Abstract
Metastatic cancer cells invade surrounding tissues by forming dynamic actin-based invadopodia, which degrade the surrounding extracellular matrix and allow cancer cell invasion. Regulatory RNAs, including circular RNA, have been implicated in this process. By microarray, we found that the circular RNA circSKA3 was highly expressed in breast cancer cells and human breast cancer tissues. We further found that the invasive capacity of breast cancer cells was positively correlated with circSKA3 expression, through the formation of invadopodia. Mechanistically, we identified Tks5 and integrin β1 as circSKA3 binding partners in these tumor-derived invadopodia. Ectopic circSKA3 expression conferred increased tumor invasiveness in vitro and in vivo. We further identified the RNA-protein binding sites between circSKA3, Tks5 and integrin β1. In tumor formation assays, we found that circSKA3 expression promoted tumor progression and invadopodium formation. Mutation of the circSKA3 binding sites or transfection with blocking oligos abrogated the observed effects. Thus, we provide evidence that the circular RNA circSKA3 promotes tumor progression by complexing with Tks5 and integrin β1, inducing invadopodium formation.
Keywords: circular RNA, circSKA3, tumorigenesis, invadopodia, invasion
Graphical Abstract
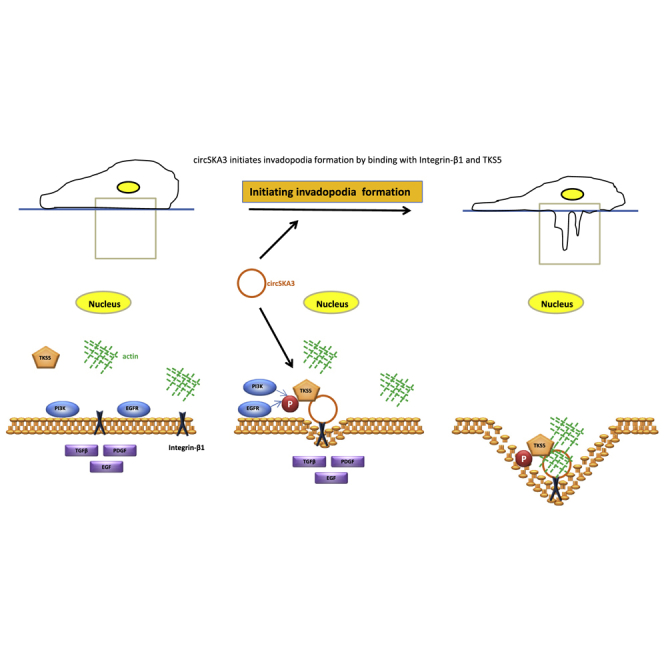
We found that the circular RNA circSka3 was highly upregulated in human breast cancer specimens and cancer cells. circSka3 plays important roles in tumorigenesis and invasiveness of breast cancer cells in vitro and in vivo through the formation of podosome/invadopodia via binding integrin β1 and the podosome marker Tks5.
Introduction
Circular RNAs (circRNAs) are a subclass of non-coding RNAs that form covalently closed loops. Although broadly grouped as a subclass of non-coding RNAs, some circRNAs have been reported to code for protein peptides.1,2 circRNAs can be generated from exons and introns of pre-mRNAs3, 4, 5 and have been detected extensively in mammalian cells.6, 7, 8 Given their abundance and evolutionary conservation, circRNAs may possess regulatory roles.9, 10, 11, 12 Some circRNAs, similar to other non-coding RNAs, have been identified as microRNA (miRNA) sponges that can inhibit miRNA functions.13, 14, 16, 17 We have previously reported that circAmotl1 binds PDK1 and AKT1, inducing nuclear translocation and antagonizing apoptosis.18 In binding to proteins, circRNAs could form three-dimensional (3D) complexes and acquire various structural conformations. This allows circRNAs to impact gene expression and protein activity through a mechanism distinct from their analogous linear mRNA counterparts. Recent studies suggest that circRNAs are involved in the development of different types of cancers, including breast,18, 19, 20, 21, 22 prostate,23,24 liver,25 lung,26 among others.27,28 These circRNAs may regulate cancer cell proliferation,18 migration,29 invasion,30 apoptosis,31 autophagy,32 wound repair,33 and protein synthesis.34
Podosomes are actin-rich structures located on the outer surface of the cell membrane and play essential roles in cell motility by coordinating degradation of the extracellular matrix to facilitate cellular movement. In cancer cells, these are called invadopodia that contain actin, Tks5, and integrins.35,36 Invadopodia are responsible for cancer invasiveness and metastasis, and can be used as markers to quantify the invasiveness of cancer cells. Thus, invadopodia play critical roles in the development of many cancers. However, the roles of circRNAs in the formation of invadopodia remain poorly understood. Our study demonstrated that the circRNA circSKA3 was highly expressed in breast cancer cell lines and human breast cancer tissue samples. Pilot in vitro experiments demonstrated a significant increase in cell motility and invasive capacity. We thus designed experiments to further interrogate the mechanism underlying this observation.
