Abstract
The presence of varied numbers of CALCINEURIN B-LIKE10 (CBL10) calcium sensor genes in species across the Brassicaceae and the demonstrated role of CBL10 in salt tolerance in Arabidopsis thaliana and Eutrema salsugineum provided a unique opportunity to determine if CBL10 function is modified in different species and linked to salt tolerance. Salinity effects on species growth and cross-species complementation were used to determine the extent of conservation and divergence of CBL10 function in four species representing major lineages within the core Brassicaceae (A. thaliana, E. salsugineum, Schrenkiella parvula, and Sisymbrium irio) as well as the first diverging lineage (Aethionema arabicum). Evolutionary and functional analyses indicate that CBL10 duplicated within expanded lineage II of the Brassicaceae and that, while portions of CBL10 function are conserved across the family, there are species-specific variations in CBL10 function. Paralogous CBL10 genes within a species diverged in expression and function probably contributing to the maintenance of the duplicated gene pairs. Orthologous CBL10 genes diverged in function in a species-specific manner, suggesting that functions arose post-speciation. Multiple CBL10 genes and their functional divergence may have expanded calcium-mediated signaling responses and contributed to the ability of certain members of the Brassicaceae to maintain growth in salt-affected soils.
Keywords: Brassicaceae, CALCINEURIN B-LIKE10, calcium sensor, functional divergence, gene duplication, salt tolerance
Duplication of the CBL10 calcium sensor within expanded lineage II of the Brassicaceae led to both conservation of and species-specific variations in CBL10 function across the family.
Introduction
Calcium has emerged as an essential component of many signaling pathways in plants, underlying growth and development by linking perception of physiological and environmental cues to cellular responses. Specificity in signaling is achieved, in part, through an array of proteins that perceive changes in cytosolic calcium levels (calcium sensors). The potential for different calcium sensors to contribute to functional specificity is found in their diverse temporal and spatial expression patterns, different affinities for calcium, and range of target proteins.
In Arabidopsis thaliana (Arabidopsis), CALCINEURIN B-LIKE10 (AtCBL10) encodes a calcium sensor that enables plants to grow in salt-affected soils by preventing the toxic accumulation of sodium ions in the cytosol (Quan et al., 2007). Upon perception of changes in cytosolic calcium levels, AtCBL10 interacts with the SALT-OVERLY-SENSITIVE2 (AtSOS2) protein kinase to activate the AtSOS1 plasma membrane sodium/proton exchanger which transports sodium out of the cell using the energy stored in the proton gradient (Quan et al., 2007; Lin et al., 2009). Mutation of AtCBL10 results in hypersensitivity when plants are grown in the presence of even low levels of salt (Quan et al., 2007). Two CBL10 genes have been found in a salt-tolerant relative of Arabidopsis, Eutrema salsugineum (EsCBL10a and EsCBL10b; Monihan et al., 2019). A comparative study of CBL10 function in Arabidopsis and E. salsugineum provided a starting point for understanding how duplication of a calcium sensor increased calcium-mediated signaling capacity in E. salsugineum and contributed to its salt tolerance. Reduced expression of EsCBL10a and EsCBL10b singly and in combination revealed that both genes function in the response of E. salsugineum to salt and probably have different functions (Monihan et al., 2019). Both EsCBL10 genes complement the Atcbl10 salt-sensitive phenotype, indicating that there is conservation of CBL10 function in the two species (Monihan et al., 2019). When the genes were expressed in a salt-sensitive strain of Saccharomyces cerevisiae (yeast) with SOS2 and SOS1 from Arabidopsis or E. salsugineum, cells expressing EsCBL10b had the greatest growth on media with salt, while cells expressing EsCBL10a had the weakest growth relative to cells expressing AtCBL10 (Monihan et al., 2019). This result suggests that EsCBL10b strongly activates the SOS pathway while EsCBL10a does so only weakly. The different expression patterns of the EsCBL10 genes provided further evidence that these two genes diverged in function. EsCBL10b, like AtCBL10, is expressed primarily in leaves, with very low expression in roots, while EsCBL10a is expressed in both leaves and roots (Monihan et al., 2019). AtSOS3, another calcium sensor in the CALCINEURIN B-LIKE family in Arabidopsis (also known as AtCBL4), is expressed in roots and also functions in plant responses to salt (Liu and Zhu, 1998; Halfter et al., 2000). The AtSOS3 and AtCBL10 proteins have non-overlapping roles during growth in the presence of salt (Quan et al., 2007). EsCBL10a, but not AtCBL10 or EsCBL10b, was able to complement the salt-sensitive Atsos3 mutant phenotype, suggesting that EsCBL10a has a function not present in AtCBL10 or EsCBL10b (Monihan et al., 2019). Together these results demonstrated that the duplication of CBL10 in E. salsugineum resulted in two calcium sensors with both shared and distinct activities expanding the response of E. salsugineum to salt.
Arabidopsis and E. salsugineum belong to the Brassicaceae, a diverse family of plants containing both economically and scientifically important species (Beilstein et al., 2006; Yang et al., 2013). The number of sequenced Brassicaceae genomes coupled with its well-established phylogeny led us to analyze the extent of conservation and divergence of CBL10 gene function in both evolutionary and genomic contexts. Specifically we determined if: (i) other species in the Brassicaceae have multiple CBL10 genes and if they are descendants of the same duplication event that resulted in two genes in E. salsugineum; (ii) the functions of the orthologous genes have been retained; and (iii) paralogous genes within selected species diverged in function. These analyses in combination with assays of plant growth in the presence of salt were used to understand how changes in CBL10 function may have contributed to Brassicaceae adaptation to soil salinity. A phylogenetic analysis of CBL10 revealed that the duplication resulting in two genes in E. salsugineum probably occurred in expanded lineage II (to which E. salsugineum belongs) of the family and resulted in several species retaining orthologs of EsCBL10a and EsCBL10b. CBL10 expression and function were examined in four species representing major lineages within the core Brassicaceae (A. thaliana, E. salsugineum, Schrenkiella parvula, and Sisymbrium irio) as well as the first diverging lineage (Aethionema arabicum) (Beilstein et al., 2006). These analyses indicate that, while portions of CBL10 function are conserved across the Brassicaceae, there are also variations in CBL10 function that are specific to each species.
Materials and methods
Phylogenetic tree
Nucleotide sequences were identified using Basic Local Alignment Search Tool (BLAST) with the coding sequence of A, thaliana CBL10 (AtCBL10; At4G33000.2) and a threshold E-score of the order of 1×10–15. The identified genes were used in a reciprocal BLAST against the Arabidopsis genome. Only those sequences that identified AtCBL10 as the closest homolog were used to generate the phylogeny. Sequences for Arabidopsis lyrata (Hu et al., 2011), Boechera stricta (Windsor et al., 2006), Brassica nigra, Brassica oleraceae (Liu et al., 2014), Brassica rapa (Wang et al., 2011), Camelina sativa (Kagale et al., 2014), Capsella grandiflora (Slotte et al., 2013), Capsella rubella (Slotte et al., 2013), Carica papaya (Ming et al., 2008), E. salsugineum Shandong (Yang et al., 2013), and Raphanus raphanistrum (Moghe et al., 2014) were retrieved from the Phytozome database (www.phytozome.net). Sequences for S. parvula (Dassanayake et al., 2011) were retrieved from the NCBI genome database, while sequences for A. arabicum, Neslia paniculata, Leavenworthia alabamica, and S. irio were provided by Dr Stephen Wright (University of Toronto; Haudry et al., 2013; Slotte et al., 2013), and a sequence for Tarenaya hassleriana (Cheng et al., 2013) was retrieved from the Comparative Genomics (CoGe) platform (Lyons and Freeling, 2008; Lyons et al., 2008). Sequences for CBL10 from the E. salsugineum accessions Cracker Creek, and Yukon were obtained by PCR using primers designed to amplify CBL10 from the Shandong accession. To strengthen the inference of the root for the gene tree within the Brassicaceae, outgroup sequences for Populus euphratica (Ma et al., 2013), Prunus mume (Zhang et al., 2012), Durio zibethinus (Teh et al., 2017), Theobroma cacao (Argout et al., 2011), and Vitis vinifera (Jaillon et al., 2007) were obtained from the GenBank nucleotide database using the CBL10 sequence from C. papaya as a query for a BLAST search (all matched with E-scores of the order of 1×10–140).
