Abstract
Genome-Wide Association Studies (GWAS) have identified several loci contributing to lung cancer and COPD risk independently; however, inflammation-related pathways likely harbor additional lung cancer risk-associated variants in biologically relevant immune genes that differ dependent on COPD. We selected single nucleotide polymorphisms (SNPs) proximal to 2069 genes within 48 immune pathways. We modeled the contribution of these variants to lung cancer risk in a discovery sample of 1932 lung cancer cases and controls stratified by COPD status and validation sample of 953 cases and controls also stratified by COPD. There were 43 validated SNPs in those with COPD and 60 SNPs in those without COPD associated with lung cancer risk. Further, 29 of 43 and 28 of 60 SNPs demonstrated a statistically significant interaction with COPD in the pooled sample. These variants demonstrated tissue-dependent effects on proximal gene expression, enhanced network connectivity, and resided together in specific immune pathways. These results reveal that key inflammatory related genes and pathways, not found in prior GWAS, impact lung cancer risk in a COPD-dependent manner. Genetic variation identified in this study supplement prior lung cancer GWAS and serve as a foundation to further interrogate risk relationships in smoking and COPD populations.
Introduction
Lung cancer is the leading cause of cancer death in the United States and the second most frequently occurring cancer type 1 Approximately 80–90% of lung cancer is attributable to smoking 2. Rates of current smoking have declined by 8% from 1990–2014; however, lung cancer incidence during this period decreased by only 2.3% 3, and the large population of at-risk former smokers in the US remains a public health concern.
Tobacco smoke exposure is closely associated with the development of a spectrum of lung diseases including emphysema, chronic bronchitis, chronic obstructive pulmonary disease (COPD) and lung cancer. Accumulating evidence suggests that prolonged exposure to cigarette smoke initiates lung and airway irritation giving rise to chronic inflammatory infiltration, closely linked to the remodeling of airway mucosa seen in airway obstruction as well as in the degradation of alveolar interstitium described in emphysema 4–6. This chronic inflammatory state is believed to underlie the 2–3 fold increase of lung cancer risk in individuals with COPD 7,8.
Although smoking accounts for a majority of lung cancers, aggregation among families, occurrence in never smokers, and variability in risk among ever smokers suggests the existence of contributing genetic factors 9. Lung cancer GWAS have identified several genotype-phenotype associations in regions such as 15q25.1 (CHRNA5, CHRNA3), 5p15.33 (CLPTM1L, TERT) and 6p21.33 (BAG6/BAT3/MSH5) that are consistent and reproducible across multiple populations 10–16. This agnostic approach is useful for identifying novel genes and generating hypotheses about biological mechanisms previously unknown to be involved in lung cancer etiology. However, with genome-wide corrections for multiple testing, these studies restrict their reporting to the few top SNPs that pass stringent statistical thresholds, potentially excluding other significant SNPs with strong biological evidence supporting a role in lung cancer susceptibility. Therefore, the incorporation of knowledge on genes and pathways relevant to lung inflammation and tumorigenesis represents a complementary approach to generate novel hypotheses regarding the genetic contributors to risk and mechanistic differences in lung carcinogenesis.
We explored the contribution of genetic variation in immune pathways to lung cancer risk, separately by COPD status, in a discovery sample of 1008 cases and 924 controls and validated findings in an independent sample of 498 cases and 455 controls from the same population of inference. This was followed by evaluating whether the impact of each validated SNP was significantly heterogeneous among those with and without COPD as well as evaluating the functional and biological significance of the identified genetic variation.
Methods
Study Participants
The Wayne State University (WSU), McLaren Health Care (MHC) and Henry Ford Health System (HFHS) Institutional Review Boards approved the procedures used in collecting and processing participant information, and written informed consent was obtained from all subjects prior to participation. The INHALE study was initiated in 2012 and has been previously described 17. Briefly, lung cancer cases were enrolled at the Karmanos Cancer Center in Detroit or its network sites, or at HFHS in Detroit or its network sites, and volunteer controls were enrolled from the same geographic areas from which cases were drawn, preferentially matched to cases on smoking status (91.7% ever-smoking cases vs. 91.1% ever-smoking controls). Participants were 21–89 years of age, and were asked to complete an interview, low-dose chest CT scan and spirometry, and provide saliva, blood and tumor tissue samples. Further eligibility was restricted to those who carried health insurance (in the event medical follow-up was required based on a clinical finding on the CT or spirometry), and never had taken Amiodarone or been diagnosed with bronchiectasis or cystic fibrosis. Additionally, controls had never been diagnosed with lung cancer nor had surgical removal of any portion of either lung.
Data collection
Age, race, gender, history of COPD, family history of lung cancer and smoking history were collected in interviews. Pack years were calculated by multiplying the number of years smoked by the average number of cigarettes smoked per day divided by 20. Pulmonary function tests with spirometry were either performed by trained technicians in accordance with ATS guidelines 18 at the time of enrollment or spirometry results from pulmonary function tests (PFTs) were abstracted from medical records if completed within 6 months of the INHALE interview date. For analysis purposes, COPD was defined based on spirometry (FEV1/FVC < 0.70); where FEV1/FVC was missing (14%), self-reported history of COPD was used.
Genotyping and selection of immune system pathways
Genotyping was performed using the Illumina Multi-Ethnic GWAS/Exome Array (MEGA), which covers 1.7 million SNPs across the genome. The chip was designed to capture variation in ethnically diverse populations including Europeans, Asians, African Americans and Hispanics. The variants originate from sequencing discoveries, other GWAS panels, and published disease association studies. Immune system related genes and pathways were obtained from either the Reactome database or a published study of inflammation pathway genes and lung cancer risk 19,20. The assembled immune system gene and pathway list contains manually curated, peer-reviewed pathway annotations cross-referenced with multiple databases including KEGG, Ensembl, and Uniprot by Reactome staff (Supplemental Table 1). Pathway annotations were included during the overrepresentation pathway analysis. MEGA SNP hg19 build 37 coordinates were cross-referenced with immune pathway gene locations according to the UCSC genome browser 21. In addition to intragenic SNPs, SNPs within flanking, proximal regulatory regions were included if contained within +/− 2kB of the gene region based upon ENCODE proximal regulatory data 22, 23. After removing invariant sites, there were 77,777 SNPs mapped to 2,015 immune pathway genes. SNPs were then filtered based on GenTrain score (<0.7), call rate <0.95 and inconsistent genotypes based on 19 CEPH sample replicates (99.88% concordance overall); 71,737 SNPs passed these quality control criteria. We required at least 15 minor allele carrier cases for each SNP evaluated (across discovery and validation samples) to avoid unstable effect estimates due to very rare SNPs. Thus, our analysis set consisted of 43,953 SNPs.
