Chlorophyll binding to the ONE-HELIX PROTEINS1/2 depends on heterodimer formation, mediated by conserved amino acids in their chlorophyll-binding motifs, and is essential for PSII biogenesis.
Abstract
Members of the light-harvesting complex protein family participate in multiple processes connected with light sensing, light absorption, and pigment binding within the thylakoid membrane. Amino acid residues of the light-harvesting chlorophyll a/b-binding proteins involved in pigment binding have been precisely identified through x-ray crystallography experiments. In vitro pigment-binding studies have been performed with LIGHT-HARVESTING-LIKE3 proteins, and the pigment-binding ability of cyanobacterial high-light-inducible proteins has been studied in detail. However, analysis of pigment binding by plant high-light-inducible protein homologs, called ONE-HELIX PROTEINS (OHPs), is lacking. Here, we report on successful in vitro reconstitution of Arabidopsis (Arabidopsis thaliana) OHPs with chlorophylls and carotenoids and show that pigment binding depends on the formation of OHP1/OHP2 heterodimers. Pigment-binding capacity was completely lost in each of the OHPs when residues of the light-harvesting complex chlorophyll-binding motif required for chlorophyll binding were mutated. Moreover, the mutated OHP variants failed to rescue the respective knockout (T-DNA insertion) mutants, indicating that pigment-binding ability is essential for OHP function in vivo. The scaffold protein HIGH CHLOROPHYLL FLUORESCENCE244 (HCF244) is tethered to the thylakoid membrane by the OHP heterodimer. We show that HCF244 stability depends on OHP heterodimer formation and introduce the concept of a functional unit consisting of OHP1, OHP2, and HCF244, in which each protein requires the others. Because of their pigment-binding capacity, we suggest that OHPs function in the delivery of pigments to the D1 subunit of PSII.
Plants possess a heterogenous family of light-harvesting-like (LIL) proteins and light-harvesting chlorophyll a/b-binding proteins (LHCPs). Within the light-harvesting complex (LHC) family, the well-characterized LHCPs are responsible for photosynthetic light harvesting and transfer of excitation energy to the photosystems as well as for energy dissipation processes collectively known as nonphotochemical quenching. The LHCPs form the outer antennae of PSI and PSII and appear in both monomeric and trimeric complexes. LHCPs contain three integral transmembrane helices. Two of these (helices 1 and 3) each contain a stretch of conserved amino acids, which together form a chlorophyll (Chl)-binding motif (Engelken et al., 2010). Similar Chl-binding motifs can also be found in the early-light-inducible proteins (ELIPs), which have three membrane helices in all, and in the PsbS subunit of PSII, which has four membrane helices. Other subsets of the LHC family comprise the two-helix proteins (stress-enhanced proteins [SEPs]) and one-helix proteins (OHPs), each of which contains only one helix with the Chl-binding motif (Engelken et al., 2010). The C-terminal segment of the plant FERROCHELATASE2 (FeCh2) isoform likewise resembles the Chl-binding motif. In contrast to the diversity of LHC and LIL protein topologies in plants, cyanobacteria only express proteins of the one-helix type (designated high-light-inducible proteins [Hlips] or small CAB-like proteins), which are considered to be the evolutionary ancestors of the entire plant LHC family (Engelken et al., 2010).
Initially, multiple functions were assigned to the cyanobacterial Hlips, ranging from the facilitation of photosystem assembly to the control of Chl biosynthesis (Komenda et al., 2012). Four different Hlip variants are encoded in the genome of Synechocystis PCC6803 (HliA–HliD). As in the plant FeCh2, the C-terminal region of the SynFeCh contains a Chl-binding motif, which is essential for dimerization but not for the catalytic activity of the enzyme (Sobotka et al., 2011). Interestingly, it was recently shown that the C termini of the homodimeric SynFeCh bind pigments in an energy-dissipating conformation (Pazderník et al., 2019).
During PSII biogenesis in cyanobacteria, the assembly of modules of the four PSII core subunits occurs in a sequential fashion (D1, D2, CP43, CP47) and ultimately leads to the formation of the PSII reaction center (Komenda et al., 2012). An HliC/HliD dimer first interacts with the D1 module and binds the assembly factor Ycf39 as well as the enzyme CHLOROPHYLL SYNTHASE (ChlG; Chidgey et al., 2014; Knoppová et al., 2014). It is therefore assumed that pigments are delivered to the nascent D1 precursor protein (pD1) via the HliC/HliD dimer. The binding of six Chl a molecules and two molecules of β-carotene (β-Car) to HliC/HliD has been confirmed, and the β-Car molecules are integrated in a twisted conformation, favoring energy dissipation (Staleva et al., 2015). These findings argue that the Hlip dimer acts primarily as a photoprotectant for the nascent D1 module and other early PSII assembly intermediates (Staleva et al., 2015; Llansola-Portoles et al., 2017). Similarly, an HliA/HliB dimer seems to be associated with the CP47 module (Boehm et al., 2012).
Recently, substantial progress has been made in the functional characterization of OHP1 and OHP2, the two OHPs found in plants. The initial analyses linked OHP2 to PSI (Andersson et al., 2003). But this hypothesis was mainly based on their apparent comigration in Suc-density gradients and is considered to be quite unlikely in light of more recent reports. One striking difference between cyanobacterial Hlips and plant OHPs is the observation that quadruple knockout strains of Synechocystis that lack all four Hlip genes are perfectly viable under normal light conditions (He et al., 2001), whereas single ohp T-DNA insertion mutants of Arabidopsis (Arabidopsis thaliana) are strongly developmentally impaired. For optimal growth, these lines have to be germinated on Murashige and Skoog (MS) medium supplemented with Suc (Beck et al., 2017). In both ohp mutant lines, the other OHP protein was partially (OHP2 in ohp1) or almost fully (OHP1 in ohp2) destabilized, and overexpression of the remaining variant in the mutant lines could not functionally compensate for depletion of the other (Beck et al., 2017). In both ohp1 and ohp2 knockout mutants, steady-state amounts of core subunits of PSI and PSII were strongly diminished, as were selected LHCPs (Beck et al., 2017; Myouga et al., 2018; Li et al., 2019). However, it is known that impairment of PSII biogenesis secondarily destabilizes PSI (Meurer et al., 1998; Armbruster et al., 2010).
This last point must be taken into consideration when ohp mutant phenotypes are evaluated, as PSII biogenesis is affected from the onset of germination in these mutants. Indeed, application of a virus-induced gene silencing (VIGS) approach to the OHP genes in 12-d-old Arabidopsis seedlings resulted in a specific reduction in amounts of PSII core subunits upon silencing of OHP2, while levels of PSI and the major LHCPs were unaltered (Hey and Grimm, 2018a). It was therefore proposed that the destabilization of PSI in the ohp mutants is a secondary effect. The low level of PSII in VIGS-OHP2 was attributable to a sharp fall in the rate of synthesis of D1, which is known to be the major bottleneck for further PSII assembly.
Interestingly, VIGS-OHP1 lines did not exhibit any macroscopic phenotype, even though a reduction in D1 synthesis could also be detected in these lines (Hey and Grimm, 2018a). However, since the two OHP proteins must interact with each other in order to perform their function, VIGS-OHP1 lines were more susceptible to elevated light intensities than VIGS-GFP control lines (Hey and Grimm, 2018b).
Moreover, the protein HIGH CHLOROPHYLL FLUORESCENCE244 (HCF244), the plant homolog of the cyanobacterial factor Ycf39, has been identified as an interaction partner of both OHPs (Hey and Grimm, 2018a; Myouga et al., 2018). HCF244 belongs to the atypical short-chain dehydrogenases, but its exact molecular function remains unclear (Link et al., 2012). The stability of HCF244 is completely dependent on the presence of OHP2, and HCF244 is attached to the stroma side of the thylakoid membrane via the OHP1/OHP2 dimer (Hey and Grimm, 2018a). OHP1 also requires OHP2 for its own stabilization, which is further increased by HCF244. Therefore, HCF244 presumably acts as a scaffold, tethering the constituents of the OHP heterodimer together (Hey and Grimm, 2018a). The intact heterotrimeric OHP1-OHP2-HCF244 complex is essential for D1 synthesis, and the OHP1/OHP2 dimer itself has been proposed to deliver pigments to pD1 (Hey and Grimm, 2018a; Myouga et al., 2018; Li et al., 2019). However, unlike the situation in cyanobacteria, no interaction of plant CHLG with OHPs has yet been detected (Hey and Grimm, 2018a; Proctor et al., 2018).
