Abstract
The study of stem cells in cnidarians has a history spanning hundreds of years, but it has primarily focused on the hydrozoan genus Hydra. While Hydra has a number of self-renewing cell types that act much like stem cells—in particular the interstitial cell line—finding cellular homologues outside of the Hydrozoa has been complicated by the morphological simplicity of stem cells and inconclusive gene expression data. In non-hydrozoan cnidarians, an enigmatic cell type known as the amoebocyte might play a similar role to interstitial cells, but there is little evidence that I-cells and amoebocytes are homologous. Instead, self-renewal and transdifferentiation of epithelial cells was probably more important to ancestral cnidarian development than any undifferentiated cell lineage, and only later in evolution did one or more cell types come under the regulation of a “stem” cell line. Ultimately, this hypothesis and competing ones will need to be tested by expanding genetic and developmental studies on a variety of cnidarian model systems.
Keywords: Cnidaria, Stem cells, Interstitial cell, Amoebocyte, Hydra, Aurelia
One could interpret [my observations] as meaning that certain interstitial cells of the ectoderm are the stem cells of the germline, and that they started to differentiate shortly before they penetrated into the endoderm. Once in the endoderm, they began to multiply by division, and then each produced one of the described clusters of germ cells … The more I learn about the hydroid organism, the more unlikely it seems to me that specifically differentiated cells, such as the endoderm cells, directly transform into germ cells.
- August Weismann (1883) pg. 46 (trans. by D.A.G)
In his book Die Entstehung Der Sexualzellen Bei Den Hydromedusen (The Origin of the Sex Cells in the Hydromedusa) August Weismann not only provides the background to his famous germ plasm theory, but also produces one of the first writings regarding the nature of the stammzellen, or stem cells. Over the last few decades, both stem cells and the “hydroids” Weismann studied—now termed hydrozoans—have been the focus of resurgent biological inquiry.
Hydrozoans are part of the phylum Cnidaria, along with the anthozoans (sea pens, corals, and sea anemones), scyphozoans (“true” jellyfish), and cubozoans (box jellies) (Fig. 1a). Together, the cnidarians are thought to represent some of the simplest living animals; they are classically considered to have “tissue-grade” complexity (lacking true organs like most animals), with radial (as opposed to bilateral) symmetry, and two embryological germ layers (as opposed to the typical three) (Brusca and Brusca 2003). Interest in cnidarians as models of developmental processes has grown substantially in recent years, partially because of this morphological simplicity, but also because of their ontogenic plasticity, remarkable regenerative capabilities, and the unique position they hold in the evolutionary tree (reviewed in Technau and Steele 2011). The majority of phylogenetic studies have placed the cnidarians as the major sister clade to the Bilateria (protostomes + deuterostomes), which encompasses 99 % of living animal species (Glenner et al. 2004; Dunn et al. 2008; Wheeler et al. 2009; Philippe et al. 2009; Pick et al. 2010; Erwin et al. 2011). This suggests that, as their simplicity indicates, cnidarians can provide valuable insight into the origins of animal complexity.
Fig. 1.
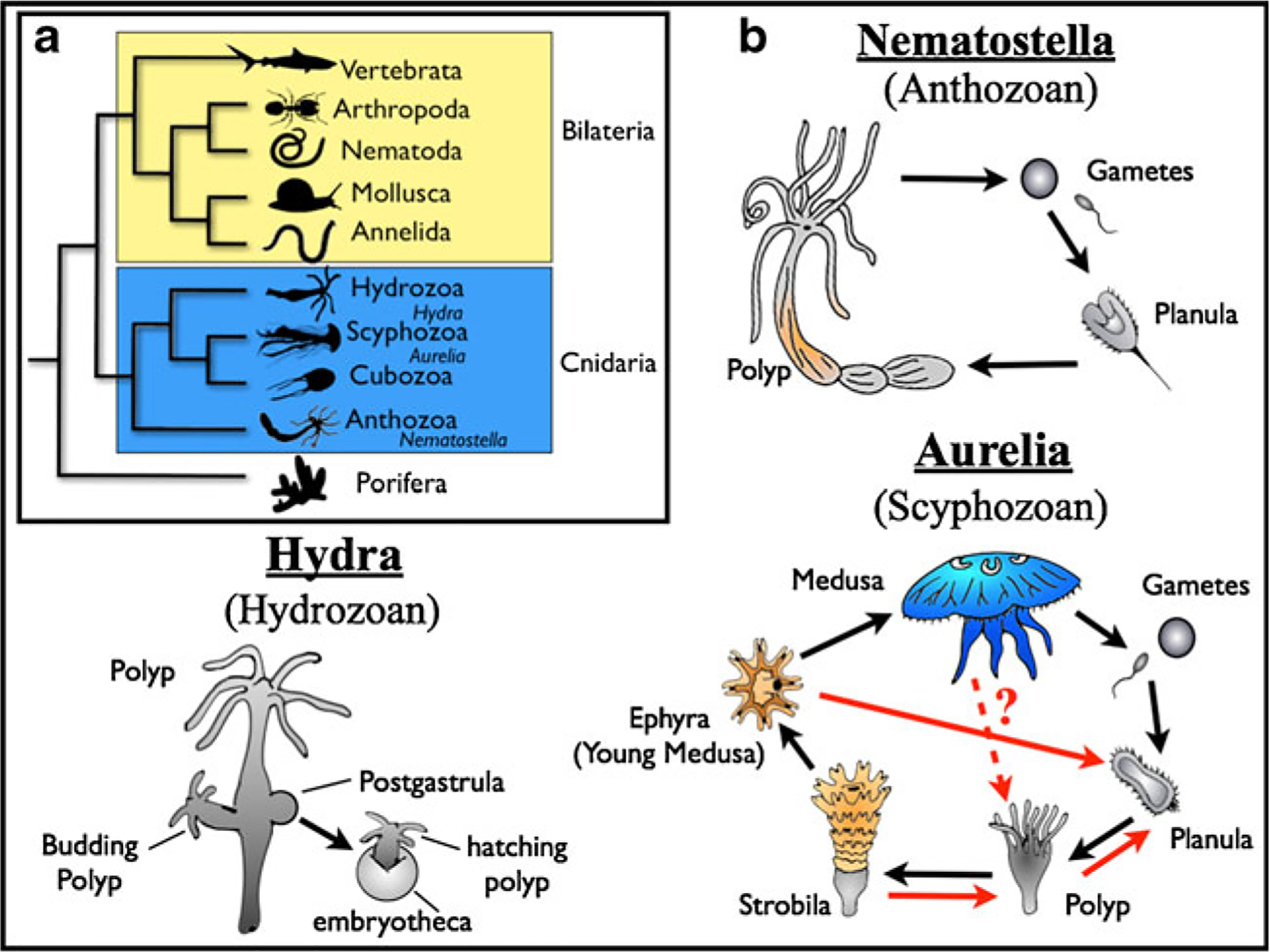
a A simple animal phylogeny, with special attention to the classes of cnidarians, based on Collins et al. (2006), Dunn et al. (2008), Philippe et al. (2009), and Pick et al. (2010). b Life cycles of several cnidarian genera. For Aurelia, derivations from the “canonical” life cycle are highlighted in red (based on Vagelli (2007) and personal observation)
However, as cnidarians have been subjected to molecular-level inquiry, study after study has found substantial cryptic complexity, challenging most of the “simple” characteristics of cnidarians mentioned previously (Kussarow et al. 2005; Lee et al. 2006; Jacobs et al. 2007, 2010; Ryan and Baxevanis 2007; Putnam et al. 2007; Matus et al. 2008). Even without these molecular insights, scientists have long recognized that the cnidarians evolved a dizzying array of diversity despite their “simple” morphology: from the clonal hydroids that build the superorganism, the Portuguese Man o’ War (Physalia physalis), to box jellies guided by complex eyes (Tripedalia cystophora), from the 2-mm-long cnidarian “worm” (Buddenbrockia plumatellae) to the 37-m-long lion’s mane jellyfish (Cyanea capillata), not to mention the corals of the great barrier reef that span over 26,000 km. Cnidarians are present in the Cambrian fossil record (Hou et al. 2005; Cartwright et al. 2007), and molecular clock analyses suggests that the origin of crown-group cnidarians—which are as genetically divergent as protostomes and deuterostomes (Erwin 2008)—might extend back 700 million years ago (Erwin et al. 2011). Thus, no single clade of cnidarians (much less any single species) can represent the cnidarians as a whole.
However, when it comes to the study of cnidarian stem cells, most research has focused on a single genus, the hydrozoan Hydra. While this research has generated significant knowledge regarding the role of stem cells in Hydra development and regeneration, fundamental questions relating to the cnidarians as a whole have yet to be clarified. Are there common principles to cnidarian stem cells? What is the relationship between cnidarian stem cells and the stem cells of other animals? A growing group of researchers are working to address these questions, but drawing broad conclusions has remained elusive.
Several factors complicate the development of unifying principles regarding cnidarian stem cells. Firstly, cnidarians have complex life cycles that include different combinations of planula larvae, benthic polyps, and free-swimming medusae (Fig. 1b). These life history stages are so disparate, that before the mid-1800 s different stages were classified as distinct taxa (recounted in Calder 1982). Cnidarians are often chosen for study based on the unique morphological aspects of their life cycle, which further complicates efforts to homologize developmental dynamics between the different model systems. Secondly, cnidarians can generate adult forms using a wide variety of developmental mechanisms, including sexual reproduction, asexual reproduction (via pinching, parthenogenesis, polarity reversal, strobilation, or the production of planuloids or propagules) and regeneration following damage (Lesh-Laurie and Suchy 1991; Van Lieshout and Martin 1992; Vagelli 2007). Even in relatively closely related species, development and sexual maturity can be fundamentally different in extremes that are uncommon to the Bilateria. Consequently, attempts to classify taxa based on developmental mechanisms have proven problematic (e.g., Fautin 1991).
To explore cnidarian stem cells and identify unifying principles or important generalizations, we start by reviewing what is known about the self-renewing cells of hydrozoans—particularly the epithelial and interstitial cells—and then present the problems faced when extending these observations to other cnidarians. While non-hydrozoans appear to lack interstitial cells, they have other undifferentiated cell types, collectively called amoebocytes, which might play a stem cell role. But there are significant differences between the morphology and cellular dynamics of interstitial cells and amoebocytes, and the later most likely do not represent stem cells. Current molecular studies of vertebrate “stem cell markers” in cnidarians suggest that the genetic control of cell pluripotency might have deep ancestry, but these studies have been largely unsuccessful in teasing out the homologies between cnidarian cell types and their vertebrate counterparts. However, comparative studies in regeneration and gametogenesis suggest that cnidarian cells are highly plastic, and that the transdifferentiation of epithelial cells and dedifferentiation of “committed” cells is probably more ancestral than any particular stem cell niche. Ultimately, this review suggests that “stem cells” are not a homologous entity shared across the animals, and caution should be made when discussing the broad evolution of stem cells or when making comparisons across animal model systems.
