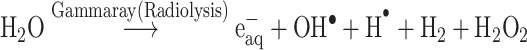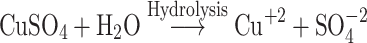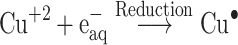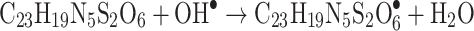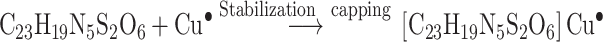Abstract
Aim
With the rapid emergence of antibiotic resistance, efforts are being made to obtain new selective antimicrobial agents. Hybridization between quinazolinone and benzenesulfonamide can provide new antimicrobial candidates. Also, the use of nanoparticles can help boost drug efficacy and lower side effects.
Materials and Methods
Novel quinazolinone-benzenesulfonamide derivatives 5–18 were synthesized and screened for their antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria, MRSA and yeast. The most potent compound 16 was conjugated with copper oxide nanoparticles 16-CuONPs by gamma irradiation (4.5 KGy). Characterization was performed using UV–Visible, TEM examination, XRD patterns and DLS. Moreover, compound 16 was used to synthesize two nanoformulations: 16-CNPs by loading 16 in chitosan nanoparticles and the nanocomposites 16-CuONPs-CNPs. Characterization of these nanoformulations was performed using TEM and zeta potential. Besides, the inhibitory profile against Staphylococcus aureus DNA gyrase was assayed. Cytotoxic evaluation of 16, 16-CNPs and 16-CuONPs-CNPs on normal VERO cell line was carried out to determine its relative safety. Molecular docking of 16 was performed inside the active site of S. aureus DNA gyrase.
Results
Compound 16 was the most active in this series against all the tested strains and showed inhibition zones and MICs in the ranges of 25–36 mm and 0.31–5.0 µg/mL, respectively. The antimicrobial screening of the synthesized nanoformulations revealed that 16-CuONPs-CNPs displayed the most potent activity. The MBCs of 16 and the nanoformulations were measured and proved their bactericidal mode of action. The inhibitory profile against S. aureus DNA gyrase showed IC50 ranging from 10.57 to 27.32 µM. Cytotoxic evaluation of 16, 16-CNPs and 16-CuONPs-CNPs against normal VERO cell lines proved its relative safety (IC50= 927, 543 and 637 µg/mL, respectively). Molecular docking of 16 inside the active site of S. aureus DNA gyrase showed that it binds in the same manner as that of the co-crystallized ligand, ciprofloxacin.
Conclusion
Compound 16 could be considered as a new antimicrobial lead candidate with enhanced activity upon nanoformulation.
Keywords: quinazolinone, benzenesulfonamide, antimicrobial, nanoparticles, nanoformulations, MBC, Staphylococcus aureus DNA gyrase, docking
Introduction
The exploration of new antimicrobial agents is essentially required in order to defeat the rapid emergence of drug resistance and toxicity.1–4 The misuse and overconsumption of antibiotics (AB) have led to the evolution of resistant strains. An example of major concern is the development of AB-resistance in S. aureus (MRSA), which is frequently linked to hospital and community-acquired infections.5,6
Quinazolinone is a privileged structure that possesses activity as anticancer, antiviral, antibacterial, antifungal, anti-tubercular and herbicidal.7–10 Some quinazolinone derivatives act as chemo-sensitizers of antibiotic activity to Enterobacter aerogenes, Klebsiella pneumoniae, Pseudomonas aeruginosa resistant strains,11 Escherichia Coli, Staphylococcus aureus, Bacillus subtilis and pathogenic fungi as Saccharomyces cerevisiae and Candida albicans.12 Other alkylamino quinazolinone derivatives were reported to restore AB-activity in Gram-negative isolates.13 Devi et al reported a series of new aza isatin derivatives containing quinazolinones (A) as potent antimicrobial agents.14,15 Suresha and his coworkers revealed a series of urea, thiourea, acetamide and sulfonamide derivatives of quinazolinone (B and C) as potent antibacterial agents.16 Al-Omary et al described some new 2-alkyl thio-quinazolinones to act as nonclassical antifolates (D).17 Also, Dinakaran et al proved that the substitution at position 3 of quinazolin-4(3H)-ones is associated with antimicrobial properties (Figure 1).18 On the other hand, sulfonamides retain its antimicrobial activity by manifesting the antagonistic properties of p-aminobenzoic acid and blocking the SH, NH2-containing enzymes and proteins.19,20
Figure 1.
Quinazolinone-based antimicrobial agents and ciprofloxacin.
The use of nanoparticles (NPs) has been widely spread in the pharmaceutical field due to the following advantages; the size and surface of the drug can be easily manipulated, it is feasible to sustain the drug release in the gastrointestinal tract, and to boost drug efficacy with lowering the side effects by controlling the clearance of the drug from the body.21 Chitosan (CS), a natural polysaccharide, is the most frequently used polymer in drug delivery systems, due to its biological, biodegradable and antibacterial properties.22 The antibacterial activity of some drugs can be effectively enhanced by its incorporation into chitosan NPs (CNPs). CNPs are active against E. coli and B. subtilis.23 It is indicated that its antibacterial activity is due to the penetration of the cell membrane causing the effusion of the cytoplasm and cell death.24 The presence of chelating sites (NH2 and OH) in chitosan structure aids in chemical reactions as metal ion sorption through electrostatic attraction and ion exchange for metal anions in acidic solutions forming the nanocomposites, which are promising NPs for reducing the bacteria-associated infections.25,26 Furthermore, the metal NPs exert the antibacterial effect by their small size and high surface to volume ratio, which allow them to closely adhere to the microbial membranes.27,28 An example of such metal NPs is copper oxide NPs (CuONPs), characterized by their low toxicity, heat resistance and broad-spectrum activity against Gram-positive bacteria, Gram-negative bacteria, MRSA and Candida albicans.25,29
Hybridization has proven to be beneficial for the preparation of new antimicrobial agents and overcoming the drawbacks of the conventionally used drugs.4,30 So, sulfonamides were integrated with quinazolin-4(3H)-ones in order to obtain more potent candidates. As a continuation of our previous efforts,12,31-35 we synthesized new quinazolinone derivatives bearing benzenesulfonamide in order to study their antimicrobial activity. The synthesized compounds were tested against Gram-positive bacteria, Gram-negative bacteria, MRSA and Candida. The most potent compound was conjugated to copper oxide nanoparticles (CuONPs) (by the aid of gamma irradiation) and incorporated into two nanoformulations; chitosan nanoparticles (CNPs) and copper oxide nanoparticles incorporated into chitosan, in order to detect the variation in antimicrobial activity. Also, an in-vitro enzyme assay was performed for the most potent compound and the nanoformulations against S. aureus DNA gyrase. DNA gyrase is considered a fundamental type II DNA topoisomerase that maneuvers DNA topology by making temporary double-strand breaks and DNA strand passage.36 It is considered as an important drug target due to its vital role in bacterial survival.37 An example of DNA gyrase poisons of commercial importance is ciprofloxacin (Figure 1). Furthermore, the cytotoxicity of the most potent compound and the nanoformulations were evaluated against VERO (African green monkey kidney epithelial cells) normal cell line. Molecular docking was used to confirm the possible binding interactions of this compound into the active site of S. aureus DNA gyrase to assert the biological activity results.
Materials and Methods
Chemistry
Uncorrected Melting points were determined on a Gallen Kamp apparatus (Sanyo Gallen Kamp, UK). Thin-layer chromatography (TLC) was performed on precoated silica gel plates (Kieselgel 0.25 mm, 60 F254, Merck, Germany) with a solvent system of chloroform/methanol (8:2). The IR spectra used to determine the functional group within the molecule were recorded using an FT-IR spectrophotometer (Perkin Elmer, USA). NMR spectra used for structural determination of molecules using the 1H and 13C atoms were scanned on an NMR spectrophotometer (Bruker AXS Inc., Switzerland), operating at 500 MHz for 1H and 125.76 MHz for 13C. Chemical shifts were expressed in δ-values (ppm) relative to TMS, using DMSO-d6 as a solvent. Mass spectra showing fragmentation patterns were characterized by their mass to charge ratios (m/z) and relative abundances and were recorded on an ISQ LT Thermo Scientific GCMS model (Massachusetts, USA). Elemental analyses were used to determine the quantity of a particular element within the molecule and were performed on a model 2400 CHNSO analyzer (Perkin Elmer, USA). All the values were within ± 0.4% of the theoretical values.
4-(2-Mercapto-4- oxoquinazolin-3(4H)-yl) Benzenesulfonamide (4)38
General Procedure for the Synthesis of 2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-substituted acetamide 5–18
A mixture of 4 (3.33 g, 0.01 mol) and 2-chloro-N-substituted acetamide (0.01 mol) in dry acetone (30 mL) and anhydrous K2CO3 (1.38 g, 0.01 mol) was stirred at room temperature for 12 hrs, filtered and the solid obtained was crystallized from ethanol to give 5–18.
2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-phenylacetamide (5)
5: Yield, 87%; m.p. 269.0°C. IR: 3447, 3319, 3211 (NH2, NH), 3098 (arom.), 2926, 2837 (aliph.), 1701, 1680 (2CO), 1612 (CN), 1334, 1161 (SO2). 1HNMR: 4.13 (s, 2H), 7.06–7.10 (m, 1H), 7.31–7.40 (m, 4H), 7.48 (dd, 1H, J= 6 & 2.5 Hz), 7.59–7.69 (m, 2H), 7.71 (d, 2H, J=8 Hz, AB), 7.84 (dd, 1H, J=6 & 2.5 Hz), 8.03 (d, 2H, J=8 Hz, AB), 8.08 (s, 2H), 10.40 (s, 1H). 13CNMR: 39.47, 119.62 (3), 126.64 (3), 127.36 (2), 129.26 (3), 130.58 (2), 139.40 (3), 147.59 (2), 156.29, 161.17, 166.39. MS m/z (%): 466 (M+) (19.43), 388 (100). Anal. Calcd. for C22H18N4O4S2 (466.53): C, 56.64; H, 3.89; N, 12.01. Found: C, 56.34; H, 3.57; N, 11.79.
2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-2-tolylacetamide (6)
6: Yield, 80%; m.p. 251.5°C. IR: 3410, 3377, 3220 (NH2, NH), 3097 (arom.), 2978, 2912 (aliph.), 1701, 1683 (2CO), 1621 (CN), 1399, 1161 (SO2). 1HNMR: 2.20 (s, 3H), 4.16 (s, 2H), 7.08 (ddd, 1H, J= 8 & 6 Hz) 7.15 (ddd, 1H, J= 8 & 6 Hz), 7.20 (dd, 1H, J= 6 & 1.5 Hz), 7.36 (dd, 1H, J= 8 & 1.5 Hz), 7.50 (dd, 1H, J= 7.5 & 2.5 Hz), 7.63–7.69 (m, 2H), 7.87 (d, 2H, J= 7.5 Hz, AB), 8.01 (dd, 1H, J= 7.5 & 2.5 Hz), 8.10 (d, 2H, J= 7.5 Hz, AB), 8.11 (s, 2H), 9.81 (s, 1H). 13CNMR: 18.32, 37.07, 125.42 (3), 125.88 (2), 126.43 (2), 127.11, 127.30, 130.50 (2), 132.33 (2), 135.57 (2), 136.56 (2), 147.62, 156.79, 161.10, 166.09. MS m/z (%): 480 (M+) (24.21), 331 (100). Anal. Calcd. for C23H20N4O4S2 (480.56): C, 57.48; H, 4.19; N, 11.66. Found: C, 57.69; H, 4.44; N, 11.92.
