Abstract
To investigate the origin and evolution of essential genes, we identified and phenotyped 195 young protein-coding genes, which originated 3 to 35 million years ago in Drosophila. Knocking down expression with RNA interference showed that 30% of newly arisen genes are essential for viability. The proportion of genes that are essential is similar in every evolutionary age group that we examined. Under constitutive silencing of these young essential genes, lethality was high in the pupal stage and also found in the larval stages. Lethality was attributed to diverse cellular and developmental defects, such as organ formation and patterning defects. These data suggest that new genes frequently and rapidly evolve essential functions and participate in development.
Essential genes are often portrayed as conserved and ancient (1, 2), whereas younger genes, which exist in only one or a few species, have been considered to be more dispensable and to perform relatively minor organismal functions (1–4). It is unclear how essential genes arise and how new genes accumulate essential functions. New genes arise continuously through various mechanisms, such as DNA-based duplication, retroposition, and de novo origination (5, 6). When they first arose, new genes were expected to be nonessential because their immediate ancestral species were able to survive without them (Fig. 1A). However, little is known about their phenotypes and degrees of essentiality.
Fig. 1.
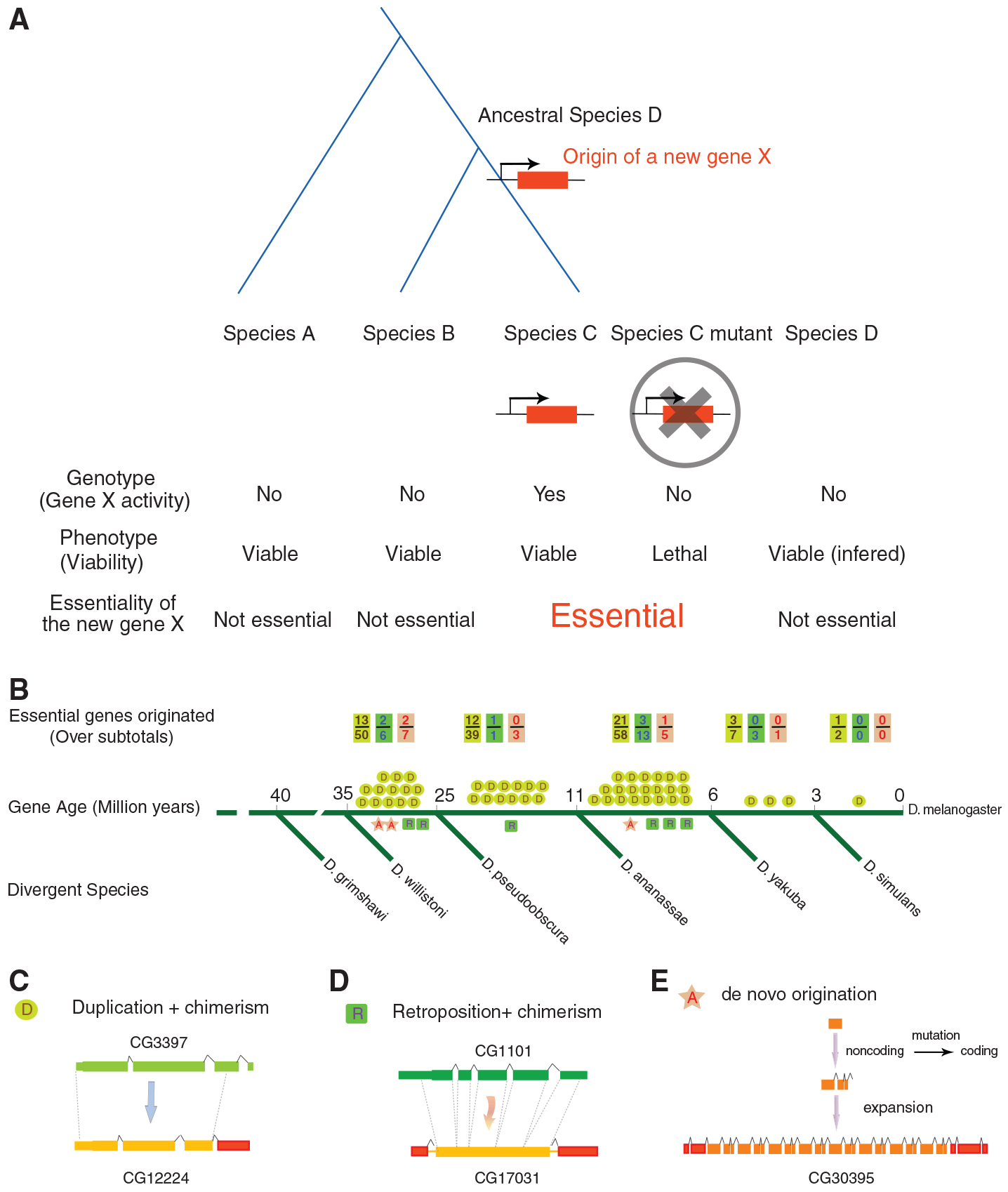
Origin of new essential genes during recent evolution in Drosophila. (A) Schematic representation for the hypothesis for the origin of a new essential gene. The ancestral species D is immediately before the new gene X originated. (B) Number of young essential genes in major evolutionary periods [D/R and A represent DNA/RNA-based duplicate genes and de novo genes with examples in (C) to (E), respectively]. The subtotal for a particular mechanism, including both essential and nonessential genes, is also shown as a denominator in B. Green, yellow, red and boxes represent exons in the parental genes, young genes, and recruited chimeric regions, respectively. Dashed lines represent paralogous duplicated regions.