The interstitial cells of hydrozoans
The stem cells of hydrozoans—particularly Hydra—have recently been reviewed in a number of excellent publications (Frank et al. 2009; Watanabe et al. 2009; Bosch 2009; Bosch et al. 2010). Subsequently, this paper will only briefly touch upon the current understanding of hydrozoan stem cells, and instead focus on the potential relationship of hydrozoan stem cells to the cells of other cnidarians.
In the Hydrozoa, stem cells are often called interstitial cells (or I-cells), because they are found “wandering” in the spaces between ectodermal cells. I-cells are generally identified by their round, undifferentiated morphology and large nucleus. The chromatin in these cells is loosely packed, allowing for easy imaging with a variety of basic nucleic acid dyes (reviewed in Frank et al. 2009). I-cells are set aside early in embryology; while older studies of I-cells trace the origin of these cells to the endoderm of planula larvae, I-cells are distinguishable earlier in Hydractinia, originating in the gastrula (Frank et al. 2009), which is similar to vertebrate embryonic stem cells.
In Hydra, it is possible to remove I-cells by chemical treatment; these “epithelial” Hydra lack stinging cells (nematocytes), gametes, neurons, secretory and sensory cells (Bode et al. 1987). Remarkably, epithelial Hydra can be kept alive in the lab by force feeding; the epithelial cells self-renew, and the animal will continue to undergo asexual reproduction through budding, even regenerating a lost head (Marcum and Campbell 1978; Holstein et al. 1991). However, both ectodermal and endodermal epithelial cells are required to keep the animal alive, and cells from one type of epithelia have not been found to differentiate into the other. Thus, it is sometimes claimed that Hydra has three distinct populations of multipotent stem cells: I-cells, ectodermal epithelial cells, and endodermal epithelial cells (Campbell and Bode 1983; Galliot et al. 2009; Watanabe et al. 2009; Bosch 2009; Technau and Steele 2011, reviewed in Fig. 2). However, it might be inappropriate to call the ectodermal and endodermal epithelial cells true stem cells. Stem cells are by most definitions undifferentiated, while Hydra’s epithelial cells are best described as differentiated, epitheliomuscular cells (see Frank et al. 2009 for a similar opinion). Hydra’s ectodermal epithelial cells are highly vacuolated, possess basal extensions composed of contractile fibers, and are connected together by gap and septate intercellular junctions (Wood 1959; West 1978; Campbell 1987). The endodermal epithelial cells share similar morphological properties, and are also capable of digesting food (Gauthier 1963; McNeil 1981, 1984; McNeil et al. 1981). The dynamics of epithelial cell division and differentiation is also distinct from most stem cells. Cellular proliferation of epithelial cells is restricted to the middle of the Hydra body column; from there, cells migrate basally towards the foot and apically into the tentacles. During their migration, epithelial cells can take on significant changes in morphology; for example, a subset of ectodermal epithelial cells that migrate into the tentacles become complex battery cells, while others migrating towards the foot become basal disk cells (Campbell and Bode 1983). However, these changes in morphology are not associated with cellular division, so it is unclear if morphological changes associated with migration should be interpreted through stem cell dynamics or as modifications of epithelial cells during their life history. Ultimately, whether or not cnidarian epithelial cells are “true” stem cells is potentially an exercise in semantics and reinforces the point that “stem cells” remain conceptually vague. Still, acknowledging the distinction between “self-renewing” epithelial cells and more conventional interstitial “stem” cells will be important to later discussions in this paper.
Fig. 2.
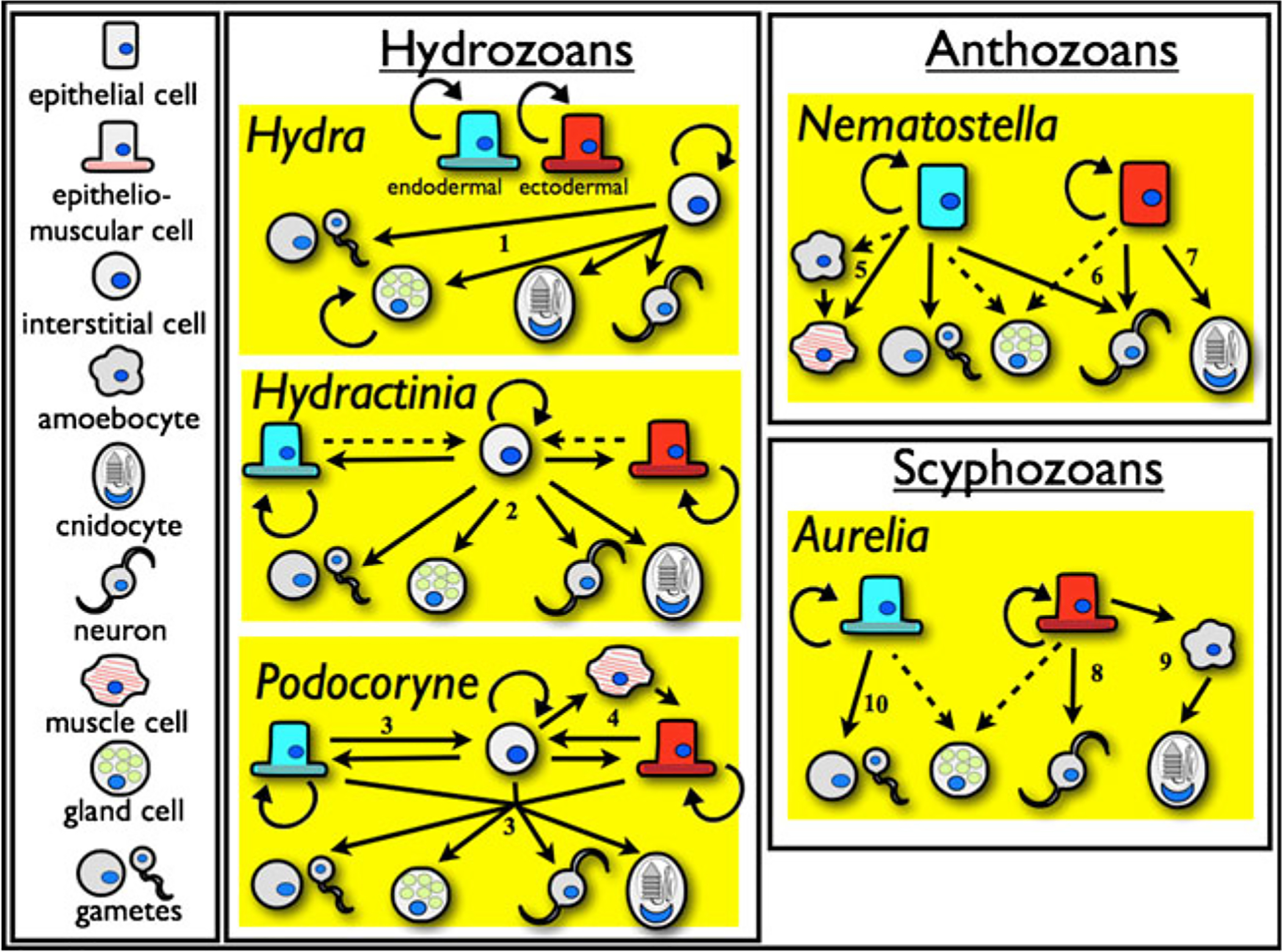
Plasticity of cell lineages in the cnidarians, based on current knowledge. A key showing major cell types is given on the left. Uncertainty is reflected by the use of dotted arrows. Numbers in the figure refer to the following references for support: (1) Bosch et al. 2010; (2) Müller and Teo 2004; (3) Schmid et al. 1982; (4) Schmid and Alder 1984; (5) Renfer et al. 2010; (6) Nakanishi et al. 2012; (7) Marlow et al. 2009; (8) Nakanishi et al. 2008; (9) Chapman 1974; (10) Eckelbarger and Larson 1988
Terminology aside, it is certainly true that in adult Hydra, all three cell lineages proliferate at levels usually exclusive to stem cells. I-cells have a cell cycle of 16–30 h; for epithelial cells, it is about 3 days, with times varying based on cell density (David and Campbell 1972; Campbell and David 1974; Holstein 1990). The majority of stem cells are generated in the middle of the animal, displacing older cells towards the aboral or oral end. Most excess cells are converted into reproductive buds, and the rest are sloughed off the ends of the animal (David and Plotnick 1980; Thomas and Edwards 1991; Bosch et al. 2010). This has two important consequences. Firstly, this constant movement implies that the cells of Hydra continually receive new positional cues, and many cell types—most noticeably neurons—appear to be receptive to such cues throughout their short lives (Koizumi and Bode 1986; Koizumi et al. 1988; Koizumi and Bode 1991; Bosch et al. 1991). Secondly, constant replacement of cells may explain the apparent immortality of Hydra polyps and the absence of cancerous tumors in these long-lived animals (Bosch 2008).
Stepping outside the genus Hydra, other hydrozoans appear to have I-cells with similar properties (Afzelius and Rosen 1965; Chapman 1974; Martin and Thomas 1981b; Denker et al. 2008; Houliston et al. 2010; Künzel et al. 2010), but there are important differences as well. Although I-cells are required for the generation of neurosensory and gland cells in Hydra, Pennaria can generate both cell types following the chemical removal of I-cells (Martin and Thomas 1981a, b). And while epithelial cells are normally self-renewing in Hydractinia, I-cells can become totipotent, capable of generating all cell types under certain conditions (such as colony fusion or chemical removal and reintroduction of I-cells) (Müller and Teo 2004; Rebscher et al. 2008; Künzel et al. 2010, reviewed in Fig. 2).
Do stem cells exist in non-hydrozoan cnidarians?
