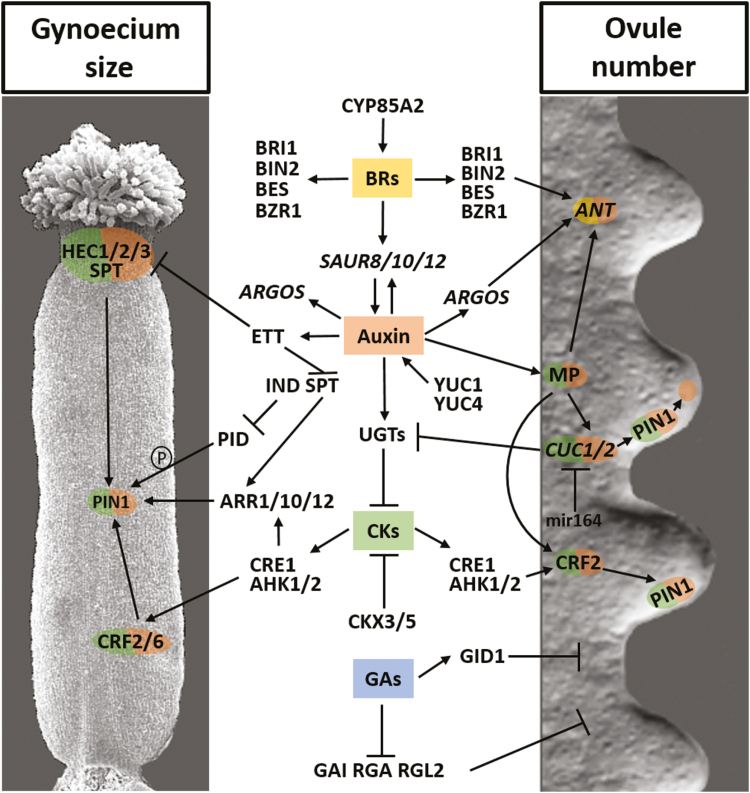
An overview of the gene and hormone regulatory network involved in determining ovule number and gynoecium size, two fundamental processes that affect seed yield.
Keywords: Gynoecium, hormones, organ boundary, ovule number, ovule primordia, pistil, seed yield
Abstract
Angiosperms form the largest group of land plants and display an astonishing diversity of floral structures. The development of flowers greatly contributed to the evolutionary success of the angiosperms as they guarantee efficient reproduction with the help of either biotic or abiotic vectors. The female reproductive part of the flower is the gynoecium (also called pistil). Ovules arise from meristematic tissue within the gynoecium. Upon fertilization, these ovules develop into seeds while the gynoecium turns into a fruit. Gene regulatory networks involving transcription factors and hormonal communication regulate ovule primordium initiation, spacing on the placenta, and development. Ovule number and gynoecium size are usually correlated and several genetic factors that impact these traits have been identified. Understanding and fine-tuning the gene regulatory networks influencing ovule number and pistil length open up strategies for crop yield improvement, which is pivotal in light of a rapidly growing world population. In this review, we present an overview of the current knowledge of the genes and hormones involved in determining ovule number and gynoecium size. We propose a model for the gene regulatory network that guides the developmental processes that determine seed yield.
Introduction
Life on earth is affected by plants in varied ways. Of the estimated 400 000 extant plant species, approximately 94% are seed plants (Govaerts, 2001; Willis, 2017). This demonstrates that seed development and dispersion strategies greatly contributed to the success of this organismal group. The vast majority of seed plants are angiosperms and only a comparatively small number are gymnosperms. Both plant divisions produce ovules; however, only angiosperm species produce flowers, and as another selective advantage, each flower produces one or more gynoecia that protect and nourish the ovules. Following fertilization, the gynoecium (or pistil) generally develops into a fruit and ovules develop into seeds.
Depending on the species, the gynoecium consists of one or more carpels, which can be fused or unfused (Endress and Igersheim, 2000). The Arabidopsis gynoecium consists of two fused carpels (Smyth et al., 1990; Alvarez-Buylla et al., 2010). Along the margins where the carpels fuse, a meristematic tissue, termed the carpel margin meristem (CMM), is formed. The CMM gives rise to the placenta, ovules, septum, and transmitting tract (Reyes-Olalde et al., 2013; Reyes-Olalde and de Folter, 2019). Inside an ovule the female gametophyte develops, comprising three antipodal cells, a central cell, two synergids, and an egg cell (Bencivenga et al., 2011; Drews and Koltunow, 2011). Therefore, ovule development is a crucial process during the plant life cycle and has been studied in many species. In recent decades, many reviews on ovule development have been written, demonstrating its importance and the degree of active research in this area (e.g. Reiser and Fischer, 1993; Angenent and Colombo, 1996; Grossniklaus and Schneitz, 1998; Gasser et al., 1998; Bowman et al., 1999; Skinner et al., 2004; Colombo et al., 2008; Shi and Yang, 2011; Endress, 2011; Cucinotta et al., 2014; Gasser and Skinner, 2019; Pinto et al., 2019; Shirley et al., 2019).
To complement existing literature, this review focuses on recent discoveries in ovule development and gynoecium size determination. An overview is provided of the genes and hormonal communication involved in the developmental programs (Fig. 1; Table 1). Understanding the regulatory networks that determine ovule number and gynoecium size is important as they hugely impact seed yield, and fine-tuning them appears to be a particularly promising strategy for enhancing crop yields.
Fig. 1.
Proposed model for the regulation of pistil growth and ovule primordium initiation. A gynoecium of Arabidopsis is shown on the left while the image on the right depicts ovule primordia; in the centre, the interconnected gene network that regulates the two processes is shown. Auxin, through ETT, regulates gynoecium fusion and elongation by repressing IND, HECs, and SPT, which in turn modulate polarization of the auxin efflux carrier PIN1 via repressing PID. CK positively regulates PIN1 expression. In particular, the CK response mediated by CRFs and ARRs is directly required for pistil elongation and indirectly affects ovule primordium initiation. CRF2 regulation by MP further integrates the auxin–CK crosstalk. Moreover, MP directly regulates CUC1 and CUC2 expression. In turn, CUCs control PIN1 expression and PIN1 protein localization, which is required for correct ovule primordium development. CUCs positively influence the CK pathway by transcriptionally repressing the CK-inactivating glycosyltransferase enzymes (UGTs). ANT, whose expression is controlled by auxin and BRs, is required for cell division in ovule primordia. ANT is also regulated by auxin via MP and ARGOS. BR signalling also positively affects pistil elongation. GA has a negative effect on ovule number, but its connection with other hormones remains to be addressed.
Table 1.
