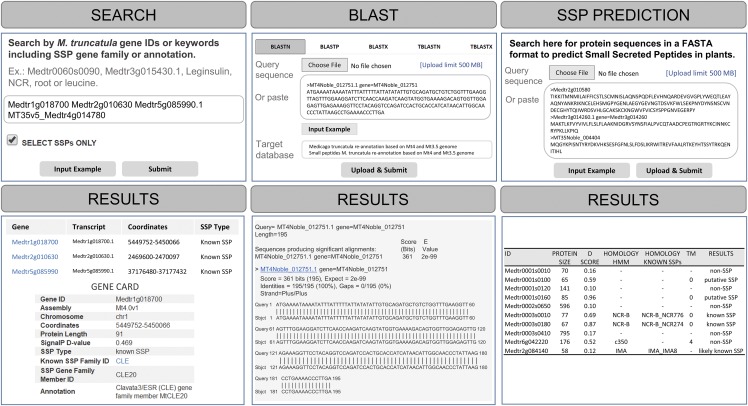
The MtSSPdb structure is categorized into three main sections: resources, Gene Expression Atlas, and tools (Fig. 2). MtSSPdb provides three analytical tools—a search function for genes, representative transcripts, functional annotations, or SSP gene families; a BLAST search function; and an online plant SSP prediction tool that identifies genes that potentially encode SSPs in the precursor proteins. These sections are described below in more detail to show their utility and usage.
Figure 2.
Framework of the MtSSPdb showing the structure, available resources, and tools.
Search Tool and Gene/Family Card Information
The search tool is a primary function to search M. truncatula genes by gene or transcript identifier (ID), keywords of annotation, or SSP gene family name (Fig. 3). There is an option to preselect “only SSP-coding genes”, which narrows down search results to SSP-coding genes. For each gene, there is a Gene Card page available with detailed information, including genomic coordinates, SignalP D-score (discrimination score.), protein length, SSP type, annotation, and sequences. Additionally, gene expression examples are shown for different experiments (Fig. 3).
Figure 3.
Workflow of the tools available on the MtSSPdb. the top row shows three different tools—Search, SSP Prediction, and BLAST—available to search genes or analyze nucleotide/protein sequences. The bottom row shows the resulting gene search displayed as a list and gene cards available for each gene. BLAST output is provided. In addition, a table is displayed as a result of the SSP Prediction tool with SSP classification from protein sequences.
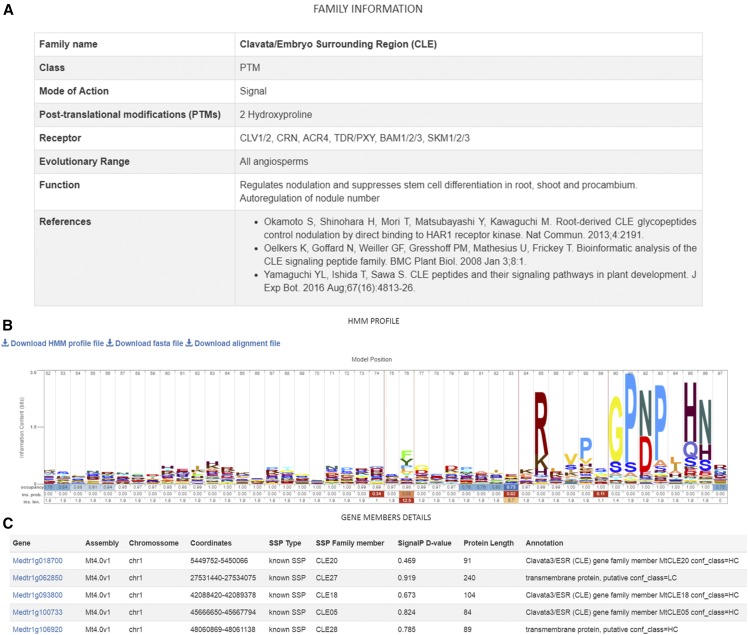
MtSSPdb contains SSP gene families divided into two groups, i.e. known and putative SSPs. There are currently 48 known SSP gene families containing 1,988 SSP-coding genes identified in M. truncatula with gene family card information for each of these (Fig. 4A). An additional 24 SSP gene families are included, although they have not yet been identified in M. truncatula. HMM profiles are available for 37 known families and 59 putative families with at least five members. HMM profiles can be downloaded or visualized by sequence logos (Fig. 4B). Users can download FASTA or alignment files and an HMM profile file for each family and visualize all gene family members (Fig. 4C). Figure 4 presents an example of a gene family card for the Clavata/Embryo Surrounding Region (CLE) with 52 gene members.
Figure 4.
MtSSPdb screenshot showing an overview of a gene family card for the Clavata/Embryo Surrounding Region (CLE) family. A, Gene family summary information. B, A HMM profile logo showing the C-terminal region of the protein. C, Five of the gene members and their associated information.
MtrunA17r5.0-ANR gene IDs (Pecrix et al., 2018) are also available on the Gene Card page. To facilitate searching SSPs in the MtrunA17r5.0-ANR genome assembly, we conducted gene mapping. The total of 44,623 MtrunA17r5.0-ANR transcripts were queried against the 70,094 Noble genome reannotation transcripts (de Bang et al., 2017). We successfully mapped 43,018 (96.4%) of the MtrunA17r5.0-ANR transcripts to the Noble M. truncatula genome reannotation, including ∼39% of small genes (<200 amino acids) and ∼77% of the SSP-coding genes (Supplemental Table S2). We further conducted SSP gene prediction on the remaining 1,605 MtrunA17r5.0-ANR genes that were not mapped to the Noble genome reannotation using our integrated online plant SSP prediction tool. Among the 1,605 MtrunA17r5.0-ANR genes, only five definitely belong to known SSP families, 11 likely belong to known SSP families, and 183 were identified as putative SSPs. These results show that the Noble M. truncatula genome reannotation was optimized for identification of small genes (de Bang et al., 2017).
