Abstract
Impaired protein homeostasis and accumulation of damaged or abnormally modified protein are common disease mechanisms in many neurodegenerative disorders including Parkinson’s disease (PD). As one of the major degradation pathways, autophagy plays a pivotal role in maintaining effective turnover of proteins and damaged organelles in cells. Several decades of research efforts led to insights into the potential contribution of impaired autophagy machinery to α-synuclein accumulation and degeneration of dopaminergic neurons, two major features of PD pathology. In this review, we summarize recent pathological, genetic, and mechanistic findings that link defective autophagy with PD pathogenesis in human patients, animal and cellular models and discuss current challenges in the field.
Keywords: α-synuclein, chaperone-mediated autophagy, macroautophagy, mitophagy, Parkinson’s disease
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder and currently remains incurable. PD is clinically characterized by the cardinal motor dysfunctions and additional non-motor symptoms including mood and cognitive changes, autonomic dysfunctions, sleep disorders, sensory symptoms and pain [1–3]. Neuropathologically, PD is characterized by progressive loss of dopaminergic neurons, which is typically accompanied by accumulation of Lewy bodies (LBs) in neuronal somata and Lewy neurites in neuronal processes with fibrillar α-synuclein aggregates as major protein component [4]. Comorbid pathological changes including tau hyperphosphorylation and resulting neurofibrillary tangles as well as amyloid-β deposition are commonly detected in PD brains within different brain regions [5]. To date, existing treatments alleviate symptoms in specific patient populations but do not stop disease progression or reverse existing disabilities. Elucidation of disease etiology and pathogenesis is crucial for the development of diagnostics and treatments for PD.
In addition to abnormal protein aggregation, malfunctioning degradation pathways such as autophagy and lysosome dysfunctions are known, early disease features that may contribute to the pathogenesis of PD, as evidenced by pathology and genetics [6–8]. The autophagy pathway is essential for the timely removal of long-lived proteins and dysfunctional organelles in eukaryotic cells to prevent subsequent toxicity and cell death. Accumulating evidence suggests that aggregation of α-synuclein and tau is a consequence of impaired autophagic-lysosomal degradation [9, 10]. In turn, α-synuclein and tau have also been shown to impact mitochondrial, autophagic, and lysosomal functions [9–13]. Dopaminergic neurons are metabolically very active with high mitochondrial energy demand and are therefore especially vulnerable towards insufficient clearance of damaged mitochondria [14, 15]. Accumulation of defective mitochondria will result in increased levels of reactive oxygen species that may damage surrounding healthy mitochondria to further accelerate disease progression in a vicious cycle.
In PD about 90–95% of cases are sporadic in nature with unknown origin and 5–10% are familial forms of the disease. Great advances have been made over the past two decades with the identification of monogenic causes of early-onset familial PD such as mutations in SNCA, LRRK2, VPS35, PRKN, PINK1, and PARK7 [16]. Other variants in genes encoding α-synuclein (SNCA), tau (MAPT), and glucocerebrosidase (GBA) were nominated among the most significant genetic risk factors known for PD [17]. In two recent large meta-analyses of genome-wide association studies (GWAS) [8, 18], previous and newly identified candidate loci were strongly enriched for lysosomal and/or autophagy functions. Despite an intriguing link between autophagic defects and PD pathogenesis (Figure 1), the precise underlying disease mechanisms remain largely unclear and their characterization requires the systematic analysis of disease-specific changes in PD brain as well as functional studies in appropriate cellular and animal models. The present review will emphasize how different PD genes/mutations affect autophagic function as well as the contribution of autophagic impairment to PD pathogenesis.
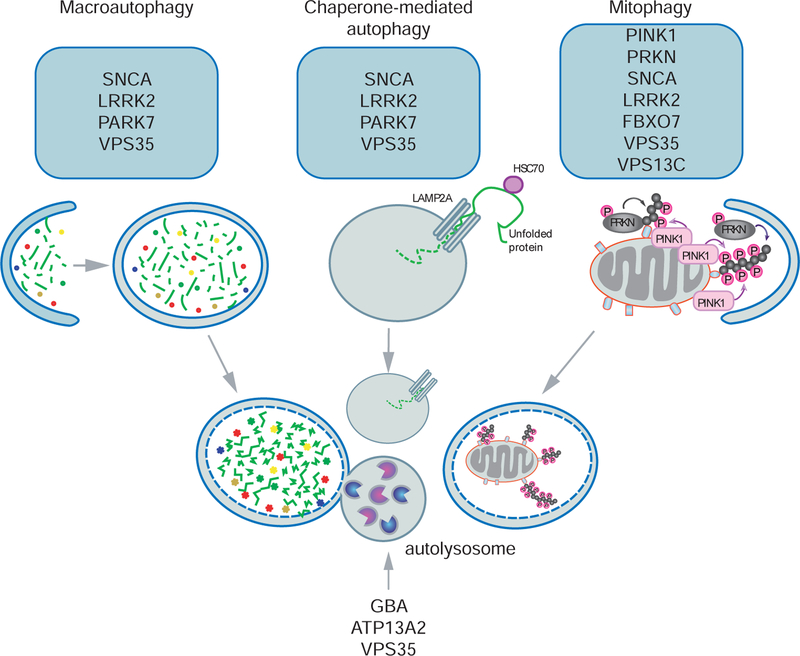
Figure 1. Schematic depiction of PD-related genes associated with different autophagy pathways.
A large group of genes associated with familial and sporadic PD are strongly linked to macroautophagy, chaperone-mediated autophagy, mitophagy, and downstream lysosomal function. Macroautophagy, the main route of cellular degradation, is initiated from an isolated membrane (phagophore) forming the double membraned autophagosome to sequester cytosolic material. Subsequent fusion of autophagosomes with lysosomes forms the autolysosome for hydrolase-mediated degradation of its contents. In chaperone-mediated autophagy, the cytosolic chaperone protein HSC70 targets and translocates unfolded proteins directly to lysosomes through binding to the lysosomal receptor LAMP2A. Compared to non-selective/bulk macroautophagy, selective mitophagy labels only damaged mitochondria for downstream autophagic degradation. See text for more detailed description of PD gene-mediated effects on various types and steps of the autophagic-lysosomal pathway.
2. Autophagy changes in autopsy PD brain
Autophagy is a degradation process to remove redundant or defective cellular components within the cell. Three different classes of autophagy are present in mammalian cells: macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy [19]. The major type of autophagy is termed macroautophagy and can be subdivided into two categories: non-selective/bulk autophagy (hereafter preferred as autophagy) and selective autophagy. Starvation causes autophagy to provide sufficient nutrients for the cell, while selective forms of autophagy remove protein aggregates (aggrephagy) as well as superfluous or damaged organelles. In either case, an isolation membrane or phagophore surrounds the entire organelle or large parts of the cytoplasm. This then initiates formation of the double membraned autophagosome. The autophagosome subsequently fuses with the lysosome to form the autolysosome, where the contents are degraded. Selective autophagy of for instance mitochondria (termed mitophagy) is regulated in a complex fashion by specific post-translational ‘tagging’ of the cargo and modification of downstream autophagy adaptors to decode the signal. CMA is a selective pathway involving translocation of unfolded proteins with a recognition sequence (KFERQ-like peptide motif) into the lysosome for degradation. The CMA pathway is mediated thought direct binding of cytosolic chaperone proteins to target substrates for lysosomal clearance [20].
Evidence of impaired autophagy, CMA, and mitophagy in vulnerable brain regions in PD patients has been provided by studies using postmortem tissue compared to age- and sex-matched controls. Early on, autophagic degeneration was observed in dopaminergic neurons in the substantia nigra in PD patients [21]. Ultrastructural examination of autophagosomes in PD brain revealed a large number of phospho-ERK labeled mitochondria, suggesting a PD-related abnormal mitophagy [7]. Our more recent studies confirmed the disease relevance of phosphorylated ubiquitin, which serves as the mitophagy tag, and demonstrated a selective increase in PD brain and a strong correlation with levels of LBs and tau tangles [22–24]. Later, the majority of LBs in the substantia nigra of PD brains were found to be immunopositive for the autophagy related protein microtubule-associated protein 1A/1B-light chain 3 (LC3) that is responsible for autophagosome formation [6, 25, 26]. The LC3 signal was present especially in the halo, where it colocalized with α-synuclein [25], and also in α-synuclein-positive Lewy neurites [25, 26]. The lipidated form of LC3 (LC3-II) is a well-characterized protein and indicator of autophagy that specifically localizes to autophagic structures [27]. LC3-II levels were also markedly elevated in nigral PD samples [25, 26], while levels and activities of lysosomal enzymes such as glucocerebrosidase (GCase) or the protease cathepsin D (CTSD) were decreased [28]. Moreover, levels of the heat shock cognate 70 (HSC70) protein, which controls the rate-limiting step of CMA, and its binding receptor, lysosomal-associated membrane protein 2A (LAMP2A), were also significantly decreased in the substantia nigra of PD cases compared to controls [26, 29]. This selective loss of CMA markers strongly correlated with α-synuclein accumulation in the same PD samples [29]. Thus, as shown in both imaging and biochemical studies, PD-related autophagic abnormalities extend to multiple autophagy pathways.
With α-synuclein being considered the major LB component, a large number of other proteins were identified in LBs, some of which are functionally associated with protein degradation systems [30, 31]. A recent postmortem study of PD brains revealed a crowded environment of organelles and lipid membranes therein, including mitochondria and structures reminiscent of lysosomes and autophagosomes [32]. These findings may provide a new dimension of understanding LB pathology in PD. It is possible that these pathological hallmarks are the common ‘dumping ground’, where impaired organellar trafficking and failed autophagicl-ysosomal clearance converges.
3. Effects of PD genes on autophagy
3.1. SNCA
PD has long been considered a non-genetic disease until the initial discovery of large families with autosomal dominant forms of PD [33, 34]. Soon after, the same group and others independently identified several PD-related mutations in SNCA: A53T [35], A30P [36], and E46K [37]. More recently, other missense mutations from familial (H50Q [38] and G51D [39]) and sporadic cases (A18T and A29S [40]) were described. Elevated α-synuclein levels in familial cases with SNCA multiplication were strongly linked to earlier disease onset, more rapid disease course, and increased disease severity [41]. α-synuclein, encoded by the SNCA gene, is a presynaptic protein in neurons of the central nervous system. Although its exact physiological function remains enigmatic, α-synuclein has been implicated in synaptic vesicle release and a variety of other functions [42–44]. As discussed later in more detail, emerging evidence suggests the convergence of PD pathways with synaptic homeostasis and autophagy [45–50]. Abnormally aggregated α-synuclein is the major component of LBs. α-synuclein is natively unfolded and can be degraded through multiple clearance routes including autophagy, CMA, and proteasome pathways.
Products from deregulated gene expression or mutant genes can misfold and start a vicious cycle. For instance, increasing amounts of wildtype or mutant α-synuclein may overwhelm or resist regular degradation and result in abnormal accumulation of aggregated α-synuclein in PD brain. Moreover, this might also impair the elimination of other cellular components by different mechanisms. Overexpression of wildtype α-synuclein compromised autophagy in mammalian cell lines and in transgenic mice by inhibiting RAB1A, a GTPase involved in the early secretory pathway, and resulted in mislocalization of the early autophagy protein ATG9 and decreased formation of omegasomes [51], an autophagic structure frequently seen in association with the endoplasmic reticulum (ER) [52]. Importantly, such defects could be rescued by RAB1A overexpression. A more recent study using rat PC12 cells demonstrated that α-synuclein bound to both cytosolic and nuclear high mobility group box 1 (HMGB1), impaired the cytosolic translocation of HMGB1, blocked HMGB1-Beclin 1 (BECN1) binding, and strengthened BECN1-BCL2 binding. Deregulation of these molecular events by α-synuclein overexpression led to autophagy inhibition and could be restored by genetic (BECN1 overexpression and HMBG1 knockdown) or pharmacological (corynoxine B, a natural autophagy inducer) manipulation [53].
However, findings using induced pluripotent stem cell (iPSC) lines or rat primary neurons showed contrary results. Differentiated iPSCs from a PD patient with SNCA triplication showed upregulated autophagic flux with greater accumulation of LC3 immunopositive puncta upon chloroquine treatment compared to control lines [54]. Similar results were reported in rat primary midbrain neurons with α-synuclein overexpression [55]. Apparent discrepancies in the literature may reflect the particular model systems employed, which vary in neuronal type, α-synuclein expression levels, post-translational modifications (PTMs), and aggregation. Upregulation of autophagy may also act as a cellular compensatory mechanism to clear accumulated monomeric α-synuclein, while this pathway may be compromised in the presence of proteolysis-resistant forms of aggregated α-synuclein. In an adeno-associated virus-mediated α-synuclein overexpression mouse model, autophagic proteins BECN1 and LC3-II were significantly increased at the presymptomatic stage but reduced in the later stage, which appeared to be associated with the expression of transcription factor EB (TFEB) [56], a key autophagy modulator [57].