Genes involved in determining gynoecium size and/or ovule number
| Gene name | Family or protein type | Gynoecium size | Ovule number | Reference |
|---|---|---|---|---|
| ANT | AP2/EREBP transcription factor | ant-9 ↓ ant-4 ↓ 35S::ANT ↑ | ant-1 ↓ ant-3 ↓ ant-4 ↓ ant-9 ↓ | Elliott et al. (1996), Liu et al. (2000), Azhakanandam et al. (2008), Krizek (2009), Wynn et al. (2014) |
| ARGOS | ARGOS protein | 35S::ARGOS ↑ | Hu et al. (2003) | |
| CRC | YABBY transcription factor | crc-1 ↓ | Gross et al. (2018) | |
| SPT | bHLH transcription factor | spt-2 ↓ | spt-2 ↓ | Heisler et al. (2001), Alvarez and Smyth (2002), Nahar et al. (2012) |
| ETT (ARF3) | ARF transcription factor | ett-1 ↓ ett-2 ↓ | Sessions et al. (1997), Nemhauser et al. (2000) | |
| HEC1, HEC2, HEC3 | bHLH transcription factor | hec1 hec2 hec3 ↓ | Gremski et al. (2007) | |
| ARR1, ARR10, ARR12 | Type-B ARR transcription factor | arr1 arr10 arr12 ↓ | arr1 arr10 arr12 ↓ | Reyes-Olalde et al. (2017) |
| CRF2, CRF3, CRF6 | ERF transcription factor | crf2 crf3 crf6 ↓ | crf2 crf3 crf6 ↓ | Cucinotta et al. (2016) |
| PIN1 | PIN auxin efflux carrier | pin1 ↓ | pin1 ↓ pin1-5 ↓ | Okada et al. (1991), Bencivenga et al. (2012), Cucinotta et al. (2016) |
| CKX3, CKX5 | CKX cytokinin oxidase/dehydrogenase protein | ckx3 ckx5 ↑ | ckx3 ckx5 ↑ | Bartrina et al. (2011) |
| UGT85A3, UGT73C1 | UDP-glucosyl transferase | 35S::UGT85A3 ↓ 35S::UGT73C1 ↓ | 35S::UGT85A3 ↓ 35S::UGT73C1 ↓ | Cucinotta et al. (2018) |
| SAUR8, SAUR10, SAUR12 | SAUR-like auxin-responsive protein family | 35S::SAUR8 ↑ 35S::SAUR10 ↑ 35S::SAUR12 ↑ | van Mourik et al. (2017) | |
| BZR1 | Brassinosteroid signalling regulatory protein | bzr1-1D ↑ | bzr1-1D ↑ | Huang et al. (2013) |
| BIN2 | ATSK (shaggy-like kinase) family | bin2 ↓ | bin2 ↓ | Huang et al. (2013) |
| DET2 | 3-Oxo-5-α-steroid 4-dehydrogenase protein | det2 ↓ | det2 ↓ | Huang et al. (2013) |
| BRI1 | Leucine-rich receptor-like protein kinase protein | bri1-5 ↓ | bri1-5 ↓ | Huang et al. (2013) |
| CYP85A2 | Cytochrome p450 enzyme | cyp85a2-1 ↓ cyp85a2-2 ↓ | Nole-Wilson et al. (2010b) | |
| SEU | Transcriptional adaptor | seu-1 ↓ | seu-1 ↓ | Nole-Wilson et al. (2010b) |
| CTR1 | RAF homologue of serine/threonine kinase | ctr1-1 ↓ | Carbonell-Bejerano et al. (2011) | |
| REV | Homeobox-leucine zipper protein | ant rev ↓ | Nole-Wilson et al. (2010a) | |
| L–UG | WD40/YVTN repeat-like-containing domain transcription factor | lug-1 ↓ lug-3 ↓ | Azhakanandam et al. (2008) | |
| PAN | bZIP transcription factor | ant pan ↓ seu pan ↓ | ant pan ↓ seu pan ↓ | Wynn et al. (2014) |
| HLL | Ribosomal protein L14p/L23e | hll ↓ | hll ↓ | Schneitz et al. (1998), Skinner et al. (2001) |
| SIN2 | P-loop containing nucleoside triphosphate hydrolase superfamily protein | sin-2 ↓ | sin-2 ↓ | Broadhvest et al. (2000) |
| YUC1, YUC4 | Flavin-binding monooxygenase protein | yuc1 yuc4 ↓ | Cheng et al. (2006) | |
| AHK2, AHK3, CRE1 | Histidine kinase | cre1-12 ahk2-2 ahk3-3 ↓ | Bencivenga et al. (2012) | |
| CUC1, CUC2 | NAC transcription factor | cuc1 cuc2 ↓ pSTK::CUC1/RNAi cuc2-1 ↓ | Galbiati et al. (2013) | |
| MIR164A | microRNA | 35S::MIR164A ↓ | Gonçalves et al. (2015) | |
| GAI, RGA, RGL2 | GRAS transcription factor | gaiT6 rgaT2 rgl2-1 ↓ | gaiT6 rgaT2 rgl2-1 ↓ | Gomez et al. (2018) |
| GID1A, GID1B | α/β-Hydrolase superfamily protein | gid1ab ↑ | Gomez et al. (2018) | |
| REM22 | B3 protein transcription factor | rem22-1 ↑ | Gomez et al. (2018) | |
| UNE16 | Homeodomain-like superfamily protein | une16-1 ↓ | Gomez et al. (2018) | |
| NERD1 | GW repeat- and PHD-finger-containing protein NERD | nerd1-2 ↓ nerd1-4 ↓ | Yuan and Kessler (2019) | |
| ONA2 | Unknown protein | ona2 ↓ | Yuan and Kessler (2019) | |
| ASHH2 | Histone-lysine N-methyltransferase | ashh2 ↓ | Grini et al. (2009) |
Up- and down-pointing arrows represent how the mutant phenotype impacts either gynoecium size or ovule number.
Placenta development and ovule primordium initiation in Arabidopsis
Periclinal cell divisions within the sub-epidermal tissue of the placenta initiate ovule primordium development at stage 9 of flower development (Roeder and Yanofsky, 2006). Subsequently, three layers of primordium cells form a finger-like structure during stage 10, which then differentiates into three regions along the proximal–distal axis: the funiculus, the chalaza, and the nucellus (Schneitz et al., 1995). These three regions undergo distinct but interdependent developmental processes. The nucellus is the site of megasporogenesis, where the megaspore mother cell differentiates and locates to the upmost, central, and subepidermal position of the digit-shaped ovule primordium (reviewed in Pinto et al., 2019). The chalaza is the region from which the inner and outer integuments develop, and these finally envelop and protect the embryonic sac. The funiculus remains attached to the gynoecium via the placental tissue and this connection is required for the transport of nutrients to the ovule (Fig. 1). For this reason, the placental tissue is fundamental for ovule primordia formation, and for determining their number and maintenance.
In Arabidopsis, placental tissue differentiates from the CMM, which is the central ridge of cells that fuse and give rise to the septum. Placental tissue differentiates along the length of the septum adjacent to the lateral walls (Alvarez and Smyth, 2002; Nole-Wilson et al., 2010a; Reyes-Olalde et al., 2013). Communication between transcription factors and hormones is essential to maintain the meristematic activity of the placenta, to determine the sites of ovule initiation and ovule identity, and to establish the distance between two adjacent ovules (Cucinotta et al., 2014). Several genes that are important for placenta development have been described in the literature (reviewed by Cucinotta et al., 2014; Reyes-Olalde and de Folter, 2019), including AINTEGUMENTA (ANT), CUP-SHAPED COTYLEDON 1 (CUC1) and CUC2, LEUNIG (LUG), MONOPTEROS (MP), and PERIANTHIA (PAN) (Fig. 1; Table 1).
