See Commentary on Page 575
Autosomal dominant polycystic kidney disease (ADPKD) is caused by loss-of-function or deleterious mutations in the PKD1 or PKD2 genes and is seen with a prevalence of 1:400 to 1:1000.1 Patients develop kidney and liver cysts that accumulate and expand to crowd out the normal organ parenchyma and lead to kidney failure in half of patients by age 60 years.2 To find targeted therapies, we need to better understand the function of the PKD1 and PKD2 gene products, polycystin-1 (PC1) and polycystin-2 (PC2). Autosomal dominant polycystic liver disease (ADPLD), also known as isolated polycystic liver disease (PCLD), is a clinical characterization of patients on the same phenotypic and mechanistic spectrum as ADPKD with regard to their liver cysts, but lacking clinically relevant kidney cysts.3 Autopsy studies suggest that PCLD has a similar prevalence to that of ADPKD.4 Approximately 50% of cases of this phenotype have been explained by mutations in any 1 of at least 7 endoplasmic reticulum protein-encoding genes, the loss of which indirectly results in insufficient PC1 functional dosage.4, 5, 6 These genes include PRKCSH, SEC63, GANAB, ALG8, SEC61B, DNAJB11, and ALG9.6 Variants in GANAB, DNAJB11, and ALG9 explain a small number of cases clinically suspected to have ADPKD.6,7 We aim to solve additional cases by improving the sensitivity of variant identification in established genes, and through implication of novel disease genes. These genes will be potential targets for treatments to upregulate the functional dosage of PC1.
Whole exome sequencing (WES) is a useful technique for gene discovery projects, as it sequences the coding regions and flanking intronic bases of all defined genes regardless of their implicated role in human homeostasis or disease.8 WES is highly sensitive at identifying small insertions, deletions, and single- or oligo-nucleotide variants. Because of its 75−base pair read length, WES is not equipped to detect large deletions. WES requires both a capture step and a polymerase chain reaction (PCR)−based amplification step that together result in significant variability of read depth across the exome. This precludes the use of read depth for a given sample to suggest areas of deletion or duplication. A statistical tool known as exome hidden Markov model (XHMM) uses comparative exome read depth across a large group of samples to predict copy number variation (CNV). Application of XHMM to the ExAC cohort of 59,898 human exomes showed that 70% of individuals carried at least one rare (<0.5%) CNV in the region of a gene.9
In this study, we investigated a cohort of 128 unrelated individuals with clinically diagnosed PCLD or ADPKD with no mutation detected (ADPKD-NMD), who remained genetically unresolved following WES. This cohort includes 115 patients enrolled at Yale or by Yale collaborators, in addition to 13 mild to moderate ADPKD-NMD patients: 9 from the Consortium of Radiologic Imaging Study of PKD (CRISP),S1 and 4 from the HALT-PKDS2 clinical cohorts obtained via the National Institute of Digestive and Kidney Diseases central repository. The established disease pathomechanism dictates that the pathogenic genotype must be a heterozygous loss-of-function or deleterious allele. We expect that the majority of these cases will be explained by either mutations in novel disease genes or mutations in established disease genes missed by standard WES analysis. We aimed to distinguish these subsets by searching for CNVs in established disease genes in our cohort.