In addition to increased levels of wildtype α-synuclein, mutant α-synuclein has also been shown to interfere with normal autophagic functions. Autophagy induction was observed in the substantia nigra of transgenic mice overexpressing A53T mutant α-synuclein via upregulating both LC3-II and BECN1 levels compared to controls [58]. This autophagy induction was further increased with aging. Ultrastructural examination of PC12 cell lines expressing A53T mutant revealed a significant accumulation of autophagic-vesicular structures, as well as impaired lysosomal hydrolysis, proteasomal function, and failure to store catecholamine with an absence of dopamine release, compared to cells expressing wildtype α-synuclein [59].
Regarding the CMA pathway, α-synuclein contains a pentapeptide sequence (95VKKDQ99) consistent with a CMA recognition motif, allowing α-synuclein to be recognized by the chaperone protein HSC70 and to bind to LAMP2A at the lysosomal membrane for degradation through CMA [60]. This has been suggested to be the primary clearance pathway for wildtype α-synuclein in its native form, while pathogenic α-synuclein mutants seem to prevent this route [60]. In PC12 cells, α-synuclein A53T and A30P mutants acted as uptake blockers by binding LAMP2A receptors on the lysosomal membrane [60]. These findings were confirmed in rat primary cortical neurons with adenoviral transduced wildtype and A53T α-synuclein with the A53T mutant causing significant reduction of CMA-dependent proteolysis [61]. Subsequent compensatory autophagy induction could not rescue the resulting perturbations of cellular homeostasis and contributed to neuronal toxicity [61].
Besides rare pathogenic missense mutations, PTMs of wildtype α-synuclein (phosphorylation, oxidation, nitrosylation, and/or dopamine-modification) impair its own CMA-mediated degradation as shown in primary neuronal mouse cultures, SH-SY5Y cells, and isolated mouse lysosomes [62]. However, unlike the strong binding of mutant α-synuclein to LAMP2A that leads to the complete block of CMA, most modified forms of α-synuclein appear to at least allow the CMA degradation of other substrates. Dopamine-modified α-synuclein is an exception and acts more similar to missense mutants [62]. Thus, dopamine-induced CMA inhibition may further contribute to the selective vulnerability observed in PD. Indeed, CMA blockage by mutant α-synuclein in transgenic mice resulted in prominent α-synuclein inclusions in brainstem and spinal cord accompanied by low CMA activities compared to more resistant brain regions [63].
Additionally, α-synuclein has been shown to both directly and indirectly interact with mitochondria and the mitophagy pathway. Stable PC12 cells expressing A53T mutant but not human wildtype α-synuclein showed marked accumulation of mitochondria-containing autophagic-vesicular structures [59]. In primary cortical neurons, expression of A53T α-synuclein led to increased colocalization between autophagosomes and healthy, polarized mitochondria. This was in contrast to the colocalization of autophagosomes with unhealthy, depolarized mitochondria in wildtype α-synuclein expressing neurons [64], which resulted in abnormal clearance of functional mitochondria and led to mitochondrial loss and a bioenergetic deficit. Mutant A53T α-synuclein-induced mitophagy activation was dependent on PRKN and mitochondrial fragmentation. Similar results were shown in an in vivo model – a transgenic mouse line with selective human α-synuclein A53T overexpression in dopamine neurons [65]. Profound mitochondrial abnormalities and increased autophagic cytoplasmic inclusions containing mitochondrial remnants were observed in the substantia nigra of these mice early on, followed by later degeneration of dopaminergic neurons. Genetic deletion of either PINK1 or PRKN in these mice exacerbated this phenotype, suggesting an important role of impaired mitophagy in the context of α-synuclein pathology. Overexpressing human wildtype and A53T mutant α-synuclein in yeast cells reduced their life span along with increased autophagy and mitophagy activities [66]. This pathway was regulated by ATG32 through sirtuin 2 (SIRT2) and may provide a possible link between α-synuclein-induced toxicity and aging. Dynamin 1 like (DNM1L/DRP1), a mitochondrial fission protein, and OPA1 mitochondrial dynamin like GTPase (OPA1), a mitochondrial fusion protein, appeared to be protective against α-synuclein-induced mitochondrial fragmentation and mitophagy activation in dopaminergic SH-SY5Y cells [67].
Despite α-synuclein’s synaptic localization under healthy, physiological conditions, cellular stress and disease conditions appeared to trigger α-synuclein translocation to the inner mitochondrial membrane where it interacted with the mitochondrial complex I. This interaction caused a reduction in complex I activity and increased free radical production in vivo [68]. Further examination of dopaminergic neurons in transgenic mice overexpressing A53T α-synuclein demonstrated monomeric and oligomeric mutant α-synuclein localized to mitochondrial membranes, which was associated with increased mitophagy upon proteasomal inhibitory stress [68]. Oligomeric, dopamine-modified, and phospho-mimetic α-synuclein species were later reported to bind with high affinity to the mitochondrial protein TOMM20 in the postmortem PD brain and in PD animal models. This strong binding prevented the interaction of TOMM20 with its co-receptor, TOMM22, and impaired mitochondrial protein import, thus leading to deficient mitochondrial respiration, enhanced production of reactive oxygen species, and loss of mitochondrial membrane potential [69]. The aberrant α-synuclein-TOMM20 interaction may further trigger the activation of mitophagy. In addition to α-synuclein missense mutations, a distinct highly neurotoxic α-synuclein species named “pα-syn*” was recently identified in primary neurons seeded with α-synuclein fibrils, in mouse brains and in autopsied brains from PD patients [70]. Pα-syn* was a truncated phosphorylated α-synuclein species resulting from incomplete autophagic degradation of α-synuclein. Pα-syn* colocalized with heat shock protein family A member 5 (HSPA5), a master regulator of the unfolded protein response, at mitochondria-associated ER membranes where it induced structural and functional damage and resulted in mitochondrial fission and enhanced mitophagy.
Collectively, emerging evidence from cell cultures, animal models, and human postmortem studies indicate strong interference of high levels of wildtype or mutant and modified α-synuclein species with autophagy, CMA, and mitophagy pathways.
3.2. LRRK2
Genetic mutations in the Leucine-rich repeat kinase 2 (LRRK2) account for the majority of autosomal dominant cases in PD. The G2019S and R1441C LRRK2 mutants are the most common pathogenic variants and responsible for more than 5% of familial PD cases [71]. The encoded protein LRRK2 has two distinct enzymatic domains: the kinase domain to catalyze phosphorylation and the Ras of complex (Roc)-GTPase domain for GTP-GDP hydrolysis. Pathogenic mutations of LRRK2 severely alter its expression levels [72] and/or kinase activity [73–75]. LRRK2 is primarily localized in membrane microdomains, multivesicular bodies, and autophagic vesicles and involved in several cellular signaling pathways including autophagy, neurite outgrowth, vesicular trafficking, and cytoskeletal maintenance [76].
Previous studies have implicated LRRK2 in the lysosomal system. Aged LRRK2 knockout rodent models exhibited increased levels of lysosomal proteins in the periphery [77] and age-dependently accumulated enlarged lysosomes containing lipofuscin in the kidney [77–79]. Recently, LRRK2 has been connected to regulate lysosomal function via its kinase activity towards several RAB GTPases [80]. Chloroquine-induced lysosomal overload led to translocation and activation of endogenous LRRK2 onto the lysosomal membrane through the mediation of upstream adaptor RAB7L1. In turn, LRRK2 stabilized its substrates RAB8A and RAB10 on lysosomes through their phosphorylation and further regulated lysosomal homeostasis [80]. Pharmacological inhibition of LRRK2 kinase resulted in reduced localization of α-synuclein in RAB35-positive compartments and might facilitate α-synuclein clearance via increased trafficking to the lysosome [81].
Autophagic dysregulation in the context of PD-associated LRRK2 mutations has been reported in different animal models in several independent studies. Immunohistochemical analysis of human brain sections revealed LRRK2 positive cytoplasmic puncta that corresponded to autophagic vacuoles [82]. LRRK2 G2019S knock-in mice showed elevated LC3-II levels [83]. Differentiated SH-SY5Y cells expressing the LRRK2 G2019S mutation exhibited a striking increase in autophagic vacuoles in both neuronal neurites and soma in addition to elevated LC3-II levels [84]. Neuritic autophagy in LRRK2 G2019S mutants was mediated via the mitogen activated protein kinase/extracellular signal regulated protein kinase (MAPK/ERK)-related signaling. Similar findings were shown in LRRK2 G2019S mutant fibroblasts, which underwent cell death likely triggered through apoptosis [85]. Compared to cells from non-mutant carriers, these fibroblasts were more sensitive to the parkinsonian toxin 1-methyl-4-phenylpyridinium ion (MPP+)-induced mTOR-dependent autophagy [86], emphasizing the synergistic contribution of genetic and environmental factors to PD etiology. In another cellular model, expression of the LRRK2 R1441C mutation caused accumulation of large autophagic vacuoles and multivesicular bodies containing incompletely degraded material along with increased levels of the autophagic receptor p62/SQSTM1 [82]. Collectively, the above experiments indicate deleterious effects of LRRK2 gain-of-function mutations on autophagy through different mechanisms.
Similar to α-synuclein, wildtype LRRK2 can be selectively degraded by CMA. One of eight putative CMA-targeting motifs in the LRRK2 protein is required for binding to the chaperone protein HSC70 [87]. This degradation pathway is compromised in the presence of pathogenic LRRK2 mutations or high concentrations of wildtype LRRK2. Yet, the mechanism of LRRK2-mediated CMA inhibition is different from that of α-synuclein. Instead of increased binding to lysosomal membranes as seen with mutant α-synuclein, both LRRK2 G2019S and R1441G blocked the formation of the CMA translocation complex at the lysosomal membrane by inducing LAMP2A and HSC70 accumulation. This was shown in neuronal cultures and LRRK2 transgenic mice as well as in human iPSC–derived dopaminergic neurons and autopsy brain from PD patients with LRRK2 mutations [87, 88]. Moreover, as a result of LRRK2-induced CMA blockade, α-synuclein degradation was inhibited and accumulation of oligomeric α-synuclein could be detected in the striatum and cortex of aged LRRK2 R1441G knockin mice compared to age-matched wildtype controls [88].
In addition to effects on general autophagy and CMA, LRRK2 mutations were also reported to affect mitophagy. LRRK2 G2019S was shown to stimulate mitophagy in several cellular models through direct interaction with ULK1, an essential regulator of autophagy [89]. Moreover, LRRK2 interacted with endogenous MKK4/7 and JIP3 and cooperatively mediated JNK signaling, which was suggested to be partially involved in LRRK2-mediated mitophagy induction. Expression of either LRRK2 G2019S or R1441C mutant in mouse cortical neurons dysregulated calcium homeostasis, leading to enhanced mitophagy and dendrite shortening [90]. The same group later showed that this pathway was mediated by transcriptional upregulation of the mitochondrial calcium uniporter (MCU) and the mitochondrial calcium uptake 1 protein (MICU1) through activation of the ERK½ pathway [91]. Both a direct binding to and phosphorylation of BCL-2, a mitochondrial anti-apoptotic protein, at threonine 56 by LRRK2 G2019S mutant was required for the mutant-induced mitochondrial depolarization and mitophagy [92]. Expression of the phospho-dead T56A BCL2 mutant corrected the autophagic damage and neuronal injury induced by LRRK2 G2019S. Furthermore, LRRK2 G2019S directly bound to and phosphorylated the fission factor DRP1 at threonine 595, leading to augmented mitochondrial fragmentation and excessive DRP1-mediated mitophagy in both cells expressing LRRK2 G2019S and in PD patient fibroblasts carrying the mutation [93]. Using live cell imaging with a pH-sensitive Rosella biosensor probe to reflect lysosomal breakdown of mitochondria, two more recent studies from the same group reported reduced mitophagy rates in fibroblasts carrying the LRRK2 G2019S mutation compared to cells isolated from healthy control subjects. Mechanistic analysis of these fibroblasts showed reduced numbers of mature autophagosomes [94] and impaired interactions between PRKN, DRP1 and their mitochondrial targets early during mitophagy activation attenuated PINK1-PRKN-directed mitophagy [95]. While wildtype LRRK2 promoted the removal of the mitochondrial transport factor RHOT1/Miro1 by forming a complex with it, LRRK2 G2019S disrupted this, slowed Miro removal and mitochondrial arrest, and delayed subsequent mitophagy [96]. However, others reported a “docking function” for Miro1 and that its reduction led to reduced PRKN recruitment to and degradation of damaged mitochondria [97]. Most recently, LRRK2 was shown to regulate PINK1-PRKN-dependent mitophagy via its substrate RAB10, which bound to the autophagy receptor optineurin (OPTN) and promoted its accumulation on depolarized mitochondria [98]. Enhanced phosphorylation of RAB10 in LRRK2 mutant patient cells impaired mitophagy, which was rescued by knockdown of LRRK2 or inhibition of its kinase activity.
