Abstract
Purpose
The retinal rod outer segment (OS) disk membranes, devoid of mitochondria, conducts oxidative phosphorylation (OxPhos). This study aimed at identifying which proteins expressed in the retinal rod OS disks determined the considerable adenosine‐5'‐triphosphate production and oxygen consumption observed in comparison with retinal mitochondria.
Procedures
Characterization was conducted by immunogold transmission electron microscopy on retinal sections. OxPhos was studied by oximetry and luminometry. The proteomes of OS disks and mitochondria purified from bovine retinas were studied by mass spectrometry. Statistical and bioinformatic analyses were conducted by univariate, multivariate, and machine learning methods.
Results
Weighted gene coexpression network analysis identified two protein expression profile modules functionally associated with either retinal mitochondria or disk samples, in function of a strikingly different ability of each sample to utilized diverse substrate for F1Fo‐ATP synthase. The OS disk proteins correlated better than mitochondria with the tricarboxylic acids cycle and OxPhos proteins.
Conclusions
The differential enrichment of the expression profile of the OxPhos proteins in the disks versus mitochondria suggests that these proteins may represent a true proteome component of the former, with different functionality. These findings may shed new light on the pathogenesis of rod‐driven retinal degenerative diseases.
Keywords: aerobic metabolism, F1Fo ATP synthase, Orbitrap Velos, oxidative phosphorylation, rod outer segment
Abbreviations
- ACOX2
acyl‐CoA oxidase 2
- Ap5A
di(adenosine)‐5‐penta‐phosphate
- ATP
adenosine‐5’‐triphosphate
- ATP synthase
F1Fo‐ATP synthase
- BSA
bovine serum albumin
- cGMP
cyclic guanosine monophosphate
- COX
cytochrome c oxidase
- DR
diabetic retinopathy
- ETC
electron transport chain
- HPR
horseradish peroxidase
- LC‐MS
liquid chromatography mass spectrometry
- EDTA
ethylene diamine‐N,N,N',N'‐tetraacetic acid
- MDS
multidimensional scaling
- MS
mass spectrometry
- NADH
reduced β‐nicotinamide adenine dinucleotide
- OS
rod outer segment
- OxPhos
oxidative phosphorylation
- PBS
phosphate buffer saline
- RHO
rhodopsin
- ROI
reactive oxygen intermediates
- TCA
tricarboxylic acid cycle
- WB
Western blot
- WGCNA
weighted gene coexpression network analysis
1. INTRODUCTION
The outer segment (OS) of the vertebrate rod is a modified cilium filled with membranous disks expressing the protein machinery for phototransduction 1 , 2 as well as for oxidative phosphorylation (OxPhos). 3 , 4 , 5 The complex of our previous biochemical, imaging, and proteomic data shows that the electron transport chain (ETC), tricarboxylic acid cycle (TCA), and F1Fo‐ATP synthase (ATP synthase) are expressed and catalytically active in vertebrate OS. 6 , 7
The provision of guanosine‐5'‐triphosphate for cyclic guanosine monophosphate (cGMP) production is a demanding process in the OS: illumination causes a fivefold increase in cGMP turnover, exceeding the OS anaerobic glycolytic capacity. 4 , 8 , 9 The possibility of a timely adenosine‐5'‐triphosphate (ATP) delivery from the IS mitochondria to the OS would be ruled out by the fact that the portion of the OS active in phototransduction is the farthest from the IS. 10 The extramitochondrial OxPhos would be linked to phototransduction, 11 as under illumination isolated OS produce reactive oxygen intermediates (ROI) in vitro. The OS of bovine, 4 , 5 mouse, 12 , 13 and Zebrafish 5 origin conduct OxPhos in the presence of classical substrates (pyruvate/malate, succinate, and glucose). 4 Activities were sensitive to ETC inhibitors and polyphenols. 14 The mismatch between the energy required by the OS for its processes in light (about 9 nmol ATP/min/mg of protein) and the amount of ATP that can be produced by glycolysis would be filled‐in by an extramitochondrial OxPhos. In this respect, the OS and the mitochondrion would share some properties, as also suggested by the fact that both selectively stain with the same vital dyes. 15 , 16 , 17
A number of proteomic studies have identified the mitochondrial OxPhos proteins in membranes other than the inner mitochondrial ones, but often considered them as contaminants. 18 We have previously conducted two proteomic analyses of purified OS disks, one by two‐dimensional electrophoresis and mass spectrometry (MS), 7 and a more recent one by liquid chromatography mass spectrometry (LC‐MS)/MS. 6 Both MS studies identified many subunits of the five complexes of respiration. In particular, our latest study 6 compared the OS disk proteins with those of liver mitochondria by statistical analyses, showing that the major discriminative proteins in the two proteomes were implied in phototransduction and lipid metabolism, respectively. Therefrom, we showed that mitochondria and OS disk do share some common features, essentially relative to OxPhos. 6 Gene Ontology (GO) analysis revealed that the proteins of the TCA, ETC, and ATP synthase were among the top gene signatures of the disk proteome. 6
Here, we asked which proteins expressed in the OS disk are responsible of its ectopic aerobic ATP production and whether these are actual component of the disk proteome. To this end, we compared the differential functionality in the presence of diverse respiring substrates and the protein expression of the OS disks and mitochondria purified from the bovine retina, by proteomics, statistical and bioinformatic analyses.
2. MATERIALS AND METHODS
2.1. Materials
All chemical compounds were of the highest chemical grade.
2.1.1. Sample preparations
Extraction of retinas
Retinas were extracted as previously described. 19 Briefly, freshly enucleated bovine eyes (from a local certified slaughterhouse) were cut to obtain the eye semi‐cup under dim red light. After vitreous and lens removal, the eye semi‐cups were incubated for 10 minutes with Mammalian Ringer (MR, 0.157 mol/L NaCl, 5 mmol/L KCl, 7 mmol/L Na2HPO4, 8 mmol/L NaH2PO4, 0.5 mmol/L MgCl2, 2 mmol/L CaCl2 pH 6.9 plus protease inhibitor cocktail [Sigma‐Aldrich]) and 50 μg/mL Ampicillin. Each free‐floating retina was then cut free of the optic nerve with scissors and collected.
