Abstract
Objective
Vibrio parahaemolyticus is a major diarrhoea-inducing pathogen in coastal areas. In this study, we analysed the pathogenic characteristics of and variation in V. parahaemolyticus isolated from acute diarrhoeal patients in seven hospitals in different areas of southeastern China from 2013 to 2017.
Methods
The fecal specimens of patients with acute diarrhoea were collected. The routine microbiological test procedure combining with MALDI Biotyper microbial identification system was carried out to identify the V. parahaemolyticus. Serum agglutination tests, PCR for the detection of virulence-related genes and the Kirby-Bauer method to test for antimicrobial sensitivity were performed.
Results
From 2013 to 2017 in southeastern China, a total of 1220 V. parahaemolyticus strains were isolated from 16,504 stool specimens collected from acute diarrhoeal patients, and the annual isolation rate fluctuated between 6.1% and 8.7%. In total, 96.7% of the V. parahaemolyticus isolates were isolated in summer and autumn, mainly in people aged 18–44. Fifty-nine serotypes were identified, and the agglutination rate of the O antigen was 98.5%. From 2014 to 2016, the dominant serotype was O3:K6, while in 2013 and 2017, it was O4:KUT. The serotypes of O3:K6, O4:KUT, O4:K8, O3:KUT, O10:K60, O1:KUT and O1:K36 appeared every year from 2013 to 2017. O4:K6 and OUT:K6 began to appear after 2014 and 2015, respectively. A total of 49.5% of the strains belonged to the pandemic group, which consisted of 26 serotypes. Most isolates were sensitive to common antibiotics, excluding ampicillin.
Conclusion
V. parahaemolyticus is still present at a high level in southeastern China. Although the pandemic O3:K6 serotype is predominant, new serotypes continue to emerge, especially the O4:KUT serotype, which exceeded O3:K6 in prevalence in some years. Long-term surveillance is necessary to prevent the outbreak or transmission of this pathogen.
Keywords: Acute diarrhoea, Vibrio parahaemolyticus, serotypes, virulence genes, antimicrobial resistance
Introduction
Diarrhoea was the eighth leading cause of death in the world in 2016, accounting for more than 1.6 million deaths.1 Vibrio parahaemolyticus, which is naturally present in coastal waters, is the leading bacterial cause of gastroenteritis associated with seafood consumption. In Southeast Asian countries, such as China, Thailand and Japan, V. parahaemolyticus is the major pathogens for foodborne illnesses.2–5 Even in the United States and Europe, V. parahaemolyticus outbreaks are frequently reported.6–8
V. parahaemolyticus is a kind of microorganism with high genetic diversity. Serological typing, pulsed field gel electrophoresis, multilocus sequence typing, and whole genome sequencing are commonly used in tracing the origin and genetic variation of V. parahaemolyticus. Serological typing is the most common method. V. parahaemolyticus can be differentiated by serotyping by the O serogroups and K serotypes. At present, there are 11 O and 69 K antigen serotypes in clinical diagnosis. In February 1996, a food poisoning event caused by a new O3:K6 serotype in India, the clone quickly spread world and became the pandemic clone.9 Group-specific PCR (GS-PCR) method can distinguish the O3:K6 strains isolated before and after 1996.9 The pandemic clone of V. parahaemolyticus had the following characteristics: tdh+, trh−, toxRS/new+ (a unique toxRS sequence detectable by GS-PCR) and orf8+/− (the orf8 sequence of f237 phage),9,10 which were found in at least 20 serotypes, including O3:K6, O4:K68, O4:K8, O1:K25 and O1:KUT (K untypeable).11 The emergence of these pandemic characteristics in other serotype strains was understood to be the result of serotype transformation.
The virulence factors of V. parahaemolyticus include thermostable direct haemolysin (TDH), TDH-related haemolysin (TRH), which can form 2 nm pores on the cell membrane, allowing water and ions to enter freely, thus making the red blood cells have osmotic dissolution and hemolytic activity.12,13 Type III secretion systems (T3SSs) are a needle like structure composed of 20–30 proteins. Their function is to secrete and inject virulence factors, cause apoptosis and produce cytotoxic effects.14 T3SS1 genes were present in all V. parahaemolyticus strains. T3SS2α genes were present and co-regulate with tdh genes, whereas T3SS2β genes were generally only found in trh strains.15–17
Southeastern China, located on the west bank of the Pacific Ocean, is a place where there are frequent outbreaks of V. parahaemolyticus. We have been monitoring the pathogen spectrum of patients with acute diarrhoea in this area since 2009. V. parahaemolyticus ranked the second most prevalent bacterial pathogen, following diarrhoeagenic Escherichia coli (DEC).18 The fluctuation of serotype, genotype and drug sensitivity of V. parahaemolyticus were also observed. In this study, we analysed the pathogenic characteristics of and variation in V. parahaemolyticus isolated from acute diarrhoeal patients in southeastern China from 2013 to 2017 in an effort to discover the epidemic trend to prevent V. parahaemolyticus outbreaks in the future.
Materials and Methods
Ethics
The study protocol was approved by the Ethics Committees of the First Affiliated Hospital, College of Medicine, Zhejiang University and all participants provided written informed consent, including children, whose parent or guardian provided informed consent on their behalf. The study was carried out in accordance with the principles of the Declaration of Helsinki.
Sample Collection and Microbiological Analysis
Stool specimens were collected from acute diarrhoeal patients (three or more watery or loose stools a day with a duration of no more than 14 days) in 6 general hospitals and 1 children’s hospital in Zhejiang Province from 2013 to 2017. Clinical information regarding the patients was collected by filling out the information questionnaire of acute diarrhoeal cases. Stool specimens were cultured in thiosulfate citrate bile salt sucrose agar culture medium after alkaline peptone water (APW) enrichment to isolate microorganisms.19 V. parahaemolyticus was identified by MALDI Biotyper microbial identification system (Bruker, Germany) according to the instructions.
Serotyping
V. parahaemolyticus isolates were serotyped using agglutination tests with 11 O (lipopolysaccharide) and 65 K (capsule) antisera (Denka Seiken Ltd., Japan) according to the instructions provided with the reagents. Serotypes were defined as a unique combination of O and K. OUT was O untypeable and KUT was K untypeable.