Exon boundaries for the Arabidopsis, E. salsugineum, S. parvula, S. irio, and A. arabicum CBL10 genes were determined by comparing the genomic sequences with cDNAs generated by PCR [AtCBL10 (Quan et al., 2007); EsCBL10a and EsCBL10b (Monihan et al., 2019); S. parvula, S. irio, and A. arabicum CBL10 genes, see cloning strategy in the complementation assay section below]. For all other CBL10 genes, exon boundaries were estimated after multiple sequence alignments to the experimentally determined annotations. Extensive length heterogeneity, indels, and low-complexity sequences within each of the eight introns resulted in poorly aligned sequences. The CBL10 coding sequences from the nine exon regions were concatenated, aligned in MUSCLE 3.8.31 (Edgar, 2004) using translated amino acids, and analyzed using IQ-TREE 1.6.7 (Nguyen et al., 2015) with standard model selection. Two sequences from L. alabamica failed the composition test, indicating statistically significant differences in composition of these two sequences relative to the rest of the alignment. The model selected by the Bayesian information criterion and the corrected Akaike information criterion was TIM3+F+I+G4, which was used in the subsequent likelihood tree search with support for branches estimated by 100 bootstrap replicates.
Synteny analysis
The genomic regions of Arabidopsis containing CBL10 and E. salsugineum containing EsCBL10a and EsCBL10b were compared with 15 genomes (A. arabicum, A. lyrata, B. stricta, B. nigra, B. oleraceae, B. rapa, C. sativa, C. grandiflora, C. rubella, C. papaya, L. alabamica, R. raphanistrum, S. parvula, S. irio, and T. hassleriana) to identify collinear regions using the SynFind feature on CoGe (Lyons and Freeling, 2008; Lyons et al., 2008). The genes located on the two identified collinear regions of the Arabidopsis genome were used as a query for a BLAST search to detect putative homologs in the genomes of the other species (Altschul et al., 1990). The species tree shown in the synteny analysis is based on Beilstein et al. (2010) and Yang et al. (2013).
Identification of transposable elements
RepeatMasker 4.0.6 was used to detect putative transposable elements in flanking regions of CBL10/CBL10a and CBL10b in Arabidopsis, E. salsugineum, S. irio, and S. parvula (A.F.A. Smit, R. Hubley, and P. Green; RepeatMasker at http://repeatmasker.org).
Plant growth
To determine the salt tolerance of each species, seeds of Arabidopsis, E. salsugineum Shangdong, S. parvula, S. irio, and A. arabicum were sown on SunGro Sunshine LC1 soil mix (SunGrow Horticulture) and stratified for 4 d at 4 °C in the dark to break dormancy. After cold treatment, plants were transferred to a growth chamber at 21 °C under a 16 h light/8 h dark photoperiod with light provided by Phillips F32T8/TL841 bulbs (135 µmol m−2 s−1) and watered every 3 d with 0.25× Hoagland’s solution (Hoagland and Arnon, 1938) with cobalt chloride in place of cobalt nitrate. When true leaves developed, plants were treated with increasing salt (sodium chloride, NaCl) in 50 mM increments every 3 d until the indicated, final concentration was reached. Three weeks after the start of treatment, photographs were taken and the fresh weight of aerial tissue was recorded. Comparisons of growth were performed by analyzing the ratio of species growth in the absence and presence of salt at each NaCl concentration using Friedman’s non-parametric, two-way ANOVA and Tukey’s honestly significant difference (HSD) tests. Statistical significance was assigned at P≤0.05.
Complementation assays
CBL10 gene function was analyzed by expressing each gene in the Atcbl10 T-DNA insertion mutant (Arabidopsis Biological Resource Center, SALK_056042; Monihan et al., 2016) and the Atsos3 ethyl methanesulfonate mutant (Dr Jian-Kang Zhu; Purdue University; Liu and Zhu, 1997). Full-length coding sequences without a stop codon were amplified from cDNA and cloned into pGEM-T Easy (Promega). All PCR amplifications were performed using Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific); primer sequences are provided in Supplementary Table S1 at JXB online. Because of the high degree of similarity between the S. parvula transcripts, forward primers were designed to anneal to the 5'-untranslated region (UTR) to enrich for a specific CBL10 gene; primer sequences are provided in Supplementary Table S1. The S. parvula PCRs were then used as a template in a second reaction with primers that anneal to the coding sequence for cloning into pGEM-T Easy (Promega). All genes were digested with XhoI and BamHI, and subcloned into the corresponding site of the plant binary vector pEZT-NL (Drs Sean Cutler and David W. Ehrhardt; Carnegie Institution of Washington) downstream of the Cauliflower mosaic virus (CaMV) 35S promoter. Agrobacterium tumefaciens LBA4404 containing the binary vector was used to transform Atcbl10 and Atsos3 via the floral dip method (Clough and Bent, 1998). T1 seed was germinated on soil for 1.5 weeks and then sprayed three times with 100 mg l–1 Basta (Rely 200 Herbicide; Bayer Crop Science) at 3 d intervals. T1 lines with antibiotic resistance were subsequently transferred to pots and grown to collect T2 seed. Single insertion lines were identified by screening T2 seed on 0.5× MS medium [Murashige and Skoog medium; PhytoTechnology Laboratories containing 2.5 mM MES, 2% sucrose (w/v), and 1% agar (w/v) (A8678; Sigma), pH 5.7 (adjusted with potassium hydroxide)] and 7.5 mg l–1 glufosinate ammonium (Santa Cruz Biotechnology). Lines with 75% resistance were selected. Homozygous lines were identified by screening T3 seed on MS plates with glufosinate ammonium and selecting lines with 100% resistance. Gene-specific primers were used to confirm the identity and expression of all transgenes (Supplementary Fig. S1).
To monitor seedling growth, transgenic Arabidopsis seeds were sown on 0.5× MS medium. Plates with seed were incubated at 4 °C in the dark for 2 d to break dormancy and transferred to a growth chamber at 21 °C under a 16 h light/8 h dark photoperiod with light provided by Phillips F32T8/TL841 bulbs (135 µmol m−2 s−1). For salt assays, seeds were germinated on medium without NaCl for 4 d, after which seedlings were transferred to medium without or with the indicated concentration of NaCl. NaCl concentrations were chosen for maximal differences in growth between the wild type and mutants. After 10 d of treatment, photographs were taken and seedling fresh weight was measured to quantify growth.
Transcript analysis
To determine expression patterns of the CBL10 genes, leaves and roots of 11-day-old seedlings grown on 0.25× MS medium were collected. RNA was isolated using the NucleoSpin RNA extraction kit (Macherey-Nagel) and used to synthesize cDNA (M-MLV Reverse Transcriptase; Promega). All PCRs were performed using recombinant Taq polymerase (Invitrogen) and were analyzed and compared in the linear range of the amplification (e.g. 23 cycles for Actin and 28 cycles for CBL10); primer sequences are provided in Supplementary Table S1.
Yeast salt screens
To monitor the ability of the CBL10 proteins to activate the SOS pathway, Saccharomyces cerevisiae strain AXT3K (ena1::HIS3::ena4, nha1::LEU2, and nhx1::KanMX4; Quintero et al., 2002) containing the pYPGE15 plasmid with AtSOS1 (Jarvis et al., 2014) and the pFL32T plasmid with AtSOS2 and AtSOS3 was modified to express the CBL10 genes. Plasmids containing AtCBL10, EsCBL10a, EsCBL10b, or without a CBL10 gene were generated as previously described (Monihan et al., 2019). The full-length coding sequences of SpCBL10a, SpCBL10b-1, SpCBL10b-2, SiCBL10a, SiCBL10b, and AaCBL10 were amplified from cDNA (Phusion High-Fidelity DNA polymerase; ThermoFisher Scientific; primer sequences are provided in Supplementary Table S1), cloned into pGEM-T Easy (Promega), digested with XhoI and NotI, and subcloned into the corresponding site of the pDR195 vector (Dr Alonso Rodriguez-Navarro; Rentsch et al., 1995). Because of the high degree of similarity between the S. parvula transcripts, forward primers were designed to anneal to the 5'-UTR to enrich for a specific CBL10 gene (Supplementary Table S1). The S. parvula PCRs were then used as a template in a second reaction with primers that anneal to the coding sequence for cloning into pGEM-T Easy (Promega). The pDR195 plasmids were digested with AgeI and NotI, and a fragment containing the CBL10 gene was subcloned into the corresponding site of the pFL32T plasmid in place of AtSOS3 to be expressed with AtSOS2. Transformed AXT3K cells were selected on synthetic dropout medium lacking both uracil and tryptophan (Clontech/TaKaRa) containing yeast nitrogen base without amino acids (VWR). Salt assays were carried out in alkali cation-free medium (AP; Rodriguez-Navarro and Ramos, 1984) containing 1 mM KCl with the designated concentrations of NaCl and cultured at 30 °C for 4 d.