By comparative genomic analysis of 12 closely related Drosophila species (7), we identified 566 new genes in the D. melanogaster genome and dated their evolutionary ages through phylogenetic distributions (8) (fig. S1). All these genes originated less than 35 million years (My) after the divergence from D. willistoni (9), so we called them young genes. To assay their phenotypic effects in viability, we obtained Drosophila RNA interference (RNAi) lines targeting these genes (10, 11) and excluded RNAi lines with predicted off-target effects and lines with detectable phenotypes by P-element insertion, resulting in a set of lines targeting 195 young genes. Crosses resulting in constitutive silencing of these genes allowed us to assay the phenotypic effects on viability in the F1 generation (8) (fig. S2).
Unexpectedly, 59 of these genes were lethal under constitutive RNAi knockdown (Table 1 and tables S1, S3, and S4). We confirmed lethality in most of the genes (93%) with different driver constructs (table S6, part I). Although the efficiency of gene knockdown by different drivers might vary, the phenotypic consistency indicated a low false-positive rate (<7%), consistent with previous estimates (10). Moreover, for the genes with multiple RNAi lines from independent upstream activating sequence–inverted repeat (UAS-IR) constructs or independent transformations that insert into different chromosomal locations, we repeated the crosses with these lines and found that 45 of 47 (96%) genes showed similar viability phenotypes between lines (table S6, part II), ruling out positional effects or construct effects. Furthermore, in deficiency libraries, lines deleting these genes are homozygous lethal, although a deletion block can be large and can contain other genes (12). Furthermore, several genes in the list (table S1)—HP6 (CG15636), CG12842, and spn2 (CG8137)—were found to be lethal using various gene disruption methods, including P-element disruption, RNAi with independent constructs, and misexpression assays (13–15). Therefore, we found 59 young genes that are essential for viability (Table 1 and table S1), a conservative number due to false negatives because RNAi does not reduce the mRNA level to zero (10). These 59 genes encode diverse protein domains with fundamental molecular and cellular functions, including putative transcription factors and/or nucleic acid–binding proteins, peptidases, G protein–coupled receptors, protease inhibitors, nicotinamide adenine dinucleotide–binding proteins, ribosomal proteins, and molecular chaperones (tables S2 and S8).
Table 1.
Summary statistics of lethal phenotypes of young genes. (I) Gene age was described in (8); age groups 0 to ~3 My and 3 to ~6 My were pooled to increase sample size. A gene was considered essential for viability if it was constitutive RNAi lethal (8); fertility is not a subject in this study. (II) Lethality stage of “pupal” includes all substage categories, such as prepupal, early pupal, late pupal, and pharate. “Before pupal” includes multiple larval stages, including early larvae and late larvae. “Other” includes mixed-stage lethal, stage unknown, or stage undefined.