AINTEGUMENTA encodes an AP2 transcription factor (Klucher et al., 1996) and positively regulates organ size via determining cell number and meristematic competence. Ant mutants have fewer and smaller floral organs than the wild-type. In particular, the ant-9 mutant is characterized by unfused carpels at the tip of the pistil (Elliott et al., 1996), whereas in ant-4, the size of floral organs is reduced (Krizek, 2009). In contrast to these mutant phenotypes, Arabidopsis plants that overexpress ANT possess larger floral organs than the wild-type (Mizukami and Fischer, 2000). Expression of ANT is controlled by AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS), an auxin-inducible gene (Hu et al., 2003). When ARGOS is overexpressed, floral organs become enlarged, resulting in longer siliques than those of wild-type (Hu et al., 2003). This was one of the first pieces of evidence that implicated a key role for auxin in pistil development.
ANT expression initiates in the placenta and is maintained throughout all stages of ovule development, in particular in the chalaza region and in the integuments. The reduced ovule number phenotype of the ant mutant is exacerbated when it is combined with other mutations that affect CMM and placenta development, such as revoluta (rev), suggesting that the activity of the REV gene, which encodes a class III homeodomain leucine zipper transcription factor, is also required for placenta formation (Nole-Wilson et al., 2010a). ANT interacts with the transcriptional repressor SEUSS (SEU) and simultaneous loss of both protein activities severely affects placenta development and leads to a complete loss of ovule formation. When a weaker ant-3 allele was combined with seu-3, placenta development was maintained but the number of ovules that initiated was reduced to approximately half of that observed in Col-0 wild-type plants (Azhakanandam et al., 2008). Another transcriptional co-regulator involved in gynoecium patterning is LEUNIG (LUG). Strong lug-1 and intermediate lug-3 alleles show a failure in ridge fusion and a reduction in the amount of placental tissue, with a consequent decrease in the number of ovules formed (Liu et al., 2000). The combination of lug and ant mutations results in gynoecia that are unable to develop ovules (Liu et al., 2000). The loss of ovules in the ant and seu backgrounds is strongly enhanced by mutations in the PERIANTHIA (PAN) gene, which encodes a bZIP transcription factor that is expressed in the gynoecium medial ridge, placenta, and ovules, where it promotes ovule formation (Wynn et al., 2014).
Similar to ANT, factors important for integument growth often affect ovule primordium formation. Two examples are HUELLENLOS (HLL) and SHORT INTEGUMENTS 2 (SIN2). HLL encodes a mitochondrial ribosomal protein and its mutation is associated with smaller gynoecia and a 10% reduction in the number of ovules (Schneitz et al., 1998; Skinner et al., 2001). Shorter gynoecia that bear fewer ovules are also observed in the sin2 mutant; however, more interestingly, the absence of SIN2 function leads to an abnormal distribution of ovules along the placenta (Broadhvest et al., 2000), in which the distance between ovules is greater than in the wild-type; thus, a reduction in ovule number is caused by a reduction in gynoecium size and by the reduced ability of the placental tissue to initiate ovule primordia. SIN2 encodes a mitochondrial DAR GTPase and, similar to HLL, is hypothesized to function in mitochondrial ribosome assembly (Hill et al., 2006). Notably, these two ribosomal proteins, which are targeted to the mitochondria, are necessary for ovule primordium formation, and it has been suggested that impaired mitochondrial function might cause cell-cycle arrest in the placenta and subsequently in the ovule integuments (Broadhvest et al. 2000).
Complex hormonal communication promotes ovule initiation and determines pistil size
Plant organogenesis requires cells to proliferate, grow, and differentiate in a coordinated way. The intercellular communication required during organ initiation is mediated by different phytohormones (Davies, 2004; Vanstraelen and Benková, 2012; Schaller et al., 2015; Marsch-Martínez and de Folter, 2016). As will be discussed in this review, auxins, cytokinins (CKs), gibberellins (GAs), and brassinosteroids (BRs) all play fundamental roles in ovule primordium formation (Fig. 1).
In most auxin-related mutants, defects in gynoecium formation lead to the reduction or absence of placental tissue and the corresponding absence of ovules (reviewed in Balanzá et al., 2006; Larsson et al., 2013; Cucinotta et al., 2014). This phenotype is common to all mutants in which auxin synthesis or transport pathways are compromised, such as yucca1 yucca4 (yuc1 yuc4) (Cheng et al., 2006) and pin1-1 (Okada et al., 1991) or is similar to that following treatment with the polar auxin transport inhibitor 1-naphthyl phthalamic acid (NPA) (Nemhauser et al., 2000).
Polar auxin transport is mediated by the PINFORMED1 (PIN1) efflux transporter and is required to create a zone with an auxin concentration maximum in the placenta, where the founder cells of the ovule primordia will be specified (Benková et al., 2003; Ceccato et al., 2013; Galbiati et al., 2013). Subsequently, the orientation of PIN1 within the membrane relocalizes and redirects auxin flow, establishing a gradient with a maximum at the apices of the formed primordia. In developing organs, auxin distribution can be monitored in vivo by imaging a synthetic auxin-inducible promoter, DR5. In plants that express green fluorescent protein (GFP) from the DR5 promoter, green fluorescence is detected at the apices of the ovule primordia, consistent with PIN1-mediated auxin flow directed to the apex (Benková et al., 2003; Galbiati et al., 2013). The weak pin1-5 mutant allele can produce some flowers in which the pistils have slightly reduced valves, which on average contain only nine ovules (Bennett et al., 1995; Sohlberg et al., 2006; Bencivenga et al., 2012).