2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-3-tolylacetamide (7)
7: Yield, 69%; m.p. 262.4°C. IR: 3427, 3313, 3188 (NH2, NH), 3053 (arom.), 2956, 2922 (aliph.), 1681, 1654 (2CO), 1612 (CN), 1332, 1165 (SO2). 1HNMR: 2.24 (s, 3H), 4.11 (s, 2H), 7.10 (ddd, 1H, J= 8 & 2 Hz), 7.47 (dd, 1H, J= 8 & 6.5 Hz), 7.48–7.61 (m, 5H), 7.76 (d, 2H, J= 7 Hz, AB), 7.85 (dd, 1H, J= 6 & 1.5 Hz), 8.03 (d, 2H, J= 7 Hz, AB), 8.10 (s, 2H), 10.21 (s, 1H). 13CNMR: 20.90, 37.80, 119.62 (3), 127.47 (3), 129.64 (3), 130.84 (3), 136.05 (2), 138.67 (2), 139.15, 146.82, 158.70, 160.32, 165.72. MS m/z (%): 480 (M+) (41.81), 373 (100). Anal. Calcd. for C23H20N4O4S2 (480.56): C, 57.48; H, 4.19; N, 11.66. Found: C, 57.21; H, 4.01; N, 11.35.
2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-4-tolylacetamide (8)
8: Yield, 91%; m.p 315.5°C. IR: 3410, 3376, 3200 (NH2, NH), 3076 (arom.), 2966, 2871 (aliph.), 1686, 1661 (2CO), 1611 (CN), 1376, 1156 (SO2). 1HNMR: 2.08 (s, 3H), 4.30 (s, 2H), 7.13–7.34 (m, 4H), 7.47–7.71 (m, 3H), 7.86 (d, 2H, J= 6.5 Hz, AB), 7.87 (dd, 1H, J= 6 & 2 Hz), 7.94 (d, 2H, J= 6.5 Hz, AB), 8.13 (s, 2H), 10.04 (s, 1H). 13CNMR: 22.13, 31.18, 117.92 (3), 122.38 (3), 126.53 (2), 126.91 (2), 130.44 (3), 134.67 (3), 142.83, 145.05, 150.96, 162.24, 175.02. MS m/z (%): 480 (M+) (7.12), 91 (100). Anal. Calcd. for C23H20N4O4S2 (480.56): C, 57.48; H, 4.19; N, 11.66. Found: C, 57.73; H, 4.31; N, 12.02.
N-(2-Ethylphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (9)
9: Yield, 68%; m.p. 170.5°C. IR: 3387, 3255, 3186 (NH2, NH), 3066 (arom.), 2966, 2927 (aliph.), 1681, 1656 (2CO), 1606 (CN), 1336, 1163 (SO2). 1HNMR: 1.06 (t, 3H, J= 10 Hz), 2.58 (q, 2H, J= 9.5 Hz), 4.16 (s, 2H), 7.15 (dd, 1H, J= 6 & 1.5 Hz), 7.22 (ddd, 1H, J= 8 & 6 Hz), 7.32–7.60 (m, 3H), 7.65–7.75 (m, 2H), 7.76 (d, 2H, J= 8.5 Hz, AB), 7.88 (dd, 1H, J= 6 & 2.5 Hz), 8.04 (d, 2H, J= 8.5 Hz, AB), 8.11 (s, 2H), 9.63 (s, 1H). 13CNMR: 14.64, 24.13, 36.99, 120.52 (3), 126.40 (2), 126.71 (2), 127.10 (2), 127.46, 128.92 (2), 130.84 (3), 139.09 (2), 146.78, 158.60, 161.11, 166.3. MS m/z (%): 494 (M+) (8.62), 148 (100). Anal. Calcd. for C24H22N4O4S2 (494.59): C, 58.28; H, 4.48; N, 11.33. Found: C, 58.51; H, 4.69; N, 11.60.
N-(3-Ethylphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (10)
10: Yield, 73%; m.p. 197.6°C. IR: 3321, 3245, 3220 (NH2, NH), 3064 (arom.), 2973, 2837 (aliph.), 1689, 1662 (2CO), 1610 (CN), 1373, 1161 (SO2). 1HNMR: 1.17 (t, 3H, J= 8.5 Hz), 2.59 (q, 2H, J= 8 Hz), 4.15 (s, 2H), 6.91 (ddd, 1H, J= 7 & 1.5 Hz), 7.22 (dd, 1H, J= 7 & 6 Hz), 7.43–7.50 (m, 2H), 7.60–7.64 (m, 3H), 7.75 (d, 2H, J= 7.5 Hz, AB), 7.84 (dd, 1H, J= 5.5 & 2 Hz), 8.07 (d, 2H, J= 7.5 Hz, AB), 8.11 (s, 2H), 10.29 (s, 1H). 13CNMR: 15.95, 28.71, 37.83, 117.11, 117.30, 119.01, 119.19, 119.97, 123.49, 123.84, 126.48, 126.65, 127.51, 129.14, 129.20, 130.83, 135.55, 138.96, 139.11, 144.83, 145.94, 156.57, 161.08, 165.03. MS m/z (%): 494 (M+) (32.18), 155 (100). Anal. Calcd. for C24H22N4O4S2 (494.59): C, 58.28; H, 4.48; N, 11.33. Found: C, 57.98; H, 4.21; N, 11.02.
N-(4-Ethylphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (11)
11: Yield, 89%; m.p. 259.4°C. IR: 3446, 3383, 3213 (NH2, NH), 3062 (arom.), 2960, 2920 (aliph.), 1701, 1658 (2CO), 1606 (CN), 1336, 1161 (SO2). 1HNMR: 1.16 (t, 3H, J= 8.5 Hz), 2.52 (q, 2H, J= 8.0 Hz), 4.12 (s, 2H), 7.13 (d, 2H, J= 9 Hz, AB), 7.15–7.50 (m, 3H), 7.58–7.71 (m, 2H), 7.83 (d, 2H, J= 8 Hz, AB), 8.02 (dd, 1H, J= 6.5 Hz, J= 2 Hz), 8.08 (d, 2H, J= 8 Hz, AB), 8.09 (s, 2H), 10.36 (s, 1H). 13CNMR: 16.16, 28.05, 37.79, 119.68, 119.95 (4), 126.46, 126.64, 127.09, 127.39 (2), 128.45 (2), 130.67, 135.58, 137.10, 138.72, 146.78, 147.58, 156.69, 161.08, 165.78. MS m/z (%): 494 (M+) (63.34), 118 (100). Anal. Calcd. for C24H22N4O4S2 (494.59): C, 58.28; H, 4.48; N, 11.33. Found: C, 58.52; H, 4.64; N, 11.64.
N-(4-Methoxyphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (12)
12: Yield, 83%; m.p. 267.3°C. IR: 3367, 3313, 3291 (NH2, NH), 3089 (arom.), 2972, 2844 (aliph.), 1694, 1668 (2CO), 1610 (CN), 1334, 1161 (SO2). 1HNMR: 3.71 (s, 3H), 4.11 (s, 2H), 6.88 (dd, 2H, J= 7 Hz, AB), 7.49 (dd, 2H, J= 7 Hz, AB), 7.51–7.62 (m, 3H), 7.75 (d, 2H, J= 6.5 Hz, AB), 7.85 (dd, 1H, J= 8.5 & 2 Hz, AB), 8.04 (d, 2H, J= 6.5 Hz, AB), 8.10 (s, 2H), 10.24 (s, 1H). 13CNMR: 27.91, 55.62, 114.38 (2), 119.95, 126.47 (2), 126.67 (2), 127.10 (2), 127.48, 130.85 (2), 132.49 (2), 135.61, 139.11, 145.89, 155.84, 156.59, 161.08, 165.47. MS m/z (%): 496 (M+) (15.66), 339 (100). Anal. Calcd. for C23H20N4O5S2 (496.56): C, 55.63; H, 4.06; N, 11.28. Found: C, 55.92; H, 4.29; N, 11.53.
N-(4-Ethoxyphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (13)
13: Yield, 90%; m.p. 262.4°C. IR: 3425, 3317, 3270 (NH2, NH), 3100 (arom.), 2976, 2833 (aliph.), 1682, 1654 (2CO), 1618 (CN), 1396, 1163 (SO2). 1HNMR: 1.30 (t, 3H, J= 5 Hz), 3.97 (s, 2H), 4.11 (q, 2H, J= 4.5 Hz), 6.86 (d, 2H, J= 6.5 Hz, AB), 7.48 (dd, 2H, J= 6.5 Hz, AB), 7.49–7.61 (m, 3H), 7.76 (d, 2H, J= 6 Hz, AB), 7.85 (dd, 1H, J= 7.5 & 1.5 Hz, AB), 8.04 (d, 2H, J= 6 Hz, AB), 8.10 (s, 2H), 10.24 (s, 1H). 13CNMR: 15.14, 31.15, 63.55, 114.89 (2), 119.95, 121.19 (2), 126.47 (2), 126.67, 127.10, 127.48, 130.84 (2), 132.38, 135.60, 139.10, 145.92, 147.58, 155.11, 156.58, 161.09, 165.46. MS m/z (%): 510 (M+) (19.47), 316 (100). Anal. Calcd. for C24H22N4O5S2 (510.59): C, 56.46; H, 4.34; N, 10.97. Found: C, 56.19; H, 4.10; N, 10.62.
N-(3,5-Dimethoxyphenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (14)
14: Yield, 92%; m.p. 283.7 °C. IR: 3450, 3366, 3170 (NH2, NH), 3097 (arom.), 2927, 2831 (aliph.), 1687, 1655 (2CO), 1602 (CN), 1336, 1159 (SO2). 1HNMR: 3.70 (s, 6H), 4.10 (s, 2H), 6.22 (dd, 1H, J= 2.5 & 1.5 Hz), 6.89 (dd, 2H, J= 2.5 & 1.5 Hz), 7.47 (dd, 1H, J= 5.5 & 2.5 Hz), 7.55–7.59 (m, 2H), 7.83 (d, 2H, J= 9 Hz, AB), 7.95 (dd, 1H, J= 5.5 & 2.5 Hz), 8.07 (m, 4H), 8.09 (s, 1H). 13CNMR: 37.83, 55.55 (2), 95.92, 97.95 (2), 119.96 (3), 126.46, 126.56, 127.04, 129.89 (2), 135.49 (2), 141.19 (2), 147.58, 157.02, 160.95 (2), 161.12, 166.25. MS m/z (%): 526 (M+) (2.94), 276 (100). Anal. Calcd. for C24H22N4O6S2 (526.58): C, 54.74; H, 4.21; N, 10.64. Found: C, 54.47; H, 4.01; N, 10.37.
2-((4-Oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(3,4,5-trimethoxyphenyl) acetamide (15)
15: Yield, 86%; m.p. 267.6°C. IR: 3315, 3224, 3113 (NH2, NH), 3072 (arom.), 2926, 2841 (aliph.), 1689, 1660 (2CO), 1610 (CN), 1342, 1165 (SO2). 1HNMR: 3.61 (s, 6H), 3.72 (s, 3H), 4.12 (s, 2H), 6.99 (d, 2H, J= 1.5 Hz), 7.49 (dd, 1H, J= 7.5 &2 Hz), 7.50–7.61 (m, 2H), 7.76 (d, 2H, J= 6 Hz, AB), 7.85 (dd, 1H, J= 7.5 & 2 Hz), 8.05 (d, 2H, J= 6 Hz, AB), 8.10 (s, 2H), 10.30 (s, 1H). 13CNMR: 31.17, 56.12 (2), 60.56, 97.20 (2), 119.96 (3), 126.49, 126.69, 127.11, 127.46 (2), 130.84, 133.91, 135.55, 135.60, 139.04, 146.02, 153.20 (2), 156.58, 161.06, 165.85. MS m/z (%): 556 (M+) (12.43), 360 (100). Anal. Calcd. for C25H24N4O7S2 (556.61): C, 53.95; H, 4.35; N, 10.07. Found: C, 53.58; H, 4.09; N, 9.84.