Statistical Analysis
The total number of INHALE participants with genotype data available was divided into a ‘discovery’ and ‘validation’ sample as follows: subjects were stratified by case-control status and randomly assigned to either the discovery or validation set by a ratio of 2:1. In this way the discovery set represented 2/3 of the total INHALE sample and the validation set represented the remaining 1/3 of the sample. There were no significant differences between the discovery and validation samples for any of the covariates used in this analysis (Supplemental Tables 2 and 3). Discovery and validation samples were further stratified by COPD status and analyzed separately.
We estimated African ancestry based on a panel of 128 ancestry informative markers (AIMs) described by Kosoy and colleagues 24; 122 AIMs were genotyped and passed QC standards in the MEGA panel. Assuming Hardy-Weinberg Equilibrium, expected genotype relative frequencies were calculated for both European (‘EUR’) and African (‘AFR’) populations based on 1000 Genomes samples. For each of the three genotypes per AIM, the proportion of African ancestry was computed as f(AFRj)/(f(AFRj)+f(EURj)), where f(AFRj) is the expected frequency for genotype j in Africans and f(EURj) is the expected frequency for genotype j in Europeans. Samples were then assigned a probability of African ancestry for each SNP corresponding to their observed genotype. These probabilities were summed and scaled for each individual to a standard uniform as the probability of African ancestry for the jth AIM, and aj and bj are the minimum and maximum possible probabilities for the jth AIM, respectively. This method of scoring ancestry was previously tested against principal component analysis (PCA) for correcting for population sub-structure and was found to perform similarly 24. We verified these findings by conducting a PCA-based ancestry estimate with EIGENSTRAT using all 43,953 immune pathway-based SNPs and then comparing this estimate with African ancestry score. The top eigenvector explained ~74% of all variance explained by significant eigenvectors (n=19). African ancestry score and this top eigenvector were highly correlated (Spearman correlation: 0.832, p = 1×10−16)(Supplemental Figure S1).
In both the discovery and validation samples, logistic regression modeling was used to estimate individual SNP effects on lung cancer risk, separately among those with and without COPD, assuming an additive (per allele effect) genetic model. SNP effects were adjusted for age, gender, African ancestry score and pack years. Due to the exploratory nature of this study, we used a threshold of α=0.05 for carrying forward SNPs in either the COPD or no COPD discovery samples for testing in the respective validation sample. We assessed race-specific SNP effects by modeling each of the validated SNPs separately in whites and African Americans (also adjusting for ancestry score), combining the (COPD or no COPD) discovery and validation samples. We also used logistic regression modeling to determine whether the effects of any of the validated SNPs were statistically dependent on COPD in the pooled sample by incorporating an interaction term, the cross-product of SNP genotype (0, 1 or 2) and COPD status (0 or 1). For interaction modeling, we analyzed the combined sample of cases and controls with and without COPD. SNP effects on tissue-specific gene expression were modeled as cis expression quantitative trait loci (eQTL) as described previously by the Genotype-Tissue Expression (GTEx) project version 7 25. Select GTEx tissues were evaluated for eQTL effects, lung and blood, to narrow the eQTL search to tissues directly involved in tobacco mediated lung injury. Multiple test corrections were computed for eQTL effects separately in each tissue. Network connectivity of genes was assessed using STRING as described previously 26. Overrepresentation analysis was conducted using the hypergeometric distribution within the 48 immune pathways containing 2015 genes.
Data A vailability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Results
Cohort description
A description of the INHALE case-control sample stratified by COPD is presented in Table 1. Lung cancer cases with COPD were more often white, ever smokers, and more likely to have a family history of lung cancer relative to controls with COPD. Cases in both strata (COPD versus no COPD) were more likely to be older and heavier smokers relative to controls, in addition to being diagnosed most often with adenocarcinoma and at later stages (stage III/IV, 72%). The total INHALE sample (N=2885) was subsequently split into a discovery (66%, n=1932/2885) and a validation (33%, n=953/2885) sample using random sampling. There were no significant differences between either cases in the discovery and validation samples or controls in the discovery and validation samples (Supplemental Tables 2 and 3).
Table 1.
Description of INHALE lung cancer case-control sample (N=2885), stratified by COPD status.
| COPD (n=1123/2885) | No COPD (n=1762/2885) | |||||
|---|---|---|---|---|---|---|
| Variable | Lung Cancer Cases (n=677) |
Controls (n=446) |
p value | Lung Cancer Cases (n=829) |
Controls (n=933) |
p value |
| Gender (n, %) | ||||||
| Male | 291 (43.0) | 208 (46.6) | 0.228 | 387 (46.7) | 421 (45.1) | 0.512 |
| Female | 386 (57.0) | 238 (53.4) | 442 (53.3) | 512 (54.9) | ||
| Race (n, %) | ||||||
| White | 435 (64.3) | 240 (53.8) | 0.001 | 545 (65.7) | 573 (61.4) | 0.060 |
| African American | 242 (35.7) | 206 (46.2) | 284 (34.3) | 360 (38.6) | ||
| Age (μ, SD) | 64.6 (9.4) | 62.5 (9.6) | <0.001 | 62.3 (11.1) | 59.4 (9.3) | <0.001 |
| <50 | 36 (5.3) | 40 (9.0) | 100 (12.1) | 131 (14.0) | ||
| 51–59 | 174 (25.7) | 127 (28.5) | 0.027 | 234 (28.2) | 353 (37.8) | <0.001 |
| 60–69 | 258 (38.1) | 167 (37.4) | 280 (33.8) | 325 (34.8) | ||
| 70+ | 209 (30.9) | 112 (25.1) | 215 (25.9) | 124 (13.3) | ||
| Smoking status (n, %) | ||||||
| Never | 19 (2.8) | 30 (6.7) | 0.002 | 119 (14.4) | 106 (11.4) | 0.060 |
| Ever | 658 (97.2) | 416 (93.3) | 710 (85.6) | 827 (88.6) | ||
| Pack years (μ, SD)* | 50.5 (29.9) | 36.9 (27.2) | <0.001 | 41.2 (29.6) | 30.4 (22.5) | <0.001 |
| Family history of lung cancer | ||||||
| No | 500 (73.9) | 384 (86.3) | <0.001 | 655 (79.1) | 764 (81.9) | 0.141 |
| Yes | 177 (26.1) | 61 (13.7) | 173 (20.9) | 169 (18.1) | ||
| Missing | 0 | 1 | 1 | 0 | ||
| Histology | ||||||
| Adenocarcinoma | 330 (49.2) | 511 (61.6) | ||||
| Squamous cell | 173 (25.8) | 139 (16.8) | ||||
| Small cell | 105 (15.7) | --- | 105 (12.7) | --- | ||
| other NSCLC | 62 (9.3) | 62 (7.5) | ||||
| unknown/missing | 7 | 12 | ||||
| Stage | ||||||
| I | 164 (24.6) | 116 (14.3) | ||||
| II | 67 (10.0) | 70 (8.6) | ||||
| III | 153 (22.9) | --- | 183 (22.5) | --- | ||
| IV | 283 (42.4) | 443 (54.6) | ||||
| Missing | 10 | 17 | ||||
Reported as the mean and SD in smoking participants only
Variant association testing
There were 43,953 immune-centric SNP association tests performed separately in samples with and without COPD (Supplementary Figures S2 and S3). SNP genotypes were modeled additively and adjusted for age, African ancestry, gender and pack years of smoking exposure. We carried forward 1,837 SNPs among cases and controls with COPD and 2,130 SNPs among cases and controls without COPD with a nominal p-value < 0.05 for evaluation in the validation sample. Upon testing these SNPs in the validation sample, 43 of 1,837 also met the α=0.05 threshold and were concordant in their direction of effect in the COPD validation sample (Supplemental Table 4). For cases and controls without COPD, 60 of 2,130 SNPs met these criteria in the validation sample (Supplemental Table 5). There were no validated SNPs in common to both strata.