BLAST
A BLAST tool was implemented with different search options (BLASTN, BLASTP, BLASTX, TBLASTN, or TBLASTX; Fig. 3). We developed a web interface for the National Center for Biotechnology Information (NCBI) program BLAST, which enables users to search their sequences against hosted sequences (Camacho et al., 2009). Users can select two different target libraries (all M. truncatula genes or only SSP-coding genes) and different output formats (Fig. 3). The BLAST tool allows users to upload up to 500 MB of data per sequence search. In the output, a link to the respective gene card page is provided for each gene.
SSP Prediction Tool
Due to a lack of SSP prediction tools in the public domain, we developed and implemented such a tool as part of MtSSPdb. The tool predicts whether a given protein sequence is likely to encompass an SSP based on several criteria (de Bang et al., 2017), including (1) protein length (≤200, 230, or 250 amino acids; Lease and Walker, 2006; Breiden and Simon, 2016; de Bang et al., 2017); (2) the presence of a signal peptide cleavage site (Petersen et al., 2011); (3) the presence of a sequence pattern characteristic of HMMs of known SSP gene families; (4) homology with known SSP-coding genes previously identified; and (5) absence of transmembrane (TM) helices. The prediction pipeline is suited to the analysis of protein sequences from multiple plant species, since its reference sequences and HMMs are built based on sequences from 35 diverse plant species, such as Arabidopsis, M. truncatula, soybean (Glycine max), maize, rice (Oryza sativa), poplar (Populus trichocarpa), tobacco, Lotus japonicus, grapevine (Vitis vinifera), Amborella trichopoda, chickpea (Cicer arietinum), common bean (Phaseolus vulgaris), tomato (Solanum lycopersicum), clementine (Citrus clementina), and others (Ghorbani et al., 2015; Grillet et al., 2018).
Preproteins comprising SSPs contain an N-terminal signal peptide that directs the preprotein to the endoplasmic reticulum for cleavage, maturation, and sorting. SignalP 4.1 has been shown to be an effective predictor of N-terminal signal peptides of proteins from a wide array of species, including prokaryotes and eukaryotes (Petersen et al., 2011). It relies on a D-score, which is a combined value from signal peptide and cleavage site prediction networks and is used to discriminate signal peptides from non-signal peptides. The default D-score cutoff for a signal peptide is 0.45 as applied in SignalP 4.1 server (Petersen et al., 2011).
After homology analysis of the sequence, prediction of TM helices is performed with putative SSP-coding genes meeting the above criteria. Any gene predicted to harbor at least one TM helix is considered not to be an SSP, since membrane anchoring is not characteristic of SSPs that act in cell–cell signaling, but note that TM predictions can vary depending on the tool used (Ganapathiraju et al., 2008; Tsirigos et al., 2015).
The final output table (Fig. 3) presents the calculated values of each of the five individual features plus a cumulative prediction that places the protein within one of three types of SSPs—“known,” “likely known,” or “putative.” A known SSP has a protein length of ≤200 amino acids, a SignalP D-score of >0.25, and homology with previous SSPs, while a putative SSP has a protein length of ≤230 amino acids, a SignalP D-score of >0.45, no TM domains, and no significant homologies with known SSPs or hits with only one type of homology. We included an additional SSP type defined as “likely known SSPs,” with significant homologies to known SSPs and a small protein length (≤250 amino acids). In this category, there are, for example, several CLE peptides, including CLE2, CLE8, CLE19, CLE27, CLE34, CLE36, CLE41, and CLE48. Details regarding criteria were described in our previously published article (de Bang et al., 2017). All details about input, output, and criteria used are provided on the Help page of MtSSPdb.
In the output result page, users can filter the results by adjusting various cutoff thresholds (“protein length,” “SignalP D-score,” “HMM homology e-value,” and “Smith-Waterman homology e-value”) using our filter function. In addition, users can filter the results by the SSP classification; for example, users can choose to display only known, likely known, or putative SSPs.
When we analyzed all M. truncatula reannotated genes (70,094) with our SSP prediction tool, our prediction generated a 98.6% matching classification with those previously produced by our group (de Bang et al., 2017). The differences are primarily due to manual curation. We recommend that SSPs predicted by our tool be further confirmed by expression evidence and subsequent experimental validation.
SSP-GEA
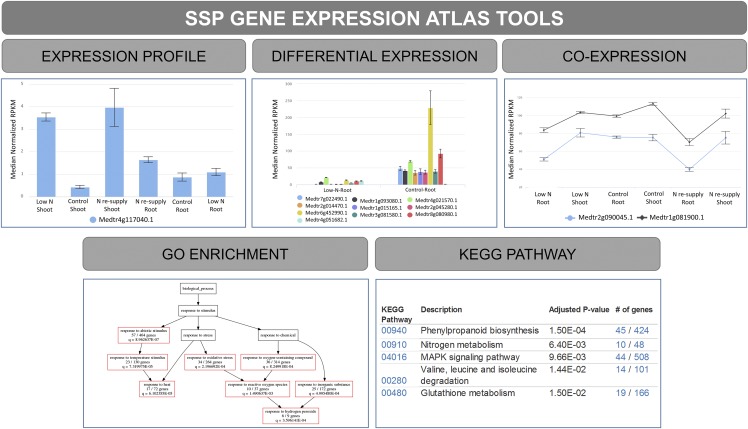
The SSP-GEA is a major component of MtSSPdb and provides several tools to analyze and display (1) an expression profile with gene search by keyword, ID, and expression pattern over conditions, (2) differential gene expression, (3) gene coexpression, (4) Gene Ontology (GO) term, and (5) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment for a selected group of genes (Fig. 5). SSP-GEA allows users to choose the RNA-seq dataset of interest, and all the above analyses are performed “on the fly.”
