Glycosyltransferase OsUGT90A1 helps to protect cellular membranes during chilling and salt stress by enhancing antioxidant enzyme activity to reduce ROS levels, and to balance the stress response with growth.
Keywords: Abiotic stress, antioxidant enzyme activity, cold tolerance, gene knockout, low temperature seedlings survivability, reactive oxygen species, salt tolerance, transgenic rice
Abstract
Due to its subtropical origins, rice (Oryza sativa) is sensitive to low-temperature stress. In this study, we identify LOC_Os04g24110, annotated to encode the UDP-glycosyltransferase enzyme UGT90A1, as a gene associated with the low-temperature seedling survivability (LTSS) quantitative trait locus qLTSS4-1. Differences between haplotypes in the control region of OsUGT90A1 correlate with chilling tolerance phenotypes, and reflect differential expression between tolerant and sensitive accessions rather than differences in protein sequences. Expression of OsUGT90A1 is initially enhanced by low temperature, and its overexpression helps to maintain membrane integrity during cold stress and promotes leaf growth during stress recovery, which are correlated with reduced levels of reactive oxygen species due to increased activities of antioxidant enzymes. In addition, overexpression of OsUGT90A1 in Arabidopsis improves freezing survival and tolerance to salt stress, again correlated with enhanced activities of antioxidant enzymes. Overexpression of OsUGT90A1 in rice decreases root lengths in 3-week-old seedlings while gene-knockout increases the length, indicating that its differential expression may affect phytohormone activities. We conclude that higher OsUGT90A1 expression in chilling-tolerant accessions helps to maintain cell membrane integrity as an abiotic stress-tolerance mechanism that prepares plants for the resumption of growth and development during subsequent stress recovery.
Introduction
Rice (Oryza sativa) is an important staple crop, feeding almost three billion people worldwide (Fairhurst and Dobermann, 2002; Shelton et al., 2002). Due to its subtropical origins, rice is susceptible to chilling stress (0–15 °C). Rice grown in temperate regions is often exposed to temperatures below 10 °C, and hence yields are severely affected (Xie et al., 2012). Only a few chilling-tolerance genes have been identified using map-based cloning strategies (da Cruz et al., 2013; Zhang et al., 2014); however, because of the large amount of genetic variation within genotyped rice accessions from around the world, numerous cold-tolerance quantitative trait loci (QTL) have been identified through genome-wide association studies (GWAS) (da Cruz et al., 2013; Fujino et al., 2015; Pan et al., 2015; Zhu et al., 2015; Lv et al., 2016; Wang et al., 2016a; Shakiba et al., 2017; Schläppi et al., 2017). Such mapping approaches are facilitated by the fact that rice has two major subspecies with considerable genetic variation. The relatively chilling-tolerant japonica subspecies is composed of the three subpopulations ‘aromatic’, ‘temperate japonica’, and ‘tropical japonica’, and the chilling-sensitive indica subspecies is composed of ‘aus’ and various ‘indica’ subpopulations.
Low-temperature stress negatively affects plant growth and development by altering the physical and chemical properties of the plasma membrane, and by disrupting ion homeostasis and overall cell metabolism (Thomashow, 1999; Ruelland and Zachowski, 2010; Ding et al. 2019). Plants utilize various strategies to protect themselves from low-temperature stress. An intact membrane is crucial for effective cold-stress response mechanisms, and the lipid compositions of plasma- and endo-membranes undergo remarkable changes during cold acclimation that help to alleviate the impact of cellular damage induced by low-temperature stress (Uemura et al., 2006; Takahashi et al., 2013; Barrero-Sicilia et al., 2017). However, this protective mechanism is simultaneously challenged by oxidative stress derived from reactive oxygen species (ROS) that are induced by cold temperatures.
ROS such as the superoxide ion (O2•–) and hydrogen peroxide (H2O2) are inevitable byproducts of cellular metabolism (Mittler et al., 2011). ROS can be categorized into metabolic and signaling, with the former originating from disruption of cellular metabolism as a consequence of stresses and the latter originating from sensors that receive the stress signals (Mittler and Blumwald, 2010; Mittler et al., 2012). The two groups are produced via different biosynthesis pathways in multiple cellular organelles, such as the plasma membrane, chloroplasts, mitochondria, peroxisomes, and the vacuole (Suzuki et al., 2011; Vaahtera et al., 2014; Gilroy et al., 2016; Mignolet-Spruyt et al., 2016). Excessive ROS can cause substantial molecular, biochemical, and physiological damage through over-oxidation of vital macromolecules such as lipids, amino acids, and DNA (Gill and Tuteja, 2010; Krasensky and Jonak, 2012). To cope with oxidative stress, higher plants have evolved mechanisms that utilize a variety of antioxidants to scavenge ROS. Antioxidants can be classified into enzymatic antioxidants, such as ascorbate peroxidase (APX), catalase (CAT), glutathione peroxidase (GPX), and superoxide dismutase (SOD), and non-enzymatic antioxidants, such as ascorbic acid and glutathione (Mittler et al., 2004). Flavonoids and anthocyanins can also scavenge ROS (Gupta et al., 2010) and the last step of their biosynthesis is catalysed by specific UDP-flavonoid/anthocyanin glycosyltransferases (UFGTs), generating various derivatives with antioxidant activity (Shi and Xie, 2014). The rice genome has 793 putative UDP-glycosyltransferases (UGTs; Cao et al., 2008), some of which are functionally characterized as UFGTs (Ko et al., 2006, 2008; Peng et al., 2017). UFGTs and other UGTs are involved in abiotic stress responses (Sun et al., 2013; Ahrazem et al., 2015; Li et al., 2017, 2018), detoxification of molecules produced by biotic stresses, and glycosylation of phytohormones (Jones and Vogt, 2001; Brazier et al., 2002; Bowles et al., 2005). Phytohormones play essential roles in the regulation of plant growth and development, and in responses to biotic and abiotic stress (Ku et al., 2018). The activity of phytohormones such as auxins, cytokinins, and abscisic acid (ABA) can be regulated by UGT-mediated glycosylation (Jackson et al., 2001; Xu et al., 2002; Hou et al., 2004), which generally serves as an inactivation mechanism for secondary plant metabolites (Jones and Vogt, 2001).
In this study, we show that the rice UGT gene OsUGT90A1 is associated with the low-temperature seedling survivability (LTSS) QTL qLTSS4-1 (Schläppi et al., 2017). Overexpression of OsUGT90A1 helps to protect the plasma membrane against cellular damage induced by abiotic stress, and is correlated with an increase in antioxidant enzyme activity and a reduction in ROS levels during abiotic stress. OsUGT90A1 has a biphasic expression pattern in rice seedlings exposed to low-temperature stress, most likely because the gene affects developmental processes such as root and shoot growth. A model for OsUGT90A1 function is proposed and discussed.
Materials and methods
Plant materials and growth condition
The USDA Rice Mini-Core (RMC) collection (Li et al., 2010) consists of 203 Oryza sativa germplasms categorized into six subpopulations: ‘aromatic’ (six accessions), ‘tropical japonica’ (33), and ‘temperate japonica’ (34) from the japonica subspecies, ‘aus’ (38) and ‘indica’ (68) from the indica subspecies, and 24 accessions that are an admixture of two or more subpopulations. The RMC collection was used as the main experimental material for both genome-wide association study (GWAS) mapping and phenotyping of chilling tolerance. Two rice reference accessions, ‘temperate japonica’ Nipponbare and ‘aus’ Kasalath, and two Arabidopsis ecotypes, Columbia-0 (Col-0) and Landsberg erecta-0 (Ler-0), were used to generate overexpression (OX) transgenic lines, and the ‘temperate japonica’ accession Zhong Hua 11 (ZH11) was used to generate knockout lines.
Rice seedlings were grown hydroponically in an AR-66L growth chamber (Percival Scientific, Inc.) under ~165 µE photon flux and 12/12 h light/dark conditions at 28/25 °C. Water was replaced by quarter-strength Murashige and Skoog (MS) liquid medium on day 7. All wild-type and transgenic Arabidopsis seeds were surface-sterilized with 10% sodium hypochlorite and germinated on agar-solidified half-strength MS medium in an AR-66L growth chamber under standard growth conditions of 16/8 h light/dark at 22/20 °C and ~165 µE photon flux.
Selection of a chilling-tolerance candidate gene associated with qLTSS4-1
Using online databases, a 1-Mb genomic region upstream and downstream from the small sequence repeat (SSR) marker RM3317 associated with the qLTSS4-1 locus was scanned for putative cold-tolerance candidate genes as follows. First, the Gene Ontology Consortium (http://www.geneontology.org) and QTARO (Yonemaru et al., 2010) databases were used to assign functions logically connected to the three potential chilling-tolerance gene classes ‘sensing/signal transduction’, ‘transcription factor’, and ‘effector’, or as to assign them as ‘hypothetical’. Second, genes that were predominantly expressed in tissues associated with LTSS, such as ‘seedling’, ‘leaf’, and also ‘root’, were selected from the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp). Third, the file ‘final_rice_v7_expression_matrix_48columns.txt’ from the MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/index.shtml; Kawahara et al., 2013) was used to select genes that were cold-induced and differentially expressed between the two reference genomes Nipponbare and 93-11 (‘indica II’), and was supplemented with information from previously published differential gene expression studies done under low-temperature stress (Yun et al., 2010; Zhang et al., 2012; Chawade et al., 2013). Finally, The SNP-Seek finder (http://snp-seek.irri.org; Alexandrov et al., 2015) and RiceVarMap (http://ricevarmap.ncpgr.cn; Zhao et al., 2015) browsers together with RMC re-sequencing data (Wang et al., 2016b) were used to select candidate genes with single-nucleotide polymorphisms (SNPs) in the coding and upstream regulatory regions of chilling-tolerant and chilling-sensitive accessions, with special emphasis on SNPs that generated non-synonymous amino acid substitutions.
Taken together, a gene was considered as a probable qLTSS4-1-associated gene if it was annotated either as ‘sensing/signal transduction’, ‘transcription factor’, or ‘effector’, if it was expressed predominantly in seedling tissues, if it was differentially expressed between japonica and indica accessions; if it was cold-temperature regulated; and if it had SNPs in both the upstream regulatory and coding regions, with the latter generating non-synonymous amino acid substitutions. A haplotype map was then generated for these genes using the RiceVarMap browser and RMC re-sequencing data. The genes for which the haplotype region was in linkage disequilibrium were then selected as the most probable candidate genes.
Cold-temperature stress treatment of rice plants
Rice seeds were germinated and grown as described above. Two-week-old seedlings from the RMC used for GWAS mapping were stressed continuously at 10 °C for 7 d. For transgenic plants, the stress conditions were 2 d continuously at 10 °C for the OsUGT90A1-OX Kasalath lines, and 4 d continuously at 4 °C for the OsUGT90A1-OX Nipponbare and osugt90a1-KO ZH11 lines. All assays related to chilling stress were done after completion of these cold-temperature treatments. See below for details of construction of the transgenic plants.
