Phosphomimetic NADH-DEPENDENT HYDROXYPYRUVATE REDUCTASE1 shows a modified cofactor preference towards NADPH.
Abstract
Photorespiration is an essential process in oxygenic photosynthetic organisms triggered by the oxygenase activity of Rubisco. In peroxisomes, photorespiratory HYDROXYPYRUVATE REDUCTASE1 (HPR1) catalyzes the conversion of hydroxypyruvate to glycerate together with the oxidation of a pyridine nucleotide cofactor. HPR1 regulation remains poorly understood; however, HPR1 phosphorylation at T335 has been reported. By comparing the kinetic properties of phosphomimetic (T335D), nonphosphorylatable (T335A), and wild-type recombinant Arabidopsis (Arabidopsis thaliana) HPR1, it was found that HPR1-T335D exhibits reduced NADH-dependent hydroxypyruvate reductase activity while showing improved NADPH-dependent activity. Complementation of the Arabidopsis hpr1-1 mutant by either wild-type HPR1 or HPR1-T335A fully complemented the photorespiratory growth phenotype of hpr1-1 in ambient air, whereas HPR1-T335D-containing hpr1-1 plants remained smaller and had lower photosynthetic CO2 assimilation rates. Metabolite analyses indicated that these phenotypes were associated with subtle perturbations in the photorespiratory cycle of HPR1-T335D-complemented hpr1-1 rosettes compared to all other HPR1-containing lines. Therefore, T335 phosphorylation may play a role in the regulation of HPR1 activity in planta, although it was not required for growth under ambient air controlled conditions. Furthermore, improved NADP-dependent HPR1 activities in peroxisomes could not compensate for the reduced NADH-dependent HPR1 activity.
The photorespiratory C-2 cycle is an essential pathway for normal plant growth and development in ambient air, representing the most important carbon metabolism process after photosynthesis (Bauwe et al., 2010; Peterhansel and Maurino, 2011). This is highlighted by the phenotypes exhibited by ambient air-grown mutants of photorespiratory proteins that include reduced growth, chlorosis, and even lethality. Whereas the severity of the phenotype depends on the affected gene, photorespiratory mutants can be rescued, or partially rescued, by high-CO2 growth conditions (Timm and Bauwe, 2013). The photorespiratory cycle involves eight core enzymes distributed in four subcellular compartments: chloroplasts, peroxisomes, mitochondria, and the cytosol (Bauwe et al., 2010). It is initiated in chloroplasts by the oxygenation reaction of Rubisco to produce one molecule of toxic 2-phosphoglycolate (2PG) and one molecule of 3-phosphoglycerate. The major roles of the photorespiratory cycle are to salvage carbon by recycling 2PG to 3-phosphoglycerate that can be used in the Calvin-Benson-Bassham cycle and at the same time to remove potentially toxic metabolites (Timm et al., 2016). However, the photorespiratory cycle also leads to the release of assimilated carbon and nitrogen as CO2 and ammonia that must be either reassimilated or lost to the atmosphere. Indeed, the important energetic cost of photorespiration and its competition with photosynthetic CO2 assimilation has a negative impact on crop yields and this is expected to become more important under changing climate conditions such as high temperature and drought (Peterhansel and Maurino, 2011; Walker et al., 2016). There have been several recent advances in improving crop growth by manipulating the photorespiratory cycle by expressing synthetic photorespiratory bypasses in chloroplasts (Nölke et al., 2014; Dalal et al., 2015; Shen et al., 2019; South et al., 2019).
The photorespiratory cycle interacts with several metabolic pathways and processes, such as photosynthesis in chloroplasts, respiration and C1 metabolism in mitochondria, cell redox homeostasis (H2O2) in peroxisomes, and nitrogen metabolism (Hodges et al., 2016). However, knowledge of the coordination of the photorespiratory cycle with these pathways and how individual enzymes are regulated is limited. Phosphoproteomics data indicate that protein phosphorylation could be a key regulatory component of the photorespiratory cycle (Hodges et al., 2013). Indeed, phosphopeptides have been associated with Arabidopsis (Arabidopsis thaliana) HYDROXYPYRUVATE REDUCTASE1 (HPR1; Aryal et al., 2012; PhosPhAt database, http://phosphat.uni-hohenheim.de/).
Hydroxypyruvate reductases catalyze the NADH/NADPH-dependent conversion of hydroxypyruvate (HP) to glycerate, and also glyoxylate to glycolate (Tolbert et al., 1970; Givan and Kleczkowski 1992; Timm et al., 2008, 2011). In Arabidopsis, three putative HPR-encoding genes have been identified: HPR1 (At1g68010), HPR2 (At1g79870), and HPR3 (At1g12550). Arabidopsis HPR1 is a 42-kD peroxisomal protein, whereas HPR2 (34 kD) is cytosolic and HPR3 (35 kD) is plastidial (Timm et al., 2011). A comparison of recombinant Arabidopsis HPR proteins indicated that HPR1 was the major NADH-dependent HPR. On the other hand, HPR2 appeared to be an NADPH-dependent HP/glyoxylate reductase and HPR3 an NADPH-dependent glyoxylate reductase. HPR2 and HPR3 showed a higher NADP-glyoxylate reductase activity compared to HPR1 (Timm et al., 2008, 2011).
HPR1 is a core enzyme of the photorespiratory cycle, since hpr1 mutants exhibit a retarded growth phenotype when grown in ambient air that is absent when grown in elevated CO2 (10,000 μL L−1; Timm et al., 2008). A dominant role for HPR1 in photorespiration was further shown by isotopic assays, where HP accumulation was 103-fold higher in hpr1 but only 3-fold higher in hpr2, compared with wild-type plants (Timm et al., 2008). The absence of HRP1 increased steady-state levels of several photorespiratory metabolites, including glycerate, glycolate, Gly, Ser, and HP (Timm et al., 2008). Based on the phenotypes of Arabidopsis hpr2 and hpr3 mutants, as well as double and triple hpr mutant lines, the cytosolic HPR2 and chloroplastic HPR3 isoforms appear to have a limited compensatory photorespiratory cycle bypass function that is mainly carried out by cytosolic HPR2 (Timm et al., 2011). A similar cytosolic HPR compensatory bypass has been suggested for a barley (Hordeum vulgare) HPR1 mutant (Murray et al., 1989) and rice (Oryza sativa) HPR RNAi lines (Ye et al., 2014). It has been reported that the cytosolic bypass does not seem to have a high enough capacity to allow the same photorespiratory carbon recycling efficiency, since hpr1 mutants show an increase in CO2 release per oxygenation, possibly from nonenzymatic decarboxylations (Cousins et al., 2011).
Although HPR1 has been identified as the major HPR-photorespiratory enzyme, little is known about how it is regulated. In order to rapidly respond to changing conditions, enzymes are often posttranslationally modified and it has been reported that pea HPR1 can be regulated by Tyr nitration (Corpas et al., 2013). In vitro, this modification led to a 65% inhibition of HPR activity, and site-directed mutagenesis showed that nitration at Y198 was responsible for the observed inhibition (Corpas et al., 2013). A major regulatory mechanism is protein phosphorylation and phosphoproteomic studies have indicated that Arabidopsis HPR1 can be phosphorylated at T335 (Aryal et al., 2012) and S229 (PhosPhAt database, http://phosphat.uni-hohenheim.de/).
In this study, the consequences of T335 phosphorylation on HPR1 enzymatic activities and kinetic properties were compared using purified recombinant wild-type, nonphosphorylatable (T335A) and phosphorylation-mimetic (T335D) HPR1 proteins. It was found that phosphorylation-mimetic HPR1-T335D exhibited an altered cofactor specificity leading to lower NADH-dependent and higher NADPH-dependent HPR activities compared to wild-type and HPR1-T335A. To evaluate the impact of these modified kinetic parameters in planta, the hpr1-1 mutant (Timm et al., 2008) was complemented with wild-type and phosphorylation-site mutated HPR1 proteins. When grown in ambient air, hpr1-1 plants expressing the phosphomimetic HPR1 form (Compl-T335D) had smaller rosettes and lower photosynthetic CO2 assimilation rates compared to the other HPR1-containing lines. Metabolic analyses suggested that the photorespiratory cycle was perturbed in the phosphomimetic HPR1 plants compared to the other complemented lines. Our data are discussed in terms of the modulation of HPR1 activities by the T335D mutation and how this impacts plant metabolism.