Detection of Virulence-Associated Genes
The species-specific marker (tlh) and haemolysin genes (tdh and trh) were tested by real-time PCR.20,21 Pandemic markers (toxRS/new and orf8) were detected by monoplex PCR.10 The type III secretion system 1 (T3SS1) genes VP1670 (vscP), VP1686 (copS), VP1689 (vscK) and VP1694 (vscF), the T3SS2α genes VP1362 (vopB2), VP1339 (vscC2), VP1335 (vscS2) and VP1327 (vopT), and the T3SS2β genes (vscC2, vopB2, vopC and vscS2) were detected by four-multiplex PCR.22 The pathogenic group was defined as tdh+ or trh+ and the non-pathogenic group was tdh−trh−. The pandemic group was defined as tdh+, trh–, toxRS/new+ and orf8+/– and all other isolates were assigned to the non-pandemic group.23
Antimicrobial Susceptibility Testing
The Kirby-Bauer method was used for in vitro antimicrobial sensitivity testing using 18 conventional antibiotics:24,25 ampicillin, piperacillin/tazobactam, piperacillin, cefazolin, cefuroxime, ceftazidime, cefotaxime, cefepime, cefoxitin, imipenem, meropenem, amikacin, gentamycin, ciprofloxacin, levofloxacin, trimethoprim/sulfamethoxazole, tetracycline, and chloramphenicol. The interpretation of susceptibility, mediation and drug resistance were determined according referred to the latest standard recommended by M45-Ed3E standards formulated by Clinical and Laboratory Standards Institute.26
Statistical Analysis
Statistical analyses were performed using the statistical package for social sciences version 18 (SPSS Inc., USA). For statistical comparisons, the Chi-square test or Fisher’s exact test was used for proportions, and t tests were used for means. All p-values were two-sided, and P <0·05 was considered statistically significant.
Results
Epidemiology
From 2013 to 2017 in southeastern China, 1220 (7.4%) V. parahaemolyticus strains were isolated from 16,504 stool specimens collected from acute diarrhoeal patients. The annual isolation rate fluctuated between 6.1% and 8.7%, and the results are shown in Table 1.
Table 1.
Detection and Distribution of Vibrio parahaemolyticus in Patients with Acute Diarrhoea in Southeastern China from 2013 to 2017
| Year | Samples | Positive | Rate % (No. of Positive/Samples) |
|---|---|---|---|
| 2013 | 3348 | 204 | 6.1 |
| 2014 | 2892 | 205 | 7.1 |
| 2015 | 1874 | 142 | 7.6 |
| 2016 | 3735 | 266 | 7.1 |
| 2017 | 4655 | 403 | 8.7 |
A total of 96.7% (1179/1220) of the V. parahaemolyticus isolates were isolated in summer and autumn, 69.6% in people aged 18–44, and there was no difference between males and females. The demographic characteristics of the patients with acute diarrhoea are listed in Table 2.
Table 2.
Demographic Characteristics of the Patients with Acute Diarrhoea from Which Vibrio parahaemolyticus Was Isolated in Southeastern China from 2013 to 2017
| Variable and Category | Total Cases | Vibrio Parahaemolyticus | P | |
|---|---|---|---|---|
| Cases (%) | Isolation Rate (%) | |||
| Gender | 0.982* | |||
| Male | 7889 | 596(48.9) | 7.6 | |
| Female | 7836 | 592(48.5) | 7.6 | |
| Unknown | 789 | 32(2.6) | 4.1 | |
| Age | <0.001* | |||
| ≤4 | 3953 | 12(1.0) | 0.3 | |
| 5–17 | 938 | 64(5.2) | 6.8 | |
| 18–44 | 6758 | 849(69.6) | 12.6 | |
| 45–59 | 1919 | 176(14.4) | 9.2 | |
| ≥60 | 2147 | 87(7.1) | 4.1 | |
| Unknown | 789 | 32(2.6) | 4.1 | |
| Time | <0.001 | |||
| March-May | 2308 | 40(3.3) | 1.7 | |
| June-August | 10,182 | 917(75.2) | 9.0 | |
| September-November | 3450 | 262(21.5) | 7.6 | |
| December-February | 564 | 1(0.1) | 0.2 | |
Notes: *The P value does not include cases of unknown sex and age.
Serotypes
Among the 1220 V. parahaemolyticus isolates, fifty-nine serotypes were identified. The agglutination rate of the O antigen was 98.5% (1202/1220), of which O3 was the highest, accounting for 49.0% (598/1220). The agglutination rate of the K antigen was 59.0% (720/1220) and the highest was K6, accounting for 45.7% (557/1220). O3:K6 was the most prevalent serotype, with 546 strains (44.8%), followed by O4:KUT (415 strains, 34.0%), O4:K8 (40 strains, 3.3%), O3:KUT (30 strains, 2.5%) and O10:K60 (25 strains, 2.0%). The serotype distribution of V. parahaemolyticus is shown in Table 3.
Table 3.
