Abstract
N6-methyladenosine (m6A) is identified as the most prevalent and abundant internal RNA modification, especially within eukaryotic mRNAs, which has attracted much attention in recent years since its importance for regulating gene expression and deciding cell fate. m6A modification is installed by RNA methyltransferases METTL3, METTL14 and WTAP (Writers), removed by the demethylases FTO and ALKBH5 (Erasers) and recognized by m6A binding proteins, such as YT521-B homology YTH domain-containing proteins (Readers). Accumulating evidence shows that m6A RNA methylation participates in almost all aspects of RNA processing, implying an association with important bioprocesses. In this review, we mainly summarize and discuss the functional relevance and importance of m6A modification in cellular processes.
Keywords: m6A modification, methylation, RNA, function
Introduction
To date >170 chemical modifications in RNA have been found in organisms 1. N6-methyladenosine (m6A) modification was firstly discovered in the early 1970s from cancer cells. This modification also exists in various species, including yeast, Arabidopsis, Drosophila, various viruses, zebrafish, plants, fruit flies, mice, and humans 2. In 2012, Dominissini et al. revealed the m6A distribution in human and mouse and identified more than 12000 methylated sites on human mRNAs by utilizing m6A-seq 3. m6A sites are highly conserved and generally enriched in the consensus motif RRACH (R = G or A and H = A, C, or U), which is more prone to be detected in the 3'-untranslated regions (3'UTRs), near stop codons and within internal long exons 3.
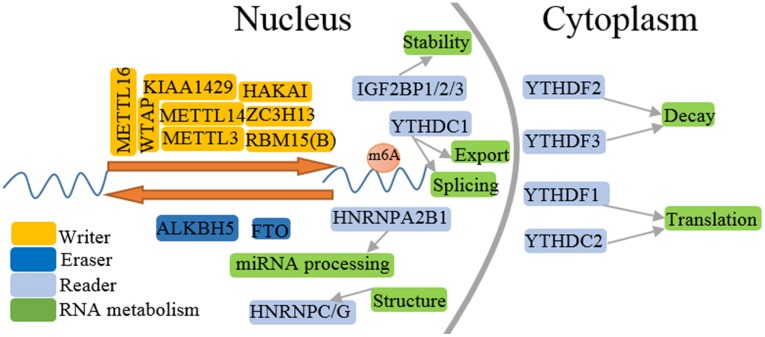
m6A methylation is introduced into RNAs by a multicomponent methyltransferase complex (m6A writers). The complex traditionally consists of methyltransferase-like 3 (METTL3), METTL14 and Wilms'tumor 1-associating protein (WTAP), which effectuates the m6A methylated group into RNAs. Subsequently, new writers, such as RBM15(B), HAKAI, METTL16, KIAA1429 (VIRMA) and ZC3H13 have been identified 1, 4. METTL3 serving as the core component and METTL14 recognizing target RNAs integrate a stable heterodimer complexes referring to interacting with other m6A cofactors to synergistically catalyze m6A methylation 5. WTAP contributes to the methyltransferase complexes anchoring in nuclear speckles. m6A methylation can be removed by RNA demethylases (m6A erasers), fat mass and obesity-associated protein (FTO) and AlkB family member 5 (ALKBH5). FTO, the first identified m6A demethylase, oxidizes m6A in RNA to N6-hydroxymethyladeosine and N6-formyladenosine 6. ALKBH5, an FTO homologue, directly abrogates m6A modification to adenosine without intermediate detected 7. Although the m6A modification is dynamically regulated by writers and erasers, the proteins (m6A readers) preferentially recognizes m6A-modified sites, influencing RNA fate and endowing distinct biological functions. m6A readers, mainly including YT521-B homology (YTH) domain family proteins (YTHDF1~3), YTH domain containing proteins (YTHDC1~2), IGF2BP1~3, HNRNPC/G/ A2B1 and eIF3, regulate RNA processing, structure, nuclear export, translation and degradation. YTH domain, as m6A binding module, shares a conserved α/β fold and can discriminate between non-modified and m6A mRNAs 8, 9. YTHDF1 can bind to the 3'UTRs and stop codon of m6A-containing RNA and promotes translation initiation by interacting with eIF3 10. Binding sites of YTHDF3 also primarily locates in 3'UTR 11. YTHDF2 associates with half-life of mRNA. YTHDC1 regulates transcription of target genes and alternative splicing of mRNA 12. HNRNPC regulates mRNA structure, while HNRNPA2/B1 involves pre-miRNA transcription 13. IGF2BPs enhances mRNA stability and storage. Reader proteins combine m6A methylation with RNA processing and biological functions.
m6A modification characterized by wide existence, unique distribution and dynamic reversibility. m6A methylation regulatory network regulates RNA processing and metabolism and participate in many cellular biological processes, such as immune modulation, fat metabolism, biological rhythm, reproductive development, and its disorders can cause various diseases. In this review, we summarized the current knowledge on the function and biology of m6A methylation.
Detection of m6A methylation
Due to technical bottlenecks, it makes the m6A methylation more mysterious and incomprehensible. m6A modification neither modulates reverse transcription nor is analogous to m7G methylation characterized by being specifically cleaved, the presence of m6A distributing and differential patterns at a particular mRNA is challenging to observe and detect 14, 15. m6A methylation has not been characterized until the availability of transcriptome-wide mapping approaches, m6A-seq and MeRIP-seq 3, 15, 16, both of which capture m6A RNA fragments through immunoprecipitation and then identify modified sequences.
Based on the theory, researchers detect an enormous amount of highly conserved m6A sites and also determine over 12,000 m6A signal peaks on 7,676 mammalian genes 3, 17. Nevertheless, the above detection analysis is insufficient to discriminate two adjacent m6A sites, and m6A mapping methods localize m6A residues to about 100~200 nucleotides, which may not accurately identify m6A sites in a whole transcript 18. Besides, both m6A-seq and MeRIP-seq may misread m6Am modification that occurs at the 3'UTR ends of mRNA and is analogous to m6A modification containing the sixth methyl group as m6A methylation. MeRIP-Seq can identify m6A-modified sites in mammalian cells, whereas the way which is complex and only separate the m6A-abundant regions. Considering the above defects, researchers have made improvements in detection techniques for m6A methylation site. Ultraviolet cross-linking immunoprecipitation technology, including miCLIP 19, PA-m6A-seq 20, m6A-CLIP (also called UV-CLIP) 14, 21, are reported to overcome the defects above, which could discriminate m6A methylation at an individual- nucleotide resolution more accurately and provide higher resolution transcriptome-wide maps of m6A methylation. Another technique, m6A-LAIC-seq, introduces spike-in RNA as an internal reference founded upon m6A-seq and then calculates the methylation level of m6A modification at each gene fragment in a full transcriptome, however, it is incapable of determining single-base resolution of m6A modification 22.