RESULTS
Recombinant Phosphorylation-Mimetic Arabidopsis HPR1-T335D Exhibits Altered Kinetic Parameters
In order to investigate the regulation of HPR1 by phosphorylation of T335, kinetic parameters of purified recombinant Arabidopsis HPR1 were measured and compared between His-tag-purified HPR1-wild-type, HPR1-T335A, and HPR1-T335D proteins (Supplemental Fig. S1). Since recombinant Arabidopsis HPR1 had been shown to catalyze both NADH- and NADPH-dependent reductase activities using either HP or glyoxylate as substrates (Timm et al., 2008), our three recombinant HPR1 proteins were tested with each substrate-cofactor combination to evaluate the effect of the nonphosphorylatable and phosphomimetic mutations on their Km and kcat (catalytic constant) properties when compared to HPR1-wild type (Tables 1 and 2). In agreement with Timm et al. (2008), recombinant HPR1-wild-type protein exhibited a preference for the NAD-HP reaction, as seen from the calculated kcat and Km values (Tables 1 and 2). Indeed, HPR1-wild type showed a 1.65-fold higher kcat for the NADH-HP reaction compared to the NADPH-HP combination, whereas this parameter was 4- and 475-fold higher when glyoxylate was used as a substrate in the presence of either NADH or NADPH, respectively (Tables 1 and 2). The Km values for substrates and cofactors of HPR1-wild type were also seen to differ according to the enzymatic reaction tested. There was a similar 8.2-fold difference in Km values for HP and cofactor when the NADH-HP and NADPH-HP activities were compared, with lower Km values observed for the NADH-HP combination. Cofactor influenced HP binding at the active site (as judged from the Km) since NADPH increased the Km HP when compared to NADH (Table 1). When glyoxylate was used as a substrate, the Km glyoxylate was not affected by cofactor; however, it was 100- and 12.6-fold higher compared to the Km HP for NADH-HP and NADPH-HP combinations, respectively. It was also found that Km NADH was lower than Km NADPH for the glyoxylate reductase activity of HPR1, whereas the difference between these two values was larger than that for the HPR activity (35- versus 8.2-fold).
Table 1. kcat and Km values of recombinant HPR1-wild-type, HPR1-T335A, and HPR1-T335D proteins in the presence of hydroxypyruvate and either NADH or NADPH.
Values represent means ± sd from three independent experiments. Statistical significance was determined by Student’s t test; values in bold are significantly different for the same recombinant protein under different reaction conditions (P < 0.05) and values marked by an asterisk are significantly different compared to HPR1-wild type under the same reaction condition (*P < 0.05 and **P < 0.01).
| Protein | HP:NADH | HP:NADPH | ||||||
|---|---|---|---|---|---|---|---|---|
| Hydroxypyruvate | NADH | Hydroxypyruvate | NADPH | |||||
| kcat | Km | kcat | Km | kcat | Km | kcat | Km | |
| s−1 | µm | s−1 | µm | s−1 | µm | s−1 | µm | |
| HPR1-wild type | 284 ± 21 | 173 ± 31 | 248 ± 9 | 18 ± 2.5 | 160 ± 13 | 1428 ± 117 | 162 ± 2 | 147 ± 5 |
| HPR1-T335D | 139 ± 26** | 167 ± 43 | 144 ± 12** | 28 ± 2.2* | 206 ± 13** | 995 ± 133* | 230 ± 5** | 102 ± 1* |
| HPR1-T335A | 253 ± 34 | 220 ± 72 | 218 ± 3 | 18 ± 4.5 | 150 ± 7 | 1579 ± 98 | 146 ± 10 | 135 ± 4 |
Table 2. kcat and Km values of recombinant HPR1-wild-type, HPR1-T335A, and HPR1-T335D proteins in the presence of glyoxylate and either NADH or NADPH.
Values are represented as means ± sd from three independent experiments. Statistical significance was determined by Student’s t test; values in bold are significantly different for the same recombinant protein under different reaction conditions (P < 0.05) and values marked by an asterisk are significantly different compared to HPR1-wild type under the same reaction condition (*P < 0.05 and **P < 0.01).
| Protein | Glyoxylate: NADH | Glyoxylate: NADPH | ||||||
|---|---|---|---|---|---|---|---|---|
| Glyoxylate | NADH | Glyoxylate | NADPH | |||||
| kcat | Km | kcat | Km | kcat | Km | kcat | Km | |
| s−1 | µm | s−1 | µm | s−1 | µm | s−1 | µm | |
| HPR1-wild type | 68 ± 10 | 17366 ± 1613 | 62 ± 2 | 10 ± 0.5 | 0.56 ± 0.03 | 17996 ± 4505 | 0.56 ± 0.04 | 353 ± 29 |
| HPR1-T335D | 75 ± 11 | 9727 ± 258* | 91 ± 10* | 5.8 ± 1.5* | 0.92 ± 0.04** | 18011 ± 5797 | 0.85 ± 0.09* | 271 ± 25* |
| HPR1-T335A | 59 ± 5 | 15195 ± 2210 | 60 ± 3 | 10.4 ± 2.2 | 0.52 ± 0.06 | 16589 ± 1985 | 0.62 ± 0.07 | 408 ± 74 |
When the kinetic parameters of HPR1-wild type were compared to those of HPR1-T335A, similar kcat and Km values were found for each of the reactions (Tables 1 and 2). However, this was not the case for the phosphomimetic HPR1-T335D protein, which exhibited significant differences in kcat and Km values (Tables 1 and 2). The kcat for the NADH-HP reaction was ∼50% lower for HPR1-T335D compared to the other two recombinant proteins, whereas the kcat for the NADPH-HP activity increased by 35% (Table 1). Opposite changes were observed for cofactor Km in the presence of HP, whereby Km NADH increased by 56%, whereas Km NADPH decreased by 31%, for the HPR activity. On the other hand, HPR1-T335D Km HP was not altered for the NADH-HP activity, whereas it was 34% reduced for the NADPH-HP activity (Table 1). The phosphomimetic mutation also led to changes in the kinetic parameters associated with glyoxylate. HPR1-T335D showed increased kcat values for both NADH-dependent (30%) and NADPH-dependent (58%) glyoxylate reductase activities when compared to HPR1-wild type and HPR1-T335A (Table 2). Km values for both glyoxylate and NADH were reduced by 50% for the NADH-glyoxylate combination, whereas only the Km NADPH decreased (25%) for the NADPH-glyoxylate reaction (Table 2).
The fact that NADH-NADPH activity balance is altered in favor of NADPH for the HPR activity while the kcat of the glyoxylate reductase activity of phosphomimetic HPR1-T335D is improved suggests that in planta phosphorylation at T335 modifies HPR1 activities and substrate specificities.
Ambient Air-Grown HPR1-T335D-Complemented hpr1-1 Plants Display Reduced Growth
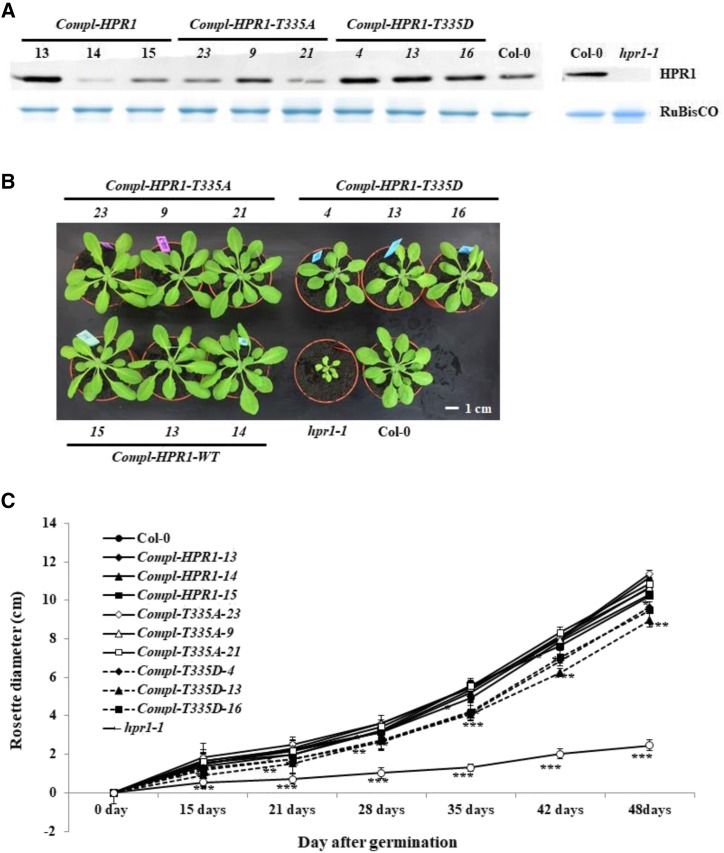
To evaluate the impact of phosphorylation-mimetic and nonphosphorylatable HPR1 on plant development, the hpr1-1 mutant (Timm et al., 2008) was transformed with constructs to express wild-type HPR1 complementary DNA and those corresponding to HPR1 with mutated phosphorylation sites. Gene expression was under the control of the AtSHMT1 promoter, which we previously used successfully to complement both amiRgox1gox2 and shmt1 mutants (Dellero et al., 2016; Liu et al., 2019). The different plant constructs (and their resulting HPR1 proteins) were as follows: Compl-HPR1 (HPR1), Compl-HPR1-T335A (HPR1-T335A), and Compl-HPR1-T335D (HPR1-T335D). Homozygous lines expressing the different HPR1 proteins were selected and analyzed. Immunoblot analysis of rosette leaf soluble proteins showed that HPR1 protein was successfully produced, albeit at varying levels, in all hpr1-1 complemented lines (Fig. 1A; Supplemental Fig, S2). All constructs complemented the photorespiratory growth phenotype of hpr1-1 in our short-day ambient-air conditions (Fig. 1B). Interestingly, Compl-T335D lines were smaller compared to Col-0, Compl-T335A, and Compl-HPR1 lines in ambient air (Fig. 1B), whereas this was not observed when plants were grown in high CO2 (3,000 μL L−1; Supplemental Fig. S3). The difference in plant size (∼20%) was quantified by measuring rosette diameter (Fig. 1C), fresh weight, and dry weight of the different plant lines (Supplemental Fig. S4, A and B). This also allowed us to show that there were no differences in leaf water content (Supplemental Fig. S4C). Taken together, our data indicate that T335D phosphorylation-mimic HPR1 in planta limits normal plant growth in controlled short-day ambient-air conditions and that this phenotype is linked to photorespiration.