Purified bovine rod OS preparations
Purified bovine rod OS were prepared under dim red light at 4°C from 20 retinas, by sucrose/Ficoll continuous gradient centrifugation, 20 in the presence of protease inhibitor cocktail (Sigma–Aldrich) and ampicillin (100 µg/mL). Rod OS preparation was characterized for integrity of plasma membrane as reported. 21 Rod OS were homogenized and disk aggregates eliminated by subjecting suspensions prior diluted 1:1 (v/v) in ultrapure water (Milli‐Q®; Millipore) to 10 passages through a needle (25 gauge) on ice.
Osmotically intact disk preparation
Osmotically intact disks were obtained from rod OS according to Smith and Litman. 22 Briefly, OS were allowed to burst for 3 hours in 5% Ficoll (Sigma‐Aldrich) and 100 μg/mL Ampicillin. Disks do not burst by hypotonic shock, due to their peculiar lipid composition. 23 Then, 2‐mL cold water was added on the top of the suspension and centrifuged for 2 hours at 25 000 rpm in a Beckman FW‐28 rotor (100 000 g). Swollen disks were collected as a pink layer at the interface between Ficoll and water.
2.1.2. Cellular subfraction preparations
All operations were carried out at 4°C.
Retinal mitochondria
Residual retinae after Rod Outer Segment preparation (5 g) were collected, washed in cold phosphate buffer saline (PBS), and homogenized in Buffer containing 0.25 mol/L sucrose, 0.15 mol/L KCl, 10 mmol/L TRIS HCl pH 7.4, 1 mmol/L ethylene diamine‐N,N,N',N'‐tetraacetic acid (EDTA), and 0.5% bovine serum albumin (BSA), and centrifuged at 800 g for 10 minutes. Supernatant was filtered and centrifuged at 12 000 g for 15 minutes. Pellet was resuspended in 0.25 mol/L sucrose, 75 mmol/L mannitol, 10 mmol/L TRIS HCl pH 7.4, 1 mmol/L EDTA (Buffer B), and centrifuged at 1000 g for 10 minutes. The final supernatant was centrifuged again at 12 000 g for 15 minutes and the mitochondrial pellet was resuspended in Buffer B. 24 Each sample was divided into aliquots, 14 one for each experimental assay plus one to determine the total proteins concentration using BCA method assay (Pierce). Sample aliquots for Western blotting and MS analysis were kept at −80°C until use.
Western Blot Analysis
Samples (50 µg proteins) were separated onto polyacrylamide gradient (8%‐16%) denaturing gels (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and transferred onto nitrocellulose. Nitrocellulose was blocked for 1 hour with 5% (w/v) BSA in PBS, incubated with rabbit polyclonal Antibody (Ab) against ACOX2 (acyl‐CoA oxidase 2), or rhodopsin (RHO) proteins (LifeSpan Bioscence) diluted in PBS‐Tween 0.15% (v/v) (PBS‐T), rinsed three times in PBS‐T and incubated with horseradish peroxidase‐conjugated anti‐rabbit IgG (Amersham Pharmacia Biothech.). Bands were detected and densitometrically quantified by the ChemiDoc Touch Imaging System and Image Lab software (Bio‐Rad).
Transmission electron microscopy
Front halves of bovine eyes were excised and the vitreous humor and lens removed. Eye cups were then filled with fixative consisting of 4% paraformaldehyde and 0.1% glutaraldehyde in PBS buffer solution. After fixation (ON at 4°C), retinas were removed from the eye capsule, cut into small pieces, washed overnight with 50 mmol/L NH4Cl, dehydrated and embedded in LR White Acrylic Resin (Sigma ‐Aldrich, St.louis, MO), and polymerized at 58°C. Ultrathin sections were placed on nickel grids and used the next day for postembedding immunogold experiments. Sections were treated with blocking solution (1% BSA, 0.1% Tween 20, PBS 1×), then incubated with mouse monoclonal anti‐RHO (1:100) (Sigma Aldrich) and rabbit polyclonal anti‐ND1 subunit of ETC I Antibody (Ab) (diluted 1:50) (Abcam) overnight at 4°C. Ab binding was detected using secondary goat anti‐mouse IgG (British BioCell International) (diluted 1:100) coupled to gold particles (5 nm) and anti‐rabbit IgG (British BioCell International) (diluted 1:100) coupled to gold particles (25) nm. Sections were analyzed at a FEI Tecnai G 2 transmission electron microscope operating at 100 kV. In negative controls, the preimmune serum was applied to the sections instead of the specific primary Ab. Images were acquired with OSIS Veleta cameras, collected and typeset in Corel Draw X6.
ATP synthesis assay
The formation of ATP from adenosine‐3',5'‐diphosfate (ADP) and inorganic phosphate (Pi) was assayed in purified OS (0.005 mg protein/mL) as reported previously. Rod OS were incubated for 10 minutes at 37°C in 50 mmol/L Tris/HCl (pH 7.4), 50 mmol/L KCl, 1 mmol/L ethylene glycol‐bis(beta‐aminoethyl ether)‐N,N,N',N'‐tetraacetic acid, 5 mmol/L MgCl2, 5 mmol/L KH2PO4, 0.6 mmol/L ouabain, and 0.25 mmol/L di(adenosine)‐5‐penta‐phosphate (Ap5A, adenylate kinase inhibitor) in the presence of 20 mmol/L fumarate; 20 mmol/L alpha‐ketoglutarate; 5 mmol/L pyruvate plus 2.5 mmol/L malate; 10 mmol/L citrate; 20 mmol/L beta‐hydroxybutyrate; 0.2 mmol/L NADH; or 20 mmol/L succinate. ATP synthesis was then induced by addition of 0.1 mmol/L ADP and an adequate volume of luciferin/luciferase reagent (Roche Diagnostics Corp.), to continuously follow ATP formation in a luminometer (Lumi‐Scint, Bioscan). ATP standard (Roche Diagnostics Corp.) solutions in the concentration range between 10−10 and 10−8 mol/L were used for calibration.
Oxygraphic measurements
Oxygen consumption from purified OS (0.04 mg of protein/1.7 mL) was measured with an amperometric electrode (Unisense‐Microsrespiration; Unisense A/S) at 22°C in a closed chamber, as previously described.4 Medium contained: 50 mMTris‐HCl pH 7.4, 50 mmol/L KCl, 2 mmol/L MgCl2, 5 mmol/L KH2PO4, 25 μg/ml ampicillin, and 0.25 mmol/L di(adenosine‐5′) penta‐phosphate Rod OS sample were diluted (1 + 4 v/v) in ultrapure water (MilliQ; Millipore) and subjected to 10 strokes through a 27‐gauge syringe to obtain homogenates. Substrates as well the OS homogenate were added by means of a Hamilton syringe. Respiring substrates were as follows: 0.2 mmol/L NADH; 20 mmol/L fumarate, 20 mmol/L alpha‐ketoglutarate; 5 mmol/L pyruvate plus 2.5 mmol/L malate; 10 mmol/L citrate; 20 mmol/L beta‐hydroxybutyrate; or 20 mmol/L succinate. MicOx software (Unisense) was used to convert data into Excel files.