To formally evaluate the context-dependency of validated SNPs in both the COPD and no COPD strata, we modeled interactions between COPD and each SNP in the pooled sample (discovery and validation samples combined). Interaction test results are presented in Table 2 for SNPs identified in individuals with COPD and Table 3 for SNPs identified in individuals without COPD. We considered an interaction test result of p<0.05 as sufficient evidence for context-dependency. We found significant COPD-dependent effects for 29 of 43 of the validated SNPs from the COPD stratum within 24 immune genes. Among the validated SNPs in samples without COPD, 28 of 60 SNPs had a significant interaction with COPD within 26 immune pathway genes. Hence, out of 103 SNPs tested for context-dependency, 57 demonstrated significant COPD-dependent effects. We observed consistent context-dependent associations among these 57 SNPs when lung cancer cases were stratified by histology (adenocarcinoma and squamous cell carcinoma, Supplemental Tables 6 and 7).
Table 2.
Tests of COPD x SNP interaction on lung cancer risk for the 43 validated SNPs in individuals with COPD
| SNP | Gene | CHR | Position | COPD × SNP interaction p value |
Pooled risk model results | ||
|---|---|---|---|---|---|---|---|
| OR* | 95% CI | p value | |||||
| rs2932538 | MOV10 | 1 | 113216543 | 0.055 | 0.73 | (0.59,0.91) | 0.004 |
| rs2901600 | DNM3 | 1 | 171835654 | <0.001 | 0.73 | (0.61,0.87) | 0.0005 |
| rs693 | APOB | 2 | 21232195 | 0.680 | 0.83 | (0.69,0.99) | 0.0437 |
| JHU_2.70774695 | TGFA | 2 | 70774696 | 0.025 | 4.34 | (1.76,10.71) | 0.0014 |
| rs10932427 | ERBB4 | 2 | 213073615 | 0.012 | 0.6 | (0.44,0.81) | 0.001 |
| rs115435003 | TRIP12 | 2 | 230629658 | 0.998 | 0.46 | (0.23,0.94) | 0.0325 |
| rs546530 | TRIP12 | 2 | 230752964 | 0.077 | 1.3 | (1.09,1.56) | 0.004 |
| rs7570061 | INPP5D | 2 | 233977318 | 0.005 | 0.7 | (0.57,0.86) | 0.0006 |
| rs79048756 | CD96 | 3 | 111323053 | 0.015 | 0.41 | (0.24,0.68) | 0.0007 |
| rs61505577 | BMPR1B | 4 | 95789665 | 0.015 | 2.48 | (1.46,4.23) | 0.0008 |
| rs73836068 | BMPR1B | 4 | 95891132 | 0.057 | 1.87 | (1.29,2.72) | 0.001 |
| JHU_6.117021274 | KPNA5 | 6 | 117021275 | 0.010 | 0.4 | (0.24,0.68) | 0.0007 |
| 6:125369362-CT | RNF217 | 6 | 125369362 | 0.022 | 0.28 | (0.11,0.7) | 0.0062 |
| rs73783372 | PARK2 | 6 | 162155477 | 0.015 | 2.54 | (1.52,4.25) | 0.0004 |
| JHU_7.54821275 | SEC61G | 7 | 54821276 | 0.070 | 4.37 | (1.6,11.93) | 0.004 |
| JHU_7.139540808 | TBXAS1 | 7 | 139540809 | 0.052 | 0.65 | (0.48,0.88) | 0.0053 |
| JHU_8.71282810 | NCOA2 | 8 | 71282811 | 0.030 | 0.24 | (0.09,0.6) | 0.0026 |
| rs4745646 | TJP2 | 9 | 71769323 | 0.011 | 1.52 | (1.18,1.95) | 0.001 |
| rs688391 | PRKCQ | 10 | 6489652 | 0.312 | 1.34 | (1.11,1.61) | 0.0024 |
| rs3793727 | PRKCQ | 10 | 6508377 | 0.005 | 1.56 | (1.26,1.93) | <0.0001 |
| rs658230 | PRKCQ | 10 | 6508563 | 0.044 | 1.39 | (1.16,1.67) | 0.0005 |
| JHU_10.32320560 | KIF5B | 10 | 32320561 | 0.080 | 0.7 | (0.55,0.88) | 0.0021 |
| rs12252698 | PRKG1 | 10 | 53608098 | 0.002 | 0.69 | (0.54,0.89) | 0.0037 |
| rs1937701 | PRKG1 | 10 | 53608977 | 0.009 | 0.7 | (0.57,0.86) | 0.0007 |
| JHU_10.75843193 | VCL | 10 | 75843194 | 0.006 | 0.44 | (0.26,0.75) | 0.0025 |
| rs3127255 | FBXW4 | 10 | 103370234 | 0.105 | 1.31 | (1.07,1.6) | 0.0086 |
| rs666432 | TRIM29 | 11 | 120003533 | 0.003 | 1.48 | (1.14,1.93) | 0.0037 |
| rs4411364 | TNFRSF19 | 13 | 24191374 | 0.029 | 1.43 | (1.12,1.82) | 0.0042 |
| rs9510787 | TNFRSF19 | 13 | 24205195 | 0.034 | 1.43 | (1.12,1.82) | 0.0042 |
| rs1630 | TNFRSF19 | 13 | 24249847 | 0.010 | 1.49 | (1.22,1.82) | 0.0001 |
| rs17446928 | FOXO1 | 13 | 41212225 | 0.001 | 0.39 | (0.25,0.6) | <0.0001 |
