Abstract
Cell-selective gene expression comprises a critical element of many adeno-associated virus (AAV) vector-based gene therapies, and to date achieving this goal has focused on AAV capsid engineering, cell-specific promoters, or cell-specific enhancers. Recently, we discovered that the capsid of AAV9 exerts a differential influence on constitutive promoters of sufficient magnitude to alter cell type gene expression in the rat CNS. For AAV9 vectors chicken β-actin (CBA) promoter-driven gene expression exhibited a dominant neuronal gene expression in the rat striatum. Surprisingly, for otherwise identical AAV9 vectors, the truncated CBA hybrid (CBh) promoter shifted gene expression toward striatal oligodendrocytes. In contrast, AAV2 vector gene expression was restricted to striatal neurons, regardless of the constitutive promoter used. Furthermore, a six-glutamate residue insertion immediately after the VP2 start residue shifted CBA-driven cellular gene expression from neurons to oligodendrocytes. Conversely, a six-alanine insertion in the same AAV9 capsid region reversed the CBh-mediated oligodendrocyte expression back to neurons without changing AAV9 capsid access to oligodendrocytes. Given the preponderance of AAV9 in ongoing clinical trials and AAV capsid engineering, this AAV9 capsid-promoter interaction reveals a previously unknown novel contribution to cell-selective AAV-mediated gene expression in the CNS.
Graphical Abstract
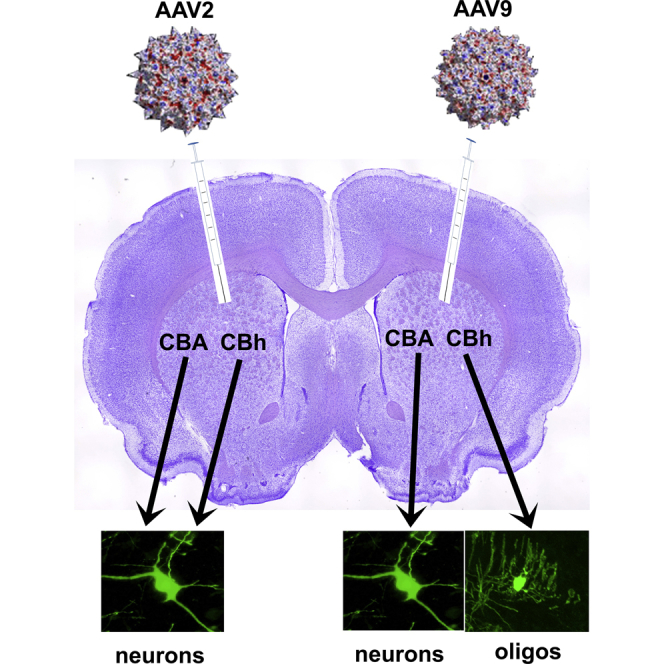
AAV9 vectors exhibit differential in vivo cellular gene expression between CBA and Cbh constitutive promoters. Six amino acid insertions into VP1/VP2 reverse the differential cellular gene expression, but no differences occur for vector cellular access. AAV9 capsids interact with cellular gene expression in vivo.
Introduction
Given highly favorable in vivo properties, adeno-associated virus (AAV) vectors have attained a prominent role in preclinical and clinical gene therapy studies. Importantly, for many applications successful outcomes depend on achieving specific properties of cellular transduction and gene expression. One means to achieve selective vector properties involves manipulation of the AAV capsid structure, based on the role of the capsid in cellular binding, endosome escape, and nuclear localization. The AAV capsid gene encodes three structural proteins (VP1, VP2, and VP3) that assemble into the capsid in a 1:1:10 ratio. VP1 and VP2 have unique N-terminal sequences that are not in VP3, whereas the VP3 sequence (C-terminal) is shared among all VP proteins.1, 2, 3 To date, manipulation of the capsid has focused on the VP3 sequence primarily through the use of rational mutagenesis or capsid DNA shuffling. In the case of capsid DNA shuffling, manipulating AAV capsid DNA by DNA shuffling or error-prone PCR creates an AAV capsid library that contains substantial diversity.4 Subsequent administration of this library either in vitro or in vivo provides the means to rescue novel capsids that fulfill the specific selection pressures exerted by directed evolution parameters. For example, novel capsids have been generated that efficiently transduce neural stem cells5 or oligodendrocytes.6
Although capsid engineering has focused on manipulating VP3, recent studies suggest that the capsid proteins, including VP1 and VP2, can influence cellular gene expression. Johnson et al.7 established that mutations in the AAV2 VP2 capsid significantly altered gene expression in vitro. Similarly, Aydemir et al.8 reported that single point mutants in the “dead zone” on the AAV2 capsid surface resulted in packaged AAV vectors that exhibited normal receptor binding, intracellular trafficking, and proper uncoating in the nucleus, but the mutants failed to transcribe the gene. These studies suggested that in addition to cellular binding, entry, and nuclear localization, AAV capsids could exert significant influence on cellular gene expression.