Figure 5.
Available tools in the SSP-GEA showing different outputs. The top shows bar charts displaying the results of the expression profile analysis of Medtr4g117040.1 transcript for different nitrogen levels in shoots and roots (left) and the DE analysis for nitrogen levels in roots (middle; low nitrogen levels and full nutrition control; adjusted P < 0.1). A line chart is displayed as a result of the coexpression analysis of the gene Medtr6g452990 for nitrogen levels in shoots and roots (right; low nitrogen levels, full nutrition control, and nitrogen resupply). The lower left shows a directed acyclic graph as a result of the GO enrichment analysis from five top GO terms from genes differentially expressed for different nitrogen levels in roots. The lower right shows a table with five top significantly (q < 0.05) enriched KEGG pathways from genes differentially expressed for nitrogen levels in roots.
SSP-GEA allows users to query expression levels of any M. truncatula gene or transcript of interest. Queries can also be made using the SSP gene family name or annotation keywords. Users begin by selecting experiment(s) of interest (Fig. 1B) and the experimental conditions. After submission, the raw read counts of selected samples are extracted from the database and normalized for further analysis, and users then can search genes by keyword or expression patterns. A bar chart or line chart is generated on the web page. Users have the option to download a “.CSV” table with all the expression values of the queried genes from the expression profile, differential expression (DE), or coexpression analyses.
Figure 5 shows the different outputs obtained from the analytical tools. In SSP-GEA, the DE analysis tool was designed for DE analysis of RNA-seq data from different experiments. Users can select the desired experiment, the two conditions to be compared (numerator and denominator), and the P-value cutoff. Users also have the option to filter out any gene with a low number of normalized mean counts or to filter the results based on log2 fold changes or adjusted P-values. For coexpression analysis, users select the relevant experiments and conditions. Once the analysis is complete, users can specify their genes of interest to extract a list of coexpressed genes. GO enrichment and KEGG pathway analyses are either available separately or are integrated with expression profile, DE, and coexpression analyses, where the output results from these upstream analyses can be directly imported into the GO/KEGG analysis module to further identify enriched GO terms or KEGG pathways in the list of genes. Additional filters are available prior to the GO and KEGG analyses, such as P- or q-value cutoff and number of top terms or pathways.
Case Studies
The following case studies demonstrate the usefulness of MtSSPdb in identifying novel candidate peptides and their biological functions.
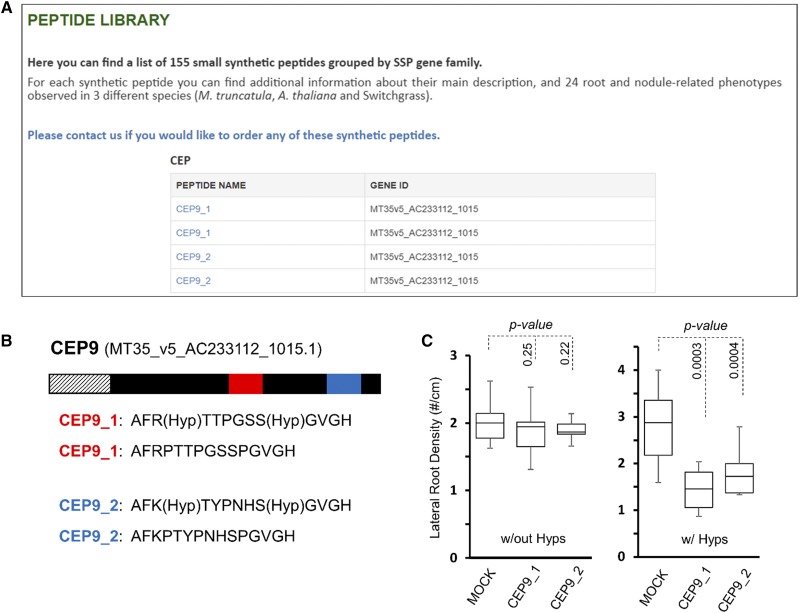
Bioactivity of CEP9 Requires Pro Hydroxylation
The C-terminally Encoded Peptide 1 (CEP1) peptide of M. truncatula was previously shown to inhibit lateral root formation when applied exogenously to germinated seedlings (Imin et al., 2013). The M. truncatula genome encodes 17 CEP genes, 15 of which are represented in the Synthetic Peptide Library of MtSSPdb (Fig. 6A). Like the CEP1 gene (MT35v5_contig_59554_1), CEP9 (MT35v5_AC233112_1015) harbors two highly similar CEP domains. Peptides representing both domains are present in the peptide library, and these peptides were synthesized with or without Hyp residues at residues 4 and 11 (Fig. 6B), which have been shown to be important for bioactivity in CEP1. The screening data in the peptide library clearly show that both CEP9 peptides reduce lateral root density, like CEP1, and that this activity depends on Hyp at residues 4 and 11 (Fig. 6C). These results indicate that the CEP9 and CEP1 peptides may have overlapping functions within M. truncatula roots.
Figure 6.
Surveying the peptide library data reveals the importance of Pro hydroxylation of CEP peptides. A, Hybrid screenshot of the peptide library page of the MtSSPdb showing the four CEP9 peptides investigated here. B, CEP9 encodes a 151-amino acid polypeptide encoding a signal peptide (hashed lines) and containing two CEP domains, designated CEP9_1 (red) and CEP9_2 (blue). The synthetic peptides used for screening are presented below, assuming the presence of Hyp. Peptides lacking Hyp held standard Pro residues at both positions. C, Synthetic peptides from both the upstream and downstream domains strongly inhibit lateral root formation, but only when Pro residues 4 and 11 are hydroxylated. Percentage values indicate the average number of secondary lateral roots in the presence of the indicated peptide relative to the mock-treated control. Plots show the median value (center lines), box limits indicate the 25th and 75th percentiles, and the bars indicate the maximum and minimum values among the data points collected (Students t test). n = 10 for peptide-treated plants and 20 for mock-treated plants.