Before the advent of molecular phylogenetics, the evolutionary relationships within the Cnidaria had been contested (Bridge et al. 1992). However, genetic studies have consistently placed the Anthozoa (sea anemones, sea pens, and corals) as sister to a medusozoan clade containing the Hydrozoa and Scyphozoa + Cubozoa (Bridge et al. 1992; Kim et al. 1999; Medina et al. 2001; Collins et al. 2006; Dunn et al. 2008; Philippe et al. 2009, Fig. 1a). This was counterintuitive to many biologists, who considered hydrozoans to be one of the most basal branches of the Cnidaria (Barnes and Harrison 1990; Brusca and Brusca 2003; see Jacobs and Gates 2003 for discussion). If this were the case, then it might be of little concern whether or not the non-hydrozoan lineages also had I-cells; their absence could easily be explained as a synapomorphic loss. But given the phylogenetic position of the hydrozoans, it becomes critical to determine whether their stem cells are derived or homologous to other cnidarian and bilaterian cell types. Some researchers have expressed hope that interstitial-like cells will be discovered outside the Hydrozoa (Frank et al. 2009), while at least one recent review suggests that the hydrozoan I-cell might have evolved convergently alongside the bilaterian stem cell (Steele et al. 2011). Given the limited taxonomic coverage, the inherent variability of cnidarians, and the ambiguous definition of cell types, it is difficult to completely discount the possibility of I-cells in non-hydrozoans. However, the available evidence suggests that I-cells, if they exist, play a much more limited role in these taxa.
I-cells have not been observed in two of the best-studied non-hydrozoan cnidarians, the sea anemone Nematostella (Marlow et al. 2009) and the jellyfish Aurelia (Steinberg 1963; Chapman 1999; Yuan et al. 2008; Nakanishi et al. 2008). Instead, these cnidarians appear to have another undifferentiated cell type, known as the amoebocyte (also spelled amebocyte). While researchers have sometimes used the terms “amoebocyte” and “interstitial cell” interchangeably, their homology is far from clear (Chapman 1974), and there are significant differences between them (Fig. 3). As the name suggests, amoebocytes take on a variety of shapes, sometimes utilizing filopodial extensions. They are often highly granulated, and are found in the epithelia and mesoglea of all life stages, although their appearance tends to be transiently associated with wound healing or regeneration (Chapman 1974, 1999; Tucker et al. 2011). This is in marked contrast to hydrozoan I-cells or vertebrate stem cells, where an undifferentiated cell population is “set aside” to generate differentiated cell types. Amoebocytes are only known from descriptive ultrastructural studies; without cell lineage-tracing experiments, it cannot be determined whether they actually play any role in developmental dynamics, as opposed to structural or immunity-related roles. In 1991, Lesh-Laurie and Suchy called the amoebocyte a “major cytologic enigma”; two decades later, this enigma has yet to be resolved.
Fig. 3.
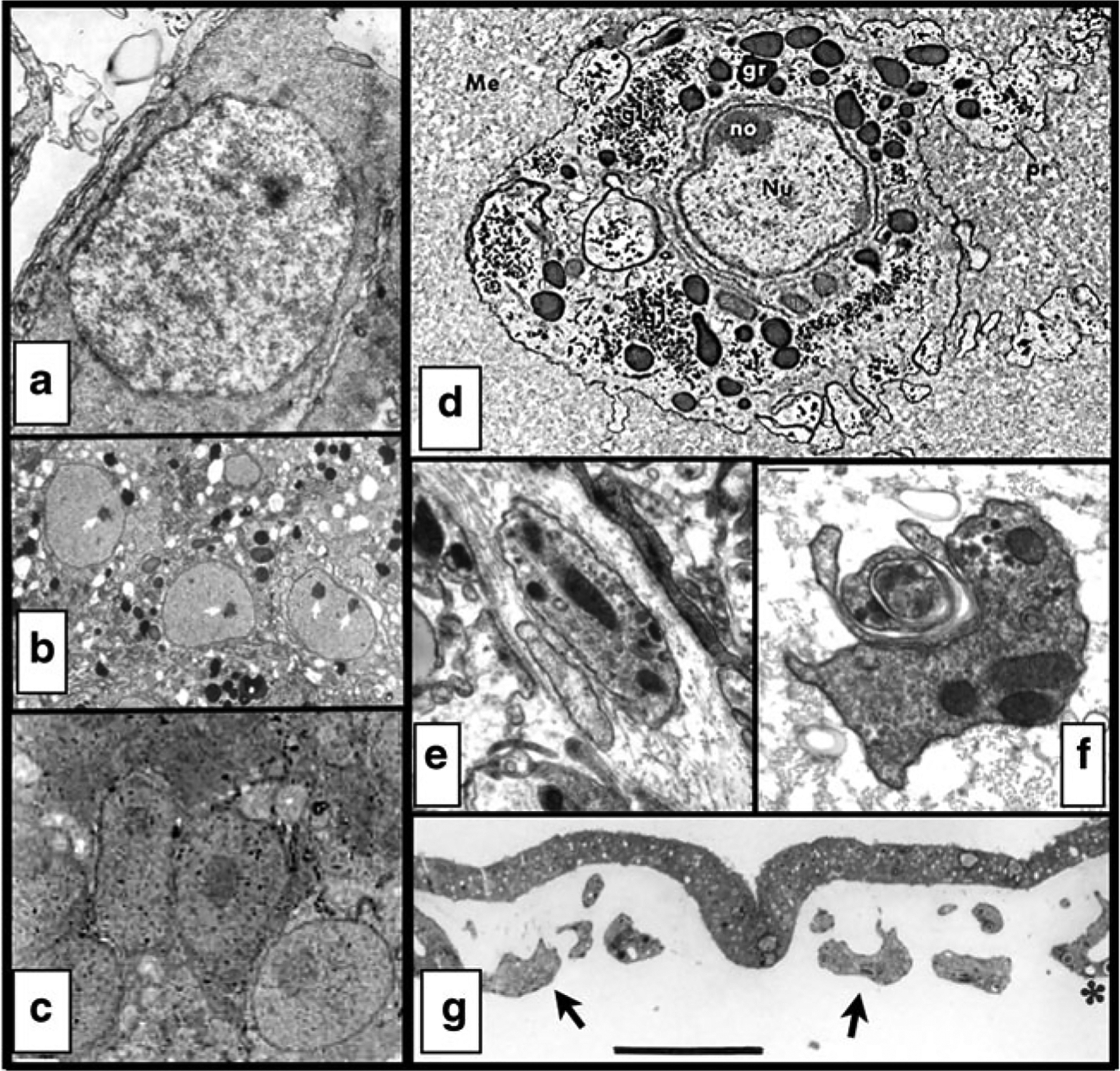
Comparisons of interstitial cells and amoebocytes in various cnidarians. a–c I-cells from hydrozoans; note the simple, rounded morphology and lack of organelles (excluding mitochondria). a Hydra (from Lentz 1965). b Halocordyle (from Martin 1988). c Clytia (from Denker et al. 2008). d–g Amoebocytes from non-hydrozoans; note the extreme variation of shapes, filopodial extensions, and abundance of granules. d Anthozoan sea anemones Actinia (from Larkman 1984b) and e–f Nematostella (from Tucker et al. 2011). g Scyphozoan jellyfish Aurelia (from Chapman 1999, arrows added for clarity)
Non-hydrozoan I-cells do exist in the literature, but they, like amoebocytes, are hypothesized from ultrastructural studies and have characteristics that appear to unite them with amoebocytes. For example, I-cells have been described in the planula larvae of the jellyfish Cassiopeia (Martin and Chia 1982) and the box jelly Tripedalia (Nordström et al. 2003). However, I-cells were not found in the polyps of either of these animals, or in regenerating fragments of Cassiopeia (Curtis and Cowden 1974; Chapman 1978). I-cells have also been described in a number of anthozoans, including the sea anemones Metridium (Westfall 1966), Renilla (Ivester 1977), and Aiptasia (Singer 1971), and in the early development of the coral Acropora (Reyes-Bermudez and Miller 2009). In both Metridium and Renilla, it is hypothesized that some I-cells develop into cnidoblasts, which are precursors to the various types of cnidocytes (or stinging cells). While this would be consistent with what is known about hydrozoan I-cells, these studies were done using transmission electron microscopy, which limits the ability to infer differentiation. It would be valuable to revisit these studies using current molecular- genetic and cell lineage-tracing techniques (e.g., Bosch 2009). In Aiptasia, Singer (1971) used radioactive thymidine labeling to suggest that undifferentiated “I-cells” proliferated during wound healing. However, differentiation was not observed. In all three anemones, several characteristics of these supposed “I-cells” (prominent granules, the spindle-shape in Aiptasia cells) suggests that they have more in common with amoebocytes than hydrozoan I-cells.
Given the limited data, it could be reasonable to hypothesize that hydrozoan I-cells are derived from the amoebocytes of other cnidarians. In this scenario, amoebocytes were “set aside” and retained throughout the hydrozoan life cycle. However, the role of amoebocytes in generating differentiated cell types is uncertain. The best understood function of amoebocytes is their role in inflammation and immunity. Based on histological evidence, amoebocyte phagocytosis in response to wounding and the introduction of foreign materials or fungal pathogens has been inferred in the gorgonian corals Swiftia (Olano and Bigger 2000), Plexaurella (Meszaros and Bigger 1999), and Gorgonia (Mydlarz et al. 2008), the scleractinian corals Montastraea (Vargas-Ángel and Peters 2007) and Porites (Palmer et al. 2011), and the sea anemones Anthopleura (Patterson 1979) and Actinia (Hutton and Smith 1996) (but not in the coral Montipora; see Work and Aeby 2010). However, not all amoebocyte “morphotypes” are involved in phagocytosis (Hutton and Smith 1996), and other cell types can play a phagocytizing role during wound healing. Sometimes, amoebocytes have been interpreted as migrating towards regions of wounding (Meszaros and Bigger 1999), and other times not (Mydlarz et al. 2008). In Gorgonia, some amoebocytes appear to have specialized functions (e.g., melanin synthesis), and because there is no evidence of amoebocyte migration, the authors conclude that specialized amoebocytes might have been generated from the proliferation of undifferentiated amoebocytes (Mydlarz et al. 2008).
There are a number of additional roles amoebocytes play in certain cnidarians, although it is not known how universal these roles are. In the gonads of anemone Actinia, amoebocytes are closely associated with oocytes and spermatogenic cysts but do not appear to differentiate into any novel cell types (Larkman 1983, 1984a, b). Amoebocytes might play a role in secreting connective mesogleal fibers (Meszaros and Bigger 1999) and the podocyst cuticle that attaches a polyp to its substrate (Chapman 1968). Perhaps most intriguingly, a histological study of Aurelia amoebocytes suggests that they can differentiate into cnidocytes and perhaps other cell types as well (Chapman 1974). If this is corroborated with experimental evidence, it would not only provide a functional link between the amoebocytes of Aurelia and the I-cells of hydrozoans, but it would also corroborate the studies of cnidocyte development from the amoebocytes/I-cells in Metridium and Renilla discussed earlier (Westfall 1966; Ivester 1977). A similar usage of amoebocytes in rebuilding tissue has also been suggested for the sea anemone Anthopleura (Patterson 1979).
