A phospholipase D is a negative regulator of plant immunity and forms complexes with regulatory receptor-like kinases.
Abstract
Plants have evolved effective strategies to defend themselves against pathogen invasion. Starting from the plasma membrane with the recognition of microbe-associated molecular patterns (MAMPs) via pattern recognition receptors, internal cellular signaling pathways are induced to ultimately fend off the attack. Phospholipase D (PLD) hydrolyzes membrane phospholipids to produce phosphatidic acid (PA), which has been proposed to play a second messenger role in immunity. The Arabidopsis (Arabidopsis thaliana) PLD family consists of 12 members, and for some of these, a specific function in resistance toward a subset of pathogens has been shown. We demonstrate here that Arabidopsis PLDγ1, but not its close homologs PLDγ2 and PLDγ3, is specifically involved in plant immunity. Genetic inactivation of PLDγ1 resulted in increased resistance toward the virulent bacterium Pseudomonas syringae pv. tomato DC3000 and the necrotrophic fungus Botrytis cinerea. As pldγ1 mutant plants responded with elevated levels of reactive oxygen species to MAMP treatment, a negative regulatory function for this PLD isoform is proposed. Importantly, PA levels in pldγ1 mutants were not affected compared to stressed wild-type plants, suggesting that alterations in PA levels are not likely the cause for the enhanced immunity in the pldγ1 line. Instead, the plasma-membrane-attached PLDγ1 protein colocalized and associated with the BAK1-INTERACTING RECEPTOR-LIKE KINASES BIR2 and BIR3, which are known negative regulators of pattern-triggered immunity. Moreover, complex formation of PLDγ1 and BIR2 was further promoted upon MAMP treatment. Hence, we propose that PLDγ1 acts as a negative regulator of plant immune responses in complex with immunity-related proteins BIR2 and BIR3.
Although plants are constantly in contact with potentially harmful microorganisms, disease development is rather an exception. This is due to an efficient plant surveillance system that detects highly conserved microbe-associated molecular patterns (MAMPs) via cell surface-located pattern recognition receptors to mount appropriate immune responses in a process called pattern-triggered immunity (PTI; Couto and Zipfel, 2016; Saijo et al., 2018). One well studied example for such a MAMP is bacterial flagellin, of which a 22-amino acid fragment is sufficient to trigger immune responses by binding to the extracellular Leu-rich repeat (LRR) domain of the receptor kinase FLAGELLIN SENSING 2 (FLS2; Gómez-Gómez and Boller, 2000; Chinchilla et al., 2006). Upon ligand-receptor interaction, activated receptor complexes recruit coreceptors of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family, most prominently BRASSINOSTEROID RECEPTOR1-ASSOCIATED KINASE1 (BAK1; Chinchilla et al., 2007; Heese et al., 2007; Sun et al., 2013). In the absence of an immunogenic stimulus, however, the coreceptor kinase BAK1 is bound to the BAK1-INTERACTING RECEPTOR-LIKE KINASES BIR2 and BIR3 to avoid inappropriate receptor activation (Halter et al., 2014; Imkampe et al., 2017). Only upon ligand-binding to the corresponding immune receptor is BAK1 released from BIR2/3 and can join the receptor-ligand complex. Subsequently, cellular signaling events are triggered, including the production of reactive oxygen species (ROS), the activation of mitogen-activated protein kinases (MAPKs), or transcriptional reprogramming of the cell (Boller and Felix, 2009; Yu et al., 2017; Saijo et al., 2018), which ultimately leads to plant resistance against the microbial invader.
The activation of MAPKs and the oxidative burst can be modulated by lipid second messengers such as phosphatidic acid (PA; Laxalt and Munnik, 2002; Testerink and Munnik, 2011), and PA levels have been shown to rapidly increase in various cell suspension cultures upon stimulation with different elicitors (van der Luit et al., 2000; Laxalt et al., 2001; den Hartog et al., 2003; de Jong et al., 2004; Bargmann et al., 2006). In plants, PA can be released from membrane phospholipids by two classes of enzymes, either directly via hydrolysis catalyzed by phospholipase D (PLD) or via the combined action of PLC and diacylglycerol kinase (Wang, 2004; Bargmann and Munnik, 2006; Arisz et al., 2009).
Arabidopsis (Arabidopsis thaliana) has 12 different genes for PLDs, and based on their gene architectures, sequence similarities, domain structures, and biochemical properties they have been grouped into six subfamilies: α, β, γ, δ, ε, and ζ (Qin and Wang, 2002). PLDs play a role in lipid metabolism but are also involved in the regulation of cellular processes, such as hormone signaling, environmental stress responses, and cellular and subcellular dynamics (Bargmann and Munnik, 2006; Wang et al., 2006). PLDα was one of the first PLD family members to have been characterized, and depletion of Arabidopsis PLDα resulted in decreased levels of PA and superoxide in leaf extracts (Sang et al., 2001). In guard cells, abscisic acid (ABA) responses rely on the production of PA via PLDα1, and this PA is required for the accumulation of ROS by directly binding to and thereby stimulating NADPH oxidase activity (Zhang et al., 2009). Moreover, PLDα1 regulates the function of the heterotrimeric G-protein GPA1, by interacting with the α-subunit and promoting the exchange of GTP to GDP, thereby mediating ABA-induced inhibition of stomatal opening (Mishra et al., 2006). A second member of this PLD subgroup, PLDα3, was also implicated in hyperosmotic stress, and pldα3 mutant plants were more sensitive to salinity and water deficiency (Hong et al., 2008). PLDs are also involved in plant growth, as exemplified by PLDε, of which mutant plants display decreased root growth and biomass accumulation (Hong et al., 2009).
In addition to the earlier observations in cell suspension cultures, PLDs have also been implicated in immunity of whole plants. Infection of Arabidopsis with the necrotrophic fungus Botrytis cinerea or the hemibiotrophic bacterial pathogen Pseudomonas syringae pv tomato (Pto) DC3000 resulted in increased PA levels, predominantly produced by PLDβ1 (Zhao et al., 2013). Genetic inactivation of PLDβ1 lead to enhanced susceptibility to Botrytis infection, but increased resistance to PtoDC3000, accompanied by higher levels of ROS and salicylic acid (SA; Zhao et al., 2013), indicating that PLDβ1 can have both positive and negative regulatory functions in plant defense, depending on the lifestyle of the pathogen. Specific PLD isoforms have also been shown to be important for penetration resistance, e.g. Arabidopsis pldδ mutants fail to restrict penetration of the two powdery mildew fungi Blumeria graminis f. sp. hordei and Erysiphe pisi (Pinosa et al., 2013). In addition, the accumulation of chitin-induced defense genes appears to be delayed in the pldδ mutant (Pinosa et al., 2013), suggesting a positive role for PLDδ in plant immunity.
We show here that Arabidopsis PLDγ1 plays a major role in immunity. Genetic inactivation of PLDγ1 leads to increased resistance toward both bacterial and fungal infection, accompanied by elevated MAMP-induced ROS levels. However, alterations in PA levels are unlikely to be the cause for this increased resistance, as they seem to be unaffected in pldγ1 mutant plants. Rather, PLDγ1 can be found in complex with the BAK1-interacting LRR receptor-like kinases (RLKs) BIR2 and BIR3, revealing a novel function for phospholipases in plant immunity.
RESULTS
PLDγ1 Is the Only PLDγ Isoform Involved in Bacterial Resistance
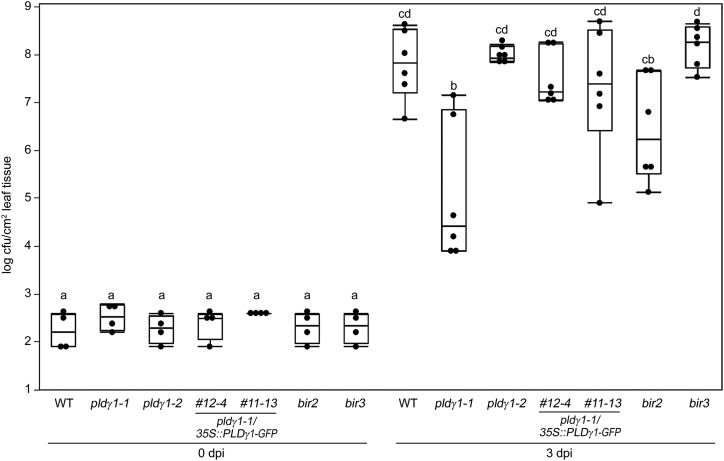
Whereas several Arabidopsis PLD family members have been shown to be involved in plant immunity (Pinosa et al., 2013; Zhao et al., 2013), little is known about the function of the PLDγ subgroup. We therefore selected transfer DNA (T-DNA) insertion lines for the three family members PLDγ1 (At4g11850), PLDγ2 (At4g11830), and PLDγ3 (At4g11840), which are closely related and share 93% to 95% similarity in their amino acid sequence (Qin and Wang, 2002). These mutant lines were previously characterized and reported to have a wild-type-like response to infection with the barley powdery mildew fungus B. graminis f. sp. hordei (Pinosa et al., 2013). To investigate whether these PLDs play a role in bacterial resistance, we infected pldγ1-1 (Salk_066687C), pldγ2 (Salk_078226), and pldγ3 (Salk_084335) plants (Pinosa et al., 2013) with the virulent bacterial strain Pseudomonas syringae pv. tomato (Pto) DC3000. Whereas pldγ2 and pldγ3 mutant plants supported bacterial growth similar to wild-type plants, genetic inactivation of PLDγ1 resulted in significantly less growth of Pto DC3000 (Fig. 1; Supplemental Fig. S1). This increased resistance phenotype could also be observed in control plants of the bir2 genotype, but not in the bir3 mutant which were reported not to be affected in their bacterial resistance (Fig. 1; Halter et al., 2014; Imkampe et al., 2017). To verify the pldγ1-1 phenotype, we infected an additional T-DNA insertion line, pldγ1-2 (GK-264A03; Supplemental Fig. S2), with Pto DC3000. However, in pldγ1-2 plants bacterial growth was not reduced compared to wild-type plants (Fig. 1). This could be explained by higher residual PLDγ1 activity in pldγ1-2 mutants compared to the pldγ1-1 line (Supplemental Fig. S2B). We therefore sequenced the genome of the pldγ1-1 mutant to determine whether this line contained secondary T-DNA insertion sites. Genome sequencing confirmed the location of the T-DNA insertion within the eighth exon of PLDγ1 (Pinosa et al., 2013) but also revealed two additional T-DNA insertion events in the pldγ1-1 genome, one in the promoter region of the gene At1g77460 and the other in the At2g31130 promoter region, which is now also indicated on the Nottingham Arabidopsis Stock Center site (http://arabidopsis.info/MultiResult?name=SALK_066687&Android=F). However, mutant lines with T-DNA insertions in the coding regions of either At1g77460 or At2g31130 (Supplemental Fig. S3, A and B) were similar to the wild type in growth of Pto DC3000 (Supplemental Fig. S3C), indicating that increased bacterial resistance of pldγ1-1 is most likely due to genetic inactivation of PLDγ1. To corroborate this hypothesis further, we generated complementation lines by expressing a PLDγ1-GFP fusion driven by the cauliflower mosaic virus 35S promoter in the pldγ1-1 mutant background. Independent transgenic lines with wild-type-like PLDγ1 transcript levels and detectable PLDγ1-GFP protein expression were selected (Supplemental Fig. S4, A and B), and T3 generation plants were infected with Pto DC3000. All tested transgenic pldγ1-1/35S::PLDγ1-GFP lines showed at least partial restoration of bacterial susceptibility (Supplemental Fig. S4C). When using T4 generation plants for infection, the two independent lines 12-4 and 11-13 showed complete restoration of the wild-type response to bacterial infection (Fig. 1), confirming that loss of PLDγ1 is the cause for decreased bacterial growth in pldγ1-1 mutants.
