Abstract
Purpose
X-linked retinitis pigmentosa can manifest in female carriers with widely variable severity, while others remain unaffected. The contribution of X-chromosome inactivation (XCI) to phenotypic variation has been postulated but not demonstrated. Furthermore, the impact of genotype and genetic modifiers has been demonstrated in affected males but has not been well established in female carriers. The purpose of this study is to describe the scope of clinical phenotype in female carriers with mutations in RPGR, and to quantify the contribution of genotype, genetic modifiers, and XCI to phenotypic severity.
Design
Cohort study.
Subjects
Seventy-seven female carriers with RPGR mutations from 41 pedigrees.
Methods
Coding single nucleotide polymorphisms were sequenced in candidate genetic modifier genes encoding known RPGR-interacting proteins. XCI ratios were determined in genomic DNA isolated from blood (n=42) and saliva (n=20) using methylation status of X-linked polymorphic repeats. These genetic data were compared to disease severity based on quantitative clinical parameters.
Main Outcome Measures
Visual acuity (VA), Humphrey visual field (HVF), full field electroretinogram (ERG), and dark adaptation.
Results
Most individuals at all ages were mildly affected or unaffected, while the individuals who had progressed to moderate or severe vision loss were over the age of 30. RPGR genotype was not associated with clinical severity. The D1264N variant in RPGRIP1L was associated with more severe disease. Skewed XCI towards inactivation of the normal RPGR allele was associated with more severe disease. The XCI ratio in both blood and saliva was a predictor of visual function as measured by HVF diameter, rod amplitude, flicker amplitude, and flicker implicit time. For carriers with extreme XCI skewing of 80:20 or more, 57% were severely affected compared to 8% for this with XCI less than 80:20 (p=0.002).
Conclusions
Female carriers with mutations in RPGR demonstrate widely variable clinical severity. XCI ratios correlate with clinical severity and may serve as a predictor of clinically significant disease. As RPGR gene therapy trials are underway, there will be a future imperative to determine which carriers require intervention and when to intervene. XCI analysis may be useful to identify candidates for early intervention.
Introduction
Retinitis Pigmentosa (RP) is a group of blinding inherited retinal degenerations with diverse genetic etiologies. Approximately 65% of RP is non-syndromic with isolated retina findings,1 and variants in over 80 genes are now known to cause non-syndromic RP, with over 70 more genes contributing to syndromic forms.2 RP can be inherited in autosomal dominant, autosomal recessive, or X-linked patterns, with 40–50% of patients presenting as simplex RP with unknown inheritance, although it is presumed that the majority of these are autosomal recessive.1 Among RP, X-linked RP (XLRP) has a more severe clinical course on average, with affected males frequently experiencing nyctalopia in childhood, peripheral vision loss in adolescence, and deterioration of visual acuity in early adulthood. The majority of XLRP (at least 70%) is caused by mutations in a single gene, retinitis pigmentosa GTPase regulator (RPGR [MIM312610]), which encodes a protein that localizes to the photoreceptor connecting cilium and likely plays a role in protein transport.3–5 RPGR also accounts for 14% of males with simplex RP and 8.5% of “autosomal dominant” pedigrees (i.e. families that were initially characterized as dominant but later found to be X-linked after screening RPGR).6, 7 This suggests that RPGR may rival USH2A as the most common cause of RP overall.
Although XLRP tends to be more severe on average than other forms, the phenotype is not uniform and there is considerable variation in disease severity across patients, sometimes even within the same family.8–10 Previous studies have shown significant genotype-phenotype correlation in affected males: mutations in exons 1–14 are associated with more severe disease than mutations in ORF15, and predicted null alleles are associated with more severe disease than variants predicted to result in a translated protein.10–15 Furthermore, a candidate modifier approach in affected males revealed 2 potential genetic modifiers in genes encoding known RPGR-interacting proteins: IQCB1 and RPGRIP1L.10 In mice, a hypomorphic allele of CEP290, another RPGR-interacting protein, was found to modify phenotype in RPGR-knockout mice.16
Although some female carriers of XLRP are asymptomatic and never come to clinical attention, XLRP is not recessive, as demonstrated by the impressive number of XLRP pedigrees which are mistaken for autosomal dominant inheritance due to the presence of affected females.7 Although males are typically more severely affected, many females are symptomatic and the phenotype can vary from unaffected to severely affected.17–21
The phenotypic variation observed in female carriers has been postulated to result from differences in X-chromosome inactivation (XCI) ratios, although reported attempts to investigate this hypothesis have not shown a correlation thus far.20, 22, 23 XCI is a gene dosage compensation mechanism used in mammals to equilibrate X-linked gene expression across genders.24 Every cell in the female embryo inactivates either the maternal or paternal X chromosome at random at the 8–16 cell stage and passes this choice down to subsequent daughter cells.25, 26 Skewing of the XCI ratio from the expected mean 50:50 ratio can occur in the early embryo at the time of XCI choice or later during development, and can occur either randomly due to the Gaussian distribution of XCI choice, or non-randomly due to X-linked genetic variants.23 Skewed XCI ratios have been shown to contribute to manifestation of X-linked disease in female carriers of Duchenne Muscular Dystrophy and Hemophilia.27, 28 Investigations of XCI in female carriers of X-linked retina disease, including XLRP, choroideremia, and X-linked retinoschisis, have included small numbers of patients, ranging from 2 to 7, and to date have not shown any relationship between XCI ratios and disease manifestation.23
Gene therapy for RPGR-associated RP has demonstrated efficacy in canine and rodent models,29–35 and clinical trials are underway in affected male patients. Any successful approved treatment will likely eventually be offered to affected female carriers as well. Understanding the range of phenotypes in females is necessary to identify the best candidates for treatment and the likelihood that a female carrier will need treatment in her lifetime. In this study we characterize the clinical phenotype in a cohort of 77 female carriers of RPGR XLRP, describe the range of clinical variation, and for the first time demonstrate and quantify the contribution of XCI to phenotype in XLRP carriers.
Methods
Subjects and clinical assessment
Female carriers with mutations in RPGR were ascertained from at-risk female family members of affected males with RPGR mutations from the Southwest Eye Registry (SER) at the Retina Foundation of the Southwest (RFSW). The SER is a database of over 3,000 patients referred to the RFSW for retinal degenerative diseases. This study was performed in accordance with the Declaration of Helsinki and informed consent was obtained from all participants. The research was approved by the Committees for Protection of Human Subjects at the University of Texas Southwestern Medical Center, the University of Texas, Health Science Center at Houston, and the Internal Review Board at the University of Michigan.
For each patient, manifest refraction and best-corrected visual acuity were assessed with the NIKON RETINOMAX using the EVA-ETDRS visual acuity chart. Humphrey visual fields were obtained using a model 740 Humphrey field analyzer. Program 30–2 was used to determine static parametric thresholds at 74 locations within the central 30 degrees. Frequency domain optical coherence tomography (OCT) retinal scans were obtained using a Heidelberg Spectralis OCT. Fundus photos were obtained with a Canon digital camera (CF-60UD) and included a posterior pole view and additional peripheral views to document pigmentary changes and vessel attenuation if present. To assess dark-adaptation, pupils were maximally dilated using 1.0% cyclopentolate hydrochloride and 2.5% phenylephrine hydrochloride, followed by 45 minutes of dark adaptation. The final dark-adapted threshold was determined using an 11-degree test stimulus located 7 degrees below fixation on a Goldmann-Weekers dark-adaptometer. Full-field electroretinograms (ERG) were obtained according to ISCEV standards using Burian-Allen contact-lens electrodes. Signals were elicited for rod response, mixed rod-cone response, and cone response, and signals were amplified and computer-averaged.