| I. Proportion of essential genes (constitutive RNAi lethal) | |||||
|---|---|---|---|---|---|
| Young genes | |||||
| Age (My) | Essential genes | Nonessential genes | Subtotal | Proportion of essential | p* |
| 0–6 | 4 | 9 | 13 | 31% | 1.00 |
| 6–11 | 25 | 51 | 76 | 33% | 0.78 |
| 11–25 | 13 | 30 | 43 | 30% | 0.60 |
| 25–35 | 17 | 46 | 63 | 27% | 0.24 |
| Total | 59 | 136 | 195 | 30% | 0.31 |
| Old genes | |||||
| Age (My) | Essential genes | Nonessential genes | Subtotal | Proportion of essential | Age (My) |
| >40 | 86 | 159 | 245 | 35% | >40 |
| II. Stage of lethality | ||||
|---|---|---|---|---|
| Young genes | Old genes | |||
| Pupal | 47 | 80% | 43 | 50% |
| Before pupal | 6 | 10% | 38 | 44% |
| Other | 6 | 10% | 5 | 6% |
| Total | 59 | 100% | 86 | 100% |
| P† | 0.0009 | |||
Two-tailed Fisher’s exact test P for essential/nonessential young genes in each age group compared with those for old genes;
Two-tailed Fisher’s exact test P for pupal/non-pupal lethals for young genes compared with those for old genes.
The proportion of essential genes in D. melanogaster is estimated at ~25 to 35% (2, 10, 16). We compared the rates of lethality between old genes and young genes using the same genesilencing methods (8). Among randomly chosen old genes, 35% (86 of 245) were essential for viability (Table 1), which was statistically similar to the 30% (59 of 195) essential young genes (two-tailed Fisher’s exact test, P = 0.3, Table 1). These data suggest that young genes are as essential as old genes in terms of viability.
We analyzed the age distribution of young essential genes by mapping the origination events of these 59 genes onto the Drosophila phylogenetic tree (8). We found that essential genes emerged throughout the evolutionary period examined (Fig. 1B and table S2). The youngest, p24-related-2 (CG33105), arose within the last 3 My and is thus D. melanogaster–specific (table S2). In each age group, the proportion of genes that are essential was around 30% (Table 1), suggesting that whether or not a gene is essential is independent of its age. These data reveal that the proportion of newly arisen essential genes reaches a plateau within a few million years. Reminiscent of the Walsh model, a new duplicate gene can quickly evolve a novel and important function by accumulating advantageous mutations (17), especially in the species with large effective population sizes, such as Drosophila (18). These observations may explain why duplicate genes are as essential as singletons (19–22), although most genes examined in these mammalian studies are relatively ancient.
We investigated the native gene expression patterns of these genes with D. melanogaster life-cycle time-course expression profiling (23). Interestingly, most of the 59 genes we identified are highly expressed at the late larval stages (L2 and L3) or during metamorphosis; some genes are also expressed during the embryonic and L1 stages (fig. S4), which suggests that their gene products are subject to transcriptional regulation during the life cycle.
We examined the developmental stages in which lethality occurs under constitutive silencing and found that lethality occurs at various developmental stages (Fig. 2). The vast majority (47 of 59, 80%) of the young essential genes consistently showed lethality during pupation; four new genes (CG11466, CG33459, CG6289, and CG8358) showed lethality at larval stage, whereas a few other genes show lethality at both larval and pupal stages, which we termed mixed-stage lethality (Table 1 and tables S1 and S5). About 50% of old genes are lethal during pupation, and the other half are lethal at earlier stages, because many early-stage developmental genes are conserved (10) (Table 1). In comparison, young genes are highly enriched in pupal lethals (Table 1; Fisher’s exact test, two-tailed, P = 9 × 10−4). These data suggest that new genes have evolved essential functions in larval and pupal development, and frequently regulate development in the pupal stage, with 10% or more regulating the development in the larval or even embryonic stages (table S1) (13).
Fig. 2.