Figure 1.
HPR1 protein content and growth phenotype of complemented hpr1 lines. A, HPR1 protein level analysis in rosette leaves of Col-0 (wild-type [WT]), hpr1-1, and HPR1-, HPR1-T335A-, and HPR1-T335D-complemented hpr1 transgenic lines. Soluble proteins were extracted from rosette leaves of 4-week-old short-day, ambient air-grown plants, and HPR1 protein levels were revealed by immunoblotting using anti-HPR1 antibodies. The large subunit of Rubisco stained with Coomassie blue was used as a loading control. B, Growth phenotype of 4-week-old short-day, ambient air-grown Col-0 (wild-type), hpr1-1, and HPR1-, HPR1-T335A-, and HPR1-T335D-complemented hpr1 plants. C, Rosette diameter of all plant lines grown under the same conditions as in B. Values are means ± sd (n = 5) of three biological repetitions. The asterisks indicate significant differences compared to Col-0 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Compl-T335D Plants Have Altered HPR Activities
In order to check whether the phenotype of Compl-T335D plants was associated with modified HPR1 activity, activity assays were performed using leaf extracts from all plant lines. Compared to Col-0, Compl-T335D lines showed a reduced maximal NADH-HP activity (40%) and higher NADPH-HP (0.5- to 1.5-fold), NADH-glyoxylate (0.3- to 2.3-fold), and NADPH-glyoxylate (2-fold, albeit not significant) activities (Table 3). However, when rosette leaf extract activities were normalized to HPR1 protein abundance estimated from immunoblots (Supplemental Fig. S2), only the NADH-HP specific activity of Compl-T335D lines was significantly different from all other HPR1-containing lines (Supplemental Fig. S5). Therefore, leaf-extracted HPR activities did not appear to follow the equivalent recombinant protein activities (compare Tables 1, 2, and 3).
Table 3. Leaf HPR1 activity of Col-0, hpr1-1, and complemented lines (Compl-HPR1, Compl-T335A, and Compl-T335D) with different substrate-cofactor combinations.
Plants were grown in ambient air for 4 weeks before rosette leaves were collected and analyzed. Mean activities ± sd (µmol min−1 mg−1 protein) from three independent experiments, each using four different plants per line (n = 12). Values in bold are significantly different from Col-0 values (Student’s t test, P < 0.05).
| Genotype | Hydroxypyruvate:NADH | Hydroxypyruvate:NADPH | Glyoxylate:NADH | Glyoxylate:NADPH |
|---|---|---|---|---|
| Col-0 | 1.02 ± 0.08 | 0.10 ± 0.03 | 0.13 ± 0.02 | 0.04 ± 0.02 |
| Compl-HPR1-13 | 1.93 ± 0.57 | 0.15 ± 0.06 | 0.21 ± 0.06 | 0.06 ± 0.03 |
| Compl-HPR1-14 | 0.66 ± 0.27 | 0.06 ± 0.01 | 0.05 ± 0.02 | 0.03 ± 0.01 |
| Compl-HPR1-15 | 0.90 ± 0.06 | 0.08 ± 0.02 | 0.09 ± 0.02 | 0.04 ± 0.02 |
| Compl-T335A-23 | 0.90 ± 0.08 | 0.07 ± 0.02 | 0.08 ± 0.03 | 0.03 ± 0.01 |
| Compl-T335A-9 | 1.21 ± 0.04 | 0.12 ± 0.04 | 0.12 ± 0.03 | 0.04 ± 0.02 |
| Compl-T335A-21 | 0.80 ± 0.15 | 0.06 ± 0.01 | 0.06 ± 0.04 | 0.03 ± 0.01 |
| Compl-T335D-4 | 0.60 ± 0.11 | 0.15 ± 0.02 | 0.18 ± 0.04 | 0.08 ± 0.06 |
| Compl-T335D-13 | 0.60 ± 0.34 | 0.25 ± 0.13 | 0.43 ± 0.23 | 0.08 ± 0.07 |
| Compl-T335D-16 | 0.57 ± 0.08 | 0.15 ± 0.01 | 0.17 ± 0.01 | 0.08 ± 0.05 |
| hpr1-1 | 0.14 ± 0.04 | 0.03 ± 0.00 | 0.03 ± 0.02 | 0.03 ± 0.01 |
Compl-T335D Lines Show Reduced Photosynthetic CO2 Assimilation
Photorespiratory perturbations alter leaf photosynthetic CO2 assimilation rate (Timm et al., 2012) and electron transfer properties (Dellero et al., 2015a, 2016). Therefore, in order to characterize the retarded growth phenotype of Compl-T335D plants, the photosynthetic properties of fully expanded rosette leaves from Col-0, hpr1-1, Compl-HPR1-13, Compl-T335A-23, and Compl-T335D-4 plants grown in ambient air were investigated under the light intensity used for plant growth. Consistent with previous reports (Timm et al., 2011, 2012), the net CO2 assimilation rate (An) of hpr1-1 was reduced by 40%, whereas dark respiration rate (Rdark) and the CO2 compensation point were increased by 37.5% and 34%, respectively, when compared to Col-0 (Table 4). Moreover, the calculated photosynthetic electron transfer rate (ETR), the photochemical chlorophyll fluorescence quenching (qP), and the relative amount of Rubisco large subunit per leaf area of hpr1-1 were reduced by 31%, 15%, and 28%, respectively, whereas nonphotochemical chlorophyll fluorescence quenching (NPQ) increased 2.7-fold (Table 4; Supplemental Fig. S6). Complemented lines showed similar parameters to Col-0, including the relative amount of Rubisco large subunit per leaf area (Table 4; Supplemental Fig. S6); however, Compl-T335D-4 exhibited a reduction in An (26%), ETR (21%), and qP (15%) when compared to the other HPR1-containing lines (Table 4). On the other hand, all plants, including hpr1-1, had similar stomatal conductance (gs), transpiration rate (Tr), and maximum potential quantum efficiency (Fv/Fm) values. These observations suggest that the altered HPR activities of Compl-T335D leaves lead to an inhibition of the net CO2 assimilation rate associated with a decreased photosynthetic ETR, which may explain the smaller plant phenotype.
Table 4. Leaf gas exchange, chlorophyll fluorescence parameters, and relative Rubisco amount per leaf surface of ambient air-grown Col-0, Compl-HPR1-13, Compl-T335A-23, Compl-T335D-4, and hpr1-1.
An, Rdark, the CO2 compensation point, gs, Tr, photosynthetic ETR, NPQ, qP, the Fv/Fm ratio, and relative Rubisco large subunit amount per leaf surface values (with the value of Col-0 set to 1) of 6-week-old Col-0, hpr1-1, Compl-HPR1-13, Compl-T335A-23, and Compl-T335D-4 rosettes. Values are means ± sd (n = 3 independent plants). Asterisks indicate significant differences from Col-0 as determined by Student’s t test (*P < 0.05 and **P < 0.01).
