Abstract
Non-alcoholic steatohepatitis (NASH) is characterized by macrovesicular steatosis with ballooning degeneration of hepatocytes, diffused lobular inflammation, and fibrosis. PPAR ligands are promising therapeutic agents in NASH; accordingly, we evaluated the effects of the first clinically available selective PPARα modulator, pemafibrate. We found that pemafibrate improves F4/80-positive macrophage accumulation, ballooning degeneration of hepatocytes, and the non-alcoholic fatty liver disease (NAFLD) activity score without affecting triglyceride (TG) accumulation in the liver of a mouse model of NASH (STAM). A global gene expression analysis indicated that pemafibrate enhances TG hydrolysis and fatty acid β-oxidation as well as re-esterification from dihydroxyacetone 3-phosphate and monoacylglycerol to TG. These changes are accompanied by the induction of genes involved in lipolysis and lipid droplet formation, along with an increased number and reduced size of lipid droplets in pemafibrate-treated livers. Pemafibrate reduced the expression of the cell adhesion molecule Vcam-1, myeloid cell markers, and inflammation- and fibrosis-related genes in STAM mice. Furthermore, pemafibrate significantly reduced VCAM-1 expression induced by high glucose in cultured human umbilical vein endothelial cells. These results suggest that pemafibrate prevents NASH development by reducing myeloid cell recruitment via interactions with liver sinusoidal endothelial cells, without altering hepatic TG accumulation.
Subject terms: Drug discovery, Diseases, Endocrinology, Molecular medicine
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most common cause of chronic liver disease and is closely linked to metabolic syndrome. Current estimates indicate that up to 30% of the general population is affected by NAFLD in industrialized countries1. Furthermore, 5–10% of patients with NAFLD can progress to non-alcoholic steatohepatitis (NASH), a more severe form of NAFLD that is broadly defined by the presence of steatosis and inflammation with ballooning, regardless of fibrosis, and eventually to cirrhosis and hepatocellular carcinoma2–4.
Hepatic TG accumulation has been suggested to play a central role in NASH development, but additional factors, such as insulin resistance, oxidative stress, ER stress, and mitochondrial dysfunction may also be involved5,6. However, the mechanisms underlying the pathogenesis of NASH for the subset of patients with steatosis has not been clarified. Additionally, no therapeutic agent has been approved for NASH. Therefore, there is an urgent need to develop an effective therapeutic approach for NASH.
Liver fatty acids and TG metabolism are tightly regulated by a balance of de novo lipogenesis (DNL), glyceroneogenesis, VLDL assembly and secretion, lipolysis, and fatty acid oxidation (FAO) at the transcriptional and post-transcriptional levels7,8. DNL is mainly transcriptionally regulated by sterol regulatory element binding protein 1c (SREBP1c) and carbohydrate response element binding protein (ChREBP), which are activated by increases in insulin signaling and glucose levels, respectively. PPARα induces hepatic FAO genes in the fasting state. Several studies have indicated that impaired PPARα function and FAO are major determinants of NASH development9,10. Therefore, PPARα ligands are considered candidate therapeutic agents for NASH.
Pemafibrate (also known as K-877), approved in Japan, is expected to replace fibrates as the first clinically available selective PPARα modulator (SPPARMα) to improve dyslipidemia and reduce macro- and micro-vascular complications11,12. Pemafibrate has greater PPARα activation potency than those of other fibrates with a lower EC50 value and a high degree of subtype selectivity (>2,000-fold subtype selectivity)13. In preclinical and clinical studies, pemafibrate shows greater plasma TG lowering and HDL-cholesterol elevating effects than those of other fibrates on the market14,15. We have reported that pemafibrate induces a series of PPARα target genes involved in TG hydrolysis, fatty acid uptake, fatty acid β-oxidation, and ketogenesis in the liver, supporting its ability to reduce plasma TG13. Recently, Honda et al. reported that pemafibrate treatment improves obesity, dyslipidemia, liver dysfunction, and the pathological condition of AMLN diet-induced NASH model16. While AMLN model showed histologic and metabolic features of human NASH with diffuse fibrosis, this model did not induce cirrhosis, hyperglycemia and hypertriglyceridemia17. In this study, we tested the therapeutic potential of pemafibrate in STAM NASH model mice, which showed diabetes-based NASH-HCC within 20 weeks of age18.
Results
Pemafibrate prevents NASH development
To investigate the protective effects of pemafibrate against NASH, we used the STAM mouse model (induced NASH in male C57BL/6J strain)18. STAM mice showed significant hyperglycemia, hypertriglyceridemia, higher NEFA and ALT levels, lower body weights, and higher liver weights and relative liver weights than those of normal C57BL/6J mice (Table 1). Obvious difference in food intake was not observed between STAM control (2.70 g/day) and pemafibrate-treated group (2.65 g/day). Pemafibrate effectively reduced serum TG and NEFA levels but did not alter the serum glucose, AST, and ALT levels. In addition, pemafibrate significantly increased liver weight and relative liver weight, which is a well-known effect in response to PPARα stimulation in rodents19.
Table 1.