Clearly, amoebocytes play a variety of roles in cnidarians, although it is still unclear whether they, as a cell type, have any shared or independent traits. It is quite possible that “amoebocyte” is a catch-all term, referring to a collection of amorphous cell types with differing degrees of specialization, which may or may not originate from one cell-type precursor. Ultimately, the stem cell role of amoebocytes, and whether or not some non-hydrozoans have true I-cells, will require much exploratory research using cell lineage-tracing techniques to verify the dynamics and potentialities of amoebocytes in various species. But are there any hypotheses we can test about the nature of cnidarian stem cells with our current information? Are there ways to determine homology when comparative histological studies are inconclusive? Recent attempts to resolve these uncertainties have used molecular markers; while these data provide some intriguing clues, the results are currently inconclusive.
The molecular control of cnidarian multipotency
A major initiative of evolutionary developmental biology is the utilization of molecular markers to establish deep homologies obscured by evolutionary divergence. Given the difficulty of establishing homologies between unspecialized cells, it would seem that the I-cells of hydrozoans, the amoebocytes of non-hydrozoans, and the stem cells of bilaterians would together represent a perfect candidate for such a comparative approach. The homology of these cell types would gain support if evidence of evolutionarily conserved transcription factors (or ideally, entire genetic regulatory pathways) were found to be used in similar ways in these disparate cell types. High-throughput sequencing (particularly RNA-Seq experiments) will likely address this issue in the near future, but currently available data for testing genetic homology largely come from candidate gene approaches. While there are no ubiquitous genetic markers among somatic stem cells, research on vertebrate embryonic stem cells has revealed a set of conserved transcription factors important to inducing pluripotency, including Nanog, Sox2, Oct4, Myc, and Klf4 (Takahashi and Yamanaka 2006; Loh et al. 2006; Huangfu et al. 2008; Feng et al. 2009). Although there is currently no evidence that these genes work together to induce pluripotency in invertebrates, it might still be instructive to look at these candidate genes in cnidarians.
Three of these candidate genes—Nanog, Klf4, and Oct4— appear to be missing from the Hydra genome (Chapman et al. 2010), but it is known that Hydra has undergone significant gene loss. A Hydra Myc homologue is expressed in I-cells, as well as differentiating nematoblasts and gland cells (Hartl et al. 2010), and changes in Myc regulation affect homeostasis between I-cell renewal and differentiation (Ambrosone et al. 2012), so there is some evidence that homologous transcription factors might regulate hydrozoan and vertebrate pluripotency. Unfortunately, data from additional genes and hydrozoans have proven less conclusive, and no evidence currently exists regarding gene expression in amoebocytes. Ultimately, while cnidarians and vertebrates show some evi- dence of shared genetic control of cell pluripotency (perhaps as part of one or more conserved gene networks), there is little molecular evidence that I-cells themselves are homologous to vertebrate stem cells.
Sox genes are present across the cnidarians, and gene expression data from the sea anemone Nematostella (Magie et al. 2005), and the coral Acropora (Shinzato et al. 2008) suggest that certain Sox orthologues play a role in neurosensory cell specification. But since gene expression is not associated with a morphological cell type, it is difficult to say whether these Sox-expressing cells are multipotent stem cells or committed but undifferentiated neurosensory cells (there are also significant differences in timing and expression of Sox genes in these two genera, see Shinzato et al. (2008) for an overview). To address this problem, Sox genes have recently been studied in the hydrozoan Clytia (Jager et al. 2011). Four paralogues (out of ten), representing three different Sox families, are expressed in the I-cells, while other paralogues are expressed in neurosensory and cnidocyte precursors, as well as larval endodermal epithelial cells. Unfortunately, Jager et al. were unable to find any pattern of commonality regarding which Sox paralogues control stem cell maintenance when comparing Clytia, Nematostella, and Acropora, or broadly between Clytia and bilaterians.
Oct4 (POU5f1) is a member of the POU transcription factor family, and the only gene known to induce pluripotency in vertebrate neural stem cells (Kim et al. 2009). In Hydractinia, the POU gene Polynem (Pln) is highly expressed in interstitial cells, and directly upregulates transcripts of Myc, as well as germ line determinants Nanos and Vasa, and to a lesser extent Piwi (Millane et al. 2011). When Pln was ectopically expressed in epithelial cells, it induced stem cell neoplasms and loss of epithelial tissue, while downregulation increased cell differentiation. Millane et al. were unable to determine if Pln was a true member of the POU-5 clade or part of the closely related POU-3 clade of paralogous POU genes. However, a more detailed analysis of the POU family suggests that the POU-5 clade is a vertebrate novelty (unpublished data), suggesting that Pln is a paralogue (as opposed to orthologue) of vertebrate Oct4. Various POU paralogues appear to be utilized in the stem cells of other invertebrates as well, such as the upregulation of POU-4 and POU-6 in planarian neoblasts (Resch et al. 2012). As in the case of the Sox genes, this suggests that different paralogues from the same gene family can play similar roles in different animals. But what this phenomenon says about the underlying homology of these structures is unclear.
So while cnidarians do deploy myc, Sox, and POU family transcription factors in control of cell pluripotency and differentiation, it is not clear that they are using the same paralogues as vertebrates, which undermines hypotheses that a homologous network exists connecting these cell types back to a common cellular ancestor. There are other genetic and cellular similarities between Hydra and bilaterian stem cells (e.g., Wnt and Notch signaling, euchromatin modification, see Khalturin et al. 2007), but it is less obvious that these characteristics are unique to stem cells, or would be difficult to evolve convergently. A growing number of studies have suggested that Sox2, Oct4, Myc, and Klf4 can all be replaced by other transcription factors in certain scenarios (reviewed in Heng et al. 2010), and that some of these genes—particularly Oct4 and Myc—are part of larger and independent networks (Ng and Surani 2011). Continued elucidation of gene regulatory networks and their functions in a wider range of animal taxa will help us determine what we might expect to be conserved between stem cells, but the current data are far from conclusive.
Cnidarian regeneration and gametogenesis supports plasticity over a conserved stem cell niche
Given the histological and molecular uncertainties described previously, the short answer to whether unifying principles exist in cnidarian stem cells is “we do not know”. The data provided in this review support the hypothesis that undifferentiated hydrozoan I-cells are derived, suggesting that the ancestral cnidarians relied on transdifferentiation and/or dedifferentiation of epithelial cells, which are conserved across the phylum. Because of the paucity of experimental data in non-hydrozoans, there is little evidence for the role of epithelial cells in differentiation during “normal” development. This is changing; recent work on transgenic Nematostella suggests that in the planula larva, both ectodermal and endodermal epithelial cells produce neurons, (Nakanishi 2012) which is consistent with our hypothesis. However, there is a much more extensive literature regarding cell dynamics during tissue regeneration, where dedifferentiation is widely hypothesized. Inferring processes of “normal” development from manipulation experiments is inherently risky because the organism is placed in conditions it might never encounter in the wild. However, with their complex life cycles and considerable regenerative abilities, cnidarians often confound normal definitions of development, and therefore cnidarian regeneration might be particularly useful in elucidating general principles of stem cell dynamics.
Animal development is often considered across a one-dimensional “arrow of time;” individuals undergo embryological change, maturation, reproduction (if they are fortunate), senescence, and death. Most of our thinking about developmental dynamics is built upon this unidirectional mode of growth. Regeneration and the repetition of developmental processes has been largely absent from modern developmental biology, in part because the model organisms of development—the mouse, the fruit fly, the nematode worm—have very limited regenerative capabilities (Sánchez Alvarado 2000). In addition, it is incompletely understood to what extent regeneration represents a true recapitulation of normal developmental, versus a unique mode of morphogenesis, although emerging research in cnidarians suggests it involves both (Burton and Finnerty 2009; Trevino et al. 2011).
In the Cnidaria, the distinction between normal development and regeneration is ambiguous. Sea anemones can undergo “fission,” where an individual pulls itself in two directions until it splits in half and then regenerates the missing material (Hand and Uhlinger 1995). In times of stress, hydrozoan medusa are known to “regress” back into polyps (reviewed in Piraino et al. 2004), and medusa of the moon jellyfish Aurelia can “degrow” parts of their bodies, including gametes, only to regrow them again when food is plentiful (Hamner and Jenssen 1974). Box jelly polyps produce larva-like propagules made from pieces of their own tissue, which swim or crawl to a new location and develop into new organisms (Fischer and Hofmann 2004), and many scyphozoan polyps produce multiple medusae at a time through a process called strobilation, after which the polyp returns to its previous life-stage and can produce more medusae in the future (e.g., Vagelli 2007). Regeneration, differentiation, and dedifferentiation produce a range of developmental processes and complex life histories across the cnidaria, which often look more like webs than cycles (Piraino et al. 2004; Vagelli 2007; the Aurelia life cycle in Fig. 1b)
In Hydra, the distinction between development and regeneration is continually blurred; new cells are constantly being generated, while old cells are repositioned across the oral/aboral axis and eventually converted into new buds or sloughed off. This ongoing cell division may explain Hydra’s unusual regenerative powers and its hypothesized immortality (Müller 1996; Watanabe et al. 2009). A piece of Hydra epithelium containing a minimum of about 300 cells (or about 0.2 mm of tissue) can regenerate into a new animal through morphallaxis, meaning that the cells do not divide, but instead change their morphology to rebuild a new, albeit tiny animal (Shimizu et al. 1993; Bosch 2007). It has been hypothesized that this minimum tissue size requirement is determined by functional mechanics; the regenerating animal needs to be able to form a spherical “shell” that creates an osmotic barrier against the environment (Shimizu et al. 1993). But it is also probably true that chunks of tissue are necessary to capture the three non-overlapping self-renewing cell lines that generate the complete Hydra (discussed in “The Interstitial Cells of Hydrozoans”). Morphallaxis is presumably possible because constant cellular repositioning during normal homeostasis means that cells are continually receptive to positional cues; this also explains how a Hydra deconstituted into individual cells can reaggregate into a complete animal (Gierer et al. 1972; Bosch 2007).