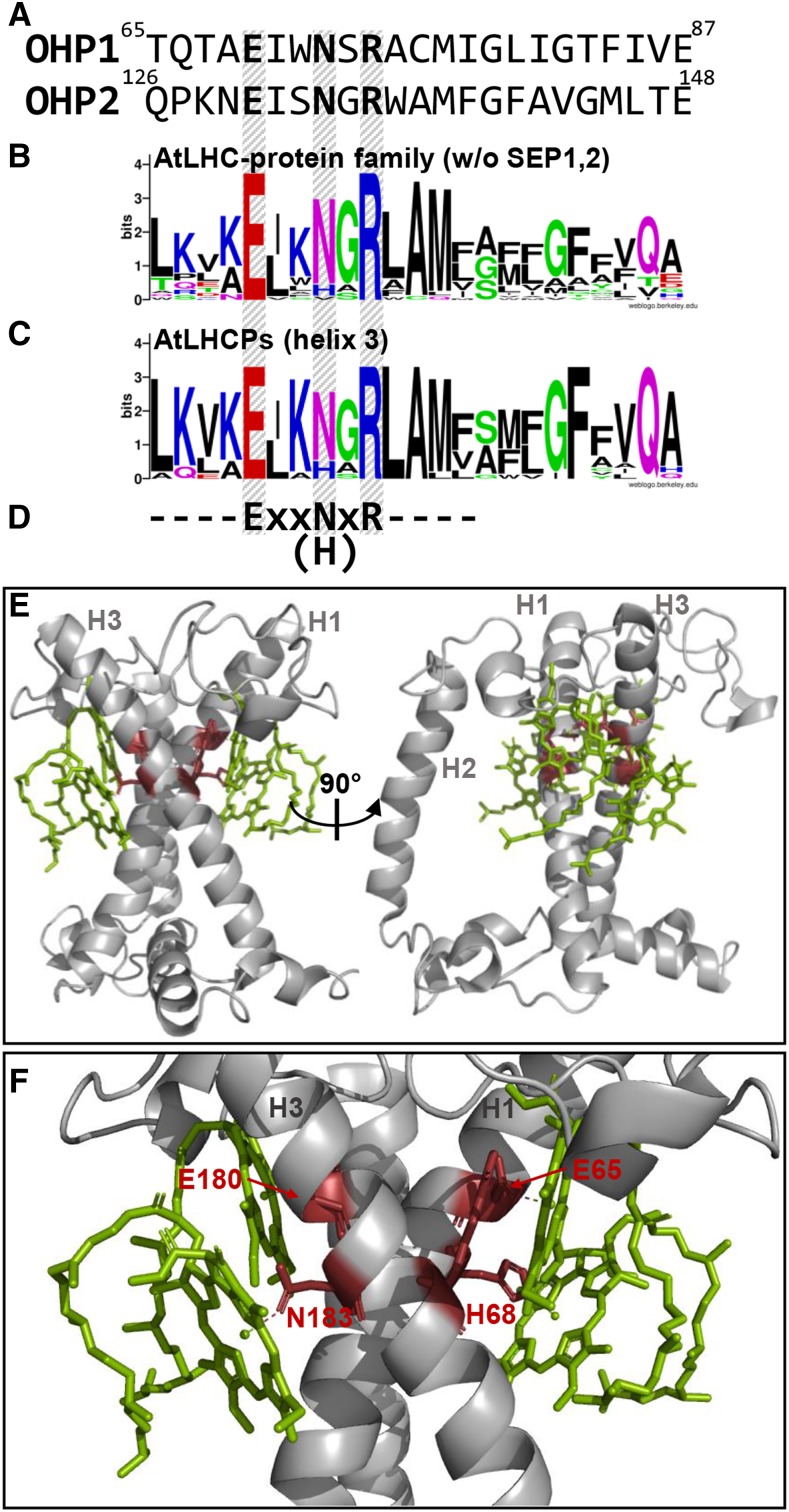
In principle, ELIPs as well as the LIL3 isoforms have been shown to possess the capacity for Chl binding (Adamska et al., 1999, 2001; Mork-Jansson et al., 2015a, 2015b; Hey et al., 2017; Mork-Jansson and Eichacker, 2018, 2019). X-ray crystallographic data for plant LHCPs provide detailed insights into the molecular organization of pigment binding mediated by the transmembrane Chl-binding motif in LHCII. Comparison of the Chl-binding motifs of all members of the LHC family identified a short stretch of highly conserved residues at the beginning of this motif (Fig. 1; Engelken et al., 2010). In this ExxN/HxR sequence, the E and R residues are conserved without exception (Fig. 1B), whereas in LHCPs, the N/H position may be occupied by either amino acid (usually N in one of the LHC helices and H in the other). In PsbS and SEP1/2, the motif is reduced to ExxxxR, but in ELIPs, LIL3s, and OHPs, the ExxNxR sequence is once again found (Fig. 1A; Engelken et al., 2010). Helices 1 and 3 of LHCII show an X-shaped arrangement in the thylakoid membrane, and the E as well as the N/H residues of both helices are in direct contact with the Mg atoms of four of the eight Chl a molecules bound by each LHCII monomer (Fig. 1, E and F; Liu et al., 2004). In addition, at least one of the R residues in LHCII makes a direct contact with one of the Chl a molecules bound by E/(N/H; Liu et al., 2004).
Figure 1.
The Chl-binding motif of the LHC protein family. A, Primary structure of the Arabidopsis OHP1 and OHP2 proteins within the region encompassing the Chl-binding motif. OHP1, Uniprot identifier O81208, amino acids 65 to 87; OHP2, Q9FEC1, amino acids 126 to 148. The conserved amino acid residues are printed in boldface. B, Conserved amino acids within the LHC protein family of Arabidopsis (apart from SEP1,2). The degree of conservation at the different positions was analyzed with WebLogo (Crooks et al., 2004). C, Conserved amino acids within the third helix of the Arabidopsis LHCPs. D, Conserved amino acids at the beginning of the helix as referenced in the text. E, Crystal structure of LHCII monomers from spinach (Protein Data Bank entry 1rwt; Liu et al., 2004). The four Chl a molecules bound by the conserved amino acids shown in D are depicted in green. The three helices are marked H1 to H3. This image was prepared with PyMol (Schrodinger, 2010). F, Closeup view of the organization of Chl binding around the conserved amino acids. The conserved residues are shown in red and are named according to their positions in the amino acid sequence of LHCII from spinach.
With regard to the other members of the LHC family, the N residue in LIL3 has been suggested to be essential for Chl binding (Mork-Jansson and Eichacker, 2019), because in vitro reconstitution assays were ineffective when this particular residue was mutated. In addition, Li et al. (2019) undertook reciprocal E/N exchanges, as well as mutating R alone in both OHPs, and successfully used these constructs for ohp complementation. These complemented seedlings phenotypically resembled wild-type seedlings but exhibited lower Fv/Fm ratios (for maximum quantum efficiency of PSII in the dark-adapted state) and decreased contents of PSII subunits. However, so far, the effects of the simultaneous mutation of all three conserved residues in this peptide motif together have not been reported.
We show here that replacement of all three conserved amino acids E-N-R in the Chl-binding motif of both OHPs by Ala (A) does not impair heterodimer formation but prevents pigment binding. Analysis of the ability of these mutants to functionally complement either of the ohp insertion mutants showed that the OHP1-AAA variant had completely lost its biological function, whereas OHP2-AAA could partially complement ohp2. Our data ultimately strengthen the idea that OHPs deliver Chl (and potentially β-Car) for at least nascent pD1 subunits. In addition, quenching of excitation energy through the carotenoids is postulated to occur in assembled OHP1/OHP2 heterodimers.
RESULTS
OHPs Can Be Reconstituted with Pigments in Vitro
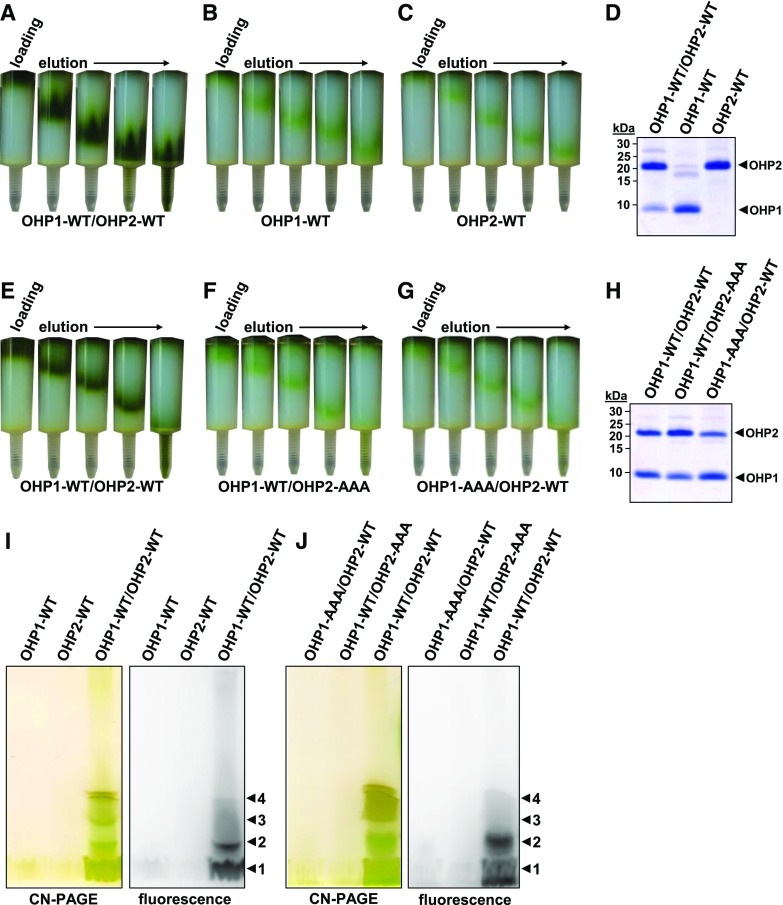
We previously reported that the formation of OHP1/OHP2 heterodimers is required for the stabilization of OHP1 as well as for the provision of adequate amounts of functional D1 in planta (Hey and Grimm, 2018a, 2018b). As LHCPs bind a large portion of their pigments by means of the two-helix pair formed by transmembrane domains 1 and 3 of these proteins (Liu et al., 2004), heterodimerization of OHP1 and OHP2 is also assumed to be an essential prerequisite for pigment binding as such. To test this hypothesis, we performed in vitro reconstitution experiments with recombinant OHPs. A protocol that was previously used for the reconstitution of pigmented LHCB (Natali et al., 2014) was adapted, and only minor changes were necessary to obtain successful reconstitution of OHPs (for details, see “Materials and Methods”). Reconstitution is followed by a purification step based on His-tag affinity chromatography on 1-mL HisTrap HP columns. After loading the column with the reconstitution mixture and extensive washing to remove unbound pigments, the green pigment-containing protein fraction became visible in the upper part of the column. Subsequent washing with 500 mm imidazole initially visualized a mobile green band consisting of pigment-protein complexes as it passed through the column before finally eluting as a pigmented fraction (Fig. 2A). Both OHP proteins were used in an equimolar ratio during the OHP1-WT/OHP2-WT (where WT represents the wild type) reconstitution experiments. Similarly, the same protein amounts were used for reconstitution assays with each individual OHP species. A dark-green protein fraction was eluted from equimolar mixtures of OHP1-WT and OHP2-WT, indicating successful reconstitution with pigments (Fig. 2A). In contrast, only a faint green protein band was detectable in the column when the individual OHP-WT isoforms were separately assayed (Fig. 2, B and C). Fractionation of equal volumes of the eluates by Tricine-SDS-PAGE revealed that all eluates contained comparable amounts of each of the respective OHP variants (Fig. 2D; Supplemental Fig. S1, top), indicating that the pigment-binding capacity of each individual OHP was drastically lower than that of the OHP1-WT/OHP2-WT combination.
Figure 2.
Reconstitution of recombinant OHPs with pigments. A to C and E to G, His-tag affinity chromatography of OHP-pigment reconstitution assays. HisTrap HP columns (1 mL; GE) are depicted following loading and washing (loading) and during elution of the bound complexes (elution). Equal molar concentrations of proteins and pigments were used for each reconstitution assay, and the intensity of the pigmented band in the column directly reflects the reconstitution efficiency. All of the pigmented eluate was collected. D and H, Tricine-SDS-PAGE analysis of the affinity chromatography eluates. Equal volumes of the eluates were fractionated on Tricine-SDS gels. The OHP bands are marked. I and J, Clear Native (CN)-PAGE analysis of the eluates. Equal volumes of the eluates were fractionated on CN gels according to Järvi et al. (2011), with 0.3% (w/v) deoxycholate in the sample and 0.05% (w/v) deoxycholate + 0.02% (w/v) DDM in the cathode buffer. The four colored bands are numbered (arrowheads 1–4).