Effects of pemafibrate on body and liver weight, biochemical parameters in the serum, immunohistochemical analysis, and NAS.
| Normal | STAM | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vehicle | Pemafibrate | ||||||||
| Body weight (g) | 23.6 | ± | 0.3** | 17.4 | ± | 0.8 | 18.4 | ± | 0.5 |
| Liver weight (g) | 1.0 | ± | 0.0** | 1.3 | ± | 0.0 | 1.8 | ± | 0.0** |
| Relative liver weight (g liver/100 g Body weight) | 4.3 | ± | 0.1** | 7.6 | ± | 0.4 | 9.7 | ± | 0.3** |
| AST (U/l) | 104.2 | ± | 6.4 | 229.2 | ± | 79.5 | 173.3 | ± | 7.5 |
| ALT (U/l) | 27.8 | ± | 2.0* | 90.7 | ± | 35.1 | 75.5 | ± | 6.2 |
| Glucose (mg/dL) | 140.4 | ± | 8.6** | 491.1 | ± | 22.7 | 511.1 | ± | 21.1 |
| NEFA (mEq/l) | 0.7 | ± | 0.0** | 2.3 | ± | 0.4 | 1.1 | ± | 0.1** |
| Triglyceride (mg/dL) | 105.4 | ± | 7.5** | 867.0 | ± | 101.6 | 270.2 | ± | 49.9** |
| Total Cholesterol (mg/dL) | 80.5 | ± | 4.5** | 178.7 | ± | 9.0 | 191.0 | ± | 9.8 |
| F4/80 positive area (%) | 2.4 | ± | 0.1** | 5.9 | ± | 0.6 | 4.3 | ± | 0.3* |
| ER-TR7 positive area (%) | 1.6 | ± | 0.1* | 3.1 | ± | 0.6 | 2.2 | ± | 0.3 |
| Sirius red positive area (%) | 0.3 | ± | 0.0** | 0.5 | ± | 0.1 | 0.4 | ± | 0.0 |
| Steatosis | 0.0 | ± | 0.0** | 1.8 | ± | 0.3 | 1.8 | ± | 0.2 |
| Inflammation | 0.0 | ± | 0.0** | 1.8 | ± | 0.2 | 1.7 | ± | 0.2 |
| Ballooning | 0.0 | ± | 0.0** | 1.5 | ± | 0.2 | 0.3 | ± | 0.2** |
| NAS | 0.0 | ± | 0.0** | 5.2 | ± | 0.6 | 3.8 | ± | 0.5* |
AST: aspartate aminotransferase, ALT: alanine aminotransferase, NEFA: non-esterified fatty acid, NAS: NAFLD activity score, n = 6 animals per group. Error bars show s.e.m. *P < 0.05; **P < 0.01: Significantly difference from STAM control group by Dunnett’s multiple comparison test.
Representative macroscopic images and microscopic H&E-stained liver sections from STAM control mice exhibited liver nodules, macro- and micro-vesicular lipid accumulation, inflammatory cell infiltration, and ballooning degeneration of hepatocytes, unlike in normal mice (Fig. 1). Pemafibrate-treated mice showed less macrovesicular lipid accumulation, less ballooning degeneration, and a significantly lower NAFLD activity score (NAS)20 than those of STAM control mice (Table 1).
Figure 1.

Pemafibrate improves macrovesicular steatosis and F4/80 positive cell accumulation in STAM mice liver. Representative gross morphology of liver, H&E stained, F4/80 stained, ER-TR7 stained, and Sirius-red stained liver section from normal, vehicle, and pemafibrate treated STAM mice.
ER-TR7 and Sirius Red staining demonstrated pericellular collagen deposition and fibrosis around central veins in STAM control mice (Fig. 1 and Table 1). Pemafibrate did not significantly improve the ER-TR7- and Sirius Red-positive areas. Marked lobular F4/80+ macrophage accumulation was observed in the STAM control livers. The pemafibrate-treated group showed a significantly smaller F4/80+ area than that of the STAM control group (Fig. 1 and Table 1).
Pemafibrate induces TG synthesis from DHAP and glycerol and re-esterification of TG in STAM mouse livers
Although pemafibrate effectively reduced macrovesicular steatosis, it did not improve the steatosis score (Table 1). To better understand the effect of pemafibrate, we used Oil Red-O staining to evaluate the TG concentration in the liver (Fig. 2A–C). The STAM control group showed a significantly increased area of fat deposition and higher TG content in the liver. There were no significant differences in the area of fat deposition and TG content between the STAM control group and pemafibrate-treated group (Fig. 2A–C).
Figure 2.
Pemafibrate induces TG synthesis in STAM mice liver. (A) Oil-Red-O stained liver section from normal, vehicle, and pemafibrate treated STAM mice. (B) Quantification of oil-red-O staining (n = 6). (C) TG contents in liver (n = 6). (D) Schematic representation of the glycolytic and TG synthesis pathways in the liver (E) qPCR validation of glycolytic and TG synthesis pathways (n = 6). (F) Immunoblots for TG metabolism-related proteins in liver extracts. Error bars show s.e.m. *P < 0.05; **P < 0.01: Significantly difference from STAM control group by Dunnett’s multiple comparison test.
To verify the effect of pemafibrate on STAM mouse livers, we performed a comprehensive transcriptome analysis by RNA-seq using liver tissues collected from normal, STAM control, and pemafibrate-treated STAM mice. We identified 187 up-regulated and 477 down-regulated genes in the pemafibrate-treated compared with the STAM control group by our stringent criteria (Supplementary Table 1). Using the web-based tool FuncAssociate 2.1 (http://llama.mshri.on.ca/funcassociate/) for a GO analysis, the upregulated genes were enriched for functions related to lipid metabolic processes and the downregulated genes were enriched for immune system processes (Supplementary Table 2). In fact, PPARα–regulated FAO-related genes were significantly induced in the pemafibrate-treated group (Supplementary Fig. 1).
To determine why pemafibrate did not reduce TG accumulation in STAM mouse livers, we further investigated its effect on lipid and carbohydrate metabolic gene expression. The expression levels of genes related to TG hydrolysis, fatty acid uptake, fatty acid activation, fatty acid binding, peroxisomal and mitochondrial oxidation, and ketogenesis were higher in the STAM control group than in the normal group (Supplementary Figs. 1 and 2). Pemafibrate apparently induced the expression of these genes. In particular, pemafibrate treatment resulted in the greatest increase in Pdk4 expression, suggesting that it mediates the suppression of glucose oxidation and preferential activation of fatty acid oxidation (Supplementary Figs. 1 and 2).