Figure 1.
Genetic inactivation of PLDγ1 results in increased resistance to bacterial infection. Wild-type (WT) plants or the indicated mutant lines for pldγ1-1, pldγ1-2, complementation lines 12-4 and 11-13 (pldγ1-1/35S:: PLDγ1-GFP, T4 generation), or bir2 and bir3 were infiltrated with 104 cfu/mL of virulent Pseudomonas syringae pv. tomato DC3000. At 0 and 3 d postinoculation (dpi), bacterial growth was quantified by counting colony-forming units. Box plots show the minimum, first quartile, median, third quartile, and a maximum of log cfu/cm2 leaf tissue (n = 4 for 0 dpi; n = 6 for 3 dpi). Lowercase letters indicate homogenous groups according to post hoc comparisons following one-way ANOVA (Tukey-Kramer multiple comparison analysis at a probability level of P < 0.05). The entire experiment was repeated three times with similar results.
Fungal Resistance Is Enhanced in pldγ1 Mutants
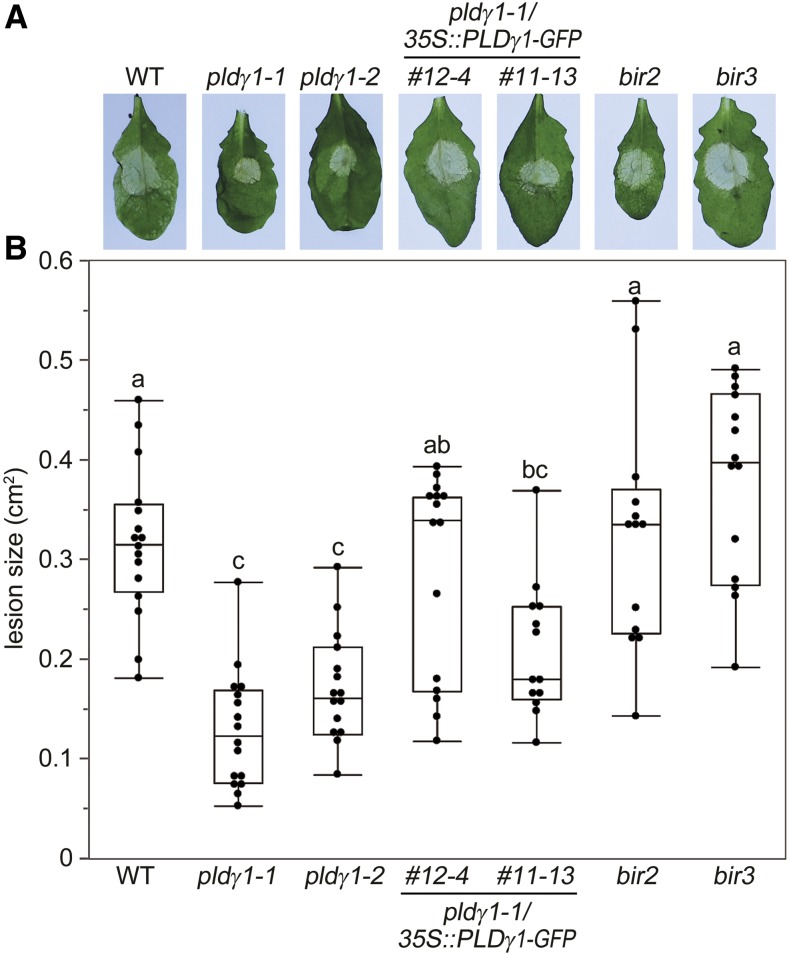
We next investigated whether pldγ1 mutants are also affected in their resistance toward the necrotrophic fungal pathogen B. cinerea. Spore inoculation of wild-type, pldγ2, and pldγ3 plants resulted in comparable lesions within 3 d postinfection, with an elevated fungal DNA content in infected leaves (Supplemental Fig. S1B). However, lesion sizes were highly reduced in pldγ1-1 and to a lesser extent in pldγ1-2 mutants (Fig. 2; Supplemental Fig. S1B). Again, wild-type responses could be restored in the two independent complementation lines (Fig. 2). Interestingly, both the bir2 and bir3 mutant lines did not display altered resistance to Botrytis infection (Fig. 2), although bir2 mutants have previously been shown to have an increased susceptibility to infection with the necrotrophic fungus Alternaria brassicicola (Halter et al., 2014). In summary, loss of functional PLDγ1 resulted in increased bacterial, but also increased fungal resistance.
Figure 2.
pldγ1-1 mutants are more resistant to B. cinerea infection. Leaves of 6-week-old plants were inoculated with 5 × 106/mL B. cinerea spores. A, After 2 d, symptom development was documented and one representative leaf per line is shown. B, Lesion sizes were determined with a pixel-based approach and then calculated using a 1-cm2 standard. Box plots show the minimum, first quartile, median, third quartile, and a maximum (n ≥ 12). Lowercase letters indicate homogenous groups according to post hoc comparisons following one-way ANOVA (Tukey-Kramer multiple comparison analysis at a probability level of P < 0.05). The experiment was performed three times with similar results. WT, Wild type.
pldγ1 Mutants Show an Elevated Oxidative Burst
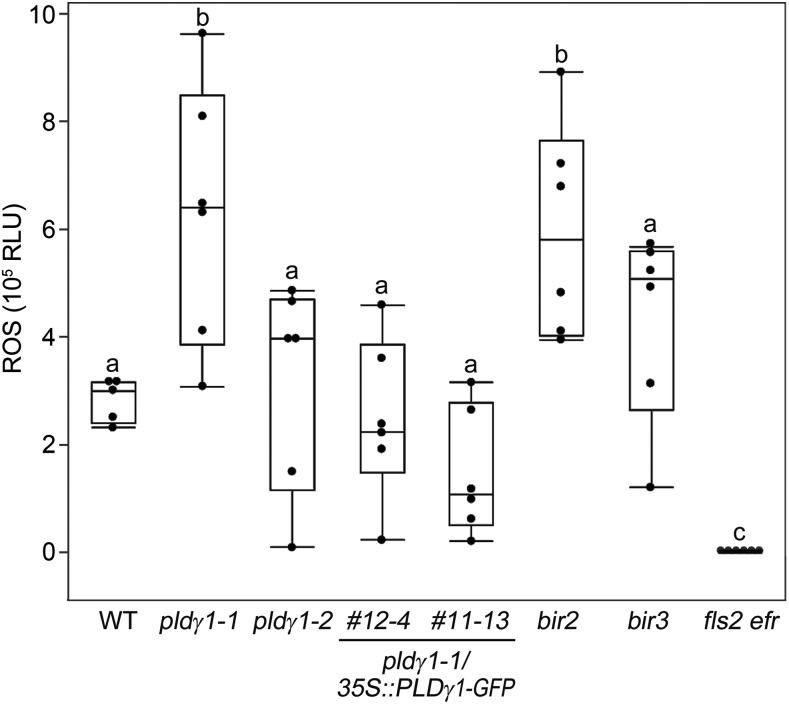
Increased resistance to bacterial infection has been associated with the activation of basal immunity (Zipfel et al., 2004, 2006). Hence, we tested whether pldγ1 mutants are also altered in PTI responses. We treated the pldγ1 mutants, the complementation lines, and wild-type plants with the MAMP flg22, a 22-amino acid peptide of the conserved N-terminal part of flagellin that is recognized by the FLS2 immune receptor (Gómez-Gómez and Boller, 2000). We then investigated the activation of MAPKs and the accumulation of ROS as two distinct branches of PTI signaling (Bigeard et al., 2015). flg22-induced activation of particular MAPKs is an early event and believed to contribute to transcriptional reprogramming and resistance (Bethke et al., 2012). Posttranslational MAPK activation was determined in Arabidopsis seedlings by western blot analysis using a p44/42 antibody raised against phosphorylated MAPKs. flg22-induced activation of immunity-related MAPKs MPK6, MPK3, and MPK4/11 was indistinguishable in pldγ1 mutants, complementation lines, and wild-type plants (Supplemental Fig. S5), indicating that a loss of PLDγ1 does not impinge on the activation of this signaling cascade. Since the accumulation of ROS after flg22 stimulation is independent of MAPK activation (Segonzac and Zipfel, 2011; Xu et al., 2014), we next studied the oxidative burst in our pldγ mutant lines. Treatment of pldγ2 and pldγ3 mutants, as well as the T-DNA insertion lines for At1g77460 and At2g31130, with flg22 resulted in ROS responses similar to those observed in wild-type plants, whereas plants mutated in the FLS2 receptor were completely unresponsive to flg22 treatment (Fig. 3; Supplemental Figs. S1C and S3D). However, ROS levels in pldγ1-1 were significantly increased, reaching about double the amount for wild-type, pldγ2, and pldγ3 plants, as well as pldγ1/35S::PLDγ1-GFP complementation lines (Fig. 3; Supplemental Figs. S1C and S3D). Increased ROS levels were also observed in the bir2 mutant, whereas bir3 mutants only showed the described weak MAMP phenotype (Fig. 3; Halter et al., 2014; Imkampe et al., 2017). Together with the increased bacterial resistance observed in pldγ1-1 mutants, these results suggest that PLDγ1 acts as a negative regulator of plant immunity.