Distribution of Serotypes of Vibro parahaemolyticus Isolates from Patients with Acute Diarrhoea in Southeastern China from 2013 to 2017
| O Serotypes (No. of Isolates*) |
Serotype (No. of Isolates) | ||||
|---|---|---|---|---|---|
| 2013 (n=204) | 2014 (n=205) | 2015 (n=142) | 2016 (n=266) | 2017 (n=403) | |
| O1(42) | O1:K36(4) O1:K56(1) O1:K63(1) O1:K9(1) O1:KUT†(8) |
O1:K36(1) O1:KUT(1) |
O1:K17(1) O1:K36(1) O1:K41(1) O1:KUT(2) |
O1:K25(2) O1:K36(2) O1:K56(1) O1:KUT(5) |
O1:K1(1) O1:K36(1) O1:K4(2) O1:K5(1) O1:K8(1) O1:KUT(4) |
| O2(9) | O2:K3(2) | – | O2:K3(1) | O2:K3(2) | O2:K28(3) O2:K3(1) |
| O3(598) | O3:K21(1) O3:K29(4) O3:K57(2) O3:K6(52) O3:KUT(3) |
O3:K29(4) O3:K46(1) O3:K53(1) O3:K57(2) O3:K6(110) O3:KUT(1) |
O3:K6(91) O3:K68(1) O3:KUT(1) |
O3:K54(1) O3:K6(141) O3:KUT(4) |
O3:K1(2) O3:K19(1) O3:K48(1) O3:K5(1) O3:K6(152) O3:KUT(21) |
| O4(491) | O4:K13(5) O4:K4(1) O4:K60(1) O4:K68(1) O4:K8(12) O4:K9(2) O4:KUT(89) |
O4:K13(1) O4:K4(1) O4:K42(1) O4:K48(1) O4:K60(1) O4:K68(1) O4:K8(4) O4:K9(2) O4:KUT(56) |
O4:K25(1) O4:K44(1) O4:K48(1) O4:K6(1) O4:K8(6) O4:KUT(25) |
O4:K55(1) O4:K6(1) O4:K60(1) O4:K68(1) O4:K8(4) O4:KUT(80) |
O4:K10(1) O4:K33(1) O4:K4(1) O4:K53(2) O4:K6(4) O4:K8(14) O4:KUT(165) |
| O5(14) | O5:K17(1) O5:KUT(2) |
O5:K52(1) O5:KUT(3) |
O5:K6(1) O5:K60(1) |
O5:KUT(2) | O5:K17(1) O5:KUT(2) |
| O6(3) | – | O6:KUT(1) | O6:K18(1) | O6:K18(1) | – |
| O8(6) | O8:K41(2) O8:KUT(1) |
O8:K41(1) | – | – | O8:K41(1) O8:KUT(1) |
| O10(32) | O10:K60(5) | O10:K60(5) O10:K8(1) O10:KUT(1) |
O10:K60(2) | O10:K60(7) O10:KUT(2) |
O10:K60(6) O10:K8(2) O10:KUT(1) |
| O11(7) | O11:KUT(2) | O11:KUT(2) | O11:KUT(2) | O11:KUT(1) | – |
| OUT† (18) | OUT:K59(1) | OUT:KUT(1) | OUT:KUT(1) | OUT:K6(3) OUT:KUT(4) |
OUT:K38(1) OUT:K6(1) OUT:KUT(6) |
Notes: *No. of isolates per serotype; †UT, untypeable; - indicates that this serotype was not detected in this year.
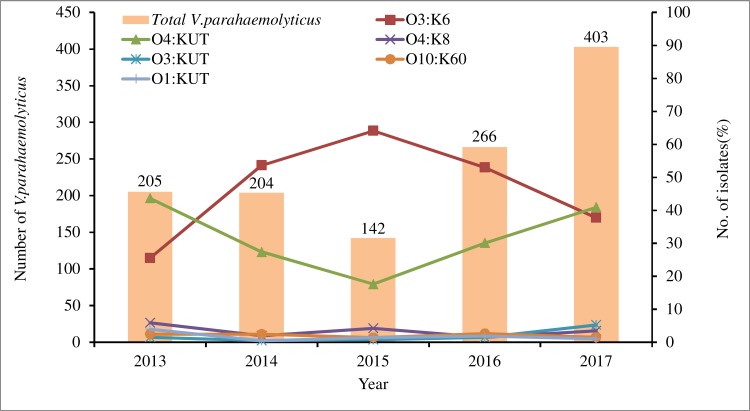
There were differences in serotype distribution between 2013 and 2017 (Table 3, Figure 1). From 2014 to 2016, the dominant serotype was O3:K6, accounting for 53.7%, 64.1% and 53.0% of the strains in those years, respectively. The O4:KUT serotype was predominant in 2013 and 2017, accounting for 43.6% and 40.9% respectively. The serotypes O3:K6, O4:KUT, O4:K8, O3:KUT, O10:K60, O1:KUT and O1:K36 appeared every year from 2013 to 2017. O4:K6 and OUT:K6 serotypes began to appear after 2014 and 2015 respectively, while O3:K29, O4:K13, O3:K57 and O4:K9 serotypes have not been detected since 2013.
Figure 1.
Distribution of the main Vibrio parahaemolyticus serotypes from patients with acute diarrhoea in southeastern China from 2013 to 2017.
Distribution of Virulence-Associated Genes
A total of 1169 pathogenic strains (95.8%, 1169/1220) were detected, of which 1150 strains (94.3%) were tdh+trh−, 13 strains were tdh+trh+ and 6 strains were tdh−trh+. Fifty-one strains (4.2%) did not carry pathogenic virulence genes (tdh−trh−). From 2013 to 2017, tdh+trh− pathogenic strains were dominant, accounting for more than 90%. There was no significant difference between different years (P=0.878), and the results are shown in Table 4. In 1169 pathogenic strains, there were 51 serotypes. In O3:K6 serotype isolates, 97.5% (14/557) were pathogenic strains, while the rate of pathogenic strains was 99.5% (413/415) for the O4:KUT serotype isolates. There were 27 serotypes of 51 non-pathogenic strains, of which O1:KUT and O5:KUT were the most abundant serotypes, with 5 strains (5.9%) each.
Table 4.
Haemolysin Genes of Vibro parahaemolyticus Isolates from Patients with Acute Diarrhoea in Southeastern China from 2013 to 2017
| Year | Total (n=1220) | Haemolysin Genes (%) | |||
|---|---|---|---|---|---|
| tdh+trh− (n= 1150) | tdh+trh+ (n= 13) | tdh−trh+ (n=6) | tdh−trh− (n=51) | ||
| 2013 | 204 | 194(95.1) | 1(0.5) | 2(1.0) | 7(3.4) |
| 2014 | 205 | 192(93.7) | 1(0.5) | 2(1.0) | 10(4.9) |
| 2015 | 142 | 136(95.8) | 3(2.1) | 1(0.7) | 2(1.4) |
| 2016 | 266 | 250(94.0) | 3(1.1) | 0(0.0) | 13(4.9) |
| 2017 | 403 | 378(93.8) | 5(1.2) | 1(0.2) | 19(4.7) |
Notes: + positive, presence of the gene; − negative, absence of the gene.