Increased glucose uptake in hepatocytes promotes glycolysis and lipogenesis to generate TG. In eukaryotes, the glycerolipid synthesis pathway (glyceroneogenesis) and the monoacylglycerol pathway play central roles in TG synthesis (Fig. 2D)21,22. The STAM control group showed higher levels of glycolysis-related gene expression than those in the normal group (Supplementary Fig. 3). In addition, we found that levels of Pck1, which encodes a key enzyme that catalyzes the conversion of oxaloacetate to phosphoenolpyruvate23, were increased in the STAM group, indicating that enhanced glyceroneogenesis contributes to lipogenesis in STAM mouse livers (Fig. 2E,F and Supplementary Fig. 3). Moreover, re-esterification of 2-monoacylglycerol was induced in STAM control livers in addition to simultaneous TG uptake and lipolysis. Pemafibrate did not influence glycolysis and Pck1 expression but significantly induced a series of genes involved in TG synthesis from DHAP and glycerol (Fig. 2E,F). Pemafibrate had the greatest effect on Mogat1, which has key roles in TG re-esterification from monoacylglycerols and diacylglycerols generated by TG hydrolysis24 in STAM mouse livers (Fig. 2E,F). These results suggest that pemafibrate enhances TG synthesis from DHAP and glycerol and the re-esterification of glycerol generated by TG hydrolysis in STAM mouse livers.
Pemafibrate increases microvesicular lipid droplets in STAM mouse livers
Because pemafibrate improved macrovesicular steatosis without reducing hepatic TG accumulation (Fig. 2), we next evaluated lipid droplet counts and size distributions. Pemafibrate treatment increased the droplet number and decreased the lipid droplet area (Fig. 3A,B). Pemafibrate increased the percentage of cells expressing small lipid droplets (<1 μm) from 34% in the STAM control to 40% and decreased large lipid droplets (>3 μm) from 18% in the STAM control to 11% (Fig. 3C). Lipid droplets consist of an inner core of neutral lipids including TG and sterol esters, a phospholipid monolayer, and lipid droplet-associated proteins (LDAPs)25,26. LDAPs influence lipid droplet function and dynamics; accordingly, we evaluated the effect of pemafibrate on LDAPs expression (Fig. 3D). The STAM control group showed increases in the expression of genes related to lipid droplet inner core lipid synthesis (Agpat6, Dgat1, and Acat1), formation (Agpat6, Acsl3, and Plin2), lipolysis (Pnpla2, Hsd17b11, and Abhd5), budding (Fitm2 and Bscl2), expansion (Arf1, Copa, Copb1, Copb2, Rab18, and Elmod2), and fusion (Cidea and Cidec). Pemafibrate further induced the Agpat6, Plin2, Pnpla2, Hsd17b11, Fitm2, Cidea, and Cidec expression. Importantly, changes in Pnpla2 mRNA expression level were reflected at the protein level in mice liver (Fig. 2F).
Figure 3.
Pemafibrate induces lipid droplets formation. (A) Quantification of lipid droplet number of vehicle and pemafibrate treated STAM mice. (B) Median lipid droplet area of vehicle and pemafibrate treated STAM mice. (C) Investigation of hepatic lipid droplet sizes in vehicle and pemafibrate treated STAM mice. (D) Heatmap of hierarchical clustering of LDAP and formation-related genes. Error bars show s.e.m. *P < 0.05; **P < 0.01: Significantly difference from STAM control group by Bonferoni’s multiple comparison test.
Pemafibrate reduces macrophage interactions with liver sinusoidal endothelial cells
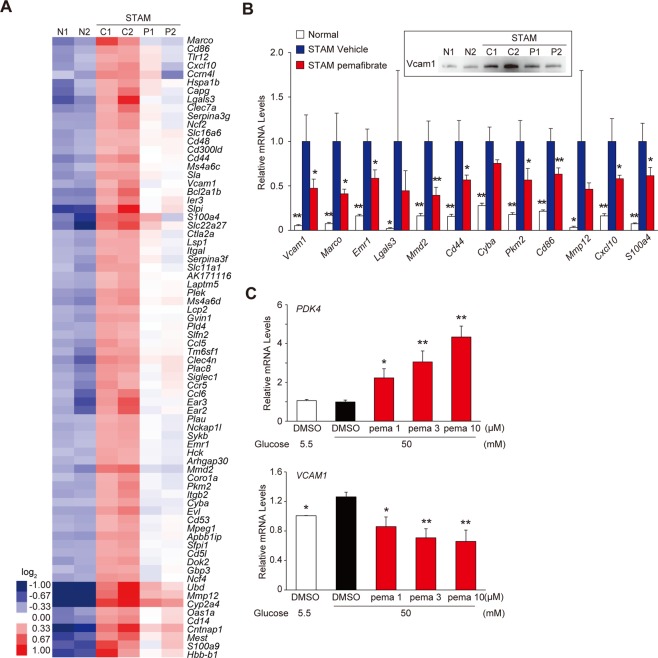
We further evaluated 74 of 473 genes that fulfilled more stringent criteria (FPKM of STAM control ≥3; STAM control/normal ratio ≥3; pemafibrate/STAM control ratio <2−0.6), as presented in a heat map in Fig. 4A. Livers from the STAM control group showed enhanced macrophage recruitment and inflammation. They expressed a number of polarization markers, including Marco, Emr1, Mmd2, Cd44, Pkm2, and Cd86. Oxidative stress and inflammatory responses have critical roles in NASH development. The NADPH oxidase components Cyba and Ncf4 and inflammatory factors Mmp12, Cxcl10, S100a4, and Lgals3 were highly induced in the STAM control mice and were significantly reduced in the pemafibrate-treated group. Resident tissue macrophages and monocyte-derived macrophages are important in chronic inflammatory processes. During inflammation, the induction of vascular cell adhesion molecule- 1 (VCAM-1) and CD31 is reported to promote the transendothelial migration of leucocytes27. Indeed, our transcriptome analysis indicated that Vcam1 levels are elevated in STAM control livers and are significantly reduced by pemafibrate treatment (Fig. 4B). These data suggested that pemafibrate prevents inflammatory monocyte recruitment and differentiation.