In addition to high-throughput sequencing, monitoring of the m6A modification at an exact position of mRNA is also very essential. SCARLET, as the most common strategy, accurately maps a single m6A site in both lncRNAs and mRNAs and then determine the m6A methylation content in total RNA 23, 24. Albeit SCARLET with low-throughput and fancy features, the feasibility and accuracy allow SCARLET to be a frequently-used strategy for examining the accuracy of high-throughput detection of m6A modification. And SCARLET can also be allowed to screen other types of epigenetic modifications in RNA, such as m5C and Ψ modifications 25. Golovina et al. reported a strategy based on high-resolution melting analysis for monitoring m6A sites in a specific RNA position 26. Mechanistically, m6A methylation alters the melting temperature, which is indirectly measured by qPCR 26; however, whether it can be popularized remains to be verified. The recently proposed m6A-REF-seq provides a novel perspective for the identification of m6A single site, which eliminates the requirement for m6A specific antibody and makes it more suitable for the detection of minute and precious samples 27. In recent years, new auxiliary detection technologies continue to emerge, which further improve the accuracy, sensitivity and application range of the traditional high-throughput sequencing. The improvement of detection strategies contributes to profoundly comprehend the m6A modification and its associations with various pathophysiologies.
Prediction of m6A methylation (site)
Since the identification of m6A methylation requires plenty of workforce and material resources, and it is rather dynamic and tissue-specific, bioinformatics prediction greatly improving research efficiency for the distribution and pattern of m6A methylation is developed. Zhang et al. firstly proposes the HIDDEN MARKOV MODEL to predict residual sites around known sites 28. Subsequently, pRNAm-PC and iRNA-Methyl (http://lin.uestc.edu.cn/server/iRNA-Methyl) for predicting m6A sites are developed, and the common characteristics of both are the application of support vector machine model 29, 30. According to this philosophy, Jia et al. develop RNA-methylPred method, a highly accurate predictor for identifying m6A sites, which is more efficient than before 31. Li et al. propose a better TargetM6A method, which can only predict the m6A sites in pri-miRNAs(http://csbio.njust.edu.cn/bioinf/TargetM6A) 32. Zhou et al. synthesize a variety of mathematical models and reports SRAMP predictor that is more effectively to predict m6A sites in mammalian RNAs 29. In 2018, a database website named RMBaseV2.0 was established, and it includes sequencing data of multiple epigenetic modifications of RNAs in 13 species (http://rna.sysu.edu.cn/rmbase/), including mass data on m6A methylation sites 33.
Biological functions of m6A modification
m6A functional enzymes cooperatively effectuates the dynamic balance of intracellular regulatory network m6A modification. Formidable evidence has revealed the important roles of m6A modification in various biological functions (Figure 1 and 2), such as RNA metabolism, adipogenesis (Figure 3), spermatogenesis, circadian rhythm, immune response, stem cell maintenance and tumorigenicity (Figure 4). Next, we mainly introduce several relatively well-studied roles of m6A modification in pathophysiological process.
Figure 1.
Regtory process of m6A methylation. m6A RNA methylation is installed by writers, reversed by erasers, and functionally facilitated by readers. m6A modification is involved in the life cycle of RNA metabolism including RNA splicing, structure, nuclear export, translation, degradation and miRNA processing.
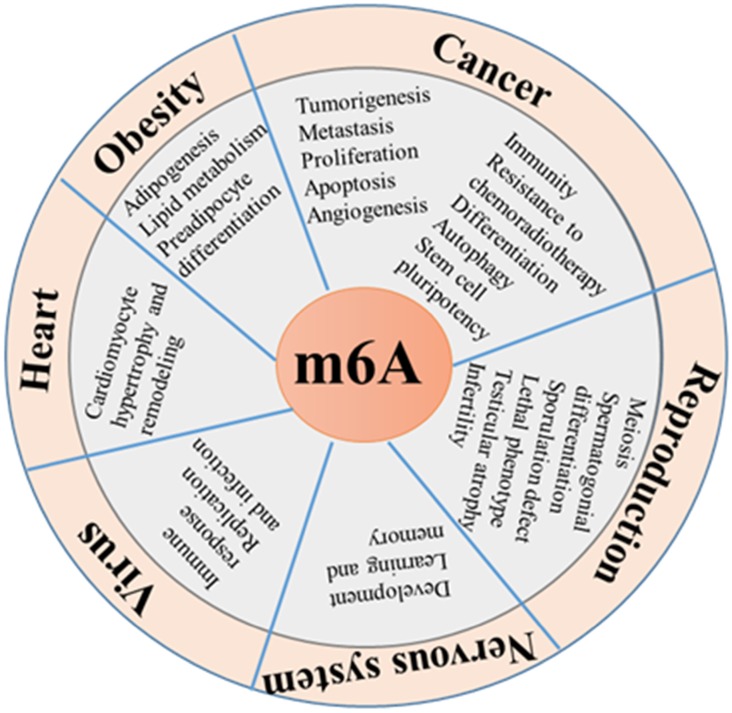
Figure 2.
Functional roles of m6A modification. m6A modification plays important role in diverse biological processes, such as tumorigenesis, fat metabolism, biological rhythm, reproductive development, immune modulation, virus replication, cardiomyocyte remodeling and stress response.
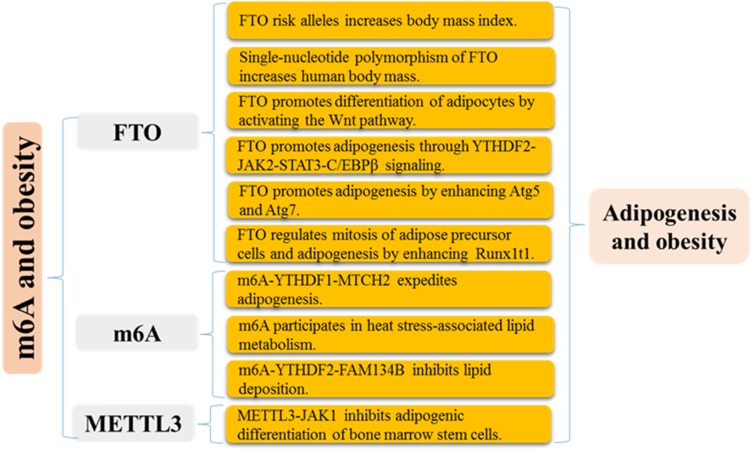
Figure 3.
Mechanisms of m6A modification in adipogenesis and obesity.