Assessment of low-temperature seedling survivability (LTSS) in rice
Seedlings at 2 weeks old were cold-temperature stressed as described above and allowed to recover for 1 week at 28/25 °C day/night cycles with a 12/12 h photoperiod at ~165 µE photon flux. Mean survivability was calculated as %LTSS, which was the number of green and healthy-looking seedlings after 1 week of recovery growth divided by the number of initial healthy-looking seedlings before treatment, expressed as a percentage. Nipponbare and Krasnodarskij 3352 (‘temperate japonica’) were used as chilling-tolerant checks, and Kasalath and Carolino 164 (‘aus’) were used as chilling-sensitive checks.
Measurement of electrolyte leakage
Immediately after the low-temperature stress treatments, four equally sized tissue sections from the middle part of different leaf blades were collected from the rice seedlings, and four leaves were removed from Arabidopsis seedlings. The leaf samples were washed in ddH2O, transferred into screw-cap glass tubes filled with 5 ml of ddH2O, and placed on a rotary shaker at 200 rpm for 1 h to release cellular electrolytes from damaged cells. Electrolyte leakage (EL) of each of the four replicate samples was measured twice in separate 120-µl aliquots of the solution using a hand-held LAQUAtwin B-771 conductivity meter (Horiba Scientific, Japan). Care was taken to ensure that the glass tubes were free of ions and that the ddH2O used had no significant conductivity. After the initial EL measurement, the tissue samples were incubated in boiling water for 10 min to release the total electrolyte contents of all cells. After cooling to room temperature, the samples were shaken at 200 rpm for 1 h and EL was measured again. Electrolyte leakage for each treatment was then determined as %EL = [(initial EL)/(total EL)] ×100.
Assessment of tolerance to freezing stress in Arabidopsis
Col-0 and the Ler-0 wild-type and OsUGT90A1-OX transgenic lines were subjected to freezing stress using a refrigerated circulating bath (Polyscience, Niles, IL, USA). Individual seedlings at 2 weeks old were removed from agar plates and transferred onto a stack of pre-wetted Light-Duty Tissue Wipes (VWR, Radnor, PA, USA) in 50-ml conical tubes and covered with pre-wetted Tissue Wipes to maintain humidity. The tubes were incubated in the circulating bath at –2 °C for 15 min followed by addition of ~50 µl of ddH2O ice chips to promote ice nucleation in the plant tissues. After further incubation at –2 °C for 1 h, the seedlings were allowed to thaw inside the tubes overnight in the dark at 4 °C. The thawed plant tissues were then subjected to different phenotypic assays.
For assessment of whole-plant survival, tubes containing 2-week-old seedlings were placed in the circulating bath as described above, and after 1 h at –2 °C the temperature in the bath was reduced at a rate of –2 °C h–1. Tubes were taken out after 1 h (i.e. –2 °C), and when the temperature reached –4 °C and –6 °C. The seedlings were thawed overnight in the dark at 4 °C and were then transferred to pre-wetted filter papers and placed for 1 week under the standard conditions in the growth chamber described see above.
Assessment of tolerance to salt stress in Arabidopsis
Salt germination assays were done as described previously (Xu et al., 2011). One hundred surface-sterilized seeds of the Ler-0 wild-type and of two of the OsUGT90A1-OX homozygous transgenic lines were positioned in a grid onto agar-solidified half-strength MS medium supplemented with various concentrations of NaCl (0, 50, 100, 150, and 200 mM) and germinated for 2 weeks under standard growth conditions. Each plate had 33–35 seeds of each line, and the position of seeds from each line was different on each of the three plates used to account for potential positional effects. Germination was defined as protrusion of the radicle and was assessed each day during the 2-week incubation period. For each line it was expressed as a percentage for each replicate plate: %germination = [(number of germinated seeds)/(total number of seeds)] × 100. Mean germination percentages were calculated from replicates.
Cloning of rice genomic DNA and plant transformation
OsUGT90A1 was PCR-amplified from genomic DNA of the accessions Krasnodarskij 3352 and Carolino 164. A genomic region of ~500 bp upstream from the coding region was also PCR-amplified from these accessions (for all primers see Supplementary Table S1 at JXB online). PCR amplifications were done using Invitrogen Platinum SuperFi DNA polymerase (ThermoFisher Scientific), with the addition of 0.5 µl of ExTaq DNA polymerase (Takara) to add adenines to the 3´-ends of the products, which were ligated into the pGEM-T vector (Promega) and sequenced. To generate the overexpression (OX) constructs, OsUGT90A1 was ligated into the BamHI and EcoRI sites between the strong MAC promoter and mannopine synthase terminator within the binary vector pPZP211. To generate the OsUGT90A1 promoter::LUC (luciferase) reporter gene constructs, the 500-bp genomic sequence was ligated into the KpnI and BamHI sites in front of the LUC reporter gene within the binary vector pPZP211. To generate OsUGT90A1::eGFP (enhanced green fluorescent protein) fusion protein constructs, genomic DNA was amplified and ligated into the XhoI and BglII sites of pA7-eGFP. The construct was removed from pA7-eGFP and ligated into the HindIII and EcoRI sites of the binary vector pPZP211. Binary vectors were introduced into Agrobacterium tumefaciens strain ABI1 and used for plant transformation.
For Arabidopsis transformation, the floral dip method was used (Clough and Bent, 1998). For rice transformation, embryogenic callus derived from scutellum tissue of the rice reference lines Nipponbare and Kasalath were co-cultivated with A. tumefaciens ABI1 and transgenic plants were regenerated as described previously (Toki et al., 2006).
To generate OsUGT90A1-knockout (-KO) lines in rice, the GGCGACGACATGGTCCGGGA sequence was inserted into the pRGRB31 vector as the sgRNA target for Cas9 cleavage of OsUGT90A1 and introduced into Agrobacterium strain AGL1. As described previously (Ma and Liu, 2015), the construct was used for Agrobacterium-mediated transformation of embryogenic callus derived from scutellum tissue of the accession ZH11. The target gene was PCR-amplified in regenerated plants and two KO lines were confirmed by sequencing.
Gene expression analysis
Quantitative reverse-transcription PCR (qPCR) analyses were done to measure the relative abundance of mRNA in rice and Arabidopsis plants. Different rice tissues or whole Arabidopsis plants were collected immediately after the various treatments and total RNA was extracted using Invitrogen Trizol reagent (ThermoFisher). First-strand cDNA was synthesized using M-MLV reverse-transcriptase (Promega), and 2× universal SYBR green supermix (BioRad) was used to perform the qPCR reaction in a CFX96 Touch™ Real-Time PCR Detection System using the following conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s; 55 °C for 30 s; and 72 °C for 1 min. The OsUGT90A1 and ubiquitin primers are listed in Supplementary Table S1.
Visualization of in vivo luciferase activity and transient transfections assays
Visualization of firefly luciferase (fLUC) activity in live plant tissues was done as described previously (Ishitani et al., 1997). Two-week-old Arabidopsis Col-0 seedlings homozygous for the 500-bp OsUGT90A1-promoter::LUC reporter gene construct were incubated in the dark at either room temperature or 4 °C for 3 h, and then seedlings were evenly sprayed three times with a solution of 1 mM luciferin (GoldBio, St. Louis, MO, USA) in 0.01% Triton X-100 and incubated for 5 min in the dark. The resulting bioluminescence signal was recorded in complete darkness using an AutoChemi Darkroom system (UVP, Upland, CA, USA) with an acquisition time of 10 min. The UVP software was used to visualize in vivo fLUC activity in transgenic plants, and area density analysis was used to quantify the activity.
To determine fLUC activity in transient transfection assays, rice protoplasts made from young seedling of the 93-11 accession were used, as described previously (Bart et al., 2006; Liu et al., 2018). pPZP211 plasmids containing different 500-bp OsUGT90A1-promoter::LUC reporter gene constructs were co-transfected into protoplasts with the pGreenII 0800-LUC vector containing the Renilla LUC gene (rLUC) to normalize transfection efficiencies. Protoplasts were incubated in the dark at 28 °C for 12 h, collected by centrifugation at 450 g for 3 min, and immediately used for LUC assays. Quantification was done using a dual-luciferase reporter assay kit (Promega). Five independent transformations were done for each construct, and relative LUC activity was calculated as the ratio of fLUC to rLUC.
Determination of total content of phenolics and anthocyanins in rice
Phenolic and anthocyanin compounds were isolated from 100 mg of 2-week-old leaf tissues using 1 ml of 70:30:1 methanol:water:trifluoroacetic acid extraction buffer and incubated in the dark at room temperature for at least 1 h. After centrifugation (16 000 g for 20 min), the supernatant was filtered through a 0.2-µm PTFE membrane using a 13-mm syringe. The total anthocyanin concentration was calculated as [(OD520–OD700) × MW × DF × 103]/(e × L) where MW is the molecular weight (449.2 g mol–1 for cyanidin-3-glucoside, Cyd-3-glu), DF is the dilution factor, L is the pathlength (1 cm), e is the molar extinction coefficient (26 900 mol–1 cm–1 for Cyd-3-glu), and 103 is the factor for conversion from g to mg. Extraction buffer alone was used as the negative control.
HPLC-MS analysis of the composition of total phenolics/anthocyanins
The mixture of total phenolics/anthocyanins in extraction buffer was vacuum-dried using a Savant UVS400 Universal Vacuum System (ThermoFisher) and resuspended in 500 μl of pure ddH2O. The solvents were H2O (A) and CH3CN (B) and the gradient elution program was as follows (v/v A in B): start, 10%; 0–30 min, 10–25%; 30–50 min, 25–45%; 50–55 min, 45–100%; 55–60 min, 100%. A 3-μm, 75 × 4.6 mm Gemini 3u C18 110a column was used (Phenomenex, Torrance, CA, USA) with an operating temperature of 40 °C and a flow rate of 1 ml min–1. A Shimadzu LC-MS-2020 system was used for mass spectrometry.
Determination of ROS content and antioxidant activity
Staining with Nitro Blue Tetrazolium (NBT) was used to visualize the O2•–. Tissues of rice and Arabidopsis were collected from 2-week-old plants before and after chilling stress and were incubated overnight in the dark at room temperature in a solution containing 0.2% NBT (Alfa Aesar, Haverhill, MA, USA) dissolved in 50 mM sodium phosphate buffer (pH 7.5). To remove chlorophyll, the tissues were incubated in 100% ethanol at ~75–80 °C for 4 h before images were taken.
For the reduction assays, 25 μl of undiluted total phenolic/anthocyanin extracts were mixed with 200 μl of 0.1 mM 2,2-diphenyl-1-picrylhydrazyl (DPPH; Sigma) solution and incubated at room temperature for 30 min on a shaker. For the blank controls, 25 μl of the phenolic/anthocyanin extraction buffer were used. Absorbance at 517 mm (OD517) was monitored using a spectrophotometer and antioxidant activity was calculated as: {[(OD517 blank – OD517 sample)/OD517 blank] × 100}/total anthocyanin content in mg.
Propidium iodide staining
Tissue samples of rice or Arabidopsis were submerged for at least 3 h in a solution of 10 μg ml–1 propidium iodide (PI; Sigma). The fluorescent signal of PI was visualized using a confocal laser-scanning microscope (Nikon A1R system), with 543 nm excitation and 615 nm emission signals.