RPGR mutation detection
Carrier DNA was sequenced for the known familial RPGR mutation in each case at the University of Texas, Health Science Center in Houston, TX using a previously described RPGR sequencing protocol.36 Genomic DNA was amplified for 35 cycles with AmpliTaq Gold® 360 Master Mix (Applied Biosystems, Foster City, CA) and M13-tailed primers designed to flank exon 1–19. RPGR ORF15 was amplified for 40 cycles using AmpliTaq Gold® 360 Master Mix and the following primers: 5’GACTAAACCATAATATCCAAATCCA3’ and 5’GCCAAAATTTACCAGTGCCTCCTAT3’. PCR product was treated with ExoSAPIT (Affymetrix, Santa Clara, CA) and sequenced using BigDye v1.1 (Applied Biosystems). Exons 1–19 were sequenced in 2 directions using M13 primers, and ORF15 was sequenced unidirectionally using a set of 7 nested primers. Sequence reactions were purified using BigDye Xterminator, run on a 3500 Genetic Analyzer, and analyzed using SeqScape v2.7 (Applied Biosystems).
XCI ratios
XCI ratios were determined using DNA extracted from blood and saliva samples using amplification of polymorphic repeats in the androgen receptor (AR) locus or the retinitis pigmentosa 2 (RP2) locus according to previously published and validated methods.37, 38 In summary, 300ng of genomic DNA was digested overnight for 16 hours with a methylation-sensitive restriction enzyme HpaII, which digests unmethylated DNA. The polymorphic repeat in AR was amplified in all samples (digested DNA in duplicate and an undigested control), using the following primers: 5’TCCAGAATCTGTTCCAGAGCGTGC3’ (conjugated to 6-Carboxyfluorescein, a.k.a. 6-FAM, on the 5’ end) and 5’GCTGTGAAGGTTGCTGTTCCTCAT3’. The sizes of the 2 alleles were then determined by fragment analysis with ROX 1000 size standards on a 3730XL Genetic Analyzer (Applied Biosystems) at the University of Michigan DNA Sequencing Core. The ratio of alleles was quantified by measuring peak heights using GeneMapper Software Version 5.0 (Applied Biosystems) and was normalized using the peak heights in the undigested control DNA. Parental DNA samples were obtained when possible, and amplification of the AR polymorphism in parental DNA was used to determine phase in the subjects, to determine which AR allele was in phase with the normal vs mutant RPGR allele.
In subjects who were homozygous at the AR polymorphism or did not have parental DNA samples available, the AR locus was uninformative for XCI ratios. In these subjects, a similar assay was used looking at methylation status of a polymorphic trinucleotide repeat in the RP2 locus, which was more recently described and validated as an alternative marker of XCI.38 The RP2 locus is only 8.5 megabases away from the RPGR locus, and the chance of crossover during meiosis is approximately 8.5%. Therefore, if parental DNA was not available, male offspring DNA samples were used to help determine phase, with a <10% chance of a crossover event confounding results. This method was used for 18 subjects, 3 of whom had 2 male offspring DNA samples available, lowering the chance of confounding crossovers to 0.7%.
A total of 42 XCI ratios were determined from blood. Of these 42 subjects, 19 also had XCI ratio determined from a saliva sample in the same manner. A 20th subject had XCI ratio determined from saliva only, as no additional blood sample was available.
Genotyping Candidate Modifier Loci
Candidate single nucleotide polymorphisms (SNPs) were genotyped as previously described.10 Briefly, DNA was extracted from blood using the Gentra Puregene blood kit (Qiagen, Valencia, CA). DNA sequences containing SNPs of interest were amplified from 37.5 ng genomic DNA with the PyroMark PCR Kit (Qiagen), using a primer with an M13 tail and a universal biotinylated M13 primer. PCR product was captured on a PyroMark Vacuum Prep Tool (Qiagen) by aspiration and washed in 70% ethanol, Pyromark Denaturation Solution (Qiagen), and PyroMark Wash Buffer (Qiagen). PCR product was released into 15 uL of 0.3 uM sequencing primer diluted in PyroMark Annealing Buffer (Qiagen), heated at 70°C for 15 minutes, and cooled at room temperature for 10 minutes before running on a PSQ hs 96A Instrument (Qiagen).
Statistical Analysis
Distributions of XCI ratios in blood and saliva samples were highly skewed and thus analyzed after log transformation. Blood and saliva means and variances were compared with a paired t-test and general least squares estimation, respectively. Interocular association for each of 7 clinical parameters was measured with Pearson’s correlation. Person-level clinical parameters were computed by averaging when more than one eye was measured. Associations between pairs of 7 clinical parameters, and the association between XCI ratio and spherical equivalent, were measured with Pearson’s correlations. Linear regression was used to determine association between XCI ratio and each clinical parameter. Multinomial regression was used to measure the probability of each disease severity group as a function of XCI ratio. Genotype-phenotype relationships, as well as associations between XCI ratio and phenotypic categories (i.e. unaffected/mild, moderate, and severe) were analyzed using chi-square tests. Associations of SNP genotypes with clinical severity were measured with Goodman-Kruskall’s gamma. No adjustments for pedigree structure were made. Analyses were conducted in R (The R Foundation for Statistical Computing, Vienna, Austria). Age was not significantly correlated with XCI ratio in our cohort. Controlling for age using partial correlations had minimal effect on correlation of XCI with clinical parameters. Therefore, no age-adjusted models are reported.
Results
Subjects and Mutations
A cohort of 77 carriers with confirmed RPGR mutations was ascertained from among family members of affected males with XLRP. The 77 carriers came from 41 pedigrees, ranging from 1–12 carriers per pedigree. The age at examination ranged from 9 to 69 years (average 43). The cohort included 62 obligate carriers. Out of 77 subjects, 58 (75.3%) were Caucasian of non-Hispanic origin, 10 (13.0%) were Hispanic, 5 (6.5%) were African American, and 1 (1.3%) was Pacific Islander, while 4 (5.2%) were not specified.
The 41 pedigrees tested positive for 35 different mutations in RPGR, including 2 missense mutations, 6 nonsense mutations, 4 splice site variants, 21 small indels with frame shift, and 2 large deletions (Table 1). Eighteen mutations were in exons 1–14 (including all mutation types), and 17 mutations were in ORF15 (nonsense and frameshift mutations only). Other than the 2 large deletions, all mutations have been previously reported and are referenced in Table 1.
Table 1. RPGR mutations in cohort of 77 XLRP carriers.