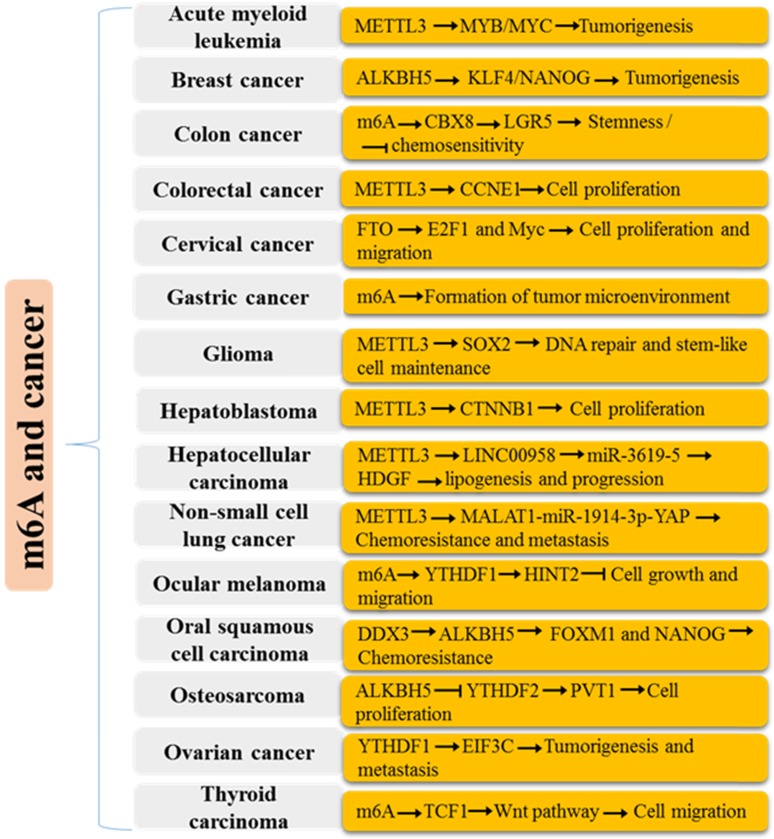
Figure 4.
Mechanisms of m6A modification in various human cancers.
m6A in mRNA metabolism
The interaction between m6A modification and its regulators reconstructs dynamic regulatory networks, which is responsible for the mRNA processing and metabolism (Figure 1), from mRNA transcription, splicing and export in the nucleus to translation, localization and degradation in the cytoplasm, and further influencing gene expression, leading to various pathophysiologic processes.
m6A and mRNA splicing
The process of pre-mRNA maturating into mRNA consists of 5' capping, 3' polyadenylation and splicing. Pre-mRNA splicing responsible for precise intron excision and exon connection is a necessary procedure for gene expression and enriches the diversity of gene products. m6A modification was initially considered as a splicing regulator, and prior research has revealed that it is more abundant in pre-mRNA than in mature mRNA 34. m6A modification is frequently enriched in the intron region of pre-mRNA 35, 36, its regulators primarily locate in nucleus speckle, a well-known location of pre-mRNA processing 37, and little methylation or demethylation occurs in cytoplasmic mRNA 38. Therefore, it can be inferred that m6A modification responsible for mRNA splicing occurs in the nucleus. Different effects of mRNA splicing accompany knockout of WTAP or METTL3, and WTAP is a known splicing factor 39. Inhibiting WTAP impairs mRNA levels and leads to abnormally alternative splicing of mRNA. METTL3 causes mis-splicing and instability of the mRNA 40. FTO silencing potentiates the splicing ability of SRSF2 and augments the number of target exons 41, 42. In addition, ALKBH5 has also been proved to affect splicing rates of mRNA 7. YTHDC1 can bind with SRSF3 and promote pre-mRNA splicing to mature mRNA 37. HNRNPC and HNRNPG are also responsible for pre-mRNA processing and mRNA maturation 43, 44. Remarkably, Ke et al. reported that m6A modification is not completely essential for the most mRNA splicing but acts as a determinant of cytoplasmic mRNA stability 38. Recently, it has been reported that m6A-assisted polyadenylation signals protect transcriptome integrity via exhibiting chimera formation in Arabidopsis thaliana RNA 45.
m6A and mRNA localization
Nuclear export of mRNA is quite an important process to connect mRNA transcription and processing in nucleus with gene expression in cytoplasm. m6A modification contributes to nuclear export of mRNA, and attenuation of METTL3 reverses m6A modification and then jeopardizes the mRNA export 46. ALKBH5 depletion accelerates the mRNA translocation from the nucleus to the cytoplasm 7, whereas ectopic expression of ALKBH5 causes nuclear retention of mRNA of antiviral transcripts, thereby inhibiting transcription of mRNA transcripts 47. YTHDC1 recognizes m6A-modified mRNA and promotes its combining with nuclear RNA export factor NXF1 and transport adaptor SRSF3 to promote nuclear export 48.
m6A and mRNA translation
METTL3 regulates mRNA translation through a different mechanism. By interacting with eIF3h, METTL3 promotes formation of densely packed polyribosomes and enhances translation of target mRNAs independently of its catalytic activity and m6A readers 48, 49. Stated differently, METTL3 localizing to the transcriptional start site frequently accompanied by CAATT-box binding protein CEBPZ of downstream genes induces m6A mRNA modification and augments translation via alleviating ribosome stalling 50. Through interacting ribosome and initiation factor, YTHDF1 binds to m6A -containing mRNAs and drives translation in an m6A-dependent manner 10, 51. On the other hand, YTHDF3 can also promote translation efficiency by combining with YTHDF1 and eIF4A3 52, indicating that YTHDF3 and YTHDF1 cooperate in the process of mRNA translation 53. Besides, IGF2BP1/2/3 can also accelerate mRNA translation 54.
m6A and mRNA degradation
Reversing m6A with the blockade of METTL3 or METTLE14 alleviates degradation efficiency of mRNA and enhances gene expression 55. ALKBH5 regulates the stability of CYR61 mRNA and elevates its expression in trophoblast, which is deciphered by the mechanism that silenced ALKBH5 enhances the half-life of CYR61 mRNA 56. YTH domain in C-terminal of YTHDF2 binds to m6A-containing mRNA, while domains in N-terminal locate the mRNA to “RNA degrader” for further degradation 57. Recent studies reported that YTHDF2 destabilizes modified mRNA mainly by two distinct pathways, YTHDF2-CCR4/NOT complex and YTHDF2-HRSP12-RNase P/MRP complex 58. And YTHDF2-HRSP12-RNase P/MRP is also scalable for m6A-modified circRNA 58. HuR binds to the U-enriched region of 3'UTR of mRNA and jeopardizes the interaction the binding of miRNA to the Ago complex, preventing mRNA degradation 59. m6A modification impairs the binding ability of HuR and destabilizes m6A-containing transcripts. Similarly, blockade of METTL3 impedes the interaction between Ago2 and target mRNA, enhancing mRNA stability 10. What is of noteworthiness is that IGF2BP-mediated m6A “Reading” recently has been reported the promotive effect on mRNA stability, and IGF2BP may combine with mRNA-stabilizing proteins, such as HuR, MATR3 and PABPC1 to elevate the mRNA stability 52. In conclusion, m6A modification can regulate gene expression through modulating mRNA stability and degradation.
m6A and mRNA structure
m6A modification can alter the mRNA structure and make it prone to combine with HNRNPC and HNRNPG. m6A methylation contributes unwind partial mRNA double-strand of MALAT1 and CDS2 to exposure single-stranded U sites, and HNRNPC can recognize specific sites to regulate gene expression 13. m6A methylation of MALAT1 hairpin strengthens accessibility of HNRNPG binding by its C-terminal low-complexity region rather than the conventional RNA binding domain 60. The effect of structural remodeling partly caused by m6A modification on the binding of mRNA to proteins is called “m6A switch”, which is ubiquitous in the transcriptome, and plays biological function by regulating the interaction between mRNAs and binding proteins 13.