Based on the relatively weak mutant phenotypes of lines expressing OHP substitution mutants in which one or two of the conserved residues in the Chl-binding motif had been replaced (Fig. 1; Li et al., 2019), we substituted Ala for all three conserved amino acids and analyzed the effects of the triple substitution on the pigment-binding capacity of the mutant OHPs. The OHP1-AAA and OHP2-AAA variants were expressed in Escherichia coli and used for in vitro reconstitution assays. Using the OHP1-WT/OHP2-WT combination as a positive control, successful pigment binding was consistently displayed by the formation of a dark-green band (Fig. 2E), but reconstitution experiments with both the OHP1-AAA/OHP2-WT pair and the OHP1-WT/OHP2-AAA combination resulted in faintly pigmented eluates (Fig. 2, F and G). Again, in all cases, the eluates contained equal amounts of both OHP variants (Fig. 2H; Supplemental Fig. S1).
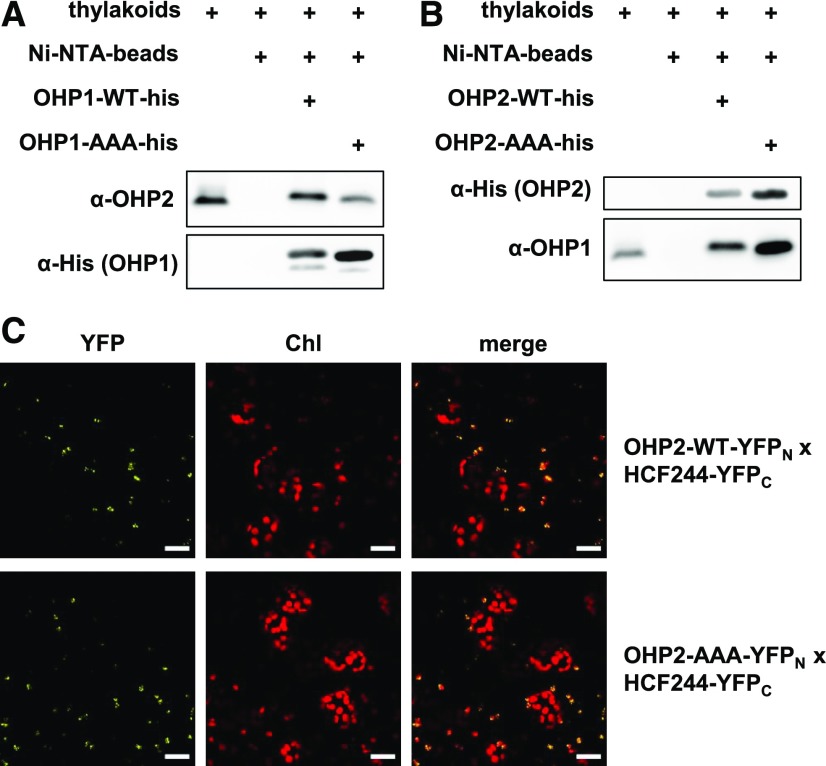
Recently it was reported that replacement of any one of the three conserved amino acids in the LIL3 protein by an Ala residue inhibited dimerization of the mutated variant with the wild-type protein (Mork-Jansson and Eichacker, 2019). To test whether the lack of heterodimer formation was responsible for the weak pigment-binding ability observed in the OHP-WT/AAA reconstitution assays, we performed pulldown experiments. Purified recombinant His-tagged OHP1 or OHP2 (100 µg) was bound to Ni-NTA agarose beads and incubated with wild-type Arabidopsis thylakoids solubilized with 0.5% (w/v) β-dodecyl maltoside (DDM). After intensive washing, bound proteins were eluted with 250 mm imidazole and fractionated by Tricine-SDS-PAGE. Both His-tagged OHP1-WT and OHP1-AAA were capable of precipitating OHP2 from thylakoids (Fig. 3). Similarly, His-tagged OHP2-WT and OHP2-AAA interacted with OHP1 from thylakoids (Fig. 3B; Supplemental Fig. S1, bottom). It is possible that the mutations slightly mitigate the binding between both heterogenous OHP partners. But it can be concluded that OHP1 and OHP2 interact in spite of simultaneous mutation of all three conserved residues in one of the proteins in each heterodimeric complex. Accordingly, OHP1-OHP2 heterodimer formation can be expected to occur in reconstitution assays containing a combination of OHP-WT and OHP-AAA.
Figure 3.
Interaction studies with the OHP-AAA variants. A and B, His-tag pulldown assays performed with recombinant OHP proteins. Purified proteins (100 µg) were allowed to bind to Ni-NTA agarose beads and incubated with solubilized Arabidopsis thylakoids (100 µL of Chl). After incubation and thorough washing of the column, the proteins bound to the OHP bait were eluted with 250 mm imidazole and fractionated by Tricine-SDS-PAGE. Recombinant and endogenous OHPs were detected with His tag-specific and OHP-specific antibodies, respectively. C, Bimolecular fluorescence complementation (BiFC) assay for verification of the interaction between OHP2-AAA and HCF244. Proteins fused to the N- or C-terminal halves of split-YFP (YFPN/C) were expressed in N. benthamiana leaves, and fluorescence was detected with an LSM800 confocal microscope (Zeiss). Bars = 20 µm.
Equal volumes of the reconstitution eluates were also fractionated by nondenaturing CN-PAGE. As expected, the OHP1-WT/OHP2-WT-containing eluate exhibited considerable amounts of pigmented protein bands on CN gels (Fig. 2, I and J). Usually, four distinct bands with different electrophoretic mobilities were visible in the low-Mr range on the gels (Fig. 2, I and J). In contrast, fractionation of eluted proteins on CN gels after reconstitution assays with the single OHPs or the OHP-WT/AAA combinations only revealed the band with the lowest Mr (band 1), which likely corresponded to free pigments, although the presence of most likely monomeric OHP proteins cannot be excluded. Interestingly, visualization of the Chl fluorescence in the OHP1-WT/OHP2-WT eluates with a PAM imager showed that only the protein bands with the lowest Mr (bands 1 and 2) were fluorescent, whereas the protein bands with higher Mr (bands 3 and 4) did not exhibit Chl fluorescence (Fig. 2, I and J). In these complexes, the fluorescence seems to be quenched, possibly due to the formation of aggregates of pigment-protein complexes of higher molarity.
OHP1/OHP2 Heterodimers Contain a Specific Combination of Pigments
We extracted the pigments from the reconstitution eluates with alkaline acetone and quantified them by HPLC. For the cyanobacterial HliC/HliD dimer, a pigment content of six Chl a and two β-Car molecules has been reported (Staleva et al., 2015). An absolute quantification (i.e. a molar quantification of the pigments per microgram of protein) was not possible in our experiment. Due to the His-tag purification step following the reconstitution procedure, a significant amount of pigment-free, nonreconstituted proteins was also expected to be present in the eluate. Thus, it should be noted that the lowest full number for all pigment molecules was obtained by normalization to six Chl a molecules.
The OHP1-WT/OHP2-WT control assays of the different reconstitution experiments exhibited comparable pigment compositions (Table 1). Besides the six Chl a molecules, one molecule each of Chl b, β-Car, and lutein was bound. Compared with the total pigment mixture used for the reconstitution, this implies a significant enrichment for β-Car as well as a depletion of Chl b. In addition, the carotenoids violaxanthin and neoxanthin, which were present in significant amounts in the pigment mixture, were not detectable in the OHP1-WT/OHP2-WT eluates (Table 1). In contrast to OHP1-WT/OHP2-WT, both OHP-WT/AAA eluates exhibited a pigment composition that was very similar to that of the total pigment mixture, with the exception of an increased amount of lutein (Table 1), indicating the presence of unspecifically bound pigments. In addition, it should be noted that the pigment contents of the single OHP-WT reconstitution eluates were always extremely low, which hampered their precise quantification. The low pigment content suggests that the protein species formed in these reconstitution assays had a very low capacity for unspecific pigment binding.
Table 1. Relative pigment contents of the OHP reconstitution eluates.
Pigments were extracted with alkaline acetone from the eluates of His-tag affinity chromatography assays and quantified by HPLC. Values were normalized to six molecules of Chl a. n.d., Not detectable; –, no informaiton available.
| Experiment | Pigment | |||||
|---|---|---|---|---|---|---|
| Chl a | Chl b | β-Car | Lutein | Neoxanthin | Violaxanthin | |
| OHP1-WT/OHP2-WT (Experiment 1) | 6 | 1.0 | 0.9 | 1.0 | n.d. | n.d. |
| OHP1-WT/OHP2-WT (Experiment 2) | 6 | 0.8 | 0.8 | 1.3 | n.d. | n.d. |
| OHP1-WT/OHP2-WT (Experiment 3) | 6 | 1.2 | 0.7 | 1.1 | n.d. | n.d. |
| Mean ± sd | 6 | 1.0 ± 0.2 | 0.8 ± 0.1 | 1.1 ± 0.1 | n.d. | n.d. |
| OHP1-WT (Experiment 2) | 6 | 0.8 | 0.4 | 1.1 | 0.6 | 0.8 |
| OHP2-WT | 6 | 1.6 | 0.4 | 1.5 | 1.0 | 1.0 |
| OHP1-AAA/OHP2-WT (Experiment 3) | 6 | 2.5 | 0.2 | 0.3 | 0.2 | 0.1 |
| OHP1-WT/OHP2-AAA | 6 | 2.0 | 0.2 | 0.4 | 0.4 | 0.4 |
| Pigment solution | 6 | 2.1 | 0.2 | 1.0 | 0.3 | 1.1 |
| HliC/HliD (Staleva et al., 2015) | 6 | – | 2 | – | – | – |
| HliC/HliC (Shukla et al., 2018) | 4 | – | 2 | – | – | – |
To summarize, the pigment composition of the OHP1-WT/OHP2-WT dimer resembles that of HliC/HliD (Staleva et al., 2015): the cyanobacterial and plant homologs contained two carotenoids per six molecules of Chl a. Whereas two β-Car molecules were bound to HliC/HliD, OHP1/OHP2 contained one molecule each of β-Car and lutein (Table 1). A natural variability in the carotenoid composition of OHPs/Hlips cannot yet be excluded, and it currently remains open whether the binding of two different carotenoids can be confirmed in planta and, if so, whether both are required for the in vivo function of the OHP1/OHP2 dimer. In addition, it is currently unclear whether one molecule of Chl b per six molecules of Chl a in OHP1/OHP2 is specifically bound to the heterodimer in planta and, if so, whether this stoichiometry is also of functional relevance.