All previously reported mutations are referenced.
| Category | Mutation | Number of Pedigrees | Number of Carriers |
|---|---|---|---|
| Exon 1–14 Missense | c.194G>A (p.Gly65Asp)15 | 1 | 3 |
| c.617C>T (p.Thr206Ile)10 | 1 | 4 | |
| Exon 1–14 Splice | c.155–1G>A (IVS2–1G>A)61 | 1 | 1 |
| c.301_310+3del (IVS4–10_+3del)10 | 1 | 1 | |
| c.1573–8A>T (IVS13–8A>G)62 | 1 | 2 | |
| c.1573–1_1576delGAAACinsAA (IVS13–1_+3delGAAACinsAA)10 | 1 | 3 | |
| Exon 1–14 Nonsense | c.838_842del (p.Leu280X)13 | 1 | 1 |
| c.851C>G (p.Ser284X)10 | 1 | 1 | |
| c.1126G>T (p.Glu376X)10 | 1 | 4 | |
| Exon 1–14 Frameshift | c.101del (p.Asn34Metfs*34)63 | 1 | 5 |
| c.219del (p.Ala74Profs*11)10 | 1 | 1 | |
| c.356del (p.Leu119Trpfs*14)64 | 1 | 1 | |
| c.1092dupA (p.Ala365Cysfs*12)10 | 1 | 3 | |
| c.1243_1244del (p.Arg415Glyfs*37)10 | 1 | 2 | |
| c.1377_1378del (p.Leu460Ilefs*2)65 | 1 | 3 | |
| c.1662_1665del (p.Glu555Glyfs*14)10 | 1 | 1 | |
| Exon 1–14 Deletion | del exons 14–19 | 1 | 1 |
| c.310+147_620–1345del (p.Glu104_Thr207delinsAla) | 1 | 1 | |
| ORF15 Nonsense | g.ORF15+327A>T (p.ORF15+Lys109X)13 | 1 | 1 |
| g.ORF15+393G>T (p.ORF15+Glu129X)66 | 1 | 1 | |
| g.ORF15+423G>T (p.ORF15+Glu141X)10 | 1 | 3 | |
| ORF15 Frameshift | g.ORF15+481_484del (p.ORF15+Arg160Lysfs*69)13 | 1 | 1 |
| g.ORF15+483_484del (p.ORF15+Glu161Argfs*23)42 | 3 | 3 | |
| g.ORF15+521_524del (p.ORF15+Gly174Lysfs*55)10 | 1 | 1 | |
| g.ORF15+587del (p.ORF15+Ala196Argfs*34)13 | 1 | 1 | |
| g.ORF15+652_653del (p.ORF15+Glu217Glyfs*32)42 | 3 | 4 | |
| g.ORF15+673_674del (p.ORF15+Glu224Glyfs*25)42 | 2 | 14 | |
| g.ORF15+689_692del (p.ORF15Gly232fs*2)42 | 1 | 1 | |
| g.ORF15+720del (p.ORF15+Glu240Argfs*264)10 | 1 | 1 | |
| g.ORF15+730_743del (p.ORF15+Glu828Glyfs*2)10 | 1 | 1 | |
| g.ORF15+763_767del (p.ORF15+Glu254Glyfs*238)13 | 1 | 1 | |
| g.ORF15+848_849del (p.ORF15+Glu283Glyfs*210)67 | 1 | 1 | |
| g.ORF15+872dupA (ORF15G291Rfs*203)10 | 1 | 1 | |
| g.ORF15+1038del (p.ORF15+Arg346fs*158)10 | 1 | 1 | |
| g.ORF15+1114del (p.ORF15+Glu371Glyfs*133)15 | 2 | 3 |
Phenotypic Variation
Sixty-eight of 77 carriers were clinically affected based on abnormalities in visual field, scotopic, or photopic ERG (88.3%). Visual acuity was obtained for all subjects in both eyes and ranged from LogMAR −0.2 to 2.0 (Snellen 20/13 to 20/2000), with an average of LogMAR 0.27 (20/37), and a median of LogMAR 0.1 (20/25). Based on visual acuity alone, 3 carriers (3.9%) were legally blind, and 7 carriers (9.1%) did not meet requirements for unrestricted driving in most states (20/40 in the better seeing eye). Refraction data was available for 50 subjects (98 eyes). Another 15 subjects reported emmetropia, although no refraction was performed. Using spherical equivalent, myopia of at least −0.5 diopters was present in 64 eyes (50%), high myopia of at least −6.0 diopters was present in 21 eyes (16%), hyperopia of at least 0.5 diopters was present in 27 eyes (21%), high hyperopia of at least 5.0 diopters was present in 6 eyes (5%), and emmetropia within 0.5 diopters was present in 37 eyes, including those that were self-reported (29%).
Humphrey visual field was obtained for 73 subjects (145 eyes). Horizontal visual field diameter using a size III spot size ranged from 0–54 (maximum) degrees, with a mean of 37 degrees, and a median of 45 degrees. Based on visual field alone, 5 carriers were legally blind. After combining visual acuity and visual field data, 5 subjects total out of 77 (6.5%) were legally blind. These individuals ranged in age from 41 to 69 (mean 56 years).
Cone function, as measured by the ERG 30hz flicker, was assessed in 77 subjects (153 eyes) and ranged from barely recordable (<1 μV) to normal (147 μV), with mean and median amplitudes mildly reduced below normal (49 and 48 μV). Twenty-five out of 153 eyes (16.3%) had severely reduced 30 Hz flicker amplitudes below 10 μV. Rod function was assessed with scotopic ERG in the same 153 eyes of 77 subjects, and ranged from non-recordable to normal, with mean and median amplitude of 131 and 144 μV (approximately 70% normal). Twenty-three eyes (15.0%) had severely reduced function below 20 μV.
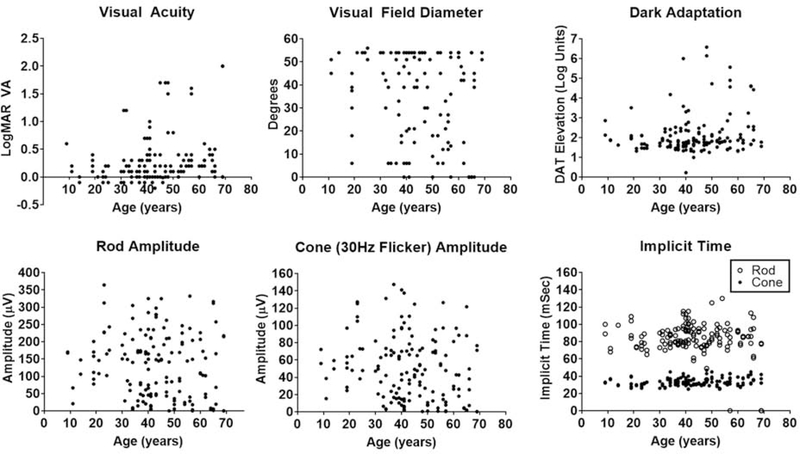
Scatter plots of 7 clinical parameters (VA, HVF diameter, dark adaptation, 30Hz flicker ERG amplitude and implicit time, and scotopic rod ERG amplitude and implicit time) by age for all subjects demonstrate the scope of variation in visual function (Figure 1). The plots show that most individuals in all age groups had retained good vision, while the subset of individuals who had progressed to moderate or severe vision loss were over the age of 30.