Results
circSKA3 Promotes Breast Cancer Cell Migration and Invasion
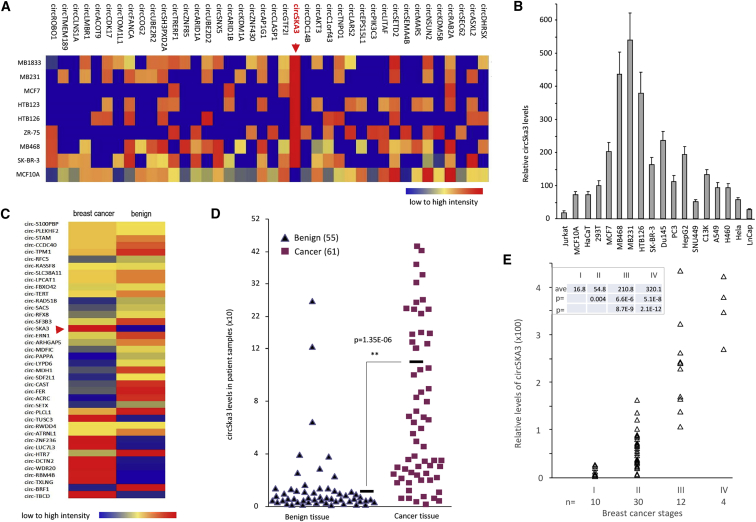
Cell lysates prepared from eight human breast cancer cell lines and a control non-tumorigenic breast epithelial cell line MCF-10A were subjected to microarray analysis to detect circRNA expression. The circSKA3 (has-circ-0000467; Figure S1A also contains sequences used in this study) was found to be highly expressed in all eight breast cancer cell lines relative to control (Figure 1A). To validate this result, we performed real-time PCR using lysates prepared from human non-tumor cell lines (HaCaT, 293T, Jurkat, and MCF10A), breast cancer cell lines (MCF-7, MDA-MB-468, MDA-MB-231, HTB126, and SK-BR-3 from ATCC, Manassas, VA, USA), and other tumor cell lines (H460, LnCap, Du145, PC3, SNU449, HepG2, CI3K, A549, and HeLa). MDA-MB-231 had the highest expression level among all cell lines tested (Figure 1B). To test the clinical relevance of increased circSKA3 expression, we performed a microarray analysis using human breast cancer specimen and adjacent benign tissues. Three pairs of human breast cancer specimen and benign tissues were pooled and subjected to microarray analysis, which demonstrated increased circSKA3 expression (Figure 1C). Real-time PCR demonstrated significantly higher levels of circSKA3 in 61 human breast cancer specimens relative to 55 benign tissue specimens (Figure 1D). circSKA3 was positively correlated with disease staging (Figure 1E).
Figure 1.
Upregulation of circSKA3 in Human Breast Cancer
(A) Microarray showed increased expression of circSKA3 in breast cancer cell lines compared with normal breast cell line MCF-10A. (B) RT-PCR showed expression of circSKA3 in human non-tumor cell lines (HaCaT, 293T, Jurkat, and MCF10A), breast cancer cell lines (MCF-7, MDA-MB-468, MDA-MB-231, HTB126, and SK-BR-3), and other tumor cell lines (H460, LnCap, Du145, PC3, SNU449, HepG2, CI3K, A549, and HeLa). (C) Microarray showed increased expression of circSKA3 in human breast cancer compared with the adjacent benign tissues. (D) Breast cancer tissues expressed higher levels of circSKA3 than the adjacent benign tissues. (E) Correlation of breast cancer stages and circSKA3 levels was analyzed.
To explore the effects of endogenous circSKA3, we designed two small interfering RNAs (siRNAs) specifically targeting the junction sequence of circSKA3 (siRNAs and control oligo provided by Gene Universal, Newark, DE, USA). We observed decreased cell invasion and migration (Figure 2A; typical images are provided in Figures S2A and S2B) after silencing circSKA3 that was confirmed on northern blotting (Figure 2B). To study the function of circSKA3, we generated a circSKA3 expression construct (Figure 2C) and stably transfected MCF-7 cells with either this construct or a vector. circSKA3 expression was confirmed by real-time PCR (Figure 2D) and northern blot (Figure 2E). By real-time PCR, we further confirmed that RNase R treatment decreased SKA3 linear mRNA, but not circSKA3 levels (Figure 2F), suggesting circularization of the circSKA3 transcript. RT-PCR products were cloned by TA cloning and subjected to DNA sequencing. We confirmed the expected circSKA3 junctional sequence (Figure S2C). Functional analysis demonstrated that stable circSKA3 expression promoted invasion and migration in MCF-7 cells (Figure 2G; typical images are provided in Figures S2D and S2E). Transfection with a linear construct (linSKA3), in which the intron responsible for circularization was mutated, did not impact cell motility or cell invasion. Silencing circSKA3 decreased but ectopic circSKA3 increased cell proliferation, whereas the linear counterpart had little effect (Figure 2H).
Figure 2.
Effect of circSKA3 on Breast Cancer Cell Invasion
(A) Left: silencing circSKA3 repressed MB-231 cell invasion. Right: silencing circSKA3 repressed cell migration in wound healing assay. ∗∗p < 0.01. Error bars, SD (n = 6). (B) Northern blotting showed decreased circSKA3 levels by transfection with circSKA3 siRNA. (C) Structure of circSKA3 expression construct and the control vector. (D) Left: RT-PCR showed that circSKA3-transfected MCF-7 cells expressed higher levels of circSKA3 compared with vector control and WT cells. Right: transfection with circSKA3 did not change linear SKA3 expression. ∗∗p < 0.01. Error bars, SD (n = 4). (E) Northern blot showed expression of SKA3 mRNA and circSKA3 in the vector control and circSKA3-transfected MCF-7 cells, without or with RNase R treatment. Biotin-labeled DNA probes reversely transcribed from the circSKA3 were used. (F) Total RNA from mock- or circSKA3-transfected MCF-7 cells was incubated with or without RNase R at 37°C for 10 min, followed by RNA spike-in using mouse total RNA and procession to real-time PCR. Although RNase R treatment decreased linear SKA3 levels, it did not affect circSKA3 levels. ∗∗p < 0.01. Error bars, SD (n = 4). (G) Left: overexpression of circSKA3 in MCF-7 cells enhanced cell invasion. Right: circSKA3-transfected MCF-7 cells showed enhanced cell migration in transwell assay. ∗∗p < 0.01. Error bars, SD (n = 6). (H) Transfection with circSKA3 siRNAs decreased (left) but overexpression of circSKA3 increased (right) cell proliferation.