Despite the lack of I-cells, some non-hydrozoan cnidarians appear to have equal—or in certain cases, superior— regenerative abilities compared to Hydra. At the polyp stage, the jellyfish Aurelia (Steinberg 1963), Cassiopea, (Curtis and Cowden 1974), and Chrysaora (Black and Riley 1985), can regenerate complete animals from isolated ectodermal fragments. As in Hydra, Chrysaora polyps can reaggregate after being disassociated (Black and Riley 1985), and unlike Hydra, Aurelia can regenerate animals from isolated polyp tentacles (Lesh-Laurie et al. 1991). As opposed to explaining this phenomenon through the presence of a tentacle stem cell niche, these differences appear to be a function of where epithelial cell proliferation occurs, for example, EdU labeling shows that cell division occurs in the tentacles of mature Aurelia polyps, which is not the case for Hydra (Fig. 4a, b).
Fig. 4.
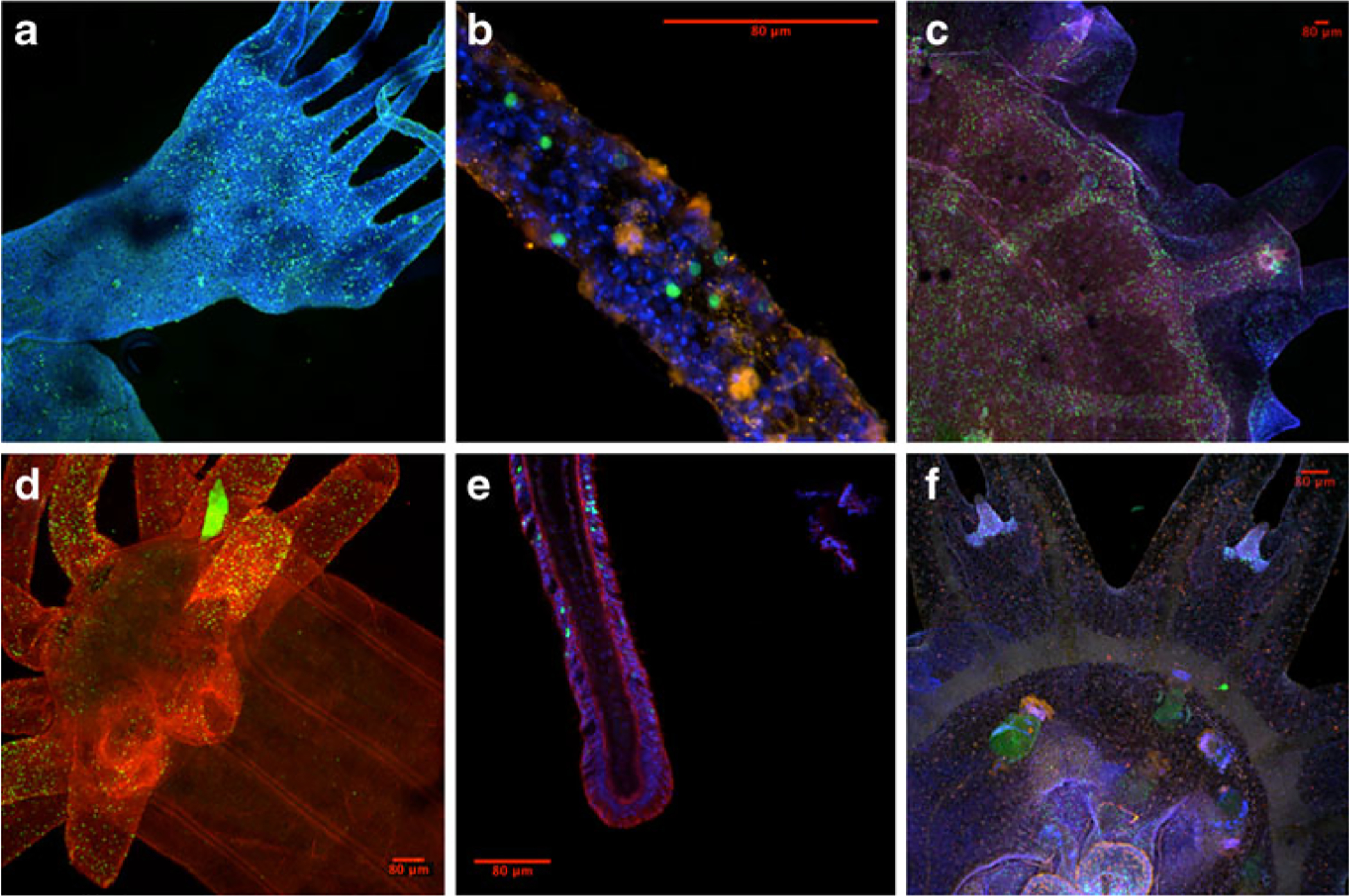
Cell proliferation (in green) in various cnidarians, labeled using 5-ethynyl-2′-deoxyuridine (EdU). a Polyp of Aurelia sp. 1 (nuclear staining in blue); b Close-up of a polyp tentacle (nuclear staining in blue); c Juvenile medusa stage of Aurelia sp. 1 (tyrosinated tubulin in red, nuclear staining in blue); d Polyp of Nematostella vectensis; and e close-up of tentacle (tyrosinated tubulin in red, nuclear staining in blue). f Ephyrae of Chrysaora colorata (fmrf-positive neurons in red, nuclear staining in blue)
Intriguingly, cnidarian morphological complexity may play a more important role in predicting regenerative capabilities than the presence or absence of I-cells. While the polyps of many jellyfish show remarkable regenerative abilities (as discussed above), the morphologically complex medusa exhibits markedly less regenerative potential (Black and Riley 1985) (however, medusa can still regenerate significant amounts of lost tissue, see Stockard’s (1908) work on Cassiopea, or Hargitt’s (1904) work on Rhizostoma). In medusa, cell division appears more spatially restricted than in the polyp (Fig. 4c, f), perhaps suggesting that the increased complexity requires more orderly cell proliferation. Anthozoans have the most complex polyp morphologies, and they exhibit less regenerative potential than scyphozoan or hydrozoan polyps. There is no evidence that anemones are capable of reaggregation after being disassociated into single cells (e.g., Schmid et al. 1981). In corals, regeneration is generally limited, decreases with time, and requires tissue from a developmental “organizing center” around the mouth (reviewed in Henry and Hart 2005). Still, many anthozoans can regenerate substantial pieces of missing tissue (Hahn 1905; Singer 1971; Reitzel et al. 2007) which again is probably linked to constant cellular proliferation of epithelial tissues. The sea anemone Nematostella, which is capable of regeneration following bisection (e.g., Burton and Finnerty 2009) shows cellular proliferation across the animal, particularly towards the oral end and into the tentacles (Fig. 4d, e), and similar results have been found in the anemone Haliplanella (Minasian 1980).
Perhaps the most convincing evidence of dedifferentiation during regeneration comes from the hydrozoan jelly Podocoryne. In Podocoryne, isolated striated muscle removed from its associated extracellular matrix dedifferentiates into smooth epitheliomuscular “stem” cells, which divide asymmetrically to produce one daughter cell and one specialized FMRFamide-positive nerve cell (Schmid et al. 1982; Schmid and Reber-Muller 1995; Reber-Müller et al. 2006). If Podocoryne striated muscle is cultured with endodermal epithelial cells, a partial organism regenerates, with differentiated nematocytes, gland cells, and even gametes and I-cells (Schmid et al. 1982). This is remarkably similar to regeneration as hypothesized in Aurelia, where epitheliomuscular cells act as a stem-like lineage, dedifferentiating into amoebocytes or directly transdifferentiating into new cell types (Steinberg 1963; Chapman 1974).
The plasticity of cell types evidenced in the above examples complicate the application of the stem cell concept to these systems, but there is evidence that Hydra and other cnidarians share broad morphogenic principles regarding regeneration. Hydra is known to emit a head-inhibition morphogen from its oral end, meaning that a decapitated Hydra will develop a new head, but a Hydra head will not necessarily develop a new body (Bode 2009). Similarly, aboral fragments of polyps in the jellyfish Cassiopeia can regenerate complete animals, while oral fragments only generate head structures (Neumann 1977). Oblique cuts to polyps, leaving portions of the head region, retard or inhibit regeneration of missing head structures in the scyphozoan Haliclystus, and, to a lesser extent, in Aurelia (Child 1951). Polyps of the cubozoan box jelly Carybdea can also regenerate new oral structures after their removal (Fischer and Hofmann 2004). Finally, both Hydra and Nematostella display Wnt signaling in their oral ends, possibly setting up the oral/aboral axis and playing a role in the head organizer region (Kussarow et al. 2005; Lengfeld et al. 2009).
When comparing gametogenesis between Hydra and other cnidarians, Hydra again shows stem cell-like regulation where other species show plasticity. Researchers have been able to create Hydra that lack the I-cells involved in somatic differentiation, while retaining I-cells involved in spermatogenesis (Littlefield 1985; Littlefield and Bode 1986; Nishimiya-Fujisawa and Sugiyama 1993). But Hydra might be unusual in having distinct somatic and germ line-restricted I-cells. The expression of the genes Nanos, Piwi, and Vasa have been studied in a number of cnidarians because of their involvement in germ line specificity in bilaterians (Extavour and Akam 2003), as well as nonbilaterians such as cetnophores (Alié et al. 2011). The expression of Vasa in Hydra is strongest in germ line cells; a weaker signal also occurs in other I-cells and ectodermal epithelial cells (Mochizuki et al. 2001). But in most other cnidarians, these molecular markers show no obvious distinction between germ line cells and multipotent cells. In the hydrozoan, Clytia, Nanos, Piwi, and Vasa are maternally inherited in the I-cell population, but are also induced in new I-cells via inductive signaling (Leclère et al. 2012). Similarly, although the maternal Vasa protein accumulates during oogenesis in Hydractinia, Vasa mRNA is a broad I-cell marker (Rebscher et al. 2008). And as mentioned earlier, the POU homologue in Hydractinia regulates Nanos, Piwi, and Vasa (Millane et al. 2011). In Podocoryne, a Piwi homologue is expressed in the germ line, as well as somatic I-cells and even cells undergoing transdifferentiation (Seipel et al. 2004). Finally, in the sea anemone Nematostella, Nanos and Vasa orthologues are restricted to germ line cells late in development but are expressed in developing somatic cells earlier on (Extavour et al. 2005). This suggests a significant flexibility in how the germ line is determined, even in cnidarians that have I-cells.
Conclusion
To review, there is considerable variation regarding what sort of cell types I-cells control in different hydrozoans. While non-hydrozoans also have undifferentiated cells (amoebocytes), they seem to largely be involved in immunity and tissue maintenance; in only a few cases have they been hypothesized to differentiate into novel cell types, and it is not clear that all amoebocytes are homologous to each other. It is possible that at least some amoebocytes represent non-dividing intermediates in the dedifferentiation process from epithelial cell to additional cell types, but only cell cycle studies or cell lineages experiments can unravel this function. Finally, many cnidarians appear to utilize transdifferentiation from a variety of cell types, particularly epithelial cells, to rebuild lost structures.