| rs76294435 | PPP2R5C | 14 | 102274571 | 0.077 | 0.4 | (0.25,0.65) | 0.0002 |
| JHU_14.103934653 | MARK3 | 14 | 103934654 | 0.099 | 0.73 | (0.6,0.89) | 0.0018 |
| rs55986634 | DAPK2 | 15 | 64275645 | 0.180 | 0.71 | (0.58,0.88) | 0.0018 |
| rs75395345 | PIAS1 | 15 | 68373718 | 0.993 | 1.26 | (1,1.57) | 0.0485 |
| rs2071501 | CSK | 15 | 75095157 | 0.033 | 0.54 | (0.37,0.79) | 0.0015 |
| JHU_16.4014963 | ADCY9 | 16 | 4014964 | 0.049 | 1.46 | (1.18,1.82) | 0.0006 |
| rs933392 | ADCY9 | 16 | 4032716 | 0.036 | 1.44 | (1.16,1.79) | 0.001 |
| exm1358199 | UBE2O | 17 | 74387284 | 0.023 | 1.39 | (1.14,1.69) | 0.0012 |
| JHU_18.49961949 | DCC | 18 | 49961950 | 0.020 | 0.68 | (0.53,0.88) | 0.0028 |
| rs10414006 | SPTBN4 | 19 | 41001921 | 0.010 | 0.62 | (0.49,0.79) | 0.0001 |
| rs11879349 | NLRP4 | 19 | 56364210 | 0.041 | 0.62 | (0.47,0.82) | 0.0008 |
| exm2262720 | PAK3 | 23 | 110379807 | 0.034 | 0.68 | (0.52,0.88) | 0.0033 |
Logistic model adjusted for age, gender, African ancestry score and pack years
Table 3.
Tests of COPD x SNP interaction on lung cancer risk for the 60 validated SNPs in individuals without COPD
| SNP | Gene | CHR | Position | COPD × SNP interaction p value |
Pooled risk model results | ||
|---|---|---|---|---|---|---|---|
| OR* | 95% CI | p value | |||||
| exm69478 | ASB17 | 1 | 76397972 | 0.156 | 1.29 | (1.1,1.5) | 0.0014 |
| JHU_1.108497389 | VAV3 | 1 | 108497390 | 0.029 | 0.72 | (0.58,0.89) | 0.0022 |
| rs3754293 | LAMTOR2 | 1 | 156024373 | 0.219 | 0.8 | (0.69,0.93) | 0.0028 |
| exm113346 | SPTA1 | 1 | 158645965 | <0.001 | 6.46 | (2.37,17.63) | 0.0003 |
| rs2230779 | TRAF5 | 1 | 211533352 | 0.534 | 1.59 | (1.18,2.14) | 0.0024 |
| rs10929693 | ATP6V1C2 | 2 | 10863267 | 0.435 | 0.8 | (0.69,0.93) | 0.0032 |
| exm175467 | APOB | 2 | 21225281 | 0.187 | 1.2 | (1.02,1.43) | 0.0316 |
| newrs676210 | APOB | 2 | 21231524 | 0.183 | 1.21 | (1.02,1.43) | 0.0299 |
| rs3749096 | EDAR | 2 | 109512428 | 0.044 | 1.34 | (1.1,1.63) | 0.0033 |
| rs13418730 | WIPF1 | 2 | 175540594 | 0.048 | 0.68 | (0.52,0.89) | 0.0046 |
| rs7583875 | AP1S3 | 2 | 224665694 | 0.062 | 0.8 | (0.7,0.92) | 0.0016 |
| JHU_3.18396523 | SATB1 | 3 | 18396524 | 0.127 | 0.35 | (0.2,0.6) | 0.0001 |
| rs80069959 | KCNH8 | 3 | 19223049 | 0.069 | 0.58 | (0.4,0.84) | 0.0040 |
| JHU_3.119275362 | CD80 | 3 | 119275363 | 0.019 | 0.53 | (0.36,0.77) | 0.0009 |
| rs953239 | TRPC1 | 3 | 142446205 | 0.003 | 1.23 | (1.07,1.41) | 0.0036 |
| rs7623154 | PIK3CA | 3 | 178921158 | 0.020 | 1.25 | (1.07,1.46) | 0.0055 |
| JHU_5.16912953 | MYO10 | 5 | 16912954 | 0.029 | 2.24 | (1.27,3.94) | 0.0051 |
| JHU_5.35873123 | IL7R | 5 | 35873124 | 0.187 | 0.6 | (0.43,0.84) | 0.0034 |
| rs7726469 | CAMK4 | 5 | 110586438 | 0.020 | 0.76 | (0.65,0.9) | 0.0015 |
| rs12153148 | KLHL3 | 5 | 136964764 | 0.066 | 0.74 | (0.62,0.88) | 0.0006 |
| rs3777376 | KLHL3 | 5 | 136965249 | 0.042 | 0.73 | (0.61,0.87) | 0.0005 |
| rs7774142 | LY86 | 6 | 6642058 | 0.027 | 1.3 | (1.12,1.5) | 0.0007 |
| exm-rs3827784 | LY86 | 6 | 6642405 | 0.031 | 1.31 | (1.12,1.52) | 0.0005 |
| JHU_6.137043810 | MAP3K5 | 6 | 137043811 | 0.016 | 2.19 | (1.23,3.89) | 0.0075 |
| rs56247201 | PARK2 | 6 | 162702092 | 0.191 | 0.5 | (0.33,0.74) | 0.0006 |
| rs35537854 | RPS6KA2 | 6 | 167072030 | 0.128 | 0.6 | (0.42,0.85) | 0.0041 |
| JHU_7.30352063 | ZNRF2 | 7 | 30352064 | 0.001 | 0.15 | (0.04,0.53) | 0.0033 |
| JHU_7.30393775 | ZNRF2 | 7 | 30393776 | 0.062 | 0.29 | (0.14,0.62) | 0.0014 |
| exm689348 | TNFRSF10A | 8 | 23049292 | 0.298 | 0.75 | (0.62,0.92) | 0.0047 |
| rs73241640 | NRG1 | 8 | 31932616 | 0.031 | 0.39 | (0.24,0.63) | 0.0002 |
| rs11776203 | NRG1 | 8 | 32419119 | 0.003 | 0.76 | (0.63,0.92) | 0.0039 |