In summary, LRRK2 pathogenic mutations can block CMA, stimulate autophagy or stall the flux of autophagy and mitophagy at several steps along the pathways. LRRK2 also influences lysosomes and recent studies further indicate a critical involvement of LRRK2 in the emerging field of synaptic autophagy as discussed below.
3.3. PINK1 and PRKN
Homozygous or compound heterozygous mutations in the PTEN-induced kinase 1 (PINK1) [99] and in PRKN (aka PARK2 or Parkin) [100] have been identified as the most common causes of autosomal recessive early-onset PD [101], with overall frequencies of 1–9% for PINK1 depending on the ethnicity [16] and nearly 50% for PRKN in young PD patients (≤ 40 years old) [102, 103]. Importantly, both proteins are functionally connected and jointly regulate selective mitochondrial autophagy [104]. PINK1 is a mitochondrial targeted kinase and constantly surveilles mitochondrial health. Upon mitochondrial depolarization, PINK1 is stabilized on the outer membrane of damaged mitochondria and then phosphorylates both the small modifier protein ubiquitin (Ub) and the Ub E3 ligase PRKN each at their conserved serine 65 (Ser65). This activates and recruits PRKN to damaged mitochondria where it provides additional Ub moieties for PINK1-dependent phosphorylation in a feedforward loop that further amplifies PINK1 and PRKN activities [105–107]. As a result, phosphorylated Ub chains (p-Ser65-Ub) decorate the outer membrane of damaged mitochondria and serve as a selective tag for the recognition by autophagy receptors and subsequent degradation via the autophagy-lysosomal pathway [104, 108–110].
PINK1 mutations with complete loss of function and heterozygous PINK1 or PD-related PRKN mutations abolish or hamper the activation of the PINK1-PRKN dependent mitophagy pathway. This is in line with our recent study showing absence or reduced mitophagy tag p-Ser65-Ub in the substantia nigra of postmortem PD brains with PINK1 or PRKN mutations [22, 23]. Reduced mitophagy leads to accumulation of damaged mitochondria and possibly to neuronal death. Since dopaminergic neurons are particularly vulnerable to mitochondrial dysfunction [14, 15], PINK1 or PRKN mutations that result in reduced mitophagy function may contribute to the selective neurodegeneration in the substantia nigra. In addition to early-onset PD, reduced activities of either enzyme may also contribute to later onset PD or may at least modify rates of progression and clinical phenotypes [111, 112]. Interestingly, neuropathological examination of PINK1 and PRKN mutation carriers often report the absence of LBs, the hallmark of PD pathology[113]. Earlier studies have shown reduced PRKN solubility in postmortem brains of aged individuals and PD patients [114–116], which could add to mitophagy impairment in sporadic PD. However, instead of decreased levels of p-Ser65-Ub, we detected selective increases of the mitophagy tag in the substantia nigra and hippocampus of sporadic PD cases [22, 23]. p-Ser65-Ub positive granules co-localized with mitochondrial and lysosomal markers and were frequently found adjacent to LBs and phospho-tau tangles. Numbers of p-Ser65-Ub positive cells increased with age and PD and strongly correlated with levels of LBs and tau tangles. In the absence of PINK1 and PRKN mutations, accumulation of the otherwise transient mitophagy tag p-S65-Ub in PD brains suggests either increased mitochondrial damage and/or decreased autophagic flux, both of which likely contribute to a generally overwhelmed mitochondrial turnover.
While studies of PINK1-PRKN directed mitophagy in human PD brain remain scarce, rigorous efforts in cellular or animal models have elegantly detailed the mechanisms underlying this pathway. Initial studies using PINK1 or PRKN mutant Drosophila exhibited obvious phenotypes including mitochondrial morphological abnormalities, locomotor defects, loss of dopaminergic neurons, muscle degeneration, and reduced life span [117, 118]. A later proteomic study in Drosophila revealed significantly reduced turnover rates of mitochondrial proteins and mitochondrial respiratory chain subunits in PRKN and PINK1 mutants [119]. The mitochondrial axonal transport machinery mediated through Miro protein was also impaired in PINK1 mutant flies [120]. Other substrates of PINK1 or PRKN, such as the mitochondrial chaperone TNF receptor-associated protein 1 (TRAP1) [121] and the mitochondrial protein BCL2 interacting protein 3 like (BNIP3L) [122], were found to act downstream to induce mitophagy and may be associated with PD pathogenesis. In contrast to flies, PINK1 or PRKN deletion in mouse models exhibited only subtle behavioral phenotypes [123–127]. Yet, PRKN deletion still resulted in reduced striatal synaptic excitability and plasticity, impaired dopamine release, and nigrostriatal deficits [125, 127]. PINK1 deletion also caused failure of striatal mitochondrial respiration and increased sensitivity to oxidative stress [128]. Mitochondrial proteins responsible for energy metabolism and membrane potential were significantly altered in PINK1 knockout mice [129]. Similar to the human condition, midbrain from these mice was most susceptible to the defective mitochondrial turnover and oxidative stress, whereas cortex and striatum could compensate for mitophagy dysfunction by feedback stimulation of other catabolic programs [129]. As PD-like phenotypes were absent in PRKN knockout mice at baseline, the importance of PRKN as an essential mitophagy regulator was further addressed in a mouse model with accumulation of dysfunctional mitochondria due to accelerated generation of mtDNA mutations (mutator mice) [130]. Consistent with the idea of a stress-induced pathway, selective loss of dopaminergic neurons in the substantia nigra was only found in aged PRKN-deficient mutator mice, but not in either single model alone. Noteworthy, the motor deficit in PRKN-mutator mice could be reversed by L-DOPA treatment.
With a better understanding of the PINK1-PRKN mitophagy pathway, emerging studies focus on how PD-associated mutations and PTMs of PINK1 and PRKN affect mitophagy and induce a PD phenotype. Depending on the exact mutation and their localization within the PRKN protein, expression of mutants in SH-SY5Y or HeLa cells led to mitophagy defects due to disruption at recognition, transport, aggregation or ubiquitination of damaged mitochondria [131–138], thereby implicating dysfunctional mitophagy in the development of PRKN-related parkinsonism. Meanwhile, several PD-linked PINK1 mutations showed impaired stabilization, enzymatic activity or binding to PRKN, thereby interfered with PRKN translocation and mitophagic execution [111, 135, 139–141]. Upon mitochondrial depolarization, iPSC derived dopaminergic neurons from PD patients with nonsense or missense mutations in the PINK1 gene showed impaired recruitment of overexpressed PRKN to mitochondria, increased mitochondrial copy number, and upregulated PPARG coactivator 1 alpha (PPARGC1A), an important regulator of mitochondrial biogenesis. These alterations could be corrected by lentiviral expression of wild-type PINK1 in mutant iPSC-derived PINK1 neurons [142]. In both mammalian cells and Drosophila, PINK1 mutations in the kinase domain failed to induce PRKN translocation to mitochondria and to induce mitochondrial aggregation [111, 140, 143]. Dopaminergic neurons derived from iPSCs with PRKN or PINK1 mutations also showed reduced p-Ser65-Ub signal and compromised mitophagy flux [144]. iPSC-derived dopamine neurons with mt-Keima confirmed neuronal mitophagy impairment in PRKN mutation carriers [145]. In addition, specific posttranslational modifications on PINK1 and PRKN can greatly alter their normal function and mimic genetic mutations [115, 146, 147]. Mitochondrial insults simulated age- or environmental-related stress and led to increased S-nitrosylated PINK1, thereby inhibiting its kinase activity [146]. S-nitrosylated PINK1 decreased PRKN translocation to mitochondrial membranes, disrupted mitophagy and subsequently led to cell death in cell lines and human iPSC-derived neurons. This modified PINK1 was also present in brains of α-synuclein transgenic mice, indicating a pathophysiological relevance for PD [146]. In addition, the neurotransmitter dopamine covalently modifies PRKN in living dopaminergic cells. This modification was shown to decrease PRKN solubility and inactivate its E3 ubiquitin ligase function [115]. More importantly, catechol-modified PRKN was only detected in the substantia nigra but not in other regions of normal human brain, suggesting a mechanism for the progressive loss of PRKN function in dopaminergic neurons during aging and sporadic PD [115]. In all, PINK1-PRKN dependent mitophagy pathway is essential for maintaining the timely turnover of damaged mitochondria and its impairment likely contributes to the pathogenesis of both familial and sporadic PD.
3.4. GBA
The initial observation of higher incidence of PD in families with Gaucher disease, a lysosomal storage disorder (LSD) caused by deficiency of the glucocerebrosidase enzyme (GCase), has led to the identification of heterozygous mutations in GBA as important and common risk factors for sporadic PD [16]. The encoded lysosomal enzyme is strongly involved in sphingolipid metabolism [148]. This discovery further strongly links autophagy-lysosomal dysfunction with PD pathogenesis. Furthermore, there is a bidirectional effect of GCase and α-synuclein in the form of a positive feedback loop [149]. Functional loss of GCase induced by PD-related GBA mutations in primary cultures or human iPSC neurons compromised lysosomal protein degradation, caused accumulation of α-synuclein, and resulted in neurotoxicity through aggregation-dependent mechanisms [149, 150]. The GCase substrate glucosylceramide (GlcCer), influenced the aggregation of α-synuclein in vitro by stabilizing soluble oligomeric intermediates. In turn, overexpression of α-synuclein inhibited the lysosomal activity of normal GCase in neurons and idiopathic PD brain, further propagating this pathogenic loop.
Intact lysosomal function and proper degradative capacity is critical for autophagic flux, thus PD-related pathogenic GBA mutations also disrupt the autophagy machinery. In primary mouse neurons and PD patient derived fibroblasts, GBA mutations compromised autophagic lysosome reformation, a cellular process to recover functional lysosomes from autolysosomes formed during autophagy, possibly due to reduced mTOR activity [151]. This defect was accompanied by elevation of PD-related disease markers including total and phosphorylated (Ser129) monomeric α-synuclein as well as amyloid oligomers [151]. To further study the molecular mechanisms in a more disease-relevant cellular model, ten independent iPSC lines from controls and unrelated PD patients heterozygous for the GBA N370S mutation were used [152]. In differentiated dopaminergic neurons, two ER-resident chaperones, HSPA5 and calreticulin (CALR) were significantly upregulated in GBA N370S cells compared to controls, suggesting the activation of ER stress. This likely contributed to the observed increase of LC3-II, the accumulation of autophagosomes, and the upregulation of key autophagy regulator BECN1 in GBA N370S mutant neurons. Increased basal levels of LC3-II were also found in GBA-PD iPSC-derived dopaminergic neurons when compared to regular and isogenic controls [153]. Inhibition of lysosomal degradation by leupeptin and ammonium chloride revealed a significant reduction in autophagic flux in GBA-PD neurons. Moreover, the decreased colocalization between LC3- and LAMP1-positive vacuoles in GBA-PD neurons suggested an impaired autophagosome-lysosome fusion in GBA mutant neurons.
A functional autophagy pathway is one of the key determinants for the effective turnover of damaged mitochondria via mitophagy. This protective pathway plays an important role in maintaining mitochondrial health. Several studies reported both mitochondrial dysfunction and autophagy defects in GBA knockout mice and in cellular models of Gaucher disease [154–157]. Therefore, PD-associated heterozygous GBA mutations may as well induce aberrant mitophagy in PD. A dual mechanism of the GBA L444P mutation-induced mitophagy defect has been demonstrated in primary hippocampal neurons from heterozygous GBA L444P knockin mouse brains and corroborated in postmortem brain tissue from PD patients carrying heterozygous GBA mutations [155]. Compared to controls, heterozygous GBA L444P neurons and tissue showed both impaired mitochondrial priming and reduced efficacy of autophagy-lysosomal degradation, which together led to mitophagy defect and accumulation of mitochondrial content. Impaired mitochondrial function and mitophagy was also found in a recent study using iPSC-derived neurons from GBA-PD patients compared to isogenic controls, along with a significant reduction of the mitophagy adaptor protein BNIP3L/NIX and increased ER stress [158]. Moreover, NAD+ might play a role in maintaining mitochondrial biogenesis and quality control. The NAD+ precursor nicotinamide riboside significantly ameliorated this cellular abnormality and increased mitophagy in patient neurons [158].