Figure 4.
Pemafibrate improves inflammatory genes expression in STAM mice liver. (A) Heatmap showing changes in expression of selected 74 genes. (B) qPCR validation of myeloid cell marker and inflammatory genes in the liver (n = 6). Immunoblot of Vcam1 protein is shown in upper boxed panel. Error bars show s.e.m. *P < 0.05; **P < 0.01. (C) HUVECs were cultured and treated with DMSO or pemafibare for 24 h in the presence of 50 mM Glucose. qPCR validation of PDK4 and VCAM1 expression (n = 3). Error bars show s.e.m. *P < 0.05; **P < 0.01: Significantly difference from STAM control group by Dunnett’s multiple comparison test.
Dysfunction of liver sinusoidal endothelial cells (LSECs) and the recruitment of inflammatory cells promote liver injury and inflammation27,28. In particular, high levels of glucose induce adhesion molecules, the transendothelial migration of monocytes, and monocyte-endothelial adhesion in human coronary artery endothelial cells and HUVECs29,30. We asked whether pemafibrate treatment prevents high-glucose-induced VCAM-1 expression in HUVECs. In accordance with previous reports, high glucose treatment induced VCAM1 expression in HUVECs (Fig. 4C). Pemafibrate dose-dependently reduced VCAM1 expression. This effect paralleled the induction of the well-known PPAR target PDK4. These data suggest that PPARα activation by pemafibrate in LSECs is pivotal for the prevention of NASH in STAM mouse livers.
Discussion
During NAFLD/NASH progression, lipotoxicity and interactions between hepatic myeloid cells and LSECs play pivotal roles31. Therefore, the STAM control of lipid flux to prevent hepatic TG accumulation and reduce hepatic inflammation is a potential therapeutic strategy for the prevention and reversion of NASH. We hypothesized that the activation of the SPPARMα pemafibrate would reduce hepatic TG accumulation and have therapeutic effects in NASH. Using the STAM mouse model, we found that therapeutic intervention with pemafibrate significantly improved the histological severity (ballooning and NAS) as well as inflammatory and fibrosis marker gene expression, without altering the hepatic TG content. A comprehensive transcriptome analysis revealed that pemafibrate decreases numerous immune-responsive genes in STAM mouse livers. In addition, pemafibrate reduced immune cell infiltration in diseased liver and high glucose-induced VCAM1 expression in HUVECs. These results suggest that PPARα activation by pemafibrate has therapeutic potential for NASH. In addition, the results emphasize the necessity of combinations of two or more drugs to modulate hepatic lipid flux and inflammation.
Under high fat and/or carbohydrate intake, lipogenesis is stimulated and excess fat is stored as TG. Excess TG accumulation in the liver results in hepatic steatosis, causing cellular lipotoxicity, fibrosis, and NASH8. This arises from an imbalance between TG accumulation (i.e., FA uptake and DNL) and removal (i.e., lipolysis and VLDL secretion)32. As indicated in Table 1, the STAM model was characterized by hyperglycemia and reduced body weight with a nearly complete loss of insulin production. Therefore, SREBP1c-mediated DNL is unlikely to contribute to TG accumulation in this model. In fact, a consistent increase in DNL was not observed in STAM model livers despite a substantial increase in genes encoding glycolytic enzymes (Supplementary Fig. 3). TG acquisition also involves fatty acid esterification pathways. The process of fatty acid esterification into TG initiates from the transfer of Acyl-CoA to the hydroxyl groups of glycerol-3-phosphate (G3P). G3P is derived from glucose via glycolysis, plasma glycerol, and de novo synthesis from pyruvate, lactate, or amino acids by glyceroneogenesis (a truncated version of gluconeogenesis)8. Recently, glyceroneogenesis has been recognized as an important pathway for G3P supply33 and a therapeutic target. Several reports have indicated that the liver utilizes mainly glycerol from glyceroneogenesis for TG synthesis in humans and rodents23,34,35. Our transcriptome analysis suggests that glyceroneogenesis is a G3P source for TG synthesis, as evidenced by the induction of higher levels of Pck1 (a rate-limiting enzyme in glyceroneogenesis) than Aqp9 or Gyk. In addition, we observed the substantial induction of genes related to TG hydrolysis (Lipe and Mgll) and re-esterification (Mogat1). Therefore, the enhancement of glyceroneogenesis and TG re-esterification are likely to play significant roles in TG accumulation in STAM mouse livers. PPARα is a well-established nutrient sensor for the induction of FAO and gluconeogenesis in the fasted liver36. In this study, PPARα activation by pemafibrate further induced FAO, TG hydrolysis, and re-esterification but did not influence glycolysis or the glyceroneogenesis rate-limiting enzyme Pck1 (Fig. 2E,F). Interestingly, various genes involved in TG synthesis from DHAP were significantly induced in pemafibrate-treated STAM mouse livers (Fig. 2E). As shown in Fig. 2F and Supplementary Figs. 1 and 2, pemafibrate had the greatest effect on Pdk4. Since Pdk4 is an inhibitor of glucose oxidation, PPARα activation by pemafibrate preferentially enhances fatty acid utilization for FAO and ketogenesis in the liver11,13. In contrast, our results suggested that the blocking of glucose oxidation by pemafibrate has a compensatory effect by enhancing the conversion of the excess glycolytic intermediate DHAP to TG synthesis in STAM mouse livers. Therefore, these results indicated that PPARα activation by pemafibrate simultaneously induces TG hydrolysis, FAO, TG synthesis from DHAP, and TG re-esterification under hyperglycemia and hyperlipidemia and thereby could not reduce TG accumulation in the STAM mouse model.