Figure 1. Scatter plots of 7 clinical parameters by age in 77 carriers of RPGR-associated XLRP.
Most individuals at all ages retained good vision, while the subset of individuals who had progressed to moderate or severe vision loss was over the age of 30. DAT= dark adaptation threshold, μV= microvolt, mSec= millisecond, VA= visual acuity.
Many clinical parameters were strongly correlated with each other, with the strongest correlation between the scotopic rod ERG amplitude and the photopic 30 Hz flicker amplitude (Table 2). Rod ERG implicit time showed poor correlation with all other parameters.
Table 2. Correlation between clinical parameters in female carriers.
VA= LogMAR visual acuity, HVF= Humphrey visual field, DAT= dark adaptation threshold, Amp= amplitude, IT= implicit time.
| VA | 1.00 | ||||||
| HVF | −0.61 | 1.00 | |||||
| DAT | 0.73 | −0.71 | 1.00 | ||||
| 30Hz Amp | −0.47 | 0.75 | −0.57 | 1.00 | |||
| 30Hz IT | 0.42 | −0.59 | 0.66 | −0.72 | 1.00 | ||
| Rod Amp | −0.47 | 0.78 | −0.56 | 0.88 | −0.66 | 1.00 | |
| Rod IT | −0.50 | −0.19 | −0.19 | 0.00 | −0.03 | −0.06 | 1.00 |
| VA | HVF | DAT | 30Hz Amp | 30Hz IT | Rod Amp | Rod IT |
Interocular Symmetry
Reports of female carriers of XLRP with asymmetric disease between eyes,39 and even more rarely with apparent unilateral disease,14 suggest that female carriers have lower interocular symmetry than affected males. To address this conjecture quantitatively, interocular correlations were calculated for measured clinical parameters for the entire cohort of 77 carriers (Table 3). For comparison, interocular correlations were also calculated for a cohort of 67 affected male patients with RPGR-associated RP from the University of Michigan (Table 3). Differences in visual testing between cohorts were most notable for visual field: Goldmann visual field was performed in the male cohort, and measured by area of the III4e isopter, compared to Humphrey visual field in the female carriers, measured by diameter of the field in degrees. However, the comparison between cohorts was restricted to a comparison of between-eye correlations, or symmetry, and the clinical measurements themselves were not compared between cohorts. With the exception of LogMAR VA, which had modest correlation of only 0.57, all interocular correlation values in the female cohort were 0.74 or greater. The correlation values were lower in females than males for each clinical parameter except for rod implicit time, which had poor correlation in the males of only 0.54.
Table 3. Interocular symmetry in affected males and female carriers.
The Pearson correlation (r) tor each of the listed clinical parameters between eyes is shown for a cohort of 67 affected males compared to the cohort of 77 female carriers. VF=visual field, Amp= amplitude, IT= implicit time.
| Males | Females | |
|---|---|---|
| LogMAR | 0.77 | 0.57 |
| VF | 0.94 | 0.79 |
| Flicker amp | 0.94 | 0.76 |
| Flicker IT | 1.00 | 0.74 |
| Rod amp | 0.96 | 0.87 |
| Rod IT | 0.54 | 0.91 |
Genotype-Phenotype Correlation
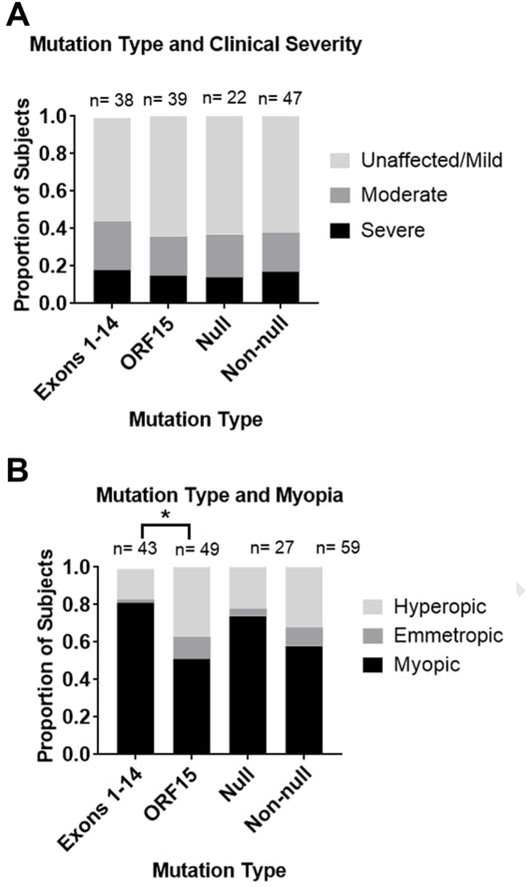
Subjects were categorized as unaffected/mild, moderate, or severe, based on visual field and ERG according to the guidelines in Table 4. Out of 77 carriers, 9 were unaffected, 37 were mildly affected, 18 were moderately affected, and 13 were severely affected. Equal numbers of subjects had mutations in ORF15 vs exons 1–14. There was no difference in clinical severity based on mutation location (p=0.73) (Figure 2). Patients were regrouped according to whether the mutation was a predicted null allele. Nonsense mutations and frameshift mutations leading to premature stop codons in non-terminal exons (exons 1–14) were predicted null, while missense mutations and all mutations in ORF15 (the terminal exon) were predicted non-null, as these mutations are not predicted to cause nonsense mediated decay of transcript. Splice site mutations were considered unpredictable and were excluded. There was no difference in clinical severity between these mutation categories (p=0.94) (Figure 2a).
Table 4. ERG and HVF features of carriers in 3 categories of phenotypic severity.
For discrepancies between eyes, averages were used. For discrepancies between parameters, 2 out of 3 determined the category. μV= microvolt.
| Category | Rod amplitude range (μV) | Cone amplitude range (μV) | HVF horizontal diameter (°) |
|---|---|---|---|
| Unaffected/Mild | >100 | >50 | >30 |
| Moderate | 30–100 | 20–50 | 10–30 |
| Severe | <30 | <20 | <10 |
Figure 2. Allelic heterogeneity is associated with myopia but not with clinical severity.
(A) The distribution of clinical severity, and (B) the distribution of myopia and hyperopia of greater than 0.5 diopters, was compared for mutations in exons 1–14 vs. ORF15 and mutations that are predicted to be null vs. non-null. Using chi-squared tests, the distributions of clinical severity (A) were not significantly different between mutation types (p= 0.728 and p= 0.937, respectively). Myopia (B) was significantly associated with mutations in Exons 1–14 compared to mutations in ORF15 (*p=0.01), but was not associated with null compared to non-null variants (p=0.30).
Since myopia is a common phenotypic feature in X-linked RP and in carrier females, refractive phenotype was also compared between genotype categories. When comparing eyes with either myopia or hyperopia of greater than 0.5 diopters, mutations in exons 1–14 were significantly associated with myopia compared to mutations in ORF15 (p=0.01, Figure 2b). There was no difference in refractive phenotype between predicted null vs. non-null variants (p=0.30).
