Abstract
Dorsal root ganglia (DRG), which contain the somata of primary sensory neurons, have increasingly been considered as novel targets for clinical neural interfaces, both for neuroprosthetic and pain applications. Effective use of either neural recording or stimulation technologies requires appropriate spatial position relative to the target neural element, whether axon or cell body. However, the internal three-dimensional spatial organization of human DRG neural fibers and somata has not been quantitatively described. In this study, we analyzed 202 cross-sectional images across the length of 31 human L4 and L5 DRG from 10 donors. We used a custom semi-automated graphical user interface to identify the locations of neural elements in the images and normalize the output to a consistent spatial reference for direct comparison by spinal level. By applying a recursive partitioning algorithm, we found that the highest density of cell bodies at both spinal levels could be found in the inner 85% of DRG length, the outer-most 25–30% radially, and the dorsal-most 69–76%. While axonal density was fairly homogeneous across the DRG length, there was a distinct low density region in the outer 7–11% radially. These findings are consistent with previous qualitative reports of neural distribution in DRG. The quantitative measurements we provide will enable improved targeting of future neural interface technologies and DRG-focused pharmaceutical therapies, and provide a rigorous anatomical description of the bridge between the central and peripheral nervous systems.
Keywords: Cell density, Cross-sectional anatomy, Dorsal root ganglion, Humans, RRID:SCR_005043, RRID:AB_2149620, RRID:SCR_001622, RRID:SCR_014242, RRID:SCR_003070
Dorsal root ganglia (DRG) are novel targets for neural interface technologies that treat neurological disorders, such as chronic pain and spinal cord injury. The three-dimensional cellular anatomy of DRG are not well-mapped, particularly in humans, limiting the effectiveness of neurotechnology. We developed a semi-automated algorithm to quantify the three-dimensional distribution of neural elements in histologically-processed tissue. We applied this algorithm to sequential NF200-stained histology slices obtained from human lumbar DRG, and demonstrated that cell bodies typically congregate around the dorsal edge of the ganglia. These results are crucial to the development of safe and effective clinical neural interface technologies.
Graphical Abstract
1. Introduction
Dorsal root ganglia (DRG) are regions of the posterior spinal roots which contain the cell bodies of all primary sensory neurons innervating a specific dermatome of the body or end organ (Devor, 1999). Recently, DRG have been increasingly investigated as sources from which to record control signals for neuroprosthetic devices, for applications ranging from treating bladder dysfunction to providing limb prosthetic control and sensory feedback (Bruns, Wagenaar, Bauman, Gaunt, & Weber, 2013; Holinski, Everaert, Mushahwar, & Stein, 2013; Ouyang, Sperry, Barrera, & Bruns, 2019; Umeda et al., 2012; Weber, Stein, Everaert, & Prochazka, 2007). While most studies have used penetrating electrode arrays to record from neurons in the interior of the DRG, recent work has demonstrated that afferent signals can be recorded from neurons near the exterior of DRG using surface recording arrays (Kashkoush, Gaunt, Fisher, Bruns, & Weber, 2019; Sperry et al., 2018) and that microelectrodes can be used for low-current microstimulation of DRG neurons (Ayers, Fisher, Gaunt, & Weber, 2016; Bruns, Weber, & Gaunt, 2015; Fisher et al., 2014). Additionally, dorsal root ganglion stimulation (DRGS) is an emerging therapy for chronic neuropathic pain (Deer et al., 2017), providing a non-addictive alternative to opioids for pain management. Although these technologies have the potential to provide dramatic improvements in quality of life for a diverse range of patients, preliminary clinical studies have demonstrated limited success thus far. Commonly reported issues include inconsistencies in the quality of elicited sensations, and variable amounts of pain relief across patients (Harrison, Epton, Bojanic, Green, & FitzGerald, 2018). These technologies rely on either the accurate recording or delivery of electric currents to specific types of DRG neurons (e.g. recording from bladder afferents, stimulating sensory inputs to reflex circuits). However, we have a limited understanding of the three-dimensional (3D) spatial distribution of primary afferents throughout human DRG. Describing the 3D cellular anatomy of DRG could inform the clinical implementation of these technologies (e.g. DRGS electrode placement relative to the ganglion, selection of penetrating or surface recording arrays to target different cells), which may lead to improved and more consistent clinical outcomes.
DRG contain both the cell bodies and portions of the axons of primary sensory afferents. Neural recording technologies detect neural activity through the voltages generated by the transmembrane currents generated by neural signal propagation (Aoyagi, Stein, Branner, Pearson, & Normann, 2003). The largest transmembrane currents, which would contribute to the bulk of the recorded signal, are thought to originate from the cell body and axon initial segment of recordable cells (Kneist, Kauff, Schröder, Koch, & Lang, 2014). In contrast, extracellular stimulation technologies are thought to primarily induce neural activation in axons (McIntyre & Grill, 1999), with modeling studies suggesting that DRGS electrodes primarily activate the axons of large myelinated tactile afferents (Graham, Bruns, Duan, & Lempka, 2019). Although the specific roles of subgroups of primary afferents are complex, DRG cells can be roughly broken into functional groups based on cell body and typically conduct thermal or noxious sensation (Dubin & Patapoutian, 2010; Winter, 1971). Myelinated A-fibers, which are typically mechanoreceptive or proprioceptive, are larger than C-fibers, typically have thinly- to thickly-myelinated axons. A-fibers can be further stratified into Aα-, Aβ-, and Aδ-fiber classes which are listed in order of descending conduction velocity and fiber size (Gardner & Johnson, 2014). Although this classification scheme is commonly used throughout the literature, there is a dearth of studies examining the spatial distribution of these fibers throughout the DRG.
The effectiveness of both recording and stimulating neural interface technologies relies heavily on the spatial relationship between the interface electrode and particular neuron types of interest. However, few studies have examined the spatial organization of neurons in DRG, and to our knowledge, no studies exist in the human literature. Various studies have discussed a weak functional somatotopy for the DRG in animal studies based on nerve tracing, with rostral-caudal, medial-lateral, and ventral-dorsal divisions all present to a certain degree (Burton & McFarlane, 1986; Kausz & Rethelyi, 1985; Peyronnard, Messier, Dubreuil, Charron, & Lebel, 1990; Prats-Galino, Puigdellívol-Sánchez, Ruano-Gil, & Molander, 1999; Puigdellívol-Sánchez, Prats-Galino, Ruano-Gil, & Molander, 1998; Wessels, Feirabend, & Marani, 1990b, 1990a). Few studies have examined the distribution of neural structures (e..g cell bodies, axons) in DRG. Only a few anecdotal references exist, including studies showing superficial cell bodies or a “central fiber stream” in the DRG (Bossy, 1970; Sato & Austin, 1961), and a neural recording study in feline DRG reported the largest density of cell bodies at a depth between 0 and 100 μm (Miletic & Lu, 1993). There is a general consensus that most cell bodies are found around the circumferential edge of the ganglion. That idea was supported by our previous study, in which a quantitative analysis of feline lumbosacral DRG showed that the highest density of cell bodies in medial sections could be found in the outer 24% radially and on the dorsal-most side of the ganglion (Ostrowski, Sperry, Kulik, & Bruns, 2017). However, in this previous study, we only examined neuronal distribution in medial sections of the DRG, while clinical neurotechnologies would likely act upon neurons distributed along the entire DRG.
In this work, we sectioned human lumbar DRG specimens at 1 mm increments along the nerve root axis. We stained these sections with neurofilament and imaged the sections for high-contrast identification of neural cell bodies and axons. We developed software to automatically segment and characterize the spatial location and size of neural elements. Finally, we used a recursive partitioning algorithm to model the 3D spatial densities of the neural elements. We also analyzed high-level donor demographic and anatomical trends in our data set. These analyses provide key insights for future neural interface technologies and therapies with human DRG.
2. Materials and Methods
2.1. Gross Tissue Processing
This study utilized human tissue donated by de-identified deceased individuals, and was therefore determined to be “Not Regulated” by the University of Michigan Institutional Review Board [HUM00109152]. We received bilateral human lumbar spinal roots (RRID:SCR_005043) from the L4 and/or L5 spinal levels from the National Disease Research Interchange (NDRI, Philadelphia, PA) from deceased donors within 72 hours of aortic cross-clamp. We chose to study L4 and L5 DRG because these DRG are common DRGS targets for managing chronic pain in the feet and lower legs (Deer et al., 2019). We excluded donors that had a history of diabetes, cancer, herpes zoster, a peripheral nerve condition, spinal cord injury, previous spine surgery, or opiate abuse. We stored the tissue in 10% neutral-buffered formalin and shipped at ambient temperature. Upon receipt, we used calipers to measure the DRG portion of each root at 1 mm increments to determine the gross tissue dimensions. We then used a razor blade to cut the DRG in half and removed two 1 cm segments constituting the approximate proximal (i.e. nearest the spinal cord) and distal (i.e. nearest the peripheral nerve) DRG halves (Figure 1, Step 1). We placed the cut sides of each DRG half face-down in a histology cassette to preserve DRG orientation across slides. If visual inspection indicated that the remaining tissue contained gray matter, we removed additional segments to obtain samples of the entire DRG region. We stored the cassettes in 70% ethanol to halt fixation, and shipped them an external histopathology lab (Histowiz Inc., Brooklyn, NY) for histological processing.