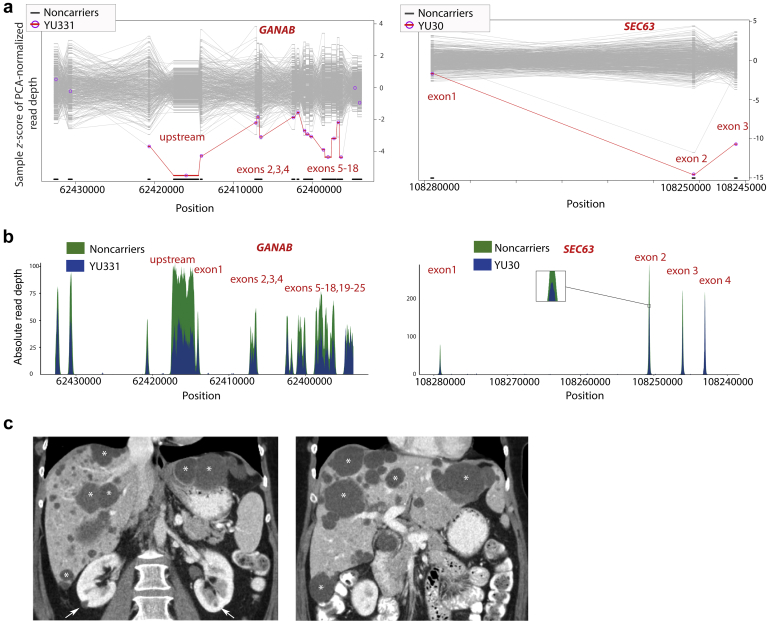
We applied XHMM to the existing WES raw data and that of healthy controls sequenced at our institution to detect possible CNVs.S3 We aligned these CNV calls to regions of known genes for ADPKD and PCLD (PKD1, PKD2, PRKCSH, SEC63, GANAB, ALG8, SEC61B, PKHD1, DNAJB11, ALG9) to identify candidate CNVs of interest. Two cases, YU331 and YU30, showed evidence of deletions overlapping GANAB and SEC63, respectively (Figure 1a). Among the approximately 20,000 alleles in the gnomAD structural variant data set, the affected GANAB region has no reported deletions and a sole deletion of 345kb spanned the SEC63 gene.S3,S4 Figure 1b shows the raw data input for the CNV call. These gene regions in the respective individuals lacked heterozygous variant calls consistent with representation from a single allele. YU331 is a 52-year-old woman who presented with right upper quadrant pain and was found to have an 8-cm liver cyst that required aspiration to relieve her symptoms. She had 5 additional large cysts (>5 cm) and numerous smaller liver cysts, as well as a small number of kidney cysts (Figure 1c). YU30 is a 36-year-old woman who presented with worsening abdominal fullness following her third pregnancy. She was found to have innumerable cysts throughout her liver, particularly severe in the right lobe, causing marked liver enlargement. There were no cysts in the kidneys or pancreas. She had no known family history of kidney or liver cysts; however, her parents and brothers had never been screened.
Figure 1.
Polycystic kidney and liver disease cases YU331 and YU30 have significantly reduced exome sequencing read depth in regions of GANAB and SEC63, respectively. (a) Z-Score plots of normalized exome read depth for deletion calls made by XHMM overlaping polycystic liver disease (PCLD) genes. Comparison between cases YU331 (left panel) or YU30 (right panel) and the remainder of cases and controls sequenced with same capture reagent show significantly reduced read depth suggesting a deletion. (b) Absolute read depth plotted at each hg19 genomic position for YU331 on chr:11 (left panel) and YU30 on chr:6 (right panel). (c) Computed tomography scan showing liver cysts (∗) and a small number of kidney cysts (arrows) in YU331.