In the present studies we show that when packaged in AAV9, two different, but similar, constitutive promoters exhibit divergent cellular gene expression within the rat striatum even though all other aspects of the transgenes were identical. Subsequent studies revealed that a region at the junction of VP1 and VP2 alters in vivo cellular gene expression in the context of AAV9 and the constitutive promoters. These findings reveal a previously unknown, novel interaction between the AAV9 capsid and constitutive promoters that can determine selective cellular gene expression in vivo.
Results
In the Context of AAV9, Striatal Cell Transgene Expression Is Promoter-Dependent
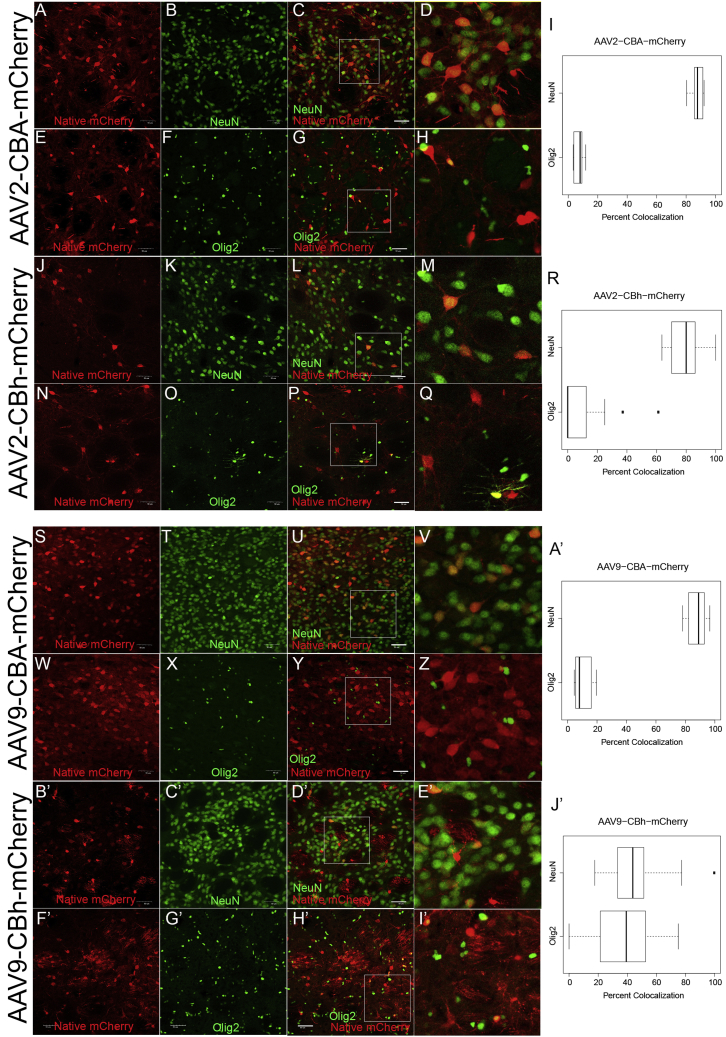
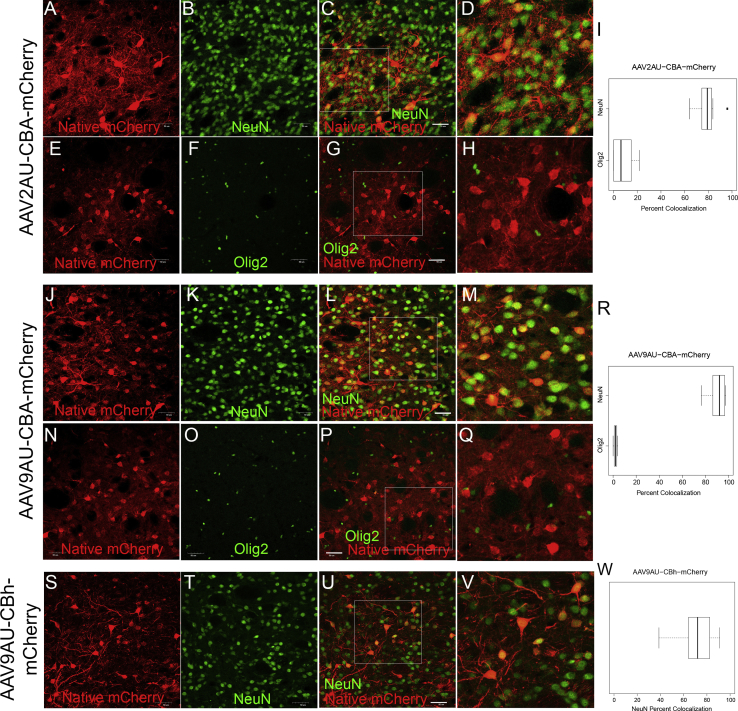
Previously, we have found that AAV2 vector gene expression was restricted to striatal neurons, regardless of whether gene expression was driven by a full-length chicken β-actin (CBA) promoter or the truncated constitutive CBA hybrid (CBh) promoter that we created for self-complimentary AAV vectors.9 However, preliminary studies with AAV9 suggested in vivo differences in cellular gene expression patterns between the two promoters. Thus, in order to make a valid comparison, we constructed identical reporter gene cassettes where mCherry gene expression was driven by either the CBA (1.6 kb) or CBh (0.8 kb) promoter (Figure 1A), but contained an identical length and sequence DNA stuffer, an identical poly(A), and identical inverted terminal repeats (ITRs) (Figure 1B). All vectors were directly infused into wild-type rat striata (Figure S1) at equal titers and volumes. Two weeks post-vector infusion, the identity of transduced cell types was determined by co-localization with known cellular-specific markers using immunofluorescence. AAV2-CBA-mCherry and AAV2-CBh-mCherry vectors exhibited a dominant neuronal gene expression regardless of the promoter (Figures 2A–2R). mCherry-positive cells co-localized with the neuronal marker, NeuN (Figures 2A–2D and 2J–2M), in 87.5% ± 1.7% and 80.4% ± 3.2% of cells when gene expression was driven by CBA or CBh, respectively (Figures 2I and 2R). Conversely, only 7.3% ± 1.3% and 9.7% ± 4.1% of mCherry-positive cells co-localized with an