3.5. Other PD Genes
Several other PD genes have been implicated in aberrant autophagy regulation and may contribute to PD phenotype and pathogenesis. Mutations in vacuolar protein sorting 35 (VPS35) are linked to autosomal dominant PD [159, 160]. VPS35 is a component of the retromer complex. It mediates endosome-to-Golgi retrograde protein sorting [161] and is critical for the endosomal recruitment of the WASH complex [162], a protein complex that facilitates protein sorting [163, 164]. Reduced VPS35 mRNA levels were shown in the nigra dopaminergic neurons of PD patients [165, 166]. Several animal models have linked VPS35 deficiency or mutation to PD-related behavior and pathological changes, including α-synuclein accumulation in dopaminergic neurons, reduced dopamine release, nigral neurodegeneration, and locomotor deficits [162, 166, 167]. Additionally, the expression of D620N VPS35, the most common PD mutation, or VPS35 down-regulation induced robust hyperphosphorylated and conformation-specific tau pathology in mouse brain [168, 169]. VPS35 deficiency induced behavior impairment and tau accumulation could be rescued by VPS35 overexpression in a CTSD activity-dependent manner [169]. The critical role of VPS35-mediated lysosomal function in tau metabolism and neuropathology also extends to atypical parkinsonism [169], where reduced VPS35 levels were detected in brains of Progressive Supranuclear Palsy cases. On the molecular level, compared to wildtype VPS35, cells with D620N mutant restricted WASH complex recruitment to endosomes and caused abnormal trafficking of the autophagy protein ATG9A and defective autophagosome formation [162]. VPS35 deficiency or mutation was further suggested to reduce α-synuclein degradation and promote PD pathogenesis through impairing CMA in dopaminergic neurons via restricting endosome-to-Golgi retrieval of LAMP2A and accelerating LAMP2A degradation [167]. Interestingly, VPS35 expression reversed phenotypes of PRKN, but not PINK1 knockout in Drosophila [170].
The autosomal recessive PD gene PARK7 encodes the protein deglycase DJ-1 that has several important functions in mitochondrial physiology, protein translation, proteasome regulation, and chaperone activity [171, 172]. DJ-1 knockdown in microglia not only increased the microglia-induced toxicity towards dopaminergic neurons but also impaired autophagy-dependent degradation of p62/SQSTM1 and LC3 proteins and reduced α-synuclein phagocytosis by microglia [173]. DJ-1 deficiency in SH-SY5Y cells exacerbated PD-toxin-induced autophagic inhibition and apoptotic cell death [174]. In contrast, DJ-1 overexpression in cultured dopaminergic cells or in the substantia nigra of rat brains protected against another PD toxin, rotenone, and apoptotic cell death. This protective function was mediated through enhancing ERK-dependent autophagy [175]. High levels of DJ-1 immunoreactivity were reported in astrocytes-surrounding neurons with pathology in idiopathic PD [176], possibly reflecting the glial response to oxidative damage. Along this line, overexpression of astrocytic DJ-1 protected rats from rotenone-induced neurodegeneration through enhancing CMA [176]. Consistently, DJ-1 deficiency in SH-SY5Y cells or PD animal models aggravated α-synuclein aggregation through CMA inhibition. This provides further evidence of the molecular interaction between DJ-1 and the CMA pathway [177]. Other studies also demonstrated that DJ-1 acted downstream of, or in parallel to, PINK1-PRKN mitophagy to maintain mitochondrial integrity [178–180].
Proteins encoded by other parkinsonism-associated genes such as F-box protein 7 (FBXO7) [181], ATPase cation transporting 13A2 (ATP13A2) [182], and vacuolar protein sorting 13 homolog C (VPS13C) also play roles in regulating autophagic functions. The ATP13A2 protein is a lysosomal type 5 P-type ATPase. ATP13A2 mutations or reduced ATP13A2 levels acted as negative regulator of the autophagic-lysosomal pathway, impaired lysosomal function in dopaminergic neurons, and induced α-synuclein accumulation and subsequent neurotoxic effects [183–185]. This inhibitory function of ATP13A2 was mediated through regulating another PD-associated gene (SYT11) at both transcriptional and post-translational levels by a mechanism dependent on MYCBP2-induced ubiquitination of TSC2. This in turn led to mTORC1 activation, decreased TFEB-mediated transcription of SYT11, and increased SYT11 protein turnover regulated by ubiquitination and degradation [186]. A recent study showed that ATP13A2 also recruited the histone deacetylase 6 [HDAC6] to lysosomes to deacetylate cortactin and promote autophagosome-lysosome fusion and autophagy [187]. FBXO7 deficiency or mutations inhibited mitophagy, while FBXO7 overexpression facilitated PRKN translocation and mitophagy through a direct interaction with PRKN [188, 189]. Homozygous or compound heterozygous truncating mutations in VPS13C have recently been associated with a distinct form of early-onset parkinsonism that was characterized by rapid and severe disease progression and early cognitive decline. Partially localizing on the outer mitochondrial membrane, VPS13C closely regulated mitochondrial membrane potential, mitochondrial fragmentation as well as PINK1-PRKN dependent mitophagy and transcriptional regulation of PRKN [190].
More recently, a GWAS study showed that several common variants in sporadic PD were components of the autophagic-lysosomal pathway [8]. Another independent study demonstrated an excessive burden of rare, likely damaging LSD gene variants associated with PD risk [191]. 56% of PD patients carried at least one putative damaging variant in a LSD gene, and 21% carried multiple alleles. As described earlier, autophagic defects were prominent in PD postmortem brain. The general dysfunction in cellular clearance in PD brain also extends to impairment of different lysosomal components [192]. The collective data suggest remarkable similarities between PD and LSD at both genetic and biochemical levels, indicating a pathological overlap between both disorders. Homozygous mutation carriers in LSDs develop disease phenotype very early in life with rapid disease progression. PD cases with heterozygous lysosomal-related variants may represent a form of mild LSD with slower disease progression and mild disease phenotypes that do not reveal themselves unless additional stressors including age are present. The lysosomal proteases CTSD and cathepsin B (CTSB) were among the recent discovered new candidate PD susceptibility genes [8, 191]. A mouse model of CTSD deficiency exhibited impaired macroautophagy and reduced proteasome activity as well as extensive endogenous α-synuclein accumulation in neurons [193]. Conversely, overexpression of CTSD, but not CTSB or cathepsin L (CTSL), was protective against α-synuclein aggregation and α-synuclein-induced dopaminergic neurodegeneration in C. elegans. Similar results were also observed in neuroglioma and neuroblastoma cells [193]. A later study suggested that partial loss of CTSD activity is already sufficient to impair the degradation of internalized exogenous α-synuclein aggregates as well as to accelerate their transcellular transmission and secretion compared to wild type cells [194]. Although the aspartyl protease CTSD plays an important role in α-synuclein metabolism and clearance, purified lysosomal extracts combined with liquid chromatography–mass spectrometry analysis revealed that CTSD alone could not degrade α-synuclein and prevent fibril formation in vitro [195]. In contrast, the cysteine proteases CTSB and CTSL cleaved α-synuclein within its amyloid region to prevent protein aggregation.
Together with the genetic evidence from GWAS, the above described functional studies in cell and animal models as well as postmortem PD brains confirmed the involvement of these PD-related genes in regulating various autophagy pathways and lysosomal functions.
4. Synaptic homeostasis and autophagy in PD
It is noteworthy that a link between autophagic and synaptic vesicle trafficking is emerging in PD [45–50]. Although the normal functions of α-synuclein remain unclear, recent studies suggest a role as a physiologic attenuator of synaptic vesicle (SV) recycling through interactions with synapsins and vesicle associated membrane protein 2 (VAMP2) [196, 197], where α-synuclein facilitated SV clustering and restricted SV mobility [198, 199]. α-synuclein was also shown to promotes SNARE-complex assembly [200], which required its both N-terminal and C-terminal sequences for this physiological function but not for its neuropathological effects [201].
As mentioned above, multiple members of the RAB family of small GTPases, including RAB8, RAB10, RAB12, are physiological substrates of LRRK2 [202]. These RAB GTPases are critical regulators of membrane trafficking and have important roles in autophagy, synaptic vesicle recycling, and sorting [203–205], which are crucial for local autophagy at the presynaptic compartment (synaptic autophagy) as well as to sort and deliver synaptic membrane proteins to lysosomes [46, 47]. In fact, synaptic autophagy is essential for the timely removal of aggregated proteins at the presynaptic compartment, which is especially important for the survival of the extremely ramified dopaminergic neurons [45]. PD models with pathogenic LRRK2 mutations showed both impaired autophagy and synaptic defects [206], which may result from the abnormal phosphorylation of RAB GTPases. Additionally, LRRK2 phosphorylated endophilin A1 (EndoA) at threonine 73 and serine 75, acting as a switch to control EndoA to form high curvature membrane zones in synaptic autophagy [207]. Moreover, LRRK2 was also shown to phosphorylate a binding partner of EndoA, synaptojanin 1 (SYNJ1) [208, 209]. PD-related pathogenic LRRK2 mutations significantly increased the phosphorylation at these sites, interfered with its interaction with Endo A and disrupted synaptic vesicle endocytosis [208, 209]. SYNJ1 itself also played a role in the regulation of synaptic autophagy, which required its SAC1 domain to dephosphorylate PI(3)P/PI(3,5)P2 and release WIPI2/ATG18A for autophagosome formation at presynaptic terminals [210]. Moreover, LRRK2 has been shown to bind Dynamin 1 (DNM1) [211], another presynaptic protein involved in regulating clathrin-mediated endocytosis, via its C-terminal WD40 domain [212] and phosphorylate DNM1 in vitro [211]. Auxilin and RME-8 are co-chaperones that affect later steps of endocytosis and HSC70-dependent autophagy processes at synapses [213–215]. A recent study has demonstrated that LRRK2 mediated phosphorylation of auxilin in its clathrin-binding domain at serine 627 [216]. Disrupted synaptic vesicle endocytosis and decreased synaptic vesicle density in LRRK2 mutant iPSC-derived dopaminergic neurons were partially attenuated by restoring auxilin function [216]. LRRK2 has also been functionally linked to VPS35 involved in protein recycling [217]. Compare to the wildtype, VPS35 D620N strikingly elevated LRRK2 kinase activity towards RABs in mouse fibroblasts tissues as well as in PD patients [218]. In a Drosophila model, VPS35 and LRRK2 were shown to cooperatively regulate synaptic endocytosis and SV regeneration through the RAB-mediated endocytic pathway [219].
Also PRKN has been implicated in various aspects of synaptic function [220]. PRKN was shown to bind to the EndoA SH3 domain through its Ub-like domain with a similar affinity as EndoA binds to SYNJ1 [221]. Of note, EndoA, SYNJ1, DNM1, and VPS35 are all ubiquitinated by PRKN at nerve terminals [221–223], which potentially may modulate presynaptic dopamine release and reuptake [220] as well as SV endocytosis [47]. Given the synaptic localization of PRKN and mitochondria, mitophagy may as well occur in the synaptic compartment [224]. While controversial results were reported, one study described local mitophagy following selective damage of a subset of mitochondria in hippocampal axons. This axonal mitophagy required PINK1-dependent PRKN and LC3 recruitment to damaged mitochondria. Moreover, mutations or variants in many genes encoding synaptic proteins SNCA, EndoA, SYNJ1, auxilin, and RME-8 are all risk factors for PD [47], suggesting a central role of impaired synaptic homeostasis in PD pathogenesis. All of the above suggest that the normal function of many PD-related genes play a critical role in maintaining synaptic autophagy.
5. Impact of impaired autophagy on PD-related phenotypes
As described above, PD-associated mutations strongly impair various steps of several autophagy signaling pathways. In turn, autophagy deficiency or blockade of flux not only dysregulates functions of proteins encoded by PD genes, but also exacerbates α-synuclein and tau pathology, forming a bidirectional pathogenic loop (Figure 2). In several studies, mice with cell-specific deletion of the essential autophagy gene ATG7 in midbrain dopamine neurons developed p62/SQSTM1 and Ub labeled inclusions, dystrophic dendrites, and reduced striatal dopamine levels as well as dopamine neuron degeneration and locomotor impairments [225–227]. In CNS-specific ATG7 knockout mice, accumulation of α-synuclein and LRRK2 proteins was found in presynaptic terminals [225]. In a zebrafish model treated with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), levels of another autophagy-related protein ATG5 were decreased and autophagy flux was blocked, which led to upregulation of β-synuclein, PRKN, and PINK1 [228]. In addition, these zebrafish exhibited locomotor deficits and dopamine neuron loss. ATG5 overexpression was able to rescue these PD phenotypes. A recent report demonstrated opposing effects of autophagy impairment in dopamine neurons on PD pathology and behavior [229]. Similarly, conditional knockout of ATG7 only in dopamine neurons of mice led to p62/SQSTM1-positive LB-like aggregates and selective neuronal loss in the nigra. Surprisingly, α-synuclein overexpression-induced PD-like behavior impairment were improved in these ATG7 deficiency mice, which was likely due to the reduced dopamine uptake and increased extracellular dopamine release as a possible compensatory response [229].