m6A in adipogenesis and obesity
Early studies revealed that ectopic expression of FTO enhances food intake and fat mass in mice 61, while blockade of FTO protects against fat accumulation 62, 63. FTO has been identified as a positive and essential regulator of preadipocyte differentiation 64-66. And accumulating evidence highlights the necessity for FTO/m6A/YTHDF2 axis in adipogenesis regulation 64, 65. FTO silencing accelerates the decay of JAK2 mRNA in an m6A-YTHDF2 dependent mechanism, which restricts STAT3 phosphorylation and C/EBPβ transcription and expression, suppressing adipogenesis, and treating with specific FTO inhibitor also eliminates the accumulation of lipid droplets 67. Loss of FTO impedes adipogenesis of 3T3-L1 preadipocyte via the prolongation of cell cycle progression in which YTHDF2 recognizes and destabilizes m6A-modified CCNA2 and CDK2 mRNAs, crucial cell cycle regulator 64. Consistent with this, epigallocatechin gallate, as upstream regulators, negatively regulates FTO/ m6A/YTHDF2 axis and induces adipogenesis inhibition in 3T3-L1 preadipocyte 65. The latest research indicated that METTL3 knockdown decreases m6A mRNA modification of JAK1, rather than JAK2, and impairs YTHDF2-mediated mRNA degradation, which activates JAK1/STAT5/C/EBPβ pathway and expedites adipogenesis of bone marrow stem cells in porcine 68. ZFP217 directly activates the FTO transcription by binding to its promoter and interacts with YTHDF2 to maintain FTO-induced m6A demethylation, thereby facilitating adipogenesis of 3T3-L1 cells 66. m6A methylation expedites adipogenesis of intramuscular preadipocyte through enhancing MTCH2 expression in a YTHDF1-dependent mRNA translation 69. Besides, m6A participates in heat stress-associated lipid metabolism, and its regulators including METTL14, WTAP, FTO, and YTHDF2 and m6A levels are increased in the liver and abdominal fat of neonatal piglets following heat stress exposure 70. In summary, m6A modification and its modulators, especially FTO, functionally regulate adipogenesis (Figure 3), which provides a new understanding for pathogenesis and treatment of obesity-associated diseases from the RNA epitranscriptome modification.
m6A in embryonic development and sex determination
m6A modification can regulate the mRNA process in the initial stage of life, and its aberrant alterations will inevitably cause abnormal embryonic development. Abnormal m6A modification affects the formation of male and female gametes, or restricts the activation of zygote genes, leading to abnormal embryonic development, and influence the normal function of offspring.
METTL3 modulates the alternative splicing of spermatogenesis-related genes, which are responsible for male fertility and spermatogenesis, and deletion of which influences spermatogonial differentiation and hinders meiosis initiation in mice 40. In Saccharomyces cerevisiae, the knockdown of IME4, a homologous gene of METTL3, causes sporulation defects 71. In Arabidopsis thaliana, the inactivation of MTA, a homologous gene of METTL3, causes the formation of lethal phenotypes or the stagnation of seed development in the spherical phase 72. Analogously, in Drosophila melanogaster, IME4, is critical for the regulation of Notch signaling in follicular development 73. Furthermore, IME4 regulates meiotic DNA replication and promotes meiosis in yeast through downregulating RME1 (a transcriptional repressor of meiosis) mRNA levels in an m6A-dependent manner 74. Upon METTL3 knockdown in the immature embryonic stem cells of mice, the immature phenotype cannot be fully terminated because of the inability of m6A modification, leading to early embryonic death 75.
The expression of ALKBH5 in mouse testis is considerably higher than that in other tissues. ALKBH5 silencing in mice results in testicular atrophy, lighter weight, reduced sperm quantity and quality and also causes reproductive dysfunction, such as apoptosis and fertility decline. Drosophila melanogaster lacking m6A modification induces fly failure and fertility decline, accompanied by the deficiencies in nerve, sex determination and dose compensation 73, 76. ALKBH5 amplification in testis is indispensable for the correct splicing of long 3'UTR of mRNA in spermatocytes, which is involved in spermatogenesis and mouse reproduction 7, 77. Inhibiting ALKBH5 results in aberrant splicing and shorter transcripts of mRNAs, ultimately developing meiosis abnormity of spermatocytes and male infertility in mice 77, 78. Meclofenamic acid suppresses cell viability and blocks G1/S progression in spermatogonia of mice through accelerating the degradation of CDK2 mRNA via specifically decreasing FTO and elevating the m6A content of 3'UTR 79.
YTHDF2-mediated mRNA degradation exerts essential functions in sexual reproduction of mammalian 80, 81. YTHDF2 deletion in an embryo of zebrafish decelerates the degradation of m6A- modified maternal transcripts and impedes the activation of zygote genome, which prevents the maternal-to-zygotic transition, thereby delaying the development of zebrafish larvae 82. YTHDF2 acts as an intrinsic determinant for early zygotic development and mammalian egg quality 81. Due to the failure of regulating the transcription of genes involved in oocyte maturation, female mice with YTHDF2 knockout are deprived of fertility 81. YTHDC1 is essential for oocyte growth and maturation in female mice and spermatogonia survival in male mice, and m6A methylation can compromise the alternative splicing defects caused by YTHDC1 silencing in mouse oocytes 37. In the testis of wild male mice, YTHDC2 expression is up-regulated at the beginning of meiosis, while in the testis with YTHDC2 knockout, healthy sperm could not be produced 82.
m6A in virus replication and infection
m6A methylation not only works a crucial role in host cells; but also in diverse viral RNAs. It was initially described in IAV and adenovirus 83, 84. Subsequently, it was discovered in HSV-1, KSHV, Rous sarcoma virus, avian sarcoma virus B77, feline leukemia virus, HIV-1, Zika virus (ZIKV), simian virus 40 (SV40), HBV, HCV, flavivirus and plant virus (alfalfa mosaic and cucumber mosaic virus) 85-89. m6A modification may present an entirely opposite pattern in the replication of different viruses. Mechanism dissection revealed that m6A methylation regulates virus replication and infection, which are mainly achieved by affecting the stability of virus mRNAs or genomic RNAs. And m6A modification in host-viral interactions is indispensable for modulating virus replication and infection 87, 90, 91.