oligodendrocyte marker (Olig2) (Figures 2E–2H and 2N–2Q) when gene expression was driven by a CBA or CBh promoter, respectively (Figures 2I and 2R). Although for AAV2, both promoters exhibit a preferential neuronal gene expression, note that in the context of single-stranded AAV vectors, the CBh promoter does not drive gene expression to the same extent as the full-length CBA promoter.
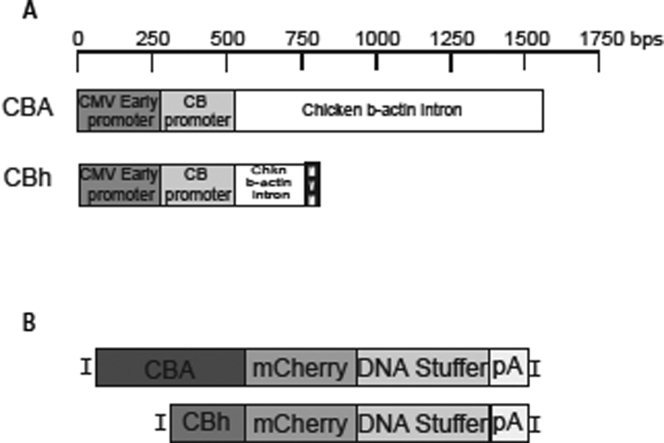
Figure 1.
Diagram of the Components of Two Constitutive Promoters CBA and CBh Used to Drive Reporter Gene Expression
(A) Shared elements of the CBA (1.6 kb) and CBh (0.8 kb) promoters: CMV early promoter, chicken β-actin promoter, and chicken β-actin intron. The CBh promoter has a truncated chicken β-actin intron and an MVM intron (minute virus of mouse). (B) Diagrams of the AAV transgenes constructed to directly compare the CBA and CBh promoters with the identical backbone, AAV2 ITRs, mCherry transgene, and DNA stuffer, to increase transgene size and human growth hormone poly(A). The transgenes are single stranded.
Figure 2.
Representative Confocal Images of AAV2 and AAV9 with mCherry Expression Driven by a CBA or CBh Promoter
Vectors were infused directly into the rat striatum at equal titers and equal volumes. mCherry transgene expression was compared with a neuronal cell marker (NeuN) and an oligodendrocyte marker (Olig2), and co-localization was quantified. Representative confocal images of AAV2-CBA-mCherry (N = 4) illustrate NeuN localization (A–D) and Olig2 localization (E–H) with subsequent quantification over several images (I). Representative confocal images of AAV2-CBh-mCherry (N = 6) illustrate NeuN localization (J–L) and Olig2 localization (N–Q) with subsequent quantification over several images (R). Representative confocal images of AAV9-CBA-mCherry (n = 6) illustrate NeuN localization (S–V) and Olig2 localization (W–Z) with subsequent quantification over several images (A′). Representative confocal images of AAV9-CBh-mCherry (N = 6) illustrate NeuN localization (B′–E′) and Olig2 localization (F′–I′) with subsequent quantification over several images (J′). Scale bars, 50 μm. The white box indicates the zoomed-in portion of the image that is shown to right of the image.