Interaction of circSKA3 with Integrin β1 and Tks5
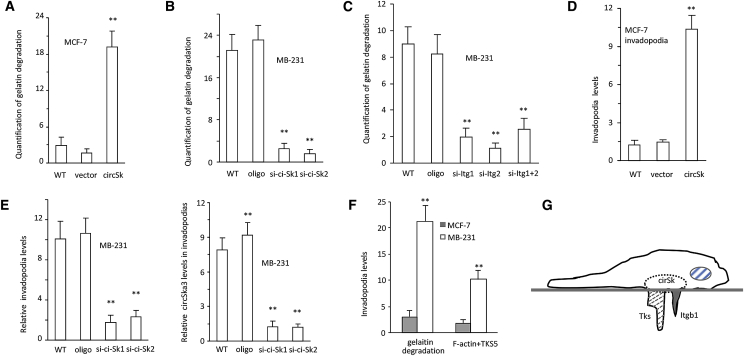
Morphological analysis detected formation of podosome/invadopodia-like core structures in the filopodia of circSKA3-transfected cells (Figure 3A). We thus examined potential interactions of circSKA3 with proteins associated with formation of invadopodia. We subjected cell lysates prepared from MB-231 cells to immunoprecipitation with relevant antibodies against invadopodia-related proteins, including Tks5, integrin β1 (Itgb1), Itgb3, epidermal growth factor (EGF), extracellular signal-regulated kinase (ERK), matrix metalloproteinase 2 (MMP2), MMP9, MMP11, Cdc42, vinculin, and F-actin. Although Tks5, Itgb1, ERK, MMP2, MMP11, and Cdc42 were shown to interact with circSKA3, we found specifically that Tks5 and Itgb1 antibodies pulled down significantly higher levels of circSKA3 (Figure 3B). We further designed a probe specific to circSKA3 and found significantly increased pull-down of circSKA3, Tks5, and Itgb1 (Figure 3C). Silencing endogenous circSKA3 abrogated this effect (Figure 3D), decreasing detected levels of Itgb1 and Tks5 (Figure 3E). Importantly, silencing circSKA3 significantly decreased detection of only Itgb1 and Tks5 (Figure 3F). Other known circSKA3-interacting proteins were globally reduced, although this effect was not statistically significant.
Figure 3.
circSKA3 Interacts with Tks5 and Integrin β1
(A) Photographs showed that expressing circSKA3 enhanced MCF-7 cell invadopodium formation. (B) Left: cell lysates from circSKA3- or vector-transfected cells were subjected to immunoprecipitation with antibodies against proteins as shown. Anti-Tks5, integrin β1 (Itgb1), ERK, MMP2, MMP11, and Cdc42 antibodies pulled down high levels of circSKA3 in circSKA3-overexpressing cells compared with the control. Right: cell lysates were subjected to immunoprecipitation followed by real-time PCR with primers specific for linear SKA3 mRNA or circSKA3. Antibodies against Tks5, Itgb1, EGF, MMP2, MMP11, and F-actin pulled down circSKA3, but not the linear SKA3 mRNA. ∗∗p < 0.01. Error bars, SD (n = 4). (C) Left: real-time PCR showed expression of circSKA3 in the inputs. Middle: PCR showed that circSKA3 probe pulled down higher levels of circSKA3 in the circSKA3-transfected cells relative to controls. Right: in the circSKA3-transfected cell lysate, circSKA3 probe pulled down more Itgb1 and Tks5 relative to controls. ∗∗p < 0.01. Error bars, SD (n = 4). (D) Left: real-time PCR showed expression of circSKA3 in the inputs. Right: PCR showed that circSKA3 pulled down lower levels of circSKA3 in the circSKA3 siRNA-transfected cells relative to controls. ∗∗p < 0.01. Error bars, SD (n = 4). (E) circSKA3 probe precipitated Itgb1 and Tks5, which were decreased by circSKA3 siRNAs. (F) Immunoprecipitation assay showed that anti-Tks5 and -Itgb1 antibodies pulled down higher levels of circSKA3 in controls compared with the siRNA-transfected cells. ∗∗p < 0.01. Error bars, SD (n = 4).
We fractionated nuclear, cytosolic, and cell membranes of MB-231 cells and isolated total RNA from these fractions to analyze circSKA3 levels. Linear SKA3 and GAPDH mRNAs were mainly detected in the cytosol, whereas circSKA3 was detected in both cytosolic and membrane fractions (Figure 4A). In MCF-7 cells transfected with circSKA3, the increased expression of circSKA3 was mainly detected in cytosol and membrane. By gelatin culture assay used to measure invadopodium levels, subcellular fractions and total invadopodia were isolated for real-time PCR. We found that circSKA3 was mainly detected in the invadopodia, whereas SKA3 mRNA was detected only in the cytosol (Figure 4B). Silencing circSKA3 decreased both circSKA3 detected in the invadopodium fraction (Figure 4C) and membrane translocation of Tks5 (Figure 4D). These results were contrasted by ectopic circSKA3 expression, which increased membrane translocation of Tks5 (Figure 4E). Breast cancer cells expressing higher levels of circSKA3 (MB-231) showed increased Tks5 membrane translocation compared with MCF-7 cells, which expressed lower levels of circSKA3 (Figures 1B and 4F). To test the specificity of the interaction, cell lysates prepared from MB-231 cells were subjected to immunoprecipitation with anti-rabbit immunoglobulin G (IgG), Tks5, and Itgb1 antibodies, followed by real-time PCR with primers specific for circSKA3, circDNAJA1, circMRPL47, circNDUF53, circRPS5, and circRPL5. We found that antibodies against Tks5 and Itgb1 pulled down circSKA3 but did not pull down the unrelated circRNAs (Figure 4G).
Figure 4.