Yeast two-hybrid assays
To determine if EsCBL10a and the SpCBL10b proteins might complement Atsos3 by interaction with a similar CBL-interacting protein kinase (CIPK), yeast-two hybrid assays were performed between both SpCBL10b proteins and the four EsCBL10a-interacting AtCIPK proteins. Cloning of AtCBL10, EsCBL10a, EsCBL10b, and AtCIPK genes was described previously (Monihan et al., 2019). SpCBL10b-1 and SpCBL10b-2 genes were PCR amplified using Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific); primer sequences are provided in Supplementary Table S1. The PCR products were digested with EcoRI and BamHI, and cloned into the corresponding site of the pGADT7 and pGBKT7 vectors (Clontech/TaKaRa) which allows for expression of the gene as a fusion protein with the GAL4 DNA activation domain (AD) or the GAL4 DNA-binding domain (BD), respectively. The pGADT7 clones were transformed into S. cerevisiae strain Y2HGold (Clontech/TaKaRa), while the pGBKT7 clones were transformed into S. cerevisiae strain Y187 (Clontech/TaKaRa). Yeast were mated and grown on synthetic defined medium (SD) minus leucine and tryptophan (SD-LW) to select for diploid yeast expressing both constructs. To determine interaction, serial dilutions of yeast colonies were grown on SD-LW and without histidine (H), and incubated for 5 d. Interaction is shown in only one orientation because the SpCBL10b proteins fused to the GAL4 BD self-activated, causing strains expressing only these fusion proteins to grow on all selection media, masking any interaction with the CIPK proteins.
Statistical analysis
To determine significant differences in growth, experiments were organized and analyzed as a randomized complete block design with genotypes and salt concentrations as treatments, and individual experiments as replicates. Treatment effects were assessed using a full-factorial mixed-model ANOVA in JMP, Version 11 (SAS Institute; 1989–2007). In these analyses, treatments were considered fixed effects and replicates random effects. The normality of the distributions of all dependent variables was analyzed by examining a plot of the residuals from a full-factorial ANOVA of untransformed data. A Shapiro–Wilk test (Shapiro and Wilk, 1965) was performed to assess normality, and Bartlett’s (Bartlett, 1937) and Levene’s (Levene, 1960) tests were performed to evaluate the homogeneity of variance. Based on the pattern of distribution and the results of these tests, a non-parametric approach was used to analyze the data throughout. Data were rank transformed using Microsoft Excel (function: RANK) followed by an ANOVA and Tukey’s HSD test for multiple comparisons of means (Conover and Iman, 1981). The HSD values from rank-based ANOVA were then applied to the actual means for each measurement (i.e. not the ranks used in ANOVA). Statistical significance was assigned at P≤0.05 throughout, and all tests of significance were two sided.
Results
CBL10 duplicated within expanded lineage II of the Brassicaceae
Previous studies have shown that there are two CALCINEURIN B-LIKE10 (CBL10) calcium sensors in E. salsugineum, a salt-tolerant relative of Arabidopsis. Reduced expression of the duplicated E. salsugineum CBL10 genes demonstrated that both genes function in the response of the plant to salt (Monihan et al., 2019). Cross-species complementation assays demonstrated that the two E. salsugineum CBL10 proteins have both shared and distinct activities (Monihan et al., 2019), suggesting that duplication of CBL10 increased calcium-mediated signaling capacity in E. salsugineum.
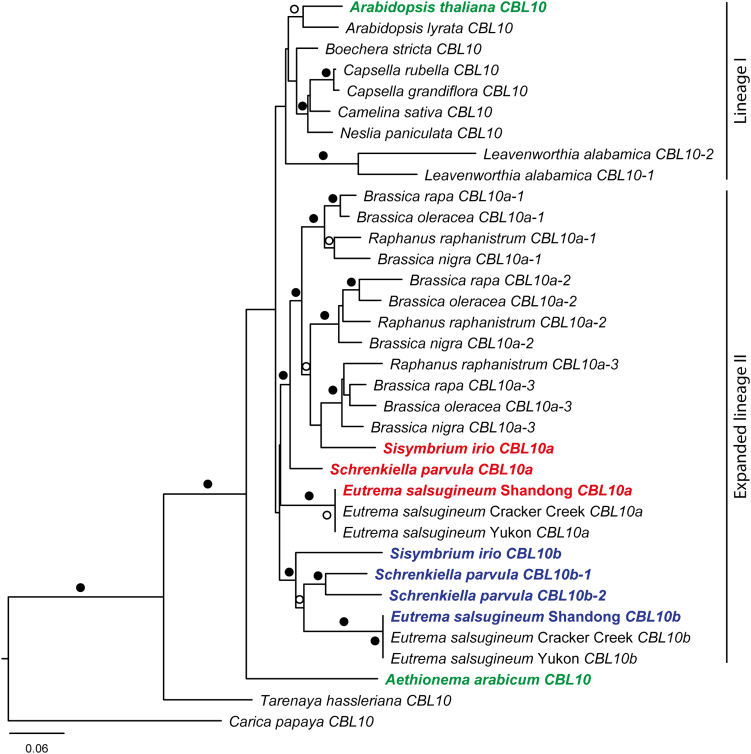
To understand CBL10’s role in calcium-mediated signaling in an evolutionary context and determine when in the evolutionary history of the Brassicaceae the CBL10 duplication arose, CBL10 sequences were identified from species within and outside of the Brassicaceae and a phylogeny was inferred based on maximum likelihood. Of eight species examined within lineage I of the Brassicaceae, seven have a single CBL10 gene: A. thaliana, A. lyrata, B. stricta, C. rubella, C. grandiflora, C. sativa, and N. paniculata (Fig. 1). Leavenworthia alabamica was the only lineage I species examined, with multiple CBL10 genes probably due to a whole-genome triplication event; however, only two CBL10 genes were identified (Fig. 1; Haudry et al., 2013). CBL10 sequences from B. rapa, B. nigra, B. oleracea, R. raphanistrum, S. irio, and S. parvula (lineage II species) were compared with the CBL10 genes in E. salsugineum (which belongs to expanded lineage II; Beilstein et al., 2006; Yang et al., 2013). Sisymbrium irio, S. parvula, and E. salsugineum have both CBL10a and CBL10b paralogs (Fig. 1), with an additional CBL10b gene in S. parvula due to a tandem duplication (Fig. 2; Dassanayake et al., 2011; Oh et al., 2014). Raphanus raphanistrum and all three Brassica species have three CBL10a genes, consistent with their whole-genome triplication events (Fig. 1; Lagercrantz and Lydiate, 1996; Moghe et al., 2014), but no CBL10b genes. Aethionema arabicum, which is a member of the first diverging lineage of the Brassicaceae, contains only a single CBL10 gene (Fig. 1). Outside of the Brassicaceae, T. hassleriana and C. papaya, members of the Cleomaceae and Caricaceae, respectively, both have a single CBL10 gene (Fig. 1). Sequences from P. euphratica, P. mume, D. zibethinus, T. cacao, and V. vinifera were included to provide additional data for the tree which was rooted on the V. vinifera sequence (data not shown). The tree is consistent with a single duplication in the ancestor of lineage II, leading to all known CBL10b genes which form a strongly supported clade (Fig. 1). The Brassica and R. raphanistrum CBL10 genes descended from a triplication in the ancestor of just those species, not from the duplication that led to CBL10b, indicating that the ancestor of the Brassica species and R. raphanistrum lost CBL10b. Sisymbrium irio CBL10a was placed with weak support among the Brassica species, suggesting that additional duplications (or other phenomena such as lineage sorting) must have occurred. While it is not possible to unequivocally determine the timing of the duplication due to low branch support, the presence of both CBL10a and CBL10b paralogs in multiple members of expanded lineage II and the absence of the paralogs in members of lineage I, A. arabicum, T. hassleriana, and C. papaya, suggest that the duplication of CBL10 occurred within expanded lineage II of the Brassicaceae.
Fig. 1.
The duplication of CBL10 probably occurred within expanded lineage II in the Brassicaceae. Exons from CBL10 nucleotide sequences were aligned and analyzed using maximum likelihood. Sequences from P. euphratica, P. mume, D. zibethinus, T. cacao, and V. vinifera were included to provide additional data for the tree which was rooted on the V. vinifera sequence. Circles above branches represent the percentage of 100 bootstrap replicates that support the topology; closed circles, 90–100; open circles, 70–89. CBL10 genes from five species were chosen for further analysis and color coded. Green, CBL10 genes from two species with a single gene; red, CBL10a genes from three species with multiple genes; blue, CBL10b genes from three species with multiple genes. Species within lineage I and expanded lineage II are indicated.