Surprisingly, the CBA and CBh promoters exhibited divergent cellular gene expression patterns in otherwise identical AAV9 vectors. When AAV9-CBA-mCherry vectors were infused into the rat striatum, 88.4% ± 1.6% of the mCherry-positive cells co-localized with NeuN (Figures 2S–2V and 2A′), and only 10.4% ± 2.1% of mCherry-positive cells co-localized with Olig2 (Figures 2W–2Z and 2A′). In marked contrast, AAV9-CBh-mCherry transgene expression co-localized with NeuN (Figures 2B′–2E′) in 46.3% ± 4.6% of the mCherry-positive cells (Figure 2J′), while 37.8% ± 4.9% of the remaining mCherry-positive cells co-localized with Olig2 (Figures 2F′–2J′). Given that in both instances the capsid was identical, the AAV9 vectors inevitably gained identical cellular access and were trafficked to the nucleus of neurons and oligodendrocytes in the rat striatum. Also, with regard to the promoter activity with AAV2, the CBh promoter supports gene expression in striatal neurons (Figures 2J–2M), while in the context of an oligodendrocyte preferring AAV capsid, previous studies have shown that the CBA promoter supports gene expression in striatal oligodendrocytes.6,10 Because both promoters can support gene expression in striatal neurons and oligodendrocytes, the divergence in cellular gene expression patterns suggests that the AAV9 capsid exerted a significant influence on promoter activity across different CNS cell types.
AAV9-CBA Striatal Transgene Expression Is Shifted to Oligodendrocytes by a Six-Glutamate Insertion in VP1/VP2
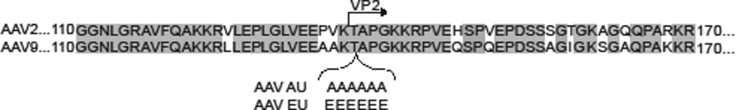
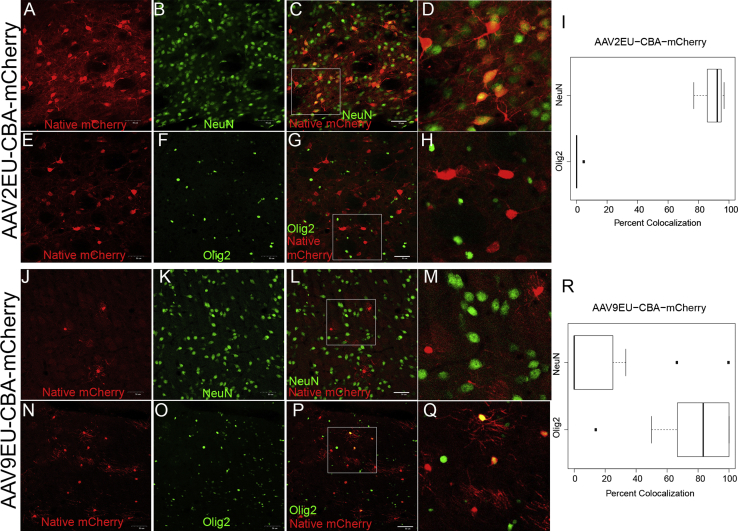
Mutations in VP2 capsids can alter gene expression in vitro,7 so we sought to disrupt a positively charged/basic region of VP1/VP2 intersection within the AAV capsid by inserting six glutamates after amino acid 138 into AAV2 (AAV2EU) and AAV9 (AAV9EU) (Figure 3). The mutant capsids were packaged with the CBA promoter construct, directly infused into rat striatum, and cellular expression was determined. The insertion of six glutamates into AAV2 (AAV2EU-CBA-mCherry) did not alter the cellular transgene expression from that of AAV2 shown in Figure 2. In the striatum the mCherry-positive cells predominantly co-localized with NeuN (89.7% ± 2.7%) and rarely co-localized with Olig2 (0.83% ± 0.8%) (Figures 4A–4I). However, AAV9EU-CBA-mCherry exhibited little co-localization with NeuN (14.2% ± 3.6%) (Figures 4J–4M and 4R), a marked departure to findings with AAV9-CBA (Figures 2S–2V and 2A′). This reduction of mCherry-positive neurons in AAV9EU-CBA was significantly lower compared to AAV9-CBA (p < 0.001). In marked contrast, AAV9EU-CBA mCherry expression co-localized with Olig2 (79.9% ± 4.6%) (Figures 4N–4R). These results suggest that the AAV9EU capsid exerted a significant influence on in vivo cellular gene expression in a manner that previously has not been described.
Figure 3.
Alignment of AAV2 and AAV9 Capsid Residues Surrounding the Site of Amino Acid Insertions
Matching residues are gray; differing residues are black and white.
Figure 4.