Determination of catalase and peroxidase enzyme activities
Tissue samples (100 mg) of rice or Arabidopsis were extracted with 800 µl of phosphate buffer (pH 7.0) to isolate total proteins. After centrifugation (16 000 g for 5 min), the supernatant was diluted 1:1 (v:v) with phosphate buffer. Total protein concentration was determined using Bradford reagent (Fisher Scientific). For the catalase (CAT) assays, discs of filter paper (diameter 8 mm) were soaked for 5 s in protein extract that had been diluted 5-fold, after which the edges of the discs were blotted for 5 s onto a paper towel to absorb excessive liquid. Each disc was then immediately placed at the bottom of a 50-ml glass beaker containing 30 ml of a freshly made 1% H2O2 solution. The time taken for the disc to reach the surface of the solution was recorded (30 mm distance). CAT activity was calculated as mm s–1 mg–1 total protein content. The peroxidase assays were done using a QuantiChromTM kit (BioAssay Systems, Hayward, CA, USA) according to the manufacturer’s instructions, using 10 µl of 20-fold diluted protein extract for each reading.
Subcellular localization of GFP
Subcellular localization of OsUGT90A1 proteins was determined in protoplasts of ‘indica II’ 93-11 using eGFP fluorescence as described previously (Liu et al., 2018). pPZP211 plasmids containing different OsUGT90A1::eGFP fusion constructs were transiently transfected into rice protoplasts using the polyethylene glycol method and incubated in the dark at 28 °C for 12 h. eGFP fluorescence was recorded using a confocal laser-scanning TCS SP8 microscope (Leica). To determine potential co-localization with the plasma membrane, FMTM 4–64 dye [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinum dibromide; ThermoFisher] was added as a plasma membrane marker. Internal membranes such as the tonoplast were not stained by FMTM 4–64. Chlorophyll autofluorescence was visualized using 480–515 nm excitation and 650–750 nm detection wavelengths.
Results
Identification of LOC_Os04g24110 as a chilling-tolerance candidate gene associated with qLTSS4-1
We previously identified a novel LOW TEMPERATURE SEEDLING SURVIVABILITY (LTSS) QTL, qLTSS4-1, using the RMC collection and a GWAS mapping approach (Schläppi et al., 2017). To identify qLTSS4-1 candidates, we screened non-transposon genes near qLTSS4-1. LOC_Os04g24110, annotated to encode the putative anthocyanin 3-O-beta-UDP-glycosyltransferase (UGT) enzyme UGT90A1, was identified as the most probable candidate gene that met all the screening criteria (see Methods), namely temporal and spatial expression patterns, differential expression between chilling-tolerant Nipponbare (‘temperate japonica’) and chilling-sensitive 93-11 (‘indica II’), evidence for low-temperature regulation, and numerous single-nucleotide polymorphisms (SNPs) in both the promoter and coding regions between the three rice reference genomes Nipponbare, 93-11, and Kasalath (‘aus’, indica).
A haplotype analysis of 2000 bp of genomic DNA of LOC_Os04g42110, including 500 bp upstream of the coding region, identified 37 SNPs and 14 haplotypes of which seven were major ones (I, II, III, IV, V, IX, and X) found in at least 100 out of the population of 4402 accessions analysed (Fig. 1a, Supplementary Fig. S1). I and V were predominantly found in accessions of the japonica subspecies, including ‘japonica admixtures’, with frequencies of 83.77% and 93.63%, respectively. In contrast, II, III, IV, and IX were predominantly found in accessions of the indica subspecies, including ‘indica/aus admix’, with frequencies of 96.90% for II, 98.41% for III, 84.96% for IV, and 97.39% for IX. Haplotype X was equally distributed between accessions of the japonica and indica subspecies (43.94% and 35.99%, respectively). Haplotypes II, III, and IX were phylogenetically similar as they clustered together, while IV clustered with I and V.
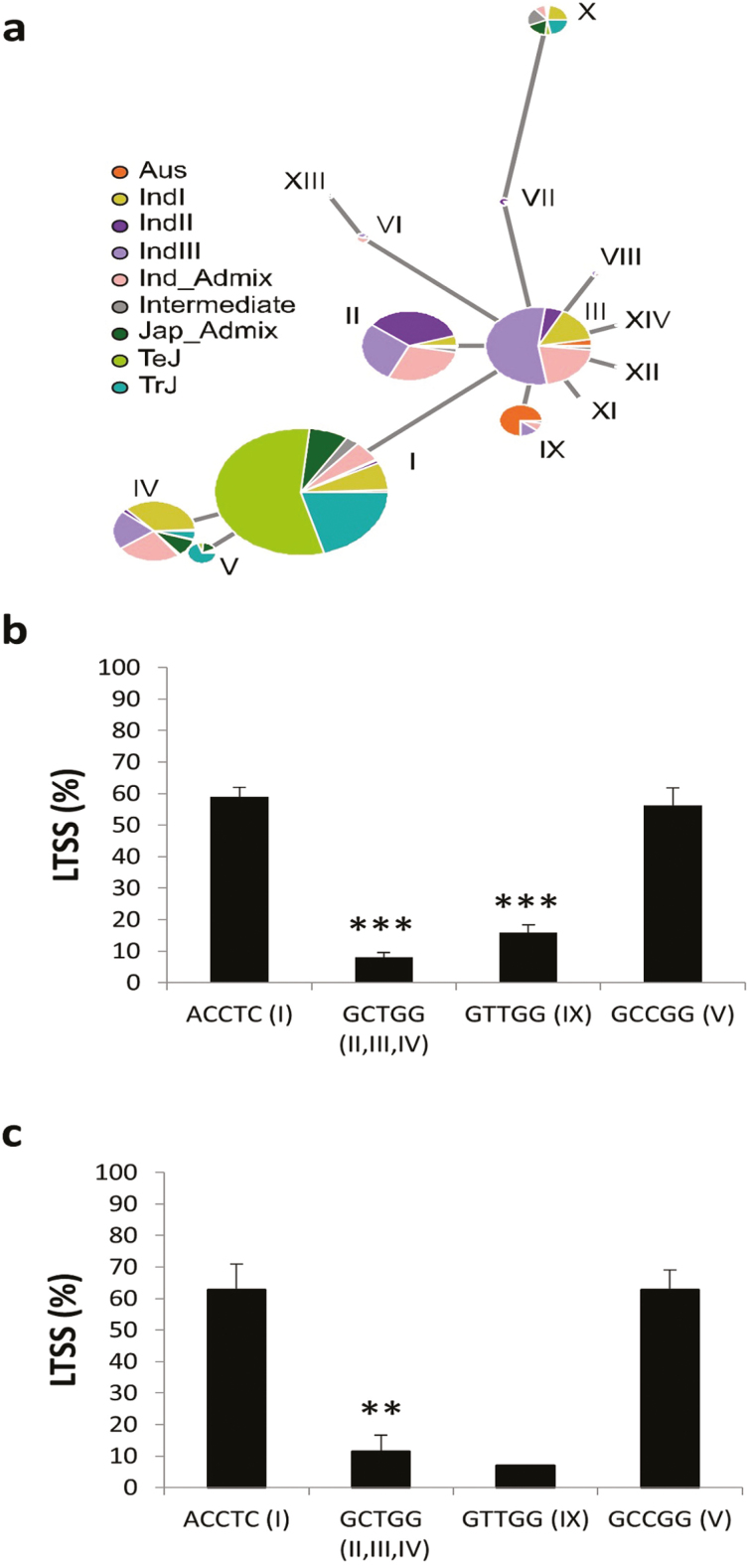
Fig. 1.
Correlation of LOC_Os04g24110 haplotypes with percent low-temperature seedling survival (LTSS) in different subpopulations of the Rice Mini-Core (RMC) collection. (a) Haplotype analysis based on 37 single-nucleotide polymorphisms (SNPs) of a 2000-bp genomic DNA fragment spanning LOC_Os04g24110 using RiceVarMap data from a population of 4402 accessions. Haplotypes (Roman numerals) are representative for the subpopulations ‘temperate japonica’ (TeJ), ‘tropical japonica’ (TrJ), ‘various indica’ (Ind), ‘aus’ (Aus), ‘indica admixtures’ (Ind_Admix), ‘japonica admixtures’ (Jap_Admix), and various intra-subspecies ‘admixtures’ (Intermediate). (b) Haplotype–LTSS correlation analysis based on five SNPs using all 203 Oryza sativa accessions of the RMC. Accessions with the ACCTC haplotype belong to the 37-SNP-based haplotype I; accessions with GCTGG belong to haplotypes II, III, and IV; accessions with GTTCC belong to haplotype IX; and accessions with GCCGG belong to haplotype V. (c) Haplotype–LTSS correlation analysis based on five SNPs using only the 24 ‘admixture’ accessions of the RMC. RMC accessions at 2 weeks old were exposed to 10 °C for 7 d and allowed to recover at warm temperatures for 1 week (28/25 °C day/night, 12-h photoperiod), after which LTSS was determined. Data are means (±SE), n=5. Significant differences compared to the ACTCC accessions were determined using two-tailed Student’s t-tests: **P<10–4; ***P<10–20.
By interrogating the RMC re-sequencing data (Wang et al., 2016b), we reduced the haplotype complexity from seven to four groups by using five SNPs, the first three of which corresponded to the last three SNPs in the 5´ upstream region whilst the fourth and fifth corresponded to the first and last SNPs in the coding region (Supplementary Fig. S1). These four haplotypes were found in different RMC subpopulations as follows. ACCTC (haplotype I; Fig. 1a) was found in 34 out of 34 (100%) ‘temperate japonica’ accessions, and in 28 out of 33 (85%) ‘tropical japonica’ accessions; GCTGG (haplotypes II, III, IV) was found in 51 out of 68 (75%) ‘indica’ accessions; GTTGG (haplotype IX) was found in 29 out of 38 (76%) ‘aus’ accessions, and in 1 out of 68 (1.5%) ‘indica’ accessions; and GCCGG (haplotype V) was exclusively found in 5 out of 33 (15%) ‘tropical japonica’ accessions. A correlation analysis between all RMC accessions having either of these haplotypes and their respective LTSS scores (Fig. 1b) indicated that those containing GCTGG and GTTGG had, as a group, significantly lower scores at P-values of 2.59×10–32 and 1.55×10–21, respectively, than those containing ACTCC, while there was no difference between those containing the ACTCC and GTTGG haplotypes (P=0.6595). When all 67 japonica accessions were removed from the analysis, ‘indica’, ‘aus’, and ‘admix’ accessions with the ACCTC haplotype had significantly higher LTSS scores than those containing GCTGG (P=1.38×10–5) and GTTGG (P=0.0017). An analysis using only the 24 RMC ‘admix’ accessions showed that 11 and eight accessions contained the ACCTC and GCTGG haplotypes, respectively, and their LTSS score distributions were significantly different with a P-value of 6.80×10–5 (or P=5.02×10–5 if the lone accession with the GTTGG sequence was included) while there was no difference between accessions containing the ACCTC and GCCGG haplotypes (P=0.9906; Fig. 1c). These results indicated that differences in either gene expression and/or protein structure of LOC_Os04g24110 (=OsUGT90A1) correlated with the observed differences in chilling tolerance between the japonica and indica accessions of the RMC collection.