Colocalization of circSKA3 with Tks5 and Itgb1
(A) Left: RNAs extracted from nuclei, cytosol, and membrane were subjected to real-time PCR. GAPDH and linear SKA3 were mainly expressed in the cytosol, whereas circSKA3 was highly expressed in the cell membrane. Right: the membrane fraction had high levels of circSKA3 in circSKA3-overexpressing cells compared with those in WT and vector-transfected cells. ∗∗p < 0.01. Error bars, SD (n = 4). (B) MB-231 cells were cultured on gelatin-coated culture dish for 24 h. Subcellular fractions and total invadopodia were isolated for real-time PCR. High levels of circSKA3 were detected in the invadopodia and cell membrane (left). SKA3 mRNA was mainly detected in cytosol (right). ∗∗p < 0.01. Error bars, SD (n = 4). (C) MB-231 cells transfected with circSKA3 siRNAs or control oligos were cultured on gelatin-coated dishes for 24 h. Real-time PCR showed decreased levels of circSKA3 in the gelatin of siRNA-transfected cells. ∗∗p < 0.01. Error bars, SD (n = 4). (D) Silencing circSKA3 increased Tks5 levels in the cytosol, but decreased Tks5 in the cell membrane. (E) Overexpression of circSKA3 promoted Tks5 translocation from cytosol to cell membrane. (F) The membrane fraction of MB-231 showed higher levels of Tks5 than MCF-7 cells. (G) Cell lysates were subjected to immunoprecipitation, followed by real-time PCR with primers specific for circRNAs as shown. Antibodies against Tks5 and Itgb1 pulled down circSKA3, but not the others. ∗∗p < 0.01. Error bars, SD (n = 4).
circSKA3 Induced Invadopodium Formation
We then analyzed the co-localization of circSKA3, Tks5, and Itgb1. In MCF-7 cells, Tks5 was mainly detected in cytosol, whereas Itgb1 was mainly located at the cell membrane. Ectopic expression of circSKA3 promoted invadopodium formation, co-localized with Tks5 and Itgb1 (Figure 5A). In MB-231 cells, silencing circSKA3 decreased the amount of invadoppdia co-localized with Tks5 and Itgb1 (Figure 5B). We isolated membrane fractions from the vector- and circSKA3-transfected MCF-7 cells. Real-time PCR showed that circSKA3 levels were significantly higher in the lipid raft than in the non-lipid raft (Figure 5C). Tks5 and Itgb1 were also highly distributed in the membrane of the circSKA3-transfected cells (Figure 5D, left). Relative to the nuclear and cytosolic fractions, the membrane-derived fractions contained much higher levels of circSKA3, Itgb1, Tks5, and Cav2 than in the lipid rafts. The precipitated Tks5 was phosphorylated (Figure 5D, right). Ectopic circSKA3 increased Itgb1 pull-down of Tks5 and vice versa (Figure 5E). Silencing circSKA3 abolished this effect (Figure 5F). Treatment with RNase A degraded circRNA and abolished the interaction of Itgb1 with Tks5 (Figure 5G). Silencing Itgb1 increased cytosolic Tks5 (Figure 5H), inhibiting cell invasion and migration (Figure S3A). To further confirm these findings, we performed in situ hybridization and found that circSKA3 was associated with Itgb1 and Tks5 (Figures S3B and S3C), resulting in increased invadopodium formation (Figure S3D). Silencing endogenous circSKA3 decreased circSKA3 levels and the associated proteins (Figures S4A and S4B). Silencing Itgb1 also decreased co-localization of F-actin and Tks5 (Figures S4C and S4D). Collectively, these results suggested the formation of a circSKA3, Itgb1, and Tks5 complex underlying invadopodium formation.
Figure 5.
Complex Formation of circSKA3 with Tks5 and Itgb1
(A) Left: Tks5 was mainly detected in cytosol, whereas Itgb1 mainly located at the cell membrane. Right: expression of circSKA3 promoted invadopodium formation with high levels of Tks5 and Itgb1 collected from gelatin. (B) Left: subcellular fractions (10 μg) were subjected to western blotting. Both Tks5 and Itgb1 were highly distributed in the cell membrane. Right: silencing circSKAs repressed invadopodium formation with less Tks5 and Itgb1 collected from gelatin. (C) Membrane fractions were isolated from vector control- and circSKA3-transfected cells, and processed to RT-PCR. circSKA3 was detected in high levels in the lipid raft compared with the non-lipid raft. (D) Left: analysis of membrane fractions revealed co-localization of Itgb1, Tks5, and Cav2 in the lipid rafts. Ectopic circSKA3 increased the formation of this complex. Right: in the lipid rafts, Tks5 tyrosine phosphorylation was promoted by ectopic circSKA3. (E) Western blotting showed that anti-Itgb1 antibody precipitated higher levels of Tks5 relative to the controls and vice versa. (F) Silencing circSKA3 did not change expression of Itgb1 and Tks5. Itgb1 precipitation pulled down more Tks5 in the control cells than in the siRNA-transfected cells and vice versa. (G) Upper: Itgb1 precipitation pulled down increased levels of Tks5, and Tks5 precipitation pulled down increased levels of Itgb1 in the circSKA3-transfected cells, which were abolished by RNase A treatment. Lower: RNase A treatment abolished anti-Itgb1 antibody precipitating Tks5 and vice versa. (H) Western blot showed that silencing Itgb1 repressed Itgb1 expression in total lysate, and decreased Itgb1 and Tks5 levels in the cell membrane.
Functionally, we quantified gelatin degradation using confocal microscopy. The vector- and circSKA3-transfected MCF-7 cells were cultured on OG 488-gelatin-coated slides for 48 h and stained with DAPI for nuclear observation and red fluorescence to detect F-actin. OG 488-gelatin staining was performed to show inversed mode with black dots to monitor invadopodia. We confirmed that circSKA3 overexpression increased invadopodium formation (Figure 6A; Figure S5A), whereas silencing endogenous circSKA3 inhibited this process (Figure 6B; Figure S5B). Silencing Itgb1 also inhibited invadopodium formation (Figure 6C; Figure S5C). The invadopodia were harvested from gelatin, and we confirmed that ectopic circSKA3 increased invadopodium formation (Figure 6D). Silencing circSKA3 decreased circSKA3 levels in the invadopodia (Figure 6E). Consistently, circSKA3 (MB-231) expression levels were correlated to invadopodium formation and in vitro markers of tumor aggressiveness (Figures 6F and 6G).