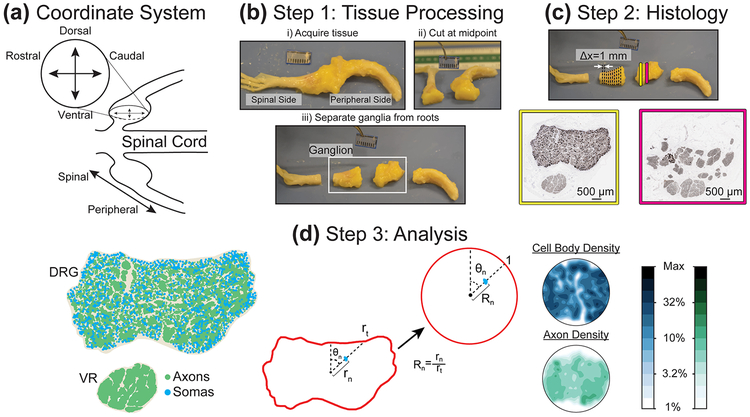
Figure 1:
(a) Illustration of coordinate system for DRG cross-section images. (b) Initial tissue processing. (c) Representation of sample locations and NF200-stained samples (dark brown). We counterstained each sample with hematoxylin, resulting in blue cell nuclei (not visible at displayed image size). (d) Analysis of histological data. (Left) DRG cross-section after automated processing, with identified axon and soma locations highlighted. (Center) Diagram of polar normalization, demonstrating transformation from perimeter and internal features to unit circle. (Right) Contour plots generated from polar normalization of DRG at right, with logarithmic color scale at right.
2.2. Histological Processing & Imaging
The histopathology lab embedded the tissue samples in paraffin (Leica EG 1150 H&C) and cut 4–5 μm thick sections at 1 mm steps (~20 sections per DRG) (Leica RM2235 microtome). They mounted the sections on slides and stained with a 1:3000 dilution of rabbit monoclonal antibody for neurofilament heavy polypeptide (NF200, ab40796, Abcam, Cambridge, UK, RRID:AB_2149620) which stains both A- and C-type fibers in humans (Rostock, Schrenk-Siemens, Pohle, & Siemens, 2018; Vega, Humara, Naves, Esteban, & Valle, 1994). They also counterstained for hematoxylin to identify nuclei. They imaged the slides at 40x with a brightfield slide scanner, with a resolution of four pixels per micron (Leica Aperio AT2). The resulting images had neural cell bodies and axons visible in dark brown and cell nuclei visible in blue (Figure 1, Step 2). The nuclei of other DRG cellular types were also visible, including satellite cells and some endothelial vascular cells. The neural cellular elements were typically arranged into at least two fascicular regions. We considered the regions containing cell bodies (or continuous with cell-body containing regions across images) as DRG. We considered all other regions as ventral root (VR). To the extent analyzed, the VR was considered separately from the DRG. We screened images for quality issues, including excessive tearing or folding, and poor quality regions were excluded from analysis.
2.3. Image Processing Software
Lumbar DRG contain on the order of 35,000 neurons (Liu, Zhou, Ma, Ge, & Cao, 2015). Therefore, to identify each neural element we developed a custom semi-automated graphical user interface (GUI) using MATLAB and its Image Processing Toolbox (MathWorks Inc., Natick, MA, RRID:SCR_001622). After loading the image, the GUI allows the user to identify the DRG, VR, and non-neural outer tissue, then calculates an optimal rotation to align the DRG above the ventral root in the upper half of the image. The GUI white-balances the image and converts it to binary using a series of thresholding steps and morphological operations. We extracted the shapes and locations of all neural elements in the original image from the resulting binary image for analysis.
In order to validate the GUI results, we selected four validation images pseudo-randomly from the final image set, to balance for inclusion from different donors, spinal levels, and sidedness (i.e. right DRG or left DRG). We randomly selected a 1 mm square region containing both axons and cell bodies from each validation image. Following consultation from a neuropathologist, the first three authors each traced the neural cell elements in the validation sub-images using the ImageJ (RRID:SCR_003070, https://imagej.nih.gov/ij/) plug-in ObjectJ (https://sils.fnwi.uva.nl/bcb/objectj/index.html, Vischer, N. & Nastase, S. University of Amsterdam).
For automated processing, we first converted all validation images to grayscale, and white balanced each image against background luminescence for each color channel (red=R, green=G, and blue=B). These grayscale images were converted to binary (black and white) using a threshold determined to minimize the difference between the initially detected object number and the number of objects determined by each human validator. A threshold of 0.78 minimized the difference between the computer- and human-identified objects, therefore we used a threshold of 0.78 for all black-white thresholding steps. We discovered that a simple threshold tended to split large lightly-stained cell bodies into many smaller objects, but that intact cell bodies could be recovered by utilizing the ratio between R and B channels. Specifically, R/B-B/R threshold of 0.5 consistently returned an image with intact cell bodies. This step was effective because the neurofilament stain tended to have a high R value and a low B value, while hematoxylin had a strong B value and weak R value. This RB ratio transformation was noisy outside of the cell bodies, so only objects with area greater than 600 μm2 were retained (roughly corresponds to circular objects with diameter 14 μm). The logical disjunction of the RB ratio image and the original binary image (RB Ratio OR Binary), denoised with a morphological “open” and with holes filled, served as the final image for automated neural element identification with MATLAB’s Image Processing (MathWorks, Natick, MA, United States) toolbox.
To compare the automated system and human performance, we found the pixel overlap between the output images and calculated the precision and recall of the automated system, using the human segmented images as ground truth. Precision is the ratio between true positives and total positives (true positives + false positives), and is a measure of how often the program incorrectly labeled a pixel as belonging to a neuron. Recall is the ratio between true positives and ground truth positives (true positives + false negatives), and is a measure of how often the program missed a neuron pixel it should have labeled. Both ratios range from 0 to 1, with a higher value being a better score (Taha & Hanbury, 2015; Wienert et al., 2012).
The precision of the algorithm was 0.92 ± 0.04 and the recall was 0.81 ± 0.05. While this is short of perfection, these values are consistent with other well-performing cell segmentation programs (Al-Kofahi, Lassoued, Lee, & Roysam, 2010; Peikari & Martel, 2016; Wienert et al., 2012). In comparison, the inter-human precision was 0.84 ± 0.06 (significantly lower than the human versus algorithm, Student’s t-test; p=0.0021), and inter-human recall was 0.86 ± 0.07 (Student’s t-test; p=0.1075). These values suggest that the algorithm is behaving overall like an average human validator, and on the basis of precision and recall would likely be indistinguishable. The absolute difference in object count between human and algorithm was 5.43 ± 4.21 % (non-absolute difference: 1.45 ± 6.89 %, not significantly different than zero (Student’s t-test; p=0.4806)). The standard deviation of inter-human count for a given image as percentage of the mean was 5.43 ± 4.21 %. Ultimately, the goal of the program was not to perfectly label all cells but to reduce the time to adequately label an image so that trends could be quantified. Each human annotation of a 1 mm2 image took approximately 6–10 hours depending on the number of objects, and there was considerable variation in duration between annotators. The same images took only ~1 minute for the program. Considerable time saving, along with the positive performance metrics, suggests that the program was a significant improvement over manual image labeling.
Figure 2 shows a portion of a raw validation image between the automated and human segmentation for comparison. In the full version of this image, the program identified 5277/5359 objects identified by the average human, a difference of −1.53%. Precision in this image was 0.95 and recall was 0.80. A note on human versus automated performance is the tendency of human segmenters to label an object with a border larger than the actual object (oversegmentation). This explains the evident difference in some object sizes between the two images.
Figure 2:
(Left) Raw portion of validation image. (Center) Binarization of human segmented image. (Right) Binarization of automatically segmented image.
2.4. DRG Reconstruction
We reconstructed the 3D profiles of individual ganglia regions by converting manually-traced 2D profiles of DRG, VR, and compact outer tissue to .svg files in MATLAB, then stacking and interpolating (lofting) between the curves using the computer-aided design (CAD) program Audodesk Fusion 360 (Autodesk, San Rafael, CA), slightly adjusting image alignment as needed (Figure 3). We used these 3D reconstructions to analyze the mean geometric properties (e.g. aspect ratio, length, width) of root regions (e.g. dorsal root, ventral root).
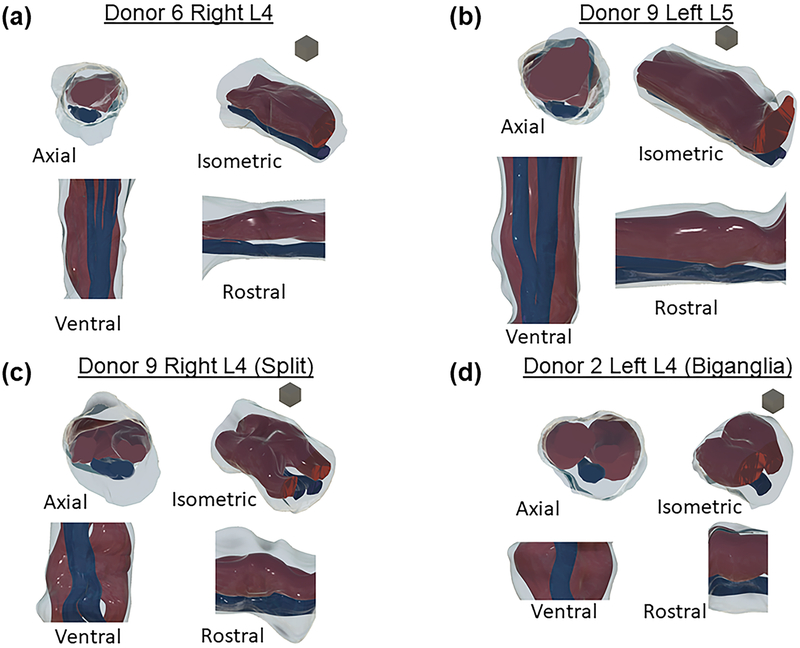
Figure 3:
Four views of reconstructed specimens. Dark red is dorsal root ganglia region, blue is ventral root, grey is epineurial tissue. In each group views are (clockwise from top left): Axial (face closest to spine [spinal face] visible), isometric (spinal face right), rostral (spinal face right), and ventral (spinal face down). Blocks shown for scale have 1 mm edges. (a) L4 monoganglia (b) L5 monoganglia (c) Near-biganglia (d) Full biganglia.