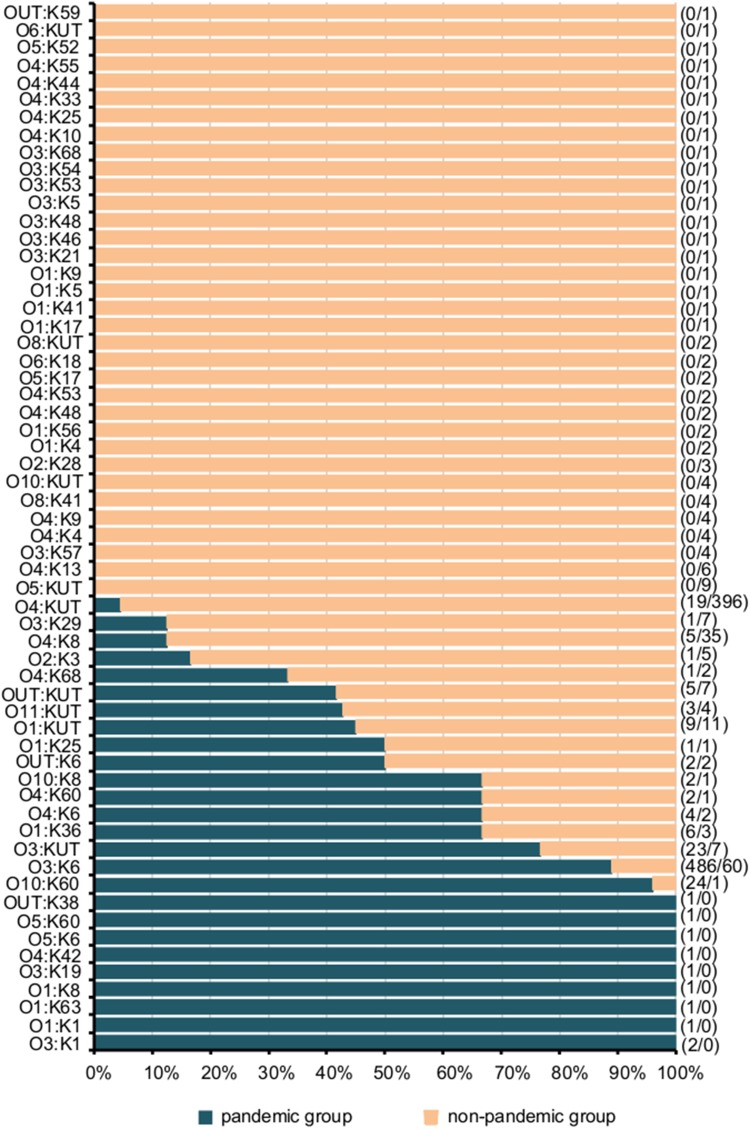
Among 1220 strains of V. parahaemolyticus, 49.5% (604/1220) were pandemic strains. The proportion of pandemic strains varied in different years (P<0.05), with pandemic strains predominating in 2015 and 2016, and non-pandemic strains predominating in 2013, 2014 and 2017 (Table 5). A total of 604 pandemic strains consisted of 26 serotypes, mainly O3:K6 (80.5%, 486/604), followed by O10:K60 (4.0%, 24/604) and O3:KUT (3.8%, 23/604), while 616 non-pandemic strains consisted of 51 serotypes, mainly O4:KUT (64.3%, 396/616), followed by O3:K6 (9.7%, 60/616) and O4:K8 (5.7%, 35/616), as shown in Figure 2. From 2013 to 2017, the proportions of O3:K6 in pandemic strains were 69.1%, 88.2%, 88.2%, 83.4% and 74.7%, respectively.
Table 5.
Distribution of Virulence Genes and Pandemic Markers in Vibrio parahaemolyticus Isolates from Patients with Acute Diarrhoea in Southeastern China from 2013 to 2017
| Haemolysin Genes | Pandemic Markers | 2013 (n=204) | 2014 (n=205) | 2015 (n=142) | 2016 (n=266) | 2017 (n=403) | |||
|---|---|---|---|---|---|---|---|---|---|
| tlh | tdh | trh | toxRS/new | orf8 | |||||
| Pandemic (49.5%, 604/1220) | 68(33.3) | 93(45.4) | 85(59.9) | 160(60.2) | 198(49.1) | ||||
| + | + | – | + | + | 60(29.4) | 70(34.1) | 76(53.5) | 131(49.2) | 182(45.2) |
| + | + | – | + | – | 8(3.9) | 23(11.2) | 9(6.3) | 29(10.9) | 16(4.0) |
| Non-pandemic (50.5%, 616/1220) | 136(66.7) | 112(54.6) | 57(40.1) | 106(39.8) | 205(50.9) | ||||
| + | + | - | - | - | 124(60.8) | 82(40.0) | 41(28.9) | 85(32.0) | 177(43.9) |
| + | + | - | - | + | 2(1.0) | 17(8.3) | 10(7.0) | 5(1.9) | 3(0.7) |
| + | + | + | + | + | 1(0.5) | 0(0.0) | 0(0.0) | 1(0.4) | 1(0.2) |
| + | + | + | + | - | 0(0.0) | 0(0.0) | 0(0.0) | 1(0.4) | 0(0.0) |
| + | + | + | - | - | 0(0.0) | 1(0.5) | 3(2.1) | 1(0.4) | 4(1.0) |
| + | - | - | + | - | 0(0.0) | 0(0.0) | 0(0.0) | 4(1.5) | 2(0.5) |
| + | - | - | - | - | 7(3.4) | 10(4.9) | 1(0.7) | 9(3.4) | 17(4.2) |
| + | - | - | - | + | 0(0.0) | 0(0.0) | 1(0.7) | 0(0.0) | 0(0.0) |
| + | - | + | - | - | 2(1.0) | 2(1.0) | 1(0.7) | 0(0.0) | 1(0.2) |
Figure 2.
Distribution of different serotypes of Vibrio parahaemolyticus isolates in pandemic and non-pandemic strains. The number on the left side of the “/” in the parenthesis on the right side of the figure is the number of pandemic strains, and the number on the right side is the number of non-pandemic strains.
A total of 1128 strains (92.5%, 1128/1220) of V. parahaemolyticus carried the four T3SS1 genes. Among the tdh+ strains, 95.1% (1106/1163) carried the four T3SS2α genes. All four T3SS2α genes were negative in three strains. Ten of 19 trh+ strains carried the four T3SS2β genes. No tdh−trh− strain carried all four T3SS2α or T3SS2β genes. From 2013 to 2017 years, the positive rates of the four T3SS1 genes were 88.7%, 86.3%, 97.2%, 91.0% and 96.8% respectively (p=0.000). Among the tdh+ strains, 97.4%, 94.3%, 88.0%, 98.0% and 95.8% respectively carried the four T3SS2α genes (p=0.001). The results of type III secretion system related genes are shown in Table 6.
Table 6.