The translation of HIV-1 mRNA is dominated by cap-dependent and independent manners 92, and the m6A modification in stem-loop II region of HIV-1 Rev response element (RRE) contributes to HIV-1 Rev protein binding to RRE, thereby affecting nuclear export of HIV-1 mRNA 86. Accumulating studies have identified the specific sites of m6A and methylation peaks in HIV-1 mRNAs 86, 93-95, all of which suggests that m6A modification performs a significant role in the HIV-1 genome. Altering m6A regulators, METTL3, METTL14, FTO and ALKBH5, affect HIV-1 related protein expressions, such as GP120, p24, p55 Gag 93. p24 and Nef are down-regulated expression in CD4+ T cells infected with HIV-1 after CRISPR-Cas-mediated deletion of YTHDF2 87. HIV-1 infection is also amplified following the knockout of reader protein DF1-3 93. On the contrary, the enforced expression of these m6A readers inhibits HIV-1 replication and decrease Gag protein level. Drugs targeting m6A modification can be used as the development direction of antiviral drugs, such as 3-deazaadenosine (3-DAA), which has been shown to block the hydrolysis of S-Adenosylhomocysteine to degrade SAM, thus inhibiting the addition of m6A group to the mRNA substrate. Kennedy et al. treated HIV-1-infected CEM-SS cells with 3-DAA and found that the replication of HIV-1 is deprived, which proves the inhibitory effect of 3-DAA on HIV-1 replication 95. The 3-DAA, too, has an excellent inhibitory effect on the replication of RSV, IAV and other viruses 96, 97.
m6A writers and erasers are found to shuttle to the cytoplasm to modulate the m6A methylation of flavivirus RNAs 98, 99, and m6A modification regulates the virus-like particle production and the infections of HCV and ZIKV 98, 99. m6A methylation enriched in the KSHV genome might regulate alternative splicing of the ORF50 pre-mRNA and control governing switch of KSHV replication 85, 100. And knockdown of METTL3 abrogates the expression of KSHV lytic genes and halts virus production 85. m6A methylation accelerates SV40 replication by potentiating the translation and export of viral transcripts 101. HBV related HBs and Hbc proteins increases followed by METTL3 or METTL14 silencing; on the contrary, inhibiting FTO or ALKBH5 enforces their expression 87, 102. m6A methylation participates in the regulation of HBV life cycle via affecting transcript stability 87, 102. The lower m6A modification makes some viruses successfully evade the immune surveillance, so whether there is some mechanism that can detect these abnormal methylations and then feedback to the immune system for treatment, which needs further study.
m6A in stress response
Following the heat shock, METTL3 locates in chromatin of the heat shock gene and catalyze m6A methylation. Nuclear YTHDF2 competes with FTO to prevent the reversal of m6A modification, thereby enhancing mRNA translation in a cap-dependent manner and improving the formation of related proteins 103. YTHDF1 is responsible for confining stress particles induced by protein-encapsulated mRNAs and protein particles to the nucleus, somehow abrogating protein release and mRNA expression 103. When the stress source disappears, the stress particles are re-depolymerized and released, and then encapsulated mRNAs re-express 103, which contributes to avoid the inflammation occurrence induced by stress sources whenever possible and is feasible and beneficial for injury repair. Similarly, due to be subjected to UV damage to DNA, METTL3-catalyzed m6A is quickly located at the injury site, and DNA polymerase κ is also recruited to ameliorate injury repair 104. Recent research has demonstrated that multivalent m6A motifs enhances phase separation of YTHDF1/2/3 proteins through juxtaposing low-complexity domain of YTHDFs, which causes the presence of phase-separated compartments, such as P-body, neuronal RNA and stress granules 105, 106.
m6A in immune response
By regulating the IL-7/STAT5/SOCS pathway, m6A methylation affects T cell state, while the absence of METTL3 can destroy the homeostasis and differentiation of T cells 107. Blockade of METTL3 in immature T cells augments the expression of SOCS1, SOCS3 and CISH, which inhibit the activation STAT5 mediated by IL-7 and the proliferation and differentiation of T cells 107. Due to the loss of differentiation ability in METTL3-deficient T cells, it is insufficient to induce autoimmune diseases 107. After virus infection, DDX46 recruits ALKBH5 to erase m6A modification of antiviral transcripts, resulting in the inability of antiviral transcripts translating into IFN-I and the failure of initiating natural antiviral immune responses 47. m6A modification also participates in tumor antigen- specific immune response through modulating the translation efficiency of lysosome cathepsin in dendritic cells 108. Knockout of YTHDF1 in mice can enhance the immune response of CD8+ T cells 108. These findings are expected to provide innovative ideas for the treatment of autoimmune diseases and understanding the molecular mechanism of innate immunity in various diseases, especially virus infection 109.
m6A in differentiation of stem cells
m6A methylation is involved in regulating the fate-determining process of stem cells 75, 110. m6A modification maintains the balance of gene expression between endothelial cells and hematopoietic cells during endothelial-hematopoietic transformation by regulating Notch1a mRNA stability via YTHDF2, which then affects the fate of hematopoietic stem cells 111. Reversing m6A modification treated with METTL3 or METTL14 blockade contributes to the amplification of transcription factors maintaining cell pluripotency and inhibits the self-renewal of embryonic stem cells 112, 113. METTL3 knockout interferes with differentiation and self-renewal of mouse embryonic stem cells via decreasing m6A methylation of NANOG, leading to embryonic death 75. However, ZC3H13 depletion impairs self-renewal of mouse embryonic stem cells due to the cells cultured in metastable naive condition 114. METTL3 depletion potentiates both calcium deposition and alkaline phosphatase activity of mesenchymal stem cells by modulating the expression of MYD required for activating NF-κB, as a repressor of osteogenesis, which uncovers the inhibitory role of METTL3 in osteogenic differentiation 115. ALKBH5 deteriorates the m6A modification of NANOG mRNA to promote its expression in breast cancer stem cells following exposure to hypoxia, which furtherly accelerates cell proliferation and regulates cell differentiation 116. It has further been proved that FTO stimulates the transformation of normal hematopoietic stem cells to pathological cells in leukemia 117, and can influence the selective splicing of key genes related adipogenesis, thereby affecting the preadipocyte differentiation 41. In conclusion, m6A methylation plays a key role in regulating the differentiation and development of stem cells.
m6A in nervous system
Brain is one of the organs with the most abundant of m6A modification 118, 119. m6A regulatory proteins are implicated in the cerebral cortex and synaptic function, axonal regeneration, self-renewal of neural stem cells and cerebellar development 118-122. m6A methylation effectuates the region-specific gene regulation in the mouse brain 118. Knockdown of METTL14 in the central nervous system of mice affect the development of cerebral cortex 120, METTL3 silencing seriously affects the development of cerebral cortex and cerebellum, leading to obvious motor dysfunction, even death during lactation 123. Mechanically, m6A modification modulates the stability of transcripts associated with cerebellar development and affects pre-mRNA splicing in synapse 123.