2.5. Image Analysis
Following parameter optimization and comparison of GUI results to human performance, we analyzed each image containing at least 10 visible soma (Offord, Ohta, Oenning, & Dyck, 1974) in the GUI to determine neural element size, shape, and location. We identified in-plane axons as elements exceeding a geometric eccentricity of 0.9 (i.e. their shape dramatically deviated from circularity), and excluded them from population size analysis. We defined mean diameter as the average of the major and minor axis. Based on values previously reported (Gardner & Johnson, 2014), we used a mean diameter cutoff of 20 μm to distinguish larger axons from smaller cell bodies. We found that objects larger than 110 μm in diameter were typically detritus, and we removed them from the data set.
Note that for the purposes of this study, we will refer to the three spatial dimensions of the DRG as follows: the axial position refers to the location of an image along the nerve trajectory, and may be either spinal (close to the spinal cord) or peripheral (away from the spinal cord); cross-sections are oriented with the dorsal side up and the ventral side down; the axis perpendicular to both these axes, and horizontal in cross-sections, is the rostral-caudal axis (Figure 1a). We determined this final label by considering the in situ anatomy of lower lumbar nerve roots, which exit the spinal cord slightly more perpendicular than parallel (Silverstein et al., 2015), and therefore to align the third axis with the rostral-caudal bodily axis. If the nerve roots were slightly more parallel to the spinal cord, it would be appropriate to refer to our rostral-caudal axis as lateral-medial respectively. A previous computational modelling study of lower lumbar DRG also used this nomenclature, shown visually in the Coordinate System section of Figure 1 (Graham et al., 2019).
2.6. Cell Density Mapping
In order to quantify neural cell densities across DRG, we normalized the location of each identified cell body and axon within an image to polar coordinates using a method similar to our prior work (Ostrowski et al., 2017). We set the centroid of the user-defined DRG region as the origin for determining the polar angle of neuron n (θn). We defined θn to equal 0 at the vertical (i.e. dorsal) normal vector calculated from the user-defined DRG region. We determined radial location (rn) by calculating the ratio of the distance from the DRG centroid of neuron n and the distance from the DRG centroid to the DRG edge along θn. The end result is a circular map with the circumference representing the DRG edge and all neurons located inside the circle. In cases where we observed multiple ganglionic regions (i.e. multiple cell body-containing fascicles) in a single slice, we combined all regions into a single region using built-in MATLAB image processing functions. To combine all ganglionic regions into a single region, we first found the perimeter of each ganglionic region (MATLAB command: bwperim). Then, we calculated the outline of a shape which formed a boundary encompassing the entirety of each ganglionic region’s perimeter (MATLAB command: boundary). Lastly, we converted the resulting boundary into a mask (MATLAB command: poly2mask), which represented the entire cell body-containing region of the slice.
We used the normalized circular maps described above to calculate cellular densities. First, we divided the circular map into concentric annular sectors of equal width and increasing radii. Next, we divided each annular sector into wedges of constant area, to create a circular grid of wedge sectors defined by a radius from the center of the circular map, and an angle from the vertical normal vector. To calculate cellular density of a wedge sector, we divided the number of cellular elements (i.e. somas or axons) in the wedge by the non-normalized area of that wedge (i.e. the area of the original image represented by the wedge sector). A shortcoming of the method used previously was a very high variance in calculated densities in sectors at the center of normalized DRG, the result of very small sectors produced by using wedges of constant angular size (Ostrowski et al., 2017). Holding wedge sector area constant, and varying the number of wedge sectors in each annulus overcame that issue (i.e. annuli with smaller circumferences closer to the center of the circular map have fewer wedge sectors, but each wedge sector has the same area). Given that the largest expected cell bodies in human and other large mammal DRG have a diameter of about 100 μm (Josephson et al., 2001; Lee et al., 1986), we chose annulus width and sector area to accommodate approximately four large cell bodies packed in each sector. We chose this size after trial and error as a compromise between fine and coarse resolution for the final maps. Figure 1d (Step 3) shows a summary of image analysis steps.
To determine axial position of each image, we fit a 3rd degree polynomial curve to the DRG fascicle areas considering the known distance between sequential images. We took the axial position corresponding to the maximum value of the fit curve as 0 μm (i.e. the middle of the DRG). We assigned a negative axial position value to images on the spinal side of the midpoint, and a positive value to images on the peripheral side. We did not assign an axial position to images from ganglia without a clear center (i.e. fascicle areas with no peak value). These images were included only in analyses that did not rely on axial position.
2.7. Donor Demographic Trend Analysis
We performed all statistical analyses for this study using JMP Pro 14 statistical software (SAS Institute, Cary, NC, RRID:SCR_014242). We tested the effect of various donor demographic and anatomical characteristics on mean neuron count and density in each DRG. We evaluated the two-factor effects (sex [male/female], side [left/right], and spinal level [L4/L5]) with a Student’s t-test. For all tests, we set the significance threshold at p=0.01. We evaluated the effect of donor size by calculating body-mass index (BMI), sorting donors into “normal,” “overweight,” and “obese” categories (group divisions at 25 and 30 (Flegal, Carroll, Kit, & Ogden, 2012)), then performing an analysis of variance (ANOVA) test for significance followed by a Tukey honest significant difference (HSD) test. We evaluated the effect of donor age by fitting a line to the density (or count) versus donor age data and assessed the significance of the slope estimate (Student’s t-test, H0: slope=0).
2.8. Spatial Density Modeling
The goal of spatial density modeling is to provide a partitioned map of neural elements throughout a DRG. We used JMP to fit separate partition models for normalized densities by spinal level (L4 and L5), and neural element type (soma and axons) for a total of four models. We further stratified the axon models by size (large [≥5 μm diameter] and small [<5 μm diameter]). Partition models split a distribution of dependent variables (e.g. cell body density) into sub-populations of distribution based on the values of one or more independent variables (e.g. axial location). This splitting operation is performed recursively, and splits distributions at values of independent variables which maximizes model fit. The result of a partition model is a decision tree that maximally captures sub-populations of distributions within a dependent variable (e.g. identifying regions of large or small cell body density based on axial location in the DRG). We used ten-fold cross-validation of the model at each split to avoid overfitting. We terminated splitting at the stage when 10 additional splits failed to improve the model R2 by at least 0.05. We additionally restricted the model to only produce groups containing more than 5% of data points, leading to a theoretical maximum of 20 groups in the model. As in Ostrowski et al. (Ostrowski et al., 2017), we used radial location and vertical angle as model inputs, defining vertical angle as the angle measured from the top (dorsal-most) point of the circle reflected across the dorsal-ventral midline of each image. For our partition models, we also added axial position as a model input. We assessed the output models by performing an ANOVA and Tukey HSD test on the measured densities in the identified spatial regions. Where relevant, results are presented as mean ± standard deviation.
3. Results
All DRG images, software, summary data, and CAD models referenced in this study are available online at the Open Science Framework (Peck-Dimit et al., 2019).
3.1. Donors and Specimens
We collected a total of 34 DRG from 10 human donors. Table 1 provides demographics for each donor. This included 7 males and 3 females, of which 7 were Caucasian, 2 Hispanic and 1 Black. Ages ranged from 25 to 59 yrs (47.2 ± 10.3 yrs). BMI ranged from 24 to 38 kg/m2 (30.1 ± 4.5 kg/m2). Based on typically-reported ranges, two of the donor BMIs were considered “Normal,” four were considered “Overweight,” and four were considered “Obese”. Seven of the donors died from natural causes (cardiovascular or stroke), and three died from external causes (trauma or asphyxiation).
Table 1:
Detailed donor information.
| Donor | Age (yr) | Sex | Wt. (kg) | Ht. (m) | BMI (kg/m2) | Race | Cause of Death | DRG |
|---|---|---|---|---|---|---|---|---|
| D1 | 25 | F | 70 | 1.64 | 24.2 | C | Cardiovascular | LL5(4), RL5(0) |
| D2 | 46 | M | 81 | 1.67 | 27.2 | C | Arrhythmia | LL4(5), RL4(5-i), LL5(6), RL5(5-i) |
| D3 | 57 | M | 91 | 1.62 | 32.4 | C | ICH/Stroke | LL4(6-i), RL4(5-i), LL5(5-i), RL5(5) |
| D4 | 49 | M | 112 | 1.69 | 36.5 | H | Head Trauma | LL4(3-i), RL4(8), LL5(7), RL5(4-i) |
| D5 | 50 | F | 94 | 1.52 | 37.9 | C | ICH/Stroke | LL5(6), RL5(6) |
| D6 | 54 | M | 97 | 1.76 | 29.0 | C | ICH/Stroke | LL4(6), RL4(8), LL5(8), RL5(8) |
| D7 | 59 | M | 93 | 1.74 | 28.6 | C | Head Trauma | LL4(5), RL4(5), LL5(3), RL5(7) |
| D8 | 46 | M | 88 | 1.81 | 24.9 | C | Head Trauma | LL5(10), RL5(9) |
| D9 | 51 | F | 92 | 1.64 | 31.2 | B | Cardiovascular | LL4(5), RL4(7), LL5(8), RL5(9) |
| D10 | 35 | M | 87 | 1.67 | 29.2 | H | Asphyxiation | XL4(4-i), XL4(5), XL5(8-i), XL5(7) |
F=female, M=male, C=Caucasian, H=Hispanic, B=black. ICH=intracranial hemorrhage. All DRG collected are listed in “DRG” column, first letter is side (L=left, R=right, X=not specified). Number of images included from each DRG is included in parentheses, with a “-i” indicating inclusion of an incomplete DRG (i.e. DRG image set did not include a non-soma-containing sample at the extreme axial positions).
We collected bilateral L5 DRG from all donors, and bilateral L4 DRG from all but three donors (D3, D5, and D8). In all but D10, the DRG were identifiable as either left or right upon receipt. Therefore, D10 was excluded from all left-right analyses, but was used when comparing features across levels (i.e. L4 vs. L5). All but one of the DRG had images suitable for inclusion in this study. Tissue slices in all images from Donor 1’s Right L5 DRG were torn and therefore unusable. Table 1 reports the DRG collected and used in this study.