CKs occupy a central role in the regulation of cell division and cell differentiation. They are positive regulators of ovule formation, as demonstrated by the phenotype of mutants in which CK pathways are altered. In the ckx3 ckx5 double mutant, the degradation of CKs is compromised and the consequent increase in the levels of these hormones leads to an increased activity of the reproductive meristem (Bartrina et al., 2011). Moreover, the longer than normal gynoecia of ckx3 ckx5 double mutants contain about twice as many ovules as those of the wild-type, indicating an increase in the meristematic capacity of placental tissue (Bartrina et al., 2011). By contrast, reduced ovule formation is observed in mutants in which the synthesis or perception of CKs is compromised. Plants that carry mutations in genes that encode all three CK receptors, cytokinin response 1 (cre1-12), histidine kinase2 (ahk2-2), and ahk3, develop five ovules per pistil on average, in addition to showing pleiotropic growth defects (Higuchi et al., 2004; Bencivenga et al., 2012). The AHK2 and AHK3 receptors are expressed throughout ovule development, from the early stages until maturity, whereas CRE1/AHK4 is expressed in the chalaza region and subsequently in the integuments, suggesting that AHK2 and AHK3 preferentially contribute to ovule primordium formation (Bencivenga et al., 2012). The ovule and gynoecium phenotype of the cre1-12 ahk2-2 ahk3-3 triple mutant resembles that of the weak pin1-5 mutant allele (Bencivenga et al., 2012). This similarity is due to the downregulation of PIN1 expression in the triple mutant, suggesting that during the early stages of ovule development, CK activates PIN1 expression. Bencivenga et al. (2012) showed that treating inflorescences with the synthetic CK 6-benzylaminopurine (BAP) increases PIN1 expression in the gynoecium. Strikingly, treatment with BAP causes the formation of on average 20 additional ovule primordia in each gynoecium, which are positioned between the existing primordia formed before the treatment. This suggests that placental tissue at the boundaries between ovules maintains meristematic competence. During root development, CK affects auxin polar transport via PIN1 both at the transcriptional and post-transcriptional levels. In contrast to the situation in the gynoecium, CK negatively regulates the expression of PIN1 in the root and controls the endorecycling of PIN1 from the membrane to direct it to vacuoles for lytic degradation (Ruzicka et al., 2009; Marhavý et al., 2011). In roots, CYTOKININ RESPONSE FACTORS (CRFs), especially CRF2, CRF3, and CRF6, transcriptionally regulate PIN1 by binding to its promoter at the cis-regulatory PIN CYTOKININ RESPONSE ELEMENT (PCRE) (Šimášková et al., 2015) and modulate its expression in response to CK. Similarly, CRFs also mediate PIN1 expression in ovules in response to CK (Cucinotta et al., 2016). Indeed, PIN1 expression is reduced in the crf2 crf3 crf6 (crf2/3/6) triple mutant and cannot be increased by CK treatment. The placenta in crf2/3/6 is also shorter, but this is not sufficient to explain the 30% decrease in ovule number as ovule density is lower in crf2/3/6 than in the wild-type (Cucinotta et al., 2016). Because PIN1 expression in crf2/3/6 was unresponsive to CK application, the mutant was significantly less sensitive to CK treatment than the wild-type with regard to an increase in ovule number and pistil length. Auxin also regulates CRF2, which is a direct target of the auxin response factor (ARF) AUXIN RESPONSE FACTOR 5/MONOPTEROS (ARF5/MP) (Schlereth et al., 2010), highlighting another convergence point between auxin and CK.
Another ARF family member that is required for appropriate apical–basal gynoecium patterning is ARF3/ETTIN (ETT). The ett mutant is characterized by a shorter ovary with an elongated style and gynophore (Sessions et al., 1997). A similar gynoecium phenotype resulted from treatment with the auxin transport inhibitor (NPA), suggesting that ETT plays a key role in auxin signalling along the apical–basal gynoecium axis (Nemhauser et al., 2000). Moreover, ETT restricts the expression domain of SPATULA (SPT), which encodes a basic helix–loop–helix (bHLH) transcription factor (Heisler et al., 2001). Mutations in SPT causes a split-carpel phenotype in the apical part of the gynoecium, leading to a slight reduction in ovule number (Alvarez and Smyth, 1999; Nahar et al., 2012). SPT dimerizes with another bHLH transcription factor, INDEHISCHENT (IND), to repress the expression of PINOID (Girin et al., 2011), which encodes a serine/threonine kinase that regulates PIN1 polarization via phosphorylation (Friml et al., 2004). The repression of PID by SPT and IND allows the formation of a radially symmetric auxin ring in the upper part of the gynoecium that is required for correct style and stigma development (Moubayidin and Østergaard, 2014).
Furthermore, SPT interacts with the three closely related bHLH transcription factors, HECATE1 (HEC1), HEC2, and HEC3 (Gremski et al., 2007), and similar to ett, hec-1 hec-2 hec-3 triple mutants possess an elongated style and shorter ovaries. The HEC proteins and SPT promote auxin transport in concert by activating PIN1 and PIN3 expression (Schuster et al., 2015) and also transcriptionally activate the type-A ARABIDOPSIS RESPONSE REGULATORS (ARR-As), which are negative regulators of CK signalling (Schuster et al., 2015). Via this dual action on auxin transport and CK response, HECs and SPT regulate wild-type gynoecium fusion at the apex, and style and stigma development. Furthermore, SPT alone in the medial domain activates the type-B ARRs, especially ARR1, which are positive regulators of CK signalling. The arr1 arr10 arr12 triple mutant possesses a shorter gynoecium and significantly fewer ovules than the wild-type (Reyes-Olalde et al., 2017).
In addition to auxin localization, correct auxin signalling is also required for wild-type gynoecium development, as confirmed by a recent study on members of the Small Auxin-Upregulated RNA (SAUR) family, which were initially identified as short transcripts that were rapidly upregulated in response to auxin (McClure and Guilfoyle, 1987). When SAUR8, SAUR10, and SAUR12 are ectopically overexpressed in Arabidopsis, the gynoecium and resulting siliques are longer than in wild-type, suggesting that auxin positively regulates gynoecium length and, probably indirectly, silique length (van Mourik et al., 2017). Notably, SAUR gene expression increased 100-fold following combined auxin and BR treatment (van Mourik et al., 2017). BRs are clearly involved in pistil growth and ovule number specification; gynoecia of the enhanced BR-signalling mutant brassinazole-resistant 1-1D (bzr1-1D) not only contained more ovules than wild-type but they were also longer. By contrast, BR-deficient mutants such as de-etiolated 2 (det-2), brassinosteroid insensitive 1 (bri1-5) and brassinosteroid-insensitive 2 (bin2-1) developed shorter pistils with fewer ovules (Huang et al., 2013).
The involvement of BRs in gynoecium and ovule development was also confirmed by Nole-Wilson et al. (2010b), who observed that a reduction in the expression of CYP85A2, which encodes an enzyme involved in the final step of brassinolide biosynthesis (Nomura et al., 2005), enhances the seuss mutant phenotypic disruptions in ovules and gynoecia (Nole-Wilson et al., 2010b).
CUP-SHAPED COTYLEDON 1 and 2 function synergistically with auxin and cytokinins
During ovule primordium formation, CK homeostasis requires two NAC-domain transcription factors, CUP-SHAPED COTYLEDON 1 (CUC1) and CUC2. These are expressed in lateral organ boundaries and function redundantly during organ boundary determination. CUC1 and CUC2 are expressed in the septum and placenta, and following the emergence of ovule primordia, CUC2 expression is restricted to the borders between the ovules (Ishida et al., 2000; Galbiati et al., 2013; Gonçalves et al., 2015). The CUC1 and CUC2 genes are both post-transcriptionally regulated by miR164 microRNAs (Laufs et al., 2004; Mallory et al., 2004). Gynoecia of the in vitro regenerated cuc1 cuc2 mutant as well as of cuc2-1 pSTK::CUC1_RNAi plants have reduced ovule numbers. The cuc1 cuc2 double mutant has on average fewer than 10 ovules per pistil (Ishida et al., 2000), whereas cuc2-1 pSTK::CUC1_RNAi plants, in which CUC1 was specifically silenced in the placenta and ovules, showed a 20% reduction in ovule number, but gynoecium length was not affected. In pistils of these plants, ovules were more widely spaced when compared with the wild-type (Galbiati et al., 2013). This result was confirmed by silencing CUC1 and CUC2 by overexpressing MIR164A, which strongly reduced ovule number, indicating a major contribution of CUC1 and CUC2 to ovule initiation (Gonçalves et al., 2015). The analysis of PIN1–GFP expression in cuc2-1 pSTK::CUC1_RNAi plants revealed that CUC1 and CUC2 redundantly promote PIN1 expression and PIN1 membrane localization in ovules. Treatment with BAP increased PIN1 expression and complemented the reduced ovule number phenotype of cuc2-1 pSTK::CUC1_RNAi plants (Galbiati et al., 2013). Therefore, CKs act downstream from or in parallel with CUC1 and CUC2 to induce the expression of PIN1. Recently, it has been demonstrated that CUC1 and CUC2 induce CK responses in vivo and function upstream of CK by transcriptionally repressing UGT73C1 and UGT85A3, which encode two enzymes involved in CK inactivation (Cucinotta et al., 2018). Consistent with this result, the concentration of inactive CK glucosides was higher in cuc2-1 pSTK::CUC1_RNAi inflorescences than in wild-type plants.