Macrovesicular steatosis (large droplet steatosis) is associated with the development of lobular inflammation and fibrosis in NASH8. Recent several reports have shed light on lipid droplet biology and its role in the pathogenesis of steatosis. A core of neutral lipids (i.e. TG and sterol esters) is surrounded by a phospholipid monolayer and LDAPs, and these have a key role in lipid droplet biology37. Neutral lipids of lipid droplets (i.e., TG and cholesterol ester) are synthesized by DGATs and ACATs residing primarily in the ER38. In addition, perilipins, seipin (encoded by Bscl2), and FITM2 are involved in lipid droplet formation and budding to the cytoplasm25,26. Furthermore, large lipid droplets are formed by coalescence or through cell death-inducing DEF45-like effector (CIDE) family protein-mediated enhancement of neutral lipids diffusion from one lipid droplet to another39. In this study, pemafibrate resulted in microvesicular steatosis with an increased number of small lipid droplets compared with those in STAM mouse livers. Our transcriptome analysis indicated that pemafibrate enhances inner core lipid synthesis and formation (Agpat6 and Dgat1), budding (Fitm2), and fusion (Cidea, and Cidec) genes expression (Fig. 3D). Recent evidence suggests that DGAT1 is responsible for the esterification of excess fatty acids40 and prevent lipotoxicity41. In addition, several reports indicated that CIDEA and CIDEC are both involved in the formation of large lipid droplet and induced in the hepatic steatosis42. As seen in adipocytes, the CIDE family proteins have been proposed to have a similar role in hepatocytes to promote lipid droplet formation43. CIDEA and CIDEC are strongly expressed in brown adipocytes and white adipocytes, and there multiolocular small lipid droplet formation by CIDEA and unilocular lipid droplet formation by CIDEC are linked with lipolysis and lipid storage, respectively. Thus, pemafibrate-induced Cidea expression may modulates lipid droplet size and basal lipolysis. In fact, pemafibrate also induced Pnpla2 which catalyze TG hydrolysis on the lipid droplet surface to generate diacylglycerol and free fatty acids44. These results suggest that pemafibrate induces DGAT1-dependent TG re-esterification, initial lipid droplet formation and budding may explain the increased number of lipid droplets. Moreover, pemafibrate-induced Cidea and Pnpla2 expression may explain a part of the prevention of larger lipid droplet formation in STAM mouse livers.
Lipotoxicity of hepatocytes initiate inflammatory cascades via the secretion of CXCL10-enriched extracellular vesicles, thereby inducing macrophage chemotaxis45. In addition, hepatic macrophage infiltration under steatohepatitis is induced by the interaction between myeloid cells and LSECs46. This interaction is mediated by cell adhesion molecules, such as VCAM1 and ICAM1, on LSECs47–49. In response to exposure to excess fatty acids, Kupffer cells and recruited macrophages undergo polarization to the M1 phenotype, characterized by increased production of cytokines, such as TNFα, IL6, IL-1β, and CCL547,49,50. These cytokines further activate hepatic stellate cells (HSCs), which in turn secrete proinflammatory cytokines, such as IL34, CCL5, and CCL2051–53. Furthermore, activated HSCs cause LSEC capillarization and ROS production46,54. Thus, a feed-forward activation loop of macrophages, HSCs, and LSECs plays a critical role in the liver fibrosis. Importantly, recent findings suggest that LSEC injury precedes hepatic inflammation and fibrosis in NASH31. In this study, we found that pemafibrate reduces the expression of polarization, inflammation, and fibrosis marker genes along with Vcam1 in STAM mouse livers (Fig. 4A,B). In addition, pemafibrate reduced high glucose-induced VCAM1 expression in HUVECs (Fig. 4C). Therefore, although mechanisms underlying LSEC dysfunction are still unknown, anti-inflammatory effects of pemafibrate may be explained its direct and/or indirect protective effect on LSECs.
The STAM model of NASH18 is characterized by the nearly complete loss of pancreatic insulin production with severe hyperglycemia indicating this model represents diabetes-based NASH patients. It may not completely reflect the hepatic nutrient status of human NASH livers because human NASH is closely linked to type 2 diabetes. In fact, 3-fold higher levels of DNL have been detected in patients with NAFLD than in healthy individuals55, but apparent induction of DNL was not observed in STAM mouse liver. Therefore, additional studies using other NASH models with obesity and insulin resistance are warranted to elucidate the mechanism by which pemafibrate effects NAFLD/NASH. Interestingly, pemafibrate has been reported to improve NASH development in another diet-induced NASH model, AMLN mice16. AMLN mice showed pronounced hepatomegaly and intrahepatic lipid accumulation, but not hypertriglyceridemia possibly due to suppressed hepatic triglyceride secretion by high dietary cholesterol intake56. Pemafibrate significantly improves not only liver TG accumulation, but also fasting plasma glucose and insulin levels in AMLN mice. Although precise mechanism of this action is still largely unknown, these observations suggest that pemafibrate potential to improve steatosis in type 2 diabetes patients. Furthermore, these differences on drug responses can be observed in clinically, which reflects on the background differences among patients. Overall, our data combined with previous observations suggest that although PPARα activation reduced macrovesicular steatosis and lipotoxicity via the reduction of excess free fatty acids by enhancing re-esterification and lipid droplet formation, PPARα agonists may not result in a sufficient TG reduction in diabetes-based NAFLD/NASH livers because PPARα is mainly involved in the regulation of nutrient flux to supply glucose and ketone bodies to peripheral tissues, rather than to provide an energy source in the liver. Furthermore, this study suggests that a multidrug intervention against TG accumulation and NAFLD/NASH progression is necessary. In particular, a combination of pemafibrate and drugs that enhance the excretion or inhibit the absorption of carbohydrates and lipids (e.g., an SGLT2 inhibitor, α-glucosidase inhibitor, or pancreatic intestinal lipase inhibitor) has the potential to improve TG accumulation and inflammation in NASH livers.
Methods
Chemicals
Pemafibrate was kindly provided from Kowa Co., Ltd. (Nagoya, Japan). Streptozotocin (STZ) was purchased from Sigma Aldrich (MO, USA). Arabic gum was obtained from Wako Pure Chemical Industries (Osaka Japan).