The majority of DRG in our data set (28/33) could be defined as monoganglia, that is, at some point we observed an image containing a single compact DRG fascicle. All of these DRG split at either the spinal or peripheral side into multiple fascicles, and in some cases were split into two primary fascicles throughout most of the region of interest. The remaining five DRG were biganglia, that is, the minimum number of observed DRG fascicles was two. It is possible that these DRG had a merged region in between images not captured by our sectioning, but the maximum length of this region would necessarily be less than 1 mm due to our sampling method. Of the biganglia, three were L4 DRG and two were L5 DRG. Figure 3 shows four samples of donor specimens reconstructed from histological images (one each of monoganglia L4 and L5, a near-biganglia, and a full biganglia).
The number of images used from each DRG ranged from 3 to 10 (6.1 ± 1.8; 202 total), varying due to anatomical differences and the quality of the final images. Considering both image quality and inclusion of only cell-body containing images, this represented approximately 25% of the overall images acquired. We imaged 17 complete DRG, identified based on a distinct continuous axial region with soma-containing images bounded by images with only axon-containing images. An additional seven DRG were considered “nearly complete,” with the region of soma-containing images bounded on one side by axon-only images and on the other by highly fascicular images with few cell bodies. The remaining nine DRG were incomplete (i.e. the DRG image set did not include a nonsoma-containing sample at the outer axial positions), but still had images useable for the study. Completeness and number of images used is also reported by DRG in Table 1.
3.2. Measurements and Reconstructions
Within the complete DRG, there was a non-significant trend toward L5 being longer than L4 (p=0.0244). L4 DRG had length 5.2 ± 1.9 mm while L5 DRG had length 7.1 ± 2.4 mm, measured as the length of the soma-containing region. Within the DRG containing a peak area, there was no apparent difference between L4 and L5 in maximum rostral-caudal width, and the overall width was 5.8 ± 1.0 mm. In these same DRG, there was a non-significant trend toward L5 having a larger maximum dorsal-ventral width than L4 (p=0.0228). L5 had a maximum dorsal-ventral width of 4.5 ± 1.2 mm while L4 was 3.7 ± 0.8 mm.
Our polar model transforms the DRG fascicular area to a perfect circle of radius 1, but it is useful to understand the typical shape of our actual samples. We found that the actual mean aspect ratio of the DRG fascicular regions (as rostral-caudal width over dorsal-ventral width) was 1.52 ± 0.47, significantly different than the unit aspect ratio of a perfect circle (Student’s t-test, p<0.0001). When considering the number of DRG fascicles, we found that the mean aspect ratio of biganglia DRG fascicle cross sections (1.78 ± 0.44) was significantly higher (p=0.0004) than for monoganglia DRG (1.47 ± 0.45). This feature of the DRG fascicle cross sections does not extend to the full tissue cross-section including VR, which had an aspect ratio of 1.04 ± 0.21 which, while significantly greater than one (Student’s t-test, p=0.0077), is an aspect ratio close to the aspect ratio of a perfect circle. Our polar model does not rely on any assumptions of aspect ratio, but these geometric trends may be of interest for future models of DRG.
3.3. Neural Counts and Densities
We observed several trends in cell body and axon count related to spinal level. L5 DRG had significantly higher mean axon count in each image (33643 ± 11007) as compared to L4 (27868 ± 10846), with p=0.0002. However, the mean density of axons did not differ significantly by spinal level, and the combined mean was 2.36×10−3 ± 8.50×10−4 axons per μm2. There was no significant difference in the number of cell bodies in each image by spinal level, though there was a slightly higher mean cell body density in L4 (3.78×10−5 ± 1.98×10−5 cells per μm2) than in L5 (3.24×10−5 ± 1.71×10−5 cells per μm2), with p=0.0492. The combined mean number of cells per image was 469 ± 303. The apparent disparity between a difference in fiber count and a non-difference in by-image cell count appears to be the result of the additional length of L5 to contain the additional cell bodies. From this point, all figures and models will consider L4 and L5 separately.
Comparing the spinal and peripheral sides of each DRG (splitting at the center axial location), we observed that the peripheral side had significantly more axons than the spinal side at both spinal levels (difference at L4: 6002 ± 5917 axons, p=0.0033; L5: 7758 ± 6398 axons, p=0.0002). This trend was true of 24/29 individual DRG where axial location was determined. For large fibers, this represents a mean increase of 4007 ± 4118 fibers (or 62.16 ± 76.24%) from the spinal to peripheral side, and an increase of 2964 ± 2825 small fibers (15.4 ± 15.5%). This trend was not accompanied by a significant increase in per-image cell body count. However, when considering cell bodies by size, there was a significant increase in confirmed (nucleated) large cell bodies on the peripheral side versus the spinal side (9 ± 15 cells, or 51.5 ± 77.7 %). The large cell body trend was true of 21/29 DRG, although there was not enough data to determine if this trend was related to DRG level.
It should be noted that large cell bodies represented only about 32.4 ± 18.0% of nucleated cell bodies, and that nucleated cells represented 27.3 ± 8.6% of all cell bodies identified in each image. Medium and small cells respectively represented 43.7 ± 12.6% and 23.9 ± 11.3% of nucleated cells identified. The mean axon diameter was 4.37 ± 1.79 μm. The mean cell body diameter was 44.94 × 15.13 μm. No axial trend was observed in either axon or cell body density (rather than count) at any size. Possible reasons for these observations are enumerated in the Discussion section.
The most significant trend related to patient demographics was that obese donors had a higher L5 DRG axon count (38298 ± 8783) than normal patients (30934 ± 8522), p=0.0047. Overweight donors overlapped both groups (33113 ± 9609). This trend was also observed in L4, although there were no normal BMI L4 donors (obese: 31171 ± 10383; overweight 26499 ± 9280; p=0.0219). Obese donors had a higher cell body count in each L4 image than overweight donors (p=0.0367), and higher than overweight and normal donors at L5 (p=0.0073 and p=0.0404 respectively). There was no significant cell body count or density trend related to BMI (i.e. the mean cell body counts and densities were not significantly different between donors categorized with different BMIs). Female donors had both higher mean L5 cell body count and cell body density than males in each image (p=0.0179 and p=0.0019, respectively).
Figure 4 shows the mean polarized densities of cell bodies and axons for each spinal level with axial position divisions at 1 mm intervals. While a full spatial description of VR axons is outside of the primary scope of this study, a few observations are relevant in comparison with the DRG. We counted 5983 ± 1474 axons in L4 VR, and 6417 ± 1564 axons in L5 VR, although the difference was not significant. There was no relationship between axon count and axial location in the VR. In each image, the number of large axons in the VR was significantly fewer (p<0.0001) than the number of small axons by 37.4 ± 35.8%, with no relationship to spinal level. The ratio of large-to-small axons was significantly higher (p<0.0001) in VR as compared to the DRG, in which the same proportion was 42.4 × 16.5% (p<0.0001). This matched our qualitative observation that it was possible to visually discern a VR fascicle from a DRG fascicle based solely on the different axon population size.
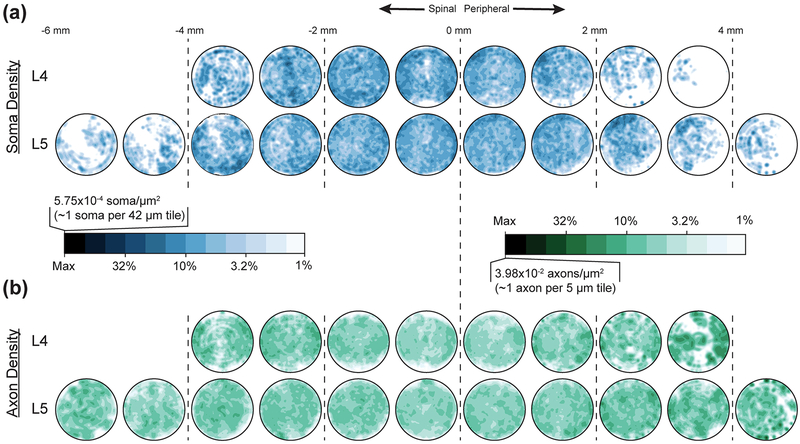
Figure 4:
Polarized mean (a) cell body (blue) and (b) axon (green) spatial densities in each millimeter of L4 (upper) and L5 (lower) dorsal root ganglia fascicular region. Inset scale left provides reference for color, which is on a logarithmic scale decreasing from maximum density to 1% of maximum.
The axon and cell densities for each level were modeled using a recursive partition algorithm which had radial, axial, and angular position as inputs. Table 2 summarizes the performance of the partition models and the contribution of each input variable to the overall model as a proportion of the total R2. For the cell body models, axial position was the most important variable, while the radial position was most important for the axon models. Both the L4 cell body and axon models had a higher R2 than the corresponding L5 models, although as in Ostrowski et al., the trends described by the model represent a relatively small portion of the overall variation (Ostrowski et al., 2017). The axon models also had higher R2 than the associated cell body models.
Table 2:
Summary of DRG density models by neural element and spinal level.
| Neural Element | Spinal Level | R2 | Axial Proportion | Radial Proportion | Angular Proportion |
|---|---|---|---|---|---|
| Cell Bodies | L4 | 0.0659 | 0.57 | 0.21 | 0.22 |
| L5 | 0.0453 | 0.60 | 0.28 | 0.12 | |
| Axons | L4 | 0.1399 | 0.04 | 0.61 | 0.35 |
| L5 | 0.0942 | 0.14 | 0.81 | 0.05 | |
| Small Axons | L5 | 0.0835 | 0.17 | 0.76 | 0.07 |
| Large Axons | 0.1003 | 0.26 | 0.70 | 0.04 |
R2 values indicate the amount of variance captured by the partition model for a neural element (i.e. cell bodies or axons) at a given spinal level. Proportion columns indicate the relative contribution of a given variable (e.g. axial position, radial distance from center) to the overall model on a scale from 0–1; proportions for a given model sum to 1.