| Parameter | Col-0 | Compl-HPR1-13 | Compl-T335A-23 | Compl-T335D-4 | hpr1-1 |
|---|---|---|---|---|---|
| An (µmol CO2 m−2 s−1) | 8.05 ± 1.20 | 8.62 ± 0.48 | 8.55 ± 0.63 | 6.22 ± 0.90* | 4.85 ± 1.33** |
| Rdark (µmol CO2 m−2 s−1) | −0.72 ± 0.10 | −0.72 ± 0.37 | −0.59 ± 0.12 | −0.61 ± 0.15 | −0.45 ± 0.11** |
| CO2 compensation point (µmol mol−1 CO2) | 72.9 ± 3.0 | 68.1 ± 5.3 | 67.0 ± 3.7 | 73.1 ± 2.8 | 110.9 ± 11.8** |
| gs (mmol H2O m−2 s−1) | 0.23 ± 0.06 | 0.21 ± 0.04 | 0.16 ± 0.05 | 0.16 ± 0.5 | 0.16 ± 0.06 |
| Tr (mmol H2O m−2 s−1) | 2.17 ± 0.84 | 3.38 ± 0.40 | 2.29 ± 0.51 | 2.43 ± 0.51 | 2.55 ± 1.02 |
| ETR | 56.4 ± 4.6 | 55.8 ± 4.7 | 56.8 ± 1.4 | 44.31 ± 5.4* | 38.9 ± 6.6** |
| NPQ | 0.56 ± 0.10 | 0.47 ± 0.06 | 0.75 ± 0.36 | 0.83 ± 0.27 | 1.53 ± 0.34** |
| qP | 0.75 ± 0.03 | 0.73 ± 0.04 | 0.70 ± 0.08 | 0.62 ± 0.04** | 0.64 ± 0.07* |
| Fv/Fm | 0.796 ± 0.012 | 0.790 ± 0.019 | 0.801 ± 0.008 | 0.791 ± 0.009 | 0.779 ± 0.008 |
| Relative Rubisco amount per leaf surface (arbitrary units) | 1 ± 0.05 | 0.94 ± 0.09 | 0.96 ± 0.03 | 0.90 ± 0.08 | 0.72 ± 0.07* |
Compl-T335D Rosettes Show Photorespiratory Perturbations
The impact of the altered enzymatic and photosynthetic activities of HPR1-T335D-complemented lines on leaf metabolism was investigated by comparing gas chromatography-mass spectrometry (GC-MS) metabolite profiles of leaf extracts from Col-0, hpr1-1, and our complemented hpr1 lines grown in either ambient-air or high-CO2 (3,000 µL L−1) conditions (Supplemental Table S1). When plants were grown in high-CO2 conditions, few significant differences in relative metabolite levels were observed between the different plant lines (Supplemental Table S1). However, in accordance with Timm et al. (2008), ambient air-grown hpr1-1 leaves displayed accumulation of photorespiratory metabolites (glycolate, glyoxylate, Gly, Ser, and glycerate) compared to Col-0 (Fig. 2). In ambient air, Compl-T335D-4 leaves also exhibited significant differences in certain photorespiratory metabolite contents when compared to either Col-0 (Fig. 2) or other HPR-containing lines (Table 5). There was an under-accumulation of Gly, glyoxylate, and glycerate (31%, 64%, and 14%, respectively), whereas glycolate accumulated more (30%) in Compl-T335D-4 compared to Col-0 (Fig. 2). These differences were not observed in high-CO2 growth conditions (Fig. 2). This suggests that the low peroxisomal NADH-HP activity of Compl-T335D-4 leaves impacts the functioning of the photorespiratory cycle in planta. GC-MS analyses also revealed a limited number of unique metabolic changes between phosphomimetic HPR1 plants and each of the other HPR1-containing lines (Table 5). In air, Compl-T335D-4 rosettes accumulated more 2-hydroxypyridine (2.3-fold), myristic acid (2.2-fold), and glycolate (1.2-fold), but less glycerate (14%), glycine (27%), glyoxylate (55%), lactose (43%), threonic acid (51%), succinate (29%), and sucrose (34%) compared to other HPR1-containing lines (Table 5).
Figure 2.
Leaf photorespiratory metabolite contents of high CO2- and ambient air-grown Col-0, Compl-HPR1-13, Compl-T335A-23, Compl-T335D-4, and hpr1-1. Plants were grown either in ambient-air or high-CO2 (3,000 μL L−1) conditions for 4 weeks before rosette leaves of four individual plants of each line were analyzed by GC-MS. Histograms show the relative metabolite contents (mean ± sd, n = 4 independent plants) from rosette leaves of Col-0, Compl-HPR1-13, Compl-T335A-23, Compl-T335D-4, and hpr1-1 with the corresponding content of Col-0 in ambient air set to 1. The complete GC-MS metabolite data set is available in Supplemental Table S1. Statistical significance was determined by Student’s t test. The asterisks indicate significant differences between Col-0 and other lines grown in the same conditions (*P < 0.05, **P < 0.01, and ***P < 0.001).
Table 5. Differences in metabolite levels between Compl-T335D rosettes and those of other HPR1-containing lines grown in ambient air.
Relative metabolite contents are shown with the Col-0 levels arbitrarily set to 1. Asterisks indicate significant differences compared to Col-0 as determined by Student’s t test (*P < 0.05). Numbers in bold indicate significant differences from all other HPR1-containing lines as determined by Student’s t test (*P < 0.05).
| Metabolite | Col-0 | Compl-HPR1-13 | Compl-HPR1-T335A-23 | Compl-HPR1-T335D-4 |
|---|---|---|---|---|
| 2-Hydroxypyridine | 1.00 ± 0.07 | 0.94 ± 0.08 | 0.88 ± 0.03 | 2.18 ± 0.66* |
| Glycerate | 1.00 ± 0.05 | 1.07 ± 0.06 | 1.13 ± 0.16 | 0.86 ± 0.06* |
| Gly | 1.00 ± 0.10 | 0.91 ± 0.06 | 0.94 ± 0.15 | 0.69 ± 0.05* |
| Glycolate | 1.00 ± 0.03 | 1.12 ± 0.05 | 1.11 ± 0.09 | 1.30 ± 0.05* |
| Glyoxylate | 1.00 ± 0.10 | 0.62 ± 0.07* | 0.78 ± 0.07 | 0.36 ± 0.03* |
| Lactose | 1.00 ± 0.05 | 0.92 ± 0.13 | 0.94 ± 0.17 | 0.54 ± 0.07* |
| Myristic acid | 1.00 ± 0.04 | 0.95 ± 0.14 | 0.73 ± 0.05 | 2.00 ± 0.63* |
| Succinate | 1.00 ± 0.05 | 0.99 ± 0.05 | 1.34 ± 0.13* | 0.79 ± 0.06* |
| Suc | 1.00 ± 0.09 | 1.33 ± 0.02* | 0.78 ± 0.07* | 0.69 ± 0.03* |
| Threonic acid | 1.00 ± 0.07 | 0.68 ± 0.07 | 0.99 ± 0.06 | 0.44 ± 0.05* |
DISCUSSION
Photorespiratory cycle regulation remains poorly understood, although there is recent evidence of the involvement of thioredoxin-dependent redox modifications of mitochondrial Gly decarboxylase l-protein activity (Reinholdt et al., 2019; Pereira et al., 2020). Furthermore, phosphoproteomics studies infer that many photorespiratory enzymes can be phosphorylated (see Hodges et al., 2006). In this study, the role of HPR1 phosphorylation at T335 was explored by analyzing enzymatic activities of phosphorylation-mimetic recombinant HPR1 and its capacity to complement the photorespiratory growth phenotype of the Arabidopsis hpr1-1 mutant.
The Enzymatic Properties of Recombinant Arabidopsis HPR1 Have Been Previously Underestimated
HPR isoforms are known to have relaxed substrate-cofactor specificities and are able to use two substrates (HP and glyoxylate) and two cofactors (NADH and NADPH; Tolbert et al., 1970; Husic and Tolbert, 1987; Kleczkowski and Randall, 1988; Timm et al., 2008, 2011). As expected, recombinant Arabidopsis HPR1-wild-type protein exhibited measurable in vitro activities for each of the four substrate-cofactor combinations, although it showed a higher NADH-HP activity compared to the other reactions tested (as seen from calculated kcat values; Tables 1 and 2), as previously reported (Timm et al., 2008). However, whereas the NADH-HP/NADPH-HP activity ratio was reported as 20 for recombinant Arabidopsis HPR1 (Timm et al., 2008), in our hands this ratio was only 1.65 due to a high NADPH-HP activity. Similarly, NADH-glyoxylate activity was only 4-fold lower than the NADH-HP activity (Tables 1 and 2), which contrasted to the 35-fold difference reported by Timm et al. (2008). Such discrepancies could be partly explained by the nonsaturating concentrations of HP and NADPH used previously for HPR1 activity measurements (see the experimental conditions of Tolbert et al., 1970; Husic and Tolbert, 1987; Timm et al., 2008), which were discovered in this work during experiments to calculate Km substrate and cofactor values for HPR1 activities. Substrate Km values were found to be similar to those already published for peroxisomal HPR proteins from other plant and algal species (for a review, see Givan and Kleczkowski, 1992). Surprisingly, no cofactor Km values for plant HPR enzymes had been reported until now. It can be seen that the Km NADH was 8-fold lower than that of NADPH for the HP reaction (Table 1), whereas there was a 35-fold difference between the Km NADH and Km NADPH for glyoxylate reductase activities (Table 2). Interestingly, the Km HP was cofactor dependent with Km HP, increasing 8-fold in the presence of NADPH compared to NADH, thus mirroring the 8-fold difference between Km NADPH and Km NADH for the HPR activity. This might reflect the catalytic process model of Lassalle et al. (2016), where cofactor binding induced domain movements and the formation of a tunnel, allowing substrate entry to the active site. Surprisingly, there was no cofactor effect on the Km glyoxylate (Table 2). Taken together, the kcat and Km values of HPR1-wild type confirmed that this enzyme would act preferentially as an HPR in planta with a preference for NADH, although this assumption would depend on substrate and cofactor concentrations within the peroxisomes. Similar conclusions can be made for the HPR1-T335A form of the enzyme, since it exhibited similar kcat and Km properties when compared to HPR1-wild type (Tables 1 and 2).