Figure 3.
ROS levels are affected in pldγ1-1 mutants. Arabidopsis leaf pieces of the indicated mutant line were treated with 1 μm flg22 or water as a control, and ROS production was monitored over time. Shown are relative light units (RLU). Box plots show the minimum, first quartile, median, third quartile, and a maximum of peak values minus background values (n = 6). Water-treated samples had no detectable ROS production and are therefore not displayed in the figure. Lowercase letters indicate homogenous groups according to post hoc comparisons following one-way ANOVA (Dunnet’s multiple comparison analysis with the wild type [WT] as a control at a probability level of P < 0.05). The experiment was performed at least three times and one representative result is shown.
pldγ1 Mutation Does Not Affect Phytohormone Levels
Plant hormones play an important role in immunity (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012; Ku et al., 2018), especially SA and jasmonic acid (JA), which are biosynthetically induced after pathogen attack, resulting in the activation of downstream pathways (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). SA is often correlated with biotrophic infection while JA is typically associated with resistance toward necrotrophic pathogens (Pieterse et al., 2012). We therefore determined SA and JA levels in untreated pldγ1-1 mutant or complemented plants. Neither basal SA nor JA levels in pldγ1-1 lines were distinguishable from those obtained from wild-type plants (Supplemental Fig. S6, A and B). Likewise, gene expression levels of SA and JA marker genes after treatment with flg22 were not affected by genetic inactivation of PLDγ1 (Supplemental Fig. S6C). Hence, PLDγ1 is unlikely to exert its function in immunity via SA- or JA-signaling pathways.
PA Levels Are Unaltered in pldγ1 Mutants
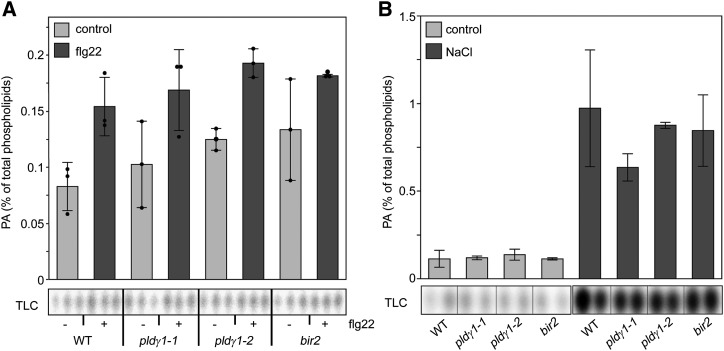
PLDs can cleave membrane phospholipids to produce PA as a lipid second messenger (Testerink and Munnik, 2011). Hence, we analyzed basal PA levels in the pldγ1 mutants. To this end, phospholipids in Arabidopsis seedlings were metabolically labeled with 32P-Pi, and 32P-PA levels were determined after extraction and thin-layer chromatography (TLC). No difference in basal PA levels was found between wild-type and pldγ1 or bir2 seedlings that would explain their increased bacterial resistance (Fig. 4). Upon flg22 treatment, a small increase in PA was observed but this was the same for both wild-type, pldγ1, and bir2 plants (Fig. 4A). Salt stress resulted in a stronger PA accumulation, reaching about 1% of total phospholipids (Bargmann et al., 2009; Yu et al., 2010), but no difference between wild type and mutants was observed (Fig. 4B). These results indicate that, at least in our experimental setup, the genetic inactivation of PLDγ1 or BIR2 does not impinge on PA levels, which are therefore not likely to be the cause for the observed increase in bacterial resistance of the pldγ1 and bir2 mutants.
Figure 4.
PA levels are not affected by PLDγ1 depletion. Five-day-old seedlings of wild-type (WT), pldγ1-1, pldγ1-2, or bir2 lines were labeled with 32Pi for 16 h and then treated for 15 min with either 1 μm flg22 (A) or 300 mm NaCl (B) or cell-free medium (control) as indicated. Lipids were extracted and separated by EtAc-TLC, and the radioactivity incorporated into PA was quantified by phosphoimaging (bottom). Data (top) represent the average ± sd of two to three biological replicates and are expressed in relation to the radioactivity of the total phospholipids. The experiment was repeated twice with similar results.
PLDγ1 Associates with BIR2 and BIR3
Since PA levels did not appear to be affected by genetic inactivation of PLDγ1, we searched for an alternative molecular mechanism as to how PLDγ1 negatively regulates the plant immune response. Intriguingly, the pldγ1 phenotype (i.e. elevated resistance to bacteria and increased production of ROS upon flg22 treatment; Figs. 1 and 3) is reminiscent of the behavior of bir2 mutants (Halter et al., 2014). The BAK1 interacting-receptor kinase BIR2 and its close homolog BIR3 act as negative regulators of plant immune responses by constitutively interacting with the coreceptor BAK1, thereby preventing its interaction with ligand-binding LRR-RLKs, such as FLS2 or the brassinolide-receptor BRI1 (Halter et al., 2014; Imkampe et al., 2017).
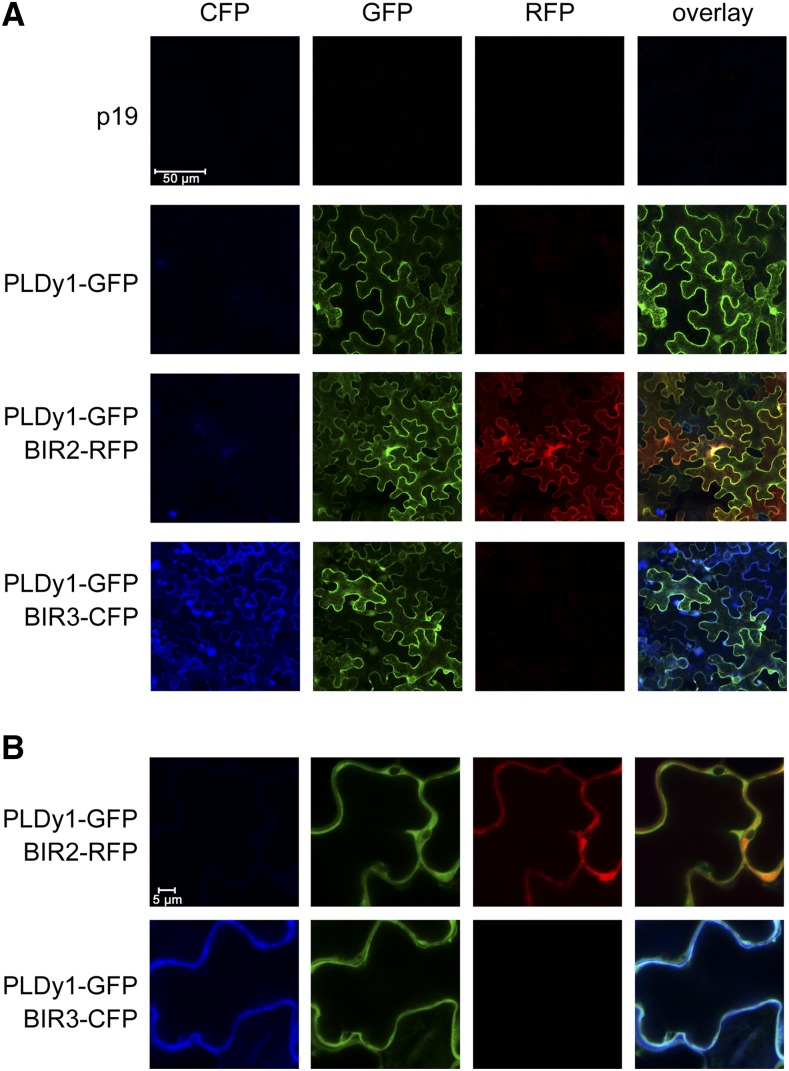
Therefore, we first investigated whether the subcellular protein localization would permit PLDγ1 to associate with BIR2/3 proteins at the plasma membrane (pm). As the fluorescence signal in the pldγ1-1/35S::PLDγ1-GFP complementation lines was too weak for microscopic detection, we transiently expressed PLDγ1-GFP together with either BIR2-red fluorescent protein (RFP) or BIR3-cyan fluorescent protein (CFP) in Nicotiana benthamiana. Using confocal microscopy, we could clearly detect GFP, RFP, and CFP signals localized to the PM of leaf epidermal cells (Fig. 5). Moreover, an overlay of fluorescent signals indicated colocalization of PLDγ1 with BIR2 and BIR3, respectively, at the PM of N. benthamiana cells.
Figure 5.
PLDγ1 colocalizes with BIR2 and BIR3 at the PM. A, PLDγ1 was transiently expressed in N. benthamiana as GFP-fusion either alone or together with BIR2-RFP or BIR3-CFP in the presence of the p19 suppressor of silencing. Infiltration of p19 alone served as control. Fluorescence in epidermal cells was monitored at day 3 using confocal laser-scanning microscopy. B, Close-up view of the samples shown in A.