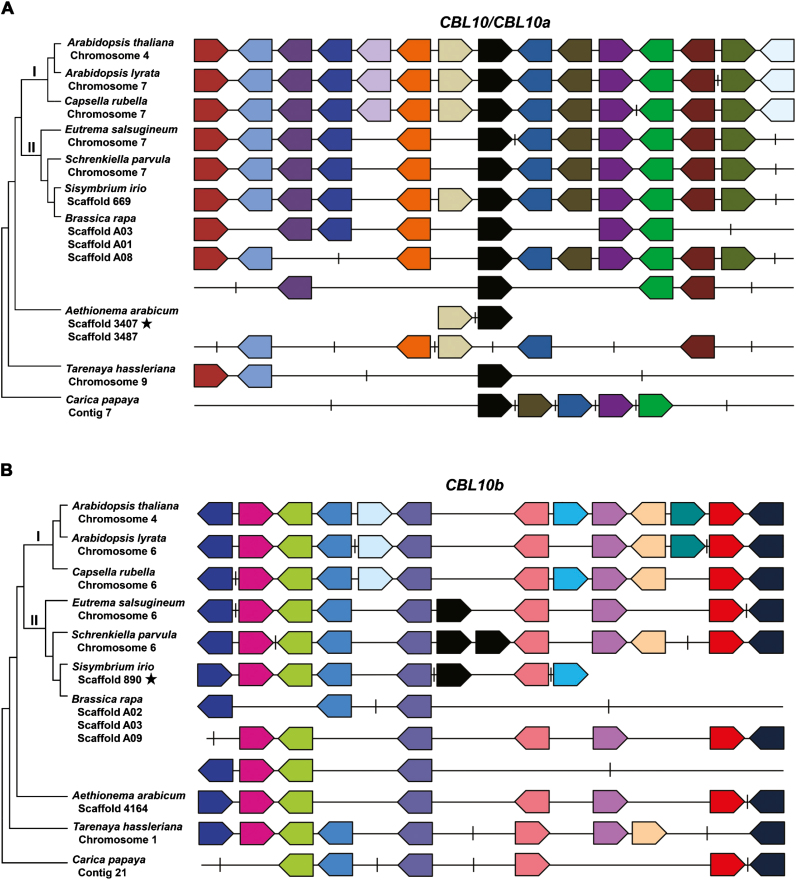
Fig. 2.
The genomic positions of AtCBL10 and the CBL10b genes differ. The organismal phylogeny is shown with the species name and the chromosome, contig, or scaffold on which CBL10 was identified. Stars, short scaffolds; horizontal line, genomic region (not drawn to scale); pentagons, genes. In Arabidopsis, CBL10 is black and the genes on either side were identified and assigned a color. In the other species, CBL10 genes were identified and the surrounding genes were colored based on ontology with genes from Arabidopsis. Vertical lines, presence of genes not syntenic to those in Arabidopsis. Tandem duplicates of the flanking genes were collapsed to one gene/pentagon to simplify the figure. (a) Regions syntenic to AtCBL10 and EsCBL10a. (b) Regions syntenic to EsCBL10b.
The region surrounding the CBL10 sequence was compared to provide additional information about the origin of the genes. EsCBL10a is syntenic with CBL10 genes from Arabidopsis, A. lyrata, both Capsella species, B. stricta, L. alabamica, C. sativa, T. hassleriana, and C. papaya, and the CBL10a genes from all Brassica species, S. parvula, and S. irio (select species shown in Fig. 2A). Synteny in N. paniculata and R. raphanistrum was difficult to determine because the CBL10 sequences were found on short scaffolds. The fact that all CBL10 genes from the Brassica species and from L. alabamica share synteny with EsCBL10a (data not shown) further supports the conclusion that these CBL10 genes arose through independent whole-genome triplication events (Lagercrantz and Lydiate, 1996; Haudry et al., 2013). The CBL10 gene from A. arabicum was found on a short scaffold with insufficient sequence to assess synteny (scaffold 3407, Fig. 2A). A second scaffold in A. arabicum sharing synteny with Arabidopsis was detected, but no CBL10 gene was identified (scaffold 3487, Fig. 2A). The genomic region syntenic to EsCBL10b was identified in all species; however, a CBL10b gene was only detected in E. salsugineum, S. parvula, and S. irio (Fig. 2B). Numerous putative transposable elements were found surrounding the CBL10b genes, suggesting that the duplication of CBL10 might have been mediated by transposons (Supplementary Fig. S2). However, it was not possible to identify a single transposon associated with all three CBL10b genes. Taken together, these results suggest that transposons may have mediated the duplication of CBL10 and that the insertion of CBL10b into a different chromosomal position took place before the divergence of expanded lineage II species within the Brassicaceae.
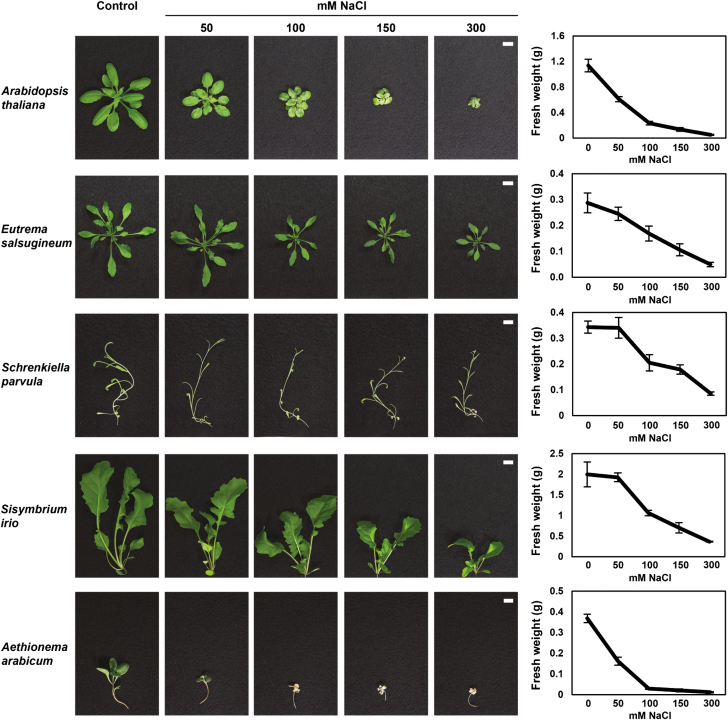
Species in expanded lineage II are salt tolerant
As a first step in linking CBL10 function to salt tolerance within the Brassicaceae, five species were selected for further analyses. In addition to Arabidopsis and E. salsugineum, S. parvula and S. irio were chosen because of their close relationship to E. salsugineum and the presence of multiple CBL10 genes in their genomes (Beilstein et al., 2006; Fig. 3). Due to its position as a member of the first diverging group within the Brassicaceae and its more distant relationship with E. salsugineum, A. arabicum was also included in the analysis (Beilstein et al., 2006; Fig. 3). The salt tolerance (FW of each species treated with increasing concentrations of salt) was measured. Schrenkiella parvula, E. salsugineum, and S. irio all maintained growth in concentrations of salt up to 300 mM, while growth of Arabidopsis and A. arabicum quickly decreased at concentrations as low as 100 mM (Fig. 3). Ratios of species growth in the absence and presence of salt at each salt concentration indicated that the salt tolerance of S. parvula, E. salsugineum, and S. irio was similar and distinct from the salt tolerance of Arabidopsis and A. arabicum which were similar (Supplementary Table S2). Because the increased salt tolerance of the expanded lineage II species analyzed might be due to an increase in the number of CBL10 genes and/or a divergence in function of those genes, four assays (cross-species complementation of the cbl10 and sos3 mutants in Arabidopsis, expression analysis, and SOS pathway activation) were used to compare the activities of the genes.
Fig. 3.
Species in expanded lineage II in the Brassicaceae are salt tolerant. Seeds from Arabidopsis, E. salsugineum, S. parvula, S. irio, and A. arabicum were germinated and grown on soil for 1 week and then treated with increasing NaCl in 50 mM increments every 3 d until the indicated final concentration was reached. Three weeks after the start of treatment, aerial portions of the plants were harvested, photographed, and weighed. Scale bar=1 cm for all images. The average fresh weight was graphed and ±SE is shown. One representative image of seven experiments is shown.