Figure 6.
Formation of Invadopodia in Gelatin
(A) The vector- or circSKA3-transfected cells were cultured on OG 488-gelatin-coated slides for 48 h and stained with DAPI (blue), red fluorescence showing F-actin. OG 488-gelatin staining showed in an inversed mode with black color. ImageJ quantified the levels of circSKA3 in the invadopodia. (B) The oligo- or circSKA3 siRNA-transfected cells were cultured on OG 488-gelatin-coated slides for 24 h. ImageJ analysis showed that silencing circSKA3 repressed invadopodium formation. (C) Control oligo- or Itgb1 siRNA-transfected cells were cultured on OG 488-gelatin-coated slides for 36 h for gelatin staining. ImageJ analysis showed that silencing Itgb1 repressed invadopodium formation. (D) ImageJ analysis showed that ectopic circSKA3 increased invadopodium formation. (E) ImageJ analysis showed that silencing circSKA3 decreased circSKA3 levels in the invadopodia and repressed invadopodium formation (colocalization of Tks5 with F-actin). (F) MB-231 cells showed higher activities in gelatin degradation and co-localization of F-actin and TKS5 forming invadopodia relative to MCF-7 cells. (G) A diagram showing that Tks5 forms invadopodia with Itgb1 and circSKA3.
Identification of the Binding Sites
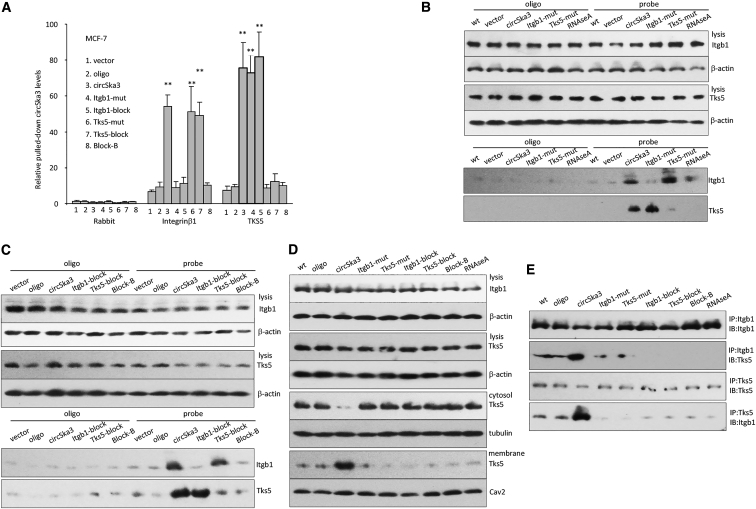
We conducted an in silico analysis of circSKA3 with Itgb1 and Tks5. Analysis of the complexes found that Itgb1 did not directly bind to Tks5. These two complexes were then combined to dock with circSKA3. The best energized conformation of more than 2,000 putative complexes was chosen as the Itgb1/Tks5/circSKA3 complex. The interaction interface was analyzed using COCOMAPS. We thus identified putative binding sites for each of these two proteins (Figure S6A). To validate this binding model, we generated mutations abolishing the interaction by site-directed mutagenesis and designed blocking oligos to inhibit circSKA3 binding to Tks5 and Itgb1 (Figure 7A). We showed that anti-Itgb1 and Tsk5 antibodies could no longer precipitate circSKA3 when the cells were transfected with their respective mutant constructs or blocking oligos. As well, the circSKA3 probe could not pull down Itgb1 and Tks5 when the binding sites were mutated (Figure 7B) or the cells were transfected with binding site blocking oligos (Figure 7C). These transfections inhibited localization of Tks5 to the cell membrane (Figure 7D) and inhibited Itgb1 pulling down Tks5 and vice versa (Figure 7E). In situ hybridization revealed that cells transfected with Itgb1 mutant or Tks5 mutant construct, or their respective blocking oligos could no longer be co-localized with circSKA3 (Figures S6B and S6C). In gelatin culture, the formation of invadopodia was inhibited by transfection with these mutants or the blocking oligos (Figure S7A), which inhibited cell invasion and migration (Figure S7B).
Figure 7.
Identification of the Binding Sites
(A) Transfection with the mutant construct harboring Itgb1 binding site or the blocking oligo masking Itgb1 binding site abolished anti-Itgb1 antibody pulling down circSKA3. Transfection with the mutant construct harboring Tks5 binding site or the blocking oligo masking Tks5 binding site abolished anti-Tks5 antibody pulling down circSKA3. (B) Transfection with these mutant constructs abolished Tks5 membrane translocation (upper) and circSKA3 probe pulling down Tks5 or Itgb1 (lower). (C) Transfection with the blocking oligos abolished Tks5 membrane translocation (upper) and circSKA3 probe pulling down Tks5 or Itgb1 (lower). (D) Transfection with these mutant constructs or the blocking oligos abolished Tks5 localization to the membrane. (E) Transfection with these mutant constructs or the blocking oligos abolished anti-Tks5 antibody pulling down Itgb1 and vice versa.
Effect of circSKA3 on Tumor Progression
We examined the effect of circSKA3 on tumor growth. Nude mice were injected with circSKA3- and vector-transfected MCF-7 cells (4 × 107 cells). The mice were injected subcutaneously with 50 μg β-estradiol 17-cypionate (in 50 μL sesame oil) every 3 days to enhance tumor development. Twenty days after the injection, the mice were sacrificed, and tumors were removed to determine their sizes. We found that nude mice injected with the circSKA3-transfected cells developed significantly larger volume tumors than the control group (Figure 8A). These tumors showed higher levels of circSKA3 than the smaller tumors (Figure 8B). The tumors were stained with DAPI, green fluorescence showing F-actin, red fluorescence showing Tks5 or Itgb1, and yellow fluorescence showing circSKA3 (Figure S8A). Expression of circSKA3 increased circSKA3 binding to Itgb1 but did not alter the percentages of the interaction (Figure 8C), suggesting that most of the ectopic-expressed circSKA3 formed complexes with Itgb1. Similarly, ectopic circSKA3 increased levels of circSKA3 in the invadopodia (Figure 8D) and total levels of invadopodia (Figure 8E).