Chl b is crucial for the stability of LHCII (Murray and Kohorn, 1991), and both Chl a and Chl b (together with carotenoids) are required for the in vitro reconstitution of LHCII with pigments (Plumley and Schmidt, 1987). Therefore, we explored the dependence of successful OHP reconstitution on Chl a and b. Purified Chl a and b were obtained from Sigma, and reconstitution assays were performed with 250 µg of Chl a + 94 µg of Chl b (+160 µg of total spinach [Spinacia oleracea] carotenoids) or 250 µg of each single Chl alone (for details, see “Materials and Methods”). Whereas reconstitution was possible with Chl a alone, no reconstituted protein complexes could be obtained when Chl b was used on its own (Supplemental Fig. S2), as deduced from the fact that reconstitution assays performed with Chl b did not result in the detection of pigmented OHP complexes on CN-PAGE gels.
Mutation of the Conserved Amino Acids in the Pigment-Binding Sites Prevents Functional Complementation of ohp Mutants
Despite OHP heterodimer formation with one mutant OHP isoform of the three conserved amino acids in the Chl-binding motifs, pigment binding in vitro was completely abolished. It was therefore tempting to explore the physiological activity of the mutated proteins in ohp complementation studies. Li et al. (2019) had already exchanged one or two residues in the Chl-binding motif, but they reported only a minor impact on growth of the corresponding transgenic lines.
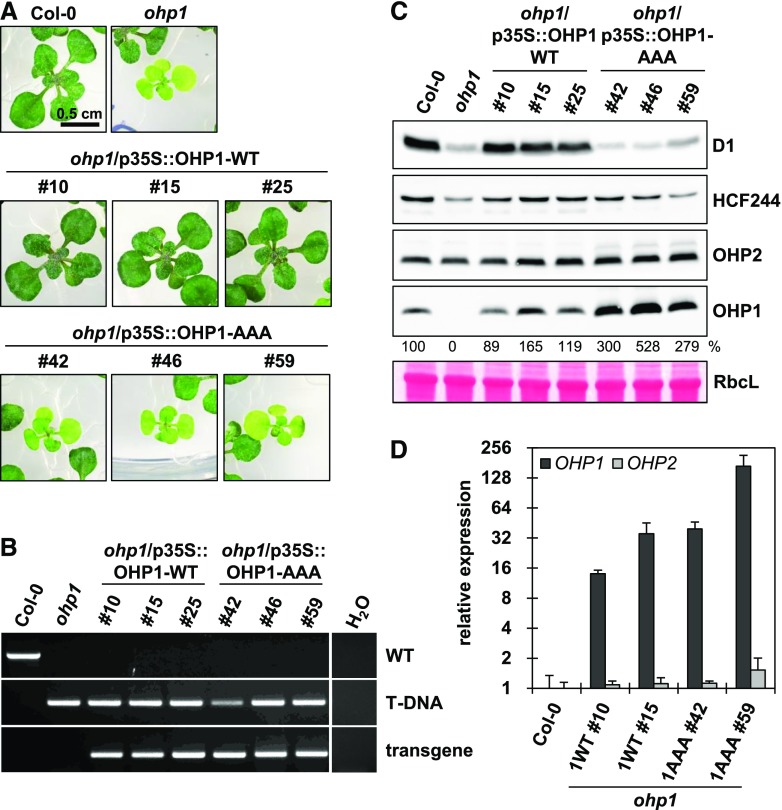
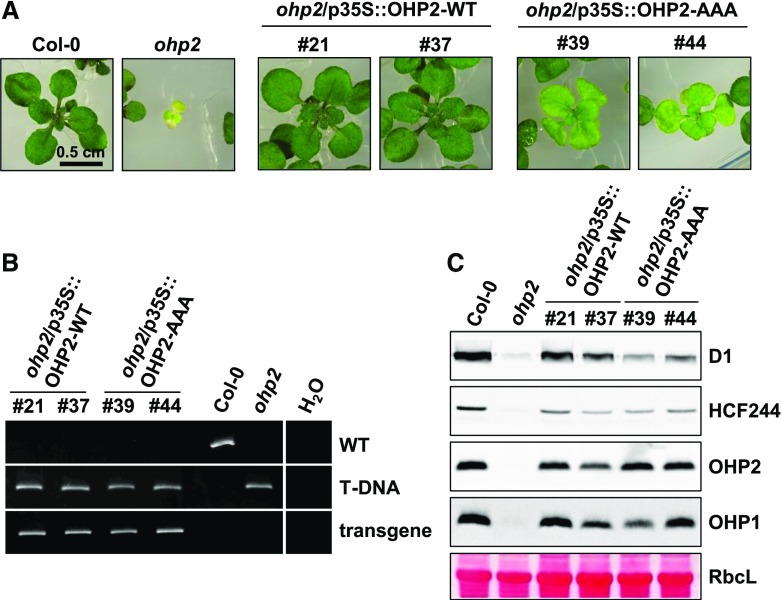
The OHP-WT- and OHP-AAA-encoding sequences were expressed under the control of the Cauliflower mosaic virus 35S promoter in the ohp1 and ohp2 mutant backgrounds. For optimal growth of the severely compromised ohp mutants, all lines were germinated on MS medium supplemented with 2% (w/v) Suc, and seedlings were incubated under continuous illumination (100 µmol photons s−1 m−2). In accordance with previous reports, even under these conditions the homozygous progeny of the ohp1 mutant were pale green and retarded in growth (Fig. 4A). Expression of the OHP1-WT cDNA in ohp1 fully complemented the mutant phenotype. In contrast, seedlings expressing the OHP1-AAA construct phenotypically resembled the ohp1 mutant (Fig. 4A). It is worth mentioning here that these plants, like ohp1, produced flowers when cultivated on Suc-containing MS medium for longer times but their seeds did not germinate. Genotyping by PCR confirmed that the lines were homozygous for the T-DNA insertion in the OHP1 gene and for the presence of the OHP1-AAA transgene, respectively (Fig. 4B).
Figure 4.
Characterization of OHP1-AAA complementation lines. A, Phenotypes of 3-week-old plants germinated on MS medium containing 2% Suc and incubated under continuous illumination (100 µmol photons s−1 m−2). Three independent lines for each construct are shown. B, Genotyping PCR for verification of homozygosity of the transgenic lines. C, Protein analysis by Tricine-SDS-PAGE and immunoblotting. Equal amounts of total leaf proteins were fractionated on Tricine-SDS gels. The large subunit of Rubisco (RbcL) served as the loading control. The band intensities of OHP1 were analyzed by densitometry and are presented as percentages of the corresponding wild-type values. D, RT-qPCR analysis of OHP gene expression in two selected complementation lines. The data were analyzed by the ΔΔCt method (Pfaffl, 2001) and normalized to the expression in the wild type. Error bars represent sd.
Protein analysis revealed that the OHP2 content remained wild-type-like in ohp1. However, in the OHP1-AAA lines, the anti-OHP2 antibody detected an additional immunoreactive band with an apparent Mr that was slightly higher than that of the mature OHP2. This protein most likely represents precursor OHP2 (i.e. it retains the chloroplast transit peptide; Fig. 4C), although posttranslational modification of full-length OHP2 cannot be excluded. Interestingly, whereas the OHP1 content was approximately wild-type-like or slightly increased in the OHP1-WT-expressing lines, large amounts of the OHP1-AAA protein (up to five times the wild-type amount) accumulated in the corresponding lines (Fig. 4C). This is surprising, as we had never observed any overaccumulation of OHP1 before, and we had assumed that OHP1 is stabilized exclusively by binding to OHP2 (Hey and Grimm, 2018a). It is important to note that the difference in protein accumulation cannot be explained by any drastic increase in mRNA level in the transgenic lines. Quantification of the OHP1 mRNA by reverse transcription quantitative PCR (RT-qPCR) indeed revealed an approximately 32-fold increase in OHP1-WT line 15 and in OHP1-AAA line 42 (Fig. 4D). However, OHP1-WT line 15 only showed a 50% increase in protein accumulation, whereas OHP1-AAA line 42 exhibited a threefold higher content of OHP1 compared with the wild type. In addition, OHP1-AAA line 59 accumulated more than 128 times as much mRNA as the amount seen in the wild type, but it still contained no more than three wild-type equivalents of the cognate protein (Fig. 4, C and D). Clearly therefore, levels of the OHP1-AAA mRNA and the corresponding protein are not directly correlated in the different complementation lines. The OHP2 mRNA remained wild-type-like in all analyzed lines.
Besides OHP1 and OHP2, we also checked the HCF244 content in the transgenic lines. The ohp1 mutant contained decreased amounts of HCF244, which was recovered in both the OHP1-WT- and OHP1-AAA-expressing complementation lines (Fig. 4C), indicating that OHP2 or the pigment-free OHP1-AAA/OHP2-WT dimer is sufficient for stabilization of HCF244. As expected from the macroscopic phenotype of the OHP1-AAA-expressing ohp1 lines, their PSII content (as indicated by levels of D1) resembled that of ohp1 (Fig. 4C). Therefore, despite formation of the OHP1-AAA/OHP2-WT heterodimer, the transgenic lines showed similarly reduced PSII contents to that seen in the ohp1 mutant (i.e. in which only OHP2 is expressed). Thus, the OHP1-AAA lines confirmed that functional OHP2 alone is not able to provide adequate support for PSII biogenesis (Hey and Grimm, 2018a, 2018b).
The ohp2 mutant was complemented by OHP2-WT and OHP2-AAA constructs expressed under the control of the 35S promoter (Fig. 5). Homozygous ohp2 seedlings grown under conditions identical to those used for the ohp1 mutant were more severely impaired than the latter (Fig. 5A). This is intuitively understandable, as the loss of OHP2 leads to the subsequent destabilization of OHP1 (Hey and Grimm, 2018a). We recently reported that OHP1 has an essential role in early developmental stages, whereas its loss can be readily tolerated once this phase has been completed (Hey and Grimm, 2018a).
Figure 5.
Characterization of OHP2-AAA complementation lines. A, Phenotypes of 3-week-old plants grown as described in the legend to Figure 4. Two independent lines for each construct are shown. B, Genotyping PCR for verification of homozygosity of the transgenic lines. C, Protein analysis by Tricine-SDS-PAGE and immunoblotting. Equal amounts of total leaf proteins were fractionated on Tricine-SDS gels. The large subunit of Rubisco (RbcL) served as the loading control.
Expression of OHP2-WT complemented the ohp2 mutant phenotype. Surprisingly, homozygous ohp2 seedlings expressing OHP2-AAA developed significantly further and reached larger sizes than the original ohp2 mutant (Fig. 5A). Nevertheless, the seedlings of OHP2-AAA lines produced fewer leaves than the wild type or OHP2-WT-expressing ohp2 complementation lines and showed weaker pigmentation. PCR-based genotyping verified that the lines were homozygous for the T-DNA in the OHP2 gene and confirmed the presence of the transgene (Fig. 5B).