2.2. Data analysis
Results are expressed as mean ± SD and n refers to the number of assays in any particular condition.
2.2.1. Protein identification by MS analysis
Sample preparation
Aliquot of purified disks and retinal mitochondria‐enriched fractions (100 µg) were solubilized, reduced, and alkylated using a solution of 6 mol/L Guanidine‐HCl, 10 mmol/L Tris(2‐carboxyethyl)phosphine hydrochloride, 40 mmol/L chloroacetamide, and 100 mmol/L Tris pH 8.5. Then, samples were overnight digested at 37°C with Lys C and Trypsin and the peptides were collected with one wash of 50% CH3OH, 45% H2O, and 5% tri‐fluoroacetic acid. Finally, the peptides were acidified and desalted on C18 StageTips before injection into MS.
LC‐MS/MS analysis and data processing
Samples were separated on a 25‐cm EASY‐Spray column (inner diameter 75 μm, PepMap C18, and 2 μm particles), using a Orbitrap Velos Pro instrument (Thermo Fisher Scientific). The instrument was operated in the data‐dependent acquisition mode and the MS survey scans were performed in the Orbitrap (mass range 350‐1650 m/z, resolution 60 000, automatic gain control 1 000 000 ions, and maximal ion injection time of 250 ms). Ten MS/MS experiments were triggered per MS scan and the minimum MS signal was set to 3000 ions using an isolation window of 2 Da [2]. The m/z values already selected for MS/MS were put on an exclusion list for 60 seconds using an exclusion window size of ±5 p.p.m.
Raw MS files were processed with MaxQuant software version 1.5.3.30. For proteins identification required a minimum length of six amino acids and false discovery rate (FDR) < 0.01 FDR < 0.01 in peptide‐spectrum match. The database used by the software was Bovine (UniProt Release 2015_09, UP000009136_9913).
Statistical and bioinformatic analysis
The sample size was determined based on biological variability of samples to allow the identification of statistical change with a probability P = .05 and power = 80%. 25 Each Label‐free quantification data were log2 converted and normalized. After normalization, unsupervised hierarchical clustering (multidimensional scaling with k‐means) and Spearman's correlation were used to identify outlier and samples similarity. Normalized datasets were used to construct the coexpression network using the weighted gene coexpression network analysis (WGCNA). After choosing the appropriate β power parameter (setting the value of independence scale at 0.8), proteins were classified into modules of coexpression profiles. Then, Spearman's correlation was used to identify the relationship between each module or intensity value of the identified proteins and the discrete (mitochondria and disk type of samples) and continuous (ATP synthesis reported as nmol ATP/min/mg in function of the respiring substrate used, ie, pyruvate/malate, citrate, alpha‐ketoglutarate, succinate, NADH, fumarate, and beta‐hydroxybutyrate) indicator traits. A heatmap was used to show the result. In the heatmap each row represents a module, and each column corresponds to a trait indicator. A pseudocolor scale depicts Pearson's coefficient of each module/trait indicator, with red indicating a perfect positive correlation (+1), white no correlation (0), and blue perfect negative correlation (−1). The tree dendrogram displays the results of an unsupervised hierarchical clustering analysis placing similar Pearson's correlation coefficients values near each other. Differentially expressed proteins between disk and mitochondria type samples were detected using a T test. P‐values for each proteins were adjusted using the method of Benjamini‐Hochberg. 25 Proteins were considered to be significantly differentially expressed with an adjusted P ≤ .05, identified in at least 70% of one of the groups and with and a fold time change ≥2. Volcano plot was used to quickly visualize the statistical differences. Cutoff lines were established using the function y = c/(x − x 0). 26 The proteome profile of the statistically significant proteins changed was visualized using the heatmap diagram. Each row represents a protein and each column corresponds to a sample. Normalized Z‐scores of protein abundance are depicted by a pseudocolor scale (with red indicating positive expression, white equal expression, and blue negative expression) compared to each protein value. The tree dendrogram displays the results of unsupervised hierarchical clustering analysis, placing similar sample/proteome profile values next to each other.
Finally, by means of Cytoscape software, the proteome profile of the identified proteins was used as a query to interrogate different databases and extract their GO annotations. The datasets obtained were used to identify a raked list of GO enrichment. These ranks were confined between −1 and 1 corresponding to minimal and maximal enrichment in the two groups. The results of this analysis are summarized by scatter plot. In the graph the points located on the straight line passing through the coordinates (1x, 1y) and (−1x, −1y) are the equally enriched signatures, while the distance from the line is proportional to the increase in signature enrichment in one of the two groups. In particular, the points above or under the straight line are the GO annotation/pathway positively enriched in disk or mitochondria, respectively. All statistical analyses were performed using software package R last version available at the time of experiments.
3. RESULTS
3.1. Transmission electron microscopy imaging of bovine retina
Characterization of bovine retinas was conducted by immunohistochemical demonstration in sections of localization of ND1, subunit of Complex I, in the mitochondria and OS, by transmission electron microscopy.
Figure 1 shows the immunogold colocalization experiment on bovine retina with antibodies against RHO (5‐nm‐diameter gold particles) and ND1 subunit of complex I (25‐nm‐diameter gold particles). The figure evidences that ND1 is expressed not only in mitochondria, as expected, but also in rod OS. Panels A and D show overviews of a labeled bovine retina, whereas details of a rod OS and of a mitochondrion are enlarged in Panels B‐C and E, respectively. Colocalization of Rh and ND1 is clearly seen in rod OS.
Figure 1.