Figure 8.
circSKA3-Containing Exosomes Potentiate Tumor Invasion
(A) Left: expression of circSKA3 formed larger tumors relative to the vector control. Right: tumor sizes at day 20 after injection. (B) Tumors collected from mice injected with circSKA3-transfected MCF-7 cells showed increased levels of circSKA3 expression. (C) Quantitation of circSKA3 levels (left), circSKA3 binding to Itgb1 (middle), and percentages of circSKA3 binding with Itgb1 (right). (D) Quantitation of circSKA3 levels (left), circSKA3 binding to invadopodia (middle), and percentages of circSKA3 complexed in the invadopodia (right). (E) Quantitation of invadopodium levels. (F) After injection with MB-231 cells, CD-1 nude mice were also injected with control oligo, circSKA3 siRNA. Delivery of circSKA3 siRNA increased mouse viability significantly relative to controls (log rank test, ∗∗p < 0.01). (G) Invadopodium formation was repressed in the tumor tissues when the mice were injected with circSKA3 siRNA. (H) Immunoprecipitation showed that overexpression of circSKA3 promoted Tks5 phosphorylation, which happened in the cell membrane. (I) Left: MB-231 cells were cultured on gelatin-coated dish for 24 h. The subcellular fractions and total invadopodia were isolated and lysed, and 10 μg of the samples was processed to immunoblot. Both Itgb1 and Tks5 were highly expressed in the invadopodia. The above fraction immunoprecipitation with antibody against Tks5 showed that the Tks5 was highly tyrosine phosphorylated in the invadopodia. Right: western blot showed that the lipid raft fractions expressed higher levels of Itgb1 and Tks5 compared with the non-raft fraction. Tks5 in the raft fractions displayed high levels of tyrosine phosphorylation. (J) Tumor sections were subjected to H&E staining. Tumor cells invading into muscle tissues were indicated by arrows.
On the other hand, we silenced circSKA3 expression and examined mouse survival. Mice bearing MB-231 tumor xenografts were intraperitoneally injected with control oligo and circSKA3 siRNA. We found that silencing circSKA3 significantly increased overall survival of the xenograft-bearing mouse (Figure 8F). By immunostaining, we found that Tks5, Itgb1, and F-actin formed invadopodia complexes in the tumor xenografts (Figure S8B). Tumors from mice injected with circSKA3 siRNA formed decreased amounts of invadopodia (Figure 8G). Given the critical role of Tks5 phosphorylation in invadopodium formation and tumor invasion, we examined Tks5 states and detected significant Tks5 phosphorylation in the invadopodia following circSKA3 expression (Figure 8H). This was reduced after silencing endogenous circSKA3 (Figure 8I). Consistently, ectopic expression of circSKA3 promoted tumor cell invasion (Figure 8J).
Discussion
By microarray analysis, we identified circSKA3 as a highly expressed circRNA in human breast cancer patient samples. We then systematically evaluated the role of this circRNA through gain- and loss-of-function approaches. We generated circSKA3 expression constructs that were validated by northern blot and real-time PCR following RNase R treatment. In vitro, we found that circSKA3 expression in breast cancer cells induced both invadopodium formation and cell invasion, as assessed by gelatin degradation assay. Tumor xenograft volume was similarly increased. These results were corroborated by siRNA-mediated silencing of circSKA3 expression. Immunoprecipitation of cell lysates revealed that circSKA3 was bound by various proteins, but most specifically Tks5 and Itgb1. circSKA3-Tks5-Itgb1 co-localization was observed by fluorescent microscopy and cellular fractionation, confirmed to be localized to the cell membrane. Site-directed mutagenesis and immunoprecipitation confirmed putative binding sites identified by bioinformatics analysis. We further isolated circSKA3-Tks5-Itgb1 in invadopodium fractions and demonstrated that this circRNA-protein complex could be potentiating cell invasion.
Thus, circSKA3 interacts with Itgb1 and Tks5, inducing invadopodium formation. The formation of actin-rich invadopodia, which are associated with basement membrane degradation, stromal migration, and intravasation, is essential for cancer invasion and metastasis.37,38 Itgb1 is a cell-surface receptor with multiple known ligands and mediated cell adhesion, detachment, and migration. Tks5 functions as an adaptor protein in src-dependent invadopodium formation.39,40 Tks5 phosphorylation is sufficient to induce invadopodium formation and tumor invasion.41 Invadopodia can be characterized by the co-localizing puncta of actin with Tks5 and can be used to quantify the invasiveness of cancer cell lines.36,42 Our results showed that circSKA3 was critical for the formation of invadopodia by binding with Itgb1 and Tks5.
Most notably, we observed that circSKA3 could recruit Tks5 to the cell membrane. In the presence of circSKA3, Tks5 formed complexes with Itgb1. Silencing endogenous circSKA3 dissociated Tks5 from the cell membrane, but Itgb1 continues to integrate in the cell membrane. Our results suggest that circSKA3 is the key molecule in the formation of invadopodia by binding and bridging both Itgb1 and Tks5, although Itgb1 and Tks5 do not directly bind with each other. Because invadopodia contain other molecules, whether or not circSKA3 also binds other invadopodia-associated molecules awaits further investigation. We demonstrated that this process led to accelerated tumor cell migration and invasion in vitro and cancer development in vivo. Our results provide a further understanding of how tumor-tumor and tumor-extracellular matrix interaction are regulated by circRNAs.