XCI Ratios and Skewing
XCI ratios were determined by assaying methylation status of a polymorphic repeat at the Androgen Receptor (AR) locus using DNA extracted from either blood or saliva samples. In subjects who were uninformative at the AR locus, XCI was assessed using a polymorphic repeat in the RP2 locus. In order to measure test-retest variability, 3 separate blood samples were drawn on 3 separate occasions at 1 week intervals from the same individual. XCI ratios were 1.17 (54:46), 1.55 (61:39), and 1.42 (59:41), with a coefficient of variation of 6.2%.
Henceforth XCI ratio will refer to the ratio of active mutant to wild-type X-chromosome with respect to RPGR. Out of 64 subjects with blood samples, a total of 42 XCI ratios were determined. Of these 42 subjects, 19 also had XCI ratio determined from a saliva sample. A 20th subject had XCI ratio determined from saliva only, as no additional blood sample was available. In 21 carriers, XCI could not be determined, either because they were homozygous at both the AR and the RP2 polymorphism, or because there were no family DNA samples available that could be used to determine phase with the RPGR alleles.
In the 42 blood samples, XCI ratios ranged from 0.02 (2:98) to 159.80 (99.4:0.6), with median 0.76 (43:57). Twenty-six out of 42 (62%) had XCI < 50:50 and 8 out of 42 (19%) had XCI < 20:80. Sixteen out of 42 (38%) had XCI > 50:50 and 7 out of 42 (17%) had XCI > 80:20. Overall, there was no apparent bias towards extreme skewing in one direction or the other, evidence that the pathogenic RPGR variants did not influence the XCI process. The same was true for the XCI ratios from the 20 saliva samples, and the median was similar to that calculated for blood samples, 0.86 (46:54). XCI ratios in blood and in saliva were highly correlated (r=0.96). Age was not significantly correlated with XCI ratio.
XCI Ratios and Phenotype
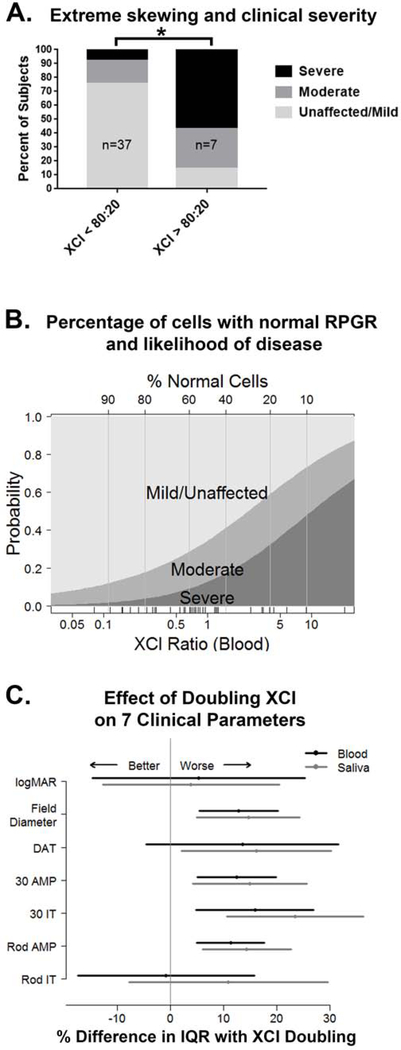
Skewed XCI towards inactivation of the normal RPGR allele was associated with more severe disease. For carriers with XCI≥80:20, the accepted standard of extreme skewing,40 57% were severely affected, compared to 8% for those with XCI<80:20 (p=0.002) (Figure 3a). The probability of a patient being unaffected/mild, moderate, or severe based on XCI ratio was plotted continuously using multinomial regression (Figure 3b) and was used to estimate what percentage of cells with normal RPGR are needed to avoid clinically significant retinal degeneration. For example, when 40% of cells have an active normal RPGR allele based on the XCI ratio, there is a 60% chance of being mildly affected or unaffected and a 40% chance of being moderately or severely affected. This question is particularly relevant in the era of gene therapy, as it is still unknown what proportion of cells need to be transduced for effective treatment.
Figure 3. XCI ratio is associated with clinical severity.
A) Extreme skewing of the XCI ratio towards inactivation of the normal RPGR allele is associated with the severe phenotype (*p= 0.002). B) The probability of being mildly, moderately, or severely affected is plotted using multinomial regression as a function of the percent of cells with normal RPGR, inferred from the XCI ratio. The vertical distance represents the probability of each disease severity category for a specific XCI blood ratio. C) The change in each clinical parameter as the XCI ratio doubles is graphed as a percentage of the interquartile range (IQR) to normalize data from different scales, and in some cases the direction is reversed so that larger values indicate worse visual function. DAT= dark adaptation threshold, AMP= amplitude, IT= implicit time.
Individual clinical parameters were also analyzed with respect to XCI ratio. Seven measurements of visual function were considered and included LogMAR VA, HVF diameter, dark adaptation threshold, scotopic ERG rod b-wave amplitude, rod b-wave implicit time, 30 Hz flicker ERG amplitude, and flicker implicit time. Each eye was measured independently, and the average value between eyes was used to determine correlation with XCI ratio.
XCI ratio in both blood and saliva was a statistically significant predictor of visual function as measured by HVF diameter, rod amplitude, flicker amplitude, and flicker implicit time. In addition, XCI ratio in saliva (but not blood) was a statistically significant predictor of dark adaptation threshold. The impact of XCI ratio is displayed in Figure 3c as a percent change in each clinical parameter. The biggest effects of XCI were on HVF diameter, rod amplitude, and flicker amplitude and implicit time. Each of these parameters demonstrated worsening by over 10% of the interquartile range as the XCI ratio doubled. In blood samples, doubling the XCI ratio was associated with a reduction of HVF by 3.8 degrees, reduction of rod amplitude by 17.4 μV, and reduction of flicker amplitude by 6.6 μV. In saliva samples, doubling the XCI ratio was associated with a reduction of HVF by 4.5 degrees, reduction of rod amplitude by 22.4 μV, and reduction of flicker amplitude by 8.0 μV. Rod implicit time, which showed poor correlation with other clinical parameters and poor interocular symmetry, also showed poor correlation with XCI.
XCI ratio was also associated with spherical equivalent. Refraction data were available for 57 eyes in 29 subjects who had XCI determined from blood samples and 26 eyes in 13 subjects who had XCI determined from saliva samples. XCI ratio (active mutant RPGR to active wild-type RPGR) had a strong negative correlation with spherical equivalent, suggesting that eyes with a higher the proportion of cells expressing mutant RPGR are more likely to be myopic. Furthermore, the correlation was stronger for saliva samples (r= −0.63 for blood samples and −0.79 for saliva samples).
Genetic Modifiers of Phenotype in Carriers of XLRP
A candidate modifier screening approach, previously described in a cohort of affected males10, was used to identify potential genetic modifiers of phenotype in female carriers of XLRP. Common coding SNPs (MAF≥2%) in genes encoding known protein binding partners of RPGR (RPGRIP1, RPGRIP1L, CEP290, and IQCB1) were sequenced in 75 female carriers from 40 families and compared to disease severity (unaffected/mild, moderate, or severe). The D1264N variant in RPGRIP1L was found to be significantly associated with disease severity (p=0.007), while the G1025S variant in RPGRIP1L demonstrated a trend towards association without statistical significance (p=0.07).