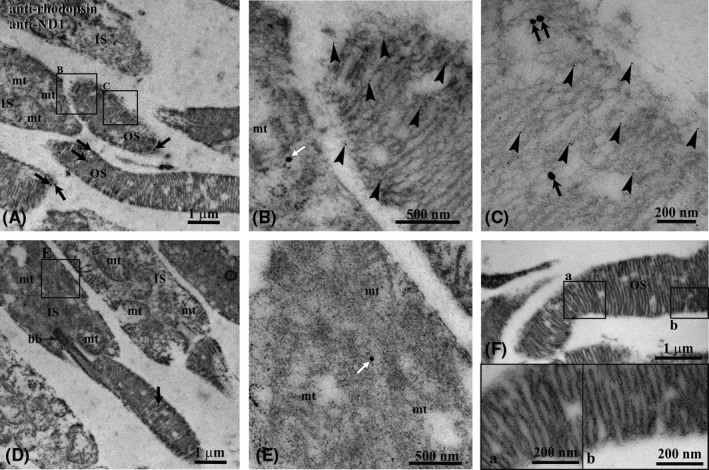
Immunogold transmission electron microscopy experiment on bovine retina. A, Retinal section showing inner (IS) and outer (OS) photoreceptor segments. B and C, Enlargements corresponding to the squared areas B and C in panel (A), to show details of an OS. Largest gold particles (25 nm width) reveal Ab against ND1 in OS (black arrows) and in IS (white arrows). The smallest gold particle (5 nm width, arrowheads) reveals Ab against Rhodopsin that, as expected, is limited to the OS. The two Abs colocalize in OS. D, Additional retinal section showing an IS with a gold particle 25 nm width (white arrow) indicating the localization of ND1 in a mitochondrion. E, Enlargement corresponding to the squared area E in panel (D). F, OS of a negative control, in which the preimmune serum was applied instead of the specific primary Ab. No gold particles are visible. bb, cilium basal body; mt, mitochondrion; a, b: enlargements of the squared areas in panel (F)
3.1.1. Oxidative Phosphorylation Assays
Both oxygen (O2) consumption (Table 1) and ATP synthesis (Table 2) by purified retinal OS or mitochondria were assayed in the presence of various substrates (20 mmol/L fumarate; 0.2 mm NADH; 20 mmol/L alpha‐ketoglutarate; 5 mmol/L pyruvate plus 2.5 mmol/L malate; 10 mmol/L citrate; 20 mmol/L beta‐hydroxybutyrate; and 20 mmol/L succinate). The O2 consumption assay can follow the transfer of electrons to O2 by the ETC. In coupled conditions the consumption of the built‐up proton gradient allows the aerobic ATP synthesis from added ADP.
Table 1.
Oxygen consumption (nmol O/min/mg). The table reports the coupled respiration rates in both retinal mitochondria‐enriched fractions (0.05 mg total protein) and isolated rod OS (0.05 mg total protein) for all sample, in the presence of TCA cycle intermediates, ie, 20 mmol/L fumarate; 20 mmol/L alpha‐ketoglutarate; 5 mmol/L pyruvate plus 2.5 mmol/L malate; 10 mmol/L citrate; 20 mmol/L beta‐hydroxybutyrate; 0.2 mmol/L NADH; and 20 mmol/L succinate. Data are reported as mean ± SD
| Retinal mitochondria | Rod OS | |
|---|---|---|
| Fumarate | 143 ± 11 | 250 ± 14 |
| Alpha‐ketoglutarate | 137 ± 9 | 284 ± 27 |
| Pyruvate/malate | 248 ± 19 | 620 ± 48 |
| Citrate | 146 ± 11 | 375 ± 31 |
| beta‐hydroxybutyrate | 316 ± 21 | 334 ± 30 |
| NADH | 42 ± 0.8 | 204 ± 18 |
| Succinate | 234 ± 17 | 540 ± 39 |
Abbreviations: NADH, reduced β‐nicotinamide adenine dinucleotide; OS, outer segment; TCA, tricarboxylic acid cycle.
Table 2.
ATP synthesis (nmol ATP/min/mg). The table reports ATP synthesis by retinal mitochondria‐enriched fractions (5 µg) or rod OS (5 µg) in the presence of TCA cycle intermediates as substrates: ie, 20 mmol/L fumarate; 20 mmol/L alpha‐ketoglutarate; 5 mmol/L pyruvate plus 2.5 mmol/L malate; 10 mmol/L citrate; 20 mmol/L beta‐hydroxybutyrate; 0.2 mmol/L NADH; and 20 mmol/L succinate. Data are reported as mean ± SD
| Retinal mitochondria | Rod OS | |
|---|---|---|
| Fumarate | 114 ± 11 | 240 ± 17 |
| Alpha‐ketoglutarate | 105 ± 9 | 333 ± 29 |
| Pyruvate/malate | 131 ± 13 | 425 ± 30 |
| Citrate | 118 ± 12 | 321 ± 29 |
| beta‐hydroxybutyrate | 142 ± 14 | 348 ± 31 |
| NADH | 5.1 ± 0.3 | 297 ± 17 |
| Succinate | 98 ± 7 | 363 ± 34 |
Abbreviation: NADH, reduced β‐nicotinamide adenine dinucleotide; OS, outer segment; TCA, tricarboxylic acid cycle.
3.1.2. Retinal disk and mitochondria proteome
Prior to the proteome characterization, purity of disk and mitochondria samples was tested by means of Western blot (WB) analysis. WB revealed that disks are positive for RHO but not for ACOX2, whereas mitochondria showed the opposite antigen profile (Figure S1).
The protein composition of disk and mitochondria, purified from bovine retinas, was determined by MS. A total of 3383 proteins were identified, 1764 (52.1%) of which were overlapping in the two groups. Instead, 1171 proteins (34.6%) or 448 (13.2%) were exclusive for disk or mitochondria, respectively (Figure S2A).
Notwithstanding the high overlapping of protein identity between the two groups, multidimensional scaling analysis and Spearman's correlogram evidenced a clear discrimination (Figure S2B,C). WGCNA was used to identify which module/proteins expression profile was functionally associated with ATP synthesis in the presence of the respiring substrates used. This analysis revealed a total of seven modules encompassing proteins with similar coexpression profiles. To distinguish between modules, we chose an arbitrary color for each module. The number of proteins included in each module ranged from 222 (magenta) to 964 (red). The blue module showed closer relationships with the mitochondrial type sample (r = .85) and its elective substrate (pyruvate/malate r = .71), whereas the red module showed closer relationships with the disk sample (r = .85) and all other substrates (alpha‐ketoglutarate, beta‐hydroxybutyrate, citrate, fumarate, NADH, and succinate with, respectively, .71, .58, .69, .72, .73, and .74 r values; Figure S3A). The proteome profile of all of the identified proteins, after Z‐score normalization (Figure S3B), or their Spearman's coefficients for each biochemical substrate or type of samples into each sample (Figure S3C) are depicted in the heatmap diagram. Visual inspection of the three heatmaps and their associated cluster analyses demonstrates the ability of the proteome profile of proteins associated with red or blue modules to distinguish between disk and mitochondria type of samples and highlight their elective biochemical substrates for the ATP synthesis.