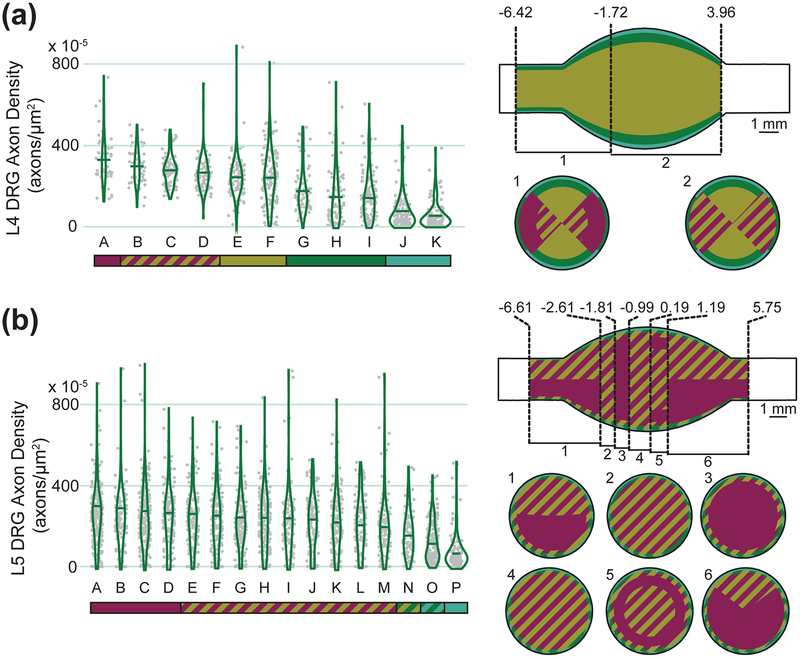
Figure 5 summarizes the measured cell body densities in each spatial region identified by the recursive partition algorithm models, as well as describing the statistical relationships between regions. Figure 6 shows the measured axon densities in each spatial region identified by the same recursive partition algorithm, and describes the statistical relationship between those regions. Each model successfully identified regions with significantly different neural densities, with a minimum ratio of ~2x between highest and lowest density regions (L5 cell bodies) and a maximum ratio of ~7x between highest and lowest density regions (L4 axons).
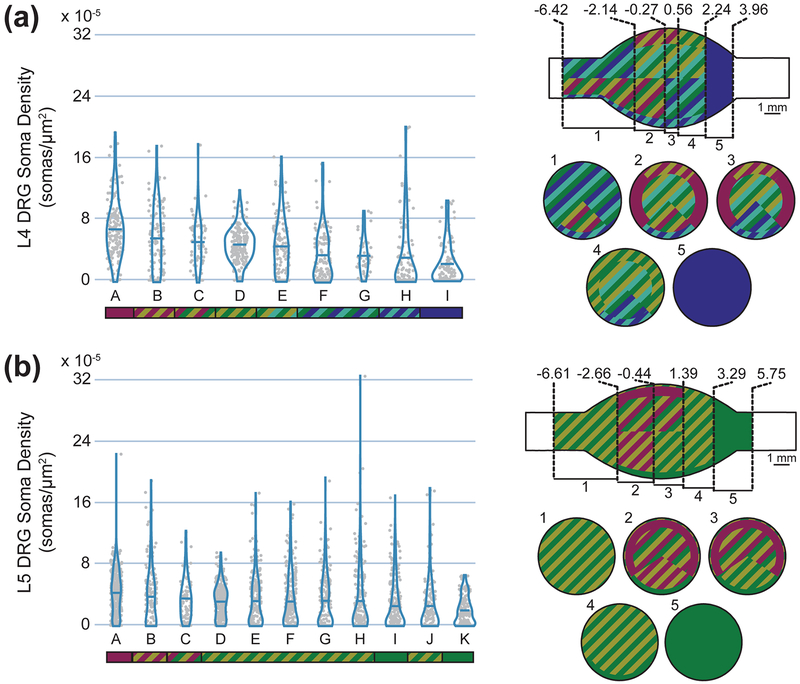
Figure 5:
Cell body densities (Left) and spatial representations of cell body density (Right) of (a) L4 and (b) L5 DRG. Density data are summarized with a violin plot and a mean bar. Letters are unique to each violin plot, although the mean in each group is descending left to right. The colored bar below each plot represents statistical significance. Groups not connected by the same color (either solid or striped) are significantly different from each other. For example, in the top left graph, group A is significantly different from all groups other than B and C, which share its magenta color. The top view of each spatial representation is a view of the DRG with the spinal side left and dorsal side up. Within each uniquely described axial region, a cross-sectional slice with corresponding number label is shown below, with the dorsal side up (e.g. for L4, slice 1 represents cell body densities at axial locations between −6.42 and −2.14 mm from the middle of the ganglion). Values along the axial axis of the DRG have units of mm.
Figure 6:
Axon densities (Left) and spatial representations of axon density (Right) of (a) L4 and (b) L5 DRG. See Figure 5 for specific information on the types of figures shown.
Axial position was the strongest contributor to the cell body models. In L4, the highest soma densities were in the inner ~4.4 mm, and in L5 the highest soma densities were within the inner ~6 mm. These values both correspond to about 85% of the mean full length of the soma-containing region. For L4, the highest soma density region was in the dorsal-most 76% angularly and the outer-most 30% radially. The lowest soma density region (in the inner axial portion of the DRG) was the ventral-most 24% angularly and the outer-most 18% radially. For L5, the highest soma density region was similarly in the dorsal-most 69% angularly and the outer-most 25% radially, although the outer-most 5% radially was characterized by slightly lower soma density. The lowest soma density region was the ventral-most 31% angularly and the outer-most 14% radially. For both levels, the interior was a more homogeneous region. In contrast to the cell body models, radial position was the most important component of the axon models at both levels. Taken together, the highest axon density regions in the model were within 89% (L5) to 93% (L4) radially. The outer 11% and 7% (respectively) had significantly lower axon densities. In L4, the medial angles (43°−136°) had the highest density, while in L5 angle played almost no role in the final model.
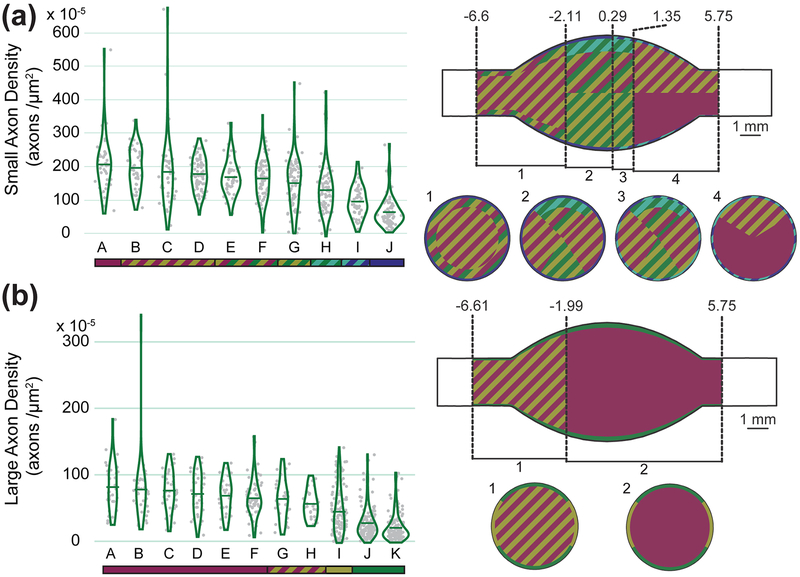
We also considered differences in modeled densities with neural element size. There were not enough nucleated cell bodies to make size models at either level. At L4, the small and large axon density models were almost identical to the overall axon density model. In L5, however, considering fiber sizes separately changed the model in a few significant ways. The contribution of each input variable to each model and the density distribution in the large axon model were fairly similar to the overall axon model (see Table 2 for variable contributions and model R2). For large axons, the model predicted low density regions throughout the outer-most 8% radially, particularly in the dorsal- and ventral-most 30–35% angularly, and a homogeneously higher inner 92% radially. The small axon model, while retaining an outer low density region (outer-most 6%), had a unique low-density region at the medial dorsal aspect (−2.11 to 1.35 mm axially, dorsal-most 25% angularly, 28% radially). This dorsal region is particularly important for DRGS applications, so a paucity of small axons in the area may be an important characteristic for ongoing research in the area. Figure 7 shows both models as violin plots and spatial models.
Figure 7:
Axon densities (Left) and spatial representations of axon density (Right) of (a) small and (b) large axon density models for L5 DRG. See Figure 5 for specific information on the types of figures shown.
4. Discussion
In this study, we analyzed 202 cross-sections across the full span of 33 human L4 and L5 DRG collected from 10 donors. To our knowledge, this represents the largest such collection of human DRG images included in a single study. Additionally, while this study was limited to a description of DRG, the full library of images produced includes over 900 images of human spinal roots, both stained and unstained, for use in potential future studies. We utilized a custom semi-automated GUI to aid in this analysis and greatly reduce processing time compared to manual image segmentation. This GUI, which relied on the high contrast neural staining provided by the use of NF200 (rather than hematoxylin & eosin), represented a significant improvement relative to our previous study of the feline DRG (Ostrowski et al., 2017).
Considering our donor demographics, the main effect we found was that the overall number of axons increases categorically with increasing BMI. To our knowledge, there is no conclusive evidence regarding the role of body size on sensory neuron counts. A previous study in frogs (St. Wecker & Farel, 1994) demonstrated that larger frogs had more DRG neurons than smaller frogs, however few studies have examined the effect of body size or weight on DRG neuron counts in mammals. One study demonstrated that in male rats, which continue to grow in body mass throughout adult life, DRG neuron count increases with age and size (Devor & Govrin-Lippmann, 1991). Other studies in rats refute this (Pover, Barnes, & Coggeshall, 1994), claiming that DRG neuron counts do not increase with age. However, that study did not report the sex of their animals, and it is understood that female rats level off in body size, unlike male rats. Future studies should examine the role of body size, and the presence of obesity, on DRG neuron counts in humans. As obesity is a common comorbidity to chronic pain, obese patients receiving DRGS may have different neuroanatomical makeups than non-obese patients. It is currently unclear if increased axon counts would have an impact on DRGS outcomes.