Identification of a Novel SSP Transcript with Support from MtSSPdb
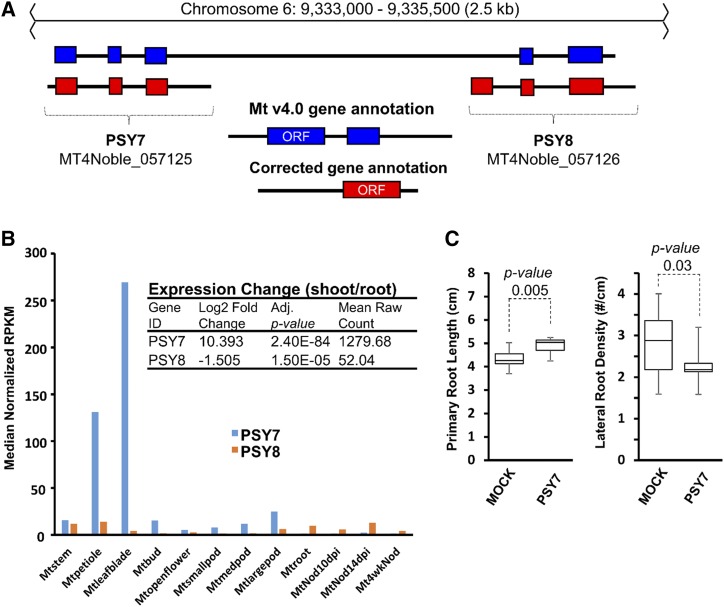
Visualization of RNA-seq data revealed that Medtr6g027320 was incorrectly annotated in Mt v4.0 (Fig. 7A). Two transcripts with widely differing expression patterns can be seen. Both corrected transcripts within this locus have ORFs with ∼60% identity, strongly predicted signal peptides, and Plant Peptide Containing Sulfated Tyr (PSY) domains near the C terminus. Accordingly, these transcripts were renamed as PSY7 and PSY8. Extracting tissue expression plots from the SSP GEA section of MtSSPdb, we found that PSY7 is the dominantly expressed transcript and is primarily expressed in aerial organs such as leaf and petiole (Fig. 7B). In contrast, PSY8 is expressed at low levels throughout much of the plant, including root, nodule, and stem. DE analysis using the SSP-GEA revealed that PSY7 transcript levels in leaves exceed those in roots by >1,000-fold, while PSY8 transcript levels were about 3-fold lower in shoots than in roots. To test for activity of PSY7, the predicted 19-residue peptide, including the expected sulfate group at Tyr-2 and Hyp at Pro-13 and Pro-16 (Amano et al., 2007), was synthesized and employed in our root phenotyping screen. Compared to the mock-treated control, the PSY7 peptide enhanced primary root growth (Fig. 7C), consistent with the characterized role of AtPSY in cell expansion (Amano et al., 2007). Additionally, a slight suppression of lateral root density is also observed (Fig. 7C). The bioactivity found for this synthetic peptide provides additional support for the identification of this novel transcript.
Figure 7.
Reannotation of a Plant Peptide Containing Sulfated Tyr (PSY) reveals a novel bioactive PSY SSP-coding gene. A, RNA-seq evidence indicates that Medtr6g027320.1 is annotated incorrectly in Mt v4.0. RNA-seq reads mapped to each locus are illustrated as a Sashimi plot representing the number of mapped reads at each nucleotide. Two distinct transcripts emerge when RNA-seq mapped reads are separated by expression in shoots or roots. Reads from one representative biological replicate are presented above. The new transcripts uncover an additional upstream PSY SSP-coding gene and have been renamed PSY7 and PSY8 accordingly. B, A plot of tissue expression levels of the newly uncovered PSY7 and PSY8 genes. PSY7 is the dominantly expressed transcript and is mostly expressed in leaves and shoots. In contrast, PSY8 demonstrates low-level expression throughout roots, nodules, and stems/petioles. The inset shows how expression changes from shoot versus root highlight the DE of the two PSY transcripts. RPKM, reads per kilobase of transcript per million mapped reads. C, Synthetic PSY7 peptide shows bioactivity when tested in the root screening platform, leading to enhanced primary root length (consistent with its characterized role in cell division/expansion), while also suppressing lateral root density. Plots show the median value (center lines), box limits indicate the 25th and 75th percentiles, and the bars indicate the maximum and minimum values among the data points collected (Students t test). n = 10 for peptide-treated plants and 20 for mock-treated plants.