Next, we applied the T test to identify the proteins that best distinguish the two types of samples. This revealed 1590 discriminative proteins. Among these, 957 and 633 proteins were enriched in disk and mitochondria sample, respectively (Figure 2A; Table S1). Their expression profiles after Z‐score normalization is shown in Figure 2B. Notably, among the proteins enriched in disks, we identified all of the proteins associated with vision process48 and most of the proteins associated with TCA and ETC biochemical pathway (68/83). A schematic representation of this last pathway is shown in Figure 2C and in the detail in Figure S4. Besides, among the 633 proteins enriched in mitochondria, 177 proteins were in close functional relationship (r > .75) with the use of pyruvate/malate for the biochemical assays. By contrast, among the 957 proteins of disk 404, 391, 268, and 264 and 6 were in close functional relationship (r > .75), respectively, with the use of citrate, NADH, fumarate, alpha‐ketoglutarate, or beta‐hydroxybutyrate for the biochemical assays.
Figure 2.
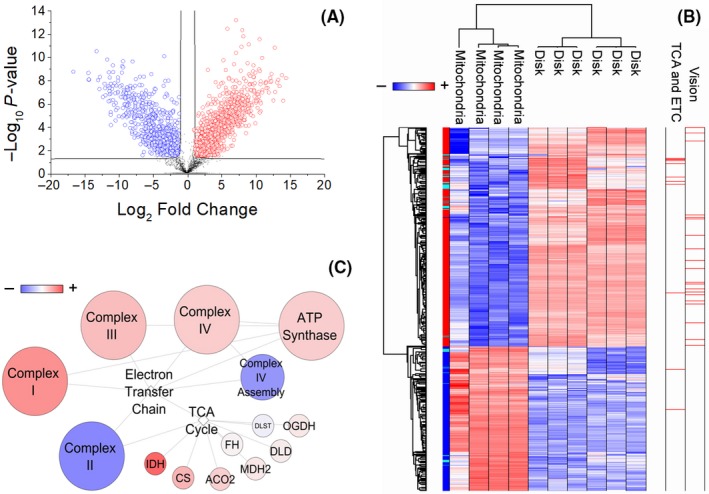
Proteins statistically significant in disk and mitochondria. A, Volcano plot of statistically significant proteins in the comparison among disk (red) and mitochondrial (blue) samples. B, Heatmap of proteome profile of the 1590 statistically significant proteins highlighted by volcano plot. In the heatmap, proteins expression profile, after Z‐score normalization, were converted into a pseudocolor scale, with red indicating positive expression; white equal expression; and blue negative expression compared with each protein values. Each row represents a protein, and each column corresponds to a sample. The tree dendrogram (top and left) displays the results of an unsupervised hierarchical clustering analysis placing similar protein expression values near each other. Visual inspection of the dendrogram and heatmap demonstrates the ability of these proteins to distinguish between the two types of samples. Notably, among the statistically significant proteins in disks, proteins associated with vision 48 and most of the proteins associated with tricarboxylic acid cycle (TCA) and electron transfer chain (ETC) (68/83) were identified. These are highlighted in red on the right of the heatmap. C, Schematic network of protein interactions in TCA and ETC pathways. In the network, circles and lines represent the proteins/complexes and their interaction, respectively. Circle colors are proportional to the fold change in protein abundance in disks vs mitochondria. The network demonstrates the enrichment of both pathways in disks
The diversity of proteins profile expression of mitochondria and disk samples and their opposite correlation in the ATP synthesis in function of the use of pyruvate/malate or all other biochemical substrates could imply a different role of these proteins in relationship with their subcellular localization. To assess this, we performed GO enrichment analysis based on cellular component, biological processes, molecular function, Kyoto Encyclopedia of Genes and Genomes, Reactome, and keywords annotation, using Cytoscape software.
Gene Ontology analysis identified 121 significant enriched gene signatures. Among these, 57 and 64 were enriched in disk or mitochondria, respectively (Table S2). These signatures were visualized using a scatter plot (Figure 2).
4. DISCUSSION
The complex of our present and previous data on the functional expression of the five Complexes of respiration in the OS disks suggests that the OS fulfills its chemical energy demand through an extramitochondrial OxPhos. 4 , 6 , 7 , 11 , 14 , 20 , 27 , 28 Aiming at identifying which proteins in the disk proteome are functionally associated with this ability, here we compared retinal mitochondria with the OS, two types of samples that would share the same origin be it either physiologic or contaminant, to be determined. OS and mitochondria behaved in a very different way in that the former utilize respiring substrates not accessible to isolated mitochondria (Tables 1 and 2).
We applied WGNCA to MS data in order to correlate the biochemical indicator traits (the different OxPhos substrates and the samples) as if they were a clinical condition to protein expression. WGCNA identified the red module as functionally associated with the disks and also to their elective substrates, the same being true for the mitochondria (blue module). The proteins clustered in each module share the expression profile and a high probability to be functionally interconnected. In our case, most of the proteins associated with TCA and ETC (68/83) (as well as those related to phototransduction, which represents a positive control) were clustered in the red module, which correlated with both the disk type of sample and the substrates on which disks are active. The GO enrichment analysis (Figure 3) demonstrated that the ETC and TCA cycle enzymes were more enriched in the disks. Mitochondria were enriched in all specific proteins that characterize this organelle, mainly involved in oxidoreduction activity (Figure 2C; Figure S4). The OxPhos proteins were not enriched in the blue module that highly correlated with the mitochondrial sample. Therefore, the OxPhos proteins in the OS would be no more contaminant than those of vision, also, their different enrichment supports the idea that the OxPhos capacity is an exquisite trait of the disk, even more so than the mitochondria.
Figure 3.