Subsequent studies showed that m6A enrichment in the prefrontal lobe of mice contributes to learning training, implying the underlying mechanisms of m6A methylation in spatial learning and memory 121. Suckling mouse with METTL3 knockout in the cortex and hippocampus displays a significant decline in learning ability 124. In YTHDF1 knockout mice, hippocampus-dependent learning and memory dysfunction occurs, and the function and plasticity of excitatory synapses in hippocampal neurons are also impaired; on the contrary, forced expression of YTHDF1 ameliorates the injury 125. The crucial role of m6A modification lays a foundation for further exploring the pathophysiology of memory formation.
m6A in cancer
Recently, emerging evidence has revealed that there are disorders of m6A components and abnormal modification of m6A in many types of cancers (Figure 4), which leads to the inactivation or overexpression of downstream carcinogenic or tumor suppressor genes and plays an important role in oncogenic transformation and tumor progression. It is worth noting that the same components in different types of tumors, or different components in the same tumor do not play the same role, or even the opposite.
m6A modification of mRNAs is significantly up-regulated during epithelial-mesenchymal transition, and it can regulate the genes related to invasion, metastasis and cell adhesion 126. Through database analysis, it is found that FTO can regulate EGFR-related pathways and angiogenic signals, and overexpression of FTO inhibits tumor growth in vivo 127. In tumor cells, METTL14 and ALKBH5 may play a cancer-promoting role by regulating the expression of angiogenesis-related genes such as TGF-β 128. METTL3 or METTL14 depletion could induce the expression of ADAM19, EPHA3 and KLF4 mRNAs and promote the growth, self-renewal and tumorigenesis of glioblastoma stem cells 128. Hypoxia stimulation-induced ALKBH5 abrogates the m6A methylation of NANOG mRNAs and enhances the mRNA stability, which in turn promotes the maintenance and metastasis of breast tumor stem cells 116. In breast cancer, targeted inhibition of FTO can reduce the production of lactic acid and ATP and inhibit the activities of pyruvate kinase and hexokinase, and lead to the inactivation of AKT, overexpression of FTO can promote the glycolysis ability of tumor cells and the activation of PI3K/AKT pathway 129. m6A modification activates Wnt/β-catenin pathway by stabilizing FZD10, which promotes BRAC deficiency and induce PARP inhibitor resistance of epithelial ovarian cancer cells 130. Blockade of METTL3 can significantly increase the sensitivity of pancreatic cancer cells to anticancer drugs 5-fluorouracil, cisplatin and radiotherapy 51. In addition, R-2-hydroxyglutarate (R-2HG) can obviate FTO activity, which augments the m6A modification in R-2HG-sensitive leukemic cells, and then reduce the stability of MYC/CEBPA transcripts, promoting cell cycle arrest and apoptosis 131. m6A modification is implicated in various aspects of multiple malignancies via regulating differentiation, proliferation, apoptosis, invasion, migration, angiogenesis, energy metabolism, autophagy and chemoradiotherapy resistance. Uncovering the underlying mechanism of m6A modification in initiation and progression of cancer extends opportunities for molecular pathological diagnosis and therapy strategies in cancer.
m6A in heart
Enhanced METTL3 induce cardiomyocyte hypertrophy without additional stimulation, and inhibition of METTL3 can effectively block the occurrence of hypertrophy in vitro 132, 133. m6A methylation can expedite adaptive growth of cardiomyocytes and induce spontaneous cardiomyocyte remodeling 132, 133. Studies have found that FTO is associated with various heart defects, including hypertrophic cardiomyopathy and interventricular defects, septal and atrioventricular defects, arrhythmias and coronary heart disease 134. Knockdown of FTO can cause the disorder of calcium homeostasis and sarcomere dynamics in heart failure and hypoxic cardiomyocytes and decrease the contractile function of cardiomyocytes 135.
Conclusions
The milestone in the study of RNA epigenetics was discovered in 2011 by He et al, who discovered the first m6A demethylase FTO, which reveals that m6A modification is dynamic and reversible, and enriches the post-transcriptional modification function of RNA and generates refueled passion in m6A research. In this review, m6A enzymes and its regulation on RNA processing and metabolism, various biological processes as well as in pathogenesis have been revealed. m6A modification can functionally regulate the transcriptome of eukaryotes, such as mRNA splicing, nucleation, localization, translation and stabilization and play crucial roles in various biological processes, such as stem cell differentiation, T cell homeostasis, brain development, biological rhythm, spermatogenesis, as well as the occurrence of a variety of diseases, including tumor, obesity and infertility. Therefore, it is particularly important to study the dynamic modification of m6A and the specific molecular mechanism of how it affects the biological function of cells.
However, the other components of m6A methyltransferase complex are not clear, the tissue-specific demethylase remains to be explored, and the binding proteins mediating distinct functions also need to be further discovered. The single-base, high-resolution and high-throughput sequencing technology for detecting m6A methylation also needs to be improved. Regulatory mechanisms and detailed biological functions of m6A modification need to be further investigated.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81872080, 81572349), Jiangsu Provincial Medical Talent (ZDRCA 2016055), the Science and Technology Department of Jiangsu Province (BK20181148), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the 333 high-level talents of Jiangsu Province (BRA2019083).
Abbreviations
- ALKBH5
AlkB homolog 5
- METTL3
methyltransferase-like protein 3
- m6A
N6-methyladenosine
- FTO
fat-mass and obesity-associated protein
- RRE
Rev response element
- R-2HG
R-2-hydroxyglutarate
- WTAP
Wilms Tumor 1 Associated Protein
- YTH
YT521-B homology
- 3-DAA
3-deazaadenosine
- 3' UTRs
3' untranslated regions
References
- 1.Huang H, Weng H, Chen J. The Biogenesis and Precise Control of RNA m(6)A Methylation. Trends in genetics: TIG. 2020;36:44–52. doi: 10.1016/j.tig.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J, Yin P. Structural Insights into N(6)-methyladenosine (m(6)A) Modification in the Transcriptome. Genomics, proteomics & bioinformatics. 2018;16:85–98. doi: 10.1016/j.gpb.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019. 363. [DOI] [PubMed]
- 5.Fitzsimmons CM, Batista PJ. It's complicated m(6)A-dependent regulation of gene expression in cancer. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862:382–93. doi: 10.1016/j.bbagrm.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature communications. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao S, Sun H, Xu C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics, proteomics & bioinformatics. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balacco DL, Soller M. The m(6)A Writer: Rise of a Machine for Growing Tasks. Biochemistry. 2019;58:363–78. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Zhan G, Chang KJ, Yang YP, Wang L, Lin J. et al. The roles of m6A RNA modifiers in human cancer. Journal of the Chinese Medical Association: JCMA. 2020;83:221–6. doi: 10.1097/JCMA.0000000000000251. [DOI] [PubMed] [Google Scholar]
- 12.Roundtree IA, He C. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends in genetics: TIG. 2016;32:320–1. doi: 10.1016/j.tig.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PJ, He C. High-Resolution Mapping of N (6)-Methyladenosine Using m(6)A Crosslinking Immunoprecipitation Sequencing (m(6)A-CLIP-Seq) Methods in molecular biology. 2019;1870:69–79. doi: 10.1007/978-1-4939-8808-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PJ, He C. Identifying the m(6)A Methylome by Affinity Purification and Sequencing. Methods in molecular biology. 2018;1649:49–57. doi: 10.1007/978-1-4939-7213-5_3. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Tang K, Zhang D, Wan Y, Wen Y, Lu Q. et al. High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PloS one. 2017;12:e0185612. doi: 10.1371/journal.pone.0185612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grozhik AV, Linder B, Olarerin-George AO, Jaffrey SR. Mapping m(6)A at Individual-Nucleotide Resolution Using Crosslinking and Immunoprecipitation (miCLIP) Methods in molecular biology. 2017;1562:55–78. doi: 10.1007/978-1-4939-6807-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nature methods. 2015;12:767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N. et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angewandte Chemie. 2015;54:1587–90. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A. et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes & development. 2015;29:2037–53. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N. et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nature methods. 2016;13:692–8. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu N, Pan T. Probing RNA Modification Status at Single-Nucleotide Resolution in Total RNA. Methods in enzymology. 