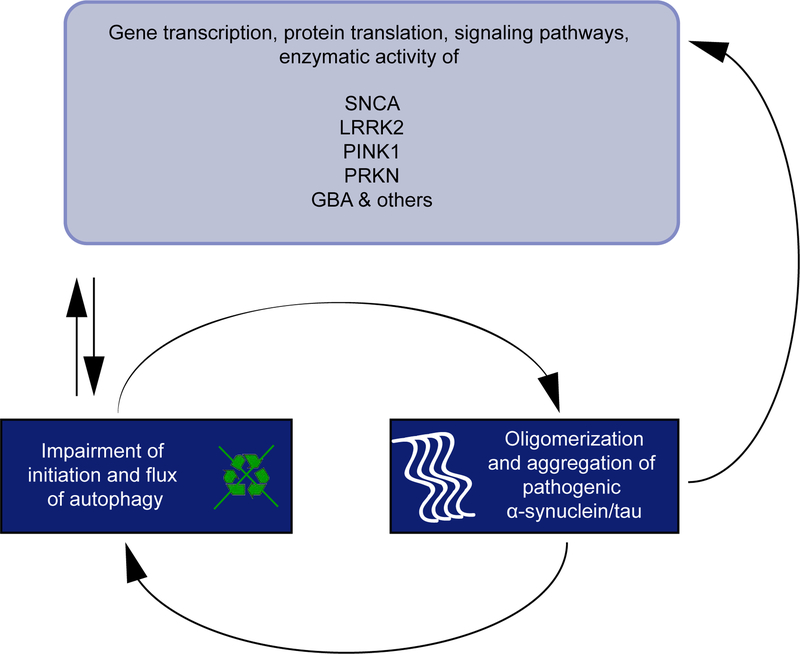
Figure 2. The multidirectional forward loop of PD-related genes, autophagy function, and PD pathology.
A variety of PD-related genes and encoded proteins are functionally linked to the autophagy pathway. Disease-associated mutations strongly impair the initiation and flux of this essential degradation process. In turn, autophagy defects dysregulate gene transcription and translation of encoded proteins as well as their downstream signaling pathways or enzymatic activities. In addition, autophagy deficiency or blockage of autophagic flux increases oligomerization or aggregation of α-synuclein and tau proteins in the PD brain. This will exacerbate levels of autophagy dysfunction and results in a pathological feedforward loop that will also interfere with physiological functions of other essential proteins.
With respect to the role of impaired CMA for PD related phenotypes, two independent studies showed that CMA inhibition led to the accumulation of soluble high molecular weight, detergent-insoluble species of α-synuclein [230] or α-synuclein and Ub double positive inclusions in rat nigra neurons [231]. This CMA defect in rats caused PD-like phenotypes such as progressive loss of dopaminergic neurons and reduced striatal dopamine levels as well as motor deficits [231]. In addition to inhibiting autophagy via knocking down autophagy-related genes, neuromelanin that usually accumulates in dopamine neurons in the human substantia nigra during aging was recently reported to induce failures in autophagy and the Ub-proteasome system [232]. This general impairment of proteostasis in neuromelanin-containing cells was associated with hypokinesia, early dopaminergic dysfunction, striatal axonal damage, LB-like formation, and nigrostriatal neurodegeneration in a unique tyrosinase-overexpressing rat model that produced human-like neuromelanin in dopamine neurons at levels similar to elderly humans. Enhancing lysosomal proteostasis by overexpressing nuclear TFEB within nigra neurons was able to reduce intracellular neuromelanin and rescue PD-like phenotypes in the animals [232].
6. Biomarkers to detect early autophagic changes in PD
Since many studies have highlighted the pivotal role of autophagy impairment in α-synuclein accumulation and neurodegeneration in PD, autophagy pathway members have great potential to serve as disease markers for early PD detection. Several studies examined autophagy markers in peripheral blood mononuclear cells (PBMC) of patients with sporadic PD and compared to healthy subjects. Reduced levels of HSC70, LAMP2 and increased levels of LC3-II have been reported [233–236], suggesting a systemic autophagy alteration in PD patients. Later, a whole-transcriptome analysis using PBMC of PD patients revealed mRNA downregulation of six core autophagy regulators (Unc-51-like kinase 3 [ULK3], ATG2A, ATG4B, ATG5, ATG6L1, and HDAC6) but upregulated protein levels of upstream autophagy markers (ULK1, BECN1, and autophagy and beclin 1 regulator 1 [AMBRA1]), possibly as a negative feedback of mRNA expression for these proteins [237]. Increased protein levels of these autophagy markers correlated with α-synuclein elevation in PBMCs. The amount of oligomeric α-synuclein in PBMCs was associated with the clinical severity of PD and the degeneration of cardiac sympathetic nerves [237]. Similar to autophagy changes in PBMCs, LC3B, LAMP2, BECN1, and ATG5 were significantly reduced in the cerebrospinal fluid (CSF) of early-stage PD patients compared to controls, with LC3B levels being the most sensitive and specific towards a diagnostic biomarker of PD [238]. Moreover, LC3B levels strongly correlated with the severity of parkinsonian motor symptoms and the asymmetry index in the caudate and putamen, one of the most representative markers for disease progression in patients with PD [238]. CMA pathway dysfunction was also detected in the CSF of female PD patients with LRRK2 mutations who had strongly reduced LAMP2A levels [239]. It is of note that recent studies emphasize a role of posttranslationally modified autophagy proteins for specific autophagy pathway. For instance, phosphorylated Ub (p-Ser65-Ub) acts as an activator of PRKN and recruits cytosolic PRKN from the cytoplasm to damaged mitochondria. PTMs of oligomerized p62/SQSTM1 at the LC3-binding site or in the Ub-binding domain regulate selective autophagy pathways such as aggrephagy (p-Ser349), mitophagy (p-Ser403), and secretory autophagy [240–243]. Thus, the development of biomarker assays to specifically target PTM signals on autophagy adaptors might provide a better understanding of PD-related autophagy defects.
7. Current challenges and future perspectives
Numerous studies have led to a better understanding of the involvement of autophagy in PD and its contribution to causes, onset, and progression of disease. Yet, it is of great importance to address the dynamic properties of this pathway. Up- or down-regulation of certain autophagy related proteins may not necessarily reflect a single directional alteration of the autophagy pathway. For instance, increases of the mitophagy tag p-Ser65-Ub can be caused by impaired downstream autophagic/lysosomal degradation in addition or alternatively to mitochondrial stress-induced overactivation of the pathway. It is thus critical to determine the flux through the entire autophagy pathway, the balance of the incoming autophagic cargo and the degradative capacity of lysosomes. Most studies that monitor autophagic flux were performed in cell culture models due to intrinsic limitations of animal and human studies. Novel transgenic animal models including mt-Keima [244], mito-QC [245], and most recently TRGL6 mice [246] have been generated to interrogate mitophagic and autophagic flux in vivo using dual-fluorescence reporters. While alterations of autophagy markers have been described before in LB disease, analyses were limited and did not necessarily consider the interconnectivity and dynamics of the system. The development of multiplexed imaging to co-label disease with various organelle markers may help to approximate the critical site of impairment. More detailed single cell or even single organelle analyses rather than whole bulk tissue/lysate analysis will reveal more accurate autophagy changes in PD. As autophagy functions may be differently regulated in neurons and glial cells, such analyses will provide additional knowledge on cell type- and cell compartment-specific autophagy alterations. For example, a recent study using multiple fluorescent probes demonstrated the presence of local degradation in the axon [247]. Soma-derived lysosomes could rapidly localize to distal axons and target autophagosomes and α-synuclein cargos for local degradation. In addition to improving the spatial resolution of autophagy research, longitudinal studies in animal and human cohorts will also benefit investigating age-related and dynamic autophagy-related changes. Nevertheless, the development of new tools/techniques is a necessary next step to investigate autophagy dysfunction in the human disease context.
Over the past decades, overexpression of PD-related genes or mutations in cells or animal models provided great opportunities to explore autophagic changes, to unravel the underlying mechanisms, and to nominate potential therapeutic targets for PD. Current and future studies with cell and animal models should attempt to mimic true physiological and pathological conditions. As other neurodegenerative diseases with abnormal protein aggregation, α-synuclein accumulation is one of the major pathological features in PD. Excessive α-synuclein impairs autophagy degradation, which in turn further exacerbate α-synuclein aggregation. It is critical to find the origin of this vicious cycle and to distinguish adaptive responses from detrimental consequences in the organism for future studies.
The autophagy system connects endosomal, lysosomal, and secretory pathways [248, 249], forming a vesicular continuum, and acting as a major hub to sort and rout cargo for recycling, degradation, or secretion. In addition to macroautophagy, CMA, and selective autophagy mentioned in this review, secretory autophagy also plays an important role in α-synuclein spreading and PD pathogenesis. Block of flux results in α-synuclein release through hybrid autophagosome/exosome vesicles and promotes spread of α-synuclein [250, 251]. Together, these data suggest a dual role for autophagy in degradation and secretion of aggregation-prone proteins. Impairments of the system at different stages contribute to processing, aggregation, and toxicity of subspecies of α-synuclein and other PD-related pathological proteins.
Due to the great advances in the field, it has become evident that aggravated PD pathology and impaired autophagy are forming a bidirectional pathogenic loop. However, different studies have shown changes of autophagic signals in PD in either direction. Therefore, an essential question to address is whether stimulation of autophagy is protective against for PD. It has been suggested that increased autophagic flux might be compensatory towards other blocked degradation pathways. However, overactivation of autophagy pathways could also be neurotoxic due to the overwhelmed lysosomal clearance that may stress the cells. To dissect pathogenic mechanisms from adaptive responses, it will be critical to identify the primary block in autophagic flux by a thorough analysis of different autophagy pathway. Future drug development targeting autophagy should not only aim at stimulating the pathway but should also ensure proper degradative flux at the same time.
Highlights:
Dysregulations of different autophagy pathways in Parkinson’s disease (PD) brain
Effects of PD-related genes on different autophagy pathways
Synaptic homeostasis and autophagy in PD
Impact of impaired autophagy on PD-related phenotype
Autophagy proteins as early PD biomarkers
Current challenges and future perspectives in the autophagy research in PD
Acknowledgements
W.S. is partially supported by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) [R01/RF1 NS085070 and U54 NS110435], the NIH/National Institute of Aging (NIA) [R56 AG062556], the Department of Defense Congressionally Directed Medical Research Programs (CDMRP) [W81XWH-17-1-0248], the Florida Department of Health - Ed and Ethel Moore Alzheimer’s Disease Research Program [9AZ10], the Michael J. Fox Foundation for Parkinson’s Research (MJFF), Mayo Clinic Foundation and the Center for Biomedical Discovery (CBD). X.H. is supported by a pilot grant from the Mayo Clinic Alzheimer Disease Research Center (ADRC) and a fellowship awarded by the American Parkinson Disease Association (APDA). F.C.F. is the recipient of fellowships from the Younkin Scholar Program and the APDA and is supported in part by the MJFF and a Gerstner Family Career Development Award from the Mayo Clinic Center for Individualized Medicine (CIM).