In contrast to previous studies, we did not find evidence that neuronal numbers increase with age (Devor & Govrin-Lippmann, 1985). However, our study was likely underpowered to determine this relationship because our sample only included two donors under the age of 35. One study in cats (H. H. Aldskogius & Risling, 1989) showed that DRG neuron count did not increase with age, however the authors reported a slight increase in axon counts with age.
A high-level anatomic trend that we observed was the significant increase in axon and cell body number in peripheral DRG compared to the spinal side. This trend is likely related to axonal branching of DRG neurons which has been observed in rat and cat studies, with branching ratios similar to those in our data (H. Aldskogius & Risling, 1981; Chung & Coggeshall, 1984; Langford & Coggeshall, 1979, 1981). Other explanations may include our underestimation of the number of small C-fiber axons present in our tissue samples, particularly considering that unmyelinated axons account for approximately two-thirds of axons in the DRG (Risling, Aldskogius, Hildebrand, & Remahl, 1983). Alternatively, it has been observed that the peripheral side of pseudounipolar DRG neuron axons are slightly larger than their spinal counterparts (Amir & Devor, 2003). Lee and colleagues demonstrated that nearly 13% of spinal axons in A-fibers have diameters less than 2 μm, suggesting they would not be detected by our algorithm (Lee et al., 1986). We found approximately 22% more fibers on the peripheral side of the DRG than the spinal side, a slightly larger portion than Lee and colleagues’ findings would suggest. It is possible that because our analysis could not detect very small fibers, there is a population of cells which can only be detected on their peripheral side.
We found a mean 33643 ± 11007 axons in L5 DRG and 27868 ± 10846 in L4 DRG. These counts are consistent with a recent study by Liu et al. which counted human dorsal root fibers and found 34455 ± 2740 in L5 and 31175 ± 2740 in L4 (Liu et al., 2015). In both cases, our mean counts are slightly lower, which may be a result of the variance in our data or our undercount of small unmyelinated fibers. However, both our study and Liu et al. stained spinal root tissue with NF200 and used computer programs to determine the number of fibers present, suggesting Liu and colleagues may have also underestimated the number of unmyelinated fibers. Davenport and Bothe reported 59,000 cells in human L3 DRG, with 25,000 unmyelinated fibers contributing to their total population (Davenport & Bothe, 1934). Davenport and Bothe’s estimation of the number of unmyelinated fibers in lumbar DRG approximately accounts for the difference in their estimate of total cellular population with the estimates made by our study and Liu et al. This suggests that both our study and Liu et al. accurately captured the population of myelinated axons in DRG, but underestimated the number of unmyelinated axons. Considering the sizes of the identified axons, there was a sharp drop-off in counted axons with mean diameter below 2 μm (only 0.018% of identified axons). We therefore concluded that our imaging and analysis method was not capable of consistently measuring unmyelinated C fiber axons, which have a typical diameter <1.6 μm (Gardner & Johnson, 2014). We also may be undercounting small myelinated fibers (Aδ), which have a minimum diameter of 1 μm (Gardner & Johnson, 2014). This was likely a result of a de-noising step in our image analysis, which could not distinguish very small axons from other cellular detritus.
For our primary quantification of human DRG spatial organization, we utilized a recursive partitioning algorithm to model axon and soma densities in normalized DRG cross-sections by axial, radial, and angular position. We found that the highest somatic densities were within about 2 mm of the widest point of L4 DRG and within 3 mm of the widest point of L5 DRG. Within these regions, both the highest and lowest cell body densities were found in the outer-most 20–25% radially, with the dorsal-most 75% having the highest overall density and the ventral-most 25% having the lowest overall density by angle. We found that axial position had a very minimal effect on axon density, which was much more affected by radial position. The outer ~10% at either level had a very low axon density, while the interior ~90% was relatively high.
Our findings related to soma density are consistent with our previous findings in Ostrowski et al. in feline lumbosacral DRG. That study found the highest density of cell bodies in the outer 25% radially and on the dorsal aspect (Ostrowski et al., 2017). As this was a study including DRG from a different spinal region (sacral), it suggests that our general characterization in human lumbar DRG can potentially be extended to other spinal levels.
4.1. Implications for Clinical Neurotechnologies
The location of different types of neurons within the DRG is vital to designing clinical neurotechnologies with the goal of treating disease. For example, dorsal root ganglion stimulation (DRGS) delivers electrical stimulation to the DRG with the goal of providing patients with pain relief. Presently, the physiological mechanisms of DRGS-induced pain relief are unknown, though previous studies have suggested that DRGS targets large myelinated DRG neurons (Graham et al., 2019), while other studies have suggested DRGS targets small unmyelinated afferents (Kent, Min, Hogan, & Kramer, 2018; Koopmeiners, Mueller, Kramer, & Hogan, 2013). As the mechanisms of DRGS become more clear, precisely targeting the cells responsible for pain relief, while avoiding cells that convey painful sensations, will be vital to the success of DRGS and to patient livelihood. Our results indicate that large axons are homogeneously distributed throughout human DRG. However, our data indicate that the density of smaller myelinated axons is lowest in the dorsal region of the middle of the DRG. Therefore, DRGS applied near the middle of the ganglion may simultaneously target the pain-relieving large axons, while minimizing the activation of small myelinated nociceptive fibers (e.g. Aδ-fibers). Furthermore, our data demonstrate the presence of biganglia in human lower lumbar DRG, supporting the findings of (Shen, Wang, Chen, & Liang, 2006). It is currently unclear if the presence of bi- or triganglia would affect the accurate delivery of electrical stimulation to DRG neurons, or if the separate ganglia are functionally distinct. We can speculate that distinct regions of connective tissue between multi-ganglia, which tend to have higher impedance than the neuron containing region (Hope, Vanholsbeeck, & McDaid, 2018), could affect the spread of current in the DRG. Shen and colleagues showed that biganglia are most commonly found at L4 (Shen et al., 2006), a common DRGS target to manage chronic pain in the foot and lower leg (Deer et al., 2019). Future studies should explore the existence of functionally-distinct biganglia, which may inform the placement of the electrode lead for DRGS.
For neurotechnologies that record signals from the DRG, the location of cell bodies is crucial to obtaining an adequate signal, as cell bodies and their initial segments produce most of the transmembrane currents that lead to recordable neural signals. Such technologies would need to be placed in close proximity to those structures to ensure obtaining a viable signal (Moffitt & McIntyre, 2005). Our results support previous findings in cats (Ostrowski et al., 2017) that cell bodies typically organize around the dorsal aspect of the ganglion. However, our results further demonstrate that this phenomenon is present not only at the most medial portion of the DRG, but over a span of several millimeters around the middle of the ganglion for human lumbar DRG. Therefore, to maximize the likelihood of achieving a viable signal, technologies seeking to record signals from the DRG should target the dorsal-middle portion of the DRG, unless future work should determine a particular somatotopy for the desired sensory modality. However, as described above, our method is likely underestimating the presence of small unmyelinated fibers in DRG. Many physiological signals of interest to neuroprosthetic technologies (e.g. bladder signals, nociception) are carried by small fibers (Dubin & Patapoutian, 2010; Fowler, 2002). Therefore, future implementations of our method must be mindful of the type of neuron carrying a signal of interest, and design their identification paradigm to successfully capture the target neural elements.
4.2. Limitations and Future Work
While our results provide a unique quantitative description of human DRG anatomy, there were several limitations to our study design. Firstly, we collected a histological slice at 1 mm increments along the nerve root axis. Human lower lumbar DRG on average range between 7.8 and 11.58 mm in length along the nerve root axis (Haberberger, Barry, Dominguez, & Matusica, 2019), meaning we were limited to between approximately 7 and 11 samples per DRG. Ideally, we would have sectioned an entire DRG with 5 μm slices, to enable a complete reconstruction of the entire cellular population of a ganglion. However, this would dramatically increase the cost of processing even a single DRG, which would have limited us from making comparisons across lumbar levels and across donors. Therefore, we believe that sacrificing slice resolution to enable statistical comparisons was an appropriate choice for our study, as we wanted to gain a rough picture of the stereotactic distribution of neurons in the DRG. Future studies should consider sectioning multiple DRG at a higher resolution, as the monetary and time costs of high-throughput histological processing decrease. Newer tissue clearing and light sheet microscopy techniques may offer opportunities to image DRG cells with minimal sectioning required.
Secondly, we collected DRG from 10 individual donors. As described above, we sacrificed spatial resolution along the nerve root axis to obtain specimens from multiple donors. However, with a sample size of 10 donors, it is unlikely that we are sufficiently powered to make strong statistical claims relating demographic data to the number and distribution of primary afferents in human DRG. Previous work in rats demonstrated an increased number of DRG neurons with increased age and body size (Devor & Govrin-Lippmann, 1985, 1991), and we were able to demonstrate body size differences (but not age differences) in our human data set. A previous study showed that sensory neuron count in human cervical DRG linearly increased with DRG volume, but DRG volume was not shown to correlate with any demographic data (e.g. height, weight) other than a significant sex difference in DRG volume (West, McKay Hart, Terenghi, & Wiberg, 2012), a parameter we did not directly quantify.
Staining our slides with only NF200 precluded us from making strong conclusions about the functional distribution of afferents in lumbar DRG. NF200 stains for cytoskeletal proteins present in all primary afferents in humans (Rostock et al., 2018; Vega et al., 1994). We believe that NF200 was the most appropriate choice in stain, as it provided sharp contrast between neural elements (which appeared brown in brightfield microscopy) and background tissue. However, the presence of NF200 did not assign a specific sensory function to a neuron (e.g. mechanoreception, nociception). Without information about sensory function, we were unable to make strong claims about the presence of functional somatotopy in human DRG. Co-staining our slides with other neurochemical markers more indicative of sensory function (e.g. TRPV1 or peripherin to stain small nociceptors (Chang et al., 2018)) would provide more convincing evidence of sensory function, and would allow us to test for the presence of functional somatotopy. Future work may utilize co-staining as an opportunity to examine out both sensory function and anatomical distribution of primary afferents.