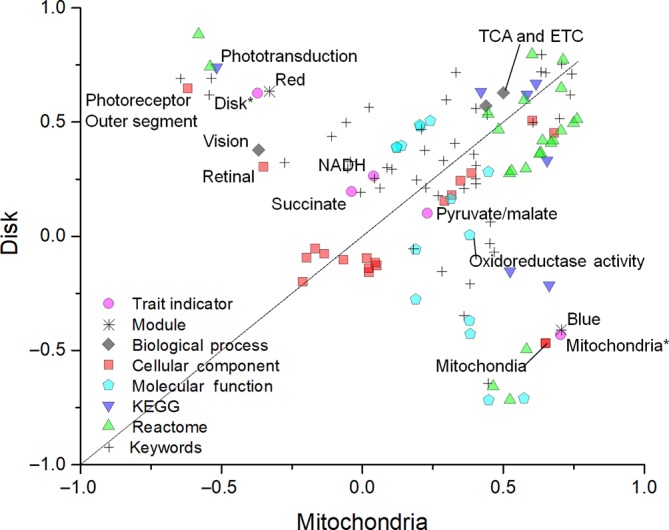
Gene ontology annotation enrichment analysis. Plot shows the enriched signatures in disk and mitochondrial samples. In the graph, the points located on the straight line drawn through the coordinates (1x, 1y) and (−1x, −1y) represent the equally enriched signatures; those above or under this line are positively enriched in disk or mitochondria, respectively. Disks are enriched in proteins involved in vision/phototransduction processes, TCA cycle/ETC, and pathways localized in the photoreceptor OS. By contrast, mitochondria are enriched in specific proteins that characterize this organelle, in particular those involved in oxidoreduction (detailed in Table S2). ETC, electron transfer chain; NADH, reduced β‐nicotinamide adenine dinucleotide; OS, outer segment; TCA, tricarboxylic acid cycle
Therefore, the main result of this work is that both mitochondria and disks perform OxPhos and express the ETC and TCA proteins, however, these are differentially enriched in function of the ability of the two types of samples to use diverse substrates. It can be supposed that the differential enrichment of the OxPhos proteins in the two types of samples depends on where those are expressed. In particular, single respiratory chain subunits were expressed in different modules, may mean that parts of the respiratory chain function differently depending on where they are expressed (ie, OS or mitochondria) and vice versa the subcellular localization would require a differential expression of the very same proteins, depending on the network they are expressed in. On the other hand, the OxPhos proteins could not be expected to be exclusive of either sample.
Proteomic data are similar to those reported previously, showing the enrichment in the disk membranes of the proteins of the TCA cycle and ETC. 6 There, liver mitochondria were utilized, as a standard reference. 6 Mitochondrial proteomes vary among tissues. 29 Our present and previous published MS studies 6 , 7 always found the expression of the mitochondrial proteins of the respiratory chain and the TCA cycle in the rod OS disks (of the 1007 proteins expressed in the mitochondria). As we have previously stated, a mere contamination would not be so selective. 18
Data confirm the existence of a cytosolic TCA cycle in the rod OS, previously reported. 3 Citrate synthase, the enzyme catalyzing the first step of the TCA cycle, and isocitrate dehydrogenase isoform 3 (IDH3) were particularly enriched in disks (Figure 2C; Figure S4). Interestingly, IDH3 is considered the isoform specific for the TCA cycle. 30 Of the five genes encoding human IDHs, IDH3 has a well‐established role in TCA cycle, where it catalyzes the first oxidative reaction. IDH3 also possesses regulatory subunits stimulated by substrate availability and inhibited by ATP and alpha‐ketoglutarate. Homozygous inactivation of IDH3 enzyme inhibits both the TCA cycle and the OxPhos. 30 NADH‐ubiquinone oxidoreductase chains 1, 3, and 5, encoded in humans by mitochondrial genes, were also enriched in disks (Figure 2C; Figure S4). Also Complex I is enriched in the OS, as confirmed by the immuno‐EM localization (Figure 1). Mutations of these subunits are implied in the Leber hereditary Optic neuropathy 31 and Leigh syndrome (subacute necrotizing encephalomyelopathy). 32 , 33 Mitochondrially encoded ATP synthase subunits 8 and 6 were both overexpressed in disks (Figure S4). Mutations in the genes encoding these subunits (MT‐ATP8 and MT‐ATP6) are related to various disorders involving the nervous system. 34 This is confirmative of previous findings showing the ectopic expression of the five complexes of respiration (reviewed in 18 ). Likely, this fact is physiological in the rod OS, where the high need of aerobically produced ATP must not trigger the inflammatory response that would follow mitochondrial damage and consequent exposure of the mitochondrial damage‐associated molecular patterns. 36
By contrast, some of the assembly proteins for Complex IV which catalyzes the limiting step of the OxPhos 37 such as SCO1 (synthesis of cytochrome c oxidase) and SURF1 (product of surf1, the first gene of the surfeit gene locus) were enriched in mitochondria (Figure S4). The COX assembly protein SCO1 is an accessory factor essential for copper delivery to the COX holoprotein, 38 typically localized in tissues with high levels of OxPhos. SURF1 is one of the early assembly factors required for the biogenesis of the COX holoenzyme. 39 SURF1 deficiency is also a frequent cause of Leigh syndrome, associated with COX deficiency. The lack of enrichment of the proteins of Complex IV assembly in disks further demonstrates that the ETC is not a contaminant in the disks, in fact assembly factors would not be necessary in the disk, as the biogenesis of the five complexes expressed inside the disk membranes would take place inside the IS mitochondria. A random contamination could not make so much sense.
Interestingly, Complex II was enriched in the mitochondria but not in the disks (Figure 2C; Figure S4). Interestingly, Complexes I, III, and IV but not II 40 , 41 , 42 are believed to assemble in a supramolecular complex (supercomplex). We have supposed that the supercomplex would be first assembled inside the IS mitochondria then transferred to the OS, possibly via the intraflagellar transport (IFT) system, 43 known to transfer proteins to the nascent disks from the IS in vesicles that travel through the cilium. 44 , 45 Defects in the IFT lead to retinal degeneration. 46 , 47
The present data may justify the fact that the retina is one of the most metabolically active tissues of the body 48 displaying the highest oxygen consumption on a weight basis. It is tempting to presume that the fall in the O2 partial pressure at the photoreceptor OS‐IS border in a whole retina in vivo 48 is due to the O2 consumption by Complex IV expressed in the disks.