Discussion
This study describes the scope of clinical severity in a large cohort of female carriers of XLRP with mutations in RPGR and demonstrates and quantifies the contribution of XCI to phenotype. This is the first report to demonstrate a significant effect of XCI skewing on disease severity in carriers of XLRP. Prior investigations have been performed in very small cohorts, and no association was found.20, 41 Although our cohort was also small, with only 42 subjects that had XCI ratios from blood samples, and even fewer (20) from saliva samples, this is substantially larger than any previous study of XCI in XLRP. Of note, this study did not include carriers with mutations in RP2, which accounts for 10–20% of XLRP and is much more rare.42 A previous investigation of 101 carriers with RPGR mutations and 20 carriers of RP2 mutations found no significant difference in phenotype.21
X-linked disease is typically classified as X-linked recessive or X-linked dominant, with the majority of conditions falling into the former category. However, the inheritance pattern in XLRP is more complex. In our cohort, 88.3% of carriers were clinically affected, and 6.5% were legally blind. Subjects were enrolled for genotyping based on potential at-risk status according to pedigree, and not based on affected status or clinical symptoms, although we cannot exclude a subject enrollment bias, as affected carriers may be more likely to enroll than unaffected carriers. In our cohort, severity did not correlate with age, and based on the scatter plots in Figure 1 this is likely due to the large background of mildly affected and unaffected subjects present at all ages. However, the scatter plots also suggest that the 30s and 40s may be a period of decline in visual function for those who do progress. Two previously published large cohorts of XLRP carriers describe different concepts of penetrance.21, 43 In a cohort of 125 subjects, either molecularly proven or obligate carriers of RPGR mutations, 40% were subjectively symptomatic (average age 34).43 In a cohort of 242 XLRP carriers, of whom 101 had known RPGR mutations, 49% had at least 1 abnormal psychophysical test (median age 38).21 In our cohort, average age 43, 38% fall into the moderate or severe category based on HVF and ERG, and 88% are affected if the mild category is included.
Fifty percent of eyes in our cohort had myopia of at least −0.5 diopters and 16% had high myopia of at least −6.0 diopters, compared to 16–26% and 2.7–4.6% respectively in the general population.44, 45 Myopia and high myopia have been reported before with a prevalence of 73% and 35% respectively in a large cohort of 125 RPGR female carriers, and a prevalence of 85% and 38% in 63 affected males.43, 46
Other systemic X-linked recessive diseases are rarely penetrant in carriers, and affected carrier status is almost always associated with XCI skewing of 80:20 or more.23, 24, 27 Given the relatively high penetrance in carriers, which is nevertheless lower than the 100% penetrance in hemizygous males, XLRP may be described as “X-linked semi-dominant,”20 or as “X-linked dominant with reduced penetrance.” In our cohort of carriers, XCI skewing was significantly associated with clinical severity. However, even in the severe group, 57% had extreme skewing, which is less than what has been reported for other non-ophthalmic X-linked diseases,23, 24, 27 suggesting that the threshold for manifesting disease is lower for XLRP. As seen in Figure 3b, even subjects with an XCI ratio of 20:80 (i.e. 80% of cells have normal RPGR) can be affected, and even subjects with 80:20 (i.e. 20% of cells have normal RPGR) can be unaffected or mild, suggesting that other factors are contributing, potentially including genetic modifiers. We report 2 variants in RPGRIP1L (D1264N and G1025S) as potential genetic modifiers of phenotype in XLRP carriers. However, no adjustment was made for multiple comparisons. We have previously reported the common protein haplotypes of RPGRIP1L and have shown that the D1264N variant mostly commonly occurs on the background of the G1025S variant.10 Other variants in RPGRIP1L were previously found to be associated with disease severity in affected males with XLRP (R744Q), and with the presence of retinopathy in syndromic ciliopathy patients (A229T), further supporting RPGRIP1L as a potential genetic modifier of phenotype in ciliopathies.10, 47
Blood and saliva were used as proxy tissues in this study, because XCI ratios cannot be directly determined in the retina. However, there is good evidence that XCI ratios can vary between tissues.48–50 It has been previously demonstrated that XCI in the blood skews with age and that XCI in buccal mucosa is more stable.25, 48, 49, 51 Our data supports the hypothesis that buccal mucosa may serve as a better proxy tissue for retina XCI ratios, as the association of XCI skewing with disease severity was stronger for the saliva samples even though there were fewer samples. However, a larger sample size is needed to confirm this.
We report here that phenotypic severity in the cohort of XLRP carriers showed no association with the location of the RPGR mutation. This contrasts with previous findings in affected males, who are more likely to be severely affected if the mutation is in exons 1–14 compared to the 3’ ORF15.10, 13 A smaller series of 4 families found the same trend in affected female carriers.52 The lack of genotype-phenotype correlation in the female carriers in our large cohort study may indicate that genetic modifiers or XCI skewing may have a more significant contribution to the observed phenotypic variation. In a previous report of 101 carriers of RPGR XLRP, a reverse genotype-phenotype correlation was found, showing more severe disease in subjects with a mutation in ORF15 compared to exons 1–14.21 Another large cohort of 125 female carriers with confirmed RPGR mutations showed no correlation between mutation location and disease severity, in agreement with the findings reported here.43
In contrast, we demonstrate a genotype-phenotype association in our cohort between myopia and mutations in exons 1–14 compared to ORF15. To our knowledge this genotype-phenotype relationship has not been previously investigated. Many previous reports have demonstrated a high penetrance of myopia in female carriers of RPGR mutations.53–57 These pedigrees show an X-linked dominant pattern of myopia but an X-linked recessive pattern of retinitis pigmentosa, and all families except 1 had mutations in ORF15.
As new therapeutic strategies for XLRP are developed, including gene therapy, we will inevitably be faced with the question of which carriers to treat and when. Previous retinal gene therapy trials have demonstrated that earlier intervention is more effective,58–60 which is clinically and biologically plausible. However, unlike male children with pathogenic RPGR variants who will invariably progress and lose vision, female carriers may be unaffected, mildly affected without functional significance, or severely affected. Our data suggest that skewed XCI ratios may be predictive of clinically significant disease. Carriers with extreme skewing of 80:20 or more had an 86% chance of being moderately or severely affected. XCI analysis may be a useful measurement in determining good candidates for early intervention.
Acknowledgments
Financial Support: Abigail Fahim acknowledges support from the NIH (2K12EY022299–06) and the University of Michigan Department of Ophthalmology. Stephen Daiger acknowledges support from the William Stamps Farish Fund, the Foundation Fighting Blindness, and the NIH (EY007142). David Birch acknowledges support from the Foundation Fighting Blindness and the NIH (EY09076). These sponsors or funding organizations had no role in the design or conduct of this research.