N-(2-Methyl-4-nitrophenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (16)
16: Yield, 76%; m.p. 255.8°C. IR: 3409, 3345, 3183 (NH2, NH), 3100 (arom.), 2972, 2848 (aliph.), 1680, 1647 (2CO), 1620 (CN), 1538, 1343 (NO2), 1398, 1163 (SO2). 1HNMR: 2.29 (s, 3H), 4.25 (s, 2H), 7.49 (dd, 1H, J= 7 Hz & 1 Hz), 7.51–7.58 (m, 2H), 7.76 (d, 2H, J= 6.5 Hz, AB), 7.83–8.10 (m, 6H), 8.11 (s, 2H), 10.23 (s, 1H). 13CNMR: 18.32, 37.33, 115.34, 117.49, 120.00, 121.78 (2), 122.32, 123.72, 124.96, 125.97, 129.80 (2), 131.76, 134.27, 135.36, 136.28, 142.81, 143.21, 147.54, 156.56, 161.69, 166.81. MS m/z (%): 525 (M+) (18.53), 118 (100). Anal. Calcd. for C23H19N5O6S2 (525.56): C, 52.56; H, 3.64; N, 13.33. Found: C, 52.85; H, 3.93; N, 13.64.
N-(2-Methyl-6-nitrophenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (17)
17: Yield, 80%; m.p. 246.5°C. IR: 3450, 3381, 3151 (NH2, NH), 3095 (arom.), 2922, 2858 (aliph.), 1695, 1658 (2CO), 1616 (CN), 1541, 1339 (NO2), 1342, 1161 (SO2). 1HNMR: 2.29 (s, 3H), 4.18 (s, 2H), 7.36 (dd, 1H, J= 7.5 & 6 Hz), 7.51 (dd, 1H, J= 7 & 1.5 Hz), 7.58 (dd, 1H, J= 7.5 & 3 Hz), 7.59–7.74 (m, 2H), 7.76 (d, 2H, J= 8.5 Hz, AB), 7.88 (dd, 1H, J= 7 & 1.5 Hz), 8.04 (d, 2H, J= 8.5 Hz, AB), 8.10 (dd, 1H, J= 6 & 3 Hz), 8.11 (s, 2H), 9.0 (s, 1H). 13CNMR: 18.35, 36.76, 119.94, 122.61 (2), 126.59 (2), 126.71 (2), 127.02, 127.49, 130.85 (2), 135.32 (2), 135.56 (2), 137.61, 139.07, 145.95, 156.33, 161.14, 166.31. MS m/z (%): 525 (M+) (20.56), 324 (100). Anal. Calcd. for C23H19N5O6S2 (525.56): C, 52.56; H, 3.64; N, 13.33. Found: C, 52.19; H, 3.28; N, 13.07.
N-(2,4-Dinitrophenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide (18)
18: Yield, 74%; m.p. 277.8°C. IR: 3428, 3361, 3313 (NH2, NH), 3100 (arom.), 2927, 2866 (aliph.), 1688, 1678 (2CO), 1606 (CN), 1537, 1345 (NO2), 1338, 1138 (SO2). 1HNMR: 4.18 (s, 2H), 7.46 (dd, 1H, J= 8 & 1.5 Hz), 7.61–7.72 (m, 2H), 7.73 (d, 2H, J= 8.5 Hz, AB), 7.82–8.04 (m, 3H), 8.06 (s, 2H), 8.10–8.62 (m, 3H), 9.91 (s, 1H). 13CNMR: 36.23, 119.10, 119.92, 120.29 (2), 121.07, 123.61, 124.80, 125.15, 128.82 (2), 129.40, 130.62, 133.71, 134.64, 136.07, 141.47, 142.72, 145.32, 160.61, 161.80, 167.92. MS m/z (%): 556 (M+) (32.82), 385 (100). Anal. Calcd. for C22H16N6O8S2 (556.53): C, 47.48; H, 2.90; N, 15.10. Found: C, 47.18; H, 2.63; N, 14.75.
Antimicrobial Activity
The in-vitro antimicrobial activity screening was accomplished at the bacteriology laboratory, Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt. The selected strains were; the Gram-positive Bacillus subtilis (ATCC 6633), Staphylococcus aureus (ATCC 29213), the Gram-negative Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), MRSA and Candida albicans (ATCC 10231). The antimicrobial potential of the target compounds was measured as the diameter of the inhibition zones using the agar plate diffusion method.39 Briefly, 100 µL of the microorganism was grown in 10 mL of fresh media until they reached a count of 108 cells/mL for bacteria or 105 cells/mL for fungi. The count of microbial suspension was determined by a serial dilution which is defined as a series of sequential dilutions used to reduce a dense culture of cells to a more usable concentration. Each dilution will reduce the concentration of bacteria by a specific amount. One mL of each compound (at 0.5 mg/mL) was added to each well (10 mm diameter holes cut in the agar gel). The plates were incubated for 24 hrs at 37°C (for bacteria) and 72 hrs at 27°C (for yeast), each test was imitated three times. Tetracycline, ciprofloxacin and vancomycin were used as standard antibacterial reference drugs, while amphotericin B was used as the antifungal. The inhibition zone diameter was measured in millimeters and used as a criterion for the antimicrobial activity. Solvent control (DMSO) was included in every experiment as a negative control. The conventional paper disk diffusion method was used in measuring the MIC of the targeted compounds by applying paper disk (266,812 W. Germany 12.7 mm in diameter). Bacteria and yeast were grown on nutrient and Sabouraud agar medium, respectively. The compounds were loaded on paper disks with different concentrations. Dried disks were loaded on the surface of agar plates inoculated with the tested organism. Growth inhibition was examined after 24 hrs at 37°C for bacteria & 72 hrs at 27°C for yeast. Each test was replicated three times.35,40-42
Synthesis and Characterization of 16-CuONPs Synthesized Using Gamma Irradiation
Gamma (γ) irradiation was performed at the National Center for Radiation Research and Technology (NCRRT), Cairo, Egypt, using 60Co-Gamma chamber 4000 A-India operating at a dose rate of 1.221 KGy/h. Compound 16 was conjugated with CuONPs by adding 16 to 4.0 mM of copper sulfate pentahydrate solution in a ratio of 1:5 (v/v) with 0.2% isopropanol as a free radical scavenger. The solution was irradiated at different doses of gamma radiation as 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5 and 5.0 kGy. Characterization of the 16-CuONPs was performed by UV–Visible spectrophotometer using a filtrate (contains 16 only) as a baseline blank. Size distribution and average particle size were determined by Dynamic Light Scattering DLS-PSS-NICOMP 380-ZLS particle sizing system (St. Barbara, California, USA). The size and morphology of 16-CuONPs were recorded using the TEM model JEOL electron microscopy JEM-100 CX. Drop coating 16-CuONPs prepared TEM studies onto carbon-coated TEM grids. X-Ray Diffraction patterns were obtained with The XRD-6000 series, including the residual austenite quantitation, stress analysis, crystallinity calculation, and crystallite size/lattice strain materials analysis by overlaying X-ray diffraction patterns using Cu-Ka target, and nickel filter Shimadzu Scientific Instruments (SSI, Tokyo, Japan).
Synthesis and Characterization of Compound 16 Nanoformulations (Chitosan Nanoparticles 16-CNPs and 16-CuONPs-CNPs)
CNPs were prepared by the ionotropic gelation method43 through the electrostatic interactions between the amine group of chitosan and the negatively charged group of Triphenyl phosphate (TPP) as a polyanion. CS was dissolved in di-distilled water (DDW) to form a stock solution of concentration 0.5% (w/v) containing 1.2% acetic acid. During the process, CS undergoes ionotropic gelation and form spherical particles that are distinguishable by the opalescence of the solution. The lyophilized compound (50 mg) was mixed with 100 mL of CS solution 0.1% (w/v) to prepare drug-loaded chitosan nanoparticles. TPP 0.1% (w/v) was added dropwise to the compound/CS mixture with stirring for 10 min. Then, the solution was centrifuged (25,000 × g, 25°C for 30 min). The 16-CuONPs-CNPs were synthesized by loading compound 16 and CuONPs into chitosan through encapsulation.
The Zeta Sizer (Malvern Instruments, Worcestershire, UK) is used to measure the particle size, size distribution (polydispersity index (PDI)), and zeta potential of nanoparticles. The mean particle size was approximated as the z-average diameter and the width of the distribution as the PDI. DLS measurements were taken at 25°C with a detection angle of 90°. All measurements were calculated as the mean ± standard deviation. Examination of the surface morphology and size distribution was performed by a transmission electron microscope (TEM) (JEOL electron microscopy JEM-100 CX). About 5 μL of the nanoparticle solution was placed on a copper grid and stained with 2% (w/v) phosphotungstic acid.
Evaluating Minimum Bactericidal Concentration (MBC)
MBC assay was conducted using the broth microdilution assay. The MBC was determined by plating 10 μL of culture volume from the MIC assay onto trypticase soy broth (TSB) plate and colony formation was examined after 24 hrs. at 37°C. MBC is defined as the lowest compound concentration resulting in a ≧ 3-log reduction in the number of colony-forming units (CFU).44
In vitro Inhibitory Activity Screening on Staphylococcus aureus DNA Gyrase
The supercoiling assay was carried out according to the manufacturer’s instructions.45 Briefly, set up a mix of assay buffer (6 μL of 5x buffer), relaxed pBR322 (0.5 μL) and water (17.5 μL) on ice. Add the aliquot 24 μL of the mix into tubes. Add 3 μL of DMSO to tubes 1 and 2. Add 3 μL of the tested compound to the other tubes then mix briefly. Add 3 μL of dilution buffer to tube 1. Dilute the enzyme in dilution buffer then add 3 μL of this to the remaining tubes. Mix and incubate for 30 mins at 37°C. Stop the reaction then centrifuge for 1 min. Load 20 μL of aqueous (upper blue) phase onto a 1% (w/v) agarose gel. Run at 75 V for approximately 2 hrs. Stain with 1 μg/mL ethidium bromide in water (15 mins), destains (5–10 mins) in water. Measurement of the concentration of the compound required for 50% inhibition of enzyme activity (IC50) was performed using GraphPad Prism 5 software and the results represent the mean of three independent measurements.
MTT Assay
The 96 well tissue culture plate was inoculated with 105 cells/mL (100 µL/well) and incubated at 37°C for 24 hrs. Two-folds dilutions of the tested sample were made in maintenance medium with 2% serum. A 0.1 mL of each dilution was tested. MTT solution was prepared (5mg/mL in phosphate-buffered saline (PBS)) (Bio Basic Canda Inc.) and 20 µL was added to each well. Incubate at 37 ºC and 5% CO2 for 1–5 h. Resuspend formazan in 200 µL DMSO. Optical density was measured at 560 nm and the IC50 was calculated using Graph-pad prism 5.
Molecular Docking
The docking study was performed using MOE 10.2008 software. The PDB file containing S. aureus DNA gyrase co-crystallized with ciprofloxacin was obtained from the protein data bank (PDB: 2XCT). Water molecules were ignored and hydrogen atoms were added. Alpha Site Finder was used to detect the active site in the enzyme. Energy minimizations were performed with an RMSD gradient of 0.001 kcal mol−1Å−1 and MMFF94X force field. Ciprofloxacin was removed from the active site and then re-docked again giving energy score (S) = −8.34 kcal mol−1 and RMSD of 1.55 Å followed by the docking of compound 16. The validated docking protocol was then used to study the ligand−target interactions in the active site to predict the binding mode and rationalize the biological activity results.