It is possible that this mechanism is not exclusive to the cancers shown in this study but may be co-opted by other malignancies or pathological processes. Furthermore, the role of circSKA3 with respect to hormone status or molecular subtyping in breast cancer remains to be explored. This circRNA is specifically expressed in human. The upstream factors regulating circSKA3 biogenesis represent a further potential target for novel cancer therapeutics. Further contextualized study of circRNAs, including circSKA3, alongside their linear RNA counterparts and their corresponding protein products is also warranted.
Materials and Methods
General Methods
Protein assays on western blot and immunohistochemistry were performed as described previously.43,44 The monoclonal antibodies against Itgb1, β-actin, tubulin, and Caveolin-2 were from Santa Cruz Biotechnology (Dallas, TX, USA). The monoclonal antibody against TKS5 was from EMD Millipore (Burlington, MA, USA). Real-time PCR was conducted as described previously.45 RNA-protein interaction assays were performed as described previously.46 Cell migration and invasion were performed as described previously.47 During the assays, the same treatments were performed in each group, and the cell number was determined at the endpoint to normalize the effect of cell proliferation in each assay. Microarray analysis for circRNAs was performed by Arraystar (Rockville, MD, USA) and KangChen BioTech (Shanghai) as described previously.22 Tumor formation was performed as described previously.19 Detection of circSKA3 in cells and tissues was performed by fluorescence in situ hybridization following the methods described previously.15
Invadopodium Detection and Gelatin Degradation Assay
The invadopodium gelatin degradation assay was performed as described previously.48 In brief, coverslips were coated with Oregon green 488-conjugated gelatin (0.2 mg/mL)/2.5% sucrose solution in PBS. Gelatin was cross-linked with 0.5% glutaraldehyde and quenched with 5 mg/mL sodium borohydride. Cells were cultured on the gelatin for 24 h, fixed with 4% paraformaldehyde (PFA), blocked with 3% BSA in PBS, and stained with DAPI (blue) for nucleus and red fluorescence for F-actin.
Invadopodium Isolation
Cells were cultured on gelatin-coated and cross-linked dishes, and separated into the cytosol fractions, cell body membranes, and invadopodia. In brief, cells were rinsed in tyrosine phosphorylated protein (YPP) buffer (10 mM 3-(N-morpholino)propanesulfonic acid, or MOPS, [pH 6.8], 100 mM KCl, 2.5 mM MgCl2, 1 mM CaCl2, 0.3M sucrose, 1× Roche protease inhibitor cocktail). Cell bodies were sheared with a glass rod into 200 μL YPP buffer. Cell body membranes were separated from the cytosol by centrifugation at 9,000 × g at 4°C for 20 min. The invadopodia, embedded in the gelatin matrix, were scraped up with the cross-linked gelatin into coimmunoprecipitation (co-IP) buffers.
Identification of Binding Sites in circSKA3
The secondary structure of circSKA3 was predicted by MC-Fold to produce 100 2D structures associated with the sequence and ranked by their free energy in kilocalories per mole (kcal/mol). The best minimum free energy (MFE) structure was chosen for 3D structure prediction. Due to the length of circSKA3, it was divided into two segments for 3D structure prediction by RNAComposer. The MFE structures of each of the producing five models were bonded together. Modeled circSKA3 was then energy minimized in two steps. Steepest decent technique was followed by conjugate gradient technique to minimize the overall modeled structure using the Discovery studio software until the structures reached the final root mean square (RMS) gradient of 0.0001 kcal/mol. All energy minimizations were done using CHARMM force field. To get appropriate conformation, we performed molecular dynamics simulation for 100 ns using CHARMM in DS.
Itgb1 structure was obtained from the B chain of integrin α5/Itgb1 complex (PDB: 4WK0). Tks5 Sh3 domain structure was downloaded from the PDB database (PDB: 2EKH). Itgb1 and Tks5 were docked onto circSKA3 respectively by Hex, and more than 2,000 poses were produced. We chose the minimum energized conformations as Itgb1/circSKA3 and Tks5/circSKA3 complexes. Analysis of the two complexes found that Itgb1 had a long distance from Tks5, and no mutual interactions were detected from the two. These two complexes were combined together. The interaction interface was analyzed using COCOMAPS.
Statistical Analysis
All experiments were performed in triplicate, and numerical data were subjected to independent sample t test. The levels of significance were set at ∗p < 0.05 and ∗∗p < 0.01.
Author Contributions
W.W.D., W.Y., and B.B.Y. were involved in project design. B.B.Y. supervised the project. W.W.D., W.Y., M.L., and B.B.Y. were involved in structuring the experiments. W.W.D., W.Y., X.L., L.F., N.W., F.L., Y.C., Q.H., E.L., and Z.Y. performed the experiments. F.M.A. performed computational analysis of the interaction between circSKA3 and the proteins. W.W.D., W.Y., and B.B.Y. wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by Canadian Institutes of Health Research, Canada (grants PJT-153105 and PJT-155962 to B.B.Y.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.03.002.