Correlation of OsUGT90A1 expression with chilling-tolerance phenotypes
Previous metadata analyses have indicated that OsUGT90A1 has higher expression in chilling-tolerant Nipponbare (‘temperate japonica’) than in chilling-sensitive 93-11 (‘indica II’) and that it is differentially expressed during exposure to low temperature (Zhang et al., 2012). To confirm these expression profiles for RMC cultivars, we selected Krasnodarskij 3352 (ACCTC haplotype I, japonica) and Carolino 164 (GTTGG haplotype IX, indica) as representatives for the two subspecies, because they are known to have consistently high and low cold-tolerance scores, respectively (Schläppi et al., 2017). Quantitative reverse-transcription PCR (qRT-PCR) analyses showed that OsUGT90A1 was more highly expressed in leaves than in roots in both accessions, more highly expressed in leaves in the japonica than indica accession at warm and cold temperatures, and expression in both tissues increased in response to low temperature in both accessions (Fig. 2a). Similar results were obtained when OsUGT90A1 expression data were averaged across three other chilling-tolerant RMC accessions containing the ACCTC haplotype (H57-3-1, WIR 911, and M202) and compared to three other chilling-sensitive accessions containing the GCTGG haplotype (Djimoron, IR 238, and Sapundali Local; Supplementary Fig. S2A). Thus, OsUGT90A1 was more highly expressed in chilling-tolerant japonica than in chilling-sensitive indica accessions in all the RMC lines that we tested.
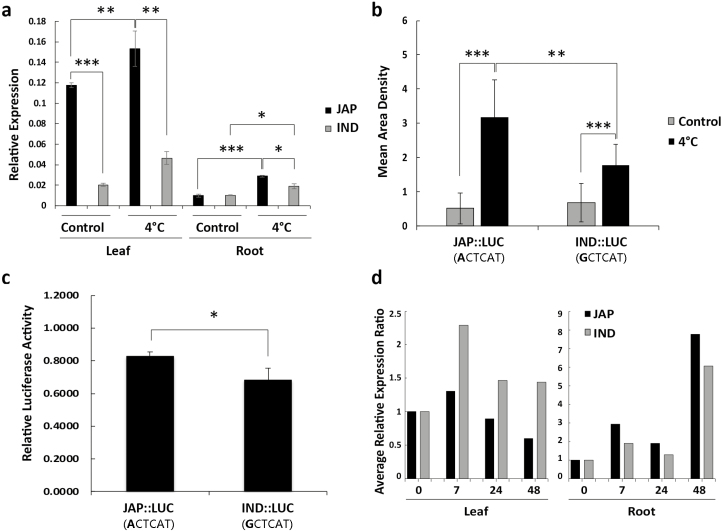
Fig. 2.
mRNA abundance of OsUGT90A1 in different rice tissues and in transgenic Arabidopsis plants at warm (control) and cold temperatures. (a) Relative expression of OsUGT90A1 in leaves and roots at control temperatures (28/25 °C day/night, 12-h photoperiod) and after continuous exposure to 4 °C for 7 h, as determined by qPCR analysis. (b) Quantification of real-time firefly luciferase (LUC) intensity in whole plants of homozygous Arabidopsis lines transformed with a 500-bp OsUGT90A1-promoter::LUC construct before (control) and after chilling treatment at 4 °C for 3 h. (c) Normalized LUC)bioluminescent activity at 28 °C in rice protoplast transiently transfected with plasmids containing a 500-bp OsUGT90A1-promoter::LUC reporter gene. (d) Time-series analysis of OsUGT90A1 expression in leaves and roots during exposure to 4 °C, as determined by qPCR analysis. Relative expression levels at 7, 24, and 48 h were normalized to the level of the warm-temperature control (time 0, set as 1). JAP, japonica accession Krasnodarskij 3352 (haplotype I; Fig. 1a); IND: indica accession Carolino 164 (haplotype IX; Fig. 1a). JAP::LUC, a 500-bp OsUGT90A1 promoter fragment from Krasnodarskij 3352 fused to LUC; IND::LUC, a 500-bp OsUGT90A1 promoter fragment from Carolino 164 fused to LUC. Data are means (±SD), n=3 (a, b, d), n=10 (c). Significant differences between means are indicated and were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001.
Because the first SNP (either A or G) of the five-SNP haplotype sequence of OsUGT90A1 is within its proximate upstream regulatory region and overlaps with a potential bZIP binding site (ACTCAT) in japonica accessions (Supplementary Fig. S1) but is lacking a binding site in indica accessions due to a G at the same position (GCTCAT), in order to test expression differences we generated firefly luciferase (fLUC) reporter gene fusions using 500 bp of genomic DNA upstream of the OsUGT90A1 coding region from Krasnodarskij 3352 (containing ACTCAT) and Carolino 164 (containing GCTCAT). Transgenic Arabidopsis plants with the ACTCAT-containing fragment (JAP::LUC) had significantly more fLUC expression after cold treatment than plants with the GCTCAT-containing fragment (IND::LUC; Fig. 2b, Supplementary Fig. S2B). Transient expression assays at room temperature in rice protoplasts showed that the ACTCA-containing fragment conferred higher fLUC expression than the GCTCA-containing fragment (Fig. 2c). Time-series analysis during cold treatment showed that expression of OsUGT90A1 in both leaves and roots was initially up-regulated (0–7 h) and then down-regulated (Fig. 2d). Expression then increased again in roots after 48 h of cold treatment. Taken together, these results indicated that OsUGT90A1 was more highly expressed in chilling-tolerant rice accessions than chilling-sensitive ones, and that SNPs in its upstream regulatory region might have been responsible for those differences in expression.
Involvement of OsUGT90A1 in protecting cell membrane integrity during abiotic stress
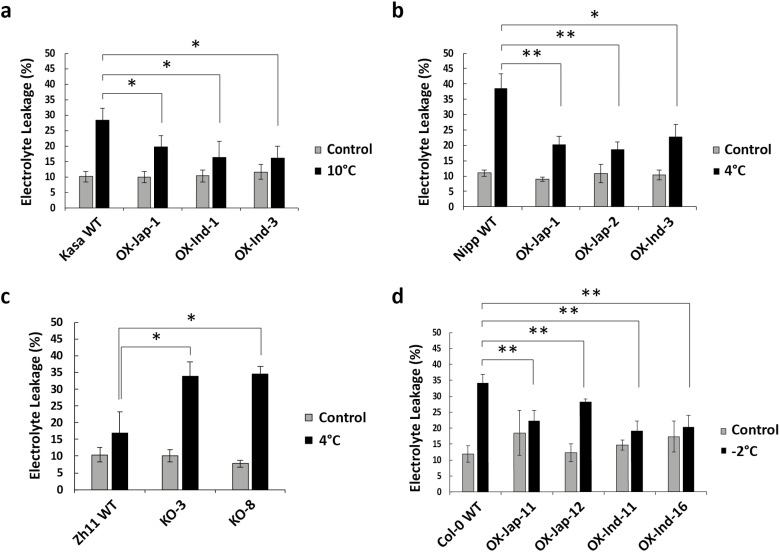
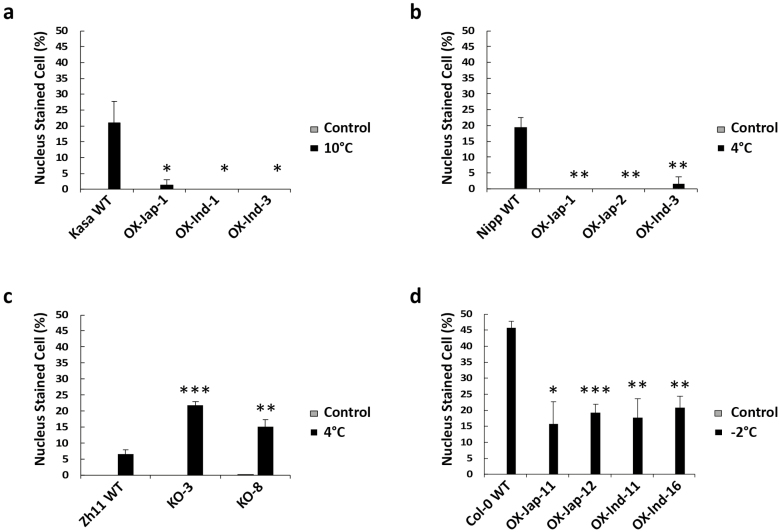
Increased OsUGT90A1 expression might contribute to the superior chilling tolerance of japonica accessions; however, it might also be due to differences in the activity of the OsUGT90A1 enzyme resulting from differences in the protein sequences (Supplementary Fig. S1C). To test these possibilities, we made transgenic rice plants overexpressing either the japonica allele of OsUGT90A1 (from Krasnodarskij 3352) or the indica allele (from Carolino 164) from a strong constitutive promoter in both the chilling-tolerant Nipponbare and chilling-sensitive Kasalath backgrounds, and knocked the gene out in the chilling-tolerant Zhong Hua 11 (ZH11) background. We also overexpressed either allele in transgenic Arabidopsis lines to assess their effects in a heterologous system. An analysis of three independent overexpression (OX) transgenic lines with constitutively high OsUGT90A1 mRNA levels for each background and two knockout (KO) lines with very low mRNA levels (Supplementary Fig. S3) showed that there were no significant differences in the LTSS scores between the wild-type (WT) controls and transgenic lines under cold-stress conditions (Supplementary Fig. S2C–E). However, overexpression of either OsUGT90A1 allele in transgenic Arabidopsis lines improved their freezing survival, and the temperature at which 50% of seedlings died shifted from –3 °C to –4 °C (Supplementary Fig. S2F). In contrast, when we determined percent electrolyte leakage (%EL) as a measure for cellular membrane damage induced by cold stress (e.g. Demidchik et al., 2014), there were significant differences between the WT and transgenic lines (Fig. 3a–c). Regardless of which allele was used, after exposure to cold the overexpression lines had lower %EL values than the WT and the knockout lines had higher values, suggesting that OsUGT90A1 helped to protect plasma membranes against cellular lesions induced by cold stress. The same membrane protection effect was observed in OsUGT90A1-OX Arabidopsis lines exposed to a mild freezing temperature (Fig. 3d). Because both the japonica and indica coding sequences of OsUGT90A1 had similar protecting effects, it is more likely that differences in expression rather than differences in enzyme activity were responsible for the observed differences in chilling-tolerance phenotypes. Consistent with this, there was a general trend for a negative correlation between OsUGT90A1 expression levels and %EL in transgenic rice and Arabidopsis plants (Supplementary Fig. S4). To validate the EL results, we stained WT, OX, and KO lines with propidium iodide, which cannot cross an intact plasma membrane and generates a fluorescent signal in the nucleus only after penetrating damaged plasma membranes (Chen et al., 2006). The results confirmed that after exposure to cold the OsUGT90A1-OX lines had fewer plasma membrane lesions compared to the WT and OsUGT90A1-KO lines had more (Fig. 4, Supplementary Fig. S5).