| JHU_8.32431713 | NRG1 | 8 | 32431714 | 0.446 | 0.42 | (0.24,0.74) | 0.0029 |
| rs1014306 | DAPK1 | 9 | 90157451 | 0.317 | 1.33 | (1.14,1.54) | 0.0002 |
| rs12378686 | DAPK1 | 9 | 90163570 | 0.547 | 1.3 | (1.1,1.52) | 0.0018 |
| JHU_9.90198587 | DAPK1 | 9 | 90198588 | 0.152 | 0.58 | (0.4,0.82) | 0.0024 |
| rs10995319 | PRKG1 | 10 | 52762887 | 0.125 | 1.32 | (1.09,1.6) | 0.0045 |
| rs7904024 | PRKG1 | 10 | 52841790 | 0.022 | 1.28 | (1.11,1.48) | 0.0007 |
| JHU_10.83841723 | NRG3 | 10 | 83841724 | 0.415 | 2.87 | (1.4,5.86) | 0.0038 |
| rs74153420 | BMPR1A | 10 | 88628433 | 0.287 | 2.4 | (1.33,4.33) | 0.0038 |
| JHU_10.93222022 | HECTD2 | 10 | 93222023 | 0.486 | 1.46 | (1,2.11) | 0.0481 |
| JHU_10.123313013 | FGFR2 | 10 | 123313014 | 0.020 | 1.49 | (1.17,1.91) | 0.0014 |
| rs548142 | DYNC2H1 | 11 | 103315520 | 0.048 | 0.75 | (0.65,0.86) | 0.0001 |
| JHU_12.6438144 | TNFRSF1A | 12 | 6438145 | 0.016 | 1.83 | (1.3,2.58) | 0.0005 |
| JHU_12.26512936 | ITPR2 | 12 | 26512937 | 0.011 | 3.4 | (1.48,7.86) | 0.0041 |
| rs61971164 | STK24 | 13 | 99190397 | 0.072 | 0.73 | (0.61,0.88) | 0.0008 |
| rs17565502 | TNFSF13B | 13 | 108954304 | 0.061 | 1.26 | (1.08,1.47) | 0.0039 |
| JHU_14.23313974 | MMP14 | 14 | 23313975 | 0.011 | 0.54 | (0.35,0.82) | 0.0038 |
| rs78656887 | PSMC1 | 14 | 90734095 | 0.018 | 0.48 | (0.31,0.76) | 0.0016 |
| rs12441042 | TLN2 | 15 | 62946064 | 0.085 | 0.79 | (0.68,0.92) | 0.0019 |
| rs74318887 | MEF2A | 15 | 100229061 | 0.027 | 0.46 | (0.29,0.74) | 0.0012 |
| rs76272325 | PSMB6 | 17 | 4699845 | 0.172 | 0.5 | (0.34,0.74) | 0.0004 |
| JHU_17.5413392 | NLRP1 | 17 | 5413393 | 0.055 | 0.79 | (0.68,0.93) | 0.0033 |
| JHU_17.40648111 | ATP6V0A1 | 17 | 40648112 | 0.003 | 1.83 | (1.33,2.51) | 0.0002 |
| rs12949223 | CD300LD | 17 | 72589264 | 0.246 | 1.24 | (1.07,1.44) | 0.0050 |
| JHU_18.21773860 | OSBPL1A | 18 | 21773861 | 0.115 | 2.78 | (1.49,5.17) | 0.0012 |
| rs11082490 | SIGLEC15 | 18 | 43412628 | <0.001 | 1.53 | (1.27,1.84) | <0.0001 |
| rs58993112 | MALT1 | 18 | 56412784 | 0.159 | 1.35 | (1.11,1.64) | 0.0023 |
| exm2253611 | PDE4A | 19 | 10546771 | 0.029 | 1.25 | (1.08,1.45) | 0.0027 |
| rs9676881 | KEAP1 | 19 | 10596780 | 0.008 | 1.32 | (1.14,1.53) | 0.0002 |
| rs2898449 | MX1 | 21 | 42814495 | 0.063 | 0.69 | (0.54,0.87) | 0.0020 |
Logistic model adjusted for age, gender, African ancestry score and pack years
Functional and biological significance
Considering our variant selection strategy, SNPs were selected for their proximity (±2kB) to immune pathway genes and each SNP was assigned to a specific gene. Through this gene assignment, we evaluated whether the genes represented by the 57 significant and context-dependent risk loci demonstrated functional or biological importance.
First, we interrogated the functional impact of risk notable SNPs on the expression of paired candidate genes using tissue-specific expression quantitative trait loci (eQTL) data from the GTEx consortium. We assessed eQTLs in two specimen types, lung tissue and whole blood leukocytes, to capture SNP functionality in the primary tissues relevant to lung inflammation (Table 4). Eleven risk significant and context-dependent SNP-gene pairs were significant eQTLs in lung, blood, or both tissues. Of variants notable for lung cancer risk with an interaction with COPD status (n=57), four SNPs demonstrated significant (p < 0.05) effects on paired candidate gene expression in lung tissue alone. Additionally, four SNPs demonstrated significant (p < 0.05) paired gene expression effects in whole blood leukocytes. Three SNP-gene pairs were significant in both tissues (CD96, NRG1 and UBE2O).
Table 4.