Animals
Pathogen-free 14-day pregnant C57BL/6J mice were purchased from CLEA Japan (Tokyo, Japan). The STAM mouse NASH model was induced in male mice by a single subcutaneous injection of 200 μg STZ (Sigma, MO, USA) at 2 days after birth and feeding with HFD32 (32%fat, CLEA Japan) ad libitum after 4 weeks of age. Mice at 6 weeks old were randomly divided into two groups: STAM control group fed a HFD32 with vehicle (3% arabic gum in distilled water) treatment and pemafibrate-treated group fed a HFD32 with pemafibrate (0.1 mg/kg in vehicle) treatment for 3 weeks (6–9 weeks). Also, a normal group fed a normal chow (CE-2; 5% fat, CLEA Japan) without STZ injection treated with vehicle for 3 weeks. Pemafibrate or vehicle were given at 5 ml/kg of body weight by oral administration between 0930 and 1000 hours57. Mice were sacrificed at 4-hour after the final administration with fasting, and parameters related to fatty liver disease were assessed. All mice were housed in a temperature-controlled (24 °C) facility with a 12-hour light/12-hour dark cycle (0800–2000 hours) and ad libitum access to food and water. The study protocol was approved in accordance with the relevant guidelines and regulations by the Animal Care and Use Committee of the University of Tokyo.
Blood parameter
Serum levels of total cholesterol, TG, glucose, free fatty acids, AST, ALT, phospholipids, and creatinine were determined using a Labospect 003 autoanalyzer (Hitachi High-Technologies Corporation, Tokyo, Japan).
Histology
For immunohistochemistry, endogenous peroxidase activity was blocked using 0.03% H2O2 in Methanol. The sections were incubated with the optimal dilutions of anti-F4/80 (AbD Serotec, Oxford, UK), ER-TR7 (Abcam, Cambridge, MA, USA) antibodies overnight at 4 °C. After incubation with appropriate secondary antibodies, substrate reaction was performed using 3,3′-Diaminobenzidine (Dojindo, Kumamoto, Japan) solution.
NAFLD activity score was calculated according to Kleiner et al.20. For qualitative assessment of Oil Red O, positive areas were scored for 5 grades by microscopy. For quantitative analysis of F4/80, ER-TR7 and Sirius red positive areas, bright field images of stained sections were captured using a digital camera (DP72, Olympus, Tokyo, Japan) around central veins at 400-fold magnification, and the positive areas in 5 fields were measured using WinROOF image processing software (Mitani, Tokyo, Japan). The results were determined as the means of five different fields of each section.
RNA-sequencing
The quality of the RNA was assessed by Nanodrop measurement (Thermo Fisher Scientific)58. RNA-sequencing (RNA-Seq) libraries were prepared by TruSeq Rapid PE Cluster Kit and TruSeq Rapid SBS kit (Illumina). The libraries were sequenced on Illumina HiSeq. 2500 using a read length of 2 × 150 bp. CASAVA v1.8.2 was conducted and RNA-seq reads were aligned to mouse transcriptome (UCSC gene) and genome (NCBI37/mm9) references respectively using Burrows-Wheeler Aligner. After transcript coordinate was converted to genomic positions, an optimal mapping result was selected either from transcript or genome mapping by comparing the minimal edit distance to the reference. Local realignment was performed within in-house short reads aligner with smaller k-mer size (k = 11). Finally, fragments per kilo base of exon per million fragments mapped (fpkm) values were calculated for each UCSC gene while considering strand-specific information.
Quantitative real-time PCR (qPCR)
First-strand cDNA was synthesized from total RNA with oligo dT primers using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific)59. The qPCR was performed by SYBR green PCR Master Mix (Thermo Fisher Scientific). The qPCR was carried out in 384-well plates using CFX384 Real-Time system (Bio-Rad). All reactions were performed in triplicate. The relative amount of all mRNAs was calculated using the comparative CT method. Ppib (mouse) and PPIA (human) mRNA were used as the invariant control. Primers used for qPCR are listed in Supplementary Tables S3 and S4.
Immunoblotting
Harvested liver tissues were homogenated in RIPA buffer (Thermo Fisher Scientific, Tokyo, Japan) supplemented with protease inhibitor cocktail (Sigma-Aldrich, MO, USA). The protein concentration was determined by the Bradford method using protein assay dye reagent concentrate (Bio-Rad, Tokyo, Japan). Whole cell samples were resolved by SDS-polyacrylamide gel electrophoresis, then electro-transferred to nitrocellulose membranes (Bio-Rad, Tokyo, Japan). Membrane were blocked with 5% bovine serum albumin (Wako pure chemical industries, Osaka, Japan) in PBS with 0.1% Tween-20 (Sigma-Aldrich, MO, USA) for 60 min at room temperature. The blot was probed with primary antibody for overnight at 4 °C and then incubated with anti-IgG horse-radish peroxidase-conjugated antibodies for 1 hour at room temperature. Proteins were detected using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific, Tokyo, Japan) according to the instructions of the manufacturer. Immunoreactive protein bands were documented using a Bio-Rad ChemiDoc XRS + system (Bio-Rad, Tokyo, Japan). Antibodies used for the immunoblot are listed in Supplementary Table S5.
Lipid droplet analysis
For hepatic lipid droplet analysis, “Image J” imaging software (https://imagej.nih.gov/ij/download.html) was used. H&E staining images were opened in Image J software, and converted into grayscale (8 bit). Then, lipid drop areas were extracted by setting threshold (Min: 220, Max: 255). After excluding blood vessels, lipid droplet areas were analyzed and quantified by “Analyze particles” function. Quantified lipid droplet area data was given by pixel and converted into μm2 (1 μm = 3 pixels: determined by scale bar size). Lipid droplet diameter was also calculated. Data were determined as mean values from three different images of each animal. Histogram was created by Microsoft Excel spreadsheet software.