The Phosphomimetic T335D Mutation of Recombinant Arabidopsis HPR1 Alters Its Kinetic Properties toward Those of Cytosolic HPR2
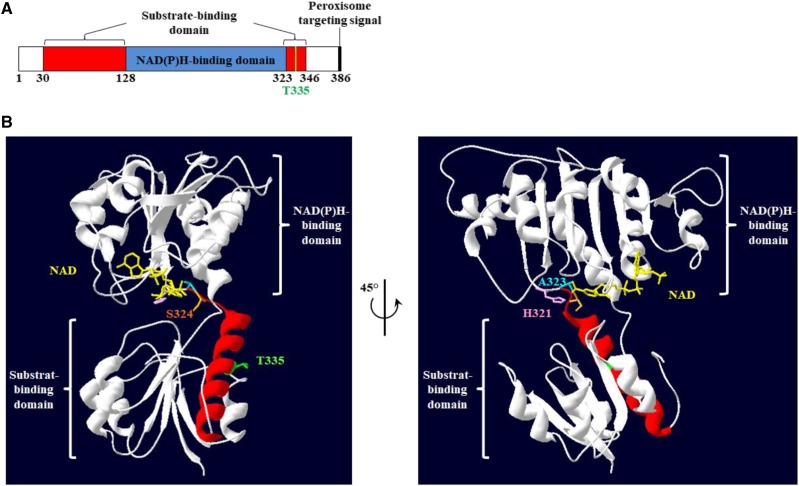
Interestingly, the phosphomimetic HPR1-T335D protein showed significant differences in both cofactor and substrate-dependent kinetic parameters when compared to HPR1-wild type and HPR1-T335A (Tables 1 and 2). Overall, these changes showed that the T335D mutation led to a shift in cofactor specificity of the HPR reaction, with a decrease in NADH-dependent activity and an increase in NADPH-dependent activity (Table 1), as well as leading to an improved glyoxylate reductase activity (Table 2). The observed alterations in activities were associated with changes in Km for substrate, cofactor, or both (Tables 1 and 2). A structural model (Fig. 3), based on the crystallized Pyrococcus furiosus recombinant glyoxylate HPR protein containing NADP and glyoxylate (PDB 5AOV; Lassalle et al., 2016), showed T335 to be located in an α-helix linking the substrate and cofactor binding domains (Yoshikawa et al., 2007). A small nonstructured region joining this α-helix to a β-sheet of the cofactor-binding fold contained three important residues, namely H321 (catalytic site), A323 (cofactor binding), and S324 (cofactor and substrate binding; https://www.ebi.ac.uk/interpro/protein/Q9C9W5). Therefore, we believe that the T335D mutation has the potential to induce structural changes that will affect both substrate and cofactor binding as well as catalytic efficiency, thus explaining the differences in kinetic parameters between HPR1-T335D and HPR1-wild type/HPR1-T335A (Tables 1 and 2). Based on our results, it is tempting to propose that a T335 phosphorylation regulatory mechanism could intervene when peroxisomal (or whole-cell) NADH/NADPH balance is shifted in favor of NADPH by either changing environmental conditions or at specific developmental stages, thus conferring cofactor flexibility to maintain the required HPR activity of HPR1. Interestingly, HPR1-T335D displayed a shift toward the substrate-cofactor preferences of cytosolic HPR2 (see Timm et al., 2008). A protein sequence alignment of Arabidopsis HPR1 and HPR2 showed that T335 of HPR1 was replaced by an Asp (D293) in HPR2 (Supplemental Fig. S7). This suggested that a negatively charged residue at this position could have an important influence on HPR cofactor specificity.
Figure 3.
Predicted 3D structure of an Arabidopsis HPR1 monomer showing the position of T335. A, Predicted domains of Arabidopsis HPR1 (https://www.ebi.ac.uk/interpro/protein/Q9C9W5). B, View of the predicted 3D structure of Arabidopsis HPR1 based on the crystallized Pyrococcus furiosus HPR1 protein structure (Lassalle et al., 2016), with T335 represented in green, H321 (catalytic site) in pink, A323 (cofactor binding) in blue, and S324 (cofactor and substrate binding) in orange. NAD is represented in yellow and the α-helix highlighted in red corresponds to the substrate-binding domain at amino acids 323 to 346 containing His-321, Ala-323, Ser-324, and Thr-335.
Major HPR1 Activities in Arabidopsis Rosettes
When the different HPR activities were compared between hpr1-1 and Col-0 (Table 3; Supplemental Fig. S5) using our saturated substrate and cofactor concentrations, it was discovered that the contribution of HPR1 to leaf total HPR activities had been previously underestimated. Indeed, HPR1 contributed to 80%, 70%, and 77% of the potential maximal NADH-HP, NADPH-HP, and NADH-glyoxylate activities, respectively, in Arabidopsis leaf extracts (as seen from Table 3) when compared to contributions reported for Arabidopsis (85%, 44%, and 30%, respectively; Timm et al., 2008) and barley (96%, 21%, and 13%, respectively; Timm et al., 2008; Kleczkowski et al., 1990).Therefore, both HPR2 and HPR3 appear to be potentially major in planta NADPH-glyoxylate reductases; however, they were recently shown to catalyze the reduction of 4-hydroxyphenylpyruvate to 4-hydroxyphenyllactic acid, a reaction involved in the biosynthesis of secondary metabolites from Tyr (Xu et al., 2018).
The Growth Phenotype of Ambient Air-Grown Compl-T335D Lines Is Associated with Reduced NADH-HP HPR1 and Photosynthetic CO2 Assimilation Activities
To evaluate the impact of HPR1 T335 phosphorylation in planta, hpr1-1 was complemented with wild-type and T335-mutated HPR1 forms. Only Compl-HPR1-T335D lines continued to display a weak photorespiration-dependent growth phenotype in ambient air (Fig. 1; Supplemental Fig. S3). When compared to other HPR1-containing lines, only Compl-T335D leaves exhibited a lower NADH-HP (44%) activity, and higher NADPH-HP (2.8-fold), NADH-glyoxylate (2.45-fold), and NADPH-glycolate (2-fold) activities (Table 3). These observations suggest that the NADH-HP activity of HPR1 is important for normal plant growth, whereas other HPR1 activities within the peroxisome are not sufficient to compensate the low NADH-HP activity and maintain normal plant growth in ambient air. Surprisingly, when activity was normalized to estimated HPR1 protein content (Supplemental Fig. S2), Compl-T335D plants exhibited only a lower specific NADH-HP activity, whereas all other specific activities were similar compared to the other HPR1-containing lines (Supplemental Fig. S5). This was surprising based on the altered kinetic properties of recombinant HPR1-T335D (Tables 1 and 2). It could be that Arabidopsis HPR1 undergoes one or more posttranslational modifications, since it has been shown that leaf HPR can undergo S-nitrosylation (Ortega-Galisteo et al., 2012; Sandalio et al., 2019). However, it might reflect a lowering of free substrate and cofactor levels below the saturation point due to enzymes in the leaf extracts that bind either HP or cofactor, thereby leading to an underestimation of maximal HPR activities for certain substrate-cofactor combinations.
As a consequence of the altered HPR1 activity in Compl-T335D rosettes, photosynthesis was negatively impacted, as seen from lower net CO2 assimilation rates, although this occurred to a lesser degree than in hpr1-1 (Table 4). Inhibition of photosynthesis has been often observed in photorespiratory mutants, including hpr1 (e.g. Chastain and Ogren, 1989; Timm et al., 2008, 2012; Dellero et al., 2016; Li et al., 2019). In contrast to hpr1, the measured decrease of An in Compl-T335D lines was not associated with a lower content of Rubisco per leaf area (Table 4; Supplemental Fig. S6). A major actor is photorespiratory 2PG, which can inhibit triose phosphate isomerase and sedoheptulose-1,7-bisphosphatase, thereby reducing ribulose-1,5-bisphosphate (RuBP) regeneration by the Calvin-Benson-Bassham cycle (see Flügel et al., 2017). This 2PG block can be overcome by a cytosolic bypass and Glc-6-phosphate shunt that requires high rates of cyclic electron flow to provide the required ATP (Li et al., 2019). These scenarios could explain why Compl-T335D rosettes exhibited lower photosynthetic CO2 assimilation rates, ETR, and qP properties (Table 4). However, this would depend on the accumulation of 2PG brought about by the low NADH-HP activity negatively affecting the functioning of the photorespiratory cycle in Compl-T335D leaves.