SSPs Regulated during Mycorrhizal and Rhizobial Symbioses
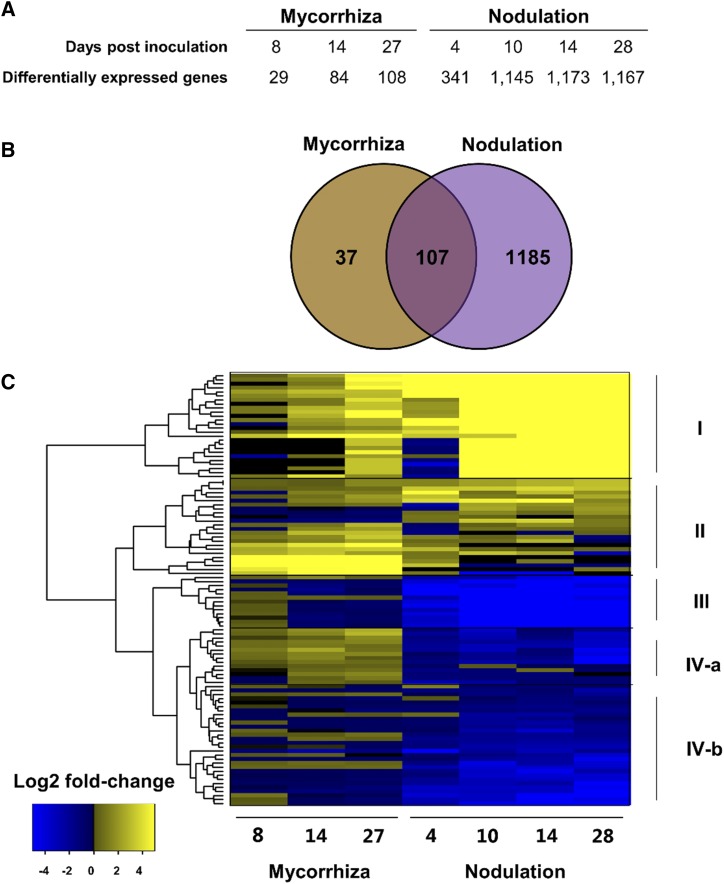
The SSP-GEA contains expression data from symbiotic interactions with nitrogen-fixing bacteria known as rhizobia and arbuscular mycorrhizal fungi (Luginbuehl et al., 2017). In legumes, these two symbionts are known to share signaling components during development of nodules (root organs hosting the rhizobia) and mycorrhizal colonization. To investigate whether any SSP gene transcripts were commonly regulated during both nodulation and mycorrhizal colonization, DE analyses were performed in SSP-GEA based on nodules at 4, 10, 14, and 28 d post rhizobia inoculation (dpi) compared to controls (de Bang et al., 2017), and mycorrhizal roots at 8, 14, and 27 dpi compared to nonmycorrhizal control roots (Luginbuehl et al., 2017). In total, 341 to 1,173 DE SSPs were identified in nodules, compared to 29 to 108 DE SSPs in mycorrhizal roots (adjusted P-value <0.1; Fig. 8A). Collectively, 1,292 individual SSPs were identified as DE during nodule development, whereas only 144 individual SSPs were found to be DE in mycorrhizal roots. Despite this large discrepancy in the number of DE SSPs between rhizobial and mycorrhizal roots, 107 SSPs were found to be shared, of which many showed a similar response to nodulation and mycorrhization, while others responded in opposite directions (Fig. 8B). Hierarchical clustering based on transcriptional changes of the 107 commonly regulated SSPs grouped the SSPs into four different clusters (Fig. 8C). SSPs in Cluster I were highly induced in nodules and during later stages of mycorrhizal symbiosis, and included 10 NCR peptides, five leginsulins, five Nodule-specific Gly Rich Peptides (NodGRPs), and three plant defensins (Supplemental Table S3). The high expression of supposedly nodule-specific NCRs and NodGRPs in mycorrhizal roots could indicate that the analyzed roots were also nodulated. However, the well-studied nodulation-marker SSP transcripts CLE12 and CLE13 were not induced, leading to the conclusion that the expression of this subset of NCRs and NodGRPs is not nodule specific. Cluster II contained SSPs generally upregulated in both mycorrhizal roots and nodules. A group of these, including five plantacyanins (PCYs), was strongly upregulated in mycorrhizal roots compared to nodules. Cluster III constituted 13 SSP transcripts with reduced abundance in nodules but moderately induced expression in mycorrhizal roots at 8 dpi. Six of these belonged to the Root Cap (RC) family. SSPs moderately upregulated by mycorrhiza and downregulated during nodulation were grouped into Cluster IV-a, which contained four SSPs from the Prorich Protein Group 669 (PRP669), and five from the subtilisin inhibitor (SubIn) families, respectively. Cluster IV-b represented 30 SSPs with significant down-regulation (adjusted P-value <0.1) during nodulation but an inconsistent response to mycorrhizal colonization. Nine of these were PCYs and four were nonspecific lipid transfer proteins.
Figure 8.
SSPs regulated by mycorrhiza colonization and nodulation. A, Number of differentially expressed SSPs during nodule development at 4, 10, 14, and 28 dpi with rhizobia, and 8, 14, and 27 dpi with mycorrhizal fungi (adjusted P < 0.1). B, Venn diagram illustrating the number of commonly regulated SSPs between mycorrhiza and nodulation. SSPs were included if they were differentially expressed in just one of the treatments. C, Hierarchical clustering of the 107 commonly regulated SSPs from log2 fold changes were calculated based on the respective control samples, i.e. nonmycorrhizal or nonrhizobial.
CAPE16 Is Implicated in Rhizobial Persistence within Nodules
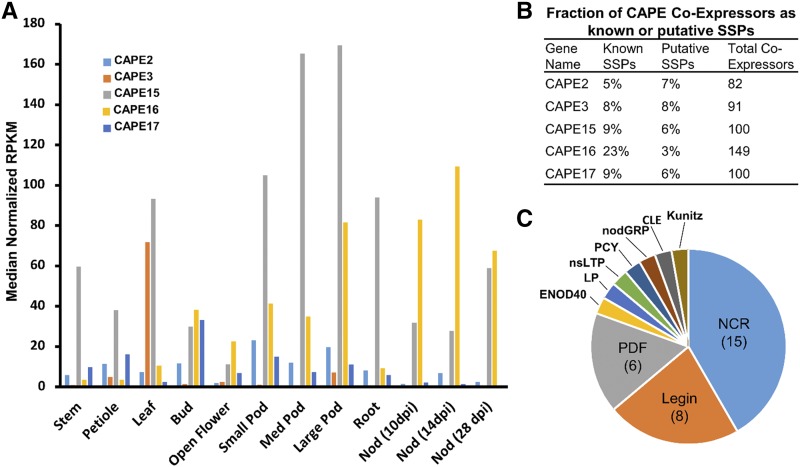
SSPs in the CAP-derived Peptide (CAPE) family are derived from functional precursor proteins involved in the pathogen defense pathway in leaves (Chen et al., 2014). CAPE peptides are embedded at the C terminus of larger Pathogenesis-Related Protein 1 proteins and are cleaved into an 11-residue peptide prior to secretion. Several CAPE peptides in Arabidopsis are induced by salt treatment (Chien et al., 2015), but no functional studies have been carried out. Analysis with SSP-GEA in MtSSPdb revealed that several CAPE gene transcripts are abundant in nodule tissue (Fig. 9A). Investigation of the coexpression patterns of selected CAPE gene transcripts revealed that CAPE16 (Medtr5g018770), in particular, was strongly enriched for coexpression with other SSP-coding genes. Filtering coexpressing genes with a >0.8 Pearson correlation coefficient showed that 23% of CAPE16 coexpressed gene transcripts were SSP-coding genes, compared to 5% to 9% of four other CAPE gene transcripts (Fig. 9B). The coexpressing SSPs were predominantly NCR, leginsulin, and plant defensin genes (Fig. 9C). NCR genes in particular encode nodule-specific SSPs with roles in development and maintenance of rhizobia within symbiotic nodules in M. truncatula (Kereszt et al., 2018). Thus, the expression patterns discerned from MtSSPdb may indicate a role in nodulation for CAPE16, but further experiments should be conducted to validate these findings.