2015;560:149–59. doi: 10.1016/bs.mie.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. Rna. 2013;19:1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rau K, Rosner L, Rentmeister A. Sequence-specific m(6)A demethylation in RNA by FTO fused to RCas9. Rna. 2019;25:1311–23. doi: 10.1261/rna.070706.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golovina AY, Dzama MM, Petriukov KS, Zatsepin TS, Sergiev PV, Bogdanov AA. et al. Method for site-specific detection of m6A nucleoside presence in RNA based on high-resolution melting (HRM) analysis. Nucleic acids research. 2014;42:e27. doi: 10.1093/nar/gkt1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z. et al. Single-base mapping of m(6)A by an antibody-independent method. Science advances. 2019;5:eaax0250. doi: 10.1126/sciadv.aax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang YC, Zhang SW, Liu L, Liu H, Zhang L, Cui X. et al. Spatially Enhanced Differential RNA Methylation Analysis from Affinity-Based Sequencing Data with Hidden Markov Model. BioMed research international. 2015;2015:852070. doi: 10.1155/2015/852070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic acids research. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen W, Feng P, Ding H, Lin H, Chou KC. iRNA-Methyl: Identifying N(6)-methyladenosine sites using pseudo nucleotide composition. Analytical biochemistry. 2015;490:26–33. doi: 10.1016/j.ab.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Jia CZ, Zhang JJ, Gu WZ. RNA-MethylPred: A high-accuracy predictor to identify N6-methyladenosine in RNA. Analytical biochemistry. 2016;510:72–5. doi: 10.1016/j.ab.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Li GQ, Liu Z, Shen HB, Yu DJ. TargetM6A: Identifying N(6)-Methyladenosine Sites From RNA Sequences via Position-Specific Nucleotide Propensities and a Support Vector Machine. IEEE transactions on nanobioscience. 2016;15:674–82. doi: 10.1109/TNB.2016.2599115. [DOI] [PubMed] [Google Scholar]
- 33.Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL. et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic acids research. 2018;46:D327–D34. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976;7:227–37. doi: 10.1016/0092-8674(76)90022-2. [DOI] [PubMed] [Google Scholar]
- 35.Carroll SM, Narayan P, Rottman FM. N6-methyladenosine residues in an intron-specific region of prolactin pre-mRNA. Molecular and cellular biology. 1990;10:4456–65. doi: 10.1128/mcb.10.9.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoltzfus CM, Dane RW. Accumulation of spliced avian retrovirus mRNA is inhibited in S-adenosylmethionine-depleted chicken embryo fibroblasts. Journal of virology. 1982;42:918–31. doi: 10.1128/jvi.42.3.918-931.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS genetics. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S. et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes & development. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF. et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell research. 2017;27:1100–14. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research. 2014;24:1403–19. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnik BC. Milk: an epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. Journal of translational medicine. 2015;13:385. doi: 10.1186/s12967-015-0746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nettersheim D, Berger D, Jostes S, Kristiansen G, Lochnit G, Schorle H. N6-Methyladenosine detected in RNA of testicular germ cell tumors is controlled by METTL3, ALKBH5, YTHDC1/F1/F2, and HNRNPC as writers, erasers, and readers. Andrology. 2019;7:498–506. doi: 10.1111/andr.12612. [DOI] [PubMed] [Google Scholar]
- 44.Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN. et al. Regulation of Co-transcriptional Pre-mRNA Splicing by m(6)A through the Low-Complexity Protein hnRNPG. Molecular cell. 2019;76(e9):70–81. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pontier D, Picart C, El Baidouri M, Roudier F, Xu T, Lahmy S, The m(6)A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life science alliance. 2019. 2. [DOI] [PMC free article] [PubMed]
- 46.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 47.Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nature immunology. 2017;18:1094–103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- 48.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Molecular cell. 2016;62:335–45. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–31. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T. et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. International journal of oncology. 2018;52:621–9. doi: 10.3892/ijo.2017.4219. [DOI] [PubMed] [Google Scholar]
- 52.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell research. 2017;27:315–28. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y. et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell research. 2017;27:444–7. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2019. [DOI] [PMC free article] [PubMed]
- 55.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP. et al. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541:371–5. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. 2019;9:3853–65. doi: 10.7150/thno.31868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y, Choe J, Park OH, Kim YK. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends in genetics: TIG. 2020;36:177–88. doi: 10.1016/j.tig.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic acids research. 2012;40:5088–100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic acids research. 2017;45:6051–63. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nature genetics. 2010;42:1086–92. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMurray F, Church CD, Larder R, Nicholson G, Wells S, Teboul L. et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS genetics. 2013;9:e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Bruning JC. et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–8. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 64.Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q. et al. FTO regulates adipogenesis by controlling cell cycle progression via m(6)A-YTHDF2 dependent mechanism. Biochimica et biophysica acta Molecular and cell biology of lipids. 2018;1863:1323–30. doi: 10.1016/j.bbalip.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 65.Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y. et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m(6)A-YTHDF2-dependent manner. International journal of obesity. 2018;42:1378–88. doi: 10.1038/s41366-018-0082-5. [DOI] [PubMed] [Google Scholar]
- 66.Song T, Yang Y, Wei H, Xie X, Lu J, Zeng Q. et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic acids research. 2019;47:6130–44. doi: 10.1093/nar/gkz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu R, Guo G, Bi Z, Liu Y, Zhao Y, Chen N. et al. m(6)A methylation modulates adipogenesis through JAK2-STAT3-C/EBPbeta signaling. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862:796–806. doi: 10.1016/j.bbagrm.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 68.Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q. et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPbeta pathway via an m(6)A-YTHDF2-dependent manner. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:7529–44. doi: 10.1096/fj.201802644R. [DOI] [PubMed] [Google Scholar]
- 69.Jiang Q, Sun B, Liu Q, Cai M, Wu R, Wang F. et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m(6)A-YTHDF1-dependent mechanism. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2019;33:2971–81. doi: 10.1096/fj.201801393RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heng J, Tian M, Zhang W, Chen F, Guan W, Zhang S. Maternal heat stress regulates the early fat deposition partly through modification of m(6)A RNA methylation in neonatal piglets. Cell stress & chaperones. 2019;24:635–45. doi: 10.1007/s12192-019-01002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic acids research. 2010;38:5327–35. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant cell. 2008;20:1278–88. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14855–60. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bushkin GG, Pincus D, Morgan JT, Richardson K, Lewis C, Chan SH. et al. m(6)A modification of a 3' UTR site reduces RME1 mRNA levels to promote meiosis. Nature communications. 2019;10:3414. doi: 10.1038/s41467-019-11232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–6. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 76.Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG. et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–4. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 77.Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E325–E33. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tong J, Flavell RA, Li HB. RNA m(6)A modification and its function in diseases. Frontiers of medicine. 2018;12:481–9. doi: 10.1007/s11684-018-0654-8. [DOI] [PubMed] [Google Scholar]