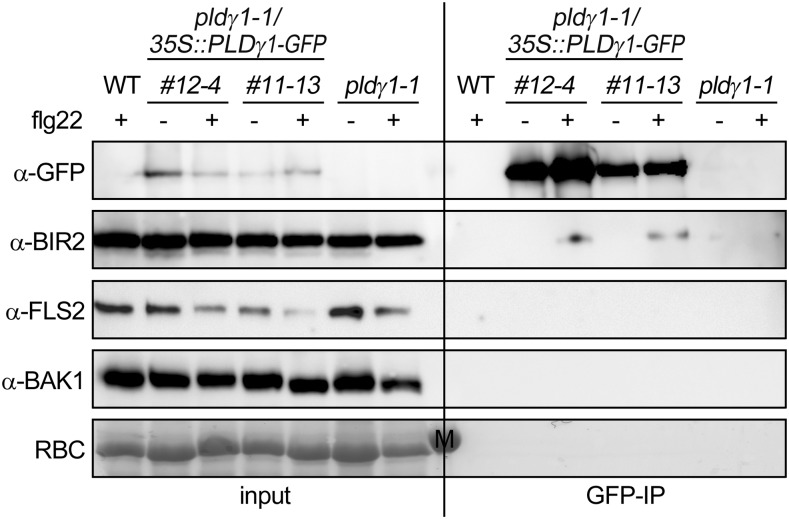
To test whether PLDγ1 could interact with BIR2, we immunopurified PLDγ1-GFP from pldγ1-1/35S::PLDγ1-GFP complementation lines using GFP-affinity beads and analyzed the immunoprecipitates for the presence of native BIR2. As shown in Figure 6, GFP immunoprecipitates from complementation lines 12-4 and 11-13 contained not only enriched PLDγ1-GFP but also BIR2 protein, though only when leaves were treated with flg22 for 5 min before harvesting. In contrast, using GFP-beads we could not copurify BIR2 protein from protein extracts from the pldγ1-1 mutant or wild-type plants (Fig. 6). As flg22-induced ROS production was altered in the pldγ1-1 mutant, we also tested whether PLDγ1 would associate with the flg22-receptor FLS2 or its coreceptor BAK1. However, using antibodies against native proteins we could not detect any FLS2 or BAK1 protein in GFP-immunoprecipitates, indicating that these proteins are not present in the same protein complex as PLDγ1-GFP (Fig. 6).
Figure 6.
PLDγ1-GFP associates with native BIR2, but not FLS2 or BAK1. Leaf material was harvested from pldγ1-1 plants, the complementation lines 12-4 and 11-13 (pldγ1-1/35S:: PLDγ1-GFP), or wild-type (WT) plants 5 min after treatment with 1 μm flg22 (+) or water (−). After protein extraction the proteins (input) were subjected to immunoprecipitation with GFP-beads (GFP-IP). Immunoprecipitated and copurified proteins were detected with GFP antibodies or specific antibodies against native proteins as indicated. Ponceau S Red-staining of the membrane served as a loading control. M indicates the marker band at 55 kD. The experiment was repeated with similar results. RBC, Ribulose-bis-phosphate-carboxylase large subunit.
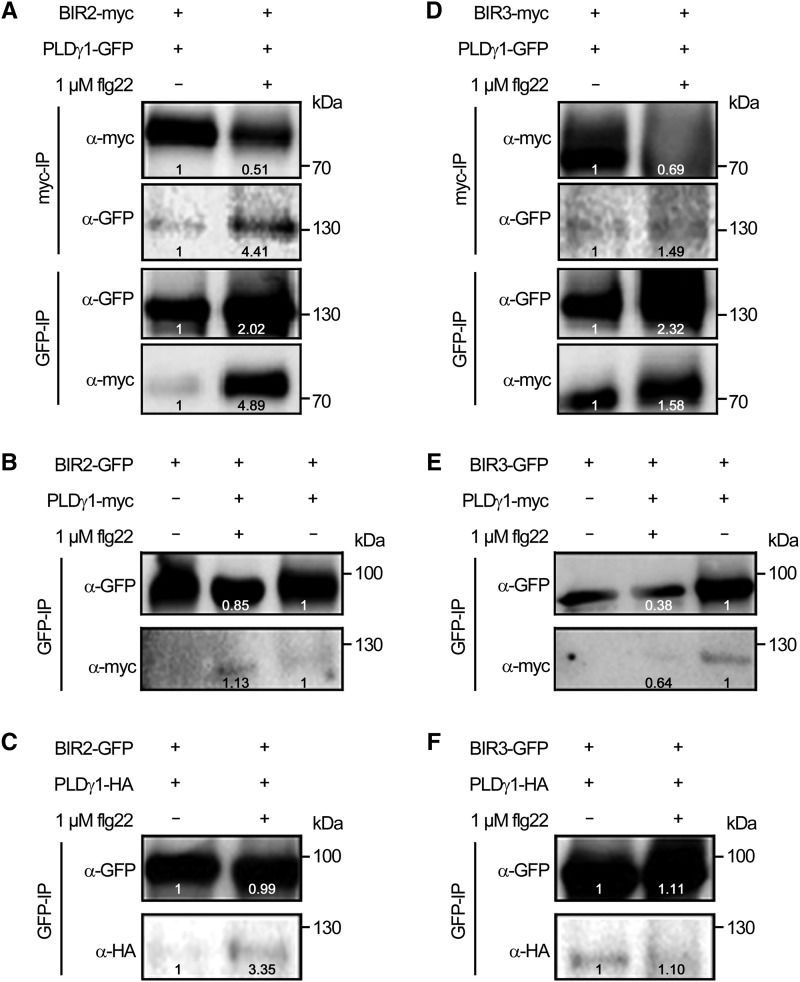
To investigate whether PLDγ1 could also be copurified when BIR2, or BIR3, would be pulled down and whether the size of the epitope tag would have an impact on protein interaction, we transiently expressed PLDγ1 fused to different C-terminal epitope tags together with either BIR2 or BIR3 fused to a C-terminal myc- or GFP-epitope in N. benthamiana leaves. Coimmunoprecipitation with protein extracts from those leaves revealed that PLDγ1 can be found in a constitutive complex with both BIR2 and BIR3 (Fig. 7), regardless of which protein partner is pulled down with the corresponding affinity beads. Again, treatment with flg22 resulted in more BIR2-myc copurified with PLDγ1-GFP compared to the control treatment, whereas the PLDγ1-BIR3 interaction seemed to be largely unaffected by elicitation (Fig. 7). Quantification of immunoprecipitated BIR2 protein levels relative to PLDγ1 signals showed that within 5 min of flg22 treatment ∼8.65 times more BIR2-myc could be found in complex with PLDγ1-GFP (when myc-beads were used for immunoprecipitation), and likewise, 2.42 times more PLDγ1-GFP could be found in complex with BIR2-myc (when GFP beads were used for immunoprecipitation), indicating a ligand-dependent enhancement of the PLDγ1/BIR2 protein interaction, as also observed in Arabidopsis (Fig. 6). PLDγ1 complex formation with BIR2 and BIR3 could also be confirmed in experiments exchanging epitope tags, and again more PLDγ1 could be found in BIR2 precipitates in the presence of flg22 (Fig. 7, B, C, E, and F). Association of transiently expressed PLDγ1 with BIR2 and BIR3 was specific, as PLDγ1 did not copurify with other LRR-RLKs, such as FLS2 or BAK1 (Supplemental Fig. S7).
Figure 7.
PLDγ1 can be found in complex with epitope-tagged BIR2 and BIR3. Western blot analysis of transiently expressed proteins in N. benthamiana 3 d after infiltration. Leaf material was harvested 5 min after treatment with 1 µm flg22 (+) or water (−). After protein extraction, the proteins were subjected to immunoprecipitation (IP) with GFP- or myc-affinity beads as indicated. For different immunoprecipitations within one experiment (A and D), the same source material was used. Immunoprecipitated and copurified proteins were detected with tag-specific antibodies as indicated. A and D, Coimmunoprecipitation of BIR2-myc (A) or BIR3-myc (D) and PLDγ1-GFP using myc-trap beads or GFP-trap beads, respectively. B and E, Copurification of BIR2-GFP (B) and BIR3-GFP (E) and PLDγ1-myc precipitated with GFP-trap beads. C and F, Copurification of BIR2-GFP (C) and BIR3-GFP (F) and PLDγ1-HA precipitated with GFP-trap beads. Numbers in the images indicate quantification of signal intensities. All experiments were repeated at least three times with similar results.
PLDγ1 Is Able to Associate with All BIR Proteins
We next investigated whether PLDγ1 would specifically colocalize and interact with BIR2 and BIR3 or also with other BIR family members. Therefore, we transiently coexpressed PLDγ1-GFP together with BIR1, BIR2, BIR3, or BIR4, each as C-terminal CFP fusion proteins in N. benthamiana. Using confocal microscopy, we could detect the CFP signal of all BIR proteins exclusively at the PM, largely overlapping with the PLDγ1-GFP signal (Supplemental Fig. S8). Moreover, we used N. benthamiana protein extracts from leaves coexpressing PLDγ1-myc with each BIR protein fused to a C-terminal yellow fluorescent protein (YFP)-tag for interaction studies with GFP-affinity beads. PLDγ1-myc could be copurified with any BIR protein used; however, only the interaction of PLDγ1-myc with BIR2-YFP and BIR4-YFP, respectively, could be enhanced by flg22 treatment (Supplemental Fig. S9). This result indicates that PLDγ1 is able to associate with any BIR protein when transiently expressed in N. benthamiana, but complex formation with BIR2 and possibly BIR4 is affected by flg22 stimulation.