Taking all of these data together, the most parsimonious explanation seems to be that early in cnidarian evolution, self-renewing epithelial cells acted like stem-like cells, generating additional differentiated cell types. However, it is also quite possible that few if any of these downstream cell types were fully “committed”, since many seemingly committed cnidarian cell types appear capable of transdifferentiation based on positional cues. Similarities in the molecular control of cnidarian I-cells and vertebrate stem cells could suggest that a gene network regulating cell pluripotency was intact in the last common ancestor, but only in certain cnidarian lineages has this pluripotency been canalized into one or more “stem” cell types. In some cases, as in Hydra, one cell (the I-cell) has come to control the development of most cell types, and in the most extreme cases—such as in Hydractinia—the I-cell can become totipotent. But in other species, such as Podocoryne, it appears that most cell types can be generated by both “stem” and “committed” cell lines. Clearly, more work is needed to assess this hypothesis relative to competing arguments that suggest one or more stem cell types are conserved across cnidarians, and perhaps between cnidarians and bilaterians. Growing interest in the emerging model anthozoans Acropora and Nematostella, as well as the scyphozoan Aurelia, will be particularly important to resolving this problem.
The plasticity of differentiated cells in Cnidaria, and the ambiguity surrounding the stem cell role of amoebocytes, leads to a number of intriguing questions for further research. Firstly, given the plasticity of cnidarian cells, it would be interesting to determine whether competition exists between growth, regeneration, and gametogenesis for a limited number of multipotent cells. In Hydra oligactis, sexual reproduction is followed by senescence, including a decline in food capture, movement, reproduction, and an increase in mortality (Yoshida et al. 2006). Bosch (2009) is suspicious of these results, suggesting they might have been caused by excessive sexual production in laboratory conditions, and that other Hydra species do not seem to show similar patterns of senescence. Yet, there is evidence that similar exhaustion might occur in Aurelia polyps that have been stimulated to strobilate (generate sexual medusa) many times (personal observation). A similar tradeoff might exist in stem cell allocation for regeneration versus sexual reproduction in corals (reviewed in Rinkevich 1996).
Furthermore, as discussed earlier, the polyps of many medusozoans appear to have regenerative abilities rivaling Hydra, while regeneration in the medusa—the gametogenic life history stage—is far more limited (Black and Riley 1985). Thus, medusae appear to have some of the more terminally differentiated attributes that are commonplace in bilaterian development. The potential relationships between cell proliferation, morphological complexity, gamete production, and senescence in cnidarians are largely unexplored.
A final question is how certain cell types come under the regulatory control of a “master” stem cell. This question is counterintuitive to how many researchers think about stem cells. It might seem logical to hypothesize that over the course of evolution, groups of cells started off more or less undifferentiated (more like stem cells) and became increasingly divergent over time. But to assume that the stem cell is ancestral simply because it occurs early in embryogenesis repeats Ernst Haeckel’s fallacy that ontogeny recapitulates phylogeny (e.g., Haeckel 1879). That differentiated choanocyte cells in sponges might be just as totipotent as undifferentiated archeocyte cells (see Funayama this issue), and given the remarkable morphological similarity between choanocytes and choanflagellates, the closest living relative to the animals (King et al. 2008), it is quite possible that the first animal cells might have been more complex than previously hypothesized.
Acknowledgments
D.A.G. would like to thank Robert Steele and Volker Hartenstein for helpful discussion while preparing this manuscript, with additional thanks to Volker Hartenstein for help in translating Weismann’s text. We gratefully thank two anonymous reviewers for valuable advice, and acknowledge funding from an NIH Training Grant in Genomic Analysis and Interpretation T32HG002536 (D.A.G.) and the National Aeronautics and Space Administration Astrobiology Program (D.K.J.).
References
- Afzelius B, Rosen B (1965) Nutritive phagocytosis in animal cells—an electron microscopical study of gastroderm of hydroid Clava squamata Mull. Z Zellforsch Mikrosk Anat 67:24. [PubMed] [Google Scholar]
- Alié A, Leclère L, Jager M, Dayraud C, Chang P, Le Guyader H, Queinnec E, Manuel M (2011) Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Dev Biol 350:183–197 [DOI] [PubMed] [Google Scholar]
- Ambrosone A, Marchesano V, Tino A, Hobmayer B, Tortiglione C (2012) Hymyc1 downregulation promotes stem cell proliferation in Hydra vulgaris. PLoS One 7(1):e30660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes R, Harrison F (1990) Introduction In: Harrison F, Westfall J (eds) Microscopic anatomy of invertebrates. Volume 2: Placozoa, Porifera, Cnidaria, and Ctenophora, 1st edn Wiley-Liss, New York [Google Scholar]
- Black RE, Riley GK (1985) Dissociation and reaggregation of cells of Chrysaora quinquecirrha (Cnidaria, Scyphozoa). J Exp Zool 233:369–375 [DOI] [PubMed] [Google Scholar]
- Bode HR (2009) Axial patterning in Hydra. Cold Spring Harb Perspect Biol 1:a000463–a000463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HR, Heimfeld S, Chow MA, Huang LW (1987) Gland cells arise by differentiation from interstitial cells in Hydra attenuata. Dev Biol 122:577–585 [DOI] [PubMed] [Google Scholar]
- Bosch T (2007) Why polyps regenerate and we don’t: towards a cellular and molecular framework for Hydra regeneration. Dev Biol 303(2):421–433 [DOI] [PubMed] [Google Scholar]
- Bosch TCG (2008) In: Bosch TCG (ed) Stem cells in immortal Hydra. Springer Netherlands, Dordrecht [Google Scholar]
- Bosch TCG (2009) Hydra and the evolution of stem cells. Bioessays 31:478–486 [DOI] [PubMed] [Google Scholar]
- Bosch TCG, Rollbhler R, Scheider B, David CN (1991) Role of the cellular environment in interstitial stem cell proliferation in Hydra. Roux’s Arch Dev Biol 200:269–276 [DOI] [PubMed] [Google Scholar]
- Bosch TCG, Anton-Erxleben F, Hemmrich G, Khalturin K (2010) The Hydra polyp: nothing but an active stem cell community. Dev Growth Differ 52:15–25 [DOI] [PubMed] [Google Scholar]
- Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW (1992) Class–level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci U S A 89:8750–8753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusca RC, Brusca GJ (2003) Invertebrates 2nd ed Sinauer Associates [Google Scholar]
- Burton PM, Finnerty JR (2009) Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Roux’s Arch Dev Biol 219:79–87 [DOI] [PubMed] [Google Scholar]
- Calder D (1982) Life history of the cannonball jellyfish, Stomolophus meleagris L. Agassiz, 1860 (Scyphozoa, Rhizostomida). Biol Bull 162:149–162 [Google Scholar]
- Campbell R (1987) Organization of the nematocyst battery in the tentacle of Hydra: arrangement of the complex anchoring junctions between nematocytes, epithelial cells, and basement membrane. Cell Tissue Res 249:647–655 [Google Scholar]
- Campbell R, Bode H (1983) Terminology for morphology and cell types In: Lenhoff H (ed) Hydra: Research methods. Plenum, New York, pp 5–14 [Google Scholar]
- Campbell R, David CN (1974) Cell cycle kinetics and development of Hydra attenuata. II. Interstitial cells. J Cell Sci 16:349–358 [DOI] [PubMed] [Google Scholar]
- Cartwright P, Halgedahl SL, Hendricks JR, Jarrard RD, Marques AC, Collins AG, Lieberman BS (2007) Exceptionally preserved jellyfishes from the Middle Cambrian. PLoS One 2:e1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DM (1968) Structure, histochemistry and formation of the podocyst and cuticle of Aurelia aurita. J Mar Biol Assoc 48: 187–208 [Google Scholar]
- Chapman DM (1974) Cnidarian histology In: Muscatine L, Lenhoff HM (eds) Coelenterate biology: Reviews and new perspectives. Academic, New York, pp 1–92 [Google Scholar]
- Chapman DM (1978) Microanatomy of the cubopolyp, Tripedalia cystophora (Class Cubozoa). Helgoländer Meeresun 31:128–168 [Google Scholar]
- Chapman DM (1999) Microanatomy of the bell rim of Aurelia aurita (Cnidaria: Scyphozoa). Can J Zool 77:34–46 [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D et al. (2010) The dynamic genome of Hydra. Nature 464: 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child CM (1951) Physiological dominance in the reconstitution of the scyphistoma of Aurelia. Physiol Zool 24:177–185 [DOI] [PubMed] [Google Scholar]
- Collins AG, Schuchert P, Marques A, Jankowski T, Medina M et al. (2006) Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol 55:97–115 [DOI] [PubMed] [Google Scholar]
- Curtis S, Cowden R (1974) Some aspects of regeneration in the scyphistoma of Cassiopea (Class Scyphozoa) as revealed by the use of antimetabolites and microspectrophotometry. Am Zool 14:851–866 [Google Scholar]
- David CN, Campbell RD (1972) Cell cycle kinetics and development of Hydra attenuata. I. Epithelial cells. J Cell Sci 11:557–568 [DOI] [PubMed] [Google Scholar]
- David CN, Plotnick I (1980) Distribution of interstitial stem cells in Hydra. Dev Biol 76:175–184 [DOI] [PubMed] [Google Scholar]
- Denker E, Manuel M, Leclère L, Le Guyader H, Rabet N (2008) Ordered progression of nematogenesis from stem cells through differentiation stages in the tentacle bulb of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev Biol 315:99–113 [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD et al. (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452:745–749 [DOI] [PubMed] [Google Scholar]
- Eckelbarger KJ, Larson RL (1988) Ovarian morphology and oogenesis in Aurelia aurita (Scyphozoa: Semaeostomae): ultra-structural evidence of heterosynthetic yolk formation in a primitive metazoan. Mar Biol 100:103–115 [Google Scholar]
- Erwin D (2008) Wonderful Ediacarans, wonderful cnidarians? Evol Dev 10:263–264 [DOI] [PubMed] [Google Scholar]