Footnotes
Declaration of interest
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet. 2009;373:2055–66. [DOI] [PubMed] [Google Scholar]
- [2].Langston JW. The Parkinson’s complex: parkinsonism is just the tip of the iceberg. Ann Neurol. 2006;59:591–6. [DOI] [PubMed] [Google Scholar]
- [3].Poewe W. Non-motor symptoms in Parkinson’s disease. Eur J Neurol. 2008;15 Suppl 1:14–20. [DOI] [PubMed] [Google Scholar]
- [4].Dickson DW, Braak H, Duda JE, Duyckaerts C, Gasser T, Halliday GM, et al. Neuropathological assessment of Parkinson’s disease: refining the diagnostic criteria. Lancet Neurol. 2009;8:1150–7. [DOI] [PubMed] [Google Scholar]
- [5].Wills J, Jones J, Haggerty T, Duka V, Joyce JN, Sidhu A. Elevated tauopathy and alpha-synuclein pathology in postmortem Parkinson’s disease brains with and without dementia. Exp Neurol. 2010;225:210–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis. 2011;43:690–7. [DOI] [PubMed] [Google Scholar]
- [7].Zhu JH, Guo F, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chang D, Nalls MA, Hallgrimsdottir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Moors T, Paciotti S, Chiasserini D, Calabresi P, Parnetti L, Beccari T, et al. Lysosomal Dysfunction and alpha-Synuclein Aggregation in Parkinson’s Disease: Diagnostic Links. Mov Disord. 2016;31:791–801. [DOI] [PubMed] [Google Scholar]
- [11].Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem Sci. 2015;40:200–10. [DOI] [PubMed] [Google Scholar]
- [12].Xilouri M, Brekk OR, Stefanis L. Autophagy and Alpha-Synuclein: Relevance to Parkinson’s Disease and Related Synucleopathies. Mov Disord. 2016;31:178–92. [DOI] [PubMed] [Google Scholar]
- [13].Caballero B, Wang Y, Diaz A, Tasset I, Juste YR, Stiller B, et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell. 2018;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bolam JP, Pissadaki EK. Living on the edge with too many mouths to feed: why dopamine neurons die. Mov Disord. 2012;27:1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haddad D, Nakamura K. Understanding the susceptibility of dopamine neurons to mitochondrial stressors in Parkinson’s disease. FEBS Lett. 2015;589:3702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Expanding Parkinson’s disease genetics: novel risk loci, genomic context, causal insights and heritable risk. bioRxiv. 2019:388165. [Google Scholar]
- [19].Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. [DOI] [PubMed] [Google Scholar]
- [20].Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–44. [DOI] [PubMed] [Google Scholar]
- [21].Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- [22].Fiesel FC, Ando M, Hudec R, Hill AR, Castanedes-Casey M, Caulfield TR, et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015;16:1114–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hou X, Fiesel FC, Truban D, Castanedes Casey M, Lin WL, Soto AI, et al. Age- and disease-dependent increase of the mitophagy marker phospho-ubiquitin in normal aging and Lewy body disease. Autophagy. 2018;14:1404–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Fiesel FC, Springer W. Disease relevance of phosphorylated ubiquitin (p-S65-Ub). Autophagy. 2015;11:2125–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dehay B, Bove J, Rodriguez-Muela N, Perier C, Recasens A, Boya P, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, et al. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67:1464–72. [DOI] [PubMed] [Google Scholar]
- [27].Yoshii SR, Mizushima N. Monitoring and Measuring Autophagy. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Moors TE, Paciotti S, Ingrassia A, Quadri M, Breedveld G, Tasegian A, et al. Characterization of Brain Lysosomal Activities in GBA-Related and Sporadic Parkinson’s Disease and Dementia with Lewy Bodies. Mol Neurobiol. 2019;56:1344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Murphy KE, Gysbers AM, Abbott SK, Spiro AS, Furuta A, Cooper A, et al. Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease. Mov Disord. 2015;30:1639–47. [DOI] [PubMed] [Google Scholar]
- [30].Xia Q, Liao L, Cheng D, Duong DM, Gearing M, Lah JJ, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wakabayashi K, Tanji K, Odagiri S, Miki Y, Mori F, Takahashi H. The Lewy body in Parkinson’s disease and related neurodegenerative disorders. Mol Neurobiol. 2013;47:495–508. [DOI] [PubMed] [Google Scholar]
- [32].Shahmoradian SH, Lewis AJ, Genoud C, Hench J, Moors TE, Navarro PP, et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat Neurosci. 2019;22:1099–109. [DOI] [PubMed] [Google Scholar]
- [33].Golbe LI, Di Iorio G, Bonavita V, Miller DC, Duvoisin RC. A large kindred with autosomal dominant Parkinson’s disease. Ann Neurol. 1990;27:276–82. [DOI] [PubMed] [Google Scholar]
- [34].Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21-q23. Science. 1996;274:1197–9. [DOI] [PubMed] [Google Scholar]
- [35].Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. [DOI] [PubMed] [Google Scholar]
- [36].Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–8. [DOI] [PubMed] [Google Scholar]
- [37].Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–73. [DOI] [PubMed] [Google Scholar]
- [38].Khalaf O, Fauvet B, Oueslati A, Dikiy I, Mahul-Mellier AL, Ruggeri FS, et al. The H50Q mutation enhances alpha-synuclein aggregation, secretion, and toxicity. J Biol Chem. 2014;289:21856–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lesage S, Anheim M, Letournel F, Bousset L, Honore A, Rozas N, et al. G51D alpha-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann Neurol. 2013;73:459–71. [DOI] [PubMed] [Google Scholar]
- [40].Hoffman-Zacharska D, Koziorowski D, Ross OA, Milewski M, Poznanski JA, Jurek M, et al. Novel A18T and pA29S substitutions in alpha-synuclein may be associated with sporadic Parkinson’s disease. Parkinsonism Relat Disord. 2013;19:1057–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rosenberg SA, Zhai Y, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, et al. Immunizing patients with metastatic melanoma using recombinant adenoviruses encoding MART-1 or gp100 melanoma antigens. J Natl Cancer Inst. 1998;90:1894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Clayton DF, George JM. The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci. 1998;21:249–54. [DOI] [PubMed] [Google Scholar]
- [43].Clayton DF, George JM. Synucleins in synaptic plasticity and neurodegenerative disorders. J Neurosci Res. 1999;58:120–9. [PubMed] [Google Scholar]
- [44].Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci U S A. 2009;106:13010–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Soukup SF, Vanhauwaert R, Verstreken P. Parkinson’s disease: convergence on synaptic homeostasis. EMBO J. 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sheehan P, Yue Z. Deregulation of autophagy and vesicle trafficking in Parkinson’s disease. Neurosci Lett. 2019;697:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nguyen M, Wong YC, Ysselstein D, Severino A, Krainc D. Synaptic, Mitochondrial, and Lysosomal Dysfunction in Parkinson’s Disease. Trends Neurosci. 2019;42:140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Luningschror P, Sendtner M. Autophagy in the presynaptic compartment. Curr Opin Neurobiol. 2018;51:80–5. [DOI] [PubMed] [Google Scholar]
- [49].Vijayan V, Verstreken P. Autophagy in the presynaptic compartment in health and disease. J Cell Biol. 2017;216:1895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Azarnia Tehran D, Kuijpers M, Haucke V. Presynaptic endocytic factors in autophagy and neurodegeneration. Curr Opin Neurobiol. 2018;48:153–9. [DOI] [PubMed] [Google Scholar]
- [51].Winslow AR, Chen CW, Corrochano S, Acevedo-Arozena A, Gordon DE, Peden AA, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Song JX, Lu JH, Liu LF, Chen LL, Durairajan SS, Yue Z, et al. HMGB1 is involved in autophagy inhibition caused by SNCA/alpha-synuclein overexpression: a process modulated by the natural autophagy inducer corynoxine B. Autophagy. 2014;10:144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Oliveira LM, Falomir-Lockhart LJ, Botelho MG, Lin KH, Wales P, Koch JC, et al. Elevated alpha-synuclein caused by SNCA gene triplication impairs neuronal differentiation and maturation in Parkinson’s patient-derived induced pluripotent stem cells. Cell Death Dis. 2015;6:e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Koch JC, Bitow F, Haack J, d’Hedouville Z, Zhang JN, Tonges L, et al. Alpha-Synuclein affects neurite morphology, autophagy, vesicle transport and axonal degeneration in CNS neurons. Cell Death Dis. 2015;6:e1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Bjorklund A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013;110:E1817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yu WH, Dorado B, Figueroa HY, Wang L, Planel E, Cookson MR, et al. Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric alpha-synuclein. Am J Pathol. 2009;175:736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5. [DOI] [PubMed] [Google Scholar]
- [61].Xilouri M, Vogiatzi T, Vekrellis K, Park D, Stefanis L. Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One. 2009;4:e5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Malkus KA, Ischiropoulos H. Regional deficiencies in chaperone-mediated autophagy underlie alpha-synuclein aggregation and neurodegeneration. Neurobiol Dis. 2012;46:732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Choubey V, Safiulina D, Vaarmann A, Cagalinec M, Wareski P, Kuum M, et al. Mutant A53T alpha-synuclein induces neuronal death by increasing mitochondrial autophagy. J Biol Chem. 2011;286:10814–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chen L, Xie Z, Turkson S, Zhuang X. A53T human alpha-synuclein overexpression in transgenic mice induces pervasive mitochondria macroautophagy defects preceding dopamine neuron degeneration. J Neurosci. 2015;35:890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues M, Tenreiro S, Franssens V, et al. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012;8:1494–509. [DOI] [PubMed] [Google Scholar]
- [67].Martinez JH, Alaimo A, Gorojod RM, Porte Alcon S, Fuentes F, Coluccio Leskow F, et al. Drp-1 dependent mitochondrial fragmentation and protective autophagy in dopaminergic SH-SY5Y cells overexpressing alpha-synuclein. Mol Cell Neurosci. 2018;88:107–17. [DOI] [PubMed] [Google Scholar]
- [68].Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486:235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Di Maio R, Barrett PJ, Hoffman EK, Barrett CW, Zharikov A, Borah A, et al. alpha-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci Transl Med. 2016;8:342ra78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Grassi D, Howard S, Zhou M, Diaz-Perez N, Urban NT, Guerrero-Given D, et al. Identification of a highly neurotoxic alpha-synuclein species inducing mitochondrial damage and mitophagy in Parkinson’s disease. Proc Natl Acad Sci U S A. 2018;115:E2634–E43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li JQ, Tan L, Yu JT. The role of the LRRK2 gene in Parkinsonism. Mol Neurodegener. 2014;9:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Skibinski G, Nakamura K, Cookson MR, Finkbeiner S. Mutant LRRK2 toxicity in neurons depends on LRRK2 levels and synuclein but not kinase activity or inclusion bodies. J Neurosci. 2014;34:418–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15:223–32. [DOI] [PubMed] [Google Scholar]
- [74].West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, et al. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–32. [DOI] [PubMed] [Google Scholar]
- [75].West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rideout HJ, Stefanis L. The neurobiology of LRRK2 and its role in the pathogenesis of Parkinson’s disease. Neurochem Res. 2014;39:576–92. [DOI] [PubMed] [Google Scholar]
- [77].Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, et al. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener. 2012;7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Herzig MC, Kolly C, Persohn E, Theil D, Schweizer T, Hafner T, et al. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum Mol Genet. 2011;20:4209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hinkle KM, Yue M, Behrouz B, Dachsel JC, Lincoln SJ, Bowles EE, et al. LRRK2 knockout mice have an intact dopaminergic system but display alterations in exploratory and motor co-ordination behaviors. Mol Neurodegener. 2012;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Eguchi T, Kuwahara T, Sakurai M, Komori T, Fujimoto T, Ito G, et al. LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci U S A. 2018;115:E9115–E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Bae EJ, Kim DK, Kim C, Mante M, Adame A, Rockenstein E, et al. LRRK2 kinase regulates alpha-synuclein propagation via RAB35 phosphorylation. Nat Commun. 2018;9:3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009;18:4022–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yue M, Hinkle KM, Davies P, Trushina E, Fiesel FC, Christenson TA, et al. Progressive dopaminergic alterations and mitochondrial abnormalities in LRRK2 G2019S knock-in mice. Neurobiol Dis. 2015;78:172–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bravo-San Pedro JM, Niso-Santano M, Gomez-Sanchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell Mol Life Sci. 2013;70:121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yakhine-Diop SM, Bravo-San Pedro JM, Gomez-Sanchez R, Pizarro-Estrella E, Rodriguez-Arribas M, Climent V, et al. G2019S LRRK2 mutant fibroblasts from Parkinson’s disease patients show increased sensitivity to neurotoxin 1-methyl-4-phenylpyridinium dependent of autophagy. Toxicology. 2014;324:1–9. [DOI] [PubMed] [Google Scholar]
- [87].Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ho PW, Leung CT, Liu H, Pang SY, Lam CS, Xian J, et al. Age-dependent accumulation of oligomeric SNCA/alpha-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: role for therapeutic activation of chaperone-mediated autophagy (CMA). Autophagy. 2019:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhu Y, Wang C, Yu M, Cui J, Liu L, Xu Z. ULK1 and JNK are involved in mitophagy incurred by LRRK2 G2019S expression. Protein Cell. 2013;4:711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Cherra SJ 3rd, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Verma M, Callio J, Otero PA, Sekler I, Wills ZP, Chu CT. Mitochondrial Calcium Dysregulation Contributes to Dendrite Degeneration Mediated by PD/LBD-Associated LRRK2 Mutants. J Neurosci. 2017;37:11151–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Su YC, Guo X, Qi X. Threonine 56 phosphorylation of Bcl-2 is required for LRRK2 G2019S-induced mitochondrial depolarization and autophagy. Biochim Biophys Acta. 2015;1852:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Su YC, Qi X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum Mol Genet. 2013;22:4545–61. [DOI] [PubMed] [Google Scholar]