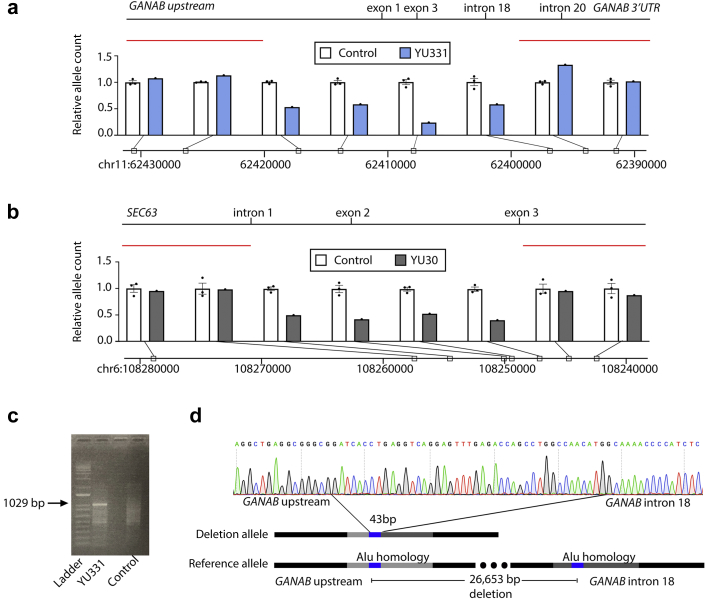
To validate the CNV calls, we performed quantitative PCR (qPCR) on genomic DNA from each putative deletion region to determine the relative normalized allele count in comparison with controls without evidence of CNV in that region. This approach showed that YU331 had relative allele counts of 24% to 58% across a minimum 25-kilobase region of GANAB, spanning exons 1 to 18, which encode the gene’s enzymatic glucosidase domain (Figure 2a). YU30 had relative allele counts of 41% to 59% across a minimum 7-kb region of SEC63, encompassing intron 1, exon 2, and intron 2 (Figure 2b). This region, which is lacking on 1 of the 2 parental alleles includes the entirety of the 100-bp exon 2 encompassing amino acids 42 to 75, which encode the first cytoplasmic loop of this triple membrane−spanning protein. Loss of this exon would not only result in loss of this domain but also result in a frameshift if exon 1 were to splice to exon 3.
Figure 2.
Confirmation and definition of GANAB and SEC63 heterozygous deletions. (a,b) Relative allele count of YU331 (a) or YU30 (b) determined by normalized genomic DNA quantitative polymerase chain reaction (qPCR) compared with the mean of 3 controls. Genomic DNA qPCR of an intronic region of GAPDH was used for normalization. Controls are 3 cases without the proposed deletion. Positions noted on hg19. (c) Agarose gel showing PCR product (arrow) using primers spanning the deletion region amplifies in YU331 but not in control. (d) Sanger sequencing of the 1029−base pair product of YU331’s deletion allele identifies the junction of GANAB upstream sequence and intron 18 sequence separated by a 43−base pair sequence present on either side of the breakpoint.
To precisely define the heterozygous deletions, we next attempted to amplify the deletion allele using a forward primer at the upstream flank and a reverse primer at the downstream flank of the deletion. In each case, these were distant enough to preclude amplification of the wild-type allele. Sanger sequencing of the YU331 PCR amplicon (Figure 2c) defined a 26,653−base pair variant: NC_000011.9:g.62395884_62422536del (Figure 2d). This variant will be available in ClinVar (SCV000995031). The upstream and downstream breakpoint each bordered an identical 43−base pair sequence on either side of the deletion region (Figure 2d). This suggests the mechanism of nonallelic homologous recombination in the generation of this mutation. Furthermore, we find this 43−base pair sequence to be present at least once on nearly every chromosome in the human reference genome. In the evaluated sequence on chromosome 11 upstream and within the GANAB genomic sequence, it is contained within approximately 300 base pairs that have high homology to the Sq and Sx families of Alu short interspersed elements known to be predisposed to recombination.S5,S6 Whether the presence of the Alu repeat in GANAB predisposes it more than other genes to large deletions cannot yet be determined.
We were unable to amplify a product for the deletion allele in YU30. We expect that this is due to PCR-related challenges, as intron 1 of SEC63 is very GC rich. However, it remains possible that there is additional structural complexity to this variant. Although the qPCR results confirm the reduced copy number, an inversion neighboring the deletion or a large insertion between the breakpoints could nonetheless prevent amplification of a product using expected primer sites.