The expression of CUC1 and CUC2 is also linked with auxin signalling: their expression pattern coincides with that of the auxin response factor ARF5/MP (see above) and both genes are downregulated in pistils of the weak mp-S319 mutant allele (Galbiati et al., 2013). During the early stages of placenta development and ovule formation, ARF5/MP directly transcriptionally activates CUC1 and CUC2, but also ANT. The observation that BAP treatment did not complement the ovule number phenotype of ant-4 suggests that ANT functions independently of CUC1 and CUC2. This is further supported by the additive effects on the reduction in ovule number observed in ant-4 cuc2-1 pSTK:CUC1_RNAi plants (Galbiati et al., 2013). Together these data suggest that ANT promotes cell proliferation, whereas CUC1 and CUC2 regulate CK homeostasis and auxin transport. Although CUC3 shares high similarity with CUC1 and CUC2, the cuc3 mutant was not affected in ovule initiation and number, but together with CUC2, CUC3 promotes ovule separation; this is reflected by the cuc2 cuc3 double mutant, which produces seeds that result from the fusion of two ovules (Gonçalves et al., 2015). These results suggest that specific CUC genes independently promote ovule initiation and ovule separation.
Lee et al. (2009) identified LATERAL ORGAN FUSION 1 (LOF1) to be involved in lateral organ separation and to functionally overlap with CUC2 and CUC3. The LOF1 gene is expressed at the base of ovule primordia and its overexpression results in a wrinkled pistil with an enlarged replum, an amorphous septum and an irregular ovule distribution (Gomez et al., 2011).
The role of gibberellins in ovule primordium formation
GAs are involved in key developmental processes throughout the plant life cycle, from seed germination in particular, to flowering time (reviewed in Hedden and Sponsel, 2015; Rizza and Jones, 2019), but their involvement in ovule initiation has only recently been demonstrated. Gomez and colleagues (2018) showed that DELLA proteins, which belong to a subfamily of the plant-specific GRAS family of transcriptional regulators that repress GA signalling, positively regulate ovule number in Arabidopsis. In addition to DELLA proteins, the GA signalling core includes the GA receptor GID1. When GID1 binds bioactive GA, the GA–GID1–DELLA complex is formed and triggers the polyubiquitination and degradation of DELLA proteins. The della triple mutant gaiT6 rgaT2 rgl2-1 produces fewer ovules than wild-type (Gomez et al., 2018). By contrast, the gain-of-function DELLA mutant gai-1, which cannot be degraded upon GA sensing, produced more ovules. Consistent with this observation, the double gid1a gid1b mutant, which cannot perceive GA, forms more ovules than the wild-type, demonstrating a negative correlation between GAs and ovule number (Gomez et al., 2018). The GAI, RGA, RGL2, GID1a, and GID1b genes are expressed in placental tissue and outgrowing ovules. The reduction in ovule number was more dramatic in the gaiT6 rgaT2 rgl2-1 triple mutant than that in ovary length, resulting in a lower ovule density, whereas the dominant gai-1 mutant has an increased ovule/placenta ratio, suggesting that GAs predominantly affect ovule initiation and not placenta elongation.
Other evidence to demonstrate that DELLA proteins promote ovule formation derives from an experiment in which the expression of the stable mutant protein rgaΔ17 under the control of the ANT promoter in the placenta resulted in the formation of 20% more ovules than in control lines (Gomez et al., 2018). This effect of GAs on the number of developing ovules was not correlated with auxin signalling or transport, and neither PIN1 localization nor DR5 expression was affected by GA treatment or DELLA activity (Gomez et al., 2018).
Confirmation of a positive role for RGL2 in determining ovule number came from the analysis of transgenic lines in which RGL2-dependent GA signalling was blocked by the expression of a dominant version of RGL2 (pRGL2:rgl2Δ17) (Gómez et al., 2019). Pistils of pRGL2:rgl2Δ17 plants contained 10% more ovules than those of the wild-type, whereas pistil length did not differ, indicating that the main function of rgl2Δ17 is to positively promote ovule primordium formation but not placenta elongation (Gómez et al., 2019). Furthermore, Gomez et al. (2018) identified REPRODUCTIVE MERISTEM 22 (REM22) and UNFERTLIZED EMBRYO SAC 16 (UNE16) via transcriptomic analysis to be DELLA targets that are positive regulators of ovule initiation. REM22 is a B3 family transcription factor that is expressed in the placenta (Mantegazza et al., 2014) and increased REM22 expression in the rem22-1 enhancer allele significantly increases ovule number. UNE16 is a transcription factor involved in embryo sac development and the knockdown allele une16-1 produces fewer ovules. Because UNE16 expression is regulated by BRs (Pagnussat et al., 2005; Sun et al., 2010), it represents a potential nexus for crosstalk between GAs and BRs in ovule initiation. The establishment of GA as an important additional component of the ovule regulatory network has introduced an additional layer of complexity to the current model for ovule initiation and it remains to be established how GAs integrate into this model. GAs might function antagonistically to CKs and BRs, which in contrast to GAs, positively regulate pistil size and ovule number.
Finally, the ctr1-1 constitutive ethylene-responsive mutant possesses a shorter gynoecium at anthesis compared with wild-type and a delay in the response to GA3 treatment that induces gynoecium senescence, suggesting that ethylene affects gynoecium size, probably by interactions with GA pathways (Carbonell-Bejerano et al., 2011).
In conclusion, there is ample evidence for complex interactions between different hormonal pathways that together determine ovule number and pistil size.