The protein analysis revealed wild-type-like levels of OHP2 as well as wild-type-like or slightly reduced OHP1 contents in all of the complementation lines (Fig. 5C). In line with previous results, the absence of OHP2 correlated with the complete loss of both OHP1 and HCF244 (Hey and Grimm, 2018a). This again highlights the crucial impact of OHP2 on the stability of its binding partners. In all other complementation lines, equal amounts of HCF244 accumulated, albeit to slightly lower levels than in the wild type (Fig. 5C). In contrast to the OHP1-AAA lines and ohp1, more D1 accumulated in the presence of OHP2-AAA than in the complete absence of OHP2 (Fig. 5C). Hence, this macroscopically discernible partial complementation of the ohp2 mutant by OHP2-AAA is correlated with modifications at the molecular level. It should be noted that OHP2-AAA represents a nonfunctional protein in terms of pigment binding, which nevertheless enables physical interaction with and stability of OHP1 and HCF244. In contrast to the rapid loss of OHP1 in ohp2 plants, OHP2-AAA has a beneficial impact on OHP1 stability in the same genetic background.
Mutation of the Chl-binding motif in LIL3 perturbs formation of the LIL3-CHLP complex (Takahashi et al., 2014). It has been suggested that the interaction of LIL3 with CHLP or POR (Tanaka et al., 2010; Hey et al., 2017) resembles the formation of the OHP-HCF244 complex (Hey and Grimm, 2018a). However, as HCF244 was found to be stable in the OHP2-AAA lines, we propose that the Chl-binding motif of OHP2 was not essential for its interaction with HCF244. BiFC experiments were performed in Nicotiana benthamiana leaves (Fig. 3C) to verify this idea. The simultaneous expression of OHP2-WT and HCF244 as well as of OHP2-AAA and HCF244 gave rise to chloroplast-localized yellow fluorescence (Fig. 3C). We therefore conclude that mutations in the Chl-binding site of OHP2 do not hamper its ability to interact with HCF244.
The C-Terminal Region of OHP2 Is Essential for Its Function
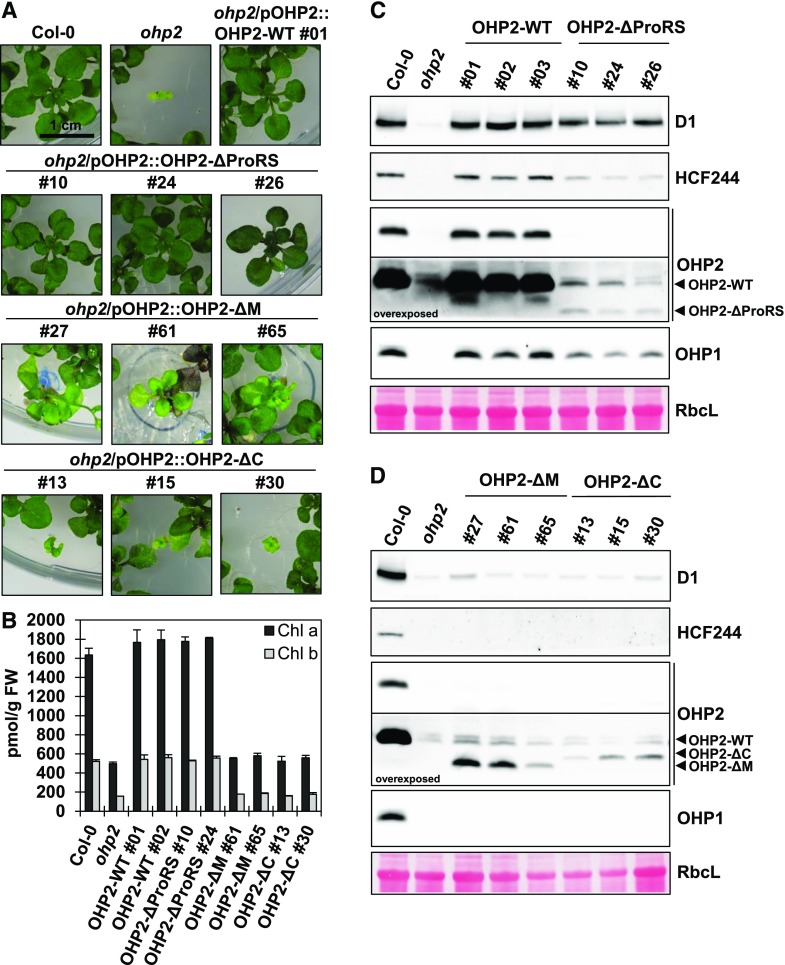
We previously reported that HCF244 specifically interacts with the middle (M) portion (amino acids 82–129) of the OHP2 protein, which is located between the N-terminal Pro-rich sequence (ProRS; amino acids 49–81) and the C-terminal transmembrane helix (TMH) harboring the Chl-binding motif (C; amino acids 130–172; Hey and Grimm, 2018a). To further characterize the three parts of OHP2, we tested truncated OHP2 variants (ΔProRS, ΔM, and ΔC; Hey and Grimm, 2018a) for their ability to complement the ohp2 mutant. The corresponding OHP2 cDNA sequences were expressed under the control of the endogenous OHP2 promoter in the ohp2 mutant background. Genotyping by PCR confirmed homozygosity for the T-DNA in the OHP2 gene, as well as the presence of the transgene, in all lines (Supplemental Fig. S3A). Seedlings were grown on Suc-containing MS medium (Fig. 6A). As expected, control expression of OHP2-WT under the control of its own promoter fully complemented the ohp2 phenotype. Surprisingly, the OHP2-ΔProRS lines also showed full phenotypic complementation (Fig. 6A). However, when the M segment was deleted from OHP2 (OHP2-ΔM), only partial complementation was achieved and loss of the OHP2 C terminus (OHP2-ΔC) completely abolished complementation of ohp2 (Fig. 6A). The different properties of the complementation lines were also reflected in their Chl contents. Whereas OHP2-WT and OHP2-ΔProRS exhibited wild-type-like amounts of Chl a/b, the Chl content of the ΔM and ΔC lines did not differ from the strongly reduced content observed in ohp2 (Fig. 6B). Therefore, partial macroscopic complementation by the ΔM lines does not reflect any significant increase in Chl levels.
Figure 6.
Complementation of ohp2 with truncated OHP2 variants. A, Phenotypes of 3-week-old plants grown as described in the legend to Figure 4. One line for the OHP2-WT construct and three independent lines for each of the deletion constructs (ΔProRS, ΔM, and ΔC) are shown. B, Quantification of the Chl content in two representative complementation lines. Pigments were extracted using alkaline acetone, separated, and quantified by HPLC using pure standards for comparison. FW, Fresh weight. Error bars represent sd. C and D, Protein analysis by Tricine-SDS-PAGE and immunoblotting. Equal amounts of total leaf proteins were fractionated on Tricine-SDS gels. The large subunit of Rubisco (RbcL) is shown as the loading control. The signals corresponding to the OHP2-WT construct and the truncated OHP2 variants are indicated.
Analysis of the contents of OHPs, HCF244, and D1 in the complementation lines revealed that surprisingly small amounts of the OHP2-ΔProRS, OHP2-ΔM, and OHP2-ΔC protein variants could be detected by immunoblotting (Fig. 6, C and D). As levels of the shortened OHP2 transcripts in the different lines were normally in the range of 0.3- to 8-fold that of the full-length OHP2 mRNA in the wild type (Supplemental Fig. S3B), we think that our antibody failed to efficiently recognize the different truncated OHP2 variants. However, because the OHP2-ΔProRS lines contained at least 50% of the wild-type contents of D1 and OHP1 (Fig. 6C), a similar amount of the OHP2-ΔProRS variant was assumed to be present. The OHP2-ΔM lines failed to accumulate either HCF244 or OHP1. Whereas the lack of HCF244 is likely to be a direct consequence of the loss of its cognate interaction site in the truncated OHP2, the lack of OHP1 confirms that HCF244 is necessary for the stabilization of OHP1 (Fig. 6D). Finally, the OHP2-ΔC variant lacking the TMH presumably cannot be integrated into the thylakoid membrane. As membrane integration is most probably crucial for the stabilization of OHP2, this would account for the failure of OHP2-ΔC to accumulate at all. Similarly, their dependence on OHP2 also explains why HCF244 and OHP1 could not accumulate in the OHP2-ΔC lines (Fig. 6D).
Both HCF244 and OHPs Are Essential for PSII Biogenesis
LIL3 is crucial for the stability of CHLP as well as POR, and lil3.1/lil3.2 mutant lines show a strong mutant phenotype (Tanaka et al., 2010). However, overexpression of a CHLP variant harboring a transmembrane domain from either LIL3 or the thylakoid-bound ascorbate peroxidase (tAPX) in the lil3.1/lil3.2 mutant background led to complementation of that strong mutant phenotype (Takahashi et al., 2014). This then prompted the proposal that LIL3 not only acts as a scaffold for CHLP but also tethers the protein to the thylakoid membrane and confers its stability.
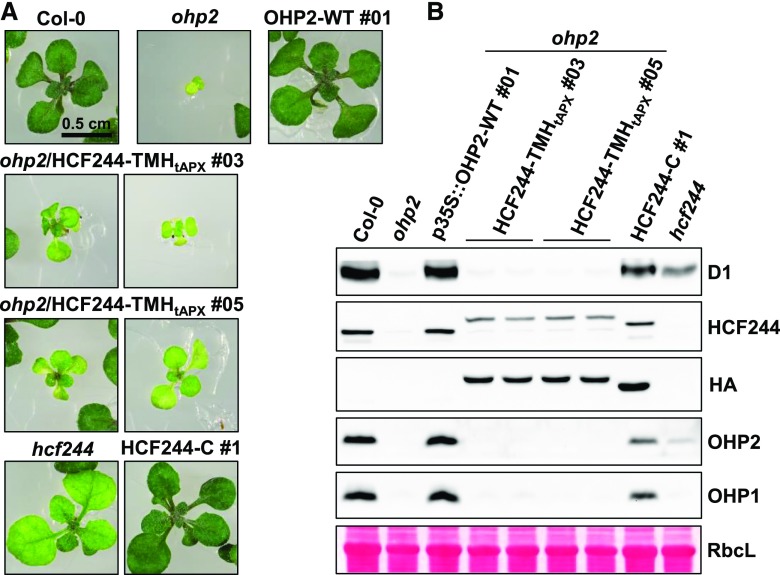
We therefore asked whether a similar strategy might restore the interaction between OHPs and HCF244. To test the idea, we used essentially the same approach as that described by Takahashi et al. (2014). The TMH of the tAPX was fused to the C terminus of HCF244 and the fusion protein was expressed in the ohp2 mutant background (ohp2/HCF244-TMHtAPX lines). In comparison with the ohp2 mutant line, the ohp2/HCF244-TMHtAPX lines showed a weak partial complementation phenotype in the seedlings. The fusion protein HCF244-TMHtAPX accumulated to at least 50% of the wild-type level in the absence of OHP2 (Fig. 7), thus corroborating the idea that formation of the OHP-HCF244 complex is ensured by OHP2-mediated membrane tethering and stabilization of HCF244.