Statistical analyses
All data are presented as mean ± s.e.m. The homogeneity in variance was evaluated by Bartlett test followed by parametric or non-parametric Dunnett’s multiple comparison test (one-side). The Bonferoni’s multiple comparison test (one-side) was used to compare the data between the STAM control and pemafibrate-treated groups. *P < 0.05, **P < 0.01.
Supplementary information
Acknowledgements
This work was supported from the Ministry of Education, Culture, Sports, Science, and Technology. This work was supported in part by a collaborative research fund from Kowa Co. Kowa Company, Ltd.
Author contributions
T.T., J.S., and T.K. planned experiments. T.T. and Y.S. wrote the manuscript. T.T., Y.S., and M.A. performed experiments. W.K. and K.M. assisted with the animal experiments and qPCR. M.A. performed the histological experiments. H.A. and S.Y. contributed to the RNA-sequencing data analysis. Y.M., T.O., M.A., and J.F. read and commented on the manuscript.
Competing interests
T.K. is an advisory board member and a recipient of support from a collaborative research fund from Kowa Company, Ltd. Kowa Co. had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. Y.S., M.A., and K.M. are employees of Kowa Company, Ltd. Rest of the authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yusuke Sasaki, Masato Asahiyama and Toshiya Tanaka.
Supplementary information
is available for this paper at 10.1038/s41598-020-64902-8.
References
- 1.Younossi Z, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018;15:11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Buzzetti E, et al. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Sutti, S. & Albano, E. Adaptive immunity: an emerging player in the progression of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 17, 81–92 (2020). [DOI] [PMC free article] [PubMed]
- 4.Suzuki A, Diehl AM. Nonalcoholic Steatohepatitis. Annu. Rev. Med. 2017;68:85–98. doi: 10.1146/annurev-med-051215-031109. [DOI] [PubMed] [Google Scholar]
- 5.Caligiuri A, et al. Molecular Pathogenesis of NASH. Int. J. Mol. Sci. 2016;17:1575. doi: 10.3390/ijms17091575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takaki A, et al. Multiple Hits, Including Oxidative Stress, as Pathogenesis and Treatment Target in Non-Alcoholic Steatohepatitis (NASH). Int. J. Mol. Sci. 2013;14:20704–20728. doi: 10.3390/ijms141020704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ress C, Kaser S. Mechanisms of intrahepatic triglyceride accumulation. World J. Gastroenterol. 2016;22:1664–1673. doi: 10.3748/wjg.v22.i4.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saponaro C, et al. The Subtle Balance between Lipolysis and Lipogenesis: A Critical Point in Metabolic Homeostasis. Nutrients. 2015;7:9453–9474. doi: 10.3390/nu7115475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montagner A, et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016;65:1202–1214. doi: 10.1136/gutjnl-2015-310798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francque S, et al. PPARα gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol. 2015;63:164–173. doi: 10.1016/j.jhep.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki Y, et al. Gene Expression Profiles Induced by a Novel Selective Peroxisome Proliferator-Activated Receptor α Modulator (SPPARMα) Pemafibrate. Int. J. Mol. Sci. 2019;20:5682–5699. doi: 10.3390/ijms20225682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fruchart JC, et al. The selective peroxisome proliferator-activated receptor α modulator (SPPARMα) paradigm: Conceptual framework and therapeutic potential: A consensus statement from the International Atherosclerosis Society (IAS) and the Residual Risk Reduction Initiative (R3i) Foundation. Cardiovasc. Diabetol. 2019;18:71. doi: 10.1186/s12933-019-0864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raza-Iqubal S, et al. Transcriptome Analysis of K-877 (a Novel Selective PPARα Modulator (SPPARMα))-Regulated Genes in Primary Human Hepatocytes and the Mouse Liver. J. Atheroscler. Thromb. 2015;22:754–772. doi: 10.5551/jat.28720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennuyer N, et al. The novel selective PPARα modulator (SPPARMα) pemafibrate improves dyslipidemia, enhances reverse cholesterol transport and decreases inflammation and atherosclerosis. Atherosclerosis. 2016;249:200–208. doi: 10.1016/j.atherosclerosis.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi S, et al. Efficacy and safety of pemafibrate (K-877), a selective peroxisome proliferator-activated receptor α modulator, in patients with dyslipidemia: Results from a 24-week, randomized, double blind, active-controlled, phase 3. trial. J. Clin. Lipidol. 2018;12:173–184. doi: 10.1016/j.jacl.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Honda Y, et al. Pemafibrate, a novel selective peroxisome proliferator-activated receptor alpha modulator, improves the pathogenesis in a rodent model of nonalcoholic steatohepatitis. Sci. Rep. 2017;7:42477. doi: 10.1038/srep42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman SL, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii M, et al. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med. Mol. Morphol. 2013;46:141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 19.Peraza MA, et al. The Toxicology of Ligands for Peroxisome Proliferator-Activated Receptors (PPAR) Toxicol. Sci. 2006;90:269–295. doi: 10.1093/toxsci/kfj062. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal AK, Garg A. Congenital generalized lipodystrophy: significance of triglyceride biosynthetic pathways. Trends Endocrinol. Metab. 2003;14:214–221. doi: 10.1016/S1043-2760(03)00078-X. [DOI] [PubMed] [Google Scholar]
- 22.Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/S0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D, et al. Early-life exposure to high-fat diet may predispose rats to gender-specific hepatic fat accumulation by programming Pepck expression. J Nutr Biochem. 2015;26:433–440. doi: 10.1016/j.jnutbio.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Lutkewitte AJ, et al. Hepatic monoacylglycerol acyltransferase 1 is induced by prolonged food deprivation to modulate the hepatic fasting response. J Lipid Res. 2019;60:528–538. doi: 10.1194/jlr.M089722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walther TC, et al. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 2017;33:491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olzamann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poisson J, et al. Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J Hepatol. 2017;66:212–227. doi: 10.1016/j.jhep.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Hammoutene A, Rautou PE. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J Hepatol. 2019;70:1278–1291. doi: 10.1016/j.jhep.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Li. Q, et al. GLP-1 Inhibits High-Glucose-Induced Oxidative Injury of Vascular Endothelial Cells. Sci. Rep. 2017;7:8008. doi: 10.1038/s41598-017-06712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, et al. Sphingosine-1-phosphate receptor 2 mediates endothelial cells dysfunction by PI3K-Akt pathway under high glucose condition. Eur. J. Pharmacol. 2016;776:19–25. doi: 10.1016/j.ejphar.2016.02.056. [DOI] [PubMed] [Google Scholar]