Compl-T335D Rosettes Show Significant Changes in Photorespiration-Associated Metabolites Compared to Fully Complemented Lines
In our growth conditions, metabolite profiling revealed metabolic differences between ambient-air and high-CO2 growth conditions similar to those previously observed for hpr1-1 (Fig. 2; Supplemental Table S1; Timm et al., 2008; 2012). Ambient-air-grown hpr1-1 accumulated photorespiratory-associated metabolites (glycolate, glyoxylate, Gly, Ser, and even glycerate), whereas our HPR1-containing lines had low levels (Fig. 2; Supplemental Table S1). However, significantly different Gly, glyoxylate, glycerate, and glycolate levels detected between Compl-T335D plants and the other HPR1-containing lines (Table 5) indicated a subtle perturbation of the photorespiratory cycle in the presence of HPR1-T335D that could explain the observed growth and photosynthetic phenotypes of Compl-T335D plants. Although 2PG was not identified in our GC-MS analyses, it is presumed that it would have accumulated in Compl-T335D rosettes, since there was an increase in glycolate and a decrease in glyoxylate, Gly, and glycerate (Table 5). In our opinion, it is difficult to correlate the reduction of glyoxylate to the observed higher maximal in vitro NADH-glyoxylate and NADPH-glyoxylate activities of Compl-TSS5D rosette extracts (Table 3), since the Km glyoxylate for recombinant HPR1-T335D remained high at 9.7 mm and 18 mm in the presence of either NADH or NADPH, respectively (Table 2). In comparison, the Km glyoxylate of two other peroxisomal enzymes using glyoxylate, namely, Glu:glyoxylate aminotransferase 1 (0.27 mm) and Ser:glyoxylate aminotransferase (0.91 mm), are much lower (Liepman and Olsen, 2003; Kendziorek and Paszkowski, 2008). We believe that leaf peroxisomal glyoxylate levels would be too low to allow a significant in planta NADH/NADPH glyoxylate activity even in the presence of HPR1-T335D. Lower glyoxylate levels might reflect a retroinhibition of Ser:glyoxylate aminotransferase activity by HP due to the low NADH-HP activity of HPR1-T335D, and this could also explain the lower Gly levels (Table 5). A low NADH-HP activity of HPR1-T335D would also explain the decreased glycerate level that in turn could affect the transport of glycolate to the peroxisomes via the plastidial glycolate glycerate translocator1 (PLGG1; Pick et al., 2013). According to this hypothesis, accumulation of glycolate, and probably 2PG, in the chloroplast would reduce photosynthesis and explain the decreased glyoxylate and glycine levels (Table 5).
Finally, Compl-T335D plants exhibited few significant metabolite differences compared to the other HPR1-containing lines grown in ambient air (Table 5), thus highlighting a subtle effect of HPR1-T335D on overall plant metabolism. Photorespiration is known to interact with mitochondrial functioning, carbohydrate metabolism, and redox homeostasis, but there were only decreases in succinate, Suc, and threonic acid (associated with oxidative stress and ascorbate metabolism; Navascués et al., 2012; Park et al., 2019), and increases of myristic acid (associated with saturated long-chain fatty acid metabolism) and 2-hydroxypyridine in Compl-T335D plants compared to the other HPR1-containing lines studied (Table 5). Surprisingly, the altered kinetic properties/activities of HPR1-T335D did not modify the levels of metabolites associated with enzymes using specific pyridine nucleotide cofactors.
In conclusion, the phosphomimetic T335D mutation of peroxisomal HPR1 led to a modification of HPR1 kinetic parameters toward those of cytosolic HPR2. Under controlled growth conditions, a reduction of NADH-HP activity could not be fully compensated for by peroxisomal NADPH-dependent HPR1 activities. We propose that T335 phosphorylation of HPR1 could be a mechanism to regulate HPR1 cofactor use at specific developmental stages and growth conditions that change peroxisomal NADH/NADPH balance in favor of NADPH.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Water Content Assays
Ecotype Columbia of Arabidopsis (Arabidopsis thaliana; Col-0) was used as the wild-type control plant in all experiments. The hpr1-1 mutant (Timm et al., 2008) was provided by Herman Bauwe (University of Rostock, Plant Physiology Department). Arabidopsis seeds were germinated on potting soil for 10 d in ambient or high-CO2 (3,000 μL L−1) air in short-day growth conditions (8 h light/16 h dark, 200 μmol photons m−2 s−1, 65% humidity, 20°C day/18°C night). Subsequently, seedlings were transplanted to individual pots for further growth. Water content was determined as previously described (Liu et al., 2019) using rosettes of 4-week-old plants. Experiments were repeated three times each with tissues from five different plants per line (n = 15), and statistical significance was determined by Student’s t test.
Plasmid Constructions, Site-Directed Mutagenesis, and Generation of Complemented Lines
To produce recombinant proteins, the pBAD-AtHPR1-WT plasmid (provided by Herman Bauwe, University of Rostock; Timm et al., 2008) was the template to introduce T335A and T335D point mutations using specific primer pairs (Supplemental Table S1), and the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) was used according to the manufacturer’s instructions to change the T335 codon (ACG) to either an Ala codon (GCG) or an Asp codon (GAT). In this way, pBAD-AtHPR1-T335A and pBAD-AtHPR1-T335D were obtained. All constructions were subsequently verified by DNA sequencing (Eurofins MWG Operon) using primers pBAD-seq-F and pBAD-seq-R (Supplemental Table S2). To generate complemented hpr1-1 lines, pBAD-AtHPR1-WT, pBAD-AtHPR1-T335A, and pBAD-AtHPR1-T335D were used as templates for two consecutive PCR reactions (using primers GW-HPR1-F/GW-HPR1-R then GateWay-F/GateWay-R; Supplemental Table S2) to add attB Gateway recombination sites to the 5′ and 3′ ends, respectively. The resulting DNAs were cloned into pDONR207 using the Gateway cloning system to generate pDONR207-AtHPR1-WT, pDONR207-AtHPR1-T335A, and pDONR207-AtHPR1-T335D. After sequencing, the different constructs were subcloned into pK7WG-promSHMT1 (Dellero et al., 2016) using the Gateway cloning system to generate pH7WG-promSHMT1::HPR1-WT, pH7WG-promSHMT1::HPR1-T335A, and pH7WG-promSHMT1::HPR2-T335D. These plasmids were transformed into Agrobacterium strain GV3101, which was used to generate complemented Arabidopsis hpr1-1 lines by floral dip (Clough and Bent, 1998). Transformed lines were selected by their resistance to hygromycin on one-half strength Murashige and Skoog medium containing 0.8% (w/v) agar, 1% (w/v) Suc, and 15 μg mL−1 hygromycin as described by Harrison et al. (2006). Homozygous lines containing a single transgene insertion were obtained by successive rounds of self-crossing and antibiotic selection on one-half strength Murashige and Skoog medium agar plates.
Expression and Purification of Recombinant HPR1 Proteins
The pBAD/His expression system (Lee et al., 1987) was used to produce recombinant N-terminal 6×His tagged HPR1 proteins. The plasmids pBAD-AtHPR1-WT and pBAD-AtHPR1-T335A/D were transferred into Escherichia coli LMG194 by electroporation. Individual transformed colonies were used to inoculate 10 mL rudic medium precultures containing 0.2% (w/v) Glc and 50 μg mL−1 ampicillin that were grown overnight at 37°C. The next day, precultures were used to inoculate 200 mL rudic medium containing 0.2% (w/v) Glc and 50 μg mL−1 ampicillin. All bacterial cultures were grown at 37°C with shaking at 200 rpm. When the OD600nm was ∼0.7, recombinant protein production was induced by adding 0.2% (w/v) l-arabinose (A3256, Sigma-Aldrich). After 20 h at 37°C, cells were harvested by centrifugation at 4,000g for 30 min at 4°C.
For His-tag protein purification, nickel affinity chromatography (His-SelectNickel Affinity, P6611, Sigma-Aldrich) was used as previously described (Dellero et al., 2015b). Bacterial pellets were resuspended in 3 mL lysis buffer (50 mm NaH2PO4 [pH 8.0], 300 mm NaCl, 5 mm imidazole, and protease inhibitor mixture [Complete Mini, EDTA-free, Roche Applied Science]). Cells were disrupted three times through a French-Press at 600 Pa. Soluble proteins were collected by centrifugation at 15,000g for 30 min at 4°C. At the same time, 2 mL of affinity resin was washed twice with 50 mL of washing buffer (lysis buffer supplemented with 20 mm imidazole), each time centrifuged at 4,000g for 10 min at 4°C and then resuspended in 10 mL of washing buffer. Then, 2.5 mL of soluble protein supernatant and 10 mL of resuspended affinity resin were mixed and incubated at 4°C for 2 h on a rotating wheel. The resin-protein mixture was poured into a syringe to make an affinity column and washed with 50 mL washing buffer. Recombinant His-tagged HPR1 was then eluted with 3 mL of elution buffer (lysis buffer supplemented with 250 mm imidazole). The eluted proteins were directly desalted using a PD10 filtration column previously equilibrated with 50 mm NaH2PO4 (pH 8.0) according to the manufacturer’s instructions (GE Healthcare). Protein concentrations were determined using Bradford reagent (B6916, Sigma-Aldrich) and bovine serum albumin as a standard before being stored at −80°C until required.