Figure 7.
Complementation of ohp2 with a membrane-bound HCF244 variant. A, Phenotypes of 3-week-old plants germinated on MS medium + 2% Suc incubated under continuous illumination (100 µmol photons s−1 m−2). Two representative seedlings of both ohp2/HCF244-TMHtAPX lines are shown. The hcf244 mutant as well as an HCF244 complementation line (HCF244-C) are shown for comparison. B, Protein analysis by Tricine-SDS-PAGE and immunoblotting. Equal amounts of total leaf proteins were fractionated on Tricine-SDS gels. The large subunit of Rubisco (RbcL) is shown as the loading control. Two independent protein samples for each of the two ohp2/HCF244-TMHtAPX lines were analyzed.
However, this partial complementation did not mitigate the loss of D1 or OHP1, neither of which could be detected (Fig. 7B), and also failed to increase the Chl content (Fig. 7A). In addition, the lack of OHP1 in the ohp2/HCF244-TMHtAPX lines supports the idea that the presence of both OHP2 and HCF244 is a prerequisite for the stabilization of OHP1.
To confirm that the C-terminal fusion of the TMHtAPX to HCF244 did not alter its activity, we expressed the variant in the hcf244 mutant background (hcf244/HCF244-TMHtAPX lines; Supplemental Fig. S4). Expression of the fusion protein indeed complemented the hcf244 mutant phenotype (Supplemental Fig. S4A). Consequently, D1 as well as OHP2 levels recovered, indicating the restoration of HCF244 activity. This also implies that the fusion construct retains the site(s) necessary for interaction with both OHPs (Supplemental Fig. S4B).
DISCUSSION
Pigment Binding of OHPs Depends on Heterodimerization
The outcome of our in vitro reconstitution assays supports the idea that the function of the two OHPs depends on their heterodimerization as a prerequisite for pigment binding. Whereas the single OHPs could not be efficiently reconstituted with pigments, the combination of OHP1 and OHP2 yielded an eluate of greenish-colored proteins after incubation with pigments (Fig. 2, A and E). That heterodimerization is necessary for pigment binding seems to be a common principle among a subset of the members of the LHC family. Thus, binding affinities measured in in vitro interaction assays comprising the two LIL3 isoforms and Chl a led to the hypothesis that LIL3.1/LIL3.2 heterodimer formation precedes the binding of Chl a (Mork-Jansson and Eichacker, 2018). However, in contrast to the two OHPs, the LIL3 isoforms show high sequence similarity and are most likely capable of binding pigments as homodimers as well (Hey et al., 2017; Mork-Jansson and Eichacker, 2019). Whereas the OHP and LIL3 dimers generate the pigment-binding site by forming an intermolecular helix pair, LHCPs possess two helices harboring the Chl-binding motif and are thus able to form the pigment-binding site by intramolecular helix pairing (Kühlbrandt et al., 1994). In vitro integration of ELIPs into etioplast membranes requires the presence of Chl a (Adamska et al., 1999, 2001), and because ELIPs are three-helix proteins (with a topology similar to that of the LHCPs), pigment binding by ELIP monomers can be confidently assumed. No details are known to date about the biological function and the pigment-binding ability of the SEP1 and SEP2 variants. However, in view of the results obtained for the OHPs and LIL3s, (hetero)dimer formation by SEP1 and SEP2 seems likely, although this hypothesis requires experimental validation.
Cyanobacterial Hlips have similarly been shown to bind pigments as dimers. Besides the preferred formation of HliA/HliB and HliC/HliD heterodimers (Boehm et al., 2012; Knoppová et al., 2014), the formation of HliC homodimers (and homooligomers) has been confirmed (Shukla et al., 2018). However, these assemblies were only detectable in Synechocystis strains that lacked HliD; hence, their biological relevance is questionable. Apart from HliC/HliD heterodimers, the formation of HliD homodimers has also been proposed, although this hypothesis may have been motivated simply by the difficulty of reliably detecting the small (5-kD) HliC protein by western blotting (Knoppová et al., 2014). In fact, detailed investigation of SynFeCh was required to reveal the pigment-binding ability of its Hlip-like C terminus in a (homo)dimeric assembly state (Pazderník et al., 2019).
The HliC/HliD dimer binds six Chl a molecules and two β-Car moieties (Staleva et al., 2015). The OHP1/OHP2 dimers reconstituted in this work comprised two carotenoids (β-Car and lutein) per six Chl a molecules (Table 1). In addition, the binding of one Chl b molecule was observed, although its biological relevance is currently unclear. As the reconstitution of an OHP-pigment complex can be achieved with Chl a alone, but not with Chl b, Chl a binding is certainly more important for OHP function (Supplemental Fig. S2).
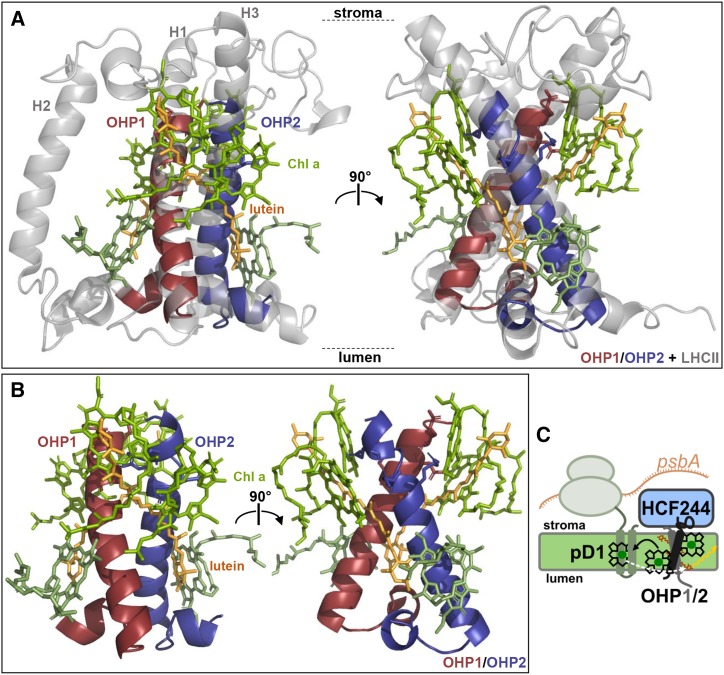
Comparisons of the LHCII crystal structure with the predicted structures of the OHP C termini gives an impression of how pigment binding might be organized in the OHP1/OHP2 dimer (Fig. 8). In LHCII, four Chl a molecules are directly coordinated by the conserved E and N/H residues of the Chl-binding motif at the stromal end of the helices, and it can be assumed that the same motif performs this function in OHPs. Two additional Chl a molecules are organized around the helix pair at the lumenal end of the helices, whereas the two carotenoids are arranged between the Chl molecules. The carotenoids are oriented perpendicular to the membrane at an angle comparable to that of the protein helices (Fig. 8, A and B). The evaluation of the structure shows that six Chl molecules can be arranged around the two helices without spatial constraints. Apparently, participation of the third helix in LHCII (helix 2) or any other stromal or lumenal loops is not essential for the uptake of Chl a. However, x-ray crystallography studies with purified OHP1/OHP2 complexes will be needed to elucidate the pigment organization in molecular detail.
Figure 8.
A model for Chl binding to the OHP1/OHP2 heterodimer. A, Three-dimensional structures of the C-terminal segments of the OHPs (OHP1, amino acids 66–110; OHP2, amino acids 127–172) were predicted using the Phyre2 server (Kelley et al., 2015) and modeled into the crystal structure of LHCII (Liu et al., 2004; Protein Data Bank entry 1rwt) in PyMol. OHP1 (red) was modeled into helix 1 and OHP2 (blue) into helix 3 of LHCII. The four Chl a molecules bound by the conserved residues, as well as two additional Chl a molecules at the lumenal end of the helices, are depicted in green. In addition, two lutein molecules from the LHCII structure are shown in orange. B, Structure of the OHP heterodimer alone with pigments. C, Working model for the function of OHPs in D1 synthesis. The OHP1/OHP2 heterodimer tethers the PSII assembly factor HCF244 to the thylakoid membrane and binds Chls as well as carotenoids. Whereas the molecular role of HCF244 is unclear, the OHP dimer delivers pigments to nascent pD1 and may also perform excitation energy quenching of light absorbed by either D1 or the OHP-bound Chls.
Pigment Binding Is Essential for the in Vivo Function of Both OHPs
The importance of the conserved amino acid residues in the Chl-binding motif was demonstrated by using mutated proteins for the reconstitution assays (Fig. 2). Characterization of the eluates on CN gels revealed that two-component (WT/AAA) combinations of the same OHP behaved exactly like each single-component OHP in reconstitution assays. In all these cases, pigment binding was highly inefficient and no pigmented protein bands were detectable on CN gels. This finding strongly supports the idea that the conserved residues of both OHP1 and OHP2 are required to establish the pigment-binding site. However, the AAA variant of either OHP formed heterodimers with the wild-type subunit of the other (Fig. 3, A and B). This contrasts with LIL3, in which single mutations of either of the conserved residues prevent dimerization with the wild-type protein (Mork-Jansson and Eichacker, 2019).
We verified the biological significance of the conserved residues in vivo by using the mutated OHP variants for complementation studies (Figs. 4 and 5). Expression of the OHP1-AAA variant failed to complement ohp1 (Fig. 4), whereas OHP2-AAA partially complemented ohp2 (Fig. 5). Therefore, it can be concluded that pigment binding of the OHP1/OHP2 heterodimer depends on the conserved amino acids in both OHP1 and OHP2 and, moreover, that pigment binding is a crucial aspect of OHP function in vivo. The partial complementation of ohp2 by OHP2-AAA can be explained if OHP1 is stabilized in these lines by its physical interaction with this (inactive) OHP2 variant. This then allows OHP1 to fulfill its essential function in early developmental stages, as reported earlier (Hey and Grimm, 2018a). In this way, some of the effects specific to the depletion of OHP1 are mitigated, so that OHP2-AAA expression effectively attenuates the ohp2 deficiency phenotype. In addition, it cannot be excluded that the OHP1-WT/OHP2-AAA dimer retains some pigment-binding capacity despite the loss of pigment-coordinating amino acids in the mutant OHP2.