Finally, the morphological operations our algorithm used to identify cell location may have individually or in tandem limited our ability to detect the location of very small axon fibers (i.e. < 2 μm in diameter). Unmyelinated C-fiber axons can be as small as 0.2 μm in diameter in cats (Lee et al., 1986). The morphology operations our algorithm used to accurately detect the location and diameter of stand-alone neural structures occasionally filtered out objects below 2 μm. However, the range of C-fiber axon diameters in humans is currently unknown. Therefore, it was difficult to estimate how many small neural structures were not captured by our framework. With algorithmic adjustments and images scanned at higher magnification, however, it may be possible to examine C-fiber population distributions using the same GUI and methods described herein.
Conclusion
The spatial distribution of neural elements in human DRG is important to clinical neurotechnologies seeking to treat neurological disease. We developed a semi-automated algorithm to identify the location of different neural elements in human DRG. We demonstrated that, at lower-lumbar levels, cell bodies preferentially organize around the dorsal-most region of the ganglion, while axons are homogeneously distributed throughout the interior 90% of the ganglion. We also presented a method to reconstruct 3D-models of human DRG based on histological images. Future studies could extend our identification algorithm to probe the presence of functional somatotopy in human DRG, or use 3D-model reconstructions as an in situ platform for developing novel DRG-interface technologies.
Acknowledgements:
We acknowledge the use of tissues procured by the National Disease Research Interchange (NDRI) with support from National Institute of Biomedical Imaging and Bioengineering award # U42OD11158. This work was also supported by funding from the National Science Foundation (CAREER award # 1653080), the National Institutes of Health (NIH R01NS089530), and the University of Michigan Rackham Graduate School (Predoctoral Fellowship; Student Research Grants). Neuropathologist Dr. Sandra Camelo-Piragua assisted in interpretation of histological slides. We also acknowledge the contribution of tissue by the anonymous donors, without whom this work would not have been possible.
Footnotes
Data Availability Statement: All DRG images, software, summary data, and CAD models referenced in this study are available online at the Open Science Framework (Peck-Dimit et al., 2019).
Disclosures: SFL holds stock options with Presidio Medical, Inc. and serves on the scientific advisory board.
References
- Al-Kofahi Y, Lassoued W, Lee W, & Roysam B (2010). Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Transactions on Biomedical Engineering, 57(4), 841–852. 10.1109/TBME.2009.2035102 [DOI] [PubMed] [Google Scholar]
- Aldskogius HH, & Risling MM (1989). Number of dorsal root ganglion neurons and axons in cats of different ages. Experimental Neurology, 106(1), 70–73. 10.1016/0014-4886(89)90145-3 [DOI] [PubMed] [Google Scholar]
- Aldskogius H, & Risling M (1981). Effect of sciatic neurectomy on neuronal number and size distribution in the L7 ganglion of kittens. Experimental Neurology, 74(2), 597–604. 10.1016/0014-4886(81)90194-1 [DOI] [PubMed] [Google Scholar]
- Amir R, & Devor M (2003). Electrical excitability of the soma of sensory neurons is required for spike invasion of the soma, but not for through-conduction. Biophysical Journal, 84(4), 2181–2191. 10.1016/S0006-3495(03)75024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi Y, Stein RB, Branner A, Pearson KG, & Normann RA (2003). Capabilities of a penetrating microelectrode array for recording single units in dorsal root ganglia of the cat. Journal of Neuroscience Methods, 128, 9–20. 10.1016/S0165-0270(03)00143-2 [DOI] [PubMed] [Google Scholar]
- Ayers CA, Fisher LE, Gaunt RA, & Weber DJ (2016). Microstimulation of the lumbar DRG recruits primary afferent neurons in localized regions of lower limb. Journal of Neurophysiology, 116(1), 51–60. 10.1152/jn.00961.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossy J (1970). Atlas of Neuroanatomy and Special Sense Organs (1st ed.). Philadelphia: WB: Saunders. [Google Scholar]
- Bruns TM, Wagenaar JB, Bauman MJ, Gaunt RA, & Weber DJ (2013). Real-time control of hind limb functional electrical stimulation using feedback from dorsal root ganglia recordings. Journal of Neural Engineering, 10(2). 10.1088/1741-2560/10/2/026020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns TM, Weber DJ, & Gaunt RA (2015). Microstimulation of afferents in the sacral dorsal root ganglia can evoke reflex bladder activity. Neurourology and Urodynamics, 34, 65–71. 10.1002/nau [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton H, & McFarlane JJ (1986). The organization of the seventh lumbar spinal ganglion of the cat. Journal of Comparative Neurology, 149, 215–232. [DOI] [PubMed] [Google Scholar]
- Chang W, Berta T, Kim YH, Lee S, Lee S-Y, & Ji R-R (2018). Expression and role of voltage-gated sodium channels in human dorsal root ganglion neurons with special focus on Nav1.7, species differences, and regulation by paclitaxel. Neuroscience Bulletin, 34(1), 4–12. 10.1007/s12264-017-0132-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, & Coggeshall RE (1984). The ration of dorsal root ganglion cells to dorsal root axons in sacral segments of the cat. The Journal of Comparative Neurology, 225(1), 24–30. [DOI] [PubMed] [Google Scholar]
- Davenport HA, & Bothe RT (1934). Cells and fibers in spinal nerves. II. A study of C2, C6, T4, T9, L3, S2, and S5 in man. The Journal of Comparative Neurology, 59(1), 167–174. 10.1002/cne.900590109 [DOI] [Google Scholar]
- Deer TR, Levy RM, Kramer J, Poree L, Amirdelfan K, Grigsby E, … Mekhail N (2017). Dorsal root ganglion stimulation yielded higher treatment success rate for complex regional pain syndrome and causalgia at 3 and 12 months: A randomized comparative trial. Pain, 158(4), 669–681. 10.1097/j.pain.0000000000000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deer TR, Pope JE, Lamer TJ, Grider JS, Provenzano D, Lubenow TR, … Mekhail N (2019). The neuromodulation appropriateness consensus committee on best practices for dorsal root ganglion stimulation. Neuromodulation, 22(1), 1–35. 10.1111/ner.12845 [DOI] [PubMed] [Google Scholar]
- Devor M (1999). Unexplained peculiarities of the dorsal root ganglion. Pain, 82(Supplement 6), S27–S35. [DOI] [PubMed] [Google Scholar]
- Devor M, & Govrin-Lippmann R (1985). Neurogenesis in adult rat dorsal root ganglia. Neuroscience Letters, 61, 189–194. 10.1016/0304-3940(85)90423-9 [DOI] [PubMed] [Google Scholar]
- Devor M, & Govrin-Lippmann R (1991). Neurogenesis in adult rat dorsal root ganglia: On counting and the count. Somatosensory & Motor Research, 8(1), 9–12. 10.3109/08990229109144724 [DOI] [PubMed] [Google Scholar]
- Dubin AE, & Patapoutian A (2010). Nociceptors: the sensors of the pain pathway. Journal of Clinical Investigation, 120(11), 3760–3772. 10.1172/JCI42843.3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LE, Ayers CA, Ciollaro M, Ventura V, Weber DJ, & Gaunt RA (2014). Chronic recruitment of primary afferent neurons by microstimulation in the feline dorsal root ganglia. Journal of Neural Engineering, 11(3). 10.1088/1741-2560/11/3/036007 [DOI] [PubMed] [Google Scholar]
- Flegal KM, Carroll MD, Kit BK, & Ogden CL (2012). Prevalence of Obesity and Trends in the Distribution of Body Mass Index Among US Adults, 1999–2010. JAMA, 307(5), 491 10.1001/jama.2012.39 [DOI] [PubMed] [Google Scholar]
- Fowler CJ (2002). Bladder afferents and their role in the overactive bladder. Urology, 59(Supplement 5A), 37–42. 10.1016/S0090-4295(02)01544-3 [DOI] [PubMed] [Google Scholar]
- Gardner EP, & Johnson KO (2014). The somatosensory system: Receptors and central pathways In Principles of Neural Science, Fifth Edition (pp. 475–497). New York, NY: McGraw-Hill Education. [Google Scholar]
- Graham RD, Bruns TM, Duan B, & Lempka SF (2019). Dorsal root ganglion stimulation for chronic pain modulates Aβ-fiber activity but not C-fiber activity: A computational modeling study. Clinical Neurophysiology, 130(6), 941–951. 10.1016/j.clinph.2019.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberberger RV, Barry C, Dominguez N, & Matusica D (2019). Human dorsal root ganglia. Frontiers in Cellular Neuroscience, 13, 1–17. 10.3389/fncel.2019.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Epton S, Bojanic S, Green AL, & FitzGerald JJ (2018). The efficacy and safety of dorsal root ganglion stimulation as a treatment for neuropathic pain: A literature review. Neuromodulation, 21(3), 225–233. 10.1111/ner.12685 [DOI] [PubMed] [Google Scholar]
- Holinski BJ, Everaert DG, Mushahwar VK, & Stein RB (2013). Real-time control of walking using recordings from dorsal root ganglia. Journal of Neural Engineering, 10(5), 056008 10.1088/1741-2560/10/5/056008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope J, Vanholsbeeck F, & McDaid A (2018). A model of electrical impedance tomography implemented in nerve-cuff for neural-prosthetics control. Physiological Measurement, 39(4), 1–17. 10.1088/1361-6579/aab73a [DOI] [PubMed] [Google Scholar]