Ovule number: the ecotype matters
It has been known for 20 years that the number of ovules varies hugely among different Arabidopsis ecotypes (diploid accessions) (Alonso-Blanco et al., 1999): for example, the Landsberg erecta accession produces 20% more ovules than the Cape Verde Islands (Cvi) accession. Recently, 189 Arabidopsis accessions from the Arabidopsis Biological Resource Center were analysed for differences in ovule number and they display a remarkable degree of variation, ranging from 39 to 82 ovules per pistil (Yuan and Kessler, 2019). The commonly used reference accession Col-0 lies in the middle of the range, with a mean ovule number of 63, which is strongly dependent on experimental growth conditions. Ovule number, in contrast to, for instance, flowering time, does not correlate with geographical origin (Stinchcombe et al., 2004; Yuan and Kessler, 2019). By conducting a genome-wide association study on these 189 accessions, two loci associated with ovule number were identified (Yuan and Kessler, 2019): NEW ENHANCER OF ROOT DWARFISM (NERD1) and OVULE NUMBER ASSOCIATED 2 (ONA2). Mutation of NERD1 or ONA2 leads to a significant reduction in ovule number, with a stronger phenotype in the nerd1-2 and nerd1-4 alleles. ONA2 encodes a protein of unknown function and was not further analysed. In addition to a reduction in ovule number, nerd mutants display additional severe male and female fertility defects. NERD1 encodes an integral membrane protein mainly localized to the Golgi. Notably, NERD1 expression is lower in Altai-5 and Kas-2 accessions, which have low ovule numbers (Yuan and Kessler, 2019), but high NERD1 expression in Altai-5 leads to a significant increase in ovule number. However, overexpression of NERD1 in Col-0 plants did not affect ovule number, indicating that NERD1 function in determining ovule number is background-dependent (Yuan and Kessler, 2019).
Considerable genetic variation in ovule number was also described for F1 triploids of different Arabidopsis genotypes by Duszynska et al. (2013), who observed differences in ovule number between genetically identical F1-hybrid offspring, after crossing parental genome excess lines (2m:1p with 1m:2p). These effects can only be explained by epigenetic mechanisms that affect genes controlling ovule number, for example DNA or histone methylation. The analysis of null alleles of ASH1 HOMOLOG 2 (ASH2), which show a remarkable 80% reduction in ovule number, provided a clear example of the involvement of histone methylation in determining ovule number (Grini et al., 2009). The transcriptional state of the ASH2 locus remains active during development via H3K36 trimethylation (Xu et al., 2008). It will be highly relevant to study the effect of epigenetic modifications induced by biotic and abiotic stresses in determining ovule number. Epigenetic responses to stress are fundamental to create the plasticity required for plant survival, especially considering that plants are sessile organisms. These epigenetic changes can be temporally transmitted, even in the absence of the original stress (Iglesias and Cerdán, 2016). Furthermore, variation in ovule number in response to fluctuations in environmental conditions, such as temperature, can be used to understand the plasticity and inheritability of (epigenetic) adaptation and response to temperature stress. Variation in ovule number under stress conditions is, of course, also highly relevant from an ecological, environmental, and evolutional perspective.
Ovule number decreases with ageing
Ovule number varies throughout inflorescence development: early flowers developing on the main inflorescence (from the fifth to the twenty-fifth flower) of Arabidopsis Ler plants produced a relatively invariable number of ovules, whereas flowers that developed later had pistils with fewer ovules (Gomez et al., 2018; Yuan and Kessler, 2019). Loss- and gain-of-function mutants of DELLA genes showed an increase in ovule number in early- and late-arising flowers (Gomez et al., 2018). To minimize age-related variation in their genome-wide association studies, Yuan and Kessler (2019) only counted ovules in flowers 6–10 from the main inflorescence.
It has been reported for other plant species that flower position as well as size influences ovule number per flower. For example, in pomegranate, the number of ovules per flower was significantly influenced by flower size, with more ovules being produced in larger flowers (Wetzstein et al., 2013).
Overall, when studying changes in ovule numbers it is important to be aware of the possible variation in the different flowers of the plant. Therefore, large numbers will have to be analysed using thorough statistical analyses, especially for genotypes that show only relatively minor changes.
A ‘gold mine’ for seed yield improvement within the Brassicaceae
Improving seed yield via the genetic manipulation of crops has historically been a central goal in agricultural research. The enormous body of data, which has been generated and shared by the scientific community over the past decades, represents a true ‘gold mine’ for translational and applied research. The determination of pistil size and ovule number may be considered one of the most straightforward traits that can be enhanced to improve overall seed yield in species characterized by multi-ovulate ovaries and the increasing amount of literature on this topic evidences an active and prolific research field. Although some questions concerning the networks controlling seed number and pistil size remain open, comprehensive knowledge of the phytohormone interactions involved in these pathways is already available and applicable (Cucinotta et al., 2014; Zúñiga-Mayo et al., 2019; Reyes-Olalde and de Folter, 2019).
Understanding these developmental processes in Arabidopsis can inform promising strategies for knowledge transfer to closely related and agronomically important crops. Rapeseed (Brassica napus), another Brassicaceae species, is an important breeding target, since it is a crop widely cultivated in Europe, Asia, Canada, and Australia. It is characterized by an oil-rich seed and its processing provides both rapeseed oil (used as edible vegetable oil or as biodiesel) and a by-product mostly used as cattle fodder (Snowdon et al., 2007).
It has recently been demonstrated that Arabidopsis and B. napus share well-conserved response mechanisms to CK treatment (Zuñiga-Mayo et al., 2018). Strikingly, exogenous CK application causes a reduction in silique length in B. napus. However, these shorter siliques contain increased ovule numbers and upon manual pollination, the plants show an increase in seed yield of 18%. Intriguingly, increases in ovule and seed number have also been observed in the offspring of the treated plants, suggesting that the mechanism has an underlying epigenetic basis (Zuñiga-Mayo et al., 2018).
An increase in CK level has also been reported to beneficially affect seed yield in transgenic B. napus lines expressing the CK biosynthetic enzyme isopentenyltransferase (IPT) under the Arabidopsis promoter of the AtMYB32 gene. An increase in seed yield of up to 23% was obtained in the transgenic lines that were analysed (Kant et al., 2015).
CK homeostasis is mediated by CYTOKININ OXIDASES/DEHYDROGENASES (CKXs) during pistil and silique development in Arabidopsis. Remarkably, the expression level of CKX genes in B. napus is associated with silique length, and RNA-sequencing and qRT-PCR analyses revealed a significantly different expression level of BnCKX5-1, 5-2, 6-1, and 7-1 in two distinct cultivated varieties with long versus short siliques (Liu et al., 2018). These findings open up promising strategies with which to modulate silique length in B. napus by manipulating CKX gene expression.
In addition to phytohormones, genetic knowledge from Arabidopsis can be successfully applied to B. napus crop improvement. Mutations in the K-box of the Arabidopsis orthologue of APETALA1 in B. napus caused a significant increase in the number of seeds per plant (Shah et al., 2018). These generated alleles could conceivably be introduced into a rapeseed breeding programme in field trials.
Germplasm of B. napus revealed substantial natural variation with respect to seed number per pod. Current rapeseed cultivars produce on average 20 seeds per pod, which is far lower than the maximum observed among the germplasm resources (Yang et al., 2017). Moreover, genetic improvement promises to deliver a massive improvement in seed yield (Yang et al., 2017). The gold mine of knowledge obtained from the closely related species Arabidopsis will certainly be fundamentally important in the exploitation of the encouraging genetic variation potential. Furthermore, it has recently been demonstrated that CRISPR–Cas9 technology can be efficiently applied to precisely induce targeted mutation in rapeseed (Braatz et al., 2017), making it a powerful tool for future genetic improvement. Similarly, existing knowledge could be used to improve other Brassicaceae species, or even non-phylogenetically related species such as soybean.