Results and Discussion
Chemistry
Scheme 1 shows the synthetic pathways adopted for the development of the quinazolinone benzenesulfonamide derivatives 5–18. The starting material 4-(2-mercapto-4-oxoquinazolin-3(4H)-yl) benzenesulfonamide 4 was prepared from the reaction of 4-isothiocyanatobenzenesulfonamide 246 and anthranilic acid 3. Coupling of 4 with the 2-chloro-N-substituted acetamide in dry acetone and anhydrous K2CO3 yielded the corresponding 2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-substituted acetamides 5–18. IR spectra of 5–18 displayed NH, CH2 aliphatic and CO bands at their assigned regions. 1H-NMR spectra of 5–18 revealed two singlets, one at the range of 3.97 to 4.30 ppm referring to the CH2 and the other at 8.09 to 10.40 ppm attributed to the NH acetamide protons. 13C-NMR of 5–18 exhibited two signals peculiar to the CH2 and CO carbons. 1H-NMR spectra of 6–8 showed singlets at 2.20, 2.24 and 2.08 ppm assigned to the CH3 group at the ortho, meta and para- positions of the phenyl ring, respectively. 13C-NMR of 6–8 showed signals at 18.32, 20.90 and 22.13 ppm for the CH3 groups. 1H-NMR spectra of 9–11 revealed triplets at 1.06, 1.17 & 1.16 ppm attributed to the CH3 ethyl, and quartet at 2.58, 2.59 and 2.52 ppm referring to the CH2 ethyl at the ortho, meta and para- positions. 13C-NMR of 9–11 showed two signals at 14.64, 15.95 & 16.16 ppm assigned to CH3 ethyl and 24.13, 28.71 & 28.05 ppm due to the CH2 ethyl groups, respectively. The 1H-NMR spectrum of 12 revealed a singlet at 3.71 ppm attributed to the OCH3 protons, while 13C-NMR of 12 showed a signal at 55.62 ppm due to the OCH3 carbon. The 1H-NMR spectrum of 13 revealed triplet at 1.30 ppm and quartet at 4.11 ppm due to the ethoxy group. The 1H-NMR spectrum of 14 revealed a singlet at 3.70 ppm attributed to the 2OCH3 protons, while 15 revealed two singlets at 3.61 and 3.72 ppm due to the 3OCH3 protons. IR spectra of 16–18 showed bands of NO2 groups at their specified regions. 1H-NMR spectra of 16 and 17 showed singlet at 2.29 ppm assigned to the CH3, while 13C-NMR of 16 and 17 showed a signal at 18.32 and 18.35 ppm attributed to the CH3 group.
Scheme 1.
Synthesis of the quinazolinone benzenesulfonamide derivatives 4–18.
Antimicrobial Activity
The synthesized compounds 4–18 were evaluated for their in vitro antibacterial potential against Bacillus subtilis and Staphylococcus aureus as examples of Gram-positive bacteria, Escherichia coli and Pseudomonas aeruginosa as examples of Gram-negative bacteria, Methicillin-resistant Staphylococcus aureus (MRSA) and antifungal potential against Candida albicans as unicellular fungi. As depicted in Table 1, the synthesized compounds 4–18 displayed potent antibacterial and antifungal activities. Compound 16 was the most potent in this series against both the bacterial and fungal strains and showed the highest inhibitory activity against the Gram-positive strains. Compound 16 showed larger inhibition zone (IZ) than that of tetracycline, ciprofloxacin, vancomycin and amphotericin B, with minimum inhibitory concentration (MIC) lower than that of tetracycline ciprofloxacin, vancomycin and amphotericin B except for MRSA, compound 16 MIC was higher than that of ciprofloxacin and vancomycin (MIC= 5.0 µg/mL versus 1.25 and 3.90 µg/mL, respectively). Regarding the antibacterial activity _in terms of the inhibition zone_ the 2-methyl-4-nitrophenyl, the 4-ethoxyphenyl and the 2-methyl-6-nitrophenyl derivative (16, 13 & 17) were the most potent compounds against Gram-positive bacteria and showed larger IZ than that of tetracycline, ciprofloxacin and vancomycin. Compounds 5, 8, 11, 13, 16–18 displayed better activity against Gram-positive bacteria compared to tetracycline. While compounds 16 and 9 showed more potent activity against Gram-negative bacteria compared to the reference drugs. Regarding the antifungal activity, compounds 7, 8, 13, 16–18 showed more potent or equipotent activity compared to amphotericin B. However, compounds 13, 16 and 17 exhibited the most potent activity towards MRSA. The MICs of all compounds are listed in Table 1. It is apparent that the majority of compounds showed lower MIC than that of tetracycline but not ciprofloxacin. The MIC value for compound 16 ranges from 0.3 to 5 µg/mL against all the tested microorganisms.
Table 1.
The In Vitro Antimicrobial Activity of Compounds 4–18 Showing the Inhibition Zones (IZ) and Minimum Inhibition Concentration (MIC) Against the Selected Strains
| Compound Code | Mean Diameter of Inhibition Zone (mm) and Minimal Inhibitory Concentrations (MIC) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. subtilis (ATCC 6633) | S. aureus (ATCC 29,213) | E. coli (ATCC 25,922) | P. aeruginosa (ATCC 27,853) | C. albicans (ATCC 10,231) | MRSA | |||||||
| IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | |
| 4 | 16 ± 0.01 | 2.50 | 13 ± 0.04 | 5.00 | NA | – | NA | – | 14 ± 0.05 | 2.50 | NA | – |
| 5 | 25 ± 0.07 | 2.50 | 22 ± 0.31 | 1.56 | 14 ± 0.23 | 10.00 | 15 ± 0.21 | 20.00 | 19 ± 0.05 | 5.00 | 20 ± 0.18 | 10.00 |
| 6 | 16 ± 0.10 | 15.63 | 14 ± 0.03 | 31.25 | 15 ± 0.31 | 5.00 | 17 ± 0.13 | 7.50 | 20 ± 0.25 | 3.91 | NA | – |
| 7 | 20 ± 0.21 | 1.25 | 15 ± 0.07 | 2.50 | 12 ± 0.15 | 20.00 | 14 ± 0.03 | 15.63 | 22 ± 0.08 | 10.00 | 10 ± 0.02 | 7.81 |
| 8 | 26 ± 0.16 | 6.31 | 23 ± 0.09 | 12.50 | NA | – | NA | – | 24 ± 0.16 | 6.31 | 19 ± 0.01 | 20.00 |
| 9 | 22 ± 0.16 | 2.50 | 18 ± 0.12 | 5.00 | 30 ± 0.28 | 1.25 | 25 ± 0.14 | 2.50 | 21 ± 0.14 | 5.00 | 14 ± 0.13 | 15.63 |
| 10 | 16 ± 0.04 | 7.81 | 12 ± 0.04 | 15.63 | NA | – | NA | – | NA | – | NA | – |
| 11 | 28 ± 0.13 | 1.25 | 26 ± 0.31 | 2.50 | 14 ± 0.14 | 5.00 | 16 ± 0.11 | 20.00 | 18 ± 0.19 | 10.00 | 22 ± 0.09 | 10.00 |
| 12 | 21 ± 0.20 | 1.95 | 24 ± 0.26 | 10.00 | 13 ± 0.12 | 10.00 | 15 ± 0.16 | 12.51 | 21 ± 0.05 | 7.81 | 20 ± 0.21 | 7.81 |
| 13 | 35 ± 0.14 | 0.49 | 33 ± 0.17 | 1.25 | 12 ± 0.17 | 1.25 | 14 ± 0.12 | 10.00 | 26 ± 0.30 | 5.00 | 28 ± 0.16 | 5.00 |
| 14 | 24 ± 0.02 | 2.50 | 27 ± 0.21 | 3.90 | 16 ± 0.01 | 5.00 | 20 ± 0.21 | 7.81 | 20 ± 0.21 | 6.31 | 24 ± 0.13 | 12.50 |
| 15 | 20 ± 0.04 | 7.81 | 16 ± 0.01 | 15.63 | 13 ± 0.04 | 15.63 | 18 ± 0.12 | 10.00 | 14 ± 0.11 | 3.80 | 12 ± 0.09 | 31.25 |
| 16 | 36 ± 0.11 | 0.31 | 35 ± 0.09 | 0.62 | 32 ± 0.20 | 1.25 | 25 ± 0.11 | 2.50 | 27 ± 0.12 | 1.25 | 29 ± 0.05 | 5.00 |
| 17 | 30 ± 0.20 | 1.25 | 28 ± 0.25 | 5.00 | 16 ± 0.23 | 5.00 | 11 ± 0.05 | 3.80 | 22 ± 0.20 | 10.00 | 25 ± 0.09 | 10.00 |
| 18 | 28 ± 0.17 | 2.50 | 26 ± 0.17 | 7.50 | 15 ± 0.12 | 7.50 | 13 ± 0.21 | 15.63 | 24 ± 0.27 | 3.13 | 23 ± 0.20 | 15.63 |
| Tetracycline | 25 ± 0.01 | 31.25 | 25 ± 0.04 | 62.50 | 23 ± 0.31 | 15.63 | 20 ± 0.10 | 62.50 | – | – | – | – |
| Ciprofloxacin | 29 ± 0.15 | 0.80 | 27 ± 0.23 | 0.78 | 27 ± 0.98 | 1.57 | 24 ± 0.47 | 3.13 | – | – | 22 ± 0.50 | 1.25 |
| Vancomycin | 21 ± 0.40 | 1.95 | 17 ± 0.11 | 1.57 | 24 ± 0.20 | 1.38 | 22 ± 0.85 | 2.50 | – | – | 14 ± 0.65 | 3.90 |
| Amphotericin B | – | – | – | – | – | – | – | – | 22± 0.11 | 15.63 | – | – |
Notes: NA indicates no activity. (-) indicates not tested. Each value represents the mean of three different experiments ± standard error. Tetracycline, ciprofloxacin and vancomycin were used as standards against the tested bacteria while Amphotericin B was used for fungi.
Synthesis and Characterization of Copper Oxide Nanoparticles (16-CuONPs)
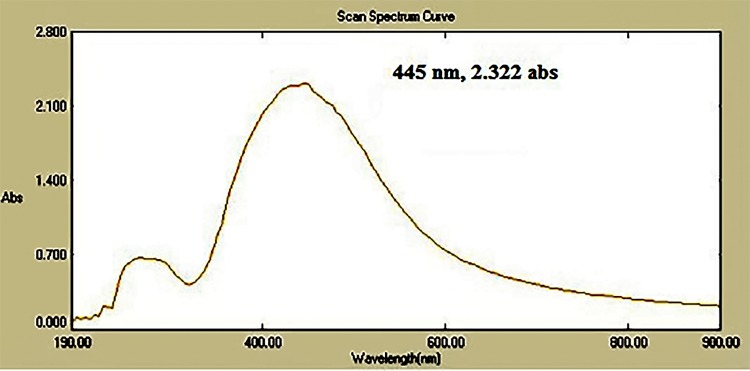
Compound 16 was mixed with copper sulfate pentahydrate solution and was irradiated at different doses of gamma radiation as shown in Table 2. The CuONPs synthesized at different gamma radiation doses showed maximum absorption (2.322) at the wavelength of 445 nm, by 4.5 kGy. In this method, hydrated electrons were produced during gamma irradiation. Compound 16 acts as a stabilizing agent by reducing the copper ions to metal oxide nanoparticles.
Table 2.