Significant eQTL and context-dependent lung cancer risk SNP-gene pairs in lung and whole blood leukocyte tissues
| SNP | Gene | CHR | Position | COPD × SNP interaction p value |
Lung cancer risk model p value |
Lung eQTL p value |
Lung eQTL FDR |
Blood eQTL p value |
Blood eQTL FDR |
|---|---|---|---|---|---|---|---|---|---|
| Significant eQTL SNP-gene pairs in individuals with COPD | |||||||||
| rs79048756 | CD96 | 3 | 111323053 | 0.015 | 0.0007 | 3.7×10−4 | 0.016 | 1.8×10−5 | 6.6×10−4 |
| rs4745646 | TJP2 | 9 | 71769323 | 0.011 | 0.0010 | >0.05 | - | 7.2×10−4 | 0.008 |
| rs3793727 | PRKCQ | 10 | 6508377 | 0.005 | 0.0000 | >0.05 | - | 0.007 | 0.049 |
| rs666432 | TRIM29 | 11 | 120003533 | 0.003 | 0.0037 | 0.036 | 0.25 | >0.05 | - |
| exm1358199 | UBE2O | 17 | 74387284 | 0.023 | 0.0012 | 4.6×10−6 | 4.0×10−4 | <0.001 | 0.002 |
| Significant eQTL SNP-gene pairs in individuals without COPD | |||||||||
| rs7623154 | PIK3CA | 3 | 178921158 | 0.020 | 0.0055 | 0.002 | 0.042 | >0.05 | - |
| rs953239 | TRPC1 | 3 | 142446205 | 0.003 | 0.0036 | 0.002 | 0.042 | >0.05 | - |
| rs3777376 | KLHL3 | 5 | 136965249 | 0.042 | 0.0005 | >0.05 | - | 0.006 | 0.045 |
| rs11776203 | NRG1 | 8 | 32419119 | 0.003 | 0.0039 | 0.044 | 0.28 | 8.1×10−10 | 6.0×10−8 |
| rs11082490 | SIGLEC15 | 18 | 43412628 | 0.001 | 0.0000 | >0.05 | - | 2.8×10−4 | 0.003 |
| rs9676881 | KEAP1 | 19 | 10596780 | 0.008 | 0.0002 | 0.004 | 0.069 | >0.05 | - |
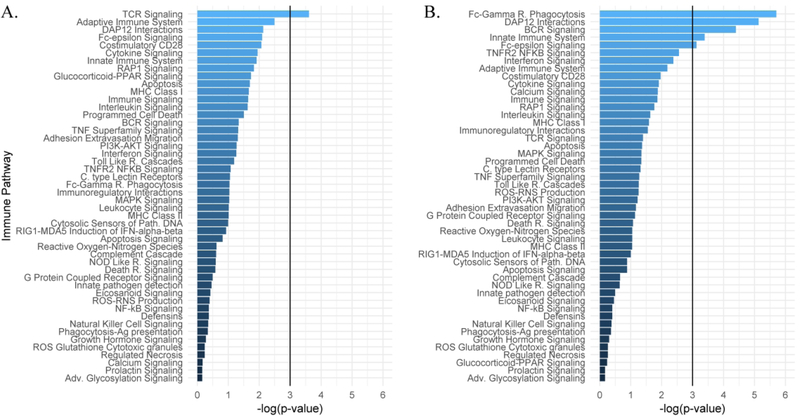
As an alternative to functional characterization, we interrogated the biological significance of immunological genes implicated by context-dependent risk loci separately by COPD. First, we assessed the network connectivity of genes demonstrating context-dependent risk associations to determine whether these genes act in cohesive biological networks. Secondly, we assessed whether any of the immune pathways contained a greater number of loci-paired genes than we would expect by chance to identify the immunological pathways involved in lung cancer risk. To limit false positive results a significance threshold of α=0.01 was used. In COPD cases and controls, eight functional protein-protein network interactions were identified among candidate genes with significant and context dependent lung cancer risk associations (n=24), which was not statistically significant (p = 0.238; Supplemental Figure 4A). The 24 gene candidates in the COPD strata were also not significantly overrepresented at α=0.01 within any of the 48 immune pathways evaluated (Figure 1A). In individuals without COPD, candidate genes (n=26) contained 20 functional protein-network associations (Supplemental Figure 4B), more than expected by chance (p = 0.0002). Two immune pathways, Fc-gamma receptor dependent phagocytosis (R-HSA-2029480) and DAP12 Interactions (R-HSA-2172127), were significantly (p<0.01) overrepresented among these significant and context dependent lung cancer risk genes (Figure 1B).
Figure 1. Pathway architecture of the significant risk candidates between COPD stratums.
Pathway analysis plots of the significant lung cancer risk and COPD interaction candidates within the designated immune pathways chosen for this study. Overrepresentation significance values are plotted on the x-axis for each pathway with a line designating the significance threshold. (A) Pathway significance values for candidates identified in individuals with COPD. (B) Pathway significance values for candidates identified in individuals without COPD.
These data demonstrate that immune-centric risk loci whose effects differ by COPD collectively reside within specific immune networks, and several directly regulate the expression of these network genes in tissues relevant to lung cancer.
Discussion
In this study we investigated the role of inflammation in lung cancer susceptibility, localized to biologically relevant immune pathways, to identify immune and inflammatory variants linked to lung cancer risk in individuals with and without COPD. Previous studies have implicated inflammation in the development of lung cancer independent of tobacco smoke exposure, and thus inflammation is thought to underlie the increased lung cancer susceptibility among those with COPD. However, prior scientific focus was limited to a narrower selection of inflammatory genes and processes 5, 27. Such studies implicated genetic variation near and within inflammatory genes in lung cancer susceptibility 28, 29, either within known candidate genes identified a posteriori utilizing gene ontology searches and customized genotyping methodology 20, 30, or by extracting inflammatory relevant variants from GWAS 31. We have complemented the efforts of previous studies, using an expanded inflammatory gene and pathway set paired with a high-density, multi-ethnic SNP array to determine which immune gene and pathway based loci confer lung cancer susceptibility in the presence and absence of COPD.
Few studies to-date assessed the role of genetic loci in lung cancer risk in individuals dual phenotyped for COPD. A study performed by Young and colleagues investigated the link between known lung cancer risk loci (n=11) and COPD in a smoking population of Caucasian New Zealanders 32. They identified differential lung cancer risk effects in two variants that differed by COPD state and reported several other loci that may act as dual risk modifiers for lung cancer and COPD. Unfortunately, only a single inflammatory locus (rs2808630, CRP) was included in the study due to the limited scope of genotyping, and we were unable to validate any COPD dependent lung cancer risk associations within the CRP gene locus, which included rs2808630. Another such study conducted by Yang et al. investigated whether known CHRNA3 lung cancer risk variants were also associated with COPD susceptibility and severity in a Chinese population of smokers, identifying a single significant association with potential mechanistic underpinnings 33. They, however, did not investigate associations between inflammatory processes, COPD, and lung cancer susceptibility.