Acknowledgements
The authors’ work is supported by the H2020 Marie Skłodowska-Curie Actions of European Commission (H2020-MSCA-RISE-2015 EXPOSEED and MSCA-RISE-2014 PROCROP projects) also acknowledges the Mexican National Council of Science and Technology (CONACyT) grants FC-2015-2/1061 and CB2017-2018 A1-S-10126. The authors would like to thank John Chandler for editing the paper.
References
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. 1999. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 96, 4710–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. 1999. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Alvarez J, Smyth DR. 2002. CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. International Journal of Plant Sciences 163, 17–41. [Google Scholar]
- Alvarez-Buylla ER, Benítez M, Corvera-Poiré A, et al. 2010. Flower development. The Arabidopsis Book 8, e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Colombo L. 1996. Molecular control of ovule development. Trends in Plant Science 1, 228–232. [Google Scholar]
- Azhakanandam S, Nole-Wilson S, Bao F, Franks RG. 2008. SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiology 146, 1165–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanzá V, Navarrete M, Trigueros M, Ferrándiz C. 2006. Patterning the female side of Arabidopsis: the importance of hormones. Journal of Experimental Botany 57, 3457–3469. [DOI] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. 2011. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. The Plant Cell 23, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S. 2011. Cross talk between the sporophyte and the megagametophyte during ovule development. Sexual Plant Reproduction 24, 113–121. [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L. 2012. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. The Plant Cell 24, 2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR. 1995. Morphogenesis in pinoid mutants of Arabidopsis thaliana. The Plant Journal 8, 505–520. [Google Scholar]
- Bowman JL, Baum SF, Eshed Y, Putterill J, Alvarez J. 1999. Molecular genetics of gynoecium development in Arabidopsis. Current Topics in Developmental Biology 45, 155–205. [DOI] [PubMed] [Google Scholar]
- Braatz J, Harloff HJ, Mascher M, Stein N, Himmelbach A, Jung C. 2017. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiology 174, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhvest J, Baker SC, Gasser CS. 2000. SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155, 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Granell A, Carbonell J, Perez-Amador MA. 2011. Ethylene is involved in pistil fate by modulating the onset of ovule senescence and the GA-mediated fruit set in Arabidopsis. BMC Plant Biology 11, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccato L, Masiero S, Sinha Roy D, Bencivenga S, Roig-Villanova I, Ditengou FA, Palme K, Simon R, Colombo L. 2013. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS ONE 8, e66148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes & Development 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo L, Battaglia R, Kater MM. 2008. Arabidopsis ovule development and its evolutionary conservation. Trends in Plant Science 13, 444–450. [DOI] [PubMed] [Google Scholar]
- Cucinotta M, Colombo L, Roig-Villanova I. 2014. Ovule development, a new model for lateral organ formation. Frontiers in Plant Science 5, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta M, Manrique S, Cuesta C, Benkova E, Novak O, Colombo L. 2018. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. Journal of Experimental Botany 69, 5169–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta M, Manrique S, Guazzotti A, Quadrelli NE, Mendes MA, Benkova E, Colombo L. 2016. Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development. Development 143, 4419–4424. [DOI] [PubMed] [Google Scholar]
- Davies PJ. 2004. Plant hormones: biosynthesis, signal, transduction, action! Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Drews GN, Koltunow AM. 2011. The female gametophyte. The Arabidopsis Book 9, e0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszynska D, McKeown PC, Juenger TE, Pietraszewska-Bogiel A, Geelen D, Spillane C. 2013. Gamete fertility and ovule number variation in selfed reciprocal F1 hybrid triploid plants are heritable and display epigenetic parent-of-origin effects. New Phytologist 198, 71–81. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. 2011. Angiosperm ovules: diversity, development, evolution. Annals of Botany 107, 1465–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK, Igersheim A. 2000. Gynoecium structure and evolution in basal angiosperms. International Journal of Plant Sciences 161, S211–S213. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306, 862–865. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Sinha Roy D, Simonini S, et al. 2013. An integrative model of the control of ovule primordia formation. The Plant Journal 76, 446–455. [DOI] [PubMed] [Google Scholar]
- Gasser CS, Broadhvest J, Hauser BA. 1998. Genetic analysis of ovule development. Annual Review of Plant Physiology and Plant Molecular Biology 49, 1–24. [DOI] [PubMed] [Google Scholar]
- Gasser CS, Skinner DJ. 2019. Development and evolution of the unique ovules of flowering plants. Current Topics in Developmental Biology 131, 373–399. [DOI] [PubMed] [Google Scholar]
- Girin T, Paicu T, Stephenson P, et al. 2011. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. The Plant Cell 23, 3641–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MD, Barro-Trastoy D, Escoms E, et al. 2018. Gibberellins negatively modulate ovule number in plants. Development 145, dev163865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez MD, Fuster-Almunia C, Ocaña-Cuesta J, Alonso JM, Pérez-Amador MA. 2019. RGL2 controls flower development, ovule number and fertility in Arabidopsis. Plant Science 281, 82–92. [DOI] [PubMed] [Google Scholar]
- Gomez MD, Urbez C, Perez-Amador MA, Carbonell J. 2011. Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS ONE 6, e18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves B, Hasson A, Belcram K, et al. 2015. A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. The Plant Journal 83, 732–742. [DOI] [PubMed] [Google Scholar]
- Govaerts R. 2001. How many species of seed plants are there? Taxon 50, 1085–1090. [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. 2007. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593–3601. [DOI] [PubMed] [Google Scholar]
- Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jørstad TS, Wilson ZA, Aalen RB. 2009. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 4, e7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross T, Broholm S, Becker A. 2018. CRABS CLAW acts as a bifunctional transcription factor in flower development. Frontiers in Plant Science 9, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K. 1998. The molecular and genetic basis of ovule and megagametophyte development. Seminars in Cell & Developmental Biology 9, 227–238. [DOI] [PubMed] [Google Scholar]
- Hedden P, Sponsel V. 2015. A century of gibberellin research. Journal of Plant Growth Regulation 34, 740–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR. 2001. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, et al. 2004. In planta functions of the Arabidopsis cytokinin receptor family. Proceedings of the National Academy of Sciences, USA 101, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Broadhvest J, Kuzoff RK, Gasser CS. 2006. Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GTPase. Genetics 174, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. 2003. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. The Plant Cell 15, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Jiang WB, Hu YW, Wu P, Zhu JY, Liang WQ, Wang ZY, Lin WH. 2013. BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Molecular Plant 6, 456–469. [DOI] [PubMed] [Google Scholar]
- Iglesias FM, Cerdán PD. 2016. Maintaining epigenetic inheritance during DNA replication in plants. Frontiers in Plant Science 7, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Aida M, Takada S, Tasaka M. 2000. Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant & Cell Physiology 41, 60–67. [DOI] [PubMed] [Google Scholar]