- Erwin D, LaFlamme M, Tweedt S, Sperling E, Pisani D, Peterson K (2011) The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334:1091. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M (2003) Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130:5869–5884 [DOI] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ (2005) vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev 7: 201–215 [DOI] [PubMed] [Google Scholar]
- Fautin DG (1991) Developmental pathways of anthozoans. Hydrobiologia 216:143–149 [Google Scholar]
- Feng B, Ng J, Heng J (2009) Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4:301–312 [DOI] [PubMed] [Google Scholar]
- Fischer AB, Hofmann DK (2004) Budding, bud morphogenesis, and regeneration in Carybdea marsupialis Linnaeus, 1758 (Cnidaria: Cubozoa). Hydrobiologia 530–531:331–337 [Google Scholar]
- Frank U, Plickert G, Müller W (2009) Cnidarian interstitial cells: the dawn of stem cell research In: Rinkevich B, Matranga V (Eds), Stem cells in marine organisms, 978-90-481-2766-5, Springer [Google Scholar]
- Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S (2009) Origins of neurogenesis, a cnidarian view. Dev Biol 332:2–24 [DOI] [PubMed] [Google Scholar]
- Gauthier G (1963) Cytological studies on gastroderm of Hydra. J Exp Zool 152:13 [Google Scholar]
- Gierer A, Berking S, Bode H, David C, Flick K (1972) Regeneration of Hydra from reaggregated cells. Nat New Biol 239:98–101 [DOI] [PubMed] [Google Scholar]
- Glenner H, Hansen AJ, Sørensen MV, Ronquist F, Huelsenbeck JP, Willerslev E (2004) Bayesian inference of the metazoan phylogeny. Curr Biol 14:1644–1649 [DOI] [PubMed] [Google Scholar]
- Haeckel E (1879) Evolution of man. Appleton, New York [Google Scholar]
- Hahn CW (1905) Dimorphism and regeneration in Metridium. J Exp Zool 2:225–235 [Google Scholar]
- Hamner WM, Jenssen RM (1974) Growth, degrowth, and irreversible cell differentiation in Aurelia aurita. Integr Comp Biol 14: 833–849 [Google Scholar]
- Hand C, Uhlinger K (1995) Asexual reproduction by transverse fission and some anomalies in the sea anemone Nematostella vectensis. Invertebr Biol 114:9–18 [Google Scholar]
- Hargitt CW (1904) Regeneration in Rhizostoma pulmo. J Exp Zool 1:73–94 [Google Scholar]
- Hartl M, Mitterstiller AM, Valovka T, Breuker K, Hobmayer B, Bister K (2010) Stem cell-specific activation of an ancestral myc proto-oncogene with conserved basic functions in the early metazoan Hydra. Proc Natl Acad Sci U S A 107:4051–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JCD, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T et al. (2010) The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6:167–174 [DOI] [PubMed] [Google Scholar]
- Henry LA, Hart M (2005) Regeneration from injury and resource allocation in sponges and corals—a review. Int Rev Hydrobiol 90:125–158 [Google Scholar]
- Holstein T (1990) Cell cycle length, cell size, and proliferation rate in Hydra stem cells. Dev Biol 142:392–400 [DOI] [PubMed] [Google Scholar]
- Holstein T, Hobmayer E, David C (1991) Pattern of epithelial cell cycling in Hydra. Dev Biol 148:602–611 [DOI] [PubMed] [Google Scholar]
- Hou XG, Stanley G, Zhao J, Ma XY (2005) Cambrian anemones with preserved soft tissue from the Chengjiang biota, China. Lethaia 38:193–203 [Google Scholar]
- Houliston E, Momose T, Manuel M (2010) Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet 26:159–167 [DOI] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA (2008) Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26:1269–1275 [DOI] [PubMed] [Google Scholar]
- Hutton D, Smith V (1996) Antibacterial properties of isolated amoebocytes from the sea anemone Actinia equina. Biol Bull 191:441–451 [DOI] [PubMed] [Google Scholar]
- Ivester M (1977) Nematocyst differentiation in the Anthozoon Renilla reniformis (Pallas). Trans Am Microsc Soc 96:238–247 [Google Scholar]
- Jacobs DK, Gates RD (2003) Developmental genes and the reconstruction of metazoan evolution—implications of evolutionary loss, limits on inference of ancestry and type 2 errors. Integr Comp Biol 43:619–646 [DOI] [PubMed] [Google Scholar]
- Jacobs DK, Nakanishi N, Yuan D, Camara A, Nichols SA, Hartenstein V (2007) Evolution of sensory structures in basal metazoa. Integr Comp Biol 47:712–723 [DOI] [PubMed] [Google Scholar]
- Jacobs DK, Gold DA, Nakanishi N, Yuan D, Camara A, Nichols SA, Hartenstein V (2010) Basal metazoan sensory evolution In: Desalle R, Scheirwater R (Eds) Key transitions in animal evolution. CRC [Google Scholar]
- Jager M, Queinnec E, Le Guyader H, Manuel M (2011) Multiple Sox genes are expressed in stem cells or in differentiating neurosensory cells in the hydrozoan Clytia hemisphaerica. EvoDevo 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Anton-Erxleben F, Milde S, Plötz C, Wittlieb J, Hemmrich G, Bosch TCG (2007) Transgenic stem cells in Hydra reveal an early evolutionary origin for key elements controlling self-renewal and differentiation. Dev Biol 309:32–44 [DOI] [PubMed] [Google Scholar]
- Kim J, Kim W, Cunningham CW (1999) A new perspective on lower metazoan relationships from 18S rDNA sequences. Mol Biol Evol 16:423–427 [DOI] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D et al. (2009) Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419 [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M et al. (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451:783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi O, Bode HR (1986) Plasticity in the nervous system of adult hydra. I. The position-dependent expression of FMRFamide-like immunoreactivity. Dev Biol 116:407–421 [DOI] [PubMed] [Google Scholar]
- Koizumi O, Bode HR (1991) Plasticity in the nervous system of adult hydra. III. Conversion of neurons to expression of a vasopressin-like immunoreactivity depends on axial location. J Neurosci 11:2011–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi O, Heimfeld S, Bode HR (1988) Plasticity in the nervous system of adult hydra. II. Conversion of ganglion cells of the body column into epidermal sensory cells of the hypostome. Dev Biol 129:358–371 [DOI] [PubMed] [Google Scholar]
- Künzel T, Heiermann R, Frank U, Müller W, Tilmann W, Bause M, Nonn A, Helling M, Schwarz RS, Plickert G (2010) Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Dev Biol 348:120–129 [DOI] [PubMed] [Google Scholar]
- Kussarow A, Pang K, Sturm C, Hrouda M, Lentfer J et al. (2005) Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433:156–160 [DOI] [PubMed] [Google Scholar]
- Larkman AU (1983) An ultrastructural study of oocyte growth within the endoderm and entry into the mesoglea in Actinia fragacea (Cnidaria, Anthozoa). J Morphol 178:155–177 [DOI] [PubMed] [Google Scholar]
- Larkman AU (1984a) An ultrastructural study of the establishment of the testicular cysts during spermatogenesis in the sea anemone Actinia fragacea (Cnidaria: Anthozoa). Gamete Res 9:303–327 [Google Scholar]
- Larkman AU (1984b) The fine structure of granular amoebocytes from the gonads of the sea anemone Actinia fragacea (Cnidaria: Anthozoa). Protoplasma 122:203–221 [Google Scholar]
- Leclère L, Jager M, Barreau C, Chang P, Le Guyader H, Manuel M, Houliston E (2012) Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev Biol 364:236–248 [DOI] [PubMed] [Google Scholar]
- Lee P, Pang K, Matus D, Martindale M (2006) A WNT of things to come: evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol 17:157–167 [DOI] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW (2009) Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330:186–199 [DOI] [PubMed] [Google Scholar]
- Lentz T (1965) The fine structure of differentiating interstitial cells in Hydra. Z Zellforsch Mikrosk Anat 67:547–560 [DOI] [PubMed] [Google Scholar]
- Lesh-Laurie G, Suchy P (1991) Cnidaria: Scyphozoa and Cubozoa In: Harrison F, Westfall J (eds) Microscopic anatomy of invertebrates. Volume 2: Placozoa, Porifera, Cnidaria, and Ctenophora; Wiley-Liss, New York [Google Scholar]
- Lesh-Laurie GE, Hujer A, Suchy P (1991) Polyp regeneration from isolated tentacles of Aurelia scyphistomae: a role for gating mechanisms and cell division. Hydrobiologia 216–217:91–97 [Google Scholar]
- Littlefield CL (1985) Germ cells in Hydra oligactis males. I. Isolation of a subpopulation of interstitial cells that is developmentally restricted to sperm production. Dev Biol 112:185–193 [DOI] [PubMed] [Google Scholar]
- Littlefield CL, Bode HR (1986) Germ cells in Hydra oligactis males. II. Evidence for a subpopulation of interstitial stem cells whose differentiation is limited to sperm production. Dev Biol 116:381–386 [DOI] [PubMed] [Google Scholar]
- Loh Y, Wu Q, Chew J, Vega V, Zhang W (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38:431–440 [DOI] [PubMed] [Google Scholar]
- Magie CR, Pang K, Martindale MQ (2005) Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev Genes Evol 215:618–630 [DOI] [PubMed] [Google Scholar]
- Marcum BA, Campbell RD (1978) Development of Hydra lacking nerve and interstitial cells. J Cell Sci 29:17–33 [DOI] [PubMed] [Google Scholar]
- Marlow HQ, Srivastava M, Matus DQ, Rokhsar D, Martindale MQ (2009) Anatomy and development of the nervous system of Nematostella vectensis, an Anthozoan Cnidarian. Dev Neurobiol 69:235–254 [DOI] [PubMed] [Google Scholar]
- Martin VJ (1988) Development of nerve cells in hydrozoan planulae: I. Differentiation of ganglionic cells. Biol Bull 174:319–329 [DOI] [PubMed] [Google Scholar]
- Martin V, Chia F (1982) Fine structure of a scyphozoan planula, Cassiopeia xamachana. Biol Bull 163:320–328 [Google Scholar]
- Martin VJ, Thomas MB (1981a) Elimination of the interstitial cells in the planula larva of the marine hydrozoan Pennaria tiarella. J Exp Zool 217:303–323 [Google Scholar]
- Martin V, Thomas M (1981b) The origin of the nervous system in Pennaria tiarella, as revealed by treatment with colchicine. Biol Bull 160:303–310 [Google Scholar]
- Matus D, Magie C, Pang K, Martindale M (2008) The Hedgehog gene family of the cnidarian, Nematostella vectensis, and implications for understanding metazoan Hedgehog pathway evolution. Dev Biol 313:501–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil PL (1981) Mechanisms of nutritive endocytosis. I. Phagocytic versatility and cellular recognition in Chlorohydra digestive cells, a scanning electron microscope study. J Cell Sci 49:311–339 [DOI] [PubMed] [Google Scholar]
- McNeil PL (1984) Mechanisms of nutritive endocytosis. III. A freeze-fracture study of phagocytosis by digestive cells of Chlorohydra. Tissue Cell 16:519–533 [DOI] [PubMed] [Google Scholar]