Fig. 3.
Determination of electrolyte leakage (EL) in leaves as a measure of cellular damage induced by low-temperature stress in wild-type (WT) and OsUGT90A1-overexpression (OX) and knockout (KO) transgenic rice and Arabidopsis plants. (a) EL in the WT indica accession Kasalath (Kasa) and in three OsUGT90A1-OX lines grown under control conditions (28/25 °C day/night, 12-h photoperiod) or after chilling treatment at 10 °C for 2 d. (b) EL in the WT japonica accession Nipponbare (Nipp) and in three OsUGT90A1-OX transgenic lines grown under control conditions or after chilling treatment at 4 °C for 4 d. (c) EL in the WT japonica accession Zhong Hua 11 (ZH11) and in two OsUGT90A1-KO lines grown under control conditions or after chilling treatment at 4 °C for 4 d. (d) EL in the Arabidopsis WT Col-0 and in four transgenic lines overexpressing OsUGT90A1 (OX) grown under control conditions (22/20 °C, 16/8 h day/night) or after freezing treatment at –2 °C for 1.5 h. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Data are means (±SD) of n=3 assays. Significant differences between means are indicated and were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001.
Fig. 4.
Determination of propidium iodide (PI) staining of nuclei in epidermal tissues of rice and Arabidopsis leaves using laser confocal microscopy. Nuclear staining indicates plasma membrane damage. (a) Percentage of cells with stained nuclei in the wild-type (WT) indica accession Kasalath (Kasa) and in three OsUGT90A1-overexpression (OX) transgenic lines grown under control conditions (28/25 °C day/night, 12-h photoperiod) or after chilling treatment at 10 °C for 2 d. (b) Percentage of cells with stained nuclei in the WT japonica accession Nipponbare (Nipp) and in three OsUGT90A1-OX transgenic lines grown under control conditions or after chilling treatment at 4 °C for 4 d. (c) Percentage of cells with stained nuclei in the WT japonica accession Zhong Hua 11 (ZH11) and in two OsUGT90A1-knockout (KO) lines grown under control conditions or after chilling treatment at 4 °C for 4 d. (d) Percentage of cells with stained nuclei in WT Arabidopsis Col-0 and in four transgenic lines overexpressing OsUGT90A1 (OX) grown under control conditions (22/20 °C, 16/8 h day/night) or after freezing treatment at –2 °C for 1.5 h. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Significant differences between the WT and the transgenic lines were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001. Note that almost no staining was observed under control conditions.
Involvement of OsUGT90A1 in abiotic stress tolerance pathways
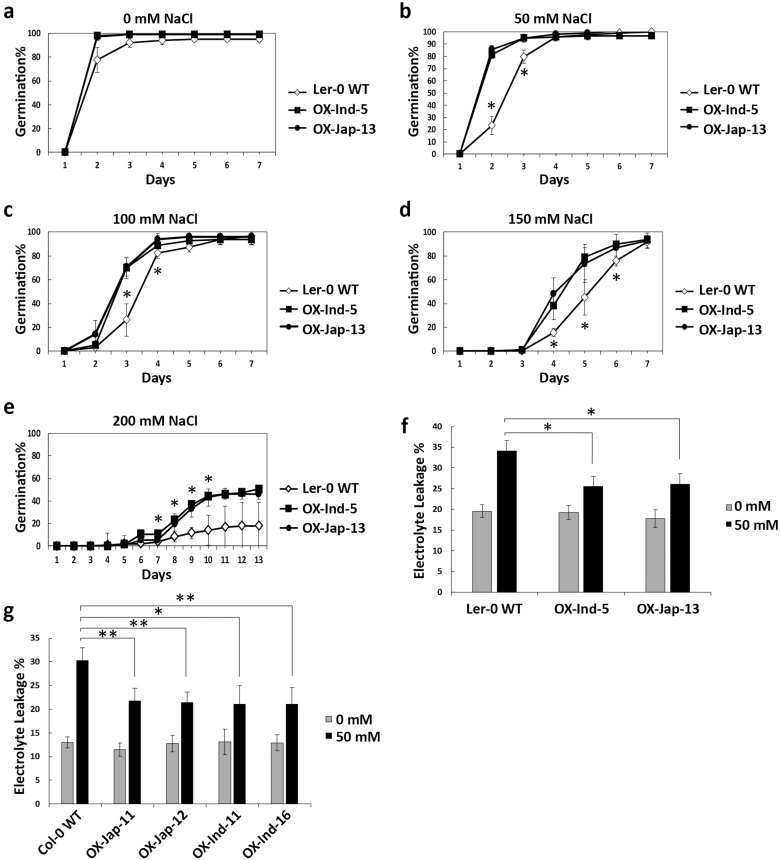
Multiple abiotic stress response mechanisms often converge in a network of coordinated biochemical pathways. Because OsUGT90A1 has previously been shown to be up-regulated by salt stress and to fall within the boundaries of a mapped salt-tolerance QTL (Saravana Kumar et al., 2014), we tested OsUGT90A1-OX Arabidopsis lines in response to salt stress using an established germination assay (Xu et al., 2011). This showed that while germination rates were similar on 0 mM NaCl control plates (Fig. 5a), there were several time-points during a 2-week germination period with significant differences on 50–200 mM NaCl for which seeds of transgenic lines germinated faster than those of the WT, regardless of which OsUGT90A1 allele was overexpressed (Fig. 5b–e). Moreover, OsUGT90A1-OX lines also experienced less membrane damage on 50 mM NaCl than WT plants (Fig. 5f, g), indicating that OsUGT90A1 also affected the mechanisms of tolerance to salt stress.
Fig. 5.
Determination of tolerance to salt stress in Arabidopsis wild-type (WT) ecotypes and in transgenic lines overexpressing OsUGT90A1 (OX). (a–e) Time-course of seed germination of WT and two OsUGT90A1-OX homozygous transgenic lines on half-strength MS medium supplemented with 0, 50, 100, 150, or 200 mM NaCl. (f, g) Electrolyte leakage in 2-week-old (f) Landsberg erecta-0 (Ler-0) and (g) Columbia-0 (Col-0) Arabidopsis ecotypes in WT and OsUGT90A1-OX transgenic lines grown with 0 mM or 50 mM NaCl. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Data are means (±SD) of n=3 assays. Significant differences between the WT and transgenic plants were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01.
Because ROS are produced during abiotic stress, we determined whether OsUGT90A1 affected their levels during cold stress. Staining with NBT, which qualitatively detects ROS in living cells (Muñoz et al., 2000; Kumar et al., 2015), showed that while there were no differences between the WT and OsUGT90A1-OX or Osugt90a1-KO transgenic rice plants at warm temperatures, at cold temperatures both OsUGT90A1-OX rice and Arabidopsis plants had fewer ROS-producing regions in the leaves than WT plants while Osugt90a1-KO rice plants had more and larger regions (Fig. 6a, Supplementary Fig. S6).
Fig. 6.
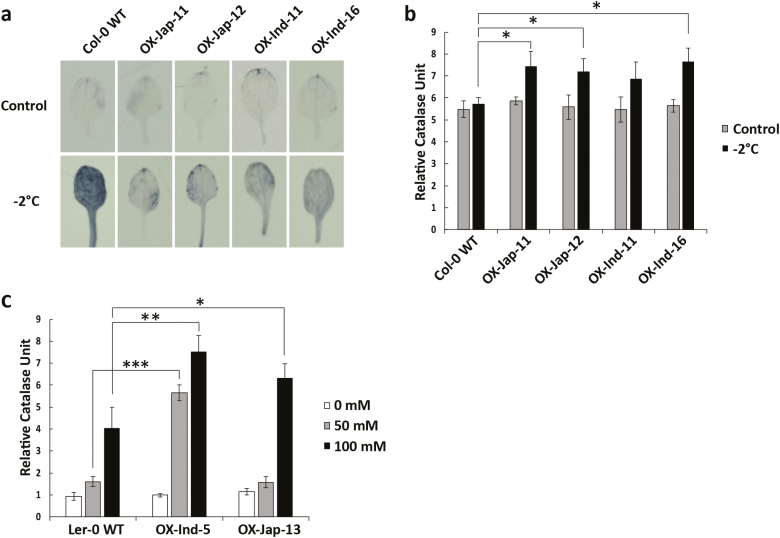
Visualization of reactive oxygen species (ROS) and activity of the antioxidant enzyme catalase (CAT) in Arabidopsis wild-type (WT) and transgenic lines overexpressing OsUGT90A1 (OX). (a) Staining with Nitro Tetrazolium Blue to visualize O2•– in WT Columbia-0 (Col-0) and in four OsUGT90A1-OX transgenic lines under control temperatures (22/20 °C, 16/8 h day/night) or after freezing stress at –2 °C for 1.5 h. (b) CAT activity in WT Col-0 plants and in four OsUGT90A1-OX transgenic lines under control conditions or after freezing stress at –2 °C for 1.5 h. (c) CAT activity in WT Landsberg erecta-0 (Ler-0) plants and in two OsUGT90A1-OX transgenic lines grown with 0, 50, or 100 mM NaCl. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Significant differences between the WT and transgenic plants were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001. (This figure is available in colour at JXB online.)
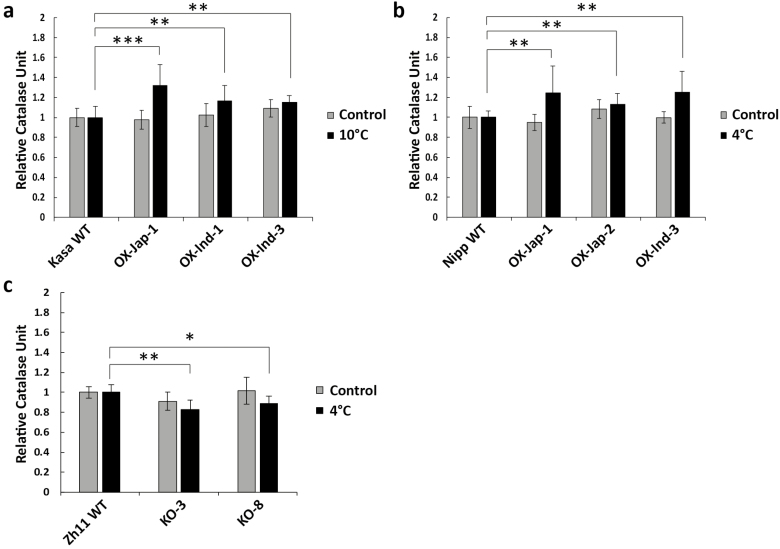
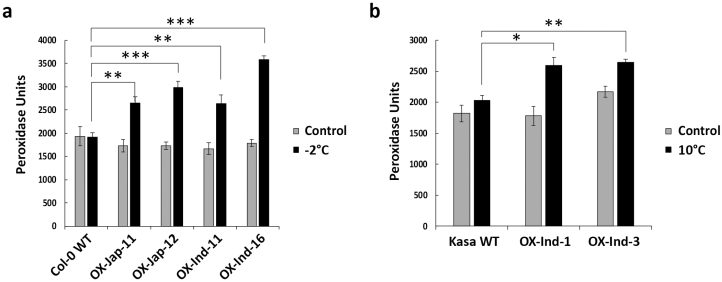
To determine whether overexpression of OsUGT90A1 affected enzymatic antioxidant activity during cold stress, we examined the activities of catalase (CAT) and peroxidase (POX) as a measure for H2O2 scavenging (Willekens et al., 1997; Mittler et al., 2004). Both assays showed that there were no differences between the WT and transgenic plants at warm temperatures (Figs 6b, 7, 8); however, under cold-stress conditions that resulted in more ROS (Fig. 6a, Supplementary Fig. S6), there was higher CAT and POX activity in both Arabidopsis and rice OsUGT90A1-OX lines (Figs 6b, 7a, b, 8), and lower CAT activity in Osugt90a1-KO lines (Fig. 7c). Moreover, while there were no differences in CAT activity between the WT and OsUGT90A1-OX Arabidopsis plants under control conditions, activity was generally higher in OsUGT90A1-OX plants grown for 2 weeks on 50 mM and 100 mM NaCl (Fig. 6c). The higher CAT and POX activities correlated well with a reduction in the leaf-bleaching (necrosis) phenotype that is observed in rice seedlings damaged by ROS during recovery growth after stress treatment (Chen et al., 2012), with OsUGT90A1-OX plants having smaller areas of necrosis than WT control plants (Supplementary Fig. S7). Hence, overexpression of OsUGT90A1 during cold stress correlated with lower levels of ROS and increased antioxidant enzyme activities, resulting in decreased membrane lesions in response to abiotic stress.