Measurement of HPR1 Activities
HPR1 enzyme activity measurements were performed based on previous reports (Tolbert et al., 1970; Husic and Tolbert 1987; Timm et al., 2008) by following cofactor oxidation at 340 nm using a Varian Cary 50 spectrophotometer. All reactions were carried out at 25°C. Recombinant HPR1 activities were carried out using 5 μg of purified protein in a final volume of 1 mL containing 50 mm phosphate buffer (pH 6.2) and variable concentrations of substrates (HP and glyoxylate) and cofactors (NADH and NADPH; see Table 6 for details). Km and Vmax values were calculated using the Michaelis-Menten curve-fitting equation of SigmaPlot 13.0. kcat = Vmax/[E], where E is the number of moles of enzyme active sites. For Arabidopsis rosette leaf HPR activity measurements, 200 mg of 4-week-old Arabidopsis leaf tissue was ground to a fine powder in liquid nitrogen before adding 1.5 mL of extraction buffer (25 mm HEPES-KOH, 1 mm MgCl2, 1 mm KCl, 10 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride, pH 7.6). Crude extracts were clarified by centrifugation at 20,000g for 20 min at 4°C. Each reaction was carried out in a final volume of 1 mL containing 50 mm phosphate buffer pH 6.2, and either 400 μm NADH (N8129, Sigma-Aldrich) or NADPH (N1630, Sigma-Aldrich) and reaction-specific substrate and protein amounts as given in Table 1. Leaf HPR activity was defined as µmol of oxidized NADH or NADPH produced per mg protein per minute. For recombinant HPR1 activities, experiments were repeated three times and for Arabidopsis rosette leaf HPR activity measurements, experiments were repeated three times each from four different plants per line (n = 12). Statistical significance was determined by a Student’s t test
Table 6. Conditions used to measure HPR activities.
| Protein Sample | Enzyme Activity | |||
|---|---|---|---|---|
| NADH-HP | NADPH-HP | NADH-glyoxylate | NADPH-glyoxylate | |
| Recombinant HPR1 | 30–2,000 μm HP | 0.5–20 mm HP | 0.625–40 mm glycolate | 5–150 mm glycolate |
| 200 μm NADH | 750 μm NADPH | 200 μm NADH | 1 mm NADH | |
| Recombinant HPR1 | 6.25–400 μm NADH | 12.5–500 μm NADPH | 25–800 μm NADH | 25–500 μm NADPH |
| 500 μm HP | 15 mm HP | 50 mm glyoxylate | 30 mm glyoxylate | |
| Leaf extract | 400 μm NADH | 400 μm NADPH | 400 μm NADH | 400 μm NADPH |
| 1.5 mm HP | 7.5 mm HP | 50 mm glyoxylate | 50 mm glyoxylate | |
| 30 μg protein | 30 μg protein | 60 μg protein | 100 μg protein | |
SDS-PAGE and Immunoblot Analyses
Two micrograms of recombinant proteins and 10 µg of leaf soluble proteins were separated by SDS-PAGE (10% [w/v] acrylamide) and detected by Coomassie blue staining (Laemmli, 1970). For immunoblot analyses, SDS-PAGE-separated proteins were transferred to nitrocellulose membranes, blocked for 1 h at room temperature in blocking buffer (150 mm NaCl, 0.1% [v/v] Tween 20, 5% [w/v] nonfat dry milk, 50 mm Tris-base [pH 7.4]), and incubated overnight at 4°C with a 1:5,000 dilution (in blocking buffer) of HPR1 antibodies (Timm et al., 2008). Membranes were washed three times with blocking buffer, then incubated with 1:5,000 diluted antirabbit antibodies (A6154, Sigma-Aldrich) for 1 h at room temperature in blocking buffer. HPR1 was revealed using a chemoluminescent peroxidase substrate-3 kit (CPS350, Sigma-Aldrich).
Leaf Gas Exchange and Chlorophyll Fluorescence Measurements
Leaf gas exchange and chlorophyll fluorescence measurements were carried out on 2 cm2 of fully expanded leaves of 6-week-old plants in a gas-exchange chamber (LCF 6400-40, LiCOR) connected to a portable measuring system (LI 6400XT, LiCOR) at 400 μmol mol−1 CO2 and 200 μmol photons m−2 s−1 of light, as in Dellero et al. (2015a). Experiments were repeated three times and statistical significance was determined by Student’s t test.
Relative Rubisco Amount
Determination of Rubisco large subunit amount by leaf area was carried out as described in Dellero et al. (2015a) using 6-week-old plants. Leaf area was measured using ImageJ software before being frozen and ground to a fine powder. Soluble protein extractions were done in 100 mm Bicine, pH 8, supplemented with an antiprotease cocktail (Complete-Mini, Roche Diagnostics) to give a leaf surface/volume ratio of 5 cm2 mL−1. After separation of 10 μL of each soluble leaf protein extract on SDS-PAGE gels (10% [w/v] acrylamide), quantification of Rubisco large subunit was performed using the Coomassie blue-stained gels and ImageJ software.
Metabolite Analyses by GC-MS
One hundred fifty milligrams of rosette leaves of 4-week-old plants were collected after 6 h of light and rapidly frozen in liquid nitrogen and stored at −80°C until extraction. Relative metabolite levels were analyzed by GC-MS using cold-methanol extraction, as described in Noctor et al. (2007). Experiment was performed on four individual plants, and statistical significance was determined by Student’s t test.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (https://www.arabidopsis.org) under the following accession numbers: At1g68010 (HPR1); At1g79870 (HPR2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. His-tag purified recombinant proteins of wild-type HPR1 (HPR1-wild type) and mutated HPR1 (HPR1-T335A and HPR1-T335D) produced in E. coli (data related to Tables 1 and 2).
Supplemental Figure S2. Rosette leaf HPR1 protein levels of ambient air-grown Col-0 (wild-type), hpr1, and complemented lines (data related to Fig. 1A).
Supplemental Figure S3. Phenotype of short-day, high-CO2-grown Col-0, hpr1-1, and complemented lines (data related to Fig. 1B).
Supplemental Figure S4. Fresh weight, dry weight, and water content of rosettes of Col-0, hpr1-1, and complemented lines (data related to Fig. 2C).
Supplemental Figure S5. Estimated specific HPR1 activities for different substrate-cofactor combinations in leaves of Col-0, hpr1-1, and complemented lines (Compl-T335D, Compl-T335A, and Compl-HPR1).
Supplemental Figure S6. Amount of Rubisco large subunit in Col-0, hpr1-1, and complemented lines (Compl-HPR1-13, Compl-T335A-23, and Compl-T335D-4; data related to Table 4).
Supplemental Figure S7. HPR1 and HPR2 protein sequence alignment.
Supplemental Table S1. Relative metabolite levels of 4-week-old Col-0, Compl-HPR1-13, Compl-T335A-23, Compl-T335D-4, and hpr1-1 rosette leaves grown in ambient air and in high CO2 conditions (data related to Fig. 2).
Supplemental Table S2. Sequence information of primers for PCR and cloning.
Acknowledgments
We thank Herman Bauwe (University of Rostock) for kindly providing hpr1-1 seeds, HPR1 antibodies, and the pBAD-HPR1 plasmid, and Dr. Stefan Timm (University of Rostock) for his help during the course of this work.
Footnotes
This work was supported by a public grant overseen by the Agence Nationale de la Recherche (ANR) as part of the “Investissement d’Avenir” program, through the “Lidex-3P” project and a French State grant (grant no. ANR–10–LABX–0040–SPS) funded by the IDEX Paris-Saclay (grant no. ANR–11–IDEX–0003–02) and a REGUL3P project grant (grant no. ANR–14–CE19–0015 to M.H.), and by the China Scholarship Council (PhD fellowship to Y.L.).