- Kant S, Burch D, Badenhorst P, Palanisamy R, Mason J, Spangenberg G. 2015. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS ONE 10, e0116349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. 1996. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. The Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. 2009. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiology 150, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Franks RG, Sundberg E. 2013. Auxin and the Arabidopsis thaliana gynoecium. Journal of Experimental Botany 64, 2619–2627. [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lee D-K, Geisler M, Springer PS. 2009. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136, 2423–2432. [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang C, Ma JQ, et al. 2018. Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus L.). Genes 9, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Franks RG, Klink VP. 2000. Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. The Plant Cell 12, 1879–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. 2004. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. The EMBO Journal 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantegazza O, Gregis V, Mendes MA, Morandini P, Alves-Ferreira M, Patreze CM, Nardeli SM, Kater MM, Colombo L. 2014. Analysis of the arabidopsis REM gene family predicts functions during flower development. Annals of Botany 114, 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Developmental Cell 21, 796–804. [DOI] [PubMed] [Google Scholar]
- Marsch-Martínez N, de Folter S. 2016. Hormonal control of the development of the gynoecium. Current Opinion in Plant Biology 29, 104–114. [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. 1987. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Molecular Biology 9, 611–623. [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. 2000. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Østergaard L. 2014. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Current Biology 24, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar MA, Ishida T, Smyth DR, Tasaka M, Aida M. 2012. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant & Cell Physiology 53, 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC. 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Azhakanandam S, Franks RG. 2010a Polar auxin transport together with AINTEGUMENTA and REVOLUTA coordinate early Arabidopsis gynoecium development. Developmental Biology 346, 181–195. [DOI] [PubMed] [Google Scholar]
- Nole-Wilson S, Rueschhoff EE, Bhatti H, Franks RG. 2010b Synergistic disruptions in seuss cyp85A2 double mutants reveal a role for brassinolide synthesis during gynoecium and ovule development. BMC Plant Biology 10, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S. 2005. The last reaction producing brassinolide is catalyzed by cytochrome P-450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. The Journal of Biological Chemistry 280, 17873–17879. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. 1991. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. The Plant Cell 3, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. 2005. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. [DOI] [PubMed] [Google Scholar]
- Pinto SC, Mendes MA, Coimbra S, Tucker MR. 2019. Revisiting the female germline and its expanding toolbox. Trends in Plant Science 24, 455–467. [DOI] [PubMed] [Google Scholar]
- Reiser L, Fischer RL. 1993. The ovule and the embryo sac. The Plant Cell 5, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, de Folter S. 2019. Control of stem cell activity in the carpel margin meristem (CMM) in Arabidopsis. Plant Reproduction 32, 123–136. [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuñiga-Mayo VM, Chávez Montes RA, Marsch-Martínez N, de Folter S. 2013. Inside the gynoecium: at the carpel margin. Trends in Plant Science 18, 644–655. [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zúñiga-Mayo VM, Serwatowska J, et al. 2017. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genetics 13, e1006726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A, Jones AM. 2019. The makings of a gradient: spatiotemporal distribution of gibberellins in plant development. Current Opinion in Plant Biology 47, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AH, Yanofsky MF. 2006. Fruit development in Arabidopsis. The Arabidopsis Book 4, e0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E. 2009. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proceedings of the National Academy of Sciences, USA 106, 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. 2010. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Baker SC, Gasser CS, Redweik A. 1998. Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125, 2555–2563. [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE. 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. The Plant Journal 7, 731–749. [Google Scholar]
- Schuster C, Gaillochet C, Lohmann JU. 2015. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. 1997. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Shah S, Karunarathna NL, Jung C, Emrani N. 2018. An APETALA1 ortholog affects plant architecture and seed yield component in oilseed rape (Brassica napus L.). BMC Plant Biology 18, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Yang WC. 2011. Ovule development in Arabidopsis: progress and challenge. Current Opinion in Plant Biology 14, 74–80. [DOI] [PubMed] [Google Scholar]
- Shirley NJ, Aubert MK, Wilkinson LG, Bird DC, Lora J, Yang X, Tucker MR. 2019. Translating auxin responses into ovules, seeds and yield: Insight from Arabidopsis and the cereals. Journal of Integrative Plant Biology 61, 310–336. [DOI] [PubMed] [Google Scholar]
- Šimášková M, O’Brien JA, Khan M, et al. 2015. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nature Communications 6, 8717. [DOI] [PubMed] [Google Scholar]
- Skinner DJ, Baker SC, Meister RJ, Broadhvest J, Schneitz K, Gasser CS. 2001. The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. The Plant Cell 13, 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DJ, Hill TA, Gasser CS. 2004. Regulation of ovule development. The Plant cell 16 Suppl, S32–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. 1990. Early flower development in Arabidopsis. The Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon RJ, Lühs W, Friedt W. 2007. Genome mapping and molecular breeding in plants. Oilseeds 6, 54–56. [Google Scholar]
- Sohlberg JJ, Myrenås M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E. 2006. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. The Plant Journal 47, 112–123. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proceedings of the National Academy of Sciences, USA 101, 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, et al. 2010. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Developmental Cell 19, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mourik H, van Dijk ADJ, Stortenbeker N, Angenent GC, Bemer M. 2017. Divergent regulation of Arabidopsis SAUR genes: a focus on the SAUR10-clade. BMC Plant Biology 17, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. 2012. Hormonal interactions in the regulation of plant development. Annual Review of Cell and Developmental Biology 28, 463–487. [DOI] [PubMed] [Google Scholar]
- Wetzstein HY, Yi W, Porter JA, Ravid N. 2013. Flower position and size impact ovule number per flower, fruitset, and fruit size in pomegranate. Journal of the American Society for Horticultural Science 138, 159–166. [Google Scholar]
- Willis KJ. 2017. State of the world’s plants 2017. Kew: Royal Botanic Gardens. [PubMed] [Google Scholar]
- Wynn AN, Seaman AA, Jones AL, Franks RG. 2014. Novel functional roles for PERIANTHIA and SEUSS during floral organ identity specification, floral meristem termination, and gynoecial development. Frontiers in Plant Science 5, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. 2008. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Molecular and Cellular Biology 28, 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang Y, Zhan J, Shi J, Wang X, Liu G, Wang H. 2017. Genetic and cytological analyses of the natural variation of seed number per pod in rapeseed (Brassica napus L.). Frontiers in Plant Science 8, 1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Kessler SA. 2019. A genome-wide association study reveals a novel regulator of ovule number and fertility in Arabidopsis thaliana. PLoS Genetics 15, e1007934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga-Mayo VM, Baños-Bayardo CR, Díaz-Ramírez D, Marsch-Martínez N, De Folter S. 2018. Conserved and novel responses to cytokinin treatments during flower and fruit development in Brassica napus and Arabidopsis thaliana. Scientific Reports 8, 6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga-Mayo VM, Gómez-Felipe A, Herrera-Ubaldo H, de Folter S. 2019. Gynoecium development: networks in Arabidopsis and beyond. Journal of Experimental Botany 70, 1447–1460. [DOI] [PubMed] [Google Scholar]