Fig. 7.
Activity of the antioxidant enzyme catalase (CAT) in rice wild-type (WT) and transgenic OsUGT90A1-overexpression (OX) lines. (a) CAT activity in WT Kasalath (Kasa) and three OsUGT90A1-OX lines at warm temperatures (control; 28/25 °C day/night, 12-h photoperiod) or after chilling stress at 10 °C for 2 d. (b) CAT activity in WT Nipponbare (Nipp) and three OsUGT90A1-OX lines under control conditions or after chilling stress at 4 °C for 4 d. (c) CAT activity in WT Zhong Hua 11 (ZH11) and two Osugt90a1-knockout (KO) lines under control conditions or after chilling treatment at 4 °C for 4 d. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Data are means (±SD) of n=3 assays and the values are expressed relative to those of the WT controls, which were set as 1. Significant differences between the WT and transgenic plants were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001.
Fig. 8.
Activity of the antioxidant enzyme peroxidase (POX) in wild-type (WT) and transgenic OsUGT90A1-overexpression (OX) Arabidopsis and rice lines. (a) POX activity in Arabidopsis WT Col-0 and in four OsUGT90A1-OX lines at warm temperatures (control; 28/25 °C day/night, 12-h photoperiod) or after freezing stress at –2 °C for 1.5 h. (b) POX activity in rice WT Kasalath (Kasa) and in two OsUGT90A1-OX lines under control conditions or after chilling stress at 10 °C for two days. Ind- and Jap- indicate that the indica or japonica allele of OsUGT90A1 was overexpressed, respectively. Data are means (±SD) of n=3 assays. Significant differences between the WT and transgenic plants were determined using two-tailed Student’s t-tests: *P<0.05; **P<0.01; ***P<0.001.
Subcellular localization OsUGT90A1 and assessment of its potential function
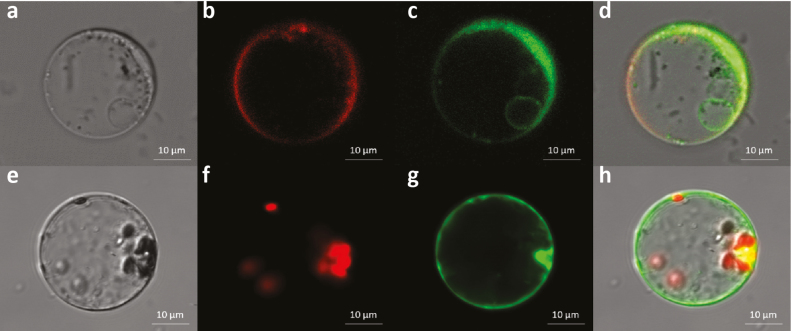
The subcellular localization of a protein can help in inferring its function. To determine the localization of OsUGT90A1, we introduced plasmids containing N-terminal fusions of the gene to the eGFP reporter into rice protoplasts. Examination of numerous eGFP-positive protoplasts using confocal laser-scanning microscopy showed that the fusion protein predominantly localized to the cytoplasm, but also had some affinity for the plasma membrane and internal membrane systems such as the tonoplast (Fig. 9c, d) but not the nucleus or chloroplasts (Fig. 9g, h). This suggested that OsUGT90A1 performs its function predominantly in the cytoplasm and/or in association with internal membranes.
Fig. 9.
Visualization of the subcellular localization of OsUGT90A1::eGFP (enhanced green fluorescent protein) in rice protoplasts at warm temperatures (28/25 °C day/night, 12-h photoperiod). (a) Bright-field image of a protoplast containing the OsUGT90A1::eGFP fusion protein. (b) Staining with FMTM 4–64 dye to visualize the plasma membrane (red). (c) Visualization of the OsUGT90A1::eGFP fusion protein (green). (d) Merged image of (b) and (c) to visualize areas of overlap (yellow). The small green ring inside the protoplast shows an affinity of OsUGT90A1::eGFP for an internal membrane, most likely the tonoplast. (e) Bright-field image of a protoplast containing the OsUGT90A1::eGFP fusion protein. (f) Visualization of chloroplasts (red). (g) Visualization of the OsUGT90A1::eGFP fusion protein (green). (h) Merged image of (f) and (g) to visualize areas of overlap (yellow).
To determine whether differential expression of OsUGT90A1 (which is annotated as a putative anthocyanin 3-O-beta-glucosyltransferase) affected the overall anthocyanin concentration, we measured the total flavonoid/anthocyanin contents in leaf tissues of plants grown with or without chilling stress, and the results indicated that there were no differences between the WT and transgenic plants (Supplementary Fig. S8A, B). To test whether OsUGT90A1 affected the abundance of particular flavonoid/anthocyanin compounds without changing the total content, we performed HPLC-MS analyses to examine their specific compositions in the leaf tissues of the WT, and OX and KO transgenic lines immediately after chilling treatment. Compared to WT plants, there was a slightly different flavonoid/anthocyanin composition profile in OsUGT90A1-OX transgenic plants (Supplementary Fig. S9), but no clear differences were detected between the WT and Osugt90a1-KO plants (Supplementary Fig. S10). To determine whether the slightly different profiles in the OX lines had an effect on the non-enzymatic antioxidant activity of the total flavonoid/anthocyanin compounds that we isolated, we performed a DPPH reduction assay. DPPH can be reduced by common antioxidants and therefore acts as an indicator for general antioxidant activity (Molyneux, 2004). We found that there were no differences in the antioxidant activity between the WT and the OsUGT90A1-OX lines (Supplementary Fig. S8C, D), suggesting that the observed increases in ROS-scavenging activity of the OX plants were not due to the slightly altered flavonoid/anthocyanin composition profiles.
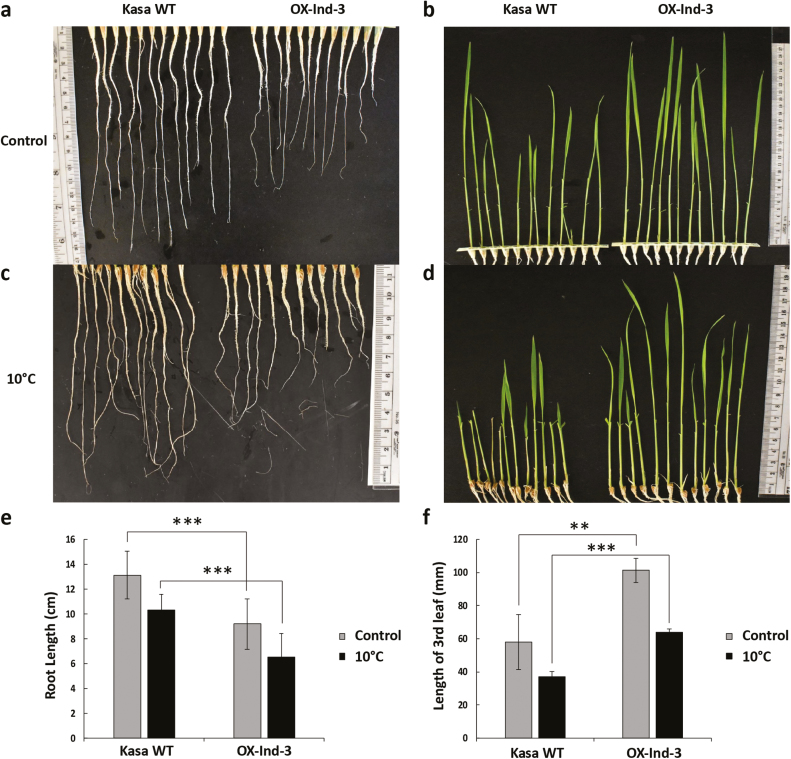
We looked next for growth and development phenotypes as indicators for other potential enzyme substrates. UGTs have previously been shown to regulate phytohormone homeostasis by glycosylation (Jackson et al., 2001; Xu et al., 2002; Hou et al., 2004). Since phytohormones are critical plant growth regulators, we determined whether root and shoot lengths were affected by differential expression of OsUGT90A1. Interestingly, OsUGT90A1-OX rice plants had longer shoots but shorter roots than control WT plants, both under warm-temperature control conditions and under cold stress (Fig. 10). This phenotype suggested that the presumed glycosylation activity of OsUGT90A1 might have a direct or indirect effect on phytohormones such as auxins and cytokinins.
Fig. 10.
Comparison of rice wild-type (WT) and a transgenic line overexpressing the indica allele of OsUGT90A1 (OX-Ind-3), showing a phenotype of short roots and long shoots in the latter. (a, c) Roots of 22-d-old WT Kasalath (Kasa) and OsUGT90A1-OX plants grown under (a) warm temperature conditions (control; 28/25 °C day/night, 12-h photoperiod), or (c) after chilling stress at 10 °C for 2 d. (b, d) The third leaf of 22-d-old Kasa WT and OsUGT90A1-OX plants grown under warm temperature growth conditions (control) or (d) after chilling stress at 10 °C for 2 d. The first and second leaves were removed for clarity. (e) Quantification of root lengths shown in (a) and (c). (f) Quantification of leaf lengths shown in (b) and (d). Ind- indicates that OsUGT90A1 was overexpressed. Data are means (±SD) of n=12 plants. Significant differences between the WT and transgenic plants were determined using two-tailed Student’s t-tests: **P<0.01; ***P<0.001.