Figure 9.
CAPE16 expression predominates in symbiotic nodules and coexpresses with many SSPs. A, RNA-seq data from the MtSSPdb. Five CAPE peptides, which have been shown previously to be induced by macronutrient deficiencies, were interrogated for expression patterns across 12 different organs. Nod, Nodule; RPKM, reads per kilobase of transcript per million mapped reads. B, CAPE16 shows a strong enrichment in coexpressing SSP-coding genes. Coexpressing SSPs were defined as those with a Pearson correlation coefficient >0.8 across 122 different experimental conditions. Thirty-five of a total 149 coexpressors of CAPE16 (23%) were SSP genes, versus 5% to 9% of the other four CAPE genes. C, Pie chart showing the distribution of the CAPE16 coexpressing SSP-coding genes, most of which are NCR peptides, leginsulins (Legin), or plant defensins (PDF). ENOD40, Early Nodulin 40; LP, LEED.PEED; nsLTP, nonspecific lipid transfer protein; nodGRP, nodule-specific Gly-rich protein.
DISCUSSION
There are only three published databases dedicated to plant small peptides (Table 2). PlantSSPdb hosts a collection of small secretory peptides from 32 plant species, including 820 M. truncatula SSP-coding genes (Ghorbani et al., 2015). Besides PlantSSPdb, there is a signal peptide database containing signal sequences of archaea, prokaryotes, and eukaryotes, but it includes only 17 SSPs described in M. truncatula (Choo et al., 2005). Also, there is a database of SSPs predicted in Arabidopsis with very limited and outdated information (Lease and Walker, 2006).
Table 2. List of available SSP and signaling peptide databases in plants.
SPdb, a signal peptide database containing signal sequences of archaea, prokaryotes, and eukaryotes.
| SSP Databases | MtSSPdb | PlantSSPdb | SPdb | Arabidopsis Unannotated Secreted Peptide Database |
|---|---|---|---|---|
| Last update | 2019 | 2015 | 2008 | 2006 |
| Species | M. truncatula (to be expanded soon to Arabidopsis and B. distachyon) | 32 species, including M. truncatula | Different species including plants and M. truncatula | Arabidopsis |
| M. truncatula SSPs (genome assembly) | 4,439 (Mt3.5v5 and Mt4.0v1) | 820 (Mt3.5v4) | 17 | Not available |
| Annotation | Available | Not available | Not available | Not available |
| Gene family | 262 available with detailed information (function, reference, HMM profile logos, genes, etc.) | 334 available for M. truncatula with limited information | Not available | Not available |
| BLAST (input sequence size) | Available (500 MB) | Available (8 MB) | Not available | Not available |
| Expression data | Available for 16 experiments and 192 conditions | Not available | Not available | Not available |
| Gene Expression Atlas | Available with multiple analyses | Not available | Not available | Not available |
| SSP Prediction tool | Available across multiple plant species and genome scale sequences | Not available | Not available | Not available |
| Synthetic Peptide Library | Available with 155 peptides tested on 3 species for root and nodule-related traits, and SSP order option available | Not available | Not available | Not available |
In PlantSSPdb, it is possible to browse SSP genes and download family HMMs and protein sequences for five pillar species (Arabidopsis, rice, poplar, grapevine, and maize). PlantSSPdb uses criteria similar to ours to identify SSPs within their reference pillar species. However, in additional species such as M. truncatula, the SSP identification relies solely on automated, unsupervised searches of each family’s HMM built from the five pillar species. Because SSPs rapidly evolve and can be species or genera specific, HMM-based searches that rely on evidence from only the five pillar species are of limited value for alternative species, in particular M. truncatula and other legumes that are known to have a number of legume-specific SSPs.
In contrast to PlantSSP, MtSSPdb relies on iterative searches and extensive manual curation throughout the M. truncatula genome (de Bang et al., 2017). These steps have resulted in high-quality SSP gene models for M. truncatula. The improved analytical procedures, described in de Bang et al. (2017), identified over 4,000 predicted SSPs, including almost 2,000 members of known SSP families and a novel legume-specific SSP family named PSN (de Bang et al., 2017). Most of the 820 M. truncatula SSP genes found in the PlantSSP database (Ghorbani et al., 2015) are included in MtSSPdb, but only ∼30% of the SSP-coding genes could be associated with the HMMs from the PlantSSP database (de Bang et al., 2017). This is likely a reflection of the extensive manual curation underlying MtSSPdb, and it highlights the greatly improved identification of putative M. truncatula SSPs.