The Effect of Different Doses of Gamma Radiation (kGy) on CuONPs Synthesis
| Radiation Doses (kGy) | Maximum Absorption (O.D) | Wavelength (nm) |
|---|---|---|
| Control (non-irradiated) | 0.065 | 425 |
| 1.0 | 0.071 | 425 |
| 1.5 | 0.085 | 430 |
| 2.0 | 1.312 | 445 |
| 2.5 | 1.125 | 465 |
| 3.0 | 1.294 | 460 |
| 3.5 | 1.125 | 465 |
| 4.0 | 1.242 | 430 |
| 4.5 | 2.322 | 445 |
| 5.0 | 1.170 | 440 |
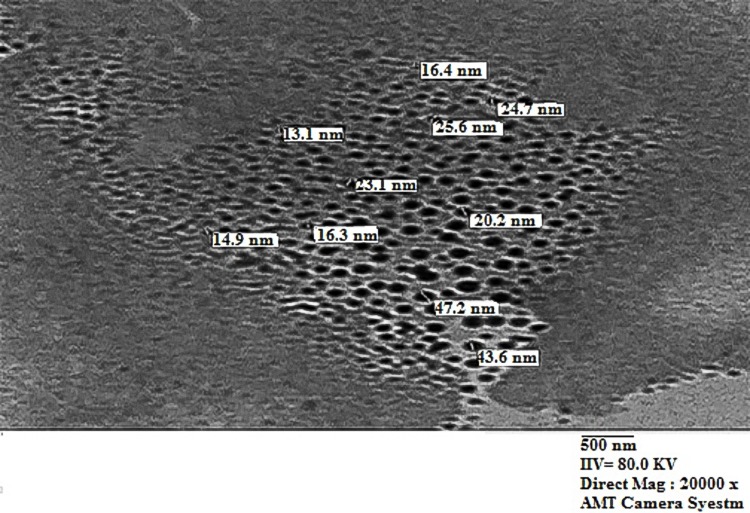
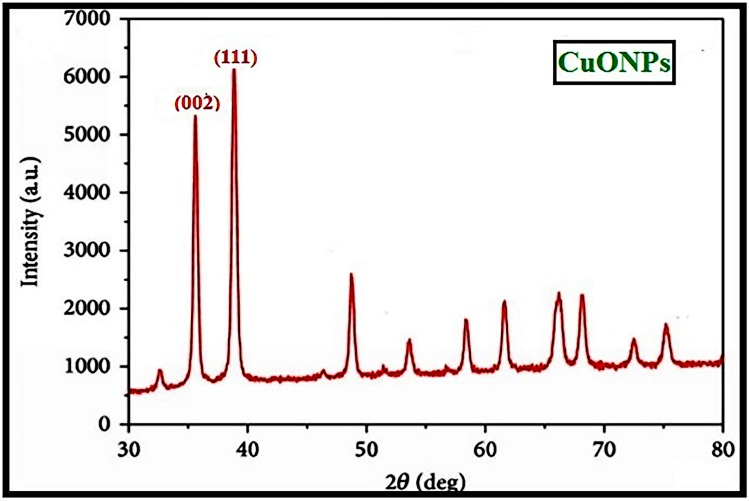
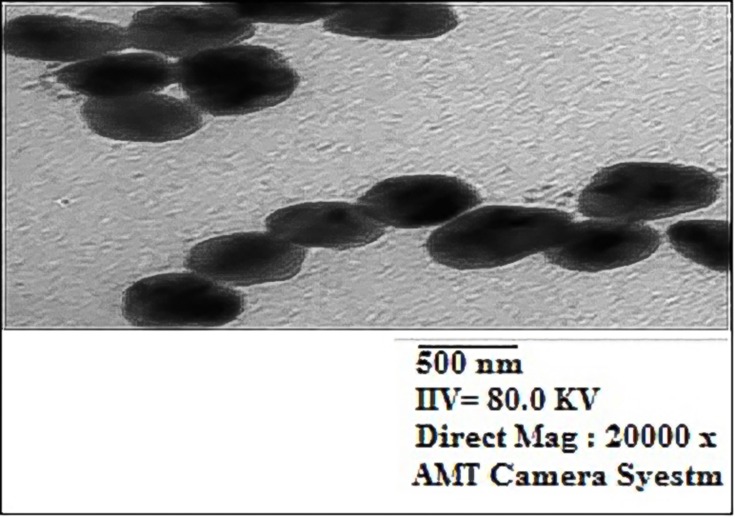
Characterization of 16-CuONPs was performed through UV–Vis spectrum, Dynamic Light Scattering (DLS), Transmission Electron Microscopy (TEM) and X-Ray Diffraction pattern (XRD). Size distribution and average particle size together with the morphology of the synthesized 16-CuONPs were figured. The colloidal solution of CuONPs displayed an intense dark green color, as a result of spectra scattered and emitted through low dimensionality. Figure 2 displays the UV–visible spectrum of CuONPs synthesized by compound 16. On the other hand, there is a slight shift in the UV–Vis peaks towards the small size. To monitor the particle size distribution, DLS was performed, and its results were compared with the TEM data. The average particle size determined by the DLS method47,48 was found to be 36.3 nm in 16-CuONPs. On the other hand, the TEM result demonstrated spherical particles within a nanoscale range from 13.1 nm to 43.6 nm with the average main diameter of 25.31 nm as shown in Figure 3. The XRD allows better visualization of the crystal structure of the observed atoms because it shows axes, shape, size and position of the atoms. The XRD pattern for the copper aggregates is shown in Figure 4, several peaks were observed. Diffraction characteristics appeared at two-theta (degree) at 35.6° and 38.8°, corresponding to the (002) and (111) planes of CuONPs, respectively, matching the conventional powder diffraction card of the Joint Committee on Powder Diffraction Standards (JCPDS).49,50
Figure 2.
The UV-visible spectrum of CuONPs synthesized by compound 16 using 4.5 KGy.
Figure 3.
TEM image of CuONPs synthesized by compound 16 using 4.5 KGy of gamma radiation.
Figure 4.
XRD image of CuONPs synthesized by compound 16 using 4.5 KGy of gamma radiation.
Mechanistic Design for Gamma-Assisted Nucleation and Germination of 16-CuONPs (Radiolysis)
The Kinetic study of the reduction method confirmed that the construction of 16-CuONPs can start without the aid of radiation; but was exceedingly enhanced by gamma irradiation particularly at 4.5 kGy, implying that radiation has a significant role in the organization of 16-CuONPs. The radicals and electrons generated in water simultaneously by gamma irradiation are e−aq, OH•, H•, H2, and H2O2 (Equ. 1). The success of gamma irradiation to form the 16-CuONPs extends the evidence that the target product of extremely reducing free electron was formed without the creation of another byproduct. The overall reaction was carried out by the use of the available electron as a reducing cause to Cu+2 ions and the effectiveness of compound 16 to act as a stabilizer towards CuONPs formation. The reaction started by dissolving CuSO4 into the water and its hydrolysis to its ions Cu+2 and SO4−2 (Equ. 2). The conversion of Cu+2 takes place by electron removal from the hydrated electrons to create zerovalent Cu• (Equ. 3). The free radicals OH• and H• are competent to discharge hydrogen from the N-(2-methyl-4-nitrophenyl)-2-((4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio) acetamide 16. Secondary radical was formed, C23 H19 N5 S2 O6• (Equ. 4). Additionally, compound 16 radical used Cu+2 to form CuONPs (Equ. 5). Eventually, the stable active compound can preserve CuONPs as referred to in Equation 6.
Synthesis of Compound 16 Nanoformulations
Synthesis and Characterization of Compound 16 Loaded Chitosan NPs (16-CNPs)
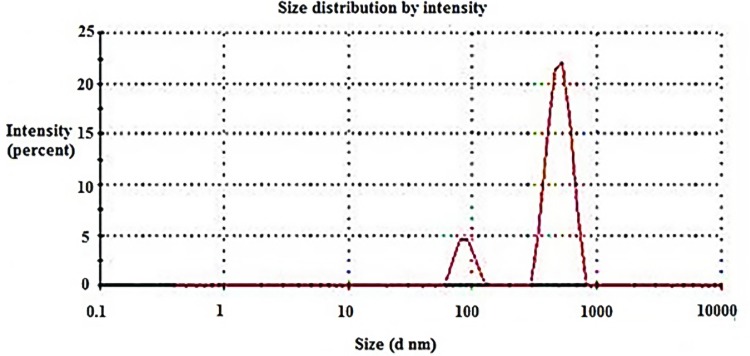
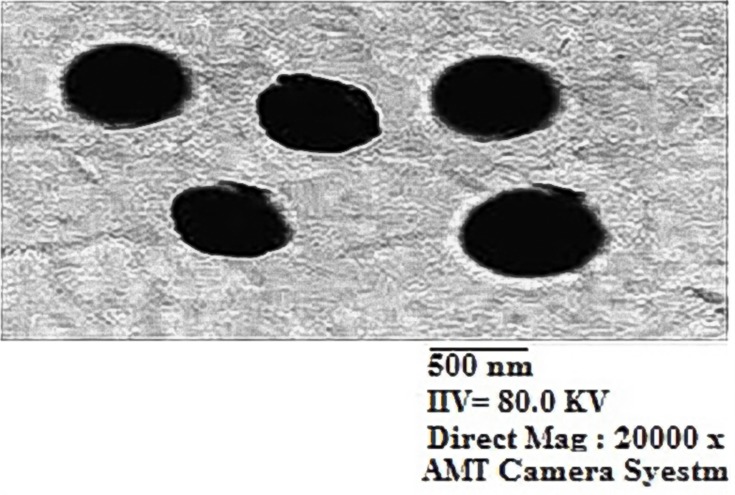
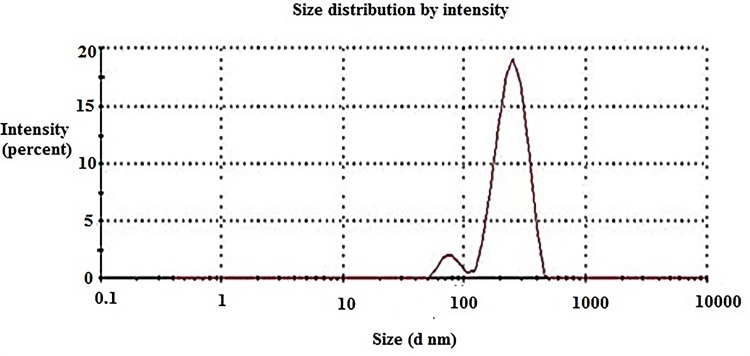
Compound 16, the most potent against all the selected bacterial and fungal strains, was further chosen to be loaded on chitosan nanoparticles in order to determine the effect of nanoformulation on its antimicrobial potency. Chitosan nanoparticles were easily obtained by means of ionic gelation between cationic chitosan and anionic triphenyl phosphate (TPP) and were applied for the encapsulation of 16-CNPs. The zeta sizer analysis of the prepared NPs revealed that the loading of compound 16 led to an increase in the NPs average diameter, to be 483.7 ± 37.83 nm in comparison to 478.4± 6.27 nm for blank CNPs. Both CNPs and 16-CNPs displayed positive Zeta potential values in SPB (sodium phosphate buffer) at pH 7.4, due to the cationic nature of chitosan. Compound 16 loading led to a decrease in the Zeta potential value from 10.0 ± 0.36 to 4.6 ± 0.88 mV (Table 3, Figures 5 & 6). The TEM examination of both CNPs and 16-CNPs demonstrated spherical shape (Figures 7 & 8).
Table 3.
The Main Characteristics of the Nanoformulations: Average Size Distribution (Nm), Particles Diameter Index (PdI) and Surface Charge (mV)
| Formulation | Size (nm ± SD) |
PdI | Zeta Potential (mV ± SD) |
|---|---|---|---|
| CNPs | 478.8 ± 6.27 | 0.502 ± 0.069 | 10.0 ± 0.36 |
| 16-CNPs | 483.7 ± 37.83 | 0.491 ± 0.023 | 4.6 ± 0.88 |
| 16-CuONPs-CNPs | 304.3 ± 83.42 | 0.424 ± 0.068 | 4.7 ± 0.58 |
Figure 5.
Average size distribution of chitosan nanoparticles (CNPs).
Figure 6.
Average size distribution of compound 16 loaded chitosan nanoparticles (16-CNPs).
Figure 7.
TEM image of chitosan nanoparticles (CNPs).
Figure 8.
TEM image of compound 16 loaded chitosan nanoparticles 16-CNPs.