CBL10 function is conserved in species across the Brassicaceae
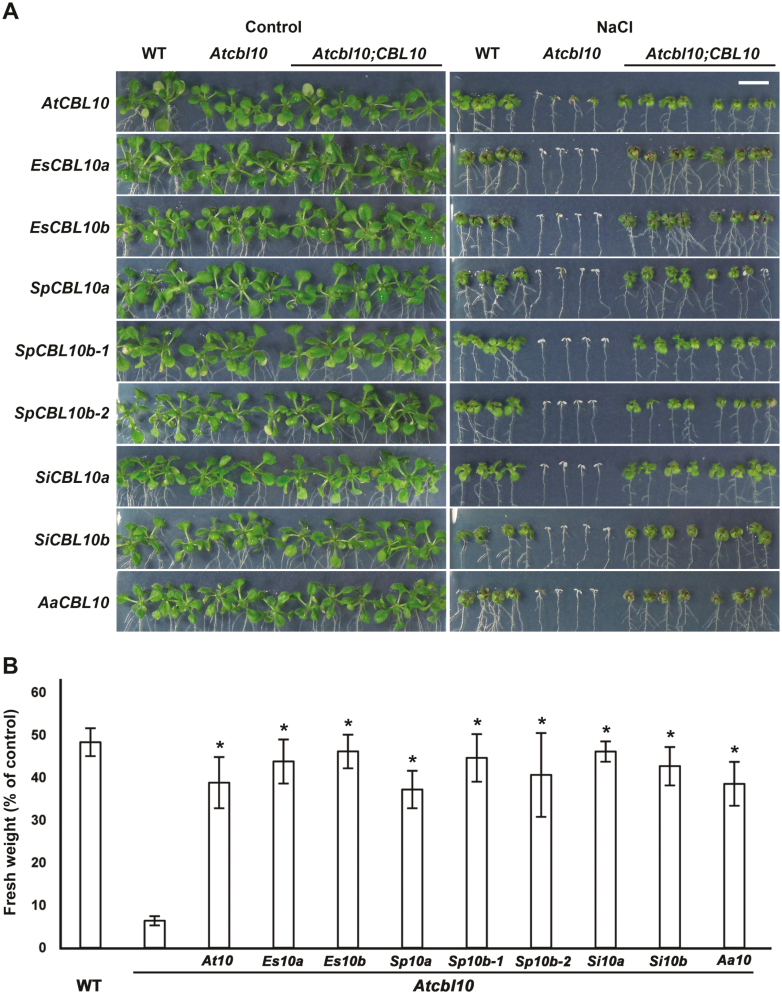
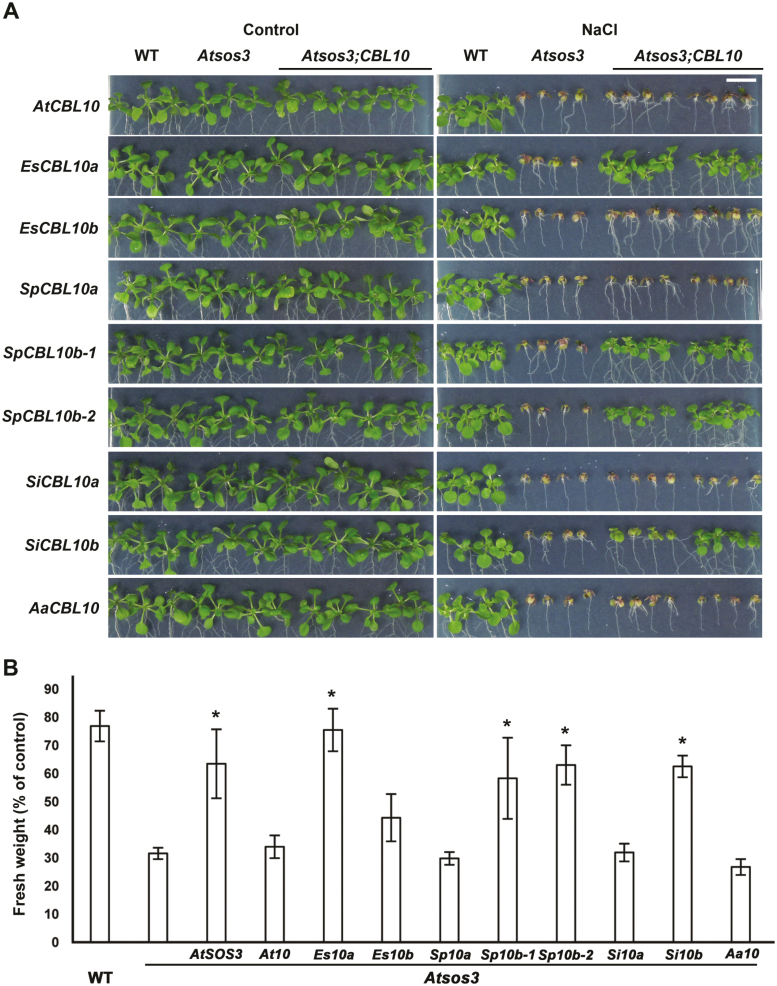
The CBL10 phylogenetic tree suggests that all the genes identified are homologs of AtCBL10 and may share activities. To determine if there is conservation of CBL10 function throughout the Brassicaceae, the CBL10 genes from S. parvula, S. irio, and A. arabicum were expressed in the Arabidopsis cbl10 mutant (Atcbl10). Our studies have shown that introns are necessary for full expression and function of AtCBL10 when using the native promoter, but can lead to alternative splicing. To avoid alternative splicing of the CBL10 genes under study and to ensure strong expression of their coding sequences, the CaMV 35S constitutive promoter was used for these cross-species complementation assays. The salt tolerance of four independently transformed, single insertion, homozygous lines was assessed for each gene. All of the tested CBL10 genes complemented the Atcbl10 salt-sensitive phenotype, indicating that there is conservation of CBL10 function throughout the Brassicaceae (Fig. 4).
Fig. 4.
CBL10 genes from all Brassicaceae species complement the Atcbl10 salt-sensitive phenotype. CBL10 genes from Arabidopsis (AtCBL10, At10), E. salsugineum (EsCBL10a, Es10a; and EsCBL10b, Es10b), S. parvula (SpCBL10a, Sp10a; SpCBL10b-1, Sp10b-1; and SpCBL10b-2, Sp10b-2), S. irio (SiCBL10a, Si10a; and SiCBL10b, Si10b), and A. arabicum (AaCBL10, Aa10) were expressed in the Atcbl10 mutant, and growth in the absence (control) and presence of salt (125 mM NaCl) was monitored. For salt assays, seeds were germinated on medium without NaCl for 4 d, after which seedlings were transferred to medium without or with the indicated concentration of NaCl. After 10 d of treatment, photographs were taken and total seedling fresh weight was measured. (A) Photographs of the wild type (WT), Atcbl10, and Atcbl10 expressing each CBL10 gene (Atcbl10;CBL10). The scale bar (1 cm, upper right panel) shows the magnification for all images. (B) Total seedling fresh weight was measured to quantify growth and is presented as a percentage of control. Data are means ±SE of at least 24 seedlings per genotype grown in three independent experiments. *Complementation of the Atcbl10 salt-sensitive phenotype (Tukey–Kramer HSD, P≤0.05).
CBL10 function diverged in a species-specific manner in the Brassicaceae
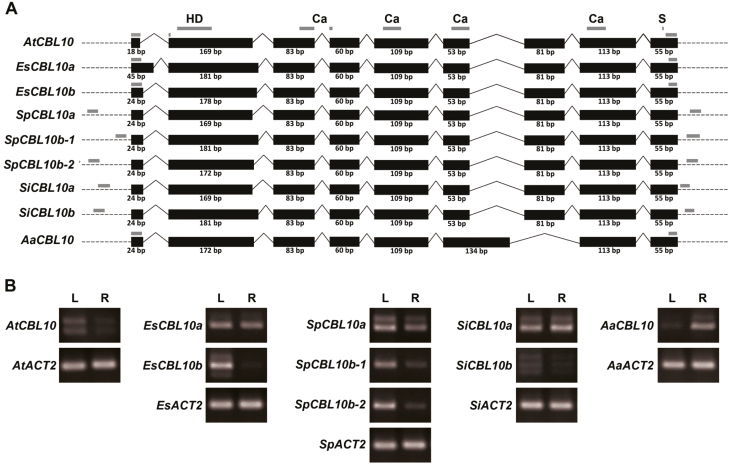
It has previously been shown that the CBL10 genes in E. salsugineum have different expression patterns. Like AtCBL10, EsCBL10b is expressed predominately in aerial tissue, whereas EsCBL10a is expressed throughout E. salsugineum (Monihan et al., 2019). To determine if CBL10 expression correlates with protein activity across the Brassicaceae, RNA was isolated from shoots and roots of each species and transcript accumulation was monitored. The exon–intron structure of the CBL10 transcripts was very similar, and protein domains known to be important for AtCBL10 function were present in all transcripts (Fig. 5). Expression patterns correlated with the phylogenetic relationship of the genes in lineage I and expanded lineage II species; CBL10b transcripts were high in leaves (similar to AtCBL10) while CBL10a transcripts accumulated in both leaves and roots (Fig. 5). The expression pattern of the CBL10 gene from A. arabicum was opposite to what was seen for the CBL10b and AtCBL10 genes; the transcript was high in roots (Fig. 5).
Fig. 5.