Distribution of T3SS Genes Among Vibrio parahaemolyticus Strains Isolated from Patients with Acute Diarrhoea in Southeastern China During 2013–2017
| Genes | Total (n=1220) | 2013 (n=204) | 2014 (n=205) | 2015 (n=142) | 2016 (n=266) | 2017 (n=403) | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tdh | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – | – |
| trh | – | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – | – | + | + | – |
| n | 1150 | 13 | 6 | 51 | 194 | 1 | 2 | 7 | 192 | 1 | 2 | 10 | 136 | 3 | 1 | 2 | 250 | 3 | 0 | 13 | 378 | 5 | 1 | 19 |
| T3SS1 | ||||||||||||||||||||||||
| VP1670[vscP] | 1144 | 13 | 6 | 51 | 192 | 1 | 2 | 7 | 191 | 1 | 2 | 10 | 136 | 3 | 1 | 2 | 248 | 3 | 0 | 13 | 377 | 5 | 1 | 19 |
| VP1686[vopS] | 1102 | 12 | 6 | 44 | 183 | 1 | 2 | 6 | 171 | 1 | 2 | 9 | 136 | 3 | 1 | 2 | 239 | 2 | 0 | 9 | 373 | 5 | 1 | 18 |
| VP1689[vscK] | 1129 | 13 | 6 | 48 | 189 | 1 | 2 | 6 | 179 | 1 | 2 | 10 | 136 | 3 | 1 | 2 | 248 | 3 | 0 | 12 | 377 | 5 | 1 | 18 |
| VP1694[vscF] | 1114 | 12 | 5 | 36 | 183 | 1 | 2 | 5 | 182 | 1 | 2 | 7 | 133 | 3 | 0 | 2 | 245 | 2 | 0 | 7 | 371 | 5 | 1 | 15 |
| All four genes | 1078 | 12 | 5 | 33 | 173 | 1 | 2 | 5 | 167 | 1 | 2 | 7 | 133 | 3 | 0 | 2 | 236 | 2 | 0 | 4 | 369 | 5 | 1 | 15 |
| T3SS2α | ||||||||||||||||||||||||
| VP1362[vopB2] | 1120 | 4 | 2 | 0 | 190 | 1 | 1 | 0 | 182 | 0 | 0 | 0 | 125 | 0 | 0 | 0 | 247 | 2 | 0 | 0 | 376 | 1 | 1 | 0 |
| VP1339[vscC2] | 1137 | 4 | 2 | 0 | 192 | 1 | 1 | 0 | 192 | 0 | 0 | 0 | 135 | 0 | 0 | 0 | 249 | 2 | 0 | 0 | 369 | 1 | 1 | 0 |
| VP1335[vscS2] | 1143 | 4 | 2 | 0 | 192 | 1 | 1 | 0 | 191 | 0 | 0 | 0 | 136 | 0 | 0 | 0 | 249 | 2 | 0 | 0 | 375 | 1 | 1 | 0 |
| VP1327[vopT] | 1132 | 4 | 2 | 0 | 192 | 1 | 1 | 0 | 183 | 0 | 0 | 0 | 136 | 0 | 0 | 0 | 248 | 2 | 0 | 0 | 373 | 1 | 1 | 0 |
| All four genes | 1108 | 4 | 2 | 0 | 189 | 1 | 1 | 0 | 182 | 0 | 0 | 0 | 125 | 0 | 0 | 0 | 246 | 2 | 0 | 0 | 366 | 1 | 1 | 0 |
| T3SS2β | ||||||||||||||||||||||||
| vscC2 | 0 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| vopB2 | 0 | 13 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 1 | 2 |
| vopC | 0 | 11 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 1 |
| vscS2 | 0 | 13 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 1 | 2 |
| All four genes | 0 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
Antimicrobial Profile
Most isolates were resistant to ampicillin, and the resistance rate fluctuated between 79.6% and 92.7% during 2013–2017. Nearly half of the isolates exhibited intermediate levels of susceptibility to cefuroxime (24.6~64.1%) and cefazolin (28.4~65.5%). More than 95% of isolates were sensitive to other antibacterial agents including fluoroquinolones, aminoglycosides, tetracyclines, trimethoprim-sulphamethoxazole, third-generation and fourth-generation cephalosporins, β-lactamase inhibitor combinations during 2013–2017. No isolate was resistance to carbapenems in five years. The antimicrobial susceptibilities of V. parahaemolyticus fluctuated with years, and the results are shown in Table 7.
Table 7.
Antimicrobial Susceptibility Testing of 1220 Vibrio Parahaemolyticus Strains Isolated from Patients with Acute Diarrhoea During 2013–2017
| Antimicrobial Agent | Total (n=1220) | 2013 (n=204) | 2014 (n=205) | 2015 (n=142) | 2016 (n=266) | 2017 (n=403) | P* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | R (%) | S (%) | ||
| Ampicillin (AMP) | 1079(88.5) | 31(2.5) | 177(86.8) | 5(2.5) | 190(92.7) | 5(2.4) | 113(79.6) | 10(7.0) | 235(88.3) | 7(2.6) | 364(90.3) | 4(1.0) | 0.002 |
| Piperacillin/tazobactam (TZP) | 5(0.4) | 1212(99.4) | 0(0.0) | 204(100.0) | 0(0.0) | 205(100.0) | 0(0.0) | 142(100.0) | 0(0.0) | 266(100.0) | 5(1.2) | 395(98.0) | 0.096 |
| Piperacillin (PRL) | 26(2.1) | 1144(93.8) | 4(2.0) | 188(92.2) | 0(0.0) | 194(94.6) | 3(2.1) | 127(89.4) | 2(0.8) | 259(97.4) | 17(4.2) | 376(93.3) | 0.002 |
| Cefazolin (KZ) | 88(7.2) | 639(52.4) | 5(2.5) | 147(72.1) | 12(5.9) | 77(37.6) | 25(17.6) | 26(18.3) | 15(5.6) | 116(43.6) | 31(7.7) | 273(67.7) | <0.001 |
| Cefuroxime (CXM) | 42(3.4) | 542(44.5) | 1(0.5) | 145(71.1) | 12(5.9) | 66(32.2) | 8(5.6) | 41(28.9) | 4(1.5) | 125(47.0) | 17(4.2) | 165(40.9) | 0.002 |
| Ceftazidime (CAZ) | 12(1.0) | 1202(98.5) | 0(0.0) | 203(99.5) | 0(0.0) | 202(98.5) | 2(1.4) | 139(97.9) | 1(0.4) | 265(99.6) | 9(2.2) | 393(97.5) | 0.018 |
| Cefotaxime (CTX) | 18(1.5) | 1145(93.8) | 4(2.0) | 195(95.6) | 1(0.5) | 183(89.3) | 3(2.1) | 131(92.3) | 1(0.4) | 259(97.4) | 9(2.2) | 377(93.5) | 0.164 |
| Cefepime (FEP) | 11(0.9) | 857(70.2) | 0(0.0) | 158(77.5) | 0(0.0) | 116(56.6) | 0(0.0) | 78(54.9) | 1(0.4) | 187(70.3) | 10(2.5) | 318(78.9) | 0.004 |