Furthermore, no gene expression or annotation data are available for the SSP genes in PlantSSPdb (Ghorbani et al., 2015). The gene expression data and related analytical functions, such as profiling, DE, coexpression, and pathway enrichment analysis, are helpful tools for exclusion of false positive predictions and identification of biological functions for the SSP genes.
Online analysis tools are a convenient way to search SSP genes from user-submitted sequences. PlantSSPdb provides a web-based BLAST search tool against SSP reference sequences with an 8 MB limit for data upload. NCBI BLAST is a heuristic search algorithm that compromises on sensitivity to sequences with lower similarity to obtain faster search performance (Camacho et al., 2009). This feature may cause the loss of SSP candidates, since SSPs rapidly evolve and conservation can be limited to short sequences. To address this issue, MtSSPdb integrates a comprehensive SSP prediction tool that utilizes HMM and Smith-Waterman searches. In addition, our database also provides the information from SignalP analysis and protein length. The SSP tool was able to predict SSPs more accurately than BLAST from PlantSSPdb (Ghorbani et al., 2015). For example, using as input M. truncatula protein sequences from Mt3.4v4 (n = 64,152), we predicted 1,218 known SSPs with the SSP tool, but using BLAST/PlantSSPdb with a stringent e-value (<1e−07), we obtained >7,000 best hits, and most of them with low identity.
The MtSSPdb prediction tool accepts user-submitted protein sequences up to 500 MB, which is enough for most genome-wide analyses. This prediction tool enables users to easily utilize the knowledge of known SSP families to identify new SSP proteins with high confidence.
In addition, MtSSPdb has gene expression information for each SSP gene, including expression profiles from common biological conditions, including various plant tissues and treatments, such as hormone and plant macro-nutrition treatments, which provide insight into the functional characterization of these SSP genes.
It is worth mentioning that MtSSPdb focuses on M. truncatula; it will be important to expand this database to other relevant model plant species and legumes such as alfalfa and soybean.
CONCLUSIONS
MtSSPdb is the first plant SSP database that integrates gene expression, an SSP prediction online tool, and synthetic peptide information. MtSSPdb hosts large-scale genomics and transcriptomics data in the model legume, M. truncatula, and provides multiple functions to search, retrieve, analyze, and visualize different datasets. It also hosts, under the synthetic peptide library, phenotyping data from synthetic peptide screens in planta. Compared to the previously published database (Ghorbani et al., 2015), MtSSPdb contains more comprehensive and up-to-date data for M. truncatula, resulting in a valuable resource for the plant research community. In addition, the integrated SSP prediction tool is the first web-based tool for the identification of plant SSPs from users’ submissions using multiple SSP characteristics. This tool also allows users to submit protein sequences on a genome scale for data analysis. To the best of our knowledge, no such comprehensive resource focusing on small peptide-coding genes, which are numerous and often still unannotated, exists for any plant species. It is worth mentioning that the database contains all known SSP family information, including 24 families which have not been identified in M. truncatula. Thus, this database can be expanded to other relevant model plant species, e.g. Arabidopsis and Brachypodium distachyon, and legume species such as alfalfa and soybean. MtSSPdb has potential to become the most comprehensive database of SSPs in plants. The MtSSPdb is available at https://mtsspdb.noble.org/.
MATERIALS AND METHODS
Medicago truncatula SSP genes and families
The M. truncatula genome (both Mt3.5v5 and Mt4.0v1 genome assemblies) was reannotated using the generic genome annotation tool MAKER pipeline (Cantarel et al., 2008). Gene model expression evidence includes 64 RNA-seq libraries that were mainly sequenced after the release of MTv4.0 and protein/EST sequences that are publicly available in legumes (de Bang et al., 2017). The SPADA pipeline (Zhou et al., 2013) and sORF Finder (Hanada et al., 2010) were used to identify short genes. The former was optimized by including HMM SSP models from PlantSSPdb (Ghorbani et al., 2015). The gene models were annotated using plant UniProt (https://www.uniprot.org/) as reference database with BLASTP for GO and KEGG (e-values < 1e−05). HMMs were established in two steps. First, we generated a multialignment file for each family using representative member sequences in M. truncatula. Second, the multialignment files were converted into HMM model files using HMMER software (Finn et al., 2011). The interactive gene family logos for HMM profiles were built using the Skylign tool (Wheeler et al., 2014). BLAT (Kent, 2002) was used to map MtrunA17r5.0-ANR genes (Pecrix et al., 2018).
Development of the MtSSPdb Web Portal
The web portal was developed using the Python Flask framework and MySQL.
RNA-Seq Data Analysis
RNA-seq data produced in-house were generated using Illumina technology representing different organs (leaves, shoot, petioles, buds, flowers, pods, roots, and nodules). More information about the experimental methods is available (de Bang et al., 2017). RNA-seq datasets were mapped against the representative transcripts of M. truncatula genes to estimate raw counts and effective transcript lengths using the Sailfish/Salmon tool (Patro et al., 2017). These results were uploaded into the database.
Normalization and DE analysis of raw counts are performed using DESeq2 (Love et al., 2014). The coexpression module was developed based on WGCNA (Langfelder and Horvath, 2008), which generates a coexpression matrix for the entire genome using biweight midcorrelation approach (also called bicor). The matrix can be processed to generate coexpressed functional gene modules (Langfelder and Horvath, 2008). The enriched GO terms or KEGG pathways in DE genes or coexpressed gene modules are detected by using P-values from hypergeometric distribution and Benjamini-Hochberg adjustment.