Expression of the CBL10a and CBL10b genes differs. (a) Transcript structure of the CBL10 genes from Arabidopsis (AtCBL10), E. salsugineum (EsCBL10a and EsCBL10b), S. parvula (SpCBL10a, SpCBL10b-1, and SpCBL10b-2), S. irio (SiCBL10a and SiCBL10b), and A. arabicum (AaCBL10). Black boxes, exons (size indicated below in bp); solid lines, introns (not drawn to scale); dotted lines, untranslated regions; gray boxes, primer annealing sites. Protein domains important for the function of AtCBL10 are indicated above the AtCBL10 transcript. HD, hydrophobic domain; Ca, calcium-binding domains; S, serine phosphorylation site. (b) CBL10 transcript accumulation in leaves (L) and roots (R) of 11-day-old seedlings grown on 0.25× MS medium. ACTIN2 (ACT2), loading control. One representative image of three replicates is shown.
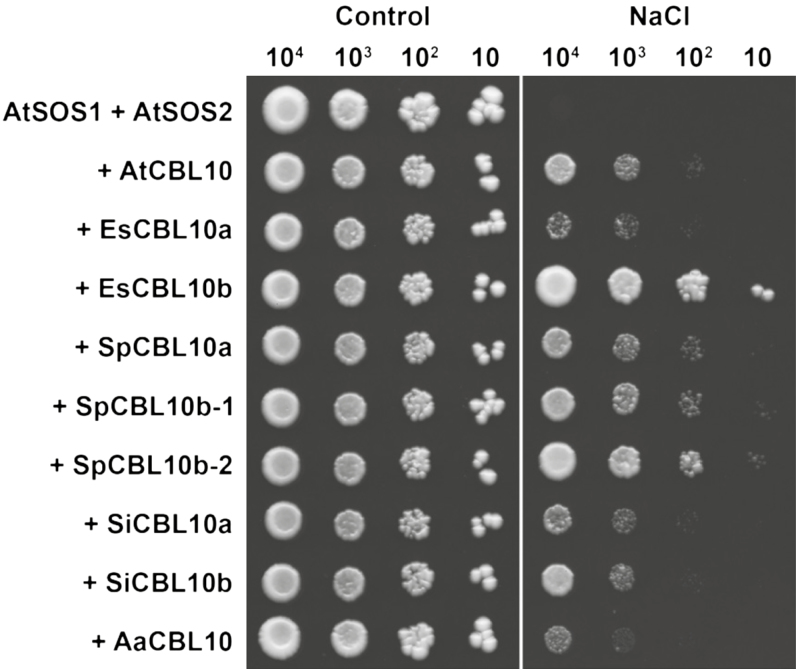
AtCBL10 has been shown to activate the SOS pathway which functions to prevent the toxic accumulation of sodium in the cytoplasm (Quan et al., 2007; Lin et al., 2009). In E. salsugineum, differences in CBL10 activation of the pathway were observed; EsCBL10b strongly activated the Arabidopsis and E. salsugineum SOS pathways, while EsCBL10a showed only weak activation (Monihan et al., 2019). To determine if the CBL10 proteins from S. irio, S. parvula, and A. arabicum function in the SOS pathway, each protein was expressed in a salt-sensitive strain of yeast (AXT3K) along with the Arabidopsis SOS2 protein kinase (AtSOS2) and SOS1 Na+/H+ exchanger (AtSOS1), and growth in the presence of salt was assessed as an indication of pathway activity. All CBL10 proteins activated the SOS pathway, but differences in the level of activation were observed. EsCBL10b had the greatest activity followed by the SpCBL10b and SpCBL10a proteins which all had greater activity than AtCBL10 (Fig. 6). The activities of SiCBL10a and SiCBL10b were similar to that of AtCBL10, while EsCBL10a and AaCBL10 had the weakest activity (Fig. 6).
Fig. 6.
SOS pathway activation is greatest with EsCBL10b. A salt-sensitive strain of S. cerevisiae (AXT3K, Δena1-4Δnha1Δnhx1) was transformed with SOS1 and SOS2 from Arabidopsis in combination with CBL10 from Arabidopsis (AtCBL10, At10), E. salsugineum (EsCBL10a, Es10a; and EsCBL10b, Es10b), S. parvula (SpCBL10a, Sp10a; SpCBL10b-1, Sp10b-1; and SpCBL10b-2, Sp10b-2), S. irio (SiCBL10a, Si10a; and SiCBL10b, Si10b), and A. arabicum (AaCBL10, Aa10). Serial decimal dilutions of yeast cells were spotted onto control medium or medium containing 125 mM NaCl. Two independently transformed colonies were assayed in three biological replicates; one representative image is shown.
EsCBL10a, but not AtCBL10 or EsCBL10b, can complement the salt-sensitive phenotype of the Arabidopsis sos3 mutant (Atsos3), suggesting that EsCBL10a has a distinct function (Monihan et al., 2019). To determine when this function arose within the Brassicaceae, the S. irio, S. parvula, and A. arabicum CBL10 genes were expressed in Atsos3 downstream of the CaMV 35S promoter and the salt tolerance of five independently transformed, single insertion, homozygous lines was determined. Four of the nine genes examined complemented Atsos3: EsCBL10a, SpCBL10b-1, SpCBL10b-2, and SiCBL10b (Fig. 7). These results indicate that at least one gene from each expanded lineage II species can perform a function that complements the Atsos3 salt-sensitive phenotype.
Fig. 7.
EsCBL10a and the CBL10b genes from S. parvula and S. irio complement the Atsos3 salt-sensitive phenotype. CBL10 genes from Arabidopsis (AtCBL10, At10), E. salsugineum (EsCBL10a, Es10a; and EsCBL10b, Es10b), S. parvula (SpCBL10a, Sp10a; SpCBL10b-1, Sp10b-1; and SpCBL10b-2, Sp10b-2), S. irio (SiCBL10a, Si10a; and SiCBL10b, Si10b), and A. arabicum (AaCBL10, Aa10) were expressed in the Atsos3 mutant, and growth in the absence (control) and presence of salt (75 mM NaCl) was monitored. For salt assays, seeds were germinated on medium without NaCl for 4 d, after which seedlings were transferred to medium without or with the indicated concentration of NaCl. After 10 d of treatment, photographs were taken and total seedling fresh weight was measured. (A) Photographs of the wild type (WT), Atsos3, and Atsos3 expressing each CBL10 gene (Atsos3;CBL10). The scale bar (1 cm, upper right panel) shows the magnification for all images. (B) Total seedling fresh weight was measured to quantify growth and is presented as a percentage of the control. Data are means ±SE of at least 24 seedlings per genotype grown in three independent experiments. *Complementation of the Atsos3 salt-sensitive phenotype (Tukey–Kramer HSD, P≤0.05).
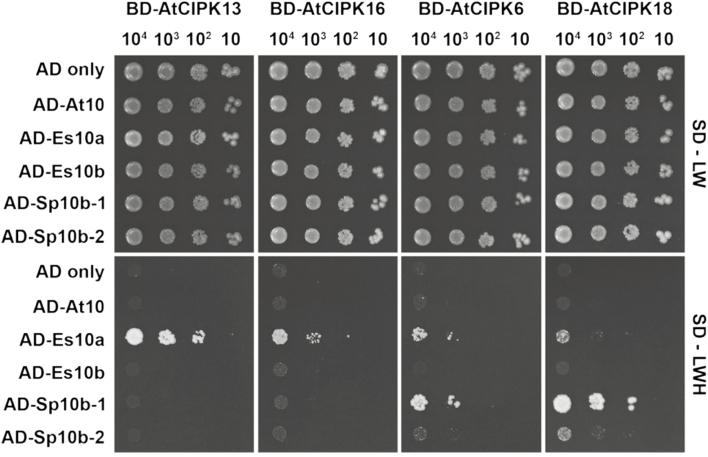
The ability of EsCBL10a to complement Atsos3 and its weak ability to activate Arabidopsis and E. salsugineum SOS2 and SOS1 in yeast suggested that it might perform its functions with a different kinase (Monihan et al., 2019). A yeast two-hybrid screen was performed with 26 kinases from the CIPK family to which AtSOS2 belongs. Four CIPKs (AtCIPK13, AtCIPK16, AtCIPK6, and AtCIPK18) were identified as interacting specifically with EsCBL10a but not AtCBL10 or EsCBL10b (Monihan et al., 2019). To determine if the CBL10b proteins from S. parvula might complement Atsos3 through a mechanism similar to EsCBL10a, interaction was tested between the SpCBL10b proteins and the four EsCBL10a-interacting CIPKs. SpCBL10b-1 interacted with two of the kinases, AtCIPK6 and AtCIPK18, while SpCBL10b-2 only interacted with AtCIPK18 (Fig. 8). In the opposite orientation, the S. parvula proteins self-activated, masking any interaction with the CIPK proteins, so interaction is only shown for the CIPK proteins fused to the GAL4 BD and the CBL10 proteins fused to the GAL4 AD.
Fig. 8.