Synthesis and Characterization of 16-CuONPs-CNPs Nanocomposites
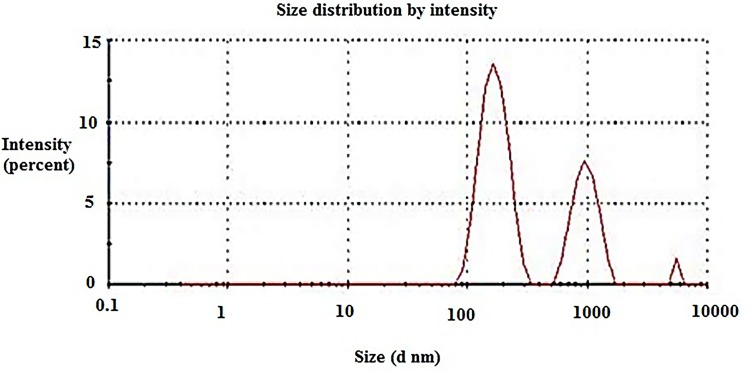
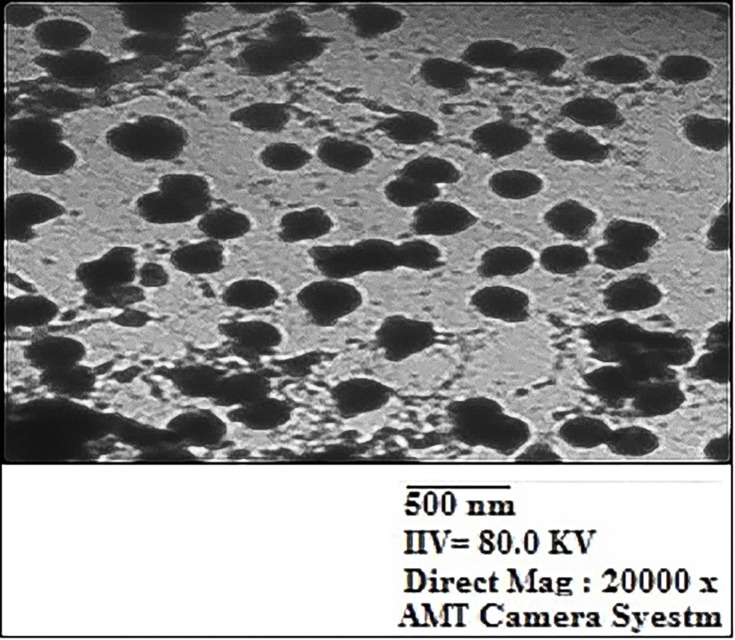
The 16-CuONPs-CNPs were synthesized by loading compound 16 and CuONPs into chitosan through encapsulation to determine the effect of nanocomposite formation on the activity. The zeta sizer analysis demonstrated that the loading of compound 16 and CuONPs has led to an increase in the average diameter, to be 304.3± 83.42 nm. Compound 16 and CuONPs loading led to a decrease of the Zeta potential values to be 4.7±0.58 mV (Table 3). The TEM demonstrated a spherical shape for these nanocomposites (Figures 9 & 10).
Figure 9.
Average size distribution of compound 16 loaded chitosan and copper oxide nanoparticles 16-CuONPs-CNPs.
Figure 10.
TEM image of compound 16 loaded chitosan and copper oxide nanoparticles 16-CuONPs-CNPs.
Antimicrobial Evaluation of the Synthesized Nanoformulations
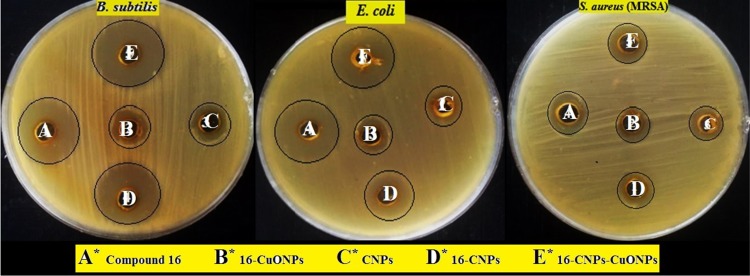
As mentioned before, CuONPs have broad-spectrum antimicrobial properties. So, compound 16 loaded CuONPs were synthesized and screened for antimicrobial activity and compared with that of CNPs and 16-CNPs. Table 4 shows the effect of different nanoformulations of compound 16 on its antimicrobial activity. It is obvious that chitosan itself possesses good antimicrobial activity and the same for compound 16 alone. However, the nano formula 16-CNPs displayed better antimicrobial activity towards all the selected strains. The inhibition zones of the nano formula 16-CNPs range from 29 to 43 mm and the MIC from 0.07 to 2.50 µg/mL. The antimicrobial activity of 16-CuONPs and 16-CuONPs-CNPs was also screened against the chosen strains. 16-CuONPs were found to exhibit moderate activity, while the nanocomposites 16-CuONPs-CNPs showed the most potent activity against all the selected strains. The 16-CuONPs-CNPs showed inhibition zones in the ranges of 34–45 mm and MIC in the range of 0.03–1.25 µg/mL, which is more potent than that of ciprofloxacin (Figure 11). In order to further study and confirm whether the above promising nanoformulations were bactericidal or bacteriostatic the relationship between MBC and MIC was applied (Table 4). The antibacterial agents are generally regarded as bactericidal if the MBC is not more than four times the MIC.51 By applying the MBC/MIC ratio it can be concluded that compound 16 and its nanoformulations exhibited bactericidal and fungicidal properties.
Table 4.
The Antimicrobial Activity of Compound 16 Nanoformulations
| Microbial Test Strains | Mean Diameter of Inhibition zone (IZ)(mm)/Minimal Inhibitory Concentration (MIC) (µg/mL)/minimal bactericidal Concentration(µg/mL). | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound 16 | 16-CuONPs | CNPs | 16-CNPs | Nanoformula 16-CuONPs-CNPs | Ciprofloxacin | |||||||||||||
| IZ | MIC | MBC | IZ | MIC | MBC | IZ | MIC | MBC | IZ | MIC | MBC | IZ | MIC | MBC | IZ | MIC | MBC | |
|
B. subtilis (ATCC 6633) |
36±0.11 | 0.31 | 0.62 | 20±0.05 | 7.81 | 15.62 | 27±0.23 | 3.91 | 7.03 | 43±0.03 | 0.07 | 0.14 | 45±0.11 | 0.03 | 0.06 | 29±0.15 | 0.80 | 1.35 |
|
S. aureus (ATCC 29,213) |
35±0.09 | 0.62 | 1.17 | 17±0.13 | 62.5 | 87.50 | 24±0.07 | 5.20 | 10.00 | 41±0.08 | 0.15 | 0.31 | 43±0.20 | 0.07 | 0.14 | 27±0.23 | 0.78 | 1.40 |
|
E. coli (ATCC 25,922) |
32±0.20 | 1.25 | 2.50 | 14±0.07 | 15.62 | 24.99 | 22±0.11 | 7.81 | 14.05 | 35±0.22 | 0.31 | 0.62 | 39±0.13 | 0.15 | 0.31 | 27±0.98 | 1.57 | 2.66 |
|
P. aeruginosa (ATCC 27,853) |
25±0.11 | 2.50 | 4.50 | 16±0.04 | 31.25 | 62.50 | 20±0.21 | 10.41 | 17.69 | 29±0.13 | 0.62 | 1.24 | 34±0.05 | 0.31 | 0.62 | 24±0.47 | 3.13 | 5.32 |
|
C. albicans (ATCC 10,231) |
27±0.12 | 1.25 | 2.12 | 18±0.18 | 62.50 | 81.25 | 21±0.02 | 12.51 | 21.26 | 33±0.04 | 0.83 | 1.66 | 37±0.04 | 0.41 | 0.82 | - | - | - |
|
S. aureus (MRSA) |
29±0.05 | 5.00 | 9.01 | 19±0.21 | 125.0 | 175.0 | 18±0.25 | 41.60 | 74.88 | 32±0.17 | 2.50 | 4.50 | 35±0.11 | 1.25 | 2.25 | 22±0.50 | 1.25 | 2.50 |
Notes: (-) indicates not tested. The results represent three independent experiments ± SE.
Figure 11.
Inhibition zones of compound 16 and its nanoformulations, A: Compound 16, B: 16-CuONPs, C: CNPs, D: 16-CNPs and E: 16-CuONPs-CNPs.
Evaluation of the DNA Gyrase Inhibitory Activity
DNA gyrase is a type II topoisomerase that is responsible for maintaining the correct level of supercoiling in bacterial DNA. It is essential in bacteria but is missing in higher eukaryotes. Ciprofloxacin is a fluoroquinolone broad-spectrum antibiotic owing to its DNA gyrase inhibitory effect. Compound 16 and the nanoformulations were screened for their inhibitory effect against Staphylococcus aureus DNA gyrase and showed IC50 values ranging from 10.57 to 27.32 µg/mL. The nano formula 16-CuONPs-CNPs showed the highest activity with IC50 = 10.57 µg/mL compared to ciprofloxacin IC50 =8.72 µg/mL.
It is clear from the results in Table 5 that compound 16 (IC50 = 16.21 µg/mL) showed more potent activity after being incorporated into nanoformulations; 16-CNPs (IC50= 14.26 µg/mL), 16-CuONPs (IC50= 12.49 µg/mL), and 16-CuONPs-CNPs. Also, chitosan NPs showed more potent inhibitory activity after being formulated with compound 16 to form 16-CNPs (IC50= 27.32 versus 14.26 µg/mL). Molecular docking was conducted to study the possible binding of 16 inside the active site of S. aureus DNA gyrase.
Table 5.
Determination of the Inhibitory Activity of Compound 16 and the Nanoformulations on S. aureus DNA Gyrase Enzyme
| Code | S. aureus DNA Gyrase IC50 (µg/mL) |
|---|---|
| Compound 16 | 16.21± 1.10 |
| CNPs | 27.32± 1.56 |
| 16-CNPs | 14.26 ± 0.68 |
| 16-CuONPs | 12.49 ± 0.24 |
| 16-CuONPs-CNPs | 10.57 ± 0.71 |
| Ciprofloxacin | 8.72 ± 0.64 |
Cytotoxicity Test
In order to determine the relative safety of compound 16, 16-CNPs and 16-CuONPs-CNPs, the cytotoxic effect on VERO cell line (ATCC CCL-81) was measured using the MTT assay.52 Compound 16 showed the least cytotoxicity with an IC50 of 927.28 µg/mL indicating its safety to normal cells. Also, 16-CNPs and 16-CuONPs-CNPs displayed a very low cytotoxic effect (IC50= 543 and 637 µg/mL, respectively) (Table 6).
Table 6.