SSP Prediction Tool
The presence of a signal peptide cleavage site is predicted by SignalP 4.1 (Petersen et al., 2011). A sequence’s homology to previously identified SSPs is determined in two ways, first by searching user-submitted protein sequences against a collection of 37 curated HMMs of known SSP families (de Bang et al., 2017) and 4,780 HMMs from PlantSSP (Ghorbani et al., 2015) using HMMER (Finn et al., 2011), and second by searching user-submitted sequences against 3,402 known plant SSP protein sequences with SSearch (Ropelewski et al., 2003), a fast implementation of the Smith-Waterman search algorithm. E-values of ≤0.01 were used for significant homologies. Prediction of TM helices is performed with TMHMM Server v.2.0 (Krogh et al., 2001), and excludes predicted N-terminal signal peptides.
Accession Numbers
RNA-seq datasets produced by Noble Research Institute can be retrieved from NCBI Sequence Read Archive at https://www.ncbi.nlm.nih.gov/sra with the Study IDs SRP110143, SRP110041, SRP109847, and SRP161571.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MtSSPdb screenshot showing an overview of the Peptide Library section.
Supplemental Table S1. Gene information for synthetic peptides tested in three species.
Supplemental Table S2. Mapping results of MtrunA17r5.0-ANR transcripts against the Noble M. truncatula genome reannotation.
Supplemental Table S3. SSPs commonly regulated between nodulation and mycorrhiza.
Acknowledgments
We thank our collaborators who provided valuable feedback during the development of MtSSPdb.
Footnotes
This work was supported by the National Science Foundation (NSF), Division of Integrative Organismal Systems (grant no. 1444549), the Oklahoma Center for the Advancement of Science and Technology (OCAST; grant no. PS18–012), the Noble Research Institute, and Novo Nordisk Fonden (grant no. NNF17OC0024884 to T.C.d.B.).
Articles can be viewed without a subscription.
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y(2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T, von Wirén N, Takahashi H(2016) CLE peptide signaling and nitrogen interactions in plant root development. Plant Mol Biol 91: 607–615 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Boschiero C, Lundquist PK, Roy S, Dai X, Zhao PX, Scheible W-R(2019) Identification and functional investigation of genome-encoded, small, secreted peptides in plants. Curr Protoc Plant Biol 4: e20098. [DOI] [PubMed] [Google Scholar]
- Breiden M, Simon R(2016) Q&A: How does peptide signaling direct plant development? BMC Biol 14: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL(2009) BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SM, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M(2008) MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR(2014) Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien PS, Nam HG, Chen YR(2015) A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J Exp Bot 66: 5301–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH, Tan TW, Ranganathan S(2005) SPdb—a signal peptide database. BMC Bioinformatics 6: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N, Yue K, Beeckman T, De Smet I(2013) Message in a bottle: Small signalling peptide outputs during growth and development. J Exp Bot 64: 5281–5296 [DOI] [PubMed] [Google Scholar]
- de Bang TC, Lundquist PK, Dai X, Boschiero C, Zhuang Z, Pant P, Torres-Jerez I, Roy S, Nogales J, Veerappan V, et al. (2017) Genome-wide identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol 175: 1669–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic MA, Mohd-Radzman NA, Imin N(2015) Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot 66: 5171–5181 [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR(2011) HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathiraju M, Balakrishnan N, Reddy R, Klein-Seetharaman J(2008) Transmembrane helix prediction using amino acid property features and latent semantic analysis. BMC Bioinformatics 9(Suppl 1): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbani S, Lin YC, Parizot B, Fernandez A, Njo MF, Van de Peer Y, Beeckman T, Hilson P(2015) Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J Exp Bot 66: 5257–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP(2003) Legumes: Importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet L, Lan P, Li W, Mokkapati G, Schmidt W(2018) IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat Plants 4: 953–963 [DOI] [PubMed] [Google Scholar]
- Hanada K, Akiyama K, Sakurai T, Toyoda T, Shinozaki K, Shiu SH(2010) sORF finder: A program package to identify small open reading frames with high coding potential. Bioinformatics 26: 399–400 [DOI] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA(2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–5409 [DOI] [PubMed] [Google Scholar]
- Kent WJ.(2002) BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A, Mergaert P, Montiel J, Endre G, Kondorosi É(2018) Impact of plant peptides on symbiotic nodule development and functioning. Front Plant Sci 9: 1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL(2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S(2008) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Walker JC(2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ(2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175–1178 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y.(2014) Posttranslationally modified small-peptide signals in plants. Annu Rev Plant Biol 65: 385–413 [DOI] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I(2012) Small signaling peptides in Arabidopsis development: How cells communicate over a short distance. Plant Cell 24: 3198–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Okamoto M, Higuchi-Takeuchi M, Yoshizumi T, Yamaguchi Y, Fukao Y, Shimizu M, Ohashi C, Tanaka M, Matsui M, et al. (2018) AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc Natl Acad Sci USA 115: 5810–5815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M, Suzaki T(2018) A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun 9: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Mohd-Radzman NA, Corcilius L, Crossett B, Connolly A, Cordwell SJ, Ivanovici A, Taylor K, Williams J, Binos S, et al. (2018) Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome. Mol Cell Proteomics 17: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C(2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecrix Y, Staton SE, Sallet E, Lelandais-Brière C, Moreau S, Carrère S, Blein T, Jardinaud MF, Latrasse D, Zouine M, et al. (2018) Whole-genome landscape of Medicago truncatula symbiotic genes. Nat Plants 4: 1017–1025 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H(2011) SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Ropelewski AJ, Nicholas HB Jr., Deerfield DW II(2003) Mathematically complete nucleotide and protein sequence searching using search. Curr Protoc Bioinformatics 4: 3.10.1-3.10.12 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, Shinozaki K(2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556: 235–238 [DOI] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A(2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43(W1): W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine AJ, Kleinert A, Benedito VA(2017) Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci 256: 46–52 [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Clements J, Finn RD(2014) Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics 15: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Udvardi M(2009) Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol 12: 193–201 [DOI] [PubMed] [Google Scholar]
- Zhou P, Silverstein KA, Gao L, Walton JD, Nallu S, Guhlin J, Young ND(2013) Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinformatics 14: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]