The SpCBL10b proteins interact with two of the four EsCBL10a-interacting CIPKs. CBL10 proteins from Arabidopsis (AtCBL10, A10), E. salsugineum (EsCBL10a, Es10a; and EsCBL10b, Es10b), and S. parvula (SpCBL10b-1, Sp10b-1; and SpCBL10b-2, Sp10b-2) were fused to the GAL4 activation domain (AD) and interaction with the Arabidopsis CIPKs fused to the GAL4 binding domain (BD) was assessed using yeast two-hybrid assays. Serial decimal dilutions of diploid yeast harboring both constructs were spotted onto synthetic defined media (SD) minus leucine (L) and tryptophan (W), or minus LW and histidine (H). Two independently mated colonies were assayed in two biological replications; one representative image is shown.
Discussion
Cross-species analysis sheds light on the evolution of CBL10 duplication in the Brassicaceae
All of the CBL10 genes examined complemented the Atcbl10 salt-sensitive phenotype, indicating that at least a portion of CBL10 function is conserved throughout the Brassicaceae (Figs 4, 9). This is consistent with studies of CBL10 genes in Poplar trichocarpa and Solanum lycopersicum which were also able to complement Atcbl10, indicating that some of CBL10 function is conserved outside of the Brassicaceae (Tang et al., 2014; Egea et al., 2018). However, within the Brassicaceae, species-specific differences in CBL10 expression and function were detected.
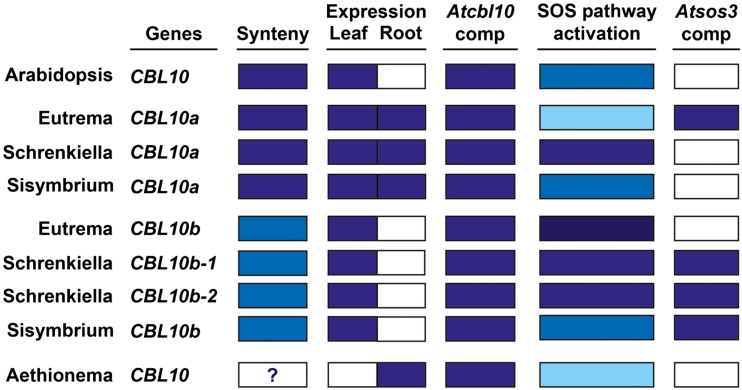
Fig. 9.
CBL10 function diverged in a species-specific manner. Synteny, genes with the same color box are syntenic (white box indicates unknown synteny; Fig. 2). Expression, presence of CBL10 transcript in leaves and roots (blue, present; white, absent; Fig. 5). Atcbl10 comp, ability to complement the Atcbl10 salt-sensitive phenotype (blue, complements; white, does not complement; Fig. 4). SOS pathway activation, ability to activate the Arabidopsis SOS pathway in yeast (strongest, strong, medium, and weak refer to the level of yeast growth correlating with strength of activation of the SOS pathway; Fig. 6). Atsos3 comp, ability to complement the Atsos3 salt-sensitive phenotype (blue, complements; white, does not complement; Fig. 7).
The absence of the CBL10a and CBL10b paralogs in members of lineage I, A. arabicum, T. hassleriana, and C. papaya in combination with the presence of CBL10a and CBL10b paralogs in multiple members of lineage II suggest that the duplication of CBL10 occurred within expanded lineage II of the Brassicaceae (Fig. 1). Lack of variation in the C-termini of the CBL10 nucleotide sequences (containing the four highly conserved EF-hand calcium-binding domains) reduced phylogenetic resolution, precluding assignment of the duplication to a specific branch of the tree.
At least one gene from each species within expanded lineage II has the ability to complement the Atsos3 salt-sensitive phenotype (Figs 7, 9). Whether the ability to complement Atsos3 evolved prior to or after subsequent speciation in the expanded lineage II species sampled remains less clear; however, there is support for the changes occurring after speciation. The ability of EsCBL10a to complement Atsos3 resides in the hydrophobic domain (Monihan et al., 2019). Conservation of amino acids in this domain in proteins that do not complement (AtCBL10, EsCBL10b, SpCBL10a, SiCBL10a, and AaCBL10) and variation in proteins that do (EsCBL10a, SpCBL10b-1, SpCBL10b-2, and SiCBL10b) suggests that sequences changed post-CBL10 duplication and after speciation (Supplementary Fig. S3). Additional studies will be required to determine if the other CBL10b proteins that complement Atsos3 do so because of changes in this or another region of the protein.
With an increase in sequenced genomes and tools to analyze those genomes, questions have emerged regarding how genomic context influences which gene changes function after a duplication event. To determine if the change in function is equally likely in both genes or more likely in the gene inserted into a new genomic position, the ratio of non-synonymous (dN) to synonymous (dS) substitutions (dN/dS) is often calculated. Studies have generally concluded that the duplicated gene in the new genomic position often has the faster rate of change and, as a result, is the one most likely to change function (Han et al., 2009; Dewey, 2011; Pegueroles et al., 2013; Rosello and Kondrashov, 2014). In keeping with these findings, the CBL10 genes in S. parvula and S. irio inserted into a new genomic position and acquired a function that complements Atsos3. However, E. salsugineum appears to be an exception to this generalization. EsCBL10a, which remained in the original genomic position, acquired the ability to complement Atsos3, while EsCBL10b, which inserted into a new genomic position, never acquired the function or acquired the function and subsequently lost it. In addition to influencing protein function, duplication and insertion of a gene into a new genomic context can lead to regulation by different cis-acting elements, suggesting that the gene that changes genomic position is more likely to have a different expression pattern (Flagel and Wendel, 2009; Pegueroles et al., 2013). Our current data do not support this model because the CBL10b genes inserted into a new genomic position yet they share a similar expression pattern with AtCBL10, while the CBL10a genes, which remained in the original genomic position, appear to have expanded expression into roots. Analysis of expression patterns from additional species will be needed to establish the major expression pattern.
Cross-species analysis sheds light on the molecular mechanism underlying CBL10 duplication in the Brassicaceae
Transposable elements have been found to replicate and integrate into genomes, leading to duplication of genes (Xiao et al., 2008; Flagel and Wendel, 2009; Panchy et al., 2016). Because numerous transposable elements were found surrounding the CBL10b genes in the species studied (Supplementary Fig. S2), replicative transposition by transposable elements is a likely mechanism underlying the duplication of CBL10. CBL10 duplication as a result of a polyploidization event is unlikely because no whole-genome duplication event is known to have occurred at the base of expanded lineage II and because, in the CBL10-containing regions of the genomes of species studied, CBL10 is the only duplicated gene (Fig. 2). Because the CBL10b genes identified contain introns and are located on a different chromosome from the CBL10a genes, the duplication probably did not arise through a retroduplication or an unequal crossover event.
In Arabidopsis, the CBL calcium sensors (10 members) and the CIPK protein kinases (26 members) form signaling networks that link changes in cytosolic calcium levels to physiological responses (Luan, 2009). The weak ability of EsCBL10a to activate the SOS pathway suggested that it might function with a CIPK protein other than SOS2 (CIPK24) to complement Atsos3 (Fig. 6; Monihan et al., 2019). Using yeast two-hybrid assays, four CIPK proteins that interact with EsCBL10a but not EsCBL10b or AtCBL10 were identified (Monihan et al., 2019). To begin to understand if EsCBL10a and the SpCBL10b proteins complement Atsos3 through a similar mechanism, yeast two-hybrid assays were used to examine interaction between the SpCBL10b proteins and the four EsCBL10a-interacting CIPK proteins. Only AtCIPK18 interacted with EsCBL10a and both SpCBL10b proteins; however, a role for AtCIPK18 in salt tolerance has not been reported and the Atcipk18 mutant does not have a salt-sensitive phenotype (Fig. 8; data not shown). SpCBL10b-1 but not SpCBL10b-2 interacted with AtCIPK6 which has been shown to interact with AtSOS3 to recruit the potassium transporter, AtAKT2, to the plasma membrane (Fig. 8; Held et al., 2011). The duplication appears to have expanded the range of CIPK interactions. Whether there is convergence on a specific CIPK remains an open question.
There are multiple ways in which CBL10 might contribute to salt tolerance in the Brassicaceae
All three Schrenkiella genes complement Atcbl10 (Figs 4, 9), but further examination of function revealed differences. In E. salsugineum, the CBL10 genes have different functions; EsCBL10b strongly activates the SOS pathway while EsCBL10a has an unknown function that allows it to complement Atsos3 (Monihan et al., 2019). In S. parvula, a different pattern is observed; SpCBL10a, like EsCBL10b, strongly activates the SOS pathway although not as well (Figs 6, 9), and does not have a function that complements Atsos3 (Figs 7, 9). SpCBL10b-1 and SpCBL10b-2 can both strongly activate the SOS pathway (like EsCBL10b although also not as well, Figs 6, 9) and complement the Atsos3 salt-sensitive phenotype (like EsCBL10a, Figs 7, 9). These results suggest that having three CBL10 genes with strong and overlapping functions has contributed to the ability of S. parvula to maintain growth in the presence of salt.