PLDγ1/BIR-Association Is Independent of N-Terminal Myristoylation or Catalytic Activity of PLDγ1
PLDγ is localized at the PM, and it was suggested that PM attachment is due to N-terminal myristoylation (Qin et al., 1997; Fan et al., 1999). We next examined whether PLDγ1 protein localization and association with BIR2 and BIR3 is dependent on the putative N-myristoylation motif or the catalytic activity of PLDγ1. To this end we generated corresponding mutants in the putative N-myristoylation motif (PLDγ1ΔMyr) or the catalytic triad (PLDγ1ΔHKD) and first tested localization of the corresponding GFP-fusion proteins. Unexpectedly, PLDγ1ΔMyr-GFP was still mainly found at the PM and this localization was not affected by coexpression of BIR2-myc or BIR3-myc (Supplemental Fig. S10A). Likewise, PLDγ1ΔHKD-GFP localized to the PM, although overall accumulation of this catalytically inactive PLDγ1 version was always much weaker, and sometimes nearly undetectable, compared to the unmutated PLDγ1-GFP or the PLDγ1ΔMyr-GFP proteins (Supplemental Fig. S10). Using ultracentrifugation, we enriched membrane fractions and could indeed find both PLDγ1-GFP and PLDγ1ΔMyr-GFP predominantly in the membrane fraction, where BIR2-myc and BIR3-myc were also exclusively detected, and to a lesser extent in the fraction of soluble proteins, where MPK6 as a control was mainly present (Supplemental Fig. S10B). Thus, neither the mutation in the putative N-myristoylation motif nor that in the catalytic site impinges on PLDγ1-GFP localization when transiently expressed in N. benthamiana.
For testing the impact of PLDγ1-mutations on the association with BIR2 or BIR3, we performed coimmunoprecipitation experiments with protein extracts from N. benthamiana leaves coexpressing the different PLDγ1 versions with a C-terminal GFP-tag together with BIR2-myc or BIR3-myc. Both myc-tagged BIR2 and BIR3 could be found in GFP-affinity-purified samples from leaves expressing unmutated PLDγ1-GFP, PLDγ1ΔMyr-GFP, or PLDγ1ΔHKD-GFP, respectively (Supplemental Fig. S11). Hence, in accordance with the mutated PLDγ1-GFP versions being localized to the PM, they were also able to interact with BIR2 and BIR3. In conclusion, the putative N-myristoylation motif is not mandatory for PLDγ1-GFP attachment to the PM and PLDγ1 catalytic activity seems dispensable for complex formation with BIR2 and BIR3.
DISCUSSION
PLDs are lipid-hydrolyzing enzymes with multiple functions during plant abiotic and biotic stress responses as well as cellular and developmental processes (Bargmann and Munnik, 2006; Zhao, 2015; Hong et al., 2016; Takáč et al., 2019). A direct product of the hydrolytic activity of PLDs toward PM lipids is PA, a typical lipid second messenger that can bind to and modulate the activity of proteins such as kinases and phosphatases, 14-3-3 proteins, transcription factors, or proteins involved in calcium signaling, oxidative burst, endocytosis, or cytoskeleton assembly (for review, see Munnik, 2001; Wang, 2004; Zhao, 2015; Pokotylo et al., 2018; Takáč et al., 2019). However, in our study, neither basal nor stimulus-induced PA levels were changed in pldγ1-1 mutants compared to wild-type plants, suggesting that this PLD isoform might not be a major PA-producing enzyme. Indeed, in addition to PLD, PA can also be produced via the combined action of PLC and diacylglycerol kinase (Arisz et al., 2009). At least in tomato cell suspensions, this is the major pathway for flg22-triggered PA production (van der Luit et al., 2000), supporting our observation that the slight flg22-induced PA accumulation was not affected by the pldγ1 mutation (Fig. 4). Nevertheless, PLDs might have a more significant role in PA production after stimulation with other MAMPs such as fungal xylanase (van der Luit et al., 2000) or in other pathways such as effector-triggered immunity (de Jong et al., 2004; Andersson et al., 2006). Moreover, Nicotiana tabacum PLDs of the subfamilies β and γ were implicated in the release of the endogenous, bioactive lipids N-acylethanolamines (NAEs) from membrane phospholipids (Chapman et al., 1998; Pappan et al., 1998). NAEs can be released upon stimulation with the fungal MAMP xylanase (Chapman et al., 1998) and were shown to inhibit ABA-mediated stomatal closure (Austin-Brown and Chapman, 2002). As phytopathogens often use stomata as entry points (Melotto et al., 2006) it can be speculated that PLD/NAE-mediated interference with stomatal closure upon pathogen attack might impact on pathogen resistance, as we observed in the pldγ1 mutant.
PLDγ1 Is Involved in Resistance to Certain Biotrophic and Necrotrophic Pathogens
We observed that pldγ1 mutants are more resistant toward infection with both biotrophic bacterial and necrotrophic fungal pathogens. This is rather intriguing, as signaling pathways mediating resistance toward biotrophic and necrotrophic pathogens are usually antagonistically regulated by phytohormones such as SA and JA (Pieterse et al., 2012). However, pldγ1 mutants did not exhibit altered SA or JA levels when compared with wild-type plants, and corresponding marker gene expression was also similar to that observed in wild-type plants. Thus, other cellular factors are most likely responsible for the increased resistance of pldγ1 mutant plants. Notably, PLDγ1 has also been implicated in ETI: PLDγ1 gene expression is up-regulated upon infection with the avirulent bacterial pathogen Pto DC3000 (avrRpm1; de Torres Zabela et al., 2002) and pldγ1 mutants display enhanced electrolyte leakage upon infection with this avirulent bacterium, however, without affecting resistance (Johansson et al., 2014). Likewise, depletion of PLDγ1 did not impinge on resistance toward the powdery mildew fungi B. graminis f. sp. hordei and E. pisi, whereas genetic inactivation of another PLD, PLDδ, resulted in decreased resistance to these nonadapted pathogens (Pinosa et al., 2013; Johansson et al., 2014). Mutants for PLDβ1 are also more susceptible to fungal infection but at the same time show an increased resistance to bacterial infection (Zhao et al., 2013), indicating that the different PLD isoforms can have both positive and negative regulatory functions in resistance to phytopathogens. It can be envisaged that the function of a certain PLD isoform is dependent on its organ-specific or cellular distribution, its requirement for certain cofactors (for instance calcium), and/or its interaction with other cellular components including other proteins.
PLDs might have a function in receptor localization and stability
Apart from producing PA or NAEs as signaling molecules, phospholipases are also involved in remodeling membrane lipids, which affects the biophysical properties and hence membrane functionality (Platre et al., 2018; Mamode Cassim et al., 2019).
A prerequisite for PLDs shaping membrane lipids is their localization to the respective membrane. We found that PLDγ1 is localized to the PM and it was previously demonstrated that PLDγ1 relocates to the PM upon activation of the intracellular immune receptor RPS2 during effector-triggered immunity (Elmore et al., 2012). Likewise, PLDδ was shown to accumulate at the entry site of fungal infection during B. graminis f. sp hordei infection and PLDδ accumulation at the PM could be enhanced by chitin treatment (Xing et al., 2019). Intriguingly, mutation of the putative N-myristoylation site did not affect PLDγ1 attachment to the PM (Supplemental Fig. S10), and PLDδ does not contain such a protein motif. Apparently, PLDs can attach to membranes without having such a lipid modification. Alternative mechanisms of PLD attachment to membranes might, for instance, simply be their high binding affinity to membrane lipids or their interaction with membrane-localized proteins.
The PM is organized in highly dynamic subcompartments, so-called micro- or nanodomains, depending on size and shape, which allow the accommodation of specific protein clusters (Malinsky et al., 2013; Ott, 2017). Protein clustering within the PM has been demonstrated for the immune receptor FLS2 (Bücherl et al., 2017; McKenna et al., 2019) as well as its coreceptor BAK1 (Hutten et al., 2017). Importantly, the stability of FLS2 protein clusters depends on the lipid composition of the surrounding membrane patch (Bücherl et al., 2017). Likewise, the mammalian receptor Tyr kinase EPIDERMAL GROWTH FACTOR RECEPTOR (EGFR) is found in PM nanodomains, and formation of these protein clusters is dependent on the integrity of PM lipid composition (Gao et al., 2015). Moreover, both FLS2 and EGFR are rapidly internalized upon ligand binding (Haigler et al., 1978; Robatzek et al., 2006). EGFR endocytosis and subsequent degradation depend on PLD activity (Shen et al., 2001), linking PLD action with receptor internalization (Donaldson, 2009). The diffusion rate of FLS2 within the PM, as well as FLS2 endocytosis, is dependent on an intact cytoskeleton assembly (Robatzek et al., 2006; McKenna et al., 2019). Cytoskeletal dynamics in turn are altered by PLDs, such as PLDβ1 and PLDδ, which bind to actin and microtubules, and PLD enzymatic activity is regulated by this protein-protein interaction (Munnik and Musgrave, 2001; Dhonukshe et al., 2003; Kusner et al., 2003; Pleskot et al., 2010; Zhao, 2015). Hence, PLD-mediated membrane lipid remodelling or PLD-cytoskeleton interactions might impinge on FLS2 (or general immune receptor) protein distribution or stability at the PM or receptor internalization upon activation. Future work will be needed to assess whether PLDγ1 also has a function in such a signaling pathway.
PLDγ1 Is a Complex Partner for the Negative Regulator BIR2
PLD proteins themselves can directly interact with a range of proteins, indicating additional PLD functions apart from lipid hydrolysis. One example is the aforementioned cytoskeleton components. PLDα1, as another example, interacts with the heterotrimeric G-protein GPA1 and promotes the exchange of GTP to GDP, thereby mediating ABA-induced inhibition of stomatal opening (Mishra et al., 2006). PLDα1 also complexes with the Cys-rich receptor-like kinase 2 (CRK2) and MPK3 (Hunter et al., 2019; Vadovič et al., 2019), indicating multiple sites of action for PLDα1 in Arabidopsis salt stress and ABA signaling. In plant immunity, PLDδ physically interacts with the immune receptor RESISTANCE TO PSEUDOMONAS SYRINGAE PV. MACULICOLA1 (RPM1) and negatively regulates its function (Yuan et al., 2019).