An oxidative metabolism in the rod OS 49 , 50 implies potential oxidative damage of the OS. In fact, the OS disks generate ROI in vitro, which implies that, in particular, conditions oxidative stress production can occur also in vivo, 11 being the ETC a major ROI producer. 51 The aerobic ATP production in the OS was also for the first time correlated with light absorption, that is, to visual transduction. 11 An increment in ATP demand due to phototransduction overwork would increment O2 consumption eliciting an increase in ROI production. In this respect, disks are a suitable model for in vitro studies on the oxidative stress produced by the functioning of the OxPhos and the ability of some natural and pharmacological antioxidants to scavenge it. 28 Oxidative stress has been implied in the onset of most of the degenerative retinal pathologies. 12 , 20 , 52 The present experimental facts may awaken the awareness that the oxidative stress may arise in part inside the rod OS. 53 The oxidative stress in the retinas from mouse models of diabetic retinopathy seems to derive from the photoreceptors, as it does not occur when retinas are deprived of these. 54 The rod OS degeneration is an early event after blue light irradiation of the eye, due to oxidative stress production and hypometabolism inside the rod OS. 13 , 55
In conclusion, data suggest that the mitochondrial OxPhos proteins may represent a true OS proteome component, outside the mitochondrion. If the OxPhos activity in the rod OS was due to a contamination by retinal mitochondria, these two types of samples should share identical biochemical activity and protein expression profiles. Further studies may lead to a deeper understanding of the mechanism of exportation of mitochondrial proteins on the plasma membranes and if these are exclusive of retina.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
I. Panfoli, M. Bruschi, and G. Candiano designed the experiments; D. Calzia, M. Bruschi, A. Petretto, M. Bartolucci, and F. Caicci performed the experiments; M. Bruschi, A. Petretto, L. Manni, and C. Traverso analyzed the data. M. Bruschi, I. Panfoli, and G. Candiano wrote the manuscript. All authors read and reviewed the article.
Supporting information
Fig S1
Fig S2
Table S2
Fig S3
Fig S4
Table S1
Supplementary Material
ACKNOWLEDGEMENTS
The work was supported by grants from Italian Ministry for Research and Technology (MIUR): FRA‐2016, Cinque per mille e Ricerca Corrente del Ministero della Salute to Istituto Giannina Gaslini, and from Renal Child Foundation.
Bruschi M, Bartolucci M, Petretto A, et al. Differential expression of the five redox complexes in the retinal mitochondria or rod outer segment disks is consistent with their different functionality. FASEB BioAdvances. 2020;2:315–324. 10.1096/fba.2020-00093
Contributor Information
Andrea Petretto, Email: AndreaPetretto@gaslini.org.
Isabella Panfoli, Email: panfoli@difar.unige.it.
REFERENCES
- 1. Molday RS, Moritz OL. Photoreceptors at a glance. J Cell Sci. 2015;128:4039‐4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palczewski K. Chemistry and biology of the initial steps in vision: the Friedenwald lecture. Invest Ophthalmol Vis Sci. 2014;55:6651‐6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Panfoli I, Calzia D, Ravera S, et al. Extramitochondrial tricarboxylic acid cycle in retinal rod outer segments. Biochimie. 2011;93:1565‐1575. [DOI] [PubMed] [Google Scholar]
- 4. Panfoli I, Calzia D, Bianchini P, et al. Evidence for aerobic metabolism in retinal rod outer segment disks. Int J Biochem Cell Biol. 2009;41:2555‐2565. [DOI] [PubMed] [Google Scholar]
- 5. Calzia D, Garbarino G, Caicci F, et al. Evidence of oxidative phosphorylation in zebrafish photoreceptor outer segments at different larval stages. J Histochem Cytochem. 2018;66:497‐509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruschi M, Petretto A, Caicci F, et al. Proteome of bovine mitochondria and rod outer segment disks: commonalities and differences. J Proteome Res. 2018;17:918‐925. [DOI] [PubMed] [Google Scholar]
- 7. Panfoli I, Musante L, Bachi A, et al. Proteomic analysis of the retinal rod outer segment disks. J Proteome Res. 2008;7:2654‐2669. [DOI] [PubMed] [Google Scholar]
- 8. Olson A, Pugh EN. Diffusion coefficient of cyclic GMP in salamander rod outer segments estimated with two fluorescent probes. Biophys J. 1993;65:1335‐1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsu SC, Molday RS. Glucose metabolism in photoreceptor outer segments. Its role in phototransduction and in NADPH‐requiring reactions. J Biol Chem. 1994;269:17954‐17959. [PubMed] [Google Scholar]
- 10. Albert AD, Boesze‐Battaglia K. The role of cholesterol in rod outer segment membranes. Prog Lipid Res. 2005;44:99‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calzia D, Degan P, Caicci F, et al. Modulation of the rod outer segment aerobic metabolism diminishes the production of radicals due to light absorption. Free Radic Biol Med. 2018;117:110–118. [DOI] [PubMed] [Google Scholar]
- 12. Calzia D, Garbarino G, Caicci F, et al. Functional expression of electron transport chain complexes in mouse rod outer segments. Biochimie. 2014;102:78‐82. [DOI] [PubMed] [Google Scholar]
- 13. Calzia D, Panfoli I, Heinig N, et al. Impairment of extramitochondrial oxidative phosphorylation in mouse rod outer segments by blue light irradiation. Biochimie. 2016;125:171‐178. [DOI] [PubMed] [Google Scholar]
- 14. Calzia D, Oneto M, Caicci F, et al. Effect of polyphenolic phytochemicals on ectopic oxidative phosphorylation in rod outer segments of bovine retina. Br J Pharmacol. 2015;172:3890‐3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Calzia D, Bianchini P, Ravera S, et al. Imaging of living mammalian retina ex vivo by confocal laser scanning microscopy. Anal Methods. 2010;2:1816‐1818. [Google Scholar]
- 16. Panfoli I, Calzia D, Ravera S, Bianchini P, Diaspro A. Immunochemical or fluorescent labeling of vesicular subcellular fractions for microscopy imaging. Microsc Res Tech. 2010;73:1086‐1090. [DOI] [PubMed] [Google Scholar]
- 17. Ravera S, Calzia D, Bianchini P, Diaspro A, Panfoli I. Confocal laser scanning microscopy of retinal rod outer segment intact disks: new labeling technique. J Biomed Opt. 2007;12:050501. [DOI] [PubMed] [Google Scholar]
- 18. Panfoli I, Ravera S, Bruschi M, Candiano G, Morelli A. Proteomics unravels the exportability of mitochondrial respiratory chains. Expert Rev Proteomics. 2011;8:231‐239. [DOI] [PubMed] [Google Scholar]