Eutrema salsugineum and S. irio have similar levels of growth in the presence of salt, the same number of CBL10 genes, similar CBL10 expression patterns, all CBL10 genes in the two species complement Atcbl10, and one of the two CBL10 genes in each species can complement Atsos3 (Figs 1–7). However, activation of the SOS pathway revealed differences in how CBL10 functions within these species. In E. salsugineum, EsCBL10b strongly activates the pathway while EsCBL10a does so only weakly. In S. irio, SiCBL10a and SiCBL10b both moderately activate the SOS pathway, suggesting that there are multiple ways to acquire salt tolerance (Figs 6, 9). As was found in E. salsugineum, the CBL10 genes in S. irio have diverged in function, probably increasing calcium-mediated signaling capacity and contributing to the ability of S. irio to grow in the presence of salt.
Aethionema arabicum is a member of the first diverging lineage of Brassicaceae and has only one CBL10 gene (Fig. 1). AaCBL10 may reside in a different genomic position; a scaffold syntenic to AtCBL10 but lacking AaCBL10 was identified in A. arabicum and AaCBL10 was found on a short scaffold (Figs 2, 9). A different genomic location for AaCBL10 might explain the altered expression of the transcript; while the CBL10 genes in the other species studied are expressed in shoots and some also in roots, AaCBL10 is the only gene expressed exclusively in roots (Figs 5, 9). Several results suggest that AaCBL10 may not play a significant role in the ability of A. arabicum to grow in the presence of salt: (i) A. arabicum is sensitive to salt, suggesting that it has few active mechanisms to deal with the presence of salt in the soil (Fig. 3); (ii) while AaCBL10 is functional in Atcbl10, it is probably complementing AtCBL10’s function in shoots (Figs 4, 9) and may not perform a similar function in A. arabicum roots; (iii) AaCBL10 is unable to complement the salt-sensitive phenotype of Atsos3 whose gene product functions in roots (Figs 7, 9; Quan et al., 2007); and (iv) AaCBL10 only weakly activates the SOS pathway (Figs 6, 9). Because calcium is an important signaling molecule in many different plant processes, AaCBL10 may function in other calcium-mediated responses in A. arabicum—a mutational analysis will be required to uncover the role of CBL10 in this species.
Taken together, results from this study have demonstrated that: the paralogous CBL10 genes within a species diverged in expression and function probably contributing to the maintenance of the duplicated gene pairs in their genomes; orthologous CBL10 genes have diverged in function in a species-specific manner, suggesting that the function of the genes was not established immediately after the duplication but after speciation; and that species studied with multiple CBL10 genes are better able to grow in the presence of salt.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Specificity of CBL10 gene expression in transgenic Arabidopsis.
Fig. S2. Transposable elements may have mediated the CBL10 duplication.
Fig. S3. Differences in CBL10 function reside in the N-terminus.
Table S1. Primers.
Table S2. Ratio of species growth in the absence and presence of salt.
Acknowledgements
We thank Brian M. Ortega and Katerina A. Oskolkoff for technical assistance, and Dr Steven E. Smith (School of Natural Resources and the Environment, University of Arizona) for statistical analyses. This work was supported by the United States National Science Foundation, Division of Integrative Organismal Systems Award 1552099 (to KSS).
Author contributions
SMM and KSS designed the research; SMM, CAM, CHR, and MMM performed the experiments; SMM, KSS, MMM, and MAB analyzed the data; and SMM and KSS wrote the manuscript with input from all co-authors.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Argout X, Salse J, Aury JM, et al. 2011. The genome of Theobroma cacao. Nature Genetics 43, 101–108. [DOI] [PubMed] [Google Scholar]
- Bartlett MS. 1937. Properties of sufficiency and statistical tests. Proceedings of the Royal Society A: Mathematical and Physical Sciences 160, 268–282. [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. 2006. Brassicaceae phylogeny and trichome evolution. American Journal of Botany 93, 607–619. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, et al. 2013. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. The Plant Cell 25, 2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conover WJ, Iman RL. 1981. Rank transformations as a bridge between parametric and nonparametric statistics. American Statistician 35, 124–129. [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, et al. 2011. The genome of the extremophile crucifer Thellungiella parvula. Nature Genetics 43, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CN. 2011. Positional orthology: putting genomic evolutionary relationships into context. Briefings in Bioinformatics 12, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Pineda B, Ortíz-Atienza A, et al. 2018. The SlCBL10 calcineurin B-like protein ensures plant growth under salt stress by regulating Na+ and Ca2+ homeostasis. Plant Physiology 176, 1676–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist 183, 557–564. [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK. 2000. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proceedings of the National Academy of Sciences, USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. 2009. Adaptive evolution of young gene duplicates in mammals. Genome Research 19, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, et al. 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nature Genetics 45, 891–898. [DOI] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, et al. 2011. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Research 21, 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. 1938. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular 347, 1–39. [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, et al. 2011. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics 43, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. [DOI] [PubMed] [Google Scholar]
- Jarvis DE, Ryu CH, Beilstein MA, Schumaker KS. 2014. Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula. Molecular Biology and Evolution 31, 2094–2107. [DOI] [PubMed] [Google Scholar]
- Kagale S, Koh C, Nixon J, et al. 2014. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Communications 5, 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U, Lydiate DJ. 1996. Comparative genome mapping in Brassica. Genetics 144, 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H. 1960. Robust tests for equality of variances. In: Olkin I, ed. Contributions to probability and statistics. Stanford, CA: Stanford University Press, 278–292. [Google Scholar]
- Lin H, Yang Y, Quan R, Mendoza I, Wu Y, Du W, Zhao S, Schumaker KS, Pardo JM, Guo Y. 2009. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. The Plant Cell 21, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. 1997. An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proceedings of the National Academy of Sciences, USA 94, 14960–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK. 1998. A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, et al. 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. 2009. The CBL–CIPK network in plant calcium signaling. Trends in Plant Science 14, 37–42. [DOI] [PubMed] [Google Scholar]
- Lyons E, Freeling M. 2008. How to usefully compare homologous plant genes and chromosomes as DNA sequences. The Plant Journal 53, 661–673. [DOI] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, et al. 2008. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiology 148, 1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang J, Zhou G, et al. 2013. Genomic insights into salt adaptation in a desert poplar. Nature Communications 4, 2797. [DOI] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, et al. 2008. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, Conner JK, Shiu SH. 2014. Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. The Plant Cell 26, 1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monihan SM, Magness CA, Yadegari R, Smith SE, Schumaker KS. 2016. Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiology 171, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monihan SM, Ryu CH, Magness CA, Schumaker KS. 2019. Linking duplication of a calcium sensor to salt tolerance in Eutrema salsugineum. Plant Physiology 179, 1176–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Hong H, Lee SY, Yun DJ, Bohnert HJ, Dassanayake M. 2014. Genome structures and transcriptomes signify niche adaptation for the multiple-ion-tolerant extremophyte Schrenkiella parvula. Plant Physiology 164, 2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Albà MM. 2013. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Molecular Biology and Evolution 30, 1830–1842. [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y. 2007. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. The Plant Cell 19, 1415–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. 2002. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proceedings of the National Academy of Sciences, USA 99, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. 1995. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters 370, 264–268. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Ramos J. 1984. Dual system for potassium transport in Saccharomyces cerevisiae. Journal of Bacteriology 159, 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselló OPI, Kondrashov FA. 2014. Long-term asymmetrical acceleration of protein evolution after gene duplication. Genome Biology and Evolution 6, 1949–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. 1965. An analysis of variance test for normality (Complete Samples). Biometrika 52, 591. [Google Scholar]
- Slotte T, Hazzouri KM, Ågren JA, et al. 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nature Genetics 45, 831–835. [DOI] [PubMed] [Google Scholar]
- Tang RJ, Yang Y, Yang L, Liu H, Wang CT, Yu MM, Gao XS, Zhang HX. 2014. Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant, Cell & Environment 37, 573–588. [DOI] [PubMed] [Google Scholar]
- Teh BT, Lim K, Yong CH, et al. 2017. The draft genome of tropical fruit durian (Durio zibethinus). Nature Genetics 49, 1633–1641. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Windsor AJ, Schranz ME, Formanová N, Gebauer-Jung S, Bishop JG, Schnabelrauch D, Kroymann J, Mitchell-Olds T. 2006. Partial shotgun sequencing of the Boechera stricta genome reveals extensive microsynteny and promoter conservation with Arabidopsis. Plant Physiology 140, 1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. 2008. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530. [DOI] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, et al. 2013. The reference genome of the halophytic plant Eutrema salsugineum. Frontiers in Plant Science 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen W, Sun L, et al. 2012. The genome of Prunus mume. Nature Communications 3, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.