We found PLDγ1 in complex with the LRR-RLKs BIR2 and BIR3, which function as negative regulators of immune receptors (Halter et al., 2014; Imkampe et al., 2017). Interestingly, we observed differences in PLDγ1 complex formation: whereas flg22 treatment resulted in further BIR2 recruitment, the PLDγ1/BIR3 complex remained largely unaffected upon elicitation (Figs. 6 and 7). Notably, although both BIR2 and BIR3 interact with BAK1 to hinder this coreceptor from joining receptor complexes without stimulus, several other behavioral differences for BIR2 and BIR3 have been reported (Halter et al., 2014; Imkampe et al., 2017): (1) BIR3, but not BIR2, directly interacts with ligand-binding receptor kinases such as FLS2 or BRI1; (2) BIR3, but not BIR2, is a negative regulator of BRI1-mediated brassinosteroid-perception; (3) bir2, but not bir3 mutants, show increased resistance toward bacterial infection; and (4) BIR2 is the better phosphorylation target of BAK1. This suggests that BIR3, although having several functions in common with BIR2, has additional distinct functions and might act at least partially via a different molecular mechanism than BIR2. When transiently over-expressed in N. benthamiana, PLDγ1 was also able to associate with BIR1 and BIR4 (Supplemental Fig. S9). BIR4 is thought to be recently derived from BIR3 by gene duplication (Halter et al., 2014); it is hardly expressed in mature tissue (www.bar.utoronto.ca), and so far, evidence for a BIR4 function in plant immunity is lacking. Mutation of BIR1 results in plants with an extreme stunted growth phenotype, extensive cell death, and constitutive defense responses (Gao et al., 2009). However, pldγ1 mutants do not display any defects in growth and do not have a constitutive activation of defense responses. Thus, we consider BIR1 and BIR4 unlikely to function in the same signaling pathway as PLDγ1. However, the pldγ1 mutant phenotype with respect to bacterial resistance and increased ROS accumulation upon flg22 stimulation mirrored the behavior of bir2, but not bir3, mutants, making BIR2 the more likely complex partner for PLDγ1. Notably, bir2 mutants display increased basal SA levels and slightly stunted growth (Halter et al., 2014) and did not show altered resistance toward infection with Botrytis (Fig. 3), phenotypes that are different from the pldγ1 mutant. Hence, PLDγ1 and BIR2 have overlapping roles, but also additional roles in separate pathways. As we did not observe any association of PLDγ1 with BAK1 or FLS2 (Fig. 6; Supplemental Fig. S7), and as PLDγ1/BIR2 interaction was enhanced by flg22 (Figs. 6 and 7), PLDγ1 is likely recruited to BIR2 after its release from BAK1.
Whether PLDγ1 directly physically interacts with BIR2 remains to be shown. However, PLDγ1 enzymatic activity does not seem to be required for PLDγ1/BIR2 association (Supplemental Fig. S11). We propose a model in which PLDγ1 and BIR2 are present in the same nanodomains in the plant PM but do not come into direct contact with each other in the resting state. Upon stimulation with flg22 (or possibly other MAMPs), BIR2 dissociates from BAK1 (which then joins the FLS2/flg22 complex) and comes into closer proximity to PLDγ1, either directly or indirectly interacting with PLDγ1. PLDγ1 might then be required for BIR2 stabilization, reassociation of BIR2 and BAK1, or recycling/relocation of these or other membrane proteins. Here, PLDγ1 might be required for membrane shaping for appropriate nanoclustering of immune receptor protein complexes at the PM. Future work will now be needed to elucidate the exact mechanism and function of PLDγ1 complex formation with BIR2.
CONCLUSION
PLDγ1 is a negative regulator of plant immune responses. We hypothesize that PLDs shape the lipid composition of membranes to allow the formation of distinct PM nanodomains to aid in immune receptor relocation and/or complex formation with positive and negative regulators (such as BIR2). Changes in lipid composition or PLD hydrolytic activity might additionally influence the association of cytoplasmic proteins with the PM or the dissociation of membrane-tethered proteins from the PM upon signaling (e.g. receptor-like cytoplasmic kinases). PLD actions on PM lipids are most likely very local (within micro- or nanodomains) and thus do not necessarily result in measurable alterations in PA levels (we did not detect any changes in PA amounts in pldγ1 mutants). Hence, in addition to being merely producers of PA, PLDs, including PLDγ1, might have novel functions in regulating cellular responses via cluster formation with immunity-related proteins.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants were grown on soil or one-half strength Murashige and Skoog medium as described (Brock et al., 2010). Plants were grown in climate chambers under short-day conditions (8 h light/16 h dark, 150 μmol cm−2 s−1 white fluorescent light, 40% to 60% humidity, 22°C). Ecotype Columbia of Arabidopsis (Col-0) was the background for all mutants used in this study, and genotyping was performed with primers listed in Supplemental Table S1. Mutant lines pldγ1-1 (Salk_066687C), pldγ2 (Salk_078226), pldγ3 (Salk_084335), bir2-1 (GK-793F12), and bir3-2 (Salk_116632) have been previously described (Pinosa et al., 2013; Halter et al., 2014; Imkampe et al., 2017) and are listed in Supplemental Table S1, together with pldγ1-2 (GK-264A03) and T-DNA insertion lines selected for At1g77460 and At2g31130. Nicotiana benthamiana plants were grown in the greenhouse (16 h light, 22°C).
Generation of Constructs and Transgenic Plants
For coimmunoprecipitation experiments, all C-terminal epitope-tag fusion constructs were made by cloning full-length coding sequences as PCR fragments first into pCR8/GW/TOPO and afterward into vectors pGWB5 (GFP tag), pGWB14 (3× hemagglutinin [HA]), pGWB17 (4× myc), or pK7YWG2.0 (YFP tag; Karimi et al., 2005; Nakagawa et al., 2007) using the Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). For colocalization, full-length coding sequences were recombined into vectors pB7RWG2.0 (BIR2-RFP), pH7CWG2.0 (BIR1-4-CFP), or pGWB5 (PLDγ1-GFP; Karimi et al., 2005; Nakagawa et al., 2007). Constructs for BIR1-4 were previously described (Halter et al., 2014).
Mutations of PLDγ1 in the putative N-myristoylation site (MGGGSS to MAAGAS from amino acids 13–18) and the catalytic triad (HQKTVIVD to AQATVIVA from amino acids 369–376) were introduced into synthetic gBlocks (Integrated DNA Technologies) that were subcloned into pCR8/GW/TOPO before cloning into pGWB5 (GFP tag).
Complementation lines were generated by the Agrobacterium-mediated floral dip method (Clough and Bent, 1998) of pldγ1-1 mutant lines using the full-length coding sequence of PLDγ1 fused to a C-terminal GFP tag in the binary vector pGWB5 (Nakagawa et al., 2007). Restored gene transcription in transformed seedlings was verified by reverse-transcription quantitative PCR (RT-qPCR) and western blot analyses, and plants were used for experiments in the T3 and T4 generation.
Plant Infections
Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) was grown overnight in King’s B medium, centrifuged, washed, and diluted in 10 mm MgCl2 to a density of 104 colony-forming units (cfu) mL−1. Bacteria were pressure-infiltrated into leaves of 5- to 6-week-old Arabidopsis plants. Leaves were harvested at days 0 and 3, surface sterilized in 70% (v/v) ethanol, and washed in double-distilled water for 1 min each. Two leaf discs (5-mm diameter) per leaf were ground in 200 μL of a 10 mm MgCl2 solution, diluted serially, and plated on Luria-Bertani plates containing rifampicin and cycloheximide. After 2 d of incubation at 28°C, colony-forming units were counted.
Spores of Botrytis cinerea isolate B0-10 were diluted to a final concentration of 5 × 106 spores mL−1 in potato dextrose broth and a 5-μL drop per leaf was used to inoculate 5- to 6-week-old Arabidopsis plants. Lesion sizes were determined after 2 d using the Photoshop CS5 Lasso tool. Selected pixels were counted and the lesion size in square centimeters was calculated using a 0.5 cm2 standard. Fungal biomass quantification was performed by extracting DNA from infected leaf material. Three infected leaves per plant were frozen in liquid nitrogen using screw-cap vials containing a ceramic bead mix (2-mm and 0.5-mm diameter). Tissue was homogenized using the Precellys 24 tissue homogenizer (Bertin Instruments) for 2 × 30 s at room temperature. After addition of 300 μL extraction buffer (2.5 mm LiCl, 50 mm Tris-HCl [pH 8], 62.5 mm EDTA, and 4% [v/v] Triton X100), samples were incubated for 10 min at room temperature, followed by purification using phenol-chloroform extraction. Genomic DNA was finally dissolved in nuclease-free water and subjected to RT-qPCR using primers listed in Supplemental Table S2.
Detection of Immune Responses
The detection of ROS in leaf pieces of 5- to 6-week-old Arabidopsis plants was measured using 96-well plates containing a final volume of 100 μL with two leaf pieces, 5 µg ml−1 horseradish peroxidase (Applichem), and 20 μm luminol l-012 (Wako Chemicals) and the elicitor to be tested per well as described (Albert et al., 2010). Light emission was measured as relative light units in a 96-well luminometer (Mithras LB 940; Berthold Technologies). Immunoblot analyses using the antiphospho p44/42 (Erk1/2) MAP kinase antibody (Cell Signaling Technology) was done with 10-d-old seedlings as described (Brock et al., 2010).
RNA Extraction and RT-qPCR
Total RNA from 100 mg leaf tissue (5- to 6-week-old Arabidopsis plants) or 10 9-d-old seedlings was purified using the NucleoSpin RNA Plus kit (Macherey-Nagel). First-strand complementary DNA synthesis was performed from 1 μg of total RNA using RevertAidTM MuLV reverse transcriptase (Thermo Scientific). RT-qPCR reactions and measurements were performed with the iQ5 Multicolor real-time PCR detection system (Bio-Rad) using the Maxima SYBR Green/Fluorescein qPCR Master Mix (Thermo Scientific) and gene specific primers listed in Supplemental Table S2. Relative gene expression was calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001) using the housekeeping gene EF-1α.