- [94].Korecka JA, Thomas R, Christensen DP, Hinrich AJ, Ferrari EJ, Levy SA, et al. Mitochondrial clearance and maturation of autophagosomes are compromised in LRRK2 G2019S familial Parkinson’s disease patient fibroblasts. Hum Mol Genet. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Bonello F, Hassoun SM, Mouton-Liger F, Shin YS, Muscat A, Tesson C, et al. LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson’s disease. Hum Mol Genet. 2019;28:1645–60. [DOI] [PubMed] [Google Scholar]
- [96].Hsieh CH, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St Lawrence E, et al. Functional Impairment in Miro Degradation and Mitophagy Is a Shared Feature in Familial and Sporadic Parkinson’s Disease. Cell Stem Cell. 2016;19:709–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Safiulina D, Kuum M, Choubey V, Gogichaishvili N, Liiv J, Hickey MA, et al. Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J. 2019;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wauters F, Cornelissen T, Imberechts D, Martin S, Koentjoro B, Sue C, et al. LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy. 2019:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. [DOI] [PubMed] [Google Scholar]
- [100].Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. [DOI] [PubMed] [Google Scholar]
- [101].Corti O, Lesage S, Brice A. What genetics tells us about the causes and mechanisms of Parkinson’s disease. Physiological reviews. 2011;91:1161–218. [DOI] [PubMed] [Google Scholar]
- [102].Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. The New England journal of medicine. 2000;342:1560–7. [DOI] [PubMed] [Google Scholar]
- [103].Puschmann A. Monogenic Parkinson’s disease and parkinsonism: clinical phenotypes and frequencies of known mutations. Parkinsonism & related disorders. 2013;19:407–15. [DOI] [PubMed] [Google Scholar]
- [104].Truban D, Hou X, Caulfield TR, Fiesel FC, Springer W. PINK1, Parkin, and Mitochondrial Quality Control: What can we Learn about Parkinson’s Disease Pathobiology? J Parkinsons Dis. 2017;7:13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. The Biochemical journal. 2014;460:127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–6. [DOI] [PubMed] [Google Scholar]
- [107].Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Molecular cell. 2014;56:360–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wong YC, Holzbaur EL. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy. 2015;11:422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60:7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Puschmann A, Fiesel FC, Caulfield TR, Hudec R, Ando M, Truban D, et al. Heterozygous PINK1 p.G411S increases risk of Parkinson’s disease via a dominant-negative mechanism. Brain. 2017;140:98–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Dawson TM, Dawson VL. Parkin plays a role in sporadic Parkinson’s disease. Neurodegener Dis. 2014;13:69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Schneider SA, Alcalay RN. Neuropathology of genetic synucleinopathies with parkinsonism: Review of the literature. Mov Disord. 2017;32:1504–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim KL, Dawson VL, et al. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J Biol Chem. 2003;278:48120–8. [DOI] [PubMed] [Google Scholar]
- [115].LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–21. [DOI] [PubMed] [Google Scholar]
- [116].Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay SP, et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet. 2005;14:3885–97. [DOI] [PubMed] [Google Scholar]
- [117].Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. [DOI] [PubMed] [Google Scholar]
- [118].Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–61. [DOI] [PubMed] [Google Scholar]
- [119].Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013;110:6400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Costa AC, Loh SH, Martins LM. Drosophila Trap1 protects against mitochondrial dysfunction in a PINK1/parkin model of Parkinson’s disease. Cell Death Dis. 2013;4:e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gao F, Chen D, Si J, Hu Q, Qin Z, Fang M, et al. The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum Mol Genet. 2015;24:2528–38. [DOI] [PubMed] [Google Scholar]
- [123].Kahle PJ, Haass C. How does parkin ligate ubiquitin to Parkinson’s disease? EMBO reports. 2004;5:681–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101:10744–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–35. [DOI] [PubMed] [Google Scholar]
- [126].Itier JM, Ibanez P, Mena MA, Abbas N, Cohen-Salmon C, Bohme GA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–91. [DOI] [PubMed] [Google Scholar]
- [127].Kitada T, Pisani A, Karouani M, Haburcak M, Martella G, Tscherter A, et al. Impaired dopamine release and synaptic plasticity in the striatum of parkin−/− mice. J Neurochem. 2009;110:613–21. [DOI] [PubMed] [Google Scholar]
- [128].Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Diedrich M, Kitada T, Nebrich G, Koppelstaetter A, Shen J, Zabel C, et al. Brain region specific mitophagy capacity could contribute to selective neuronal vulnerability in Parkinson’s disease. Proteome Sci. 2011;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, et al. Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron. 2015;87:371–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Walinda E, Morimoto D, Sugase K, Shirakawa M. Dual Function of Phosphoubiquitin in E3 Activation of Parkin. J Biol Chem. 2016;291:16879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sauve V, Lilov A, Seirafi M, Vranas M, Rasool S, Kozlov G, et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 2015;34:2492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–31. [DOI] [PubMed] [Google Scholar]
- [136].Fiesel FC, Caulfield TR, Moussaud-Lamodiere EL, Ogaki K, Dourado DF, Flores SC, et al. Structural and Functional Impact of Parkinson Disease-Associated Mutations in the E3 Ubiquitin Ligase Parkin. Hum Mutat. 2015;36:774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Yi W, MacDougall EJ, Tang MY, Krahn AI, Gan-Or Z, Trempe JF, et al. The landscape of Parkin variants reveals pathogenic mechanisms and therapeutic targets in Parkinson’s disease. Hum Mol Genet. 2019;28:2811–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–8. [DOI] [PubMed] [Google Scholar]
- [140].Ando M, Fiesel FC, Hudec R, Caulfield TR, Ogaki K, Gorka-Skoczylas P, et al. The PINK1 p.I368N mutation affects protein stability and ubiquitin kinase activity. Mol Neurodegener. 2017;12:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Siuda J, Jasinska-Myga B, Boczarska-Jedynak M, Opala G, Fiesel FC, Moussaud-Lamodiere EL, et al. Early-onset Parkinson’s disease due to PINK1 p.Q456X mutation--clinical and functional study. Parkinsonism Relat Disord. 2014;20:1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, Krainc D. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Song S, Jang S, Park J, Bang S, Choi S, Kwon KY, et al. Characterization of PINK1 (PTEN-induced putative kinase 1) mutations associated with Parkinson disease in mammalian cells and Drosophila. J Biol Chem. 2013;288:5660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014;33:2473–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Suzuki S, Akamatsu W, Kisa F, Sone T, Ishikawa KI, Kuzumaki N, et al. Efficient induction of dopaminergic neuron differentiation from induced pluripotent stem cells reveals impaired mitophagy in PARK2 neurons. Biochem Biophys Res Commun. 2017;483:88–93. [DOI] [PubMed] [Google Scholar]
- [146].Oh CK, Sultan A, Platzer J, Dolatabadi N, Soldner F, McClatchy DB, et al. S-Nitrosylation of PINK1 Attenuates PINK1/Parkin-Dependent Mitophagy in hiPSC-Based Parkinson’s Disease Models. Cell Rep. 2017;21:2171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Meng F, Yao D, Shi Y, Kabakoff J, Wu W, Reicher J, et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol Neurodegener. 2011;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Indellicato R, Trinchera M. The Link between Gaucher Disease and Parkinson’s Disease Sheds Light on Old and Novel Disorders of Sphingolipid Metabolism. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, et al. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Bae EJ, Yang NY, Lee C, Lee HJ, Kim S, Sardi SP, et al. Loss of glucocerebrosidase 1 activity causes lysosomal dysfunction and alpha-synuclein aggregation. Exp Mol Med. 2015;47:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Magalhaes J, Gegg ME, Migdalska-Richards A, Doherty MK, Whitfield PD, Schapira AH. Autophagic lysosome reformation dysfunction in glucocerebrosidase deficient cells: relevance to Parkinson disease. Hum Mol Genet. 2016;25:3432–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Fernandes HJ, Hartfield EM, Christian HC, Emmanoulidou E, Zheng Y, Booth H, et al. ER Stress and Autophagic Perturbations Lead to Elevated Extracellular alpha-Synuclein in GBA-N370S Parkinson’s iPSC-Derived Dopamine Neurons. Stem Cell Reports. 2016;6:342–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Schondorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun. 2014;5:4028. [DOI] [PubMed] [Google Scholar]
- [154].Gegg ME, Schapira AH. Mitochondrial dysfunction associated with glucocerebrosidase deficiency. Neurobiol Dis. 2016;90:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Li H, Ham A, Ma TC, Kuo SH, Kanter E, Kim D, et al. Mitochondrial dysfunction and mitophagy defect triggered by heterozygous GBA mutations. Autophagy. 2019;15:113–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Osellame LD, Rahim AA, Hargreaves IP, Gegg ME, Richard-Londt A, Brandner S, et al. Mitochondria and quality control defects in a mouse model of Gaucher disease--links to Parkinson’s disease. Cell Metab. 2013;17:941–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Ivanova MM, Changsila E, Iaonou C, Goker-Alpan O. Impaired autophagic and mitochondrial functions are partially restored by ERT in Gaucher and Fabry diseases. PLoS One. 2019;14:e0210617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Schondorf DC, Ivanyuk D, Baden P, Sanchez-Martinez A, De Cicco S, Yu C, et al. The NAD+ Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC and Fly Models of Parkinson’s Disease. Cell Rep. 2018;23:2976–88. [DOI] [PubMed] [Google Scholar]
- [159].Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, et al. VPS35 mutations in Parkinson disease. American journal of human genetics. 2011;89:162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. American journal of human genetics. 2011;89:168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Molecular biology of the cell. 2000;11:4105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, et al. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat Commun. 2014;5:3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–23. [DOI] [PubMed] [Google Scholar]
- [164].Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, et al. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77:425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tsika E, Glauser L, Moser R, Fiser A, Daniel G, Sheerin UM, et al. Parkinson’s disease-linked mutations in VPS35 induce dopaminergic neurodegeneration. Hum Mol Genet. 2014;23:4621–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Tang FL, Erion JR, Tian Y, Liu W, Yin DM, Ye J, et al. VPS35 in Dopamine Neurons Is Required for Endosome-to-Golgi Retrieval of Lamp2a, a Receptor of Chaperone-Mediated Autophagy That Is Critical for alpha-Synuclein Degradation and Prevention of Pathogenesis of Parkinson’s Disease. J Neurosci. 2015;35:10613–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Chen X, Kordich JK, Williams ET, Levine N, Cole-Strauss A, Marshall L, et al. Parkinson’s disease-linked D620N VPS35 knockin mice manifest tau neuropathology and dopaminergic neurodegeneration. Proc Natl Acad Sci U S A. 2019;116:5765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Vagnozzi AN, Li JG, Chiu J, Razmpour R, Warfield R, Ramirez SH, et al. VPS35 regulates tau phosphorylation and neuropathology in tauopathy. Mol Psychiatry. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [170].Malik BR, Godena VK, Whitworth AJ. VPS35 pathogenic mutations confer no dominant toxicity but partial loss of function in Drosophila and genetically interact with parkin. Human molecular genetics. 2015;24:6106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Ariga H, Takahashi-Niki K, Kato I, Maita H, Niki T, Iguchi-Ariga SM. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxid Med Cell Longev. 2013;2013:683920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Biosa A, Sandrelli F, Beltramini M, Greggio E, Bubacco L, Bisaglia M. Recent findings on the physiological function of DJ-1: Beyond Parkinson’s disease. Neurobiol Dis. 2017;108:65–72. [DOI] [PubMed] [Google Scholar]
- [173].Nash Y, Schmukler E, Trudler D, Pinkas-Kramarski R, Frenkel D. DJ-1 deficiency impairs autophagy and reduces alpha-synuclein phagocytosis by microglia. J Neurochem. 2017;143:584–94. [DOI] [PubMed] [Google Scholar]
- [174].Gonzalez-Polo R, Niso-Santano M, Moran JM, Ortiz-Ortiz MA, Bravo-San Pedro JM, Soler G, et al. Silencing DJ-1 reveals its contribution in paraquat-induced autophagy. J Neurochem. 2009;109:889–98. [DOI] [PubMed] [Google Scholar]
- [175].Gao H, Yang W, Qi Z, Lu L, Duan C, Zhao C, et al. DJ-1 protects dopaminergic neurons against rotenone-induced apoptosis by enhancing ERK-dependent mitophagy. J Mol Biol. 2012;423:232–48. [DOI] [PubMed] [Google Scholar]