Large deletions have not been previously reported as the germline pathogenic mutation in any of the disease genes for non-PKD1, non-PKD2 polycystic kidney and liver patients. Large deletions up to 17 kilobases have been reported in PKD1 or PKD2 detected by either the gold standard long-range PCR sequencing method for ADPKD genes or multiplex ligation-dependent probe amplification (MLPA).S7−S10 When patients have a liver-predominant or mild kidney cystic phenotype, WES is the highest-yield method of genetic testing, given the genetic heterogeneity of the non-PKD1, non-PKD2 phenotype. If traditional WES analysis results are negative, CNVs are considered, yet standard methods such as single nucleotide polymorphism genotyping, comparative genomic hybridization, or whole genome sequencing for detecting large genomic deletions, require costly additional testing. Fromer et al. describe that XHMM applied to WES has a 79% to 85% sensitivity for detection of CNVs overlapping 3 to 5 exome-targeted regions, with a high specificity when compared to alternative methods.S3,S11 The limitations of XHMM are also true of other WES-based CNV calling algorithms. Sensitivity for CNV detection is significantly lower when the CNV spans fewer exons and will be 0 for purely intronic CNVs; limitations in specificity often require human interpretation of data plots and biological confirmation for certainty. Superior sensitivity and specificity for some CNV-calls may be achieved with a combination of multiple algorithms.S12 These technologies will likely continue to improve until whole genome sequencing replaces WES as an affordable initial sequencing methodology and dramatically simplifies and broadens our ability to detect CNVs.
In summary, this is the first study to describe the identification of large deletions in autosomal dominant polycystic kidney and liver disease genes from WES data, and reports the first such clinically relevant deletions in the non-PKD1, non-PKD2 genes. We describe the phenotype of these cases and highlight the utility of CNV analysis to identify pathogenic mutations. The XHMM tool represents a cost-free analysis to identify exon-spanning CNVs in samples with clinical or research genetic testing by WES to help reduce the percentage of genetically unresolved cases. Although not investigated as part of this study, this approach could also help to identity candidate loci for additional gene discovery, which is the ultimate goal of the genetic investigation of this cohort.
Disclosure
SS is a founder, consultant, and scientific advisory board member for Goldfinch Bio, outside the submitted work. VET reports research support from Otsuka Pharmaceuticals, Palladio Biosciences, Mironid, Blueprint Medicines, and Sanofi Genzyme, outside the submitted work. All the other authors declared no competing interests.
Acknowledgments
This study was funded by the following: a PKD Foundation Grant (217G18a), a Robert E. Leet and Clara Guthrie Patterson Trust Mentored Research Award, and a NIH grant K08DK119642 to WB; NIH grants DK100592 and DK51041 to SS; the George M. O’Brien Kidney Center at Yale (P30 DK079310); the Mayo Translational PKD Center (P30 DK090728) and an endowment from Robert M. and Billie J. Pirnie for Kidney Research; and the Yale Center for Mendelian Genomics (Yale CMG; NIH M#UM1HG006504-05). The Yale Center for Mendelian Genomics (UM1HG006504) is funded by the National Human Genome Research Institute. The GSP Coordinating Center (U24 HG008956) contributed to cross-program scientific initiatives and provided logistical and general study coordination. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors would like to thank Dr. Ali Gharavi for his discussion of XHMM usage, the Yale Center for Mendelian Genomics for the whole exome sequencing, and the patients presented in this study.
Footnotes
Supplementary References.
Supplementary Methods.
Table S1. Genomic DNA qPCR primers.
Supplementary Material
References
- 1.Torres V.E., Harris P.C., Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 2.Harris P.C., Torres V.E. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian Q., Li A., King B.F. Clinical profile of autosomal dominant polycystic liver disease. Hepatology. 2003;37:164–171. doi: 10.1053/jhep.2003.50006. [DOI] [PubMed] [Google Scholar]
- 4.Besse W., Dong K., Choi J. Isolated polycystic liver disease genes define effectors of polycystin-1 function. J Clin Invest. 2017;127:3558. doi: 10.1172/JCI96729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fedeles S.V., Tian X., Gallagher A.R. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet. 2011;43:639–647. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornec-Le Gall E., Torres V.E., Harris P.C. Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J Am Soc Nephrol. 2018;29:13–23. doi: 10.1681/ASN.2017050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besse W., Chang A.R., Luo J.Z. ALG9 mutation carriers develop kidney and liver cysts. J Am Soc Nephrol. 2019;30:2091–2102. doi: 10.1681/ASN.2019030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamshad M.J., Ng S.B., Bigham A.W. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 9.Ruderfer D.M., Hamamsy T., Lek M. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat Genet. 2016;48:1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.