OHP1-AAA Shows Enhanced Protein Stability
The overaccumulation of OHP1-AAA in the ohp1 mutant is unexpected (Fig. 4C), as, to our knowledge, OHP variants have not been found to accumulate beyond the wild-type levels in complemented ohp. We conclude that this effect is related to the mutation of the conserved amino acids. Normally, OHP1 is degraded unless stabilized by binding to OHP2 (Hey and Grimm, 2018a). There are two possible functional explanations for the enhanced stability of the OHP1-AAA variant, although the formation of proteolysis-resistant OHP1 aggregates cannot be fully excluded. (1) The Chl-binding motif could serve as a general recognition site for the proteases that degrade OHP1. If so, other members of the LHC protein family should show this type of instability. This would open up the possibility of a specific degradation mechanism for these proteins. However, no experimental data on the degradation of LHCPs and LHC-like proteins are available. FtsH6 was suggested to be the protease responsible for the degradation of LHCB1 and LHCB3 under high-light conditions, but this idea was later refuted (Zelisko et al., 2005; Wagner et al., 2011). (2) The second explanation is based on the idea that, in the heterodimer, the OHP1 protein is particularly sensitive to photooxidative damage due to light absorption by the bound pigments attached to the OHP-heterodimer complex. This would imply the need for constant replacement of photodamaged OHP1 by newly synthesized copies of the protein (e.g. after the OHP heterodimer has delivered Chl molecules to pD1). As OHP1-AAA is unable to bind pigments, it should be less susceptible to such photooxidative damage in the ohp1 mutant background. Then more OHP1-AAA could accumulate as a result of ongoing synthesis of the protein concurrently with a reduced degradation rate.
Membrane-Bound HCF244 Is Stable in the Absence of OHPs
The expression of an HCF244 variant fused to the TMH of tAPX in the ohp2 mutant background revealed that membrane tethering of HCF244 is in principle sufficient for its stability, even in the complete absence of OHPs (Fig. 7B). In addition to our previous report, the results presented here also clearly support the hypothesis that OHP1 not only depends on OHP2 but also requires HCF244 for its own stabilization (Figs. 6D and 7B). Thus, anchorage of HCF244 to the thylakoid membrane does not stabilize OHP1 in the absence of OHP2. The lack of accumulation of OHP1 in various mutant lines (expression of truncated OHP2 and of HCF244-TMHtAPX in ohp2) supports the model presented in our previous report (Hey and Grimm, 2018a; i.e. that HCF244 first binds to OHP2, and OHP1 is added in a second step in vivo).
In summary, OHP1, OHP2, and HCF244 can be considered to form a single functional unit, insofar as each protein requires the other two for stabilization. In addition, the OHP1-OHP2 subcomplex and HCF244 each perform unique and indispensable functions. Identification of the precise molecular function of HCF244 is therefore essential for the further functional characterization of the OHP1-OHP2-HCF244 complex.
OHPs Mediate Pigment Delivery to pD1 as Well as Energy Quenching
The function of OHPs is undoubtedly connected to the synthesis of PSII, or more specifically, to the synthesis of the functional D1 protein (Hey and Grimm, 2018a, 2018b; Li et al., 2019). In addition, HCF244 contributes to the action of the two OHP isoforms (Link et al., 2012). In light of the Chl-binding capacity of OHP1/OHP2 heterodimers, a function of OHPs in pigment delivery for pD1 has previously been suggested (Hey and Grimm, 2018a, 2018b; Myouga et al., 2018; Li et al., 2019). However, the source of the Chl molecules remains an open question, as no direct interaction of OHPs with CHLG could be detected (Hey and Grimm, 2018a; Proctor et al., 2018). This contrasts with findings in cyanobacteria, where the Hlip-ChlG interaction is well established. An interaction of the OHPs with LIL3, which has been reported to fulfill a pigment-shuttling function within the thylakoid membrane and between the enzymes that mediate late steps in Chl biosynthesis, is conceivable. However, this needs to be explored in future research.
The fact that the OHP heterodimer, like the HliC/HliD dimer, contains both Chl and carotenoids makes the quenching of excitation energy possible. Indeed, this mode of action has been confirmed spectroscopically for the Hlip dimer (Staleva et al., 2015), and it is reasonable to assume that this functionality is conserved among the one-helix members of the LHC family. Thanks to our protocol for the reconstitution of the OHP1/OHP2 heterodimer with pigments, large amounts of this complex are now easily accessible and can be used for spectroscopic analyses in future studies.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Generation of Transgenic Lines
Arabidopsis (Arabidopsis thaliana) seeds (wild-type Columbia-0, ohp1 [GABI_362D02], ohp2 [GABI_071E10; Myouga et al., 2018], and hcf244) were surface sterilized with Meliseptol (Braun) and germinated on one-half-strength MS medium supplemented with 2% (w/v) Suc. Plants were incubated in growth chambers under continuous illumination (100 µmol photons s−1 m−2, 20°C). For stable transformation, ohp1, ohp2, and hcf244 plants were transferred onto soil and incubated in a greenhouse under long-day conditions (16-h photoperiod, 100 µmol photons s−1 m−2) until bolting. A modified floral dip method was used for Agrobacterium tumefaciens-mediated transformation. The coding sequences of both OHP genes were amplified with Phusion polymerase (New England Biolabs) using the specific primers listed in Supplemental Table S1, and point mutations were introduced by overlap-extension PCR. The resulting DNA fragments were then ligated into pJet (Thermo Fisher), and their sequences were confirmed. Subsequently, the fragments were excised by restriction digestion and subcloned into pCAMstrepII (Hey and Grimm, 2018a). The coding sequences for truncated OHP2 peptides and the HCF244 fusion protein were produced by amplification with Phusion polymerase (New England Biolabs) as described above (for primer sequences, see Supplemental Table S1) and subsequent combination of the sequences by overlap-extension PCR. Following confirmation of sequence identity as described above, the fragments were excised by restriction digestion and subcloned into pCAMBIA3301 (OHP2 variants) or pCAMstrepII (HCF244 variants).
Preparation of Total Leaf Protein, and (Tricine)-SDS-PAGE and Immunoblot Analyses
Total leaf proteins were extracted from leaf material that had been powdered in liquid N2. After resuspension in sample buffer (100 mm Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, 200 mm DTT, and 0.01% [w/v] Bromophenol Blue), samples were incubated at 95°C for 10 min. SDS-PAGE in 12% SDS-polyacrylamide (supplemented with 6 m urea for separation of photosynthetic subunits), Tricine-SDS-PAGE (for separation of OHPs), and immunoblot analyses were performed as previously described (Hey and Grimm, 2018a).
Gene Expression Analyses
Total RNA was extracted from powdered leaf material as described previously (Oñate-Sánchez and Vicente-Carbajosa, 2008). Aliquots (1 µg) of RNA were subsequently digested with DNase, and cDNA synthesis was performed with RevertAid RT (Thermo-Fisher) according to the manufacturer’s instructions. Expression of OHP genes was analyzed by RT-qPCR, which was carried out in a CFX 96 real-time system (Bio-Rad) using 2× SensiMixSYBR (Bioline) with primers listed in Supplemental Table S2. ACT2 (At3g18780) and SAND (At2g28390) were used as reference genes, and normalization was performed by the ∆∆Ct method (Pfaffl, 2001).
DNA Extraction and Genotyping
Genomic DNA was extracted from leaf material by resuspension in extraction buffer (200 mm Tris-HCl, pH 8, 100 mm NaCl, 25 mm EDTA, and 0.5% [w/v] SDS) followed by a 10-min centrifugation (20,000g). The supernatant was mixed with an equal volume of isopropanol, and the DNA was precipitated by centrifugation. Subsequently, the DNA was washed twice with 75% (v/v) ethanol, dried, and resuspended in ultrapure water. For genotyping PCRs, DreamTaq polymerase (Thermo) was used according to the manufacturer’s instructions. The primers employed are listed in Supplemental Table S1.
Expression and Purification of Recombinant OHP Proteins
Coding sequences of both OHP genes (without the putative transit peptides) were amplified from cDNA with specific primers listed in Supplemental Table S1 and cloned into the pET22b vector (Novagen). Protein expression was carried out at 37°C for 3 h in Rosetta2 cells (Novagen) grown in 2YT medium supplemented with 1% (w/v) Glc and induced by adding 1 mm isopropyl β-d-1-thiogalactopyranoside (Sigma). Cells were resuspended in lysis buffer (50 mm NaH2PO4, pH 8, and 300 mm NaCl) and lysed by adding 1 µg µL−1 lysozyme and sonicating for 5 min on ice. The lysate was cleared by centrifugation (15 min at 10,000g) and incubated with HisPur Ni-NTA Resin (Thermo-Fisher). After washing with lysis buffer plus 75 mm imidazole, proteins were eluted in wash buffer (lysis buffer plus 250 mm imidazole). The eluted proteins were dialyzed into storage buffer (50 mm NaH2PO4, pH 7.6, 150 mm NaCl, and 2% [v/v] glycerol) and stored at −80°C. OHP protein-containing Escherichia coli membranes intended for use in reconstitution assays were enriched as follows. Cells were resuspended in lysis buffer as above and mixed with 1 µg µL−1 lysozyme. After a 30-min incubation on ice, 5 µL mL−1 DNase I (New England Biolabs), 2 µL mL−1 RNase (Thermo), 10 µL mL−1 1 m MgCl2, and 10 µL mL−1 1 m NaCl were added and cells were incubated on ice for an additional 30 min. Subsequently, cells were sonicated for 5 min, and the lysate was distributed to 2-mL microcentrifuge tubes for a 10-min centrifugation (20,000g). Afterward, the pellet was resuspended by short sonication steps on ice (5 s, each followed by a break of 20 s) into TE buffer (10 mm Tris-HCl, pH 8, and 1 mm EDTA) and centrifuged again as before. This washing step was repeated, and the membranes were finally resuspended into TE buffer by sonication.