- [176].De Miranda BR, Rocha EM, Bai Q, El Ayadi A, Hinkle D, Burton EA, et al. Astrocyte-specific DJ-1 overexpression protects against rotenone-induced neurotoxicity in a rat model of Parkinson’s disease. Neurobiol Dis. 2018;115:101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Xu CY, Kang WY, Chen YM, Jiang TF, Zhang J, Zhang LN, et al. DJ-1 Inhibits alpha-Synuclein Aggregation by Regulating Chaperone-Mediated Autophagy. Front Aging Neurosci. 2017;9:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Hauser DN, Mamais A, Conti MM, Primiani CT, Kumaran R, Dillman AA, et al. Hexokinases link DJ-1 to the PINK1/parkin pathway. Mol Neurodegener. 2017;12:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Di Fonzo A, Dekker MC, Montagna P, Baruzzi A, Yonova EH, Correia Guedes L, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–5. [DOI] [PubMed] [Google Scholar]
- [182].Park JS, Mehta P, Cooper AA, Veivers D, Heimbach A, Stiller B, et al. Pathogenic effects of novel mutations in the P-type ATPase ATP13A2 (PARK9) causing Kufor-Rakeb syndrome, a form of early-onset parkinsonism. Hum Mutat. 2011;32:956–64. [DOI] [PubMed] [Google Scholar]
- [183].Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, et al. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci U S A. 2012;109:9611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Usenovic M, Tresse E, Mazzulli JR, Taylor JP, Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, alpha-synuclein accumulation, and neurotoxicity. J Neurosci. 2012;32:4240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Tsunemi T, Perez-Rosello T, Ishiguro Y, Yoroisaka A, Jeon S, Hamada K, et al. Increased Lysosomal Exocytosis Induced by Lysosomal Ca(2+) Channel Agonists Protects Human Dopaminergic Neurons from alpha-Synuclein Toxicity. J Neurosci. 2019;39:5760–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Bento CF, Ashkenazi A, Jimenez-Sanchez M, Rubinsztein DC. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat Commun. 2016;7:11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Wang R, Tan J, Chen T, Han H, Tian R, Tan Y, et al. ATP13A2 facilitates HDAC6 recruitment to lysosome to promote autophagosome-lysosome fusion. J Cell Biol. 2019;218:267–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Burchell VS, Nelson DE, Sanchez-Martinez A, Delgado-Camprubi M, Ivatt RM, Pogson JH, et al. The Parkinson’s disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat Neurosci. 2013;16:1257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Zhou ZD, Xie SP, Sathiyamoorthy S, Saw WT, Sing TY, Ng SH, et al. F-box protein 7 mutations promote protein aggregation in mitochondria and inhibit mitophagy. Hum Mol Genet. 2015;24:6314–30. [DOI] [PubMed] [Google Scholar]
- [190].Lesage S, Drouet V, Majounie E, Deramecourt V, Jacoupy M, Nicolas A, et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am J Hum Genet. 2016;98:500–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Robak LA, Jansen IE, van Rooij J, Uitterlinden AG, Kraaij R, Jankovic J, et al. Excessive burden of lysosomal storage disorder gene variants in Parkinson’s disease. Brain. 2017;140:3191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Klein AD, Mazzulli JR. Is Parkinson’s disease a lysosomal disorder? Brain. 2018;141:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain. 2008;1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Bae EJ, Yang NY, Lee C, Kim S, Lee HJ, Lee SJ. Haploinsufficiency of cathepsin D leads to lysosomal dysfunction and promotes cell-to-cell transmission of alpha-synuclein aggregates. Cell Death Dis. 2015;6:e1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].McGlinchey RP, Lee JC. Cysteine cathepsins are essential in lysosomal degradation of alpha-synuclein. Proc Natl Acad Sci U S A. 2015;112:9322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Atias M, Tevet Y, Sun J, Stavsky A, Tal S, Kahn J, et al. Synapsins regulate alpha-synuclein functions. Proc Natl Acad Sci U S A. 2019;116:11116–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Sun J, Wang L, Bao H, Premi S, Das U, Chapman ER, et al. Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling. Proc Natl Acad Sci U S A. 2019;116:11113–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Wang L, Das U, Scott DA, Tang Y, McLean PJ, Roy S. alpha-synuclein multimers cluster synaptic vesicles and attenuate recycling. Curr Biol. 2014;24:2319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Scott D, Roy S. alpha-Synuclein inhibits intersynaptic vesicle mobility and maintains recycling-pool homeostasis. J Neurosci. 2012;32:10129–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Burre J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sudhof TC. Alpha-synuclein promotes SNARE-complex assembly in vivo and in vitro. Science. 2010;329:1663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Burre J, Sharma M, Sudhof TC. Systematic mutagenesis of alpha-synuclein reveals distinct sequence requirements for physiological and pathological activities. J Neurosci. 2012;32:15227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Steger M, Tonelli F, Ito G, Davies P, Trost M, Vetter M, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Pavlos NJ, Jahn R. Distinct yet overlapping roles of Rab GTPases on synaptic vesicles. Small GTPases. 2011;2:77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–46. [DOI] [PubMed] [Google Scholar]
- [206].Roosen DA, Cookson MR. LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol Neurodegener. 2016;11:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, et al. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–21. [DOI] [PubMed] [Google Scholar]
- [208].Islam MS, Nolte H, Jacob W, Ziegler AB, Putz S, Grosjean Y, et al. Human R1441C LRRK2 regulates the synaptic vesicle proteome and phosphoproteome in a Drosophila model of Parkinson’s disease. Hum Mol Genet. 2016;25:5365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Pan PY, Li X, Wang J, Powell J, Wang Q, Zhang Y, et al. Parkinson’s Disease-Associated LRRK2 Hyperactive Kinase Mutant Disrupts Synaptic Vesicle Trafficking in Ventral Midbrain Neurons. J Neurosci. 2017;37:11366–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, Schoovaerts N, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017;36:1392–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Stafa K, Tsika E, Moser R, Musso A, Glauser L, Jones A, et al. Functional interaction of Parkinson’s disease-associated LRRK2 with members of the dynamin GTPase superfamily. Hum Mol Genet. 2014;23:2055–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Piccoli G, Onofri F, Cirnaru MD, Kaiser CJ, Jagtap P, Kastenmuller A, et al. Leucine-rich repeat kinase 2 binds to neuronal vesicles through protein interactions mediated by its C-terminal WD40 domain. Mol Cell Biol. 2014;34:2147–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Chang HC, Hull M, Mellman I. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J Cell Biol. 2004;164:1055–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Xhabija B, Vacratsis PO. Receptor-mediated Endocytosis 8 Utilizes an N-terminal Phosphoinositide-binding Motif to Regulate Endosomal Clathrin Dynamics. J Biol Chem. 2015;290:21676–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–6. [DOI] [PubMed] [Google Scholar]
- [216].Nguyen M, Krainc D. LRRK2 phosphorylation of auxilin mediates synaptic defects in dopaminergic neurons from patients with Parkinson’s disease. Proc Natl Acad Sci U S A. 2018;115:5576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Follett J, Norwood SJ, Hamilton NA, Mohan M, Kovtun O, Tay S, et al. The Vps35 D620N mutation linked to Parkinson’s disease disrupts the cargo sorting function of retromer. Traffic. 2014;15:230–44. [DOI] [PubMed] [Google Scholar]
- [218].Mir R, Tonelli F, Lis P, Macartney T, Polinski NK, Martinez TN, et al. The Parkinson’s disease VPS35[D620N] mutation enhances LRRK2-mediated Rab protein phosphorylation in mouse and human. Biochem J. 2018;475:1861–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Inoshita T, Arano T, Hosaka Y, Meng H, Umezaki Y, Kosugi S, et al. Vps35 in cooperation with LRRK2 regulates synaptic vesicle endocytosis through the endosomal pathway in Drosophila. Hum Mol Genet. 2017;26:2933–48. [DOI] [PubMed] [Google Scholar]
- [220].Sassone J, Serratto G, Valtorta F, Silani V, Passafaro M, Ciammola A. The synaptic function of parkin. Brain. 2017;140:2265–72. [DOI] [PubMed] [Google Scholar]
- [221].Trempe JF, Chen CX, Grenier K, Camacho EM, Kozlov G, McPherson PS, et al. SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol Cell. 2009;36:1034–47. [DOI] [PubMed] [Google Scholar]
- [222].Cao M, Milosevic I, Giovedi S, De Camilli P. Upregulation of Parkin in endophilin mutant mice. J Neurosci. 2014;34:16544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Williams ET, Glauser L, Tsika E, Jiang H, Islam S, Moore DJ. Parkin mediates the ubiquitination of VPS35 and modulates retromer-dependent endosomal sorting. Hum Mol Genet. 2018;27:3189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Evans CS, Holzbaur ELF. Quality Control in Neurons: Mitophagy and Other Selective Autophagy Mechanisms. J Mol Biol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Ahmed I, Liang Y, Schools S, Dawson VL, Dawson TM, Savitt JM. Development and characterization of a new Parkinson’s disease model resulting from impaired autophagy. J Neurosci. 2012;32:16503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Sato S, Uchihara T, Fukuda T, Noda S, Kondo H, Saiki S, et al. Loss of autophagy in dopaminergic neurons causes Lewy pathology and motor dysfunction in aged mice. Sci Rep. 2018;8:2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Hu ZY, Chen B, Zhang JP, Ma YY. Up-regulation of autophagy-related gene 5 (ATG5) protects dopaminergic neurons in a zebrafish model of Parkinson’s disease. J Biol Chem. 2017;292:18062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Hunn BHM, Vingill S, Threlfell S, Alegre-Abarrategui J, Magdelyns M, Deltheil T, et al. Impairment of Macroautophagy in Dopamine Neurons Has Opposing Effects on Parkinsonian Pathology and Behavior. Cell Rep. 2019;29:920–31 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Xilouri M, Brekk OR, Polissidis A, Chrysanthou-Piterou M, Kloukina I, Stefanis L. Impairment of chaperone-mediated autophagy induces dopaminergic neurodegeneration in rats. Autophagy. 2016;12:2230–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Carballo-Carbajal I, Laguna A, Romero-Gimenez J, Cuadros T, Bove J, Martinez-Vicente M, et al. Brain tyrosinase overexpression implicates age-dependent neuromelanin production in Parkinson’s disease pathogenesis. Nat Commun. 2019;10:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Wu G, Wang X, Feng X, Zhang A, Li J, Gu K, et al. Altered expression of autophagic genes in the peripheral leukocytes of patients with sporadic Parkinson’s disease. Brain Res. 2011;1394:105–11. [DOI] [PubMed] [Google Scholar]
- [234].Sala G, Stefanoni G, Arosio A, Riva C, Melchionda L, Saracchi E, et al. Reduced expression of the chaperone-mediated autophagy carrier hsc70 protein in lymphomonocytes of patients with Parkinson’s disease. Brain Res. 2014;1546:46–52. [DOI] [PubMed] [Google Scholar]
- [235].Papagiannakis N, Xilouri M, Koros C, Stamelou M, Antonelou R, Maniati M, et al. Lysosomal alterations in peripheral blood mononuclear cells of Parkinson’s disease patients. Mov Disord. 2015;30:1830–4. [DOI] [PubMed] [Google Scholar]
- [236].Prigione A, Piazza F, Brighina L, Begni B, Galbussera A, Difrancesco JC, et al. Alpha-synuclein nitration and autophagy response are induced in peripheral blood cells from patients with Parkinson disease. Neurosci Lett. 2010;477:6–10. [DOI] [PubMed] [Google Scholar]
- [237].Miki Y, Shimoyama S, Kon T, Ueno T, Hayakari R, Tanji K, et al. Alteration of autophagy-related proteins in peripheral blood mononuclear cells of patients with Parkinson’s disease. Neurobiol Aging. 2018;63:33–43. [DOI] [PubMed] [Google Scholar]
- [238].Youn J, Lee SB, Lee HS, Yang HO, Park J, Kim JS, et al. Cerebrospinal Fluid Levels of Autophagy-related Proteins Represent Potentially Novel Biomarkers of Early-Stage Parkinson’s Disease. Sci Rep. 2018;8:16866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Klaver AC, Coffey MP, Aasly JO, Loeffler DA. CSF lamp2 concentrations are decreased in female Parkinson’s disease patients with LRRK2 mutations. Brain Res. 2018;1683:12–6. [DOI] [PubMed] [Google Scholar]
- [240].Liu H, Dai C, Fan Y, Guo B, Ren K, Sun T, et al. From autophagy to mitophagy: the roles of P62 in neurodegenerative diseases. J Bioenerg Biomembr. 2017;49:413–22. [DOI] [PubMed] [Google Scholar]
- [241].Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, et al. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11:e1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Lee Y, Weihl CC. Regulation of SQSTM1/p62 via UBA domain ubiquitination and its role in disease. Autophagy. 2017;13:1615–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–24. [DOI] [PubMed] [Google Scholar]
- [244].Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015;60:685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Lee JH, Rao MV, Yang DS, Stavrides P, Im E, Pensalfini A, et al. Transgenic expression of a ratiometric autophagy probe specifically in neurons enables the interrogation of brain autophagy in vivo. Autophagy. 2019;15:543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Farfel-Becker T, Roney JC, Cheng XT, Li S, Cuddy SR, Sheng ZH. Neuronal Soma-Derived Degradative Lysosomes Are Continuously Delivered to Distal Axons to Maintain Local Degradation Capacity. Cell Rep. 2019;28:51–64 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Davis S, Wang J, Ferro-Novick S. Crosstalk between the Secretory and Autophagy Pathways Regulates Autophagosome Formation. Dev Cell. 2017;41:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Ejlerskov P, Rasmussen I, Nielsen TT, Bergstrom AL, Tohyama Y, Jensen PH, et al. Tubulin polymerization-promoting protein (TPPP/p25alpha) promotes unconventional secretion of alpha-synuclein through exophagy by impairing autophagosome-lysosome fusion. J Biol Chem. 2013;288:17313–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Lee HJ, Cho ED, Lee KW, Kim JH, Cho SG, Lee SJ. Autophagic failure promotes the exocytosis and intercellular transfer of alpha-synuclein. Exp Mol Med. 2013;45:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]