To expand upon these efforts, as well as to determine the COPD dependency of inflammatory gene and pathway variants in lung cancer risk, we conducted lung cancer association testing on 43,953 loci proximal (±2kB) to 2069 immune-centric genes in individuals separately by COPD status. We observed significant context-independent associations in the pooled discovery/validation samples for several genes (APOB, PARK2, and PRKG1). While SNPs in PRKG1 were also validated in the context-dependent analyses, none of the 57 SNPs were in common to both strata (nor were they in LD with each other). However, we did find several correlations (D’>0.9) between SNPs that were identified in samples with COPD and those without COPD. LD was estimated using both D’ and r2 separately for white and African American samples; D’ values were used to determine strong associations due to the presence of lower frequency SNPs in the MEGA panel and the sensitivity of r2 to relative allele frequency differences between pairs of SNPs. Among white samples, three SNP pairs were in strong LD: rs2901600 in DNM3 (COPD) and exm113346 in SPTA1 (No COPD), rs10932427 in ERBB4 (COPD) and rs3749096 in EDAR (No COPD) and JHU_8.71282810 in NCOA2 (COPD) and rs73241640 in NRG1 (No COPD). No SNP pairs were in strong LD among African Americans. A lack of substantial overlapping loci/gene associations between the COPD stratums provides evidence that the genetic risk profiles for lung cancer greatly differ in individuals susceptible to COPD as opposed to individuals without COPD. As such, these findings align with prior work highlighting divergent inflammatory processes in smokers who are susceptible to COPD as opposed to smokers who are not 34; and further, the divergent inflammatory processes likely contribute to the increased risk of lung carcinogenesis in individuals with COPD 35, 36.
Next, we interrogated the functional and biological significance of the immune-centric risk candidates, separately by COPD state. Functional analyses identified 19% (11/57) of the risk significant variants as significant cis-eQTLs in tissues of primary interest: lung and whole blood leukocyte tissues. This suggests that these variants may impart risk through modulating the expression patterns of lung and immunological tissues. Risk variants with no detectable eQTL signals may still play a role in regulating immunological gene expression in cases where lung inflammation is present; GTEx style studies in populations with active lung disease will be necessary to elucidate these effects.
Immune-centric gene candidates associated with lung cancer risk also demonstrated substantially more network connectivity in individuals without COPD (20 functional interactions) as opposed to individuals with COPD (8 functional interactions), despite the fact that approximately the same number of genes were represented in each stratum (24 in COPD versus 26 in no COPD). Immune-related risk signatures in smokers without COPD may reside in a narrow biological process whereas immune-related risk signatures in smokers with COPD reside across a more broad biological process. Moreover, pathway analysis revealed several pathways with an overrepresentation of risk candidates. Two classical innate immune activation schemes were represented; DAP12 Interactions (R-HSA-2172127) and Fc-gamma receptor dependent phagocytosis (R-HSA-2029480). Pathway involvement of the candidates was distinct between individuals with and without COPD, as no pathway was enriched in both stratums. These findings provide a biological context by which these variants may be contributing to differential lung cancer risk in individuals with and without COPD.
Due to known heterogeneity of minor allele relative frequencies between whites and African Americans, it is possible that race-specific effects exist among the 57 validated COPD-dependent SNPs. Indeed, six of 29 SNPs with risk effects in COPD (rs868936562, rs61505577, rs72969686, rs73783372, rs1074822 and rs61731180) and 4 of 28 SNPs in the no COPD stratum (rs867806199, rs114240594, rs4149646 and rs78466637) were restricted to either whites or African Americans, due to very low relative allele frequencies in the other race (i.e., no valid test). These variants could prove useful in understanding race related lung cancer susceptibility patterns. Conversely, the remaining variants represent a race-independent inflammatory relationship in lung cancer susceptibility in individuals with or without COPD. This set of variants could serve as useful risk stratification loci in admixed smoking populations. Furthermore, neither the race dependent or independent variants were shared between individuals with nor without COPD suggesting that the genetic risk profiles in lung cancer differ greatly in individuals susceptible to COPD as opposed to individuals without COPD regardless of an individual’s race.
The strengths of this study lie in the recruitment of a large set of racially diverse cases and controls with a well-defined COPD phenotype as well as the targeting of variation within genes directly involved in immune functions. This has allowed us to stratify the study population by COPD status to interrogate the contribution of immune-related variation to lung cancer susceptibility. There are, however, limitations to the approach we have taken. The requirement that participants carry a valid health insurance policy may limit generalizability to those who either can afford private insurance, have employer-provided benefits or qualify for Medicaid. In the state of Michigan, uninsured cancer patients can apply for Medicaid and therefore the most appropriate control group is one that is also insured. In addition, clinically actionable findings on CT could not be ignored based on ethical grounds; thus, insured participants had a mechanism through which findings could be acted upon. Another potential weakness of this study was the choice of a relatively loose significance threshold of α=0.05 with no correction for multiple testing in the discovery phase of the analysis. Among the 43,953 SNPs, none were significant at a Bonferroni-corrected threshold (p≤1.1×10−6). However, we employed a stringent step-wise approach downstream to restrict our findings to the most consistent stratum-specific SNP effects. Likewise, the selection of the multi-ethnic genotyping array, while useful for a population containing Caucasians and African Americans, limited our ability to externally validate variant findings in prior lung cancer risk datasets, ultimately leading us to use an internal validation set.
Further research is necessary to determine the mechanistic link between risk-notable immune and inflammatory variants which differ by COPD states in at-risk populations. Moreover, the variants identified in this study will serve as a foundation to further interrogate the relationship of differential lung cancer risk profiling in individuals who have COPD as opposed to individuals who do not have COPD.
Supplementary Material
Impact.
We evaluated the impact of genetic variation in immune and inflammatory genes and pathways on lung cancer in metropolitan Detroit lung cancer cases and controls with or without COPD. We demonstrate that variation in these genes and pathways impact lung cancer risk in a COPD dependent manner.