- McNeil PL, Hohman TC, Muscatine L (1981) Mechanisms of nutritive endocytosis. II. The effect of charged agents on phagocytic recognition by digestive cells. J Cell Sci 52:243–269 [DOI] [PubMed] [Google Scholar]
- Medina M, Collins AG, Silberman JD, Sogin ML (2001) Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci U S A 98:9707–9712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros A, Bigger C (1999) Qualitative and quantitative study of wound healing processes in the coelenterate, Plexaurella fusifera: spatial, temporal, and environmental (light attenuation) influences. J Invertebr Pathol 73:321–331 [DOI] [PubMed] [Google Scholar]
- Millane RC, Kanska J, Duffy DJ, Seoighe C, Cunningham S, Plickert G, Frank U (2011) Induced stem cell neoplasia in a cnidarian by ectopic expression of a POU domain transcription factor. Development 138:2429–2439 [DOI] [PubMed] [Google Scholar]
- Minasian LL Jr (1980) The distribution of proliferating cells in an anthozoan polyp, Haliplanella luciae (Actiniaria: Acontiaria), as indicated by 3H-thymidine incorporation In: Tardent P, Tardent R (eds) Developmental and cellular biology of coelenterates. New York, Elsevier North-Holland [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T (2001) Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Roux’s Arch Dev Biol 211: 299–308 [DOI] [PubMed] [Google Scholar]
- Müller WA (1996) Pattern formation in the immortal Hydra. Trends Genet 12:91–96 [DOI] [PubMed] [Google Scholar]
- Müller W, Teo R (2004) Totipotent migratory stem cells in a hydroid. Dev Biol 275:215–224 [DOI] [PubMed] [Google Scholar]
- Mydlarz LD, Holthouse SF, Peters EC, Harvell CD (2008) Cellular responses in sea fan corals: granular amoebocytes react to pathogen and climate stressors. PLoS One 3:e1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Yuan D, Jacobs DK, Hartenstein V (2008) Early development, pattern, and reorganization of the planula nervous system in Aurelia (Cnidaria, Scyphozoa). Dev Genes Evol 218:511–524 [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Renfer E, Technau U, Rentzsch F (2012) Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 139:347–357 [DOI] [PubMed] [Google Scholar]
- Neumann R (1977) Polyp morphogenesis in a scyphozoan: evidence for a head inhibitor from the presumptive foot end in vegetative buds of Cassiopeia andromeda. Roux’s Arch Dev Biol 183:79–83 [DOI] [PubMed] [Google Scholar]
- Ng HH, Surani MA (2011) The transcriptional and signalling networks of pluripotency. Nature 13:490–496 [DOI] [PubMed] [Google Scholar]
- Nishimiya-Fujisawa C, Sugiyama T (1993) Genetic analysis of developmental mechanisms in hydra. XX. Cloning of interstitial stem cells restricted to the sperm differentiation pathway in Hydra magnipapillata. Dev Biol 157:1–9 [DOI] [PubMed] [Google Scholar]
- Nordström K, Wallén R, Seymour J, Nilsson D (2003) A simple visual system without neurons in jellyfish larvae. Proc Biol Sci 270:2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano CT, Bigger CH (2000) Phagocytic activities of the gorgonian coral Swiftia exserta. J Invertebr Pathol 76:176–184 [DOI] [PubMed] [Google Scholar]
- Palmer CV, Traylor-Knowles NG, Willis BL, Bythell JC (2011) Corals use similar immune cells and wound-healing processes as those of higher organisms. PLoS One 6:e23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M (1979) Cellular reaction to injury in the anthozoan Anthopleura elegantissima. J Invertebr Pathol 33:189–196 [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E et al. (2009) Phylogenomics revives traditional views on deep animal relatioships. Curr Biol 19:706–712 [DOI] [PubMed] [Google Scholar]
- Pick KS, Philippe H, Schreiber F, Erpenbeck D, Jackson DJ, Wrede P, Wiens M, Alié A, Morgenstern B, Manuel M et al. (2010) Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol 27:1983–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piraino S, De Vito D, Schmich J, Bouillon J, Boero F (2004) Reverse development in Cnidaria. Can J Zool 82:1748–1754 [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV et al. (2007) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94 [DOI] [PubMed] [Google Scholar]
- Reber-Müller S, Streitwolf-Engel R, Yanze N, Schmid V, Stierwald M, Erb M, Seipel K (2006) BMP2/4 and BMP5–8 in jellyfish development and transdifferentiation. Int J Dev Biol 50:377–384 [DOI] [PubMed] [Google Scholar]
- Rebscher N, Volk C, Teo R, Plickert G (2008) The germ plasm component Vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev Dyn 237:1736–1745 [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Burton PM, Krone C, Finnerty JR (2007) Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: embryogenesis, regeneration, and two forms of asexual fission. Invertebr Biol 126:99–112 [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PR, Technau U (2010) A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc Natl Acad Sci U S A 107: 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch AM, Palakodeti D, Lu YC, Horowitz M, Graveley BR (2012) Transcriptome analysis reveals strain-specific and conserved stemness genes in Schmidtea mediterranea. PLoS One 7:e34447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Bermudez A, Miller D (2009) In vitro culture of cells derived from larvae of the staghorn coral Acropora millepora. Coral Reefs 28:859–864 [Google Scholar]
- Rinkevich B (1996) Do reproduction and regeneration in damaged corals compete for energy allocation? Mar Ecol Prog Ser 143:297–302 [Google Scholar]
- Ryan JF, Baxevanis AD (2007) Hox, Wnt, and the evolution of the primary body axis: insights from the early-divergent phyla. Biol Direct 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A (2000) Regeneration in the metazoans: why does it happen? Bioessays 22:578–590 [DOI] [PubMed] [Google Scholar]
- Schmid V, Alder H (1984) Isolated, mononucleated, striated muscle can undergo pluripotent transdifferentiation and form a complex regenerate. Cell 38:801–809 [DOI] [PubMed] [Google Scholar]
- Schmid V, Reber-Muller S (1995) Transdifferentiation of isolated striated muscle of jellyfish: the initiation process. Semin Cell Biol 6:109–116 [DOI] [PubMed] [Google Scholar]
- Schmid V, Stidwill R, Bally A, Marcum B, Tardent P (1981) Heat dissociation and maceration of marine Cnidaria. Roux’s Arch Dev Biol 190:143–149 [DOI] [PubMed] [Google Scholar]
- Schmid V, Wydler M, Alder H (1982) Transdifferentiation and regeneration in vitro. Dev Biol 92:476–488 [DOI] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V (2004) The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol 48:1–7 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Sawada Y, Sugiyama T (1993) Minimum tissue size required for hydra regeneration. Dev Biol 155:287–296 [DOI] [PubMed] [Google Scholar]
- Shinzato C, Iguchi A, Hayward DC, Technau U, Ball EE, Miller DJ (2008) Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol Biol 8:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer I (1971) Tentacular and oral-disc regeneration in sea anemone, Aiptasia diaphana.3. Autoradiographic analysis of patterns of tritiated thymidine uptake. J Embryol Exp Morpholog 26: 253–270 [PubMed] [Google Scholar]
- Steele RE, David CN, Technau U (2011) A genomic view of 500 million years of cnidarian evolution. Trends Genet 27:7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S (1963) Regeneration of whole polyps from ectodermal fragments of scyphistoma larvae of Aurelia aurita. Biol Bull 124:337–343 [Google Scholar]
- Stockard C III (1908). Studies of tissue growth An experimental study of the rate of regeneration in Cassiopea xamachana (Bigelow). Publication 103, Carnegie Institution of Washington; 61–102 [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- Technau U, Steele RE (2011) Evolutionary crossroads in developmental biology: Cnidaria. Development 138:1447–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Edwards N (1991) Cnidaria: Hydrozoa In: Harrison F, Westfall J (eds) Microscopic anatomy of invertebrates. Volume 2: Placozoa, Porifera, Cnidaria, and Ctenophora; Wiley-Liss, New York [Google Scholar]
- Trevino M, Stefanik DJ, Rodriguez R, Harmon S, Burton PM (2011) Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis. Dev Dyn 240:2673–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Shibata B, Blankenship TN (2011) Ultrastructure of the mesoglea of the sea anemone Nematostella vectensis (Edwardsiidae). Invertebr Biol 130:11–24 [Google Scholar]
- Vagelli A (2007) New observations on the asexual reproduction of Aurelia aurita (Cnidaria, Scyphozoa) with comments on its life cycle and adaptive significance. Invertebr Zool 4:111–127 [Google Scholar]
- Van Lieshout JS, Martin VJ (1992) Development of planuloid buds of Cassiopea xamachana (Cnidaria: Scyphozoa). Trans Am Microsc Soc 111:89–110 [Google Scholar]
- Vargas-Ángel B, Peters E (2007) Cellular reactions to sedimentation and temperature stress in the Caribbean coral Montastraea cavernosa. J Invertebr Pathol 95:140–145 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Hoang VT, Mättner R, Holstein TW (2009) Immortality and the base of multicellular life: lessons from cnidarian stem cells. Semin Cell Dev Biol 20:1114–1125 [DOI] [PubMed] [Google Scholar]
- West DL (1978) The epitheliomuscular cell of hydra: its fine structure, three-dimensional architecture and relation to morphogenesis. Tissue Cell 10:629–646 [DOI] [PubMed] [Google Scholar]
- Westfall JA (1966) The differentiation of nematocysts and associated structures in the cnidaria. Z Zellforsch Mikrosk Anat 75:381–403 [DOI] [PubMed] [Google Scholar]
- Wheeler BM, Heimberg AM, Moy VN, Sperling EA, Holstein TW, Heber S, Peterson KJ (2009) The deep evolution of metazoan microRNAs. Evol Dev 11:50–68 [DOI] [PubMed] [Google Scholar]
- Wood R (1959) Intercellular attachment in the epithelium of Hydra as revealed by electron microscopy. J Biophys Biochem Cytol 6:343–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work TM, Aeby GS (2010) Wound repair in Montipora capitata. J Invertebr Pathol 105:116–119 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Fujisawa T, Hwang JS, Ikeo K, Gojobori T (2006) Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene 385:64–70 [DOI] [PubMed] [Google Scholar]
- Yuan D, Nakanishi N, Jacobs DK, Hartenstein V (2008) Embryonic development and metamorphosis of the scyphozoan Aurelia. Dev Genes Evol 218:525–539 [DOI] [PubMed] [Google Scholar]