Supplemental Information
References
- 1.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatomer D.C., Wilusz J.E. An Unchartered Journey for Ribosomes: Circumnavigating Circular RNAs to Produce Proteins. Mol. Cell. 2017;66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 5.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 6.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.R. ncRNA-encoded peptides or proteins and cancer. Mol. Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sang M., Meng L., Sang Y., Liu S., Ding P., Ju Y., Liu F., Gu L., Lian Y., Li J. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37–46. doi: 10.1016/j.canlet.2018.03.049. [DOI] [PubMed] [Google Scholar]
- 14.Wang J.J., Liu C., Shan K., Liu B.H., Li X.M., Zhang S.J., Zhou R.M., Dong R., Yan B., Sun X.H. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics. 2018;8:3408–3415. doi: 10.7150/thno.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W., Du W.W., Li X., Yee A.J., Yang B.B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z.B., Huang G.X., Fu Q., Han B., Lu J.J., Chen A.M., Zhu L. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol. Ther. 2019;27:531–541. doi: 10.1016/j.ymthe.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu K.P., Zhang C.L., Ma X.L., Hu J.P., Cai T., Zhang L. Analyzing the interactions of mRNAs and ncRNAs to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol. Ther. 2019;27:518–530. doi: 10.1016/j.ymthe.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du W.W., Yang W., Li X., Awan F.M., Yang Z., Fang L., Lyu J., Li F., Peng C., Krylov S.N. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 20.Fang L., Du W.W., Lyu J., Dong J., Zhang C., Yang W., He A., Kwok Y.S.S., Ma J., Wu N. Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 2018;25:2195–2208. doi: 10.1038/s41418-018-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sang Y., Chen B., Song X., Li Y., Liang Y., Han D., Zhang N., Zhang H., Liu Y., Chen T. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol. Ther. 2019;27:1638–1652. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang L., Du W.W., Awan F.M., Dong J., Yang B.B. The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex suppressing cell invasion and tumorigenesis. Cancer Lett. 2019;459:216–226. doi: 10.1016/j.canlet.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 23.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843.e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Nakase I., Gallis B., Takatani-Nakase T., Oh S., Lacoste E., Singh N.P., Goodlett D.R., Tanaka S., Futaki S., Lai H., Sasaki T. Transferrin receptor-dependent cytotoxicity of artemisinin-transferrin conjugates on prostate cancer cells and induction of apoptosis. Cancer Lett. 2009;274:290–298. doi: 10.1016/j.canlet.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Ren S., Xin Z., Xu Y., Xu J., Wang G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle. 2017;16:2204–2211. doi: 10.1080/15384101.2017.1346754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu W., Bi Z.Y., Chen Z.L., Liu C., Li L.L., Zhang F., Zhou Q., Zhu W., Song Y.Y., Zhan B.T. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K., Liao X., Gong Y., He J., Zhou J.K., Tan S., Pu W., Huang C., Wei Y.Q., Peng Y. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol. Cancer. 2019;18:98. doi: 10.1186/s12943-019-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B., Zheng P., Li Z., Li H., Wang X., Shi Z., Han Q. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle. 2018;17:2644–2654. doi: 10.1080/15384101.2018.1553354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y., Zhang S., Yue M., Li Y., Bi J., Liu H. Angiotensin II inhibits apoptosis of mouse aortic smooth muscle cells through regulating the circNRG-1/miR-193b-5p/NRG-1 axis. Cell Death Dis. 2019;10:362. doi: 10.1038/s41419-019-1590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z.G., Awan F.M., Du W.W., Zeng Y., Lyu J., Wu, Gupta S., Yang W., Yang B.B. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol. Ther. 2017;25:2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu N., Yuan Z., Du K.Y., Fang L., Lyu J., Zhang C., He A., Eshaghi E., Zeng K., Ma J. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758–2773. doi: 10.1038/s41418-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddy R.J., Weidmann M.D., Sharma V.P., Condeelis J.S. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 2017;27:595–607. doi: 10.1016/j.tcb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stylli S.S., Stacey T.T., Verhagen A.M., Xu S.S., Pass I., Courtneidge S.A., Lock P. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 2009;122:2727–2740. doi: 10.1242/jcs.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz M., Yamaguchi H., Wang Y., Singer R.H., Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr. Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Marx J. Cell biology. Podosomes and invadopodia help mobile cells step lively. Science. 2006;312:1868–1869. doi: 10.1126/science.312.5782.1868. [DOI] [PubMed] [Google Scholar]
- 39.Mueller S.C., Chen W.T. Cellular invasion into matrix beads: localization of beta 1 integrins and fibronectin to the invadopodia. J. Cell Sci. 1991;99:213–225. doi: 10.1242/jcs.99.2.213. [DOI] [PubMed] [Google Scholar]
- 40.Diaz B., Shani G., Pass I., Anderson D., Quintavalle M., Courtneidge S.A. Tks5-dependent, nox-mediated generation of reactive oxygen species is necessary for invadopodia formation. Sci. Signal. 2009;2:ra53. doi: 10.1126/scisignal.2000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burger K.L., Learman B.S., Boucherle A.K., Sirintrapun S.J., Isom S., Díaz B., Courtneidge S.A., Seals D.F. Src-dependent Tks5 phosphorylation regulates invadopodia-associated invasion in prostate cancer cells. Prostate. 2014;74:134–148. doi: 10.1002/pros.22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurski L.A., Xu X., Labrada L.N., Nguyen N.T., Xiao L., van Golen K.L., Jia X., Farach-Carson M.C. Hyaluronan (HA) interacting proteins RHAMM and hyaluronidase impact prostate cancer cell behavior and invadopodia formation in 3D HA-based hydrogels. PLoS ONE. 2012;7:e50075. doi: 10.1371/journal.pone.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du W.W., Liu F., Shan S.W., Ma X.C., Gupta S., Jin T., Spaner D., Krylov S.N., Zhang Y., Ling W., Yang B.B. Inhibition of Dexamethasone-induced Fatty Liver Development by Reducing miR-17-5p Levels. Mol. Ther. 2015;23:1222–1233. doi: 10.1038/mt.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma J., Fang L., Yang Q., Hibberd S., Du W.W., Wu N., Yang B.B. Posttranscriptional regulation of AKT by circular RNA angiomotin-like 1 mediates chemoresistance against paclitaxel in breast cancer cells. Aging (Albany NY) 2019;11:11369–11381. doi: 10.18632/aging.102535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutnam Z.J., Du W.W., Yang W., Yang X., Yang B.B. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 2014;5:2914. doi: 10.1038/ncomms3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mader C.C., Oser M., Magalhaes M.A., Bravo-Cordero J.J., Condeelis J., Koleske A.J., Gil-Henn H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011;71:1730–1741. doi: 10.1158/0008-5472.CAN-10-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.