Determination of PA, SA, and JA
Analysis of phospholipids was essentially done as previously described (Munnik and Zarza, 2013). In brief, 5-d-old seedlings were labeled overnight with 32P-orthophosphate and treated the next day with buffer with or without flg22 or NaCl. Lipids were subsequently extracted and analyzed by TLC using an ethyl acetate solvent system composed of the organic upper phase of an ethyl acetate/iso-octane/acetic acid/water mixture of 13:2:3:10 (v/v; Munnik and Laxalt, 2013). Radio-labeled phospholipids were visualized and quantified using a PhosphoImager (GE Healthcare) and the program QuantityOne (Bio-Rad).
For SA and JA quantification, 200 mg leaf material of 8-week-old Arabidopsis plants was frozen. The amounts of SA and JA were measured by gas chromatography coupled to mass spectrometry as previously described (Wan et al., 2018), but using a Shimadzu TQ8040 triple-quad mass spectrometer (Shimadzu Cooperation) with splitless injection mode and a SH-Rxi-17SIL-MS column (30 m, 0.25-mm internal diameter, 0.25-μm film; Shimadzu Cooperation). The gas chromatmography oven temperature was held initially at 70°C, then ramped at 25°C min−1 to 280°C and held for 7 min, then ramped at 10°C min−1 to 300°C and afterward held for an additional 3 min at 300°C. Helium was used as the carrier gas, with a flow rate of 0.86 mL min−1. The mass spectrometer was operated in electron impact ionization and multiple-reaction monitoring mode.
Protein Interaction and Localization Studies
For transient expression of epitope-tagged proteins, plasmid constructs were transformed into Agrobacterium tumefaciens strain GV3101 and overnight cultures were used for leaf infiltration of N. benthamiana at OD600 of 0.5 as described (Brock et al., 2010). Leaf material was harvested after 3 d. For protein extraction, ground leaf tissue (200 mg from N. benthamiana plants or 1 g from Arabidopsis plants) was resuspended in 1.6 mL solubilization buffer (25 mm Tris-HCl [pH 8.0], 150 mm NaCl, 1% [v/v] NP40, 0.5% [w/v] deoxycholate (DOC), 2 mm dithiothreitol, and 1 tablet of cOmplete ULTRA Tablets, Mini, EASYpack” [Roche] per 10 mL). Proteins were solubilized for 1 h at 4°C with end-over-end shaking and the cell debris was removed by two times centrifugation at 20,000g for 10 min at 4°C. For immunoprecipitation, each supernatant was incubated with prewashed and solubilization buffer-equilibrated affinity beads (GFP-trap or myc-trap; ChromoTek) for 1 h at 4°C with head-over-head rotation at 6 rpm. Beads were washed twice with solubilization buffer and twice with washing buffer (25 mm Tris-HCl [pH 8.0], 150 mm NaCl, and 2 mm dithiothreitol) before adding SDS-PAGE loading dye.
For the enrichment of membranes, 200 mg of ground N. benthamiana leaf tissue was resuspended in 500 µL of solubilization buffer without NP40 and DOC and centrifuged at 100,000g for 1 h at 4°C. The supernatant (soluble proteins) was removed and SDS-PAGE loading dye was added. The pellet was resuspended in solubilization buffer including NP40 and DOC. After 1 h at 4°C with end-over-end shaking, the cell debris was removed by centrifugation at 100,000g for 1 h at 4°C. The supernatant (membrane proteins) was removed and SDS-PAGE loading dye was added.
Samples were analyzed by immunoblotting using the following antibody dilutions: anti-GFP (1:10,000, from goat; Sicgen), anti-myc (1:5,000, from rabbit; Sigma), anti-HA (1:5,000, from mouse; Sigma), anti-BIR2 (1:500, from rabbit; Halter et al., 2014), anti-BAK1 (1:5000, from rabbit; Agrisera), anti-FLS2 (1:5000, from rabbit; Agrisera), anti-MPK6 (1:5000, from rabbit; Sigma), anti-rabbit IgG- -conjugate (1:10,000, from goat; Sigma), anti-goat IgG-HRP-conjugate, and anti-mouse IgG-HRP-conjugate (each 1:10,000, from rabbit; Sigma).
Fluorescence images were acquired with the confocal laser scanning microscope TCS-SP8 from Leica (HC PL APO CS2 63×/1.20 water objective; Leica Application Suite X software) or Zeiss LSM 880 (C-Apochromat 40×/1.2 W objective, Corr FCS M27 and Plan-Apochromat 10×/0.45 M27; Zeiss Efficient Navigation Blue software) with the following excitation/emission wavelength: enhanced GFP, 488 nm/509 nm; enhanced CFP, 434 nm/477 nm; and RFP, 555 nm/584 nm. Overlays and contrast/brightness adjustments of images were performed with Adobe Photoshop CS5 software.
Whole Genome Sequencing
Genomic DNA of the pldγ1-1 was isolated using the DNeasy Plant Kit (Qiagen). The library was prepared using a modified Nextera protocol (Karasov et al., 2018) and was sequenced using a HiSeq3000 platform (Illumina). Forty-fold genome coverage was obtained. The raw reads were trimmed using SKEWER (v0.2.2) software (minimum quality 20 and minimum length 30; Jiang et al., 2014) and the processed reads were mapped against the expected pROK2 insertion using the BWA MEM (v0.7.15) algorithm (Li and Durbin, 2009). Downstream processing of the files was performed using SAMTOOLS (v1.3.1; Li et al., 2009) and PCR duplicates were removed using the PICARD (v2.2.1) algorithm (http://broadinstitute.github.io/picard). SAMTOOLS (v1.3.1) was used for subsetting reads that were mapped at the first or last 100 nucleotides of the pROK2 insertion (Li et al., 2009). These reads were mapped to the Arabidopsis TAIR10 reference genome using the BWA MEM (v0.7.15) algorithm (Li and Durbin, 2009). As done previously, the newly generated file was processed using SAMTOOLS (v1.3.1; Li et al., 2009). The file was evaluated for multiple integration points of the transgene and visualized using Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013).
Statistical Analysis
To test for significant differences in different experiments, the distribution of the data was first determined. For parametric data, one-way ANOVA followed by Tukey-Kramer or Dunnet’s multiple comparison analysis at a probability level of P < 0.05 was applied. For nonparametric data, Kruskal-Wallis one-way ANOVA followed by an each-pair comparison Wilcoxon rank-sum test with a probability level of P < 0.05 was used. All data analyses were performed using JMP 14 software.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource under the following accession numbers: At4g11850 (PLDγ1), At4g11830 (PLDγ2), At4g11840 (PLDγ3), At5g48380 (BIR1), At3g28450 (BIR2), At1g27190 (BIR3), At1g69990 (BIR4), At4g33430 (BAK1), At5g46330 (FLS2), and At2g43790 (MPK6). Accession numbers for all other genes/proteins used in this work are listed in Supplemental Tables S1 and S2.
Supplemental Material
The following supplemental materials are available.
Supplemental Figure S1. pldγ1 mutants are more resistant to pathogen infection compared to pldγ2 and pldγ3 mutants.
Supplemental Figure S2. Characterization of a second T-DNA insertion line for PLDγ1.
Supplemental Figure S3. Characterization of secondary mutations in the pldγ1-1 mutant.
Supplemental Figure S4. Characterization of pldγ1-1 complementation lines.
Supplemental Figure S5. Flg22-induced MAP kinase activation is not altered in pldγ1 mutants or complementation lines.
Supplemental Figure S6. PLDγ1 depletion does not affect SA and JA-signaling.
Supplemental Figure S7. PLDγ1 does not interact with FLS2 and BAK1.
Supplemental Figure S8. PLDγ1 colocalizes with all BIR proteins.
Supplemental Figure S9. PLDγ1 can be found in association with all BIR proteins.
Supplemental Figure S10. PLDγ1 mutants in the N-myristoylation motif or the catalytic site are still located in the PM.
Supplemental Figure S11. PLDγ1 mutants in catalytic triad or N-myristoylation motif still interact with BIR2 and BIR3.
Supplemental Table S1. Primers used for genotyping T-DNA insertion lines.
Supplemental Table S2. Primers used in RT-qPCR analyses.
Acknowledgments
We are grateful to Birgit Löffelhardt for excellent technical support; Mats Andersson for pldγ mutants; Birgit Kemmerling for BIR1-4 constructs, bir2 and bir3 seeds, and the antibodies raised against BIR2; Katja Fröhlich for help with statistical analysis; Sarina Schulze and Heribert Hirt for helpful discussions; Detlef Weigel for critical comments on the manuscript; and Thorsten Nürnberger for critical comments and helpful discussions about the project.
Footnotes
This work was supported by the University of Tübingen Graduate College “Of Plants and Men”, the Deutsche Forschungsgemeinschaft (grant no. SFB 766), the Max Planck Society (E.S.), and the Netherlands Organisation for Scientific Research (grant no. 867.15.020 to T.M.).
Articles can be viewed without a subscription.