Discussion
To the best of our knowledge, we present the first evidence that a gene annotated to encode a UDP-glycosyltransferase, LOC_Os04g24110 (=OsUGT90A1), plays a role in chilling tolerance of rice. OsUGT90A1 is one of the probable candidate genes associated with the chilling-tolerance QTL qLTSS4-1(Schläppi et al., 2017), and our functional assays revealed a role in protecting the plasma membrane from lesions induced by cold stress. Accessions from both the indica and japonica subspecies in which OsUGT90A1 was overexpressed experienced less membrane damage after cold stress than wild-type (WT) plants, while a generally cold-tolerant accession in which the gene was knocked out experienced more membrane damage (Fig. 3). However, although Arabidopsis lines overexpressing OsUGT90A1 had better freezing survival abilities than WT plants, transgenic rice lines did not have improved low-temperature seedling survivability (LTSS) scores; this might be, at least in part, because rice chilling tolerance is a highly quantitative trait. We have previously shown that each QTL contributes only 2–20% to any of the studied chilling-tolerance indices, indicating that these are polygenic traits (Schläppi et al., 2017). This is also in agreement with the recently proposed omnigenic theory, which suggests that almost any gene can influence a complex trait (Boyle et al., 2017). It is therefore remarkable that in the current study overexpression or knockout of a single enzyme-encoding gene had such a measurable effect on plasma membrane integrity, which is essential for plant survival during exposure to abiotic stress (Kim and Tai, 2011). Thus, OsUGT90A1 has a significant auxiliary role and could be used to improve chilling tolerance of rice without affecting major gene regulons, which is a major concern when transcription factors or signaling molecules are used to improve the performance of plants under abiotic stress. Taken together, our results indicate that OsUGT90A1 is probably a major candidate gene associated with qLTSS4-1; however, due to the polygenic nature of chilling tolerance, we cannot rule out that other genes within the qLTSS4-1 boundary contribute to this QTL.
OsUGT90A1 was more highly expressed in japonica than indica accessions (Fig. 2). A reason for this differential expression might be the A/G SNP at –289 from the start of the coding region. At this position, almost all japonica accessions tested had an A, which generates a potential ACTCAT cis-element for binding of Group-S basic leucine zipper (bZIP) factors (Satoh et al., 2002), while the majority of indica accessions had a G. It has previously been shown that the Group-S factor bZIP73 plays a major role in the superior chilling tolerance of japonica accessions, because a single amino acid substitution reduces the bZIP73 trans-activation potential in indica accessions (Liu et al., 2018). The lower abundance of OsUGT90A1 mRNA in indica than japonica accessions that we consistently observed might be in part due to the absence of a bZIP73-interacting ACTCAT element and/or the presence of a bZIP73 protein with reduced trans-activation potential. Consistent with this, in our transient expression assays the ACTCAT-containing promoter element produced only 1.2-fold higher reporter gene expression than the GCTCAT one, which may have been because the assays were done in protoplasts derived from the 93-11 accession, which contains a bZIP73 protein with reduced trans-activation potential (Liu et al., 2018).
The positive effect of OsUGT90A1 in reducing membrane lesions induced by abiotic stress was validated in the heterologous system of Arabidopsis (Fig. 3d). Thus, OsUGT90A1 had a similar auxiliary role in both a monocot and a dicot species. Arabidopsis salt germination assays suggested that the OsUGT90A1-overexpressing (-OX) lines had lower levels of abscisic acid (ABA), because they germinated faster than WT plants on salt (Fig. 5) whereas salt stress generally increases ABA levels to repress germination (Xu et al., 2011). In contrast, activities of the antioxidant enzyme catalase (CAT) were higher in the OX than WT plants when exposed to salt and during freezing stress, but levels of reactive oxygen species (ROS) were lower in the OX plants than in the WT (Fig. 6). Similarly, ROS levels were lower and CAT and peroxidase (POX) levels were higher in OX plants than in the WT during chilling stress in rice (Figs 7, 8).
How overexpression of OsUGT90A1 enhances antioxidant enzyme activities to reduce ROS levels during abiotic stress is currently unknown. It is tempting to speculate that OsUGT90A1 glycosylates metabolites that are involved in balancing abiotic stress responses with the general requirement for growth and development. Although OsUGT90A1 is annotated to encode an anthocyanin 3-O-beta-glucosyltransferase, we detected no significant alterations in either the total content or composition of flavonoid/anthocyanin metabolites (Supplementary Figs S8–S10), indicating that flavonoids and/or anthocyanins might not be the actual substrates for the OsUGT90A1 enzyme.
An alternative hypothesis is that OsUGT90A1 glycosylates phytohormones, which generally leads to the sequestration of hormone glycosides into storage organelles (Mok and Mok, 2001). Chilling-tolerant rice accessions have been shown to have lower ABA levels than chilling-sensitive ones, which balances stress responses with growth and development (Li et al., 2018). Hence, OsUGT90A1 might help to control the amount of active plant hormones by forming sugar conjugates and might help to regulate redox status through hormone-mediated pathways, which may allow harmless levels of ROS to function as signaling molecules (Xia et al., 2015; Tognetti et al., 2017). The crosstalk between plant hormones (e.g. auxin and cytokinin) and ROS plays an important role in regulating plant growth and development. When plants are exposed to environmental stresses, RESPIRATORY BURST OXIDASE HOMOLOG (RBOH) is activated to produce signaling ROS, such as H2O2, which work together with mitogen-activated protein kinases (MAPKs) and Ca2+-dependent protein kinases (CDPKs) to regulate stress tolerance responses (e.g. Abbasi et al., 2004). Phytohormones such as auxins and ABA generate positive feedback for this signaling pathway and produce more ROS via an auxin- or ABA-receptor mediated activation of the RBOH mechanism (Xia et al., 2015). This auxin/ABA–ROS positive feedback loop might be modulated by OsUGT90A1-mediated glycosylation of active auxin and/or ABA molecules.
We further showed that overexpression of OsUGT90A1 in rice inhibited root development and enhanced shoot development (Fig. 10). This further suggests that OsUGT90A1 might modulate the activity of general growth regulators. Although the effect of overexpression on root and shoot growth was different, it was consistent with the known antagonistic interactions of auxins and cytokinins, such that a low ratio of auxin:cytokinin leads to shorter roots but longer shoots (Dello Ioio et al., 2008; Schaller et al., 2015). Likewise, ABA and gibberellic acid (GA) have antagonistic effects on germination (Vishal and Kumar, 2018). Overexpression of OsUGT90A1 in Arabidopsis increased seed germination rates under salt-stress conditions (Fig. 5). This was consistent with the hypothesis that auxins and/or ABA rather than anthocyanins might be substrates for OsUGT90A1. In this model, reduced activities of auxins and/or ABA due to glycosylation-mediated sequestration into vacuoles would result in shorter roots, longer leaves, and improved germination rates.
Interestingly, although there were amino acid substitutions between japonica and indica haplotypes, overexpression of either haplotype had a similar positive effect on reducing cellular lesions induced by abiotic stress (Figs 3–5). Thus, naturally occurring ‘overexpression’ of OsUGT90A1 in japonica accessions under both warm and cold stress conditions (Fig. 2) most likely results in high OsUGT90A1 protein levels and plays a positive role in balancing abiotic stress responses with the requirement of low ROS levels and an intact cellular membrane in preparation for growth and development. However, we cannot rule out that amino acid substitutions have a subtle effect on the function of OsUGT90A1. On the other hand, OsUGT90A1 was recently assigned to the ‘down-regulated’ core transcriptome response induced by abiotic stress in rice (Cohen and Leach, 2019), most likely because after the initial up-regulation during cold stress, OsUGT90A1 may be down-regulated to fine-tune its effect on growth and development.
Conclusions
We propose the following model for the auxiliary role of OsUGT90A1 in the chilling-stress response of rice. Cold-tolerant japonica accessions have higher mRNA abundance of OsUGT90A1 than cold-sensitive indica accessions, and transcript abundance increases (at least temporarily) during exposure to cold temperatures, most likely leading to an increase in abundance of the OsUGT90A1 protein. OsUGT90A1 might glycosylate ABA and/or auxins to sequester them into the vacuole for storage. The consequence of this is a fine-tuning of the ABA- and/or auxin-mediated abiotic stress response, leading to an increase in the activities of CAT, POX, and possibly other antioxidant enzymes. This in turn reduces ROS levels and thus maintains cell membrane integrity as a general mechanism of abiotic stress tolerance to help the resumption of shoot growth and development during subsequent stress recovery.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. SNP haplotypes of the LOC_Os04g24110 promoter, coding region, and protein sequences in japonica and indica reference subspecies.
Fig. S2. In vivo OsUGT90A1 promoter activity in response to cold stress, and the effect of OsUGT90A1 expression on low-temperature seedling survivability in rice.
Fig. S3. Abundance of UGT90A1 mRNA in the individual transgenic rice and Arabidopsis plants used in Figs 3–8, 10, and Supplementary Fig. S4.
Fig. S4. Correlations of OsUGT90A1 mRNA abundance with plasma membrane integrity in cold-stressed transgenic rice and Arabidopsis plants.
Fig. S5. Propidium iodide staining of wild-type and OsUGT90A1-OX and -KO transgenic rice and Arabidopsis lines.
Fig. S6. Effect of OsUGT90A1 expression on ROS-scavenging activity under control and chilling-stress conditions in transgenic rice.
Fig. S7. Leaf-bleaching phenotype of wild-type rice and OsUGT90A1-overexpression plants.
Fig. S8. Overall concentrations and non-enzymatic antioxidant activities of total flavonoids/anthocyanins in leaves of wild-type rice and OsUGT90A1-overexpression plants.
Fig. S9. Effects of OsUGT90A1 overexpression on flavonoid/anthocyanin composition under chilling stress in rice.
Fig. S10. Effects of OsUGT90A1 knockout on flavonoid/anthocyanin composition under chilling stress in rice.
Table S1. List of primers used in this study.
Acknowledgements
We are grateful to Marquette University undergraduate student André Beaumont for helping with the determination of LUC activity in transgenic Arabidopsis plants as part of his ‘Independent Study’ (BIOL4956) project. We would like to thank Mr Gupo Li (Chinese Academy of Sciences) for management of the rice plants, Dr Wei Wang for help with the protein localization experiments, and the undergraduate students of Marquette University’s 2018 ‘Foundation in Biological Inquiry’ (BIOL1101) class who helped perform some of the Arabidopsis seed germination and CAT assays. We greatly appreciate the help of Dr Cai Sheng (Department of Chemistry, Marquette University) who performed the HPLC-MS assays. YS, HP, and MRS were supported by Bridge Funding from Marquette University and by National Institute of Food and Agriculture-Agriculture and Food Research Initiative (NIFA-AFRI) grant no. 2016-67013-24587 from the United States Department of Agriculture (USDA). ZZ, SC, and CC were supported by grant no. 31271680 and 31501283 from the National Science Foundation of China.
Author contributions
MRS and YS conceived the project and designed the experiments; YS, HP, YL, ZZ, and MRS performed the experiments; SC generated the transgenic rice plants; CC provided material and intellectual support; YS and MRS performed all data analyses and wrote the manuscript; all authors edited and approved the final version of the manuscript.