Representative Confocal Images of AAV2EU-CBA and AAV9EU-CBA with mCherry Expression Driven by a CBA Promoter
Vectors were infused directly into the rat striatum at equal titers and equal volumes. mCherry transgene expression was compared with a neuronal cell marker (NeuN) and an oligodendrocyte marker (Olig2), and co-localization was quantified. Representative confocal images of AAV2EU-CBA-mCherry (N = 2) compared with NeuN localization (A–D), Olig2 localization (E–H), and the quantification over several images (I) are shown. Representative confocal images of AAV9EU-CBA-mCherry (N = 5) compared with NeuN localization (J–M), Olig2 localization (N–Q), and the quantification over several images (R) are shown. Scale bars, 50 μm. The white box indicates the zoomed-in portion of the image that is shown to right of the image.
AAV9 Striatal Transgene Expression Is Neuronal After a Six-Alanine Insertion into VP1/VP2
Next, to assess whether the insertion itself was responsible for the change in cellular expression, six neutral alanine residues were inserted into the same VP1/VP2 site in AAV2 and AAV9 (AAV2AU and AAV9AU). The mutant capsids were packaged with the CBA promoter construct, directly infused into rat striatum, and cellular expression was determined. The insertion of six alanines into AAV2 (AAV2AU) did not alter the cellular transgene expression from that of AAV2EU-CBA or AAV2-CBA (Figures 2A–2I, 4A–4D, and 4I); the mCherry-positive cells co-localized mostly with NeuN (79.2% ± 3.8%) with limited co-localization with Olig2 (8.3% ± 3.6%) (Figures 5A–5I). Also, the insertion of six alanines into AAV9 (AAV9AU-CBA) resulted in a predominant neuronal tropism similar to that seen with AAV9-CBA (Figures 2S–2A′). AAV9AU-CBA mCherry expression co-localized with NeuN (89.8% ± 3.9%) while only 2.1% ± 0.8% of mCherry-positive cells co-localized with Olig2 (Figures 5J–5R). Notably, the increase in mCherry-positive neurons with AAV9AU-CBA is significant compared to AAV9EU-CBA (p < 0.001). Furthermore, the reduction in mCherry-positive oligodendrocytes in AAV9AU-CBA compared to AAV9-CBA was also significant (p < 0.005). Surprisingly, when gene expression was driven by the CBh promoter, the six alanine insertions into AAV9 reversed the predominant oligodendrocyte gene expression back to a predominant neuronal gene expression pattern (Figures 5S–5W). Taken together, our results suggest that the glutamates’ negative charge in AAV9EU disrupts a capsid-transgene interaction that determines neuronal transgene expression of AAV9-CBA while the neutral alanine insertions reverse the capsid interaction with the CBh promoter that supports oligodendrocyte expression. Again, these results implicate a capsid-promoter interaction in vivo.
Figure 5.
Representative Confocal Images of AAV2AU-CBA and AAV9AU-CBA with mCherry Expression Driven by a CBA Promoter
Vectors were infused directly into the rat striatum at equal titers and equal volumes. mCherry transgene expression was compared with a neuronal cell marker (NeuN) and an oligodendrocyte marker (Olig2), and co-localization was quantified. Representative confocal images of AAV2AU-CBA-mCherry (N = 2) compared with NeuN localization (A–D), Olig2 localization (E–H), and the quantification over several images (I) are shown. Representative confocal images of AAV9AU-CBA-mCherry (N = 2) compared with NeuN localization (J–M), Olig2 localization (N–Q), and the quantification over several images (R) are shown. Representative confocal images of AAV9AU-CBh-mCherry (N = 2) compared with NeuN localization (S–V) and the quantification over several images (W) are shown. Scale bars, 50 μm. The white box indicates the zoomed-in portion of the image that is shown to right of the image.
AAV9AU Can Transduce Striatal Oligodendrocytes
While there are no published reports of the VP1/VP2 region influencing AAV gene expression, it was entirely possible that the alanine insertion could possibly have shifted AAV9 capsid binding away from oligodendrocytes. Therefore, we constructed a transgene where gene expression was driven by an oligodendrocyte-specific promoter (MBP [myelin basic protein]) and then packaged the transgene into AAV9AU or AAV9. Our previous results showed that AAV9AU-CBA-mediated transgene expression was predominantly neuronal (Figures 5J–5R). However, for both the AAV9AU and AAV9, MBP-driven expression of GFP was non-neuronal (Figure S2). Thus, the alanine insertion did not alter the ability of AAV9AU to bind and transduce oligodendrocytes but did prevent oligodendrocyte localized gene expression driven by the constitutive CBh promoter.