Reconstitution of OHPs with Pigments
Pigments (total Chl and carotenoids) were extracted from spinach (Spinacia oleracea; obtained from a local market) as described before (Natali et al., 2014; for the composition of the pigment solution, see Table 1). For reconstitution of OHPs with pigments according to Natali et al. (2014), E. coli membranes containing OHP variants were mixed in equimolar ratios (i.e. OHP1, 270 µg:OHP2, 530 µg) and suspended in 400 µL of TE buffer. Four hundred microliters of reconstitution buffer (200 mm HEPES, 5% [w/v] Suc, 4% [w/v] lithium dodecylsulfate, 2 mm benzamidine, and 10 mm aminocaproic acid) and 0.6 µL of β-mercaptoethanol were then added, and the proteins were denatured by incubation at 100°C for 1 min. After incubation on ice for 2 min, 500 µg of total spinach pigments plus 80 µg of carotenoids were resuspended in 35 µL of 100% ethanol and added to the protein mixture during mixing. Afterward, 94 µL of 20% (w/v) octylglucoside (OG) was added during mixing, and the assay was incubated on ice for 10 min. Finally, 90 µL of 2 m KCl was added, and precipitation of potassium dodecylsulfate was stimulated by a 20-min incubation on ice followed by a 10-min centrifugation (20,000g). The supernatant was mixed with 5 mL of OG buffer (20 mm HEPES, pH 7.5, 200 mm NaCl, 12.5% [w/v] Suc, 10 mm imidazole, and 1% [w/v] OG) and loaded onto a 1-mL HisTrap HP column (GE Healthcare) equilibrated with OG buffer at a flow rate of 1 mL min−1. After washing with 5 mL of OG buffer, nonspecifically bound pigments were eluted from the column by washing with 3 mL of rinse buffer (40 mm HEPES, pH 8, 200 mm NaCl, and 0.06% [w/v] DDM). Finally, reconstituted complexes were eluted with 3 mL of elution buffer (40 mm HEPES, pH 8, 200 mm NaCl, 500 mm imidazole, and 0.06% [w/v] DDM). Approximately 500 µL of pigmented eluate was collected for each assay.
For the reconstitution of OHPs with purified Chls, only 400 µg of total OHPs (i.e. OHP1, 135 µg:OHP2, 265 µg) in a 400-µL assay volume was used, and all other volumes were halved. Purified Chl was obtained from Sigma, and 250 µg of Chl a and 94 µg of Chl b or 250 µg of Chl a or b (plus 160 µg of carotenoids for each assay) was used per assay.
CN-PAGE Analysis
Native electrophoresis of reconstitution eluates was performed on 4% to 12.5% native PAGE gels according to Järvi et al. (2011). The protein complexes were charged by adding 0.3% (w/v) sodium deoxycholate to the eluates. In addition, the cathode buffer contained 0.05% (w/v) DDM and 0.02% (w/v) sodium deoxycholate. In-gel Chl fluorescence was recorded in a PAM imager chamber (FluorCam 700MF, Photon Systems Instruments).
His-Tag Pulldown of Proteins
Purified recombinant proteins (100 µg) were bound to 50 µL of HisPur Ni-NTA Resin (Thermo) beads in assay buffer (50 mm NaH2PO4, pH 8, 150 mm NaCl, and 10% [v/v] glycerol) and incubated with solubilized Arabidopsis thylakoids (100 µg of Chl, solubilized at a Chl concentration of 1 µg µL−1 with 1% [w/v] DDM in the assay buffer). After incubation and intense washing with assay buffer, the proteins bound to the OHP bait proteins were eluted with assay buffer containing 250 mm imidazole and fractionated by Tricine-SDS-PAGE.
BiFC Assay
OHP2 and HCF244 cDNAs were amplified from total cDNA using specific primers carrying attB sites (Supplemental Table S1) and cloned into pDEST-GW-VYNE/-VYCE vectors (Gehl et al., 2009) via pDON207 using the Gateway system (Thermo Fisher). Plasmids were transformed into A. tumefaciens strain GV2260, and BiFC experiments were performed as described previously (Hey and Grimm, 2018a). YFP fluorescence was recorded on an LSM 800 confocal microscope (Zeiss).
Pigment Extraction and HPLC
Pigments were extracted from the reconstitution eluates by adding 9 volumes of ice-cold alkaline acetone (acetone:0.2 m NH4OH, 9:1). Pigments were separated by HPLC and quantified using pure standards. Pigments from leaf material were similarly extracted, and Chls as well as carotenoids were separated and quantified by HPLC.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers At5g02120 (OHP1), At1g34000 (OHP2), At4g35250 (HCF244), and At1g77490 (tAPX).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Raw data for Figures 2, D and H, and 3, A and B.
Supplemental Figure S2. OHP1-WT/OHP2-WT reconstitution efficiency depends on the presence of Chl a.
Supplemental Figure S3. Complementation of ohp2 with truncated OHP2 variants.
Supplemental Figure S4. Complementation of hcf244 with a membrane-bound HCF244 variant.
Supplemental Table S1. Primers used for genotyping and cloning procedures.
Supplemental Table S2. Primers used for gene expression analyses.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Gr936 14–1 and Gr936 14–1 to B.G.).
Articles can be viewed without a subscription.
References
- Adamska I, Kruse E, Kloppstech K(2001) Stable insertion of the early light-induced proteins into etioplast membranes requires chlorophyll a. J Biol Chem 276: 8582–8587 [DOI] [PubMed] [Google Scholar]
- Adamska I, Roobol-Bóza M, Lindahl M, Andersson B(1999) Isolation of pigment-binding early light-inducible proteins from pea. Eur J Biochem 260: 453–460 [DOI] [PubMed] [Google Scholar]
- Andersson U, Heddad M, Adamska I(2003) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J, Lohscheider JN, Albert S, Andersson U, Mendgen KW, Rojas-Stütz MC, Adamska I, Funck D(2017) Small one-helix proteins are essential for photosynthesis in Arabidopsis. Front Plant Sci 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Yu J, Reisinger V, Beckova M, Eichacker LA, Schlodder E, Komenda J, Nixon PJ(2012) Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: Implications for the assembly and repair of photosystem II. Philos Trans R Soc Lond B Biol Sci 367: 3444–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R(2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE(2004) WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelken J, Brinkmann H, Adamska I(2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R(2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- He Q, Dolganov N, Bjorkman O, Grossman AR(2001) The high light-inducible polypeptides in Synechocystis PCC6803: Expression and function in high light. J Biol Chem 276: 306–314 [DOI] [PubMed] [Google Scholar]
- Hey D, Grimm B(2018a) ONE-HELIX PROTEIN2 (OHP2) is required for the stability of OHP1 and assembly factor HCF244 and is functionally linked to PSII biogenesis. Plant Physiol 177: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey D, Grimm B(2018b) Requirement of ONE-HELIX PROTEIN 1 (OHP1) in early Arabidopsis seedling development and under high light intensity. Plant Signal Behav 13: e1550317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey D, Rothbart M, Herbst J, Wang P, Müller J, Wittmann D, Gruhl K, Grimm B(2017) LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiol 174: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM(2011) Optimized native gel systems for separation of thylakoid protein complexes: Novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE(2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Sobotka R, Tichy M, Yu J, Konik P, Halada P, Nixon PJ, Komenda J(2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ(2012) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y(1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367: 614–621 [DOI] [PubMed] [Google Scholar]
- Li Y, Liu B, Zhang J, Kong F, Zhang L, Meng H, Li W, Rochaix JD, Li D, Peng L(2019) OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol 179: 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link S, Engelmann K, Meierhoff K, Westhoff P(2012) The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol 160: 2202–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W(2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Llansola-Portoles MJ, Sobotka R, Kish E, Shukla MK, Pascal AA, Polívka T, Robert B(2017) Twisting a β-carotene, an adaptive trick from nature for dissipating energy during photoprotection. J Biol Chem 292: 1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P(1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork-Jansson A, Bue AK, Gargano D, Furnes C, Reisinger V, Arnold J, Kmiec K, Eichacker LA(2015a) Lil3 assembles with proteins regulating chlorophyll synthesis in barley. PLoS ONE 10: e0133145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork-Jansson AE, Eichacker LA(2018) Characterization of chlorophyll binding to LIL3. PLoS ONE 13: e0192228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork-Jansson AE, Eichacker LA(2019) A strategy to characterize chlorophyll protein interaction in LIL3. Plant Methods 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork-Jansson AE, Gargano D, Kmiec K, Furnes C, Shevela D, Eichacker LA(2015b) Lil3 dimerization and chlorophyll binding in Arabidopsis thaliana. FEBS Lett 589: 3064–3070 [DOI] [PubMed] [Google Scholar]
- Murray DL, Kohorn BD(1991) Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol Biol 16: 71–79 [DOI] [PubMed] [Google Scholar]
- Myouga F, Takahashi K, Tanaka R, Nagata N, Kiss AZ, Funk C, Nomura Y, Nakagami H, Jansson S, Shinozaki K(2018) Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol 176: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natali A, Roy LM, Croce R(2014) In vitro reconstitution of light-harvesting complexes of plants and green algae. J Vis Exp e51852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J(2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazderník M, Mareš J, Pilný J, Sobotka R(2019) The antenna-like domain of the cyanobacterial ferrochelatase can bind chlorophyll and carotenoids in an energy-dissipative configuration. J Biol Chem 294: 11131–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW.(2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley FG, Schmidt GW(1987) Reconstitution of chlorophyll a/b light-harvesting complexes: Xanthophyll-dependent assembly and energy transfer. Proc Natl Acad Sci USA 84: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor MS, Chidgey JW, Shukla MK, Jackson PJ, Sobotka R, Hunter CN, Hitchcock A(2018) Plant and algal chlorophyll synthases function in Synechocystis and interact with the YidC/Alb3 membrane insertase. FEBS Lett 592: 3062–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. (2010) The PyMOL molecular graphics system, version 1.0. https://pymol.org/2/#page-top
- Shukla MK, Llansola-Portoles MJ, Tichý M, Pascal AA, Robert B, Sobotka R(2018) Binding of pigments to the cyanobacterial high-light-inducible protein HliC. Photosynth Res 137: 29–39 [DOI] [PubMed] [Google Scholar]
- Sobotka R, Tichy M, Wilde A, Hunter CN(2011) Functional assignments for the carboxyl-terminal domains of the ferrochelatase from Synechocystis PCC 6803: The CAB domain plays a regulatory role, and region II is essential for catalysis. Plant Physiol 155: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staleva H, Komenda J, Shukla MK, Šlouf V, Kaňa R, Polívka T, Sobotka R(2015) Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat Chem Biol 11: 287–291 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Takabayashi A, Tanaka A, Tanaka R(2014) Functional analysis of light-harvesting-like protein 3 (LIL3) and its light-harvesting chlorophyll-binding motif in Arabidopsis. J Biol Chem 289: 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Rothbart M, Oka S, Takabayashi A, Takahashi K, Shibata M, Myouga F, Motohashi R, Shinozaki K, Grimm B, et al. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci USA 107: 16721–16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Pružinská A, Jänkänpää HJ, Jansson S, Funk C(2011) Fitness analyses of Arabidopsis thaliana mutants depleted of FtsH metalloproteases and characterization of three FtsH6 deletion mutants exposed to high light stress, senescence and chilling. New Phytol 191: 449–458 [DOI] [PubMed] [Google Scholar]
- Zelisko A, García-Lorenzo M, Jackowski G, Jansson S, Funk C(2005) AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc Natl Acad Sci USA 102: 13699–13704 [DOI] [PMC free article] [PubMed] [Google Scholar]