References
- Abbasi F, Onodera H, Toki S, Tanaka H, Komatsu S. 2004. OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Molecular Biology 55, 541–552. [DOI] [PubMed] [Google Scholar]
- Ahrazem O, Rubio-Moraga A, Trapero-Mozos A, Climent MF, Gómez-Cadenas A, Gómez-Gómez L. 2015. Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Science 234, 60–73. [DOI] [PubMed] [Google Scholar]
- Alexandrov N, Tai S, Wang W, et al. 2015. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Research 43, D1023–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Sicilia C, Silvestre S, Haslam RP, Michaelson LV. 2017. Lipid remodelling: unravelling the response to cold stress in Arabidopsis and its extremophile relative Eutrema salsugineum. Plant Science 263, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. 2006. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim EK, Poppenberger B. 2005. Glycosyltransferases: managers of small molecules. Current Opinion in Plant Biology 8, 254–263. [DOI] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier M, Cole DJ, Edwards R. 2002. O-Glucosyltransferase activities toward phenolic natural products and xenobiotics in wheat and herbicide-resistant and herbicide-susceptible black-grass (Alopecurus myosuroides). Phytochemistry 59, 149–156. [DOI] [PubMed] [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC. 2008. Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Molecular Plant 1, 858–877. [DOI] [PubMed] [Google Scholar]
- Chawade A, Lindlöf A, Olsson B, Olsson O. 2013. Global expression profiling of low temperature induced genes in the chilling tolerant japonica rice Jumli Marshi. PLoS ONE 8, e81729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JX, Hall DE, Murata J, De Luca V. 2006. L-alanine induces programmed cell death in V. labrusca cell suspension cultures. Plant Science 171, 734–744. [Google Scholar]
- Chen S, Yin C, Strasser RJ, Govindjee Yang C, Qiang S. 2012. Reactive oxygen species from chloroplasts contribute to 3-acetyl-5-isopropyltetramic acid-induced leaf necrosis of Arabidopsis thaliana. Plant Physiology and Biochemistry 52, 38–51. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen SP, Leach JE. 2019. Abiotic and biotic stresses induce a core transcriptome response in rice. Scientific Reports 9, 6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz RP, Sperotto RA, Cargnelutti D, Adamski JM, de FreitasTerra T, Fett JP. 2013. Avoiding damage and achieving cold tolerance in rice plants. Food Energy Security 2, 96–119. [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Straltsova D, Medvedev SS, Pozhvanov GA, Sokolik A, Yurin V. 2014. Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. Journal of Experimental Botany 65, 1259–1270. [DOI] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S. 2019. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytologist 222, 1690–1704. [DOI] [PubMed] [Google Scholar]
- Fairhurst T, Dobermann A. 2002. Rice in the global food supply. World 5, 454–349. [Google Scholar]
- Fujino K, Obara M, Shimizu T, Koyanagi KO, Ikegaya T. 2015. Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breeding Science 65, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiology and Biochemistry 48, 909–930. [DOI] [PubMed] [Google Scholar]
- Gilroy S, Białasek M, Suzuki N, Górecka M, Devireddy AR, Karpiński S, Mittler R. 2016. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiology 171, 1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, Kumira R, Garg M, Gupta M. 2010. Recent updates on free radicals scavenging flavonoids: an overview. Asian Journal of Plant Sciences 9, 108–117. [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. 2004. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. The Journal of Biological Chemistry 279, 47822–47832. [DOI] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. 1997. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. The Plant Cell 9, 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. 2001. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. The Journal of Biological Chemistry 276, 4350–4356. [DOI] [PubMed] [Google Scholar]
- Jones P, Vogt T. 2001. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Tai TH. 2011. Evaluation of seedling cold tolerance in rice cultivars: a comparison of visual ratings and quantitative indicators of physiological changes. Euphytica 178, 437–447. [Google Scholar]
- Ko JH, Kim BG, Hur HG, Lim Y, Ahn JH. 2006. Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Reports 25, 741–746. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim BG, Kim JH, Kim H, Lim CE, Lim J, Lee C, Lim Y, Ahn JH. 2008. Four glucosyltransferases from rice: cDNA cloning, expression, and characterization. Journal of Plant Physiology 165, 435–444. [DOI] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku YS, Sintaha M, Cheung MY, Lam HM. 2018. Plant hormone signaling crosstalks between biotic and abiotic stress responses. International Journal of Molecular Sciences 19, 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Singh A, Mithra SV, et al. 2015. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Research 22, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK. 2017. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. The Plant Journal 89, 85–103. [DOI] [PubMed] [Google Scholar]
- Li Q, Yu HM, Meng XF, Lin JS, Li YJ, Hou BK. 2018. Ectopic expression of glycosyltransferase UGT 76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biology 20, 10–19. [DOI] [PubMed] [Google Scholar]
- Li X, Yan W, Agrama H, Hu B, Jia L, Jia M, Jackson A, Moldenhauer K, McClung A, Wu D. 2010. Genotypic and phenotypic characterization of genetic differentiation and diversity in the USDA rice mini-core collection. Genetica 138, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Liu C, Ou S, Mao B, et al. 2018. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nature Communications 9, 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Guo Z, Li X, Ye H, Li X, Xiong L. 2016. New insights into the genetic basis of natural chilling and cold shock tolerance in rice by genome-wide association analysis. Plant, Cell & Environment 39, 556–570. [DOI] [PubMed] [Google Scholar]
- Ma X, Liu Y. 2015. CRISPR/Cas9-based genome editing systems and the analysis of targeted genome mutations in plants. Hereditas (Beijing) 38, 118–125. [DOI] [PubMed] [Google Scholar]
- Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J. 2016. Spreading the news: subcellular and organellar reactive oxygen species production and signalling. Journal of Experimental Botany 67, 3831–3844. [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E. 2010. Genetic engineering for modern agriculture: challenges and perspectives. Annual Review of Plant Biology 61, 443–462. [DOI] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P. 2012. How do plants feel the heat? Trends in Biochemical Sciences 37, 118–125. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. 2004. Reactive oxygen gene network of plants. Trends in Plant Science 9, 490–498. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends in Plant Science 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC. 2001. Cytokinin metabolism and action. Annual Review of Plant Physiology and Plant Molecular Biology 52, 89–118. [DOI] [PubMed] [Google Scholar]
- Molyneux P. 2004. The use of the stable free radical diphenylpicrylhydrazyl (DPPH) for estimating antioxidant activity. Songklanakarin Journal of Science and Technology 26, 211–219. [Google Scholar]
- Muñoz M, Cedeño R, Rodríguez J, van der Knaap WP, Mialhe E, Bachère E. 2000. Measurement of reactive oxygen intermediate production in haemocytes of the penaeid shrimp, Penaeus vannamei. Aquaculture 191, 89–107. [Google Scholar]
- Pan Y, Zhang H, Zhang D, et al. 2015. Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS ONE 10, e0120590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Shahzad R, Gul A, et al. 2017. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV tolerance. Nature Communications 8, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelland E, Zachowski A. 2010. How plants sense temperature. Environmental and Experimental Botany 69, 225–232. [Google Scholar]
- Saravana Kumar P, Al-Dhabi NA, Duraipandiyan V, Balachandran C, Praveen Kumar P, Ignacimuthu S. 2014. In vitro antimicrobial, antioxidant and cytotoxic properties of Streptomyces lavendulae strain SCA5. BMC Microbiology 14, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2002. ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiology 130, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläppi MR, Jackson AK, Eizenga GC, Wang A, Chu C, Shi Y, Shimoyama N, Boykin DL. 2017. Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-core collection. Frontiers in Plant Science 8, 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakiba E, Edwards JD, Jodari F, Duke SE, Baldo AM, Korniliev P, McCouch SR, Eizenga GC. 2017. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 12, e0172133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton AM, Zhao JZ, Roush RT. 2002. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annual Review of Entomology 47, 845–881. [DOI] [PubMed] [Google Scholar]
- Shi MZ, Xie DY. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Patents on Biotechnology 8, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YG, Wang B, Jin SH, Qu XX, Li YJ, Hou BK. 2013. Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS ONE 8, e59924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. 2011. Respiratory burst oxidases: the engines of ROS signaling. Current Opinion in Plant Biology 14, 691–699. [DOI] [PubMed] [Google Scholar]
- Takahashi D, Li B, Nakayama T, Kawamura Y, Uemura M. 2013. Plant plasma membrane proteomics for improving cold tolerance. Frontiers in Plant Science 4, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Bielach A, Hrtyan M. 2017. Redox regulation at the site of primary growth: auxin, cytokinin and ROS crosstalk. Plant, Cell & Environment 40, 2586–2605. [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. 2006. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. The Plant Journal 47, 969–976. [DOI] [PubMed] [Google Scholar]
- Uemura M, Tominaga Y, Nakagawara C, Shigematsu S, Minami A, Kawamura Y. 2006. Responses of the plasma membrane to low temperatures. Physiologia Plantarum 126, 81–89. [Google Scholar]
- Vaahtera L, Brosché M, Wrzaczek M, Kangasjärvi J. 2014. Specificity in ROS signaling and transcript signatures. Antioxidants & Redox Signaling 21, 1422–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishal B, Kumar PP. 2018. Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Frontiers in Plant Science 9, 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu J, Li C, et al. 2016. a Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu X, Vieira FG, Xiao Y, Li Z, Wang J, Nielsen R, Chu C. 2016b The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Molecular Plant 9, 975–985. [DOI] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W. 1997. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. The EMBO Journal 16, 4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XJ, Zhou YH, Sji K, Zhou J, Foyer CH, Yu JQ. 2015. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. Journal of Experimental Botany 66, 2839–2856. [DOI] [PubMed] [Google Scholar]
- Xie G, Kato H, Imai R. 2012. Biochemical identification of the OsMKK6–OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochemistry Journal 443, 95–102. [DOI] [PubMed] [Google Scholar]
- Xu D, Huang X, Xu ZQ, Schläppi M. 2011. The HyPRP gene EARLI1 has an auxiliary role for germinability and early seedling development under low temperature and salt stress conditions in Arabidopsis thaliana. Planta 234, 565–577. [DOI] [PubMed] [Google Scholar]
- Xu ZJ, Nakajima M, Suzuki Y, Yamaguchi I. 2002. Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiology 129, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemaru J-i, Yamamoto T, Fukuoka S, Uga Y, Hori K, Yano M. 2010. Q-TARO: QTL annotation rice online database. Rice 3, 194–203. [Google Scholar]
- Yun KY, Park MR, Mohanty B, Herath V, Xu F, Mauleon R, Wijaya E, Bajic VB, Bruskiewich R, de Los Reyes BG. 2010. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biology 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen Q, Wang S, Hong Y, Wang Z. 2014. Rice and cold stress: methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B. 2012. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 7, e43274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yao W, Ouyang Y, Yang W, Wang G, Lian X, Xing Y, Chen L, Xie W. 2015. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Research 43, D1018–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Chen K, Mi X, Chen T, Ali J, Ye G, Xu J, Li Z. 2015. Identification and fine mapping of a stably expressed QTL for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS ONE 10, e0145704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.