AAV9-CBh Oligodendrocyte-Preferring Gene Expression Is Consistent with Other Transgenes
Due to the size differences in CBA and CBh promoters, the final single-stranded constructs differed by approximately 1 kb. To address whether packaging size issues influenced the capsid contribution to the change in cellular gene expression, we constructed an AAV9 vector packaged with self-complementary CBh-GFP containing a different poly(A) (bovine growth hormone) and transgene (GFP) such that the entire transgene was close to the packaging capacity. This vector exhibited both in vivo oligodendrocyte and neuronal gene expression (Figure S3) as was found with the single-stranded AAV9 vectors where CBh drove gene expression.
Discussion
In the context of AAV9, divergent in vivo cellular gene expression occurred between two similar constitutive promoters, CBA and CBh, even though all other components of the vector were identical. Because in both instances the capsid was identical, differential cellular binding, intracellular trafficking, and nuclear localization appear unlikely to be contributing factors. Also, as seen in Figure 2, both promoters can support gene expression in neurons and oligodendrocytes albeit to differing degrees. Finally, the 1-kb difference in cassette size does not appear to contribute, because a self-complimentary AAV9 vector with the CBh promoter exhibited oligodendrocyte gene expression in vivo. Although, no such differences in in vivo gene expression occurred with AAV2 vectors, it is possible that other AAV serotypes could exhibit similar capsid-promoter interactions. Clearly, these initial findings suggest that unknown elements of the AAV9 capsid can influence promoter permissiveness within different CNS cellular populations. To date, cell-specific gene expression has been achieved by manipulation of the capsid, including utilization of cell-specific promoters or cell-specific enhancers.4,11, 12, 13 Thus, the observed interaction of the AAV9 capsid with constitutive promoter gene expression in vivo reveals a previously unknown, novel property of AAV9 vectors.
Subsequent investigations not only further reinforced the conclusion that the AAV9 capsid can interact with promoter activity, but they also provided evidence for a specific site that appears directly involved in this capsid/promoter interaction. The insertion of six negatively charged glutamates into the VP1/VP2 junction had no effect on cellular gene expression in the context of AAV2, even though with the MBP promoter, AAV2 supports oligodendrocyte gene expression in the rat CNS.14 For AAV9, this mutation shifted the balance of CBA-mediated gene expression from neurons to oligodendrocyte gene expression. Because the six-glutamate insertion exerted no influence on AAV2 neuronal gene expression, this amino acid insertion likely did not cause a general alteration in capsid-binding properties. With regard to the potential for some non-specific insertion effect, insertion of six neutral alanines in the same position did not alter the predominant neuronal gene expression by the CBA promoter. However, surprisingly the six-alanine insertion into AAV9 reversed the CBh-mediated cellular gene expression pattern from oligodendrocytes to a predominant neuronal transduction pattern. Importantly, this reversal in cellular gene expression did not arise from a reduction of cellular capsid binding to oligodendrocytes. When gene expression was driven by an oligodendrocyte-specific promoter, both AAV9 and AAV9AU vectors exhibited in vivo gene expression that was confined to oligodendrocytes. Clearly, both AAV9 and AAV9AU gained access to oligodendrocytes, yet in the context of the six-alanine insertions, CBh-mediated gene expression was shifted from oligodendrocytes to neurons. Thus, these amino acid insertions not only validate an AAV9 capsid-promoter interaction, but they implicate an area of the capsid that to date has not been associated with modulation of capsid binding or gene expression. Further in-depth investigations will be necessary to determine whether the capsid directly interacts with the promoters, as well as potential permissive or suppressive elements that prove uniquely expressed in the different cell types.
Although the many approaches to AAV capsid engineering focus on the VP3 capsid protein, several previous studies have provided tangential evidence that the AAV capsid can influence gene expression. Johnson et al.7 found that mutations in the VP1/VP2 junction in AAV2 caused a significant decrease in in vitro gene expression. Similarly, Aydemir et al.8 showed that mutations in the “dead zone” of the AAV2 capsid resulted in a lack of gene expression, even though no changes were found in receptor binding, endosome escape, nuclear localization, or capsid uncoating. Currently, AAV9 vectors have attained a central prominence in clinical gene therapy, particularly with regard to single-gene disorders such as RPE65 retinal mutations, spinal muscular atrophy, and giant axon neuropathy.15, 16, 17 In addition, AAV9 capsid sequences comprise a significant proportion of engineered chimeric capsids.18,19 Given the rapidly expanding generation of synthetic promoters,20 an AAV9 capsid interaction with constitutive promoter activity certainly could complicate the actual contributions to cellular gene expression profiles in the CNS. Thus, future studies will need to determine the overall mechanistic landscape by which the AAV9 capsid interacts with constitutive promoters in the CNS.