- 79.Huang T, Guo J, Lv Y, Zheng Y, Feng T, Gao Q. et al. Meclofenamic acid represses spermatogonial proliferation through modulating m(6)A RNA modification. Journal of animal science and biotechnology. 2019;10:63. doi: 10.1186/s40104-019-0361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao BS, He C. "Gamete On" for m(6)A: YTHDF2 Exerts Essential Functions in Female Fertility. Molecular cell. 2017;67:903–5. doi: 10.1016/j.molcel.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN. et al. The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Molecular cell. 2017;67(e4):1059–67. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A. et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–8. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. Journal of virology. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moss B, Koczot F. Sequence of methylated nucleotides at the 5'-terminus of adenovirus-specific RNA. Journal of virology. 1976;17:385–92. doi: 10.1128/jvi.17.2.385-392.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye F. RNA N(6)-adenosine methylation (m(6)A) steers epitranscriptomic control of herpesvirus replication. Inflammation and cell signaling. 2017. 4. [PMC free article] [PubMed]
- 86.Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y. et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nature microbiology. 2016;1:16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manners O, Baquero-Perez B, Whitehouse A. m(6)A: Widespread regulatory control in virus replication. Biochimica et biophysica acta Gene regulatory mechanisms. 2019;1862:370–81. doi: 10.1016/j.bbagrm.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arribas-Hernandez L, Brodersen P. Occurrence and Functions of m(6)A and Other Covalent Modifications in Plant mRNA. Plant physiology. 2020;182:79–96. doi: 10.1104/pp.19.01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m(6)A RNA reader essential for replication of Kaposi's sarcoma-associated herpesvirus. eLife. 2019. 8. [DOI] [PMC free article] [PubMed]
- 90.Brocard M, Ruggieri A, Locker N. m6A RNA methylation, a new hallmark in virus-host interactions. The Journal of general virology. 2017;98:2207–14. doi: 10.1099/jgv.0.000910. [DOI] [PubMed] [Google Scholar]
- 91.Fleming AM, Nguyen NLB, Burrows CJ. Colocalization of m(6)A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N (6)-Methylation. ACS central science. 2019;5:218–28. doi: 10.1021/acscentsci.8b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blissenbach M, Grewe B, Hoffmann B, Brandt S, Uberla K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. Journal of virology. 2010;84:6598–604. doi: 10.1128/JVI.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016. 5. [DOI] [PMC free article] [PubMed]
- 94.Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M. et al. N(6)-Methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. The Journal of biological chemistry. 2018;293:12992–3005. doi: 10.1074/jbc.RA118.004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB. et al. Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression. Cell host & microbe. 2016;19:675–85. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bader JP, Brown NR, Chiang PK, Cantoni GL. 3-Deazaadenosine, an inhibitor of adenosylhomocysteine hydrolase, inhibits reproduction of Rous sarcoma virus and transformation of chick embryo cells. Virology. 1978;89:494–505. doi: 10.1016/0042-6822(78)90191-5. [DOI] [PubMed] [Google Scholar]
- 97.Chiang PK. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacology & therapeutics. 1998;77:115–34. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- 98.Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C. et al. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell host & microbe. 2016;20:666–73. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA. et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell host & microbe. 2016;20:654–65. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J. et al. Viral and cellular N(6)-methyladenosine and N(6),2'-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nature microbiology. 2018;3:108–20. doi: 10.1038/s41564-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6641–6. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY. et al. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:8829–34. doi: 10.1073/pnas.1808319115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W. et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–6. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF. et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424–8. doi: 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC. et al. Multivalent m(6)A motifs promote phase separation of YTHDF proteins. Cell research. 2019;29:767–9. doi: 10.1038/s41422-019-0210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J. et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–42. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R. et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–4. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang C, Fu J, Zhou Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Frontiers in immunology. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell. 2014;15:707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D. et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–6. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 112.Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell research. 2017;27:1216–30. doi: 10.1038/cr.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Molecular cell. 2018;69(e6):1028–38. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu J, Shen L, Liu Y, Ming H, Zhu X, Chu M. et al. The m6A methyltransferase METTL3 cooperates with demethylase ALKBH5 to regulate osteogenic differentiation through NF-kappaB signaling. Molecular and cellular biochemistry. 2020;463:203–10. doi: 10.1007/s11010-019-03641-5. [DOI] [PubMed] [Google Scholar]
- 116.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Z, Weng H, Su R, Weng X, Zuo Z, Li C. et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang M, Lv H, Zhang W, Ma C, He X, Zhao S, Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open biology. 2017. 7. [DOI] [PMC free article] [PubMed]
- 119.Livneh I, Moshitch-Moshkovitz S, Amariglio N, Rechavi G, Dominissini D. The m(6)A epitranscriptome: transcriptome plasticity in brain development and function. Nature reviews Neuroscience. 2020;21:36–51. doi: 10.1038/s41583-019-0244-z. [DOI] [PubMed] [Google Scholar]
- 120.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D. et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell. 2017;171(e17):877–89. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Widagdo J, Zhao QY, Kempen MJ, Tan MC, Ratnu VS, Wei W. et al. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2016;36:6771–7. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J, Yang X, Qi Z, Sang Y, Liu Y, Xu B. et al. The role of mRNA m(6)A methylation in the nervous system. Cell & bioscience. 2019;9:66. doi: 10.1186/s13578-019-0330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang CX, Cui GS, Liu X, Xu K, Wang M, Zhang XX. et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS biology. 2018;16:e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Z, Wang M, Xie D, Huang Z, Zhang L, Yang Y. et al. METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell research. 2018;28:1050–61. doi: 10.1038/s41422-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z. et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–53. doi: 10.1038/s41586-018-0666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J. et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nature communications. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 127.Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao J. et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Frontiers in oncology. 2019;9:369. doi: 10.3389/fonc.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Science advances. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Y, Wang R, Zhang L, Li J, Lou K, Shi B. The lipid metabolism gene FTO influences breast cancer cell energy metabolism via the PI3K/AKT signaling pathway. Oncology letters. 2017;13:4685–90. doi: 10.3892/ol.2017.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S. et al. N(6)-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer research. 2019;79:2812–20. doi: 10.1158/0008-5472.CAN-18-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172(e23):90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kehat I, Davis J, Tiburcy M, Accornero F, Saba-El-Leil MK, Maillet M. et al. Extracellular signal-regulated kinases 1 and 2 regulate the balance between eccentric and concentric cardiac growth. Circulation research. 2011;108:176–83. doi: 10.1161/CIRCRESAHA.110.231514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M. et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419–37. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gustavsson J, Mehlig K, Leander K, Lissner L, Bjorck L, Rosengren A. et al. FTO genotype, physical activity, and coronary heart disease risk in Swedish men and women. Circulation Cardiovascular genetics. 2014;7:171–7. doi: 10.1161/CIRCGENETICS.111.000007. [DOI] [PubMed] [Google Scholar]
- 135.Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N. et al. FTO-Dependent N(6)-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation. 2019;139:518–32. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]