Footnotes
Conflict of Interest: Dr. Fahim reports grants from National Institutes of Health during the conduct of the study. Dr. Birch reports grants and personal fees from AGTC, grants and personal fees from Nightstar, outside the submitted work. Dr. Daiger reports grants from National Institutes of Health, grants from the Foundation Fighting Blindness, and grants from the William Stamps Farish Fund during the conduct of the study; grants and personal fees from the Foundation Fighting Blindness, and personal fees from AGTC and from Spark Therapeutics outside the submitted work. No conflicting relationships exist for other authors.
This cohort study of 77 female carriers with mutations in RPGR describes the scope of clinical phenotype and for the first time quantifies the contribution of X-chromosome inactivation to phenotypic severity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daiger SP, Bowne SJ, and Sullivan LS Perspective on genes and mutations causing retinitis pigmentosa. Arch Ophthalmol. 2007; 125: p. 151–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daiger SP RetNet. https://sph.uth.edu/retnet; 2018. Accessed 07.10.18.
- 3.Roepman R, Bernoud-Hubac N, Schick DE, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet. 2000; 9: p. 2095–105 [DOI] [PubMed] [Google Scholar]
- 4.Hong DH, Pawlyk B, Sokolov M, et al. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci. 2003; 44: p. 2413–21 [DOI] [PubMed] [Google Scholar]
- 5.Khanna H, Hurd TW, Lillo C, et al. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J Biol Chem. 2005; 280: p. 33580–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branham K, Othman M, Brumm M, et al. Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest Ophthalmol Vis Sci. 2012; 53: p. 8232–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Churchill JD, Bowne SJ, Sullivan LS, et al. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013; 54: p. 1411–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keith CG, Denton MJ, and Chen JD Clinical variability in a family with X-linked retinal dystrophy and the locus at the RP3 site. Ophthalmic Paediatr Genet. 1991; 12: p. 91–8 [DOI] [PubMed] [Google Scholar]
- 9.Walia S, Fishman GA, Swaroop A, et al. Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene. Arch Ophthalmol. 2008; 126: p. 379–84 [DOI] [PubMed] [Google Scholar]
- 10.Fahim AT, Bowne SJ, Sullivan LS, et al. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One. 2011; 6: p. e23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Peachey NS, Moshfeghi DM, et al. Mutations in the RPGR gene cause X-linked cone dystrophy. Hum Mol Genet. 2002; 11: p. 605–11 [DOI] [PubMed] [Google Scholar]
- 12.Demirci FY, Rigatti BW, Wen G, et al. X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am J Hum Genet. 2002; 70: p. 1049–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharon D, Sandberg MA, Rabe VW, et al. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003; 73: p. 1131–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebenezer ND, Michaelides M, Jenkins SA, et al. Identification of novel RPGR ORF15 mutations in X-linked progressive cone-rod dystrophy (XLCORD) families. Invest Ophthalmol Vis Sci. 2005; 46: p. 1891–8 [DOI] [PubMed] [Google Scholar]
- 15.Pelletier V, Jambou M, Delphin N, et al. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype-phenotype correlations and impact on genetic counseling. Hum Mutat. 2007; 28: p. 81–91 [DOI] [PubMed] [Google Scholar]
- 16.Rao KN, Zhang W, Li L, et al. Ciliopathy-associated protein CEP290 modifies the severity of retinal degeneration due to loss of RPGR. Hum Mol Genet. 2016; 25: p. 2005–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souied E, Segues B, Ghazi I, et al. Severe manifestations in carrier females in X linked retinitis pigmentosa. J Med Genet. 1997; 34: p. 793–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenekoop RK, Loyer M, Hand CK, et al. Novel RPGR mutations with distinct retinitis pigmentosa phenotypes in French-Canadian families. Am J Ophthalmol. 2003; 136: p. 678–87 [DOI] [PubMed] [Google Scholar]
- 19.Hong DH, Pawlyk BS, Adamian M, et al. Dominant, gain-of-function mutant produced by truncation of RPGR. Invest Ophthalmol Vis Sci. 2004; 45: p. 36–41 [DOI] [PubMed] [Google Scholar]
- 20.Banin E, Mizrahi-Meissonnier L, Neis R, et al. A non-ancestral RPGR missense mutation in families with either recessive or semi-dominant X-linked retinitis pigmentosa. Am J Med Genet A. 2007; 143A: p. 1150–8 [DOI] [PubMed] [Google Scholar]
- 21.Comander J, Weigel-DiFranco C, Sandberg MA, et al. Visual Function in Carriers of X-Linked Retinitis Pigmentosa. Ophthalmology. 2015; 122: p. 1899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vajaranant TS, Seiple W, Szlyk JP, et al. Detection using the multifocal electroretinogram of mosaic retinal dysfunction in carriers of X-linked retinitis pigmentosa. Ophthalmology. 2002; 109: p. 560–8 [DOI] [PubMed] [Google Scholar]
- 23.Fahim AT and Daiger SP The Role of X-Chromosome Inactivation in Retinal Development and Disease. Adv Exp Med Biol. 2016; 854: p. 325–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyon MF X-chromosome inactivation and human genetic disease. Acta Paediatr Suppl. 2002; 91: p. 107–12 [DOI] [PubMed] [Google Scholar]
- 25.Amos-Landgraf JM, Cottle A, Plenge RM, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006; 79: p. 493–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Berg IM, Laven JS, Stevens M, et al. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009; 84: p. 771–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pegoraro E, Schimke RN, Arahata K, et al. Detection of new paternal dystrophin gene mutations in isolated cases of dystrophinopathy in females. Am J Hum Genet. 1994; 54: p. 989–1003 [PMC free article] [PubMed] [Google Scholar]
- 28.Di Michele DM, Gibb C, Lefkowitz JM, et al. Severe and moderate haemophilia A and B in US females. Haemophilia. 2014; 20: p. e136–43 [DOI] [PubMed] [Google Scholar]
- 29.Beltran WA, Cideciyan AV, Lewin AS, et al. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci U S A. 2012; 109: p. 2132–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beltran WA, Cideciyan AV, Lewin AS, et al. Gene augmentation for X-linked retinitis pigmentosa caused by mutations in RPGR. Cold Spring Harb Perspect Med. 2014; 5: p. a017392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Hiriyanna S, Qian H, et al. A long-term efficacy study of gene replacement therapy for RPGR-associated retinal degeneration. Hum Mol Genet. 2015; 24: p. 3956–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng WT, Dyka FM, Dinculescu A, et al. Stability and Safety of an AAV Vector for Treating RPGR-ORF15 X-Linked Retinitis Pigmentosa. Hum Gene Ther. 2015; 26: p. 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawlyk BS, Bulgakov OV, Sun X, et al. Photoreceptor rescue by an abbreviated human RPGR gene in a murine model of X-linked retinitis pigmentosa. Gene Ther. 2016; 23: p. 196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer MD, McClements ME, Martinez-Fernandez de la Camara C, et al. Codon-Optimized RPGR Improves Stability and Efficacy of AAV8 Gene Therapy in Two Mouse Models of X-Linked Retinitis Pigmentosa. Mol Ther. 2017; 25: p. 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltran WA, Cideciyan AV, Boye SE, et al. Optimization of Retinal Gene Therapy for X-Linked Retinitis Pigmentosa Due to RPGR Mutations. Mol Ther. 2017; 25: p. 1866–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan LS, Bowne SJ, Birch DG, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006; 47: p. 3052–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen RC, Zoghbi HY, Moseley AB, et al. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992; 51: p. 1229–39 [PMC free article] [PubMed] [Google Scholar]