Acknowledgments
Funding
National Institutes of Health (T32CA009531, R01CA141769, P30CA022453), Herrick Foundation
Abbreviations:
- COPD
chronic obstructive pulmonary disease
- GWAS
genome-wide association study
- SNP
single nucleotide polymorphism
- MEGA
multi-ethnic genotyping array
- GTEx
genotype-tissue expression project
- PFT
pulmonary function test
- eQTL
expression quantitative trait loci
Footnotes
Disclosure
Dr. Shirish Gadgeel reports personal fees from Genentech/Roche, Astra-Zeneca, Boehringer-Ingelheim, Pharmamar, Novocure, Xcovery, and Takeda. The authors report no other conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians 2017;67: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Alberg AJ, Samet JM. Epidemiology of Lung Cancer. Chest 2003;123: 21S–49S. [DOI] [PubMed] [Google Scholar]
- 3.Statistics NCfH. Compressed Mortality File, 1999–2015 Hyattsville, Maryland, 2016. [Google Scholar]
- 4.Hogg JC. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. The Lancet 2004;364: 709–21. [DOI] [PubMed] [Google Scholar]
- 5.Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer 2013;13: 233–45. [DOI] [PubMed] [Google Scholar]
- 6.Brody JS, Spira A. State of the Art. Chronic Obstructive Pulmonary Disease, Inflammation, and Lung Cancer. Proceedings of the American Thoracic Society 2006;3: 535–7. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz AG, Lusk CM, Wenzlaff AS, Watza D, Pandolfi S, Mantha L, Cote ML, Soubani AO, Walworth G, Wozniak A, Neslund-Dudas C, Ardisana AA, et al. Risk of Lung Cancer Associated with COPD Phenotype Based on Quantitative Image Analysis. Cancer Epidemiology Biomarkers & Prevention 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koshiol J, Rotunno M, Consonni D, Pesatori AC, De Matteis S, Goldstein AM, Chaturvedi AK, Wacholder S, Landi MT, Lubin JH, Caporaso NE. Chronic Obstructive Pulmonary Disease and Altered Risk of Lung Cancer in a Population-Based Case-Control Study. PLOS ONE 2009;4: e7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musolf MA, Simpson LC, de Andrade M, Mandal D, Gaba C, Yang P, Li Y, You M, Kupert YE, Anderson WM, Schwartz GA, Pinney MS, et al. Familial Lung Cancer: A Brief History from the Earliest Work to the Most Recent Studies. Genes 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40: 616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452: 633–7. [DOI] [PubMed] [Google Scholar]
- 12.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452: 638–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, et al. A Genome-wide Association Study of Lung Cancer Identifies a Region of Chromosome 5p15 Associated with Risk for Adenocarcinoma. The American Journal of Human Genetics 2009;85: 679–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong T, Hung RJ, Amos CI, Wu X, Bickeböller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, Dienemann H, Kaaks R, et al. Replication of Lung Cancer Susceptibility Loci at Chromosomes 15q25, 5p15, and 6p21: A Pooled Analysis From the International Lung Cancer Consortium. JNCI: Journal of the National Cancer Institute 2010;102: 959–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang IA, Holloway JW, Fong KM. Genetic susceptibility to lung cancer and co-morbidities. Journal of Thoracic Disease 2013;5: S454–S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zanetti KA, Wang Z, Aldrich M, Amos CI, Blot WJ, Bowman ED, Burdette L, Cai Q, Caporaso N, Chung CC, Gillanders EM, Haiman CA, et al. Genome-wide association study confirms lung cancer susceptibility loci on chromosomes 5p15 and 15q25 in an African-American population. Lung Cancer 2016;98: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz AG, Lusk CM, Wenzlaff AS, Watza D, Pandolfi S, Mantha L, Cote ML, Soubani AO, Walworth G, Wozniak A, Neslund-Dudas C, Ardisana AA, et al. Risk of Lung Cancer Associated with COPD Phenotype Based on Quantitative Image Analysis. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2016;25: 1341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrino R, Viegi G, Brusasco V, Crapo RO, Burgos F, Casaburi R, Coates A, van der Grinten CPM, Gustafsson P, Hankinson J, Jensen R, Johnson DC, et al. Interpretative strategies for lung function tests. European Respiratory Journal 2005;26: 948. [DOI] [PubMed] [Google Scholar]
- 19.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, et al. The Reactome pathway knowledgebase. Nucleic Acids Research 2014;42: D472–D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitz MR, Gorlov IP, Dong Q, Wu X, Chen W, Chang DW, Etzel CJ, Caporaso NE, Zhao Y, Christiani DC, Brennan P, Albanes D, et al. Multistage Analysis of Variants in the Inflammation Pathway and Lung Cancer Risk in Smokers. Cancer Epidemiology Biomarkers & Prevention 2012;21: 1213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler aD. The Human Genome Browser at UCSC. Genome Research 2002;12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koudritsky M, Domany E. Positional distribution of human transcription factor binding sites. Nucleic Acids Research 2008;36: 6795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human Mutation 2009;30: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, et al. The Genotype-Tissue Expression (GTEx) project. Nature Genetics 2013;45: 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research 2017;45: D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballaz S, Mulshine JL. The Potential Contributions of Chronic Inflammation to Lung Carcinogenesis. Clinical Lung Cancer 2003;5: 46–62. [DOI] [PubMed] [Google Scholar]
- 28.Shiels MS, Engels EA, Shi J, Landi MT, Albanes D, Chatterjee N, Chanock SJ, Caporaso NE, Chaturvedi AK. Genetic variation in innate immunity and inflammation pathways associated with lung cancer risk. Cancer 2012;118: 5630–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engels EA, Wu X, Gu J, Dong Q, Liu J, Spitz MR. Systematic Evaluation of Genetic Variants in the Inflammation Pathway and Risk of Lung Cancer. Cancer Research 2007;67: 6520. [DOI] [PubMed] [Google Scholar]
- 30.Brenner DR, Brennan P, Boffetta P, Amos CI, Spitz MR, Chen C, Goodman G, Heinrich J, Bickeböller H, Rosenberger A, Risch A, Muley T, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Human Genetics 2013;132: 579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung RJ, Ulrich CM, Goode EL, Brhane Y, Muir K, Chan AT, Marchand LL, Schildkraut J, Witte JS, Eeles R, Boffetta P, Spitz MR, et al. Cross Cancer Genomic Investigation of Inflammation Pathway for Five Common Cancers: Lung, Ovary, Prostate, Breast, and Colorectal Cancer. Journal of the National Cancer Institute 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young RP, Hopkins RJ, Whittington CF, Hay BA, Epton MJ, Gamble GD. Individual and Cumulative Effects of GWAS Susceptibility Loci in Lung Cancer: Associations after Sub-Phenotyping for COPD. PLOS ONE 2011;6: e16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Qiu F, Lu X, Huang D, Ma G, Guo Y, Hu M, Zhou Y, Pan M, Tan Y, Zhong H, Ji W, et al. Functional Polymorphisms of CHRNA3 Predict Risks of Chronic Obstructive Pulmonary Disease and Lung Cancer in Chinese. PLOS ONE 2012;7: e46071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isajevs S, Taivans I, Svirina D, Strazda G, Kopeika U. Patterns of Inflammatory Responses in Large and Small Airways in Smokers with and without Chronic Obstructive Pulmonary Disease. Respiration 2011;81: 362–71. [DOI] [PubMed] [Google Scholar]
- 35.Shen H-M, Tergaonkar V. NFkB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis 2009;14: 348–63. [DOI] [PubMed] [Google Scholar]
- 36.Birru R, Kahkonon B, Di YP. Chronic Inflammation in the Pathogenesis of COPD and Lung Cancer. Proceedings of the American Thoracic Society 2012;9: 81-. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.