References
- Aryal UK, Krochko JE, Ross ARS(2012) Identification of phosphoproteins in Arabidopsis thaliana leaves using polyethylene glycol fractionation, immobilized metal-ion affinity chromatography, two-dimensional gel electrophoresis and mass spectrometry. J Proteome Res 11: 425–437 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR(2010) Photorespiration: Players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Chastain CJ, Ogren WL(1989) Glyoxylate inhibition of ribulosebisphosphate carboxylase oxygenase activation state in vivo. Plant Cell Physiol 30: 937–944 [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cousins AB, Walker BJ, Pracharoenwattana I, Smith SM, Badger MR(2011) Peroxisomal hydroxypyruvate reductase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Photosynth Res 108: 91–100 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Leterrier M, Begara-Morales JC, Valderrama R, Chaki M, López-Jaramillo J, Luque F, Palma JM, Padilla MN, Sánchez-Calvo B, et al. (2013) Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim Biophys Acta 1830: 4981–4989 [DOI] [PubMed] [Google Scholar]
- Dalal J, Lopez H, Vasani NB, Hu Z, Swift JE, Yalamanchili R, Dvora M, Lin X, Xie D, Qu R, et al. (2015) A photorespiratory bypass increases plant growth and seed yield in biofuel crop Camelina sativa. Biotechnol Biofuels 8: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellero Y, Jossier M, Glab N, Oury C, Tcherkez G, Hodges M(2016) Decreased glycolate oxidase activity leads to altered carbon allocation and leaf senescence after a transfer from high CO2 to ambient air in Arabidopsis thaliana. J Exp Bot 67: 3149–3163 [DOI] [PubMed] [Google Scholar]
- Dellero Y, Lamothe-Sibold M, Jossier M, Hodges M(2015a) Arabidopsis thaliana ggt1 photorespiratory mutants maintain leaf carbon/nitrogen balance by reducing RuBisCO content and plant growth. Plant J 83: 1005–1018 [DOI] [PubMed] [Google Scholar]
- Dellero Y, Mauve C, Boex-Fontvieille E, Flesch V, Jossier M, Tcherkez G, Hodges M(2015b) Experimental evidence for a hydride transfer mechanism in plant glycolate oxidase catalysis. J Biol Chem 290: 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügel F, Timm S, Arrivault S, Florian A, Stitt M, Fernie AR, Bauwe H(2017) The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 29: 2537–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givan CV, Kleczkowski LA(1992) The enzymic reduction of glyoxylate and hydroxypyruvate in leaves of higher plants. Plant Physiol 100: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A(2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M, Dellero Y, Keech O, Betti M, Raghavendra AS, Sage R, Zhu XG, Allen DK, Weber APM(2016) Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. J Exp Bot 67: 3015–3026 [DOI] [PubMed] [Google Scholar]
- Hodges M, Jossier M, Boex-Fontvieille E, Tcherkez G(2013) Protein phosphorylation and photorespiration. Plant Biol (Stuttg) 15: 694–706 [DOI] [PubMed] [Google Scholar]
- Husic W, Tolbert NE(1987) NADH :hydroxypyruvate reductase and NADPH :glyoxylate in algae : Partial purification and characterization from Chlamydomonas reinhardtii. Arch Biochem Biophys 252: 396–408 [DOI] [PubMed] [Google Scholar]
- Kendziorek M, Paszkowski A(2008) Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves. Acta Biochim Biophys Sin (Shanghai) 40: 102–110 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD(1988) Purification and characterization of a novel NADPH(NADH)-dependent hydroxypyruvate reductase from spinach leaves. Comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem J 250: 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Edwards GE, Blackwell RD, Lea PJ, Givan CV(1990) Enzymology of the reduction of hydroxypyruvate and glyoxylate in a mutant of barley lacking peroxisomal hydroxypyruvate reductase. Plant Physiol 94: 819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK.(1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lassalle L, Engilberge S, Madern D, Vauclare P, Franzetti B, Girard E(2016) New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases. Sci Rep 6: 20629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Francklyn C, Hamilton EP(1987) Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci USA 84: 8814–8818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Weraduwage SM, Preiser AL, Tietz S, Weise SE, Strand DD, Froehlich JE, Kramer DM, Hu J, Sharkey TD(2019) A cytosolic bypass and G6P shunt in plants lacking peroxisomal hydroxypyruvate reductase. Plant Physiol 180: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Olsen LJ(2003) Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol 131: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mauve C, Lamothe-Sibold M, Guérard F, Glab N, Hodges M, Jossier M(2019) Photorespiratory serine hydroxymethyltransferase 1 activity impacts abiotic stress tolerance and stomatal closure. Plant Cell Environ 42: 2567–2583 [DOI] [PubMed] [Google Scholar]
- Murray AJS, Blackwell RD, Lea PJ(1989) Metabolism of hydroxypyruvate in a mutant of barley lacking NADH-dependent hydroxypyruvate reductase, an important photorespiratory enzyme activity. Plant Physiol 91: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navascués J, Pérez-Rontomé C, Sánchez DH, Staudinger C, Wienkoop S, Rellán-Álvarez R, Becana M(2012) Oxidative stress is a consequence, not a cause, of aluminum toxicity in the forage legume Lotus corniculatus. New Phytol 193: 625–636 [DOI] [PubMed] [Google Scholar]
- Noctor G, Bergot G, Mauve C, Thominet D, Lelarge-Trouverie C, Prioul JL(2007) A comparative study of amino acid measurement in leaf extracts by gas chromatography-time of flight-mass spectrometry and high performance liquid chromatography with fluorescence detection. Metabolomics 3: 161–174 [Google Scholar]
- Nölke G, Houdelet M, Kreuzaler F, Peterhänsel C, Schillberg S(2014) The expression of a recombinant glycolate dehydrogenase polyprotein in potato (Solanum tuberosum) plastids strongly enhances photosynthesis and tuber yield. Plant Biotechnol J 12: 734–742 [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC(2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J Exp Bot 63: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-H, Scheffler J, Scheffler B, Cantrell CL, Pauli CS(2019) Chemical defense responses of upland cotton, Gossypium hirsutum L. to physical wounding. Plant Direct 3: e00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F, Geigenberger P, Thormählen I, Souza PVL, Nesi AN, Timm S, Fernie AR, Araújo WL, Daloso DM(2020) Thioredoxin h2 contributes to the redox regulation of mitochondrial photorespiratory metabolism. Plant Cell Environ 43: 188–208 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Maurino VG(2011) Photorespiration redesigned. Plant Physiol 155: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schulz MA, Obata T, Fernie AR, Weber AP(2013) PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA 110: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholdt O, Schwab S, Zhang Y, Reichheld JP, Fernie AR, Hagemann M, Timm S(2019) Redox-regulation of photorespiration through mitochondrial thioredoxin o1. Plant Physiol 181: 442–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandalio LM, Gotor C, Romero LC, Romero-Puertas MC(2019) Multilevel regulation of peroxisomal proteome by post-translational modifications. Int J Mol Sci 20: E4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen BR, Wang LM, Lin XL, Yao Z, Xu HW, Zhu CH, Teng HY, Cui LL, Liu EE, Zhang JJ, et al. (2019) Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Mol Plant 12: 199–214 [DOI] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR(2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Bauwe H(2013) The variety of photorespiratory phenotypes—employing the current status for future research directions on photorespiration. Plant Biol (Stuttg) 15: 737–747 [DOI] [PubMed] [Google Scholar]
- Timm S, Florian A, Fernie AR, Bauwe H(2016) The regulatory interplay between photorespiration and photosynthesis. J Exp Bot 67: 2923–2929 [DOI] [PubMed] [Google Scholar]
- Timm S, Florian A, Jahnke K, Nunes-Nesi A, Fernie AR, Bauwe H(2011) The hydroxypyruvate-reducing system in Arabidopsis: Multiple enzymes for the same end. Plant Physiol 155: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Mielewczik M, Florian A, Frankenbach S, Dreissen A, Hocken N, Fernie AR, Walter A, Bauwe H(2012) High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS One 7: e42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Nunes-Nesi A, Pärnik T, Morgenthal K, Wienkoop S, Keerberg O, Weckwerth W, Kleczkowski LA, Fernie AR, Bauwe H(2008) A cytosolic pathway for the conversion of hydroxypyruvate to glycerate during photorespiration in Arabidopsis. Plant Cell 20: 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE, Yamazaki RK, Oeser A(1970) Localization and properties of hydroxypyruvate and glyoxylate reductases in spinach leaf particles. J Biol Chem 245: 5129–5136 [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR(2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Xu JJ, Fang X, Li CY, Zhao Q, Martin C, Chen XY, Yang L(2018) Characterization of Arabidopsis thaliana hydroxyphenylpyruvate reductases in the tyrosine conversion pathway. Front Plant Sci 9: 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Yang G, Chen Y, Zhang C, Zhang J, Peng X(2014) Two hydroxypyruvate reductases encoded by OsHPR1 and OsHPR2 are involved in photorespiratory metabolism in rice. J Integr Plant Biol 56: 170–180 [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Arai R, Kinoshita Y, Uchikubo-Kamo T, Wakamatsu T, Akasaka R, Masui R, Terada T, Kuramitsu S, Shirouzu M, et al. (2007) Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate. Acta Crystallogr D Biol Crystallogr 63: 357–365 [DOI] [PubMed] [Google Scholar]