- 38.Machado FB, Machado FB, Faria MA, et al. 5meCpG epigenetic marks neighboring a primate-conserved core promoter short tandem repeat indicate X-chromosome inactivation. PLoS One. 2014; 9: p. e103714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson SG, Yagasaki K, Feuer WJ, et al. Interocular asymmetry of visual function in heterozygotes of X-linked retinitis pigmentosa. Exp Eye Res. 1989; 48: p. 679–91 [DOI] [PubMed] [Google Scholar]
- 40.Harris A, Collins J, Vetrie D, et al. X inactivation as a mechanism of selection against lethal alleles: further investigation of incontinentia pigmenti and X linked lymphoproliferative disease. J Med Genet. 1992; 29: p. 608–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berghmans LV, de Mendonca RH, Coppieters F, et al. Discordance for retinitis pigmentosa in two monozygotic twin pairs. Retina. 2011; 31: p. 1164–9 [DOI] [PubMed] [Google Scholar]
- 42.Vervoort R, Lennon A, Bird AC, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000; 25: p. 462–6 [DOI] [PubMed] [Google Scholar]
- 43.Talib M, van Schooneveld MJ, Van Cauwenbergh C, et al. The Spectrum of Structural and Functional Abnormalities in Female Carriers of Pathogenic Variants in the RPGR Gene. Invest Ophthalmol Vis Sci. 2018; 59: p. 4123–4133 [DOI] [PubMed] [Google Scholar]
- 44.Holden BA, Fricke TR, Wilson DA, et al. Global Prevalence of Myopia and High Myopia and Temporal Trends from 2000 through 2050. Ophthalmology. 2016; 123: p. 1036–42 [DOI] [PubMed] [Google Scholar]
- 45.Kempen JH, Mitchell P, Lee KE, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004; 122: p. 495–505 [DOI] [PubMed] [Google Scholar]
- 46.Talib M, van Schooneveld MJ, Thiadens AA, et al. CLINICAL AND GENETIC CHARACTERISTICS OF MALE PATIENTS WITH RPGR-ASSOCIATED RETINAL DYSTROPHIES: A Long-Term Follow-up Study. Retina. 2018 [DOI] [PubMed] [Google Scholar]
- 47.Khanna H, Davis EE, Murga-Zamalloa CA, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009; 41: p. 739–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp A, Robinson D, and Jacobs P Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000; 107: p. 343–9 [DOI] [PubMed] [Google Scholar]
- 49.Knudsen GP, Pedersen J, Klingenberg O, et al. Increased skewing of X chromosome inactivation with age in both blood and buccal cells. Cytogenet Genome Res. 2007; 116: p. 24–8 [DOI] [PubMed] [Google Scholar]
- 50.Bolduc V, Chagnon P, Provost S, et al. No evidence that skewing of X chromosome inactivation patterns is transmitted to offspring in humans. J Clin Invest. 2008; 118: p. 333–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatakeyama C, Anderson CL, Beever CL, et al. The dynamics of X-inactivation skewing as women age. Clin Genet. 2004; 66: p. 327–32 [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Yin X, Feng L, et al. Novel mutations of RPGR in Chinese retinitis pigmentosa patients and the genotype-phenotype correlation. PLoS One. 2014; 9: p. e85752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parmeggiani F, Barbaro V, De Nadai K, et al. Identification of novel X-linked gain-of-function RPGR-ORF15 mutation in Italian family with retinitis pigmentosa and pathologic myopia. Sci Rep. 2016; 6: p. 39179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez Tocino H, Diez Montero C, Villanueva Gomez A, et al. Phenotypic high myopia in X-linked retinitis pigmentosa secondary to a novel mutation in the RPGR gene. Ophthalmic Genet. 2019; 40: p. 170–176 [DOI] [PubMed] [Google Scholar]
- 55.Yokoyama A, Maruiwa F, Hayakawa M, et al. Three novel mutations of the RPGR gene exon ORF15 in three Japanese families with X-linked retinitis pigmentosa. Am J Med Genet. 2001; 104: p. 232–8 [PubMed] [Google Scholar]
- 56.Jin ZB, Liu XQ, Hayakawa M, et al. Mutational analysis of RPGR and RP2 genes in Japanese patients with retinitis pigmentosa: identification of four mutations. Mol Vis. 2006; 12: p. 1167–74 [PubMed] [Google Scholar]
- 57.Sheng X, Li Z, Zhang X, et al. A novel mutation in retinitis pigmentosa GTPase regulator gene with a distinctive retinitis pigmentosa phenotype in a Chinese family. Mol Vis. 2010; 16: p. 1620–8 [PMC free article] [PubMed] [Google Scholar]
- 58.Bemelmans AP, Kostic C, Crippa SV, et al. Lentiviral gene transfer of RPE65 rescues survival and function of cones in a mouse model of Leber congenital amaurosis. PLoS Med. 2006; 3: p. e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mowat FM, Breuwer AR, Bartoe JT, et al. RPE65 gene therapy slows cone loss in Rpe65-deficient dogs. Gene Ther. 2013; 20: p. 545–55 [DOI] [PubMed] [Google Scholar]
- 60.Bainbridge JW, Mehat MS, Sundaram V, et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med. 2015; 372: p. 1887–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demirci FY, Radak AL, Rigatti BW, et al. A presumed missense mutation of RPGR causes abnormal RNA splicing with eon skipping. Am J Ophthalmol. 2004; 138: p. 504–5 [DOI] [PubMed] [Google Scholar]
- 62.Fujita R, Buraczynska M, Gieser L, et al. Analysis of the RPGR gene in 11 pedigrees with the retinitis pigmentosa type 3 genotype: paucity of mutations in the coding region but splice defects in two families. Am J Hum Genet. 1997; 61: p. 571–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guevara-Fujita M, Fahrner S, Buraczynska K, et al. Five novel RPGR mutations in families with X-linked retinitis pigmentosa. Hum Mutat. 2001; 17: p. 151. [DOI] [PubMed] [Google Scholar]
- 64.Sharon D, Bruns GA, McGee TL, et al. X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest Ophthalmol Vis Sci. 2000; 41: p. 2712–21 [PubMed] [Google Scholar]
- 65.Zito I, Thiselton DL, Gorin MB, et al. Identification of novel RPGR (retinitis pigmentosa GTPase regulator) mutations in a subset of X-linked retinitis pigmentosa families segregating with the RP3 locus. Hum Genet. 1999; 105: p. 57–62 [DOI] [PubMed] [Google Scholar]
- 66.Breuer DK, Yashar BM, Filippova E, et al. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet. 2002; 70: p. 1545–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bader I, Brandau O, Achatz H, et al. X-linked retinitis pigmentosa: RPGR mutations in most families with definite X linkage and clustering of mutations in a short sequence stretch of exon ORF15. Invest Ophthalmol Vis Sci. 2003; 44: p. 1458–63 [DOI] [PubMed] [Google Scholar]