- 31.Miyao M, et al. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Lab. Invest. 2015;95:1130–1144. doi: 10.1038/labinvest.2015.95. [DOI] [PubMed] [Google Scholar]
- 32.Ipsen DH, et al. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 2018;75:3313–3327. doi: 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson RW, Reshef L. Glyceroneogenesis revisited. Biochimie. 2003;85:1199–1205. doi: 10.1016/j.biochi.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Kalhan SC, et al. Estimates of hepatic glyceroneogenesis in type 2 diabetes mellitus in humans. Metabolism. 2008;57:305–312. doi: 10.1016/j.metabol.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalhan SC, et al. Glyceroneogenesis and the source of glycerol for hepatic triacylglycerol synthesis in humans. J. Biol. Chem. 2001;276:12928–12931. doi: 10.1074/jbc.M006186200. [DOI] [PubMed] [Google Scholar]
- 36.Preidis GA, et al. Nutrient-sensing nuclear receptors PPARα and FXR control liver energy balance. J. Clin. Invest. 2017;127:1193–1201. doi: 10.1172/JCI88893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gluchowski NL, et al. Lipid droplets and liver disease: from basic biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2017;14:343–355. doi: 10.1038/nrgastro.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFie PJ, et al. Murine Diacylglycerol Acyltransferase-2 (DGAT2) Can Catalyze Triacylglycerol Synthesis and Promote Lipid Droplet Formation Independent of Its Localization to the Endoplasmic Reticulum. J. Biol. Chem. 2011;286:28235–28246. doi: 10.1074/jbc.M111.256008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilfling F, et al. Lipid Droplet Biogenesis. Curr. Opin. Cell Biol. 2014;29:39–45. doi: 10.1016/j.ceb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chitraju C, et al. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab. 2017;26:407–418. doi: 10.1016/j.cmet.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen TB, et al. DGAT1-Dependent Lipid Droplet Biogenesis Protects Mitochondrial Function during Starvation-Induced Autophagy. Dev. Cell. 2017;42:9–21. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao G, et al. Control of lipid droplet fusion and growth by CIDE family proteins. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids. 2017;1862:1197–1204. doi: 10.1016/j.bbalip.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Chen FJ, et al. CIDE family proteins control lipid homeostasis and the development of metabolic diseases. Traffic. 2020;21:94–105. doi: 10.1111/tra.12717. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi H, et al. Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. J. Cell Biochem. 2008;105:1430–6. doi: 10.1002/jcb.21964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibrahim SH, et al. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology. 2016;63:731–744. doi: 10.1002/hep.28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters KM, et al. Non-parenchymal hepatic cell lipotoxicity and the coordinated progression of non-alcoholic fatty liver disease and atherosclerosis. Curr. Opin. Lipidol. 2018;29:417–422. doi: 10.1097/MOL.0000000000000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyachi Y, et al. Roles for Cell-Cell Adhesion and Contact in Obesity-Induced Hepatic Myeloid Cell Accumulation and Glucose Intolerance. Cell Rep. 2017;18:2766–2779. doi: 10.1016/j.celrep.2017.02.039. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, et al. Role of sterile inflammation in fatty liver diseases. Liver Res. 2018;2:21–29. doi: 10.1016/j.livres.2018.02.003. [DOI] [Google Scholar]
- 49.Karasawa T, et al. Saturated Fatty Acids Undergo Intracellular Crystallization and Activate the NLRP3 Inflammasome in Macrophages. Arterioscler. Thromb. Vasc. Biol. 2018;38:744–756. doi: 10.1161/ATVBAHA.117.310581. [DOI] [PubMed] [Google Scholar]
- 50.Kazankov K, et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroentero.l Hepatol. 2019;16:145–159. doi: 10.1038/s41575-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 51.Shoji H, et al. Interleukin-34 as a fibroblast-derived marker of liver fibrosis in patients with non-alcoholic fatty liver disease. Sci. Rep. 2016;6:28814. doi: 10.1038/srep28814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim BM, et al. Hepatic stellate cells secrete Ccl5 to induce hepatocyte steatosis. Sci. Rep. 2018;8:7499. doi: 10.1038/s41598-018-25699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanson A, et al. Chemokine ligand 20 (CCL20) expression increases with NAFLD stage and hepatic stellate cell activation and is regulated by miR-590-5p. Cytokine. 2019;123:154789. doi: 10.1016/j.cyto.2019.154789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammoutene A, Rautou PE. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J. Hepatol. 2019;70:1278–1291. doi: 10.1016/j.jhep.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Lambert JE, et al. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014;146:726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henkel, J. et al. Induction of Steatohepatitis (NASH) with Insulin Resistance in Wild-type B6 Mice by a Western-type Diet Containing Soybean Oil and Cholesterol. Mol Med.23, 70–82 (2017). [DOI] [PMC free article] [PubMed]
- 57.Tanaka T, et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto R, et al. Overexpression of p54nrb/NONO induces differential EPHA6 splicing and contributes to castration-resistant prostate cancer growth. Oncotarget. 2018;9:10510–10524. doi: 10.18632/oncotarget.24063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka T, et al. PPARβ/δ activation of CD300a controls intestinal immunity. Sci. Rep. 2014;4:5412. doi: 10.1038/srep05412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.