Cytotoxicity of compound 16 and its nanoformulations on normal VERO cell Line
| Compound No. | IC50 (µg/mL) |
|---|---|
| 16 | 927.28±0.17 |
| 16-CNPs | 543.37±0.25 |
| 16- CuONPs-CNPs | 637.69±0.5 |
Molecular Docking
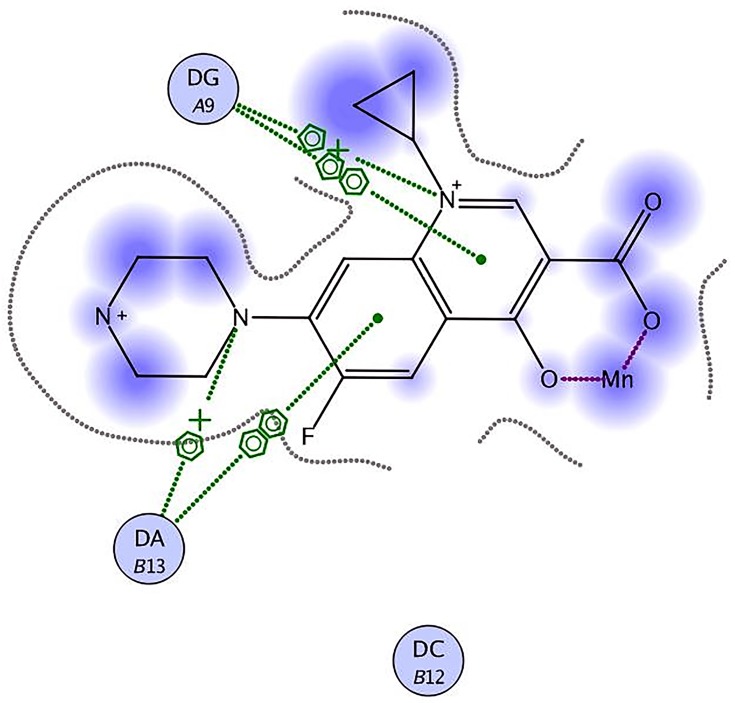
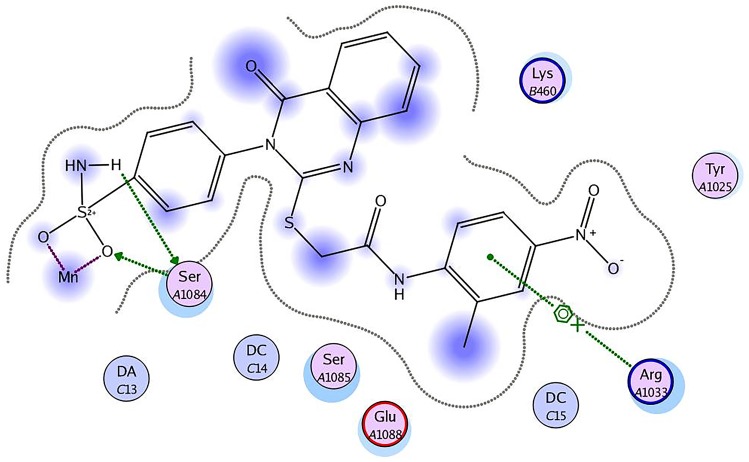
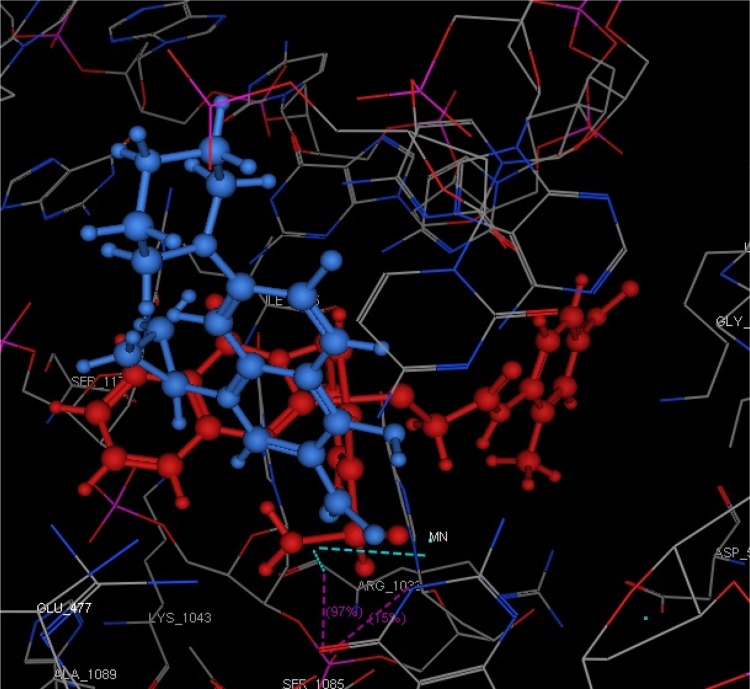
Staphylococcus aureus DNA gyrase (PDB: 2XCT) consists of four lobes with four active sites in which ciprofloxacin binds. The DNA strands intercalate with ciprofloxacin-forming interactions with DNA structure.53 The various substitutions on the quinazolinone ring can play an effective role in its binding to the receptor. The interaction energy and the binding mode of compound 16 were in agreement with that of ciprofloxacin. Ciprofloxacin binds inside the active site through two π–π and cation–π interactions to the DNA fragments DG:A9, DA:B13 and Mn2+ through its carboxylic group with binding energy S= −8.34 Kcal mol−1 (Figure 12). Compound 16 was able to adopt the same binding mode as ciprofloxacin and establish the same contact with Mn2+ through its SO2 group, which is crucial for binding and forms cation–π interaction with Arg A1033, hydrogen bonds with Ser A1084 of binding energy S= −8.82 Kcal mol−1 (Figure 13). Superimposition of ciprofloxacin & compound 16 showed that both adopt the same orientation directed towards Mn2+ by the carboxylic & SO2 groups, respectively (Figure 14 & Table 7).
Figure 12.
Ciprofloxacin binds inside the active site of 2XCT to the DNA fragments DG:A9 & DA:B13 and Mn2+.
Figure 13.
Compound 16 forms hydrogen bonds with Ser A1084, cation–π interaction with Arg A1033 and Mn2+.
Figure 14.
Superimposition of compound 16 (red) and ciprofloxacin (blue) in which the SO2 group of 16 and the carboxylic of ciprofloxacin are directed towards the Mn2+.
Table 7.
Docking Results of Compound 16 in the Active Site of S. aureus Gyrase Active Site (PDB: 2XCT)
| Compound | Energy Score (S) (Kcal/mol) | Amino Acids | Interacting Groups | Length (Å) |
|---|---|---|---|---|
| 16 | −8.82 | Ser 1084 Ser 1084 Arg 1033 |
SO2 NH2 Ph |
2.28 2.52 4.07 |
Conclusion
Novel quinazolinone benzenesulfonamide derivatives 4–18 were synthesized and screened for their antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria, MRSA and yeast. The 2-methyl-4-nitrophenyl derivative 16 was the most potent against the selected strains and showed inhibition zones and MIC in the ranges of 25–36 mm and 0.31–5.0 µg/mL, respectively. The most potent activity was against the Gram-positive Bacillus subtilis and the least potent was against Pseudomonas aeruginosa. Compound 16 was selected to be conjugated with CuONPs and synthesize two nanoformulations. The 16-CuONPs synthesized by the aid of gamma irradiation (4.5 KG), exhibited moderate activity towards the selected strains. While the 16-CNPs displayed inhibition zones and MIC in the ranges of 29–43 mm and 0.07–2.50 µg/mL, and were more potent than CNPs. The nanocomposites 16-CuONPs-CNPs synthesized from compound 16 and CuONPs and encapsulated in chitosan proved to be the most potent nano formula and showed inhibition zones and MIC in the ranges of 34–45 mm and 0.03–1.25 µg/mL, respectively. Measurement of the MIC and MBC of the nanoformulations has confirmed their bactericidal and fungicidal activity. In vitro enzyme assay screening of compound 16 and the nanoformulations revealed that 16-CuONPs-CNPs showed the most potent inhibitory activity towards S. aureus DNA gyrase that is relatively equipotent to that of ciprofloxacin (IC50=10.57 versus 8.72 µg/mL, respectively). Cytotoxic activity screening of 16, 16-CNPs and 16-CuONPs-CNPs on normal VERO cell line has proved their relative safety. Molecular docking of 16 revealed its proper binding inside the active site of S. aureus DNA gyrase, which may incorporate at least in part to its antimicrobial activity. Finally, hybridization between quinazolinone and benzenesulfonamide can provide potent antimicrobial candidates. The use of nanoformulations for the most potent compound has increased its potency against pathogenic microbes with multi-drug resistance such as MRSA. It is clear that compound 16 could be considered as a promising antimicrobial agent with enhanced activity upon nanoformulation.
Acknowledgment
M.M. Ghorab and A.M. Soliman appreciate the staff members of the gamma irradiation unit at the National Center for Radiation Research and Technology (NCRRT) for carrying out the irradiation process.
A.S. Alqahtani is thankful to the Researchers Supporting Project number (RSP-2019/132), King Saud University, Riyadh, Saudi Arabia.
Disclosure
The authors declare no conflict of interest.
References
- 1.Zhang L, Peng XM, Damu GL, Geng RX, Zhou CH. Comprehensive review in current developments of imidazole‐based medicinal chemistry. Med Res Rev. 2014;34(2):340–437. doi: 10.1002/med.21290. [DOI] [PubMed] [Google Scholar]
- 2.Peng X-M, Cai G-X, Zhou C-H. Recent developments in azole compounds as antibacterial and antifungal agents. Curr Top Med Chem. 2013;13(16):1963–2010. doi: 10.2174/15680266113139990125 [DOI] [PubMed] [Google Scholar]
- 3.Cui S-F, Addla D, Zhou C-H. Novel 3-aminothiazolquinolones: design, synthesis, bioactive evaluation, sars, and preliminary antibacterial mechanism. J Med Chem. 2016;59(10):4488–4510. doi: 10.1021/acs.jmedchem.5b01678 [DOI] [PubMed] [Google Scholar]
- 4.Zhang H-Z, Jeyakkumar P, Kumar KV, Zhou C-H. Synthesis of novel sulfonamide azoles via C–N cleavage of sulfonamides by azole ring and relational antimicrobial study. New J Chem. 2015;39(7):5776–5796. doi: 10.1039/C4NJ01932F [DOI] [Google Scholar]
- 5.Jang M-Y, De Jonghe S, Segers K, Anné J, Herdewijn P. Synthesis of novel 5-amino-thiazolo [4, 5-d] pyrimidines as E. coli and S. aureus SecA inhibitors. Bioorg Med Chem. 2011;19(1):702–714. doi: 10.1016/j.bmc.2010.10.027 [DOI] [PubMed] [Google Scholar]
- 6.Khatkar A, Nanda A, Kumar P, Narasimhan B. Synthesis, antimicrobial evaluation and QSAR studies of p-coumaric acid derivatives. Arab J Chem. 2017;10:S3804–S3815. doi: 10.1016/j.arabjc.2014.05.018 [DOI] [Google Scholar]
- 7.Manikprabhu D, Lingappa K. Antibacterial activity of silver nanoparticles against methicillin-resistant Staphylococcus aureus synthesized using model Streptomyces sp. pigment by photo-irradiation method. J Pharm Res. 2013;6(2):255–260. doi: 10.1016/j.jopr.2013.01.022 [DOI] [Google Scholar]
- 8.Durán N, Marcato PD, De Souza GI, Alves OL, Esposito E. Antibacterial effect of silver nanoparticles produced by fungal process on textile fabrics and their effluent treatment. J Biomed Nanotechnol. 2007;3(2):203–208. doi: 10.1166/jbn.2007.022 [DOI] [Google Scholar]
- 9.Panagiotopoulos A, Zafiropoulos TF, Perlepes SP, et al. Molecular structure and magnetic properties of acetato-bridged lanthanide (III) dimers. Inorg Chem. 1995;34(19):4918–4920. doi: 10.1021/ic00123a029 [DOI] [Google Scholar]
- 10.El-Nawawy M, Farag R, Sabbah I, Abu-Yamin A. Synthesis, spectroscopic, thermal studies and biological activity of a new sulfamethoxazole schiff base and its copper complexes. Int J Pharm Sci Res. 2011;2(12):3143–3148. [Google Scholar]
- 11.Chevalier J, Mahamoud A, Baitiche M, et al. Quinazoline derivatives are efficient chemosensitizers of antibiotic activity in Enterobacter aerogenes, Klebsiella pneumoniae and Pseudomonas aeruginosa resistant strains. Int J Antimicrob Agents. 2010;36(2):164–168. doi: 10.1016/j.ijantimicag.2010.03.027 [DOI] [PubMed] [Google Scholar]
- 12.Ghorab M, AbdelHamide S, ElHakim A. Synthesis of some new biologically active 2-phenyl-6-iodo-3-substituted-4 (3H) quinazolinones. Indian J Heterocy Chem. 1995;5(2):115–120. [Google Scholar]
- 13.Mahamoud A, Chevalier J, Baitiche M, Adam E, Pages J-M. An alkylaminoquinazoline restores antibiotic activity in Gram-negative resistant isolates. Microbiology. 2011;157(2):566–571. doi: 10.1099/mic.0.045716-0 [DOI] [PubMed] [Google Scholar]
- 14.Devi KA, Sarangapani M. Synthesis and antimicrobial activity of some quinazolinones derivatives. Int J Drug Dev Res. 2012;4(3):324–327. [Google Scholar]
- 15.Jafari E, Khajouei MR, Hassanzadeh F, Hakimelahi GH, Khodarahmi GA. Quinazolinone and quinazoline derivatives: recent structures with potent antimicrobial and cytotoxic activities. Res Pharm Sci. 2016;11(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- 16.Suresha G, Suhas R, Kapfo W, Gowda DC. Urea/thiourea derivatives of quinazolinone–lysine conjugates: synthesis and structure–activity relationships of a new series of antimicrobials. Eur J Med Chem. 2011;46(6):2530–2540. doi: 10.1016/j.ejmech.2011.03.041 [DOI] [PubMed] [Google Scholar]
- 17.Al-Omary FA, Hassan GS, El-Messery SM, Nagi MN, Habib E-SE, El-Subbagh HI. Nonclassical antifolates, part 3: synthesis, biological evaluation and molecular modeling study of some new 2-heteroarylthio-quinazolin-4-ones. Eur J Med Chem. 2013;63:33–45. doi: 10.1016/j.ejmech.2012.12.061 [DOI] [PubMed] [Google Scholar]