Materials and Methods
Cloning Construct
In order to directly compare the transgenes, promoters were constructed by inserting a DNA stuffer into pAAV-CMV (cytomegalovirus)-mCherry-hGH (human growth hormone)-poly(A) using SacII. The CBA promoter was digested out of an existing construct using BglII (blunted) and SalI and then ligated into pAAV-mCherry-DNA stuffer-hGH-poly(A) at MluI (blunted) and SalI sites. The CBh promoter was digested out of an existing construct using KpnI (blunted) and Age1 and then ligated into pAAV-mCherry-DNA stuffer-hGH-poly(A) at MluI (blunted) and AgeI sites.
Virus Production
The virus was produced in HEK293 cells as previously described.18 Briefly, polyethylenimine max (PEI) was used for the triple transfection of a rep/cap plasmid (pGSK2/9 [AAV9], pGSK2/9EU, pSGK2/9AU, pXR2 [AAV2], pXR2AU, and pXR2EU), the pXX6-80 helper plasmid, and a transgene plasmid (pAAV-CBA-mCherry-DNA stuffer-hGH-poly(A), pAAV-CBh-mCherry-DNA stuffer-hGH-poly(A), pTR-MBP-GFP, pTR-CBh-GFP). Cells were harvested 48 h post-transfection, and the virus was purified by cesium chloride ultracentrifugation. After identifying peak fractions by qPCR, the virus was dialyzed into PBS/NaCl/d-sorbitol. Titers were calculated by qPCR according to established procedures using a LightCycler 480 instrument and ITR primers. The individual titers were as follows: AAV2-CBA, 1.9 × 1012 vector genomes (vg)/mL; AAV2-CBh, 1.3 × 1013 vg/mL; AAV9-CBA, 6 × 1013 vg/mL; AAV9-CBh, 2.5 × 1013 vg/mL; AAV2EU-CBA, 5 × 1012 vg/mL; AAV2AU-CBA, 2 × 1012 vg/mL; AAV9AU-CBh, 3.6 × 1012 vg/mL; scAAV9-MBP, 3 × 1012 vg/mL; scAAV9AU-MBP, 1.5 × 1012 vg/mL; scAAV9-CBh-GFP, 1.4 × 1012 vg/mL.
Animals and Stereotactic Infusions
All of the animals were male Sprague-Dawley rats (Charles River Laboratories) weighing between 200 and 300 g at the time of intracranial injection. The animals were maintained on a 12-h light/12-h dark cycle and had free access to water and food. For all animal studies, care and procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and all procedures received prior approval by the University of North Carolina Institutional Animal Care and Usage Committee.
Virus vector infusions were performed as previously described.6,9 First, animals were anesthetized with 50 mg/kg pentobarbital and placed into a stereotactic frame. Using a 32G stainless steel injector and a Sage infusion pump, animals received 2 μL unilaterally of each vector 10 min into the striatum (0.5 mm anterior to bregma, 3.5 mm lateral, and 5.5 mm vertical, according to the atlas of Paxinos and Watson.19 The injector was left in place for 3 min post-infusion in order to allow diffusion from the injector. The animal numbers were as follows: AAV2-CBA-mCherry (N = 4); AAV2-CBh-mCherry (N = 6); AAV9-CBA-mCherry (N = 6); AAV9-CBh-mCherry (N = 6); AAV2EU-CBA-mCherry (N = 2); AAV9EU-CBA-mCherry (N = 5); AAV9-MBP-GFP (N = 2); AAV9-CBh-GFP (N = 2).
Immunochemistry and Confocal Microscopy
Two weeks after AAV vector infusion, animals received an overdose of pentobarbital (100 mg/kg pentobarbital intraperitoneally [i.p.]), and they were perfused transcardially with ice-cold 100 mM PBS (pH 7.4), followed by 4% paraformaldehyde in phosphate buffer (PB) (pH 7.4). After brains were post-fixed 12–48 h at 4°C in the paraformaldehyde-PB, 40-μm coronal sections were cut using a vibrating blade microtome for subsequent immunofluorescence. To determine fluorescent transgene (mCherry or GFP) cellular co-localization, tissue sections were incubated in the blocking solution with either cellular marker antibodies NeuN (1:500, Chemicon) or Olig2 (1:250, Abcam). Following incubation at 4°C for 48–72 h in primary antibodies, the sections were rinsed three times with PBS and blocked again for 45 min at room temperature. Subsequently, the tissue sections were incubated in either Alexa Fluor 488- or Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin G (IgG) or goat anti-mouse (1:500, Invitrogen) for 1 h at 4°C. Rinsed sections were mounted, and fluorescence was visualized using an Olympus FV3000RS confocal microscope in the UNC Neuroscience Center Confocal. Transgene fluorescence co-localization was determined on the z stacks and counted using an ImageJ cell counter plug-in. At least 125 individual cells were counted per vector and antibody. The cell counts were tallied and basic statistics were performed in Excel. Cell counts are reported as percent co-localization (±SEM). R21 was used to graph and perform independent t tests to test for significance.