- 19. Bianchini P, Calzia D, Ravera S, et al. Live imaging of mammalian retina: rod outer segments are stained by conventional mitochondrial dyes. J Biomed Opt. 2008;13:054017. [DOI] [PubMed] [Google Scholar]
- 20. Calzia D, Barabino S, Bianchini P, et al. New findings in ATP supply in rod outer segments: insights for retinopathies. Biol Cell. 2013;105:345‐358. [DOI] [PubMed] [Google Scholar]
- 21. Schnetkamp PP, Daemen FJ. Isolation and characterization of osmotically sealed bovine rod outer segments. Methods Enzym. 1982;81:110‐116. [DOI] [PubMed] [Google Scholar]
- 22. Gilbert Smith H, Litman BJ. Preparation of osmotically intact rod outer segment disks by Ficoll flotation. Methods Enzymol. 1982;81:57‐61. [DOI] [PubMed] [Google Scholar]
- 23. Lamba OP, Borchman D, O'Brien PJ. Fourier transform infrared study of the rod outer segment disk and plasma membranes of vertebrate retina. Biochemistry. 1994;33:1704‐1712. [DOI] [PubMed] [Google Scholar]
- 24. Fernández‐Vizarra E, Ferrín G, Pérez‐Martos A, Fernández‐Silva P, Zeviani M, Enríquez JA. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion. 2010;10:253‐262. [DOI] [PubMed] [Google Scholar]
- 25. Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. Statistical protein quantification and significance analysis in label‐free LC‐MS experiments with complex designs. BMC Bioinformatics. 2012;13:S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fabris A, Bruschi M, Santucci L, et al. Proteomic‐based research strategy identified laminin subunit alpha 2 as a potential urinary‐specific biomarker for the medullary sponge kidney disease. Kidney Int. 2017;91:459‐468. [DOI] [PubMed] [Google Scholar]
- 27. Panfoli I, Calzia D, Bruschi M, et al. Functional expression of oxidative phosphorylation proteins in the rod outer segment disc. Cell Biochem Funct. 2013;31:532‐538. [DOI] [PubMed] [Google Scholar]
- 28. Calzia D, Candiani S, Garbarino G, et al. Are rod outer segment ATP‐ase and ATP‐synthase activity expression of the same protein? Cell Mol Neurobiol. 2013;33:637‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629‐640. [DOI] [PubMed] [Google Scholar]
- 30. Al‐Khallaf H. Isocitrate dehydrogenases in physiology and cancer: biochemical and molecular insight. Cell Biosci. 2017;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sadun AA, La Morgia C, Carelli V. Leber's hereditary optic neuropathy. Curr Treat Options Neurol. 2011;13:109‐117. [DOI] [PubMed] [Google Scholar]
- 32. Man PY, Turnbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet. 2002;39:162‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carelli V, Ross‐Cisneros FN, Sadun AA. Optic nerve degeneration and mitochondrial dysfunction: genetic and acquired optic neuropathies. Neurochem Int. 2002;40:573‐584. [DOI] [PubMed] [Google Scholar]
- 34. Dautant A, Meier T, Hahn A, Tribouillard‐Tanvier D, di Rago JP, Kucharczyk R. ATP synthase diseases of mitochondrial genetic origin. Front Physiol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chi SL, Pizzo SV. Cell surface F1Fo ATP synthase: a new paradigm? Ann Med. 2006;38:429‐438. [DOI] [PubMed] [Google Scholar]
- 36. Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hroudová J, Fišar Z. Control mechanisms in mitochondrial oxidative phosphorylation. Neural Regen Res. 2013;8:363‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horng YC, Leary SC, Cobine PA, et al. Human Sco1 and Sco2 function as copper‐binding proteins. J Biol Chem. 2005;280:34113‐34122. [DOI] [PubMed] [Google Scholar]
- 39. Zhu Z, Yao J, Johns T, et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet. 1998;20:337‐343. [DOI] [PubMed] [Google Scholar]
- 40. Lenaz G, Genova ML. Structural and functional organization of the mitochondrial respiratory chain: a dynamic super‐assembly. Int J Biochem Cell Biol. 2009;41:1750‐1772. [DOI] [PubMed] [Google Scholar]
- 41. Lenaz G, Genova ML. Supramolecular organisation of the mitochondrial respiratory chain: a new challenge for the mechanism and control of oxidative phosphorylation. Adv Exp Med Biol. 2012;748:107‐144. [DOI] [PubMed] [Google Scholar]
- 42. Wittig I, Carrozzo R, Santorelli FM, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta: Bioenerg. 2006;1757:1066‐1072. [DOI] [PubMed] [Google Scholar]
- 43. Insinna C, Besharse JC. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn. 2008;237:1982‐1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilliam JC, Chang JT, Sandoval IM, et al. Three‐dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029‐1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retin Eye Res. 2013;36:24‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marszalek JR, Liu X, Roberts EA, et al. Genetic evidence for selective transport of opsin and arrestin by kinesin‐II in mammalian photoreceptors. Cell. 2000;102:175‐187. [DOI] [PubMed] [Google Scholar]
- 47. Pazour GJ, Baker SA, Deane JA, et al. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002;157:103‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wangsa‐Wirawan ND, Linsenmeier RA. Retinal oxygen: fundamental and clinical aspects. Arch Ophthalmol. 2003;121:547‐557. [DOI] [PubMed] [Google Scholar]
- 49. Albert AD, Boesze‐Battaglia K. The role of cholesterol in rod outer segment membranes. Prog Lipid Res. 2005;44(2‐3):99‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elias RV, Sezate SS, Cao W, McGinnis JF. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha‐transducin in mouse photoreceptor cells. Mol Vis. 2004;10:672‐681. [PubMed] [Google Scholar]
- 51. Dröse S, Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol. 2012;748:145‐169. [DOI] [PubMed] [Google Scholar]
- 52. Araki E, Nishikawa T. Oxidative stress: a cause and therapeutic target of diabetic complications. J Diabetes Investig. 2010;1:90‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Panfoli I, Calzia D, Ravera S, Morelli AM, Traverso CE. Extra‐mitochondrial aerobic metabolism in retinal rod outer segments: new perspectives in retinopathies. Med Hypotheses. 2012;78:423‐427. [DOI] [PubMed] [Google Scholar]
- 54. Du Y, Veenstra A, Palczewski K, Kern TS. Photoreceptor cells are major contributors to diabetes‐induced oxidative stress and local inflammation in the retina. Proc Natl Acad Sci USA. 2013;110:16586‐16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roehlecke C, Schumann U, Ader M, et al. Stress reaction in outer segments of photoreceptors after blue light irradiation. PLoS ONE. 2013;8:e71570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S2
Fig S3
Fig S4
Table S1
Supplementary Material