| Cefoxitin (FOX) | 22(1.8) | 1119(91.7) | 4(2.0) | 198(97.1) | 1(0.5) | 178(86.8) | 2(1.4) | 114(80.3) | 1(0.4) | 258(97.0) | 14(3.5) | 371(92.1) | 0.021 |
| Imipenem (IPM) | 0(0.0) | 1220(100.0) | 0(0.0) | 204(100.0) | 0(0.0) | 205(100.0) | 0(0.0) | 142(100.0) | 0(0.0) | 266(100.0) | 0(0.0) | 403(100.0) | – |
| Meropenem (MEM) | 0(0.0) | 1220(100.0) | 0(0.0) | 204(100.0) | 0(0.0) | 205(100.0) | 0(0.0) | 142(100.0) | 0(0.0) | 266(100.0) | 0(0.0) | 403(100.0) | – |
| Amikacin (AK) | 9(0.7) | 966(79.2) | 1(0.5) | 176(86.3) | 1(0.5) | 150(73.2) | 1(0.7) | 93(65.5) | 1(0.4) | 201(75.6) | 5(1.2) | 346(85.9) | 0.815 |
| Gentamycin (CN) | 8(0.7) | 1188(97.3) | 0(0.0) | 203(99.5) | 0(0.0) | 204(99.5) | 0(0.0) | 139(97.9) | 1(0.4) | 258(97.0) | 7(1.7) | 384(95.3) | 0.054 |
| Ciprofloxacin (CIP) | 15(1.2) | 1173(96.2) | 0(0.0) | 203(99.5) | 0(0.0) | 199(97.1) | 3(2.1) | 132(93.0) | 2(0.8) | 262(98.5) | 10(2.5) | 377(93.5) | 0.013 |
| Levofloxacin (LEV) | 11(0.9) | 1207(98.9) | 0(0.0) | 204(100.0) | 0(0.0) | 205(100.0) | 2(1.4) | 140(98.6) | 1(0.4) | 265(99.6) | 8(2.0) | 393(97.5) | 0.035 |
| Trimethoprim/ sulfamethoxazole (SXT) | 35(2.9) | 1179(96.6) | 2(1.0) | 202(99.0) | 2(1.0) | 203(99.0) | 6(4.2) | 136(95.8) | 3(1.1) | 262(98.5) | 22(5.5) | 376(93.3) | 0.001 |
| Tetracycline (TE) | 37(3.0) | 1177(96.5) | 4(2.0) | 200(98.0) | 0(0.0) | 204(99.5) | 5(3.5) | 135(95.1) | 5(1.9) | 261(98.1) | 23(5.7) | 377(93.5) | <0.001 |
| Chloramphenicol (C) | 14(1.1) | 1195(98.0) | 0(0.0) | 203(99.5) | 0(0.0) | 205(100.0) | 3(2.1) | 135(95.1) | 0(0.0) | 265(99.6) | 11(2.7) | 387(96.0) | 0.001 |
Note: P*, difference in resistance rates.
Discussion
V. parahaemolyticus is a common pathogen that induces diarrhoea. In our study, the annual separation rate of V. parahaemolyticus in acute diarrhoeal patients was between 6.1% and 8.7%, which was close to the 8.1% isolation rate in this region between 2009 and 2013,27 and the 6.0% isolation rate reported in southern China between 2007 and 2012.28 However, the rate was higher compared with the 4.5% rate in Shanghai between 2012 and 2016,29 and the 5.1% rate in northwestern Mexico between 2004 and 2010.30 The difference may be related to geographical location and local eating habits, or even to sample selection and laboratory testing. In our study, 96.7% of the V. parahaemolyticus isolates were isolated between June and November, mainly in people aged 18–44. This result may be attributed to the optimum growth temperature of V. parahaemolyticus and the fact that young adults are more likely to eat in restaurants, thus increasing the risk of raw or inadequately cooked seafood. In early summer the temperature is 15°C or higher, and V. parahaemolyticus is released into the water column from the sediments where it survived during winter.31
Serotyping the 1220 V. parahaemolyticus isolates revealed that the agglutination rate of the O antigen was 98.5% and that of the K antigen was 59.0% which was much lower than any previously recorded rate.27 O3:K6 was the main prevalent serotype, with 546 strains (44.8%), followed by O4:KUT (415/1220, 34.0%) and O4:K8 (40/1220, 3.3%). Since 1996, a new O3:K6 strain appeared in Kolkata, India.32 The O3:K6 strain rapidly spread throughout in the world in subsequent years and became a pandemic strain in clinical V. parahaemolyticus isolates.7,9,10,33,34 It is worth noting that from 2013 to 2017 in southeastern China, the detection rate of the O3:K6 serotype was reduced and the second dominant serotype O4:KUT reached up to 34.0%. Even in 2013 and 2017, O4:KUT surpassed O3:K6 to become the dominant serotype, accounting for 43.6% and 40.9% respectively. In addition, O4:K6 and OUT:K6 serotypes began to appear after 2014 and 2015 respectively, while O3:K29, O4:K13, O3:K57 and O4:K9 serotypes have not been detected since 2013. These changes in the serotypes of V. parahaemolyticus may be related to environmental changes, such as rising water temperature and acidification of seawater.11
Among the 1220 V. parahaemolyticus isolates, pathogenic strains (tdh+ or trh+) accounted for 95.8% and pandemic strains (tdh+, trh−, toxRS/new+) accounted for 49.5% of all strains. Non-pandemic strains were predominant in 2013, 2014 and 2017. This is a newly discovered phenomenon. The main reason was that 95.4% (396/415) of the isolates of the O4:KUT serotype which was the first dominant serotype in 2013 and 2017 did not have the toxRS/new+ characteristic of the pandemic strain. Although O4:KUT is not a pandemic serovariant, its clinical prevalence has increased and the clinical symptoms of patients infected with O4:KUT were similar to those infected with O3:K6. Whether the O4:KUT serotype strain has other pathogenic factors remains to be further studied.