Author Contributions
S.K.P. and T.J.M. designed and conducted the experiments, and S.K.P., T.J.M., and R.J.S. wrote and edited the manuscript.
Conflicts of Interest
R.J.S. is a founder and a shareholder at Asklepios Biopharmaceutical. He holds patents that have been licensed by UNC to Asklepios Biopharmaceutical, for which he receives royalties. The remaining authors declare no competing interests.
Acknowledgments
These studies were supported by NIH NINDS grant NS082289 (to T.J.M.) and a Pfizer-NCBiotech post-doctoral fellowship (to S.K.P.). Confocal microscopy was conducted in the UNC Neuroscience Center Confocal and Multiphoton Imaging Core (NINDS center grant P30 NS045892).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.03.007.
Supplemental Information
References
- 1.Buller R.M., Rose J.A. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J. Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson F.B., Ozer H.L., Hoggan M.D. Structural proteins of adenovirus-associated virus type 3. J. Virol. 1971;8:860–863. doi: 10.1128/jvi.8.6.860-863.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose J.A., Maizel J.V., Jr., Inman J.K., Shatkin A.J. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang J.H., Koerber J.T., Kim J.S., Asuri P., Vazin T., Bartel M., Keung A., Kwon I., Park K.I., Schaffer D.V. An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Mol. Ther. 2011;19:667–675. doi: 10.1038/mt.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powell S.K., Khan N., Parker C.L., Samulski R.J., Matsushima G., Gray S.J., McCown T.J. Characterization of a novel adeno-associated viral vector with preferential oligodendrocyte tropism. Gene Ther. 2016;23:807–814. doi: 10.1038/gt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson J.S., Li C., DiPrimio N., Weinberg M.S., McCown T.J., Samulski R.J. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 2010;84:8888–8902. doi: 10.1128/JVI.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aydemir F., Salganik M., Resztak J., Singh J., Bennett A., Agbandje-McKenna M., Muzyczka N. Mutants at the 2-fold interface of adeno-associated virus type 2 (AAV2) structural proteins suggest a role in viral transcription for AAV capsids. J. Virol. 2016;90:7196–7204. doi: 10.1128/JVI.00493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray S.J., Foti S.B., Schwartz J.W., Bachaboina L., Taylor-Blake B., Coleman J., Ehlers M.D., Zylka M.J., McCown T.J., Samulski R.J. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 2011;22:1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg M.S., Criswell H.E., Powell S.K., Bhatt A.P., McCown T.J. Viral vector reprogramming of adult resident striatal oligodendrocytes into functional neurons. Mol. Ther. 2017;25:928–934. doi: 10.1016/j.ymthe.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh M.S., Hong S.J., Huh Y., Kim K.S. Expression of transgenes in midbrain dopamine neurons using the tyrosine hydroxylase promoter. Gene Ther. 2009;16:437–440. doi: 10.1038/gt.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.A., Guo L., Xu Q., Liu R., Lu C., Chu J. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankvoort S., Witter M.P., Noonan J., Cotney J., Kentros C. Marked diversity of unique cortical enhancers enables neuron-specific tools by enhancer-driven gene expression. Curr. Biol. 2018;28:2103–2114.e5. doi: 10.1016/j.cub.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H., McCarty D.M., Bruce A.T., Suzuki K. Oligodendrocyte-specific gene expression in mouse brain: use of a myelin-forming cell type-specific promoter in an adeno-associated virus. J. Neurosci. Res. 1999;55:504–513. doi: 10.1002/(SICI)1097-4547(19990215)55:4<504::AID-JNR10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 15.Bennett J., Wellman J., Marshall K.A., McCague S., Ashtari M., DiStefano-Pappas J., Elci O.U., Chung D.C., Sun J., Wright J.F. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: a follow-on phase 1 trial. Lancet. 2016;388:661–672. doi: 10.1016/S0140-6736(16)30371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 17.Bailey R.M., Armao D., Nagabhushan Kalburgi S., Gray S.J. Development of intrathecal AAV9 gene therapy for giant axonal neuropathy. Mol. Ther. Methods Clin. Dev. 2018;9:160–171. doi: 10.1016/j.omtm.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojala D.S., Sun S., Santiago-Ortiz J.L., Shapiro M.G., Romero P.A., Schaffer D.V. In vivo selection of a computationally designed SCHEMA AAV library yields a novel variant for infection of adult neural stem cells in the SVZ. Mol. Ther. 2018;26:304–319. doi: 10.1016/j.ymthe.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domenger C., Grimm D. Next-generation AAV vectors—do not judge a virus (only) by its cover. Hum. Mol. Genet. 2019;28(R1):R3–R14. doi: 10.1093/hmg/ddz148. [DOI] [PubMed] [Google Scholar]
- 21.R Studio Team RStudio: Integrated Development for R (RStudio, Inc.) 2015. http://www.rstudio.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.