- Josephson A, Widenfalk J, Trifunovski A, Widmer HR, Olson L, & Spenger C (2001). GDNF and NGF family members and receptors in human fetal and adult spinal cord and dorsal root ganglia. Journal of Comparative Neurology, 440(2), 204–217. 10.1002/cne.1380 [DOI] [PubMed] [Google Scholar]
- Kashkoush AI, Gaunt RA, Fisher LE, Bruns TM, & Weber DJ (2019). Recording single- and multi-unit neuronal action potentials from the surface of the dorsal root ganglion. Scientific Reports, 9(1), 1–12. 10.1038/s41598-019-38924-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausz M, & Rethelyi M (1985). Lamellar arrangment of neuronal somata in the dorsal root ganglion of the cat. Somatosensory Research, 2(3), 193–204. 10.1080/19397030902947041 [DOI] [PubMed] [Google Scholar]
- Kent AR, Min X, Hogan QH, & Kramer JM (2018). Mechanisms of dorsal root ganglion stimulation in pain suppression: A computational modeling analysis. Neuromodulation, 21, 234–246. 10.1111/ner.12754 [DOI] [PubMed] [Google Scholar]
- Kneist W, Kauff DW, Schröder M, Koch KP, & Lang H (2014). Percutaneous nerve evaluation based on electrode placement under control of autonomic innervation. Techniques in Coloproctology, 18(8), 725–730. 10.1007/s10151-014-1160-x [DOI] [PubMed] [Google Scholar]
- Koopmeiners AS, Mueller S, Kramer J, & Hogan QH (2013). Effect of electrical field stimulation on dorsal root ganglion neuronal function. Neuromodulation, 16(4), 304–311. 10.1111/ner.12028 [DOI] [PubMed] [Google Scholar]
- Langford LA, & Coggeshall RE (1979). Branching of sensory axons in the dorsal root and evidence for the absence of dorsal root efferent fibers. Journal of Comparative Neurology, 184, 193–204. [DOI] [PubMed] [Google Scholar]
- Langford LA, & Coggeshall RE (1981). Branching of Sensory Axons in the Peripheral Nerve of the Rat, 750, 745–750. [DOI] [PubMed] [Google Scholar]
- Lee KH, Chung K, Chung JM, & Coggeshall RE (1986). Correlation of cell body size, axon size, and signal conduction velocity for individually labelled dorsal root ganglion cells in the cat. The Journal of Comparative Neurology, 243(3), 335–346. 10.1002/cne.902430305 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou X, Ma J, Ge Y, & Cao X (2015). The diameters and number of nerve fibers in spinal nerve roots. The Journal of Spinal Cord Medicine, 38(4), 532–537. 10.1179/1079026814z.000000000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre CC, & Grill WM (1999). Excitation of central nervous system neurons by nonuniform electric fields. Biophysical Journal, 76(2), 878–888. 10.1016/S0006-3495(99)77251-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletic V, & Lu GW (1993). Characteristics of action potentials recorded from cat spinal ganglion neurons in vivo. Brain Research Bulletin, 31(5), 531–538. [DOI] [PubMed] [Google Scholar]
- Moffitt MA, & McIntyre CC (2005). Model-based analysis of cortical recording with silicon microelectrodes. Clinical Neurophysiology, 116(9), 2240–2250. 10.1016/j.clinph.2005.05.018 [DOI] [PubMed] [Google Scholar]
- Offord K, Ohta M, Oenning RF, & Dyck PJ (1974). Method of morphometric evaluation of spinal and autonomic ganglia. Journal of the Neurological Sciences, 22, 65–71. 10.1016/0022-510X(74)90054-9 [DOI] [PubMed] [Google Scholar]
- Ostrowski AK, Sperry ZJ, Kulik G, & Bruns TM (2017). Quantitative models of feline lumbosacral dorsal root ganglia cell density. Neuroscience Methods, 290, 116–124. 10.1016/j.jneumeth.2017.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Sperry ZJ, Barrera ND, & Bruns TM (2019). Real-time bladder pressure estimation for closed-loop control in a detrusor overactivity model. IEEE Transactions on Neural Systems and Rehabilitation Engineering, 27(6), 1209–1216. 10.1109/TNSRE.2019.2912374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [dataset] Peck-Dimit N, Sperry ZJ, Graham RD, Lempka SF, Bruns TM (2019) Human DRG 3D Cross Section Analysis. Open Science Framework. DOI: 10.17605/OSF.IO/TQRFZ [DOI] [Google Scholar]
- Peikari M, & Martel AL (2016). Automatic cell detection and segmentation from H and E stained pathology slides using colorspace decorrelation stretching. Proc. SPIE 9791, Medical Imaging 2016: Digital Pathology, 9791 10.1117/12.2216507 [DOI] [Google Scholar]
- Peyronnard J-M, Messier J-P, Dubreuil M, Charron L, & Lebel F (1990). Three-dimensional computer-aided analysis of the intraganglionic topography of primary muscle afferent neurons in the rat. The Anatomical Record, 227(4), 405–417. 10.1002/ar.1092270404 [DOI] [PubMed] [Google Scholar]
- Pover CM, Barnes MC, & Coggeshall RE (1994). Do primary afferent cell numbers change in relation to increasing weight and surface area in adult rats? Somatosensory & Motor Research, 11(2), 163–167. 10.3109/08990229409028869 [DOI] [PubMed] [Google Scholar]
- Prats-Galino A, Puigdellívol-Sánchez A, Ruano-Gil D, & Molander C (1999). Representations of hindlimb digits in rat dorsal root ganglia. Journal of Comparative Neurology, 408(1), 137–145. [DOI] [PubMed] [Google Scholar]
- Puigdellívol-Sánchez A, Prats-Galino A, Ruano-Gil D, & Molander C (1998). Sciatic and femoral nerve sensory neurones occupy different regions of the L4 dorsal root ganglion in the adult rat. Neuroscience Letters, 251(3), 169–172. 10.1016/S0304-3940(98)00518-7 [DOI] [PubMed] [Google Scholar]
- Risling M, Aldskogius H, Hildebrand C, & Remahl S (1983). Effects of sciatic nerve resection on L7 spinal roots and dorsal root ganglia in adult cats. Experimental Neurology, 82(3), 568–580. 10.1016/0014-4886(83)90081-X [DOI] [PubMed] [Google Scholar]
- Rostock C, Schrenk-Siemens K, Pohle J, & Siemens J (2018). Human vs. mouse nociceptors - similarities and differences. Neuroscience, 387, 13–27. 10.1016/j.neuroscience.2017.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, & Austin G (1961). Intracellular potentials of mammalian dorsal root ganglion cells. Journal of Neurophysi, 24(6), 569–582. [DOI] [PubMed] [Google Scholar]
- Shen J, Wang H-Y, Chen J-Y, & Liang B-L (2006). Morphologic analysis of normal human lumbar dorsal root ganglion by 3D MR imaging. AJNR. American Journal of Neuroradiology, 27(10), 2098–2103. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/17110675 [PMC free article] [PubMed] [Google Scholar]
- Silverstein MP, Romrell LJ, Benzel EC, Thompson N, Griffith S, & Lieberman IH (2015). Lumbar dorsal root ganglia location: An anatomic and MRI assessment. International Journal of Spine Surgery, 9 10.14444/2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry ZJ, Na K, Parizi SS, Chiel HJ, Seymour J, Yoon E, & Bruns TM (2018). Flexible microelectrode array for interfacing with the surface of neural ganglia. Journal of Neural Engineering, 15(3). 10.1088/1741-2552/aab55f [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Wecker PGR, & Farel PB (1994). Hindlimb sensory neuron number increases with body size. Journal of Comparative Neurology, 342(3), 430–438. 10.1002/cne.903420309 [DOI] [PubMed] [Google Scholar]
- Taha AA, & Hanbury A (2015). Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Medical Imaging, 15(1). 10.1186/s12880-015-0068-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda T, Seki K, Sato M, Nishimura Y, Kawato M, & Isa T (2012). Population coding of forelimb joint kinematics by peripheral afferents in monkeys. PLoS ONE, 7(10). 10.1371/journal.pone.0047749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega JA, Humara JM, Naves FJ, Esteban I, & Valle M. E. Del. (1994). Immunoreactivity for phosphorylated 200-kDa neurofilament subunit is heterogeneously expressed in human sympathetic and primary sensory neurons. Anatomy and Embryology, 190, 453–459. [DOI] [PubMed] [Google Scholar]
- Weber DJ, Stein RB, Everaert DG, & Prochazka A (2007). Limb-state feedback from ensembles of simultaneously recorded dorsal root ganglion neurons. Journal of Neural Engineering, 4(3). 10.1088/1741-2560/4/3/S04 [DOI] [PubMed] [Google Scholar]
- Wessels WJT, Feirabend HKP, & Marani E (1990a). Evidence for a rostrocaudal organization in dorsal root ganglia during development as demonstrated by intra-uterine WGA-HRP injections into the hindlimb of rat fetuses. Developmental Brain Research, 54(2), 273–281. 10.1016/0165-3806(90)90150-W [DOI] [PubMed] [Google Scholar]
- Wessels WJT, Feirabend HKP, & Marani E (1990b). Somatotopic organization in the sensory innervation of the rat hindlimb during development, using half dorsal root ganglia as subsegmental units. European Journal of Morphology, 28(2–4), 394–403. [PubMed] [Google Scholar]
- West CA, McKay Hart A, Terenghi G, & Wiberg M (2012). Sensory neurons of the human brachial plexus: A quantitative study employing optical fractionation and in vivo volumetric magnetic resonance imaging. Neurosurgery, 70(5), 1183–1194. 10.1227/NEU.0b013e318241ace1 [DOI] [PubMed] [Google Scholar]
- Wienert S, Heim D, Saeger K, Stenzinger A, Beil M, Hufnagl P, … Klauschen F (2012). Detection and segmentation of cell nuclei in virtual microscopy images: A minimum-model approach. Scientific Reports, 2(503), 1–7. 10.1038/srep00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DL (1971). Receptor characteristics and conduction velocites in bladder afferents. Journal of Psychiatric Research, 8(3), 225–235. 10.1016/0022-3956(71)90021-5 [DOI] [PubMed] [Google Scholar]