T3SS is associated with the pathogenicity of V. parahaemolyticus. The T3SS1 genes are present in all V. parahaemolyticus isolates. The T3SS2α genes co-regulated with the tdh and T3SS2β genes are closely related to the trh gene.15,17 The results of this study were consistent with previous results.27 All 1220 V. parahaemolyticus isolates had at least one of the T3SS1 genes and no tdh−trh− strain carrying T3SS2α or T3SS2β gene was detected.
In this study, V. parahaemolyticus isolates were usually generally to common antibiotics, except ampicillin. A total of 1079 isolates (88.5%) showed a resistance to ampicillin. Similar results were observed in isolates from seafood.35 However, the resistance rates of amikacin and cefuroxime in this study in Zhejiang Province were 0.7% and 3.4%, which were inconsistent with the resistance rates of clinical V. parahaemolyticus isolates in Shanghai at 90.5% and 92.6%.36,37 The reason for this difference might be that the Shanghai samples came from only one hospital and clustered events might have been included, while our samples came from 7 hospitals in different regions and the results were more reliable. Although V. parahaemolyticus remained highly sensitive to fluoroquinolone (such as ciprofloxacin and levofloxacin), which were commonly used to treat infectious diarrhea in clinic, a trend of fluoroquinolone resistance had emerged. Changes in drug-resistance rate were also observed in other species in the Vibrionaceae family. According to surveillance findings in Nepal, resistance patterns in Vibrio cholerae were shown dramatic fluctuations in resistance to routinely used antibiotics. Resistance to ampicillin decreased from 93% in 2006 to 18% by 2010 and again raised to 100% by 2016. Ciprofloxacin and tetracycline resistance emerged in 2007, reached a peak during 2010–2012 and declined to 0 by 2016.38 In addition, Aeromonas spp., which is one of the main pathogens of infectious diarrhea, was uniformly resistant to penicillin, cephalosporins or carbapenems due to the production of various β-lactamases. From 2011 to 2017, Aeromonas demonstrated the high resistance rates to ceftriaxone (29.4%), ceftazidime (28.9%), cefepime (22.2%), ciprofloxacin (27.3%) and trimethoprim-sulfamethoxazole (45%), respectively, in southwest China.39 In a 10-year retrospective survey in Hungary, the resistance trends of Aeromonas were showed.40 The highest resistance levels overall was observed for ceftriaxone (20%), sulphamethoxazole/trimethoprim (18%), ciprofloxacin (14%) and cefepime (13.0%). Meropenem resistance was also above 10%, while resistance rates against doxycycline, tigecycline and gentamicin were around or lower than 5%. There was a significant increase in the number of resistant isolates corresponding to ceftriaxone, sulphamethoxazole/trimethoprim and meropenem. The variation in the antimicrobial resistance may be linked to how heavily antimicrobial drugs are used. In recent years, the consumption of antibiotics in aquaculture has increased globally to prevent or treat related diseases, leading to an increase in resistance rate of related bacteria, such as V. parahaemolyticus and Aeromonas. It is obvious that the pace of antibiotic drug discovery cannot keep up with the continuous and detrimental changes in resistance trends. Therefore, it is important to preserve the drugs that we currently have (through the development of rapid and sensitive diagnostic tools to ensure their prudent use, and antibiotic stewardship practices), in addition to facilitating the development of new ideal antibacterial drugs.41
Conclusions
In summary, V. parahaemolyticus was present at high levels in southeastern China from 2013 to 2017. Although the pandemic O3:K6 serotype was predominant, new serotypes continue to emerge, especially the O4:KUT serotype, which exceeded O3:K6 in prevalence in some years. Fluctuations in the virulence gene distribution and drug sensitivity of these isolates were obvious. Long-term surveillance is necessary to prevent the outbreak or transmission of this pathogen.
Acknowledgments
We thank Sijia Liu of the First Affiliated Hospital, College of Medicine, Zhejiang University, for his opinion on revising this manuscript.
Funding Statement
This work was supported by grants from the National Key Programs for Infectious Diseases of China (2017ZX10103008001 and 2017ZX10103008007) and the National Nature Science Foundation of China (81802067).
Disclosure
The authors declare no competing interests in this study.