- 18.Dinakaran M, Selvam P, DeClercq E, Sridhar SK. Synthesis, antiviral and cytotoxic activity of 6-bromo-2, 3-disubstituted-4 (3H)-quinazolinones. Biol Pharm Bull. 2003;26(9):1278–1282. doi: 10.1248/bpb.26.1278 [DOI] [PubMed] [Google Scholar]
- 19.Lubenets V, Karpenko O, Ponomarenko M, Zahoriy G, Krychkovska A, Novikov V. Development of New Antimicrobial Compositions of Thiosulfonate Structure. 2013:119–124. [Google Scholar]
- 20.Lubenets V, Vasylyuk S, Monka N, et al. Synthesis and antimicrobial properties of 4-acylaminobenzenethiosulfoacid S-esters. Saudi Pharm J. 2017;25(2):266–274. doi: 10.1016/j.jsps.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elgadir MA, Uddin MS, Ferdosh S, Adam A, Chowdhury AJK, Sarker MZI. Impact of chitosan composites and chitosan nanoparticle composites on various drug delivery systems: a review. J Food Drug Anal. 2015;23(4):619–629. doi: 10.1016/j.jfda.2014.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JH, Saravanakumar G, Kim K, Kwon IC. Targeted delivery of low molecular drugs using chitosan and its derivatives. Adv Drug Deliv Rev. 2010;62(1):28–41. doi: 10.1016/j.addr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 23.Wu T, Wu C, Fu S, et al. Integration of lysozyme into chitosan nanoparticles for improving antibacterial activity. Carbohydr Polym. 2017;155:192–200. doi: 10.1016/j.carbpol.2016.08.076 [DOI] [PubMed] [Google Scholar]
- 24.Mendoza G, Regiel-Futyra A, Andreu V, et al. Bactericidal effect of gold–chitosan nanocomposites in coculture models of pathogenic bacteria and human macrophages. ACS Appl Mater Interfaces. 2017;9(21):17693–17701. doi: 10.1021/acsami.6b15123 [DOI] [PubMed] [Google Scholar]
- 25.Usman MS, El Zowalaty ME, Shameli K, Zainuddin N, Salama M, Ibrahim NA. Synthesis, characterization, and antimicrobial properties of copper nanoparticles. Int J Nanomedicine. 2013;8:4467–4479. doi: 10.2147/IJN.S50837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salehi E, Daraei P, Shamsabadi AA. A review on chitosan-based adsorptive membranes. Carbohydr Polym. 2016;152:419–432. doi: 10.1016/j.carbpol.2016.07.033 [DOI] [PubMed] [Google Scholar]
- 27.Ramyadevi J, Jeyasubramanian K, Marikani A, Rajakumar G, Rahuman AA. Synthesis and antimicrobial activity of copper nanoparticles. Mater Lett. 2012;71:114–116. doi: 10.1016/j.matlet.2011.12.055 [DOI] [Google Scholar]
- 28.Khashan KS, Sulaiman GM, Abdulameer FA. Synthesis and antibacterial activity of CuO nanoparticles suspension induced by laser ablation in liquid. Arab J Sci Eng. 2016;41(1):301–310. doi: 10.1007/s13369-015-1733-7 [DOI] [Google Scholar]
- 29.Emami-Karvani Z, Chehrazi P. Antibacterial activity of ZnO nanoparticle on gram-positive and gram-negative bacteria. Afr J Microbiol Res. 2011;5(12):1368–1373. [Google Scholar]
- 30.Gong -H-H, Addla D, Lv J-S, Zhou C-H. Heterocyclic naphthalimides as new skeleton structure of compounds with increasingly expanding relational medicinal applications. Curr Top Med Chem. 2016;16(28):3303–3364. doi: 10.2174/1568026616666160506145943 [DOI] [PubMed] [Google Scholar]
- 31.Ghorab MM, Alsaid MS, Soliman AM, Al-Mishari AA. Benzo [g] quinazolin-based scaffold derivatives as dual EGFR/HER2 inhibitors. J Enzyme Inhib Med Chem. 2018;33(1):67–73. doi: 10.1080/14756366.2017.1389922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghorab MM, Alsaid MS, Soliman AM, Ragab FA. VEGFR-2 inhibitors and apoptosis inducers: synthesis and molecular design of new benzo [g] quinazolin bearing benzenesulfonamide moiety. J Enzyme Inhib Med Chem. 2017;32(1):893–907. doi: 10.1080/14756366.2017.1334650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alsaid MS, Al-Mishari AA, Soliman AM, Ragab FA, Ghorab MM. Discovery of Benzo [g] quinazolin benzenesulfonamide derivatives as dual EGFR/HER2 inhibitors. Eur J Med Chem. 2017;141:84–91. doi: 10.1016/j.ejmech.2017.09.061 [DOI] [PubMed] [Google Scholar]
- 34.Ghorab MM, Alsaid MS, El-Gaby MS, Safwat NA, Elaasser MM, Soliman AM. Biological evaluation of some new N-(2, 6-dimethoxypyrimidinyl) thioureido benzenesulfonamide derivatives as potential antimicrobial and anticancer agents. Eur J Med Chem. 2016;124:299–310. doi: 10.1016/j.ejmech.2016.08.060 [DOI] [PubMed] [Google Scholar]
- 35.Ghorab MM, Soliman AM, Alsaid MS, Askar AA. Synthesis, antimicrobial activity and docking study of some novel 4-(4, 4-dimethyl-2, 6-dioxocyclohexylidene) methylamino derivatives carrying biologically active sulfonamide moiety. Arab J Chem. 2020;13(1):545–556. doi: 10.1016/j.arabjc.2017.05.022 [DOI] [Google Scholar]
- 36.Murugavel S, Sundramoorthy S, Lakshmanan D, Subashini R, Kumar PP. Synthesis, crystal structure analysis, spectral (NMR, FT-IR, FT-Raman and UV–Vis) investigations, molecular docking studies, antimicrobial studies and quantum chemical calculations of a novel 4-chloro-8-methoxyquinoline-2 (1H)-one: an effective antimicrobial agent and an inhibition of DNA gyrase and lanosterol-14α-demethylase enzymes. J Mol Struct. 2017;1131:51–72. doi: 10.1016/j.molstruc.2016.11.035 [DOI] [Google Scholar]
- 37.Lesher GY, Froelich EJ, Gruett MD, Bailey JH, Brundage RP. 1, 8-Naphthyridine derivatives. A new class of chemotherapeutic agents. J Med Chem. 1962;5(5):1063–1065. doi: 10.1021/jm01240a021 [DOI] [PubMed] [Google Scholar]
- 38.Soliman AM, Ghorab MM. Exploration of N-alkyl-2-[(4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl) thio] acetamide derivatives as anticancer and radiosensitizing agents. Bioorg Chem. 2019;88:102956. doi: 10.1016/j.bioorg.2019.102956 [DOI] [PubMed] [Google Scholar]
- 39.Cappuccino J, Sherman N. Microbiology, Laboratory Manual. India: Pearson Education, Inc New Delhi; 2004:282–283. [Google Scholar]
- 40.Cooper K. The theory of antibiotic inhibition zones. Anal Microbiol. 1963;1–86. [Google Scholar]
- 41.Hassan AS, Askar AA, Nossier ES, Naglah AM, Moustafa GO, Al-Omar MA. Antibacterial evaluation, in silico characters and molecular docking of schiff bases derived from 5-aminopyrazoles. Molecules. 2019;24(17):3130. doi: 10.3390/molecules24173130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wikler M, Hindler J, Cockerill F, et al. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard,; CLSI Document M07-A8; ISBN: ISBN 1-56238-689-1. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 43.Kawashima Y, Handa T, Kasai A, Takenaka H, Lin S, Ando Y. Novel method for the preparation of controlled‐release theophylline granules coated with a polyelectrolyte complex of sodium polyphosphate–chitosan. J Pharm Sci. 1985;74(3):264–268. doi: 10.1002/jps.2600740308 [DOI] [PubMed] [Google Scholar]
- 44.Dias FR, Novais JS, Devillart T, et al. Synthesis and antimicrobial evaluation of amino sugar-based naphthoquinones and isoquinoline-5, 8-diones and their halogenated compounds. Eur J Med Chem. 2018;156:1–12. doi: 10.1016/j.ejmech.2018.06.050 [DOI] [PubMed] [Google Scholar]
- 45.Alt S, Mitchenall LA, Maxwell A, Heide L. Inhibition of DNA gyrase and DNA topoisomerase IV of Staphylococcus aureus and Escherichia coli by aminocoumarin antibiotics. J Antimicrob Chemother. 2011;66(9):2061–2069. doi: 10.1093/jac/dkr247 [DOI] [PubMed] [Google Scholar]
- 46.Ghorab MM, El Ella DAA, Heiba HI, Soliman AM. Synthesis of certain new thiazole derivatives bearing a sulfonamide moiety with expected anticancer and radiosensitizing activities. J Mater Sci Eng A. 2011;1:684–691. [Google Scholar]
- 47.Berne BJ, Pecora R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics. Courier Corporation; 2000. [Google Scholar]
- 48.El-Batal A, El-Sayed MH, Refaat BM, Askar AAZ. Marine streptomyces cyaneus strain alex-SK121 mediated eco-friendly synthesis of silver nanoparticles using gamma radiation. Br J Pharm Res. 2014;4(21):2525–2547. doi: 10.9734/BJPR/2014/12224 [DOI] [Google Scholar]
- 49.Etefagh R, Azhir E, Shahtahmasebi N. Synthesis of CuO nanoparticles and fabrication of nanostructural layer biosensors for detecting Aspergillus niger fungi. Sci Iran. 2013;20(3):1055–1058. [Google Scholar]
- 50.Clark J. Joint Committee on Powder Diffraction Standards (JCPDS). 1961. In Card. [Google Scholar]
- 51.Sun N, Li M, Cai S, et al. Antibacterial evaluation and mode of action study of BIMQ, a novel bacterial cell division inhibitor. Biochem Biophys Res Commun. 2019;514(4):1224–1230. doi: 10.1016/j.bbrc.2019.05.086 [DOI] [PubMed] [Google Scholar]
- 52.Keepers YP, Pizao PE, Peters GJ, van Ark-otte J, Winograd B, Pinedo HM. Comparison of the sulforhodamine B protein and tetrazolium (MTT) assays for in vitro chemosensitivity testing. Eur J Cancer Clin Oncol. 1991;27(7):897–900. doi: 10.1016/0277-5379(91)90142-Z [DOI] [PubMed] [Google Scholar]
- 53.Bax BD, Chan PF, Eggleston DS, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466(7309):935. doi: 10.1038/nature09197 [DOI] [PubMed] [Google Scholar]