References
- Albert M, Jehle AK, Mueller K, Eisele C, Lipschis M, Felix G(2010) Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J Biol Chem 285: 19035–19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M(2006) Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47: 947–959 [DOI] [PubMed] [Google Scholar]
- Arisz SA, Testerink C, Munnik T(2009) Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 1791: 869–875 [DOI] [PubMed] [Google Scholar]
- Austin-Brown SL, Chapman KD(2002) Inhibition of phospholipase Dα by N-acylethanolamines. Plant Physiol 129: 1892–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Laxalt AM, Riet BT, Schouten E, van Leeuwen W, Dekker HL, de Koster CG, Haring MA, Munnik T(2006) LePLDβ1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J 45: 358–368 [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Laxalt AM, ter Riet B, van Schooten B, Merquiol E, Testerink C, Haring MA, Bartels D, Munnik T(2009) Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol 50: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Munnik T(2006) The role of phospholipase D in plant stress responses. Curr Opin Plant Biol 9: 515–522 [DOI] [PubMed] [Google Scholar]
- Bethke G, Pecher P, Eschen-Lippold L, Tsuda K, Katagiri F, Glazebrook J, Scheel D, Lee J(2012) Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol Plant Microbe Interact 25: 471–480 [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H(2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G(2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G, Lee J, Nürnberger T, Gust AA(2010) The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 153: 1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C(2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Tripathy S, Venables B, Desouza AD(1998) N-Acylethanolamines: Formation and molecular composition of a new class of plant lipids. Plant Physiol 116: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G(2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T(2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C(2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- de Jong CF, Laxalt AM, Bargmann BO, de Wit PJ, Joosten MH, Munnik T(2004) Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J 39: 1–12 [DOI] [PubMed] [Google Scholar]
- de Torres Zabela M, Fernandez-Delmond I, Niittyla T, Sanchez P, Grant M(2002) Differential expression of genes encoding Arabidopsis phospholipases after challenge with virulent or avirulent Pseudomonas isolates. Mol Plant Microbe Interact 15: 808–816 [DOI] [PubMed] [Google Scholar]
- den Hartog M, Verhoef N, Munnik T(2003) Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol 132: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TW, Munnik T(2003) Phospholipase d activation correlates with microtubule reorganization in living plant cells. Plant Cell 15: 2666–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG.(2009) Phospholipase D in endocytosis and endosomal recycling pathways. Biochim Biophys Acta 1791: 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, Coaker G(2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics 11: M111 014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zheng S, Cui D, Wang X(1999) Subcellular distribution and tissue expression of phospholipase Dα, Dβ, and Dγ in Arabidopsis. Plant Physiol 119: 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang Y, Cai M, Pan Y, Xu H, Jiang J, Ji H, Wang H(2015) Mechanistic insights into EGFR membrane clustering revealed by super-resolution imaging. Nanoscale 7: 2511–2519 [DOI] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Haigler H, Ash JF, Singer SJ, Cohen S(1978) Visualization by fluorescence of the binding and internalization of epidermal growth factor in human carcinoma cells A-431. Proc Natl Acad Sci USA 75: 3317–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T, Imkampe J, Mazzotta S, Wierzba M, Postel S, Bücherl C, Kiefer C, Stahl M, Chinchilla D, Wang X, et al. (2014) The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr Biol 24: 134–143 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP(2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, Wang X(2009) Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J 58: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Pan X, Welti R, Wang X(2008) Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhao J, Guo L, Kim SC, Deng X, Wang G, Zhang G, Li M, Wang X(2016) Plant phospholipases D and C and their diverse functions in stress responses. Prog Lipid Res 62: 55–74 [DOI] [PubMed] [Google Scholar]
- Hunter K, Kimura S, Rokka A, Tran HC, Toyota M, Kukkonen JP, Wrzaczek M(2019) CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol 180: 2004–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten SJ, Hamers DS, Aan den Toorn M, van Esse W, Nolles A, Bücherl CA, de Vries SC, Hohlbein J, Borst JW(2017) Visualization of BRI1 and SERK3/BAK1 nanoclusters in Arabidopsis roots. PLoS One 12: e0169905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, Zhu S(2014) Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ON, Fahlberg P, Karimi E, Nilsson AK, Ellerström M, Andersson MX(2014) Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpm1 in Arabidopsis thaliana. Front Plant Sci 5: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov TL, Almario J, Friedemann C, Ding W, Giolai M, Heavens D, Kersten S, Lundberg DS, Neumann M, Regalado J, et al. (2018) Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe 24: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P(2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Ku YS, Sintaha M, Cheung MY, Lam HM(2018) Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 19: 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusner DJ, Barton JA, Qin C, Wang X, Iyer SS(2003) Evolutionary conservation of physical and functional interactions between phospholipase D and actin. Arch Biochem Biophys 412: 231–241 [DOI] [PubMed] [Google Scholar]
- Laxalt AM, Munnik T(2002) Phospholipid signalling in plant defence. Curr Opin Plant Biol 5: 332–338 [DOI] [PubMed] [Google Scholar]
- Laxalt AM, ter Riet B, Verdonk JC, Parigi L, Tameling WI, Vossen J, Haring M, Musgrave A, Munnik T(2001) Characterization of five tomato phospholipase D cDNAs: Rapid and specific expression of LePLDβ1 on elicitation with xylanase. Plant J 26: 237–247 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R(2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malinsky J, Opekarová M, Grossmann G, Tanner W(2013) Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol 64: 501–529 [DOI] [PubMed] [Google Scholar]
- Mamode Cassim A, Gouguet P, Gronnier J, Laurent N, Germain V, Grison M, Boutté Y, Gerbeau-Pissot P, Simon-Plas F, Mongrand S(2019) Plant lipids: Key players of plasma membrane organization and function. Prog Lipid Res 73: 1–27 [DOI] [PubMed] [Google Scholar]
- McKenna JF, Rolfe DJ, Webb SED, Tolmie AF, Botchway SW, Martin-Fernandez ML, Hawes C, Runions J(2019) The cell wall regulates dynamics and size of plasma-membrane nanodomains in Arabidopsis. Proc Natl Acad Sci USA 116: 12857–12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY(2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X(2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Munnik T.(2001) Phosphatidic acid: An emerging plant lipid second messenger. Trends Plant Sci 6: 227–233 [DOI] [PubMed] [Google Scholar]
- Munnik T, Laxalt AM(2013) Measuring PLD activity in vivo. Methods Mol Biol 1009: 219–231 [DOI] [PubMed] [Google Scholar]
- Munnik T, Musgrave A(2001) Phospholipid signaling in plants: Holding on to phospholipase D. Sci STKE 2001: pe42. [DOI] [PubMed] [Google Scholar]
- Munnik T, Zarza X(2013) Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol Biol 1009: 3–15 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T(2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Ott T.(2017) Membrane nanodomains and microdomains in plant-microbe interactions. Curr Opin Plant Biol 40: 82–88 [DOI] [PubMed] [Google Scholar]
- Pappan K, Austin-Brown S, Chapman KD, Wang X(1998) Substrate selectivities and lipid modulation of plant phospholipase Dα, -β, and -γ. Arch Biochem Biophys 353: 131–140 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC(2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pinosa F, Buhot N, Kwaaitaal M, Fahlberg P, Thordal-Christensen H, Ellerström M, Andersson MX(2013) Arabidopsis phospholipase Dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol 163: 896–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platre MP, Noack LC, Doumane M, Bayle V, Simon MLA, Maneta-Peyret L, Fouillen L, Stanislas T, Armengot L, Pejchar P, et al. (2018) A combinatorial lipid code shapes the electrostatic landscape of plant endomembranes. Dev Cell 45: 465–480 [DOI] [PubMed] [Google Scholar]
- Pleskot R, Potocký M, Pejchar P, Linek J, Bezvoda R, Martinec J, Valentová O, Novotná Z, Zárský V(2010) Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J 62: 494–507 [DOI] [PubMed] [Google Scholar]
- Pokotylo I, Kravets V, Martinec J, Ruelland E(2018) The phosphatidic acid paradox: Too many actions for one molecule class? Lessons from plants. Prog Lipid Res 71: 43–53 [DOI] [PubMed] [Google Scholar]
- Qin C, Wang X(2002) The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol 128: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Pappan K, Wang X(1997) Molecular heterogeneity of phospholipase D (PLD). Cloning of PLDγ and regulation of plant PLDγ, -β, and -α by polyphosphoinositides and calcium. J Biol Chem 272: 28267–28273 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T(2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD(2011) Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Loo EP, Yasuda S(2018) Pattern recognition receptors and signaling in plant-microbe interactions. Plant J 93: 592–613 [DOI] [PubMed] [Google Scholar]
- Sang Y, Cui D, Wang X(2001) Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol 126: 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Zipfel C(2011) Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol 14: 54–61 [DOI] [PubMed] [Google Scholar]
- Shen Y, Xu L, Foster DA(2001) Role for phospholipase D in receptor-mediated endocytosis. Mol Cell Biol 21: 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J(2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628 [DOI] [PubMed] [Google Scholar]
- Takáč T, Novák D, Šamaj J(2019) Recent advances in the cellular and developmental biology of phospholipases in plants. Front Plant Sci 10: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Munnik T(2011) Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot 62: 2349–2361 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP(2013) Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadovič P, Šamajová O, Takáč T, Novák D, Zapletalová V, Colcombet J, Šamaj J(2019) Biochemical and genetic interactions of phospholipase Dα1 and mitogen-activated protein kinase 3 affect Arabidopsis stress response. Front Plant Sci 10: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Luit AH, Piatti T, van Doorn A, Musgrave A, Felix G, Boller T, Munnik T(2000) Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol 123: 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan WL, Zhang L, Pruitt R, Zaidem M, Brugman R, Ma X, Krol E, Perraki A, Kilian J, Grossmann G, et al. (2018) Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol 221: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.(2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R(2006) Signaling functions of phosphatidic acid. Prog Lipid Res 45: 250–278 [DOI] [PubMed] [Google Scholar]
- Xing J, Li X, Wang X, Lv X, Wang L, Zhang L, Zhu Y, Shen Q, Baluška F, Šamaj J, et al. (2019) Secretion of phospholipase Dδ functions as a regulatory mechanism in plant innate immunity. Plant Cell 31: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S(2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W(2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, Shan L(2017) From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol 55: 109–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Wang Z, Huang J, Xuan H, Gao Z(2019) Phospholipidase Dδ negatively regulates the function of Resistance to Pseudomonas syringae pv. Maculicola 1 (RPM1). Front Plant Sci 9: 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X(2009) Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.(2015) Phospholipase D and phosphatidic acid in plant defence response: From protein-protein and lipid-protein interactions to hormone signalling. J Exp Bot 66: 1721–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Devaiah SP, Wang C, Li M, Welti R, Wang X(2013) Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytol 199: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G(2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T(2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]