References
- 1.Troeger C, Blacker BF, Khalil IA, GBD. 2016 Diarrhoeal disease collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the global burden of disease study 2016. Lancet Infect Dis. 2018;18(11):1211–1228. doi: 10.1016/S1473-3099(18)30362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiou CS, Hsu SY, Chiu SI, et al. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J Clin Microbiol. 2000;38(12):4621–4625. doi: 10.1128/JCM.38.12.4621-4625.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vuddhakul V, Chowdhury A, Laohaprertthisan V, et al. Isolation of a pandemic O3:K6 Clone of a Vibrio parahaemolyticus Strain from environmental and clinical sources in Thailand. Appl Environ Microbiol. 2000;66(6):2685–2689. doi: 10.1128/AEM.66.6.2685-2689.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubota K, Kasuga F, Iwasaki E, et al. Estimating the burden of acute gastroenteritis and foodborne illness caused by Campylobacter, Salmonella, and Vibrio parahaemolyticus by using population-based telephone survey data, Miyagi Prefecture, Japan, 2005 to 2006. J Food Protect. 2011;74(10):1592–1598. doi: 10.4315/0362-028X.JFP-10-387 [DOI] [PubMed] [Google Scholar]
- 5.Yan WX, Dai Y, Zhou YJ, et al. Risk factors for sporadic Vibrio parahaemolyticus gastroenteritis in east China: a matched case-control study. Epidemiol Infect. 2015;143(5):1020–1028. doi: 10.1017/S0950268814001599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels NA, MacKinnon L, Bishop R, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis. 2000;181(5):1661–1666. doi: 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Urtaza J, Simental L, Velasco D, et al. Pandemic Vibrio parahaemolyticus O3:K6, Europe. Emerg Infect Dis. 2005;11(8):1319–1320. doi: 10.3201/eid1108.050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scallan E, Hoekstra RM, Angulo FJ, et al. Foodborne illness acquired in the United States major pathogens. Emerg Infect Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto C, Okuda J, Ishibashi M, et al. Pandemic spread of an O3: k6clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38(2):578. doi: 10.1128/JCM.38.2.578-585.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laohaprertthisan V, Chowdhury A, Kongmuang U, et al. Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in southern Thailand. Epidemiol Infect. 2003;130(3):395–406. doi: 10.1017/S0950268803008458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nair GB, Ramamurthy T, Bhattacharya SK, et al. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20(1):39. doi: 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda S, Kodama T, Okada N, et al. Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect Immun. 2010;78(2):603–610. doi: 10.1128/IAI.00946-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raghunath P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front Microbiol. 2015;5(805):805. doi: 10.3389/fmicb.2014.00805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park KS, Ono T, Rokuda M, et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72(11):6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhoopong P, Palittapongarnpim P, Pomwised R, et al. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J Clin Microbiol. 2007;45(5):1544–1550. doi: 10.1128/JCM.02371-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meador CE, Parsons MM, Bopp CA, et al. Virulence gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J Clin Microbiol. 2007;45(4):1133–1139. doi: 10.1128/JCM.00042-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noriea NFI, Johnson CN, Griffitt KJ, et al. Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J Appl Microbiol. 2010;109(3):953–962. doi: 10.1111/j.1365-2672.2010.04722.x [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Chen Y, Yang Q, et al. Plesiomonas shigelloides infection in Southeast China. PLoS One. 2013;8(11):e77877. doi: 10.1371/journal.pone.0077877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen JH, Pfaller MA. Manual of Clinical Microbiology 2015. Washington, DC: ASM press; 2015. [Google Scholar]
- 20.West CKG, Klein SL, Lovell CR. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microbiol. 2013;9(7):2247–2252. doi: 10.1128/AEM.03792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson WB, Turner JW. The thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus: sequence variation and implications for detection and function. J Microbiol Methods. 2016;126:1–7. doi: 10.1016/j.mimet.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 22.Jones JL, Lüdeke CH, Bowers JC, et al. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol. 2012;50(7):2343–2352. doi: 10.1128/JCM.00196-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De JHL, Leonsicairos N, Velazquezroman J, et al. A pandemic Vibrio parahaemolyticus O3: k6clone causing most associated diarrhea cases in the Pacific Northwest coast of Mexico. Front Microbiol. 2015;6(221):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 25.Gajdács M, Ábrók M, Lázár A, et al. Comparative epidemiology and resistance trends of common urinary pathogens in a tertiary-care hospital: a 10-year surveillance study. Medicina (Kaunas). 2019a;55(7):E356. doi: 10.3390/medicina55070356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. Methods for Antimicrobial Dilution and Dusk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria Third Edition. CLSI Document M45. Waye, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 27.Chen Y, Chen X, Yu F, et al. Serology, virulence, antimicrobial susceptibility and molecular characteristics of clinical Vibrio parahaemolyticus strains circulating in southeastern China from 2009 to 2013. Clin Microbiol Infect. 2016;22(3):258. doi: 10.1016/j.cmi.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Xie X, Shi X, et al. Vibrio parahaemolyticus, southern coastal region of China, 2007–2012. Emerg Infect Dis. 2014;20(4):685–688. doi: 10.3201/eid2004.130744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong XH, Wu HY, Li J, et al. Epidemiology, aetiology and seasonality of infectious diarrhoea in adult outpatients through active surveillance in Shanghai, China, 2012–2016: a cross-sectional study. BMJ Open. 2018;8(9):e019699. doi: 10.1136/bmjopen-2017-019699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velazquez-Roman J, Leon-Sicairos N, Flores-Villasenor H, et al. Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl Environ Microbiol. 2012;78(6):1794–1803. doi: 10.1128/AEM.06953-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko T, Colwell RR. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J Bacteriol. 1973;113:24–32. doi: 10.1128/JB.113.1.24-32.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda J, Ishibashi M, Hayakawa E, et al. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35(12):3150–3155. doi: 10.1128/JCM.35.12.3150-3155.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ansaruzzaman M, Lucas M, Deen JL, et al. Pandemic Serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus Associated With Diarrhea In Mozambique: spread Of The Pandemic into the African continent. J Clin Microbiol. 2005;43(6):2559–2562. doi: 10.1128/JCM.43.6.2559-2562.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velazquez-Roman J, León-Sicairos N, de Jesus Hernández-díaz L, et al. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front Cell Infect Microbiol. 2014;3:110. doi: 10.3389/fcimb.2013.00110.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Chu Y, Xie G, et al. Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int J Food Microbiol. 2019;290:116–124. doi: 10.1016/j.ijfoodmicro.2018.10.005 [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Jiang Y, Yang L, et al. Molecular and conventional analysis of acute diarrheal isolates identifies epidemiological trends, antibiotic resistance and virulence profiles of common enteropathogens in Shanghai. Front Microbiol. 2018;9:164. doi: 10.3389/fmicb.2018.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Tang R, Lou Y, et al. A comprehensive epidemiological research for clinical Vibrio parahaemolyticus in Shanghai. Front Microbiol. 2017;8:1043. doi: 10.3389/fmicb.2017.01043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rijal N, Acharya J, Adhikari S, et al. Changing epidemiology and antimicrobial resistance in Vibrio cholerae: AMR surveillance findings (2006–2016) from Nepal. BMC Infect Dis. 2019;19(1):801. doi: 10.1186/s12879-019-4432-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang S, He T, Sun J, et al. Distinct antimicrobial resistance profiling of clinically important Aeromonas spp. In Southwest China: a seven-year surveillance study. Infect Drug Resist. 2019;12:2971–2978. doi: 10.2147/IDR.S216926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gajdács M. Resistance trends and epidemiology of Aeromonas and Plesiomonas infections (RETEPAPI): a 10-year retrospective survey. Infect Dis. 2019b;51(9):710–713. doi: 10.1080/23744235.2019.1640389 [DOI] [PubMed] [Google Scholar]
- 41.Gajdács M. The concept of an ideal antibiotic: implications for drug design. Molecules. 2019c;24(5):E892. doi: 10.3390/molecules24050892 [DOI] [PMC free article] [PubMed] [Google Scholar]