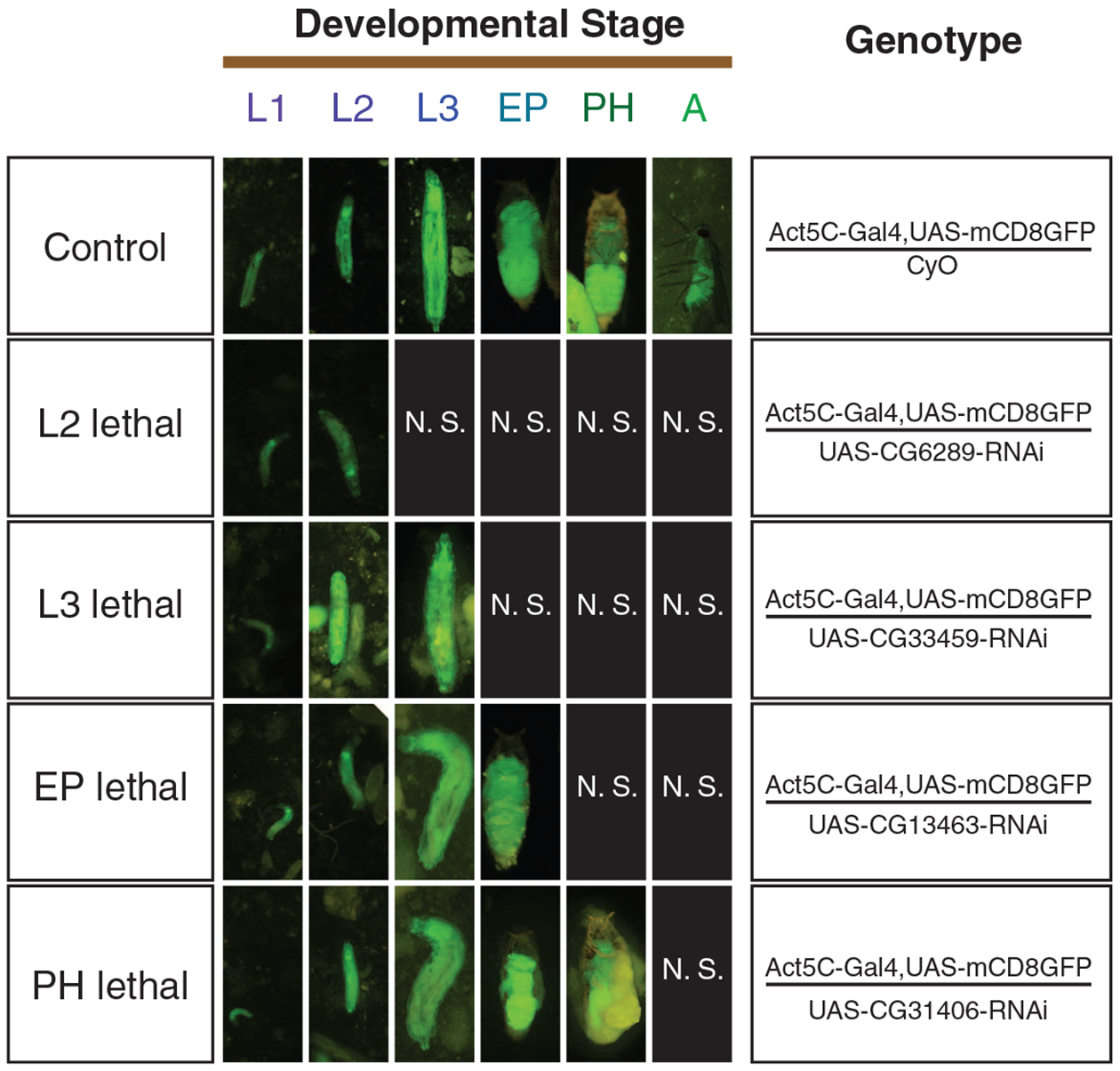
Staging lethality of gene silencing by fluorescence tracking. Living flies with RNAi–green fluorescent protein dual constructs are shown for six major D. melanogaster developmental stages: L1, first instar larva; L2, second instar larva; L3, third instar larva; EP, early pupa; PH, pharate (late pupa); A, adult. (Right) Genotypes of the flies. (Left) Stage of lethality. N.S., no flies of this genotype survived to this stage.
Examination of metamorphosis failures of pupal lethals demonstrated several distinct classes. The majorities (37 of 47, 79%) of pupal lethals were classified as class I (i.e., pharate lethal; complete pharates formed but failed in the final steps of pupal development and/or eclosion), with only a few falling into class II (pupae development aborted at the prepupal or early pupal stage, without proper formation of rudimentary heads or early leg structures) or class III (development failed over multiple stages, including prepupal, early pupal, late pupal and/or complete pharate stages) (tables S1 and S5 and fig. S3). These data suggested that young essential genes tend to play vital roles in middle or late stages of development, with a few cases in early stages.
We applied a tissue-specific loss-of-function (LOF) analysis to wing and notum development to investigate specific underlying defects (8). Under tissue-specific RNAi, almost every young essential gene we examined showed visible morphological abnormalities that were distinct in range, position, affected cell type, severity, and penetrance (Fig. 3 and table S7). Several types of canonical cellular and developmental defects were observed: (i) gross morphological defects in the overall shapes of the wing or notum (Fig. 3A and table S7); (ii) cell misdifferentiation or cell fate switching, as seen in loss of bristle cells or ectopic bristles (Fig. 3, B and E); (iii) tissue necrosis or death (Fig. 3C); (iv) tumor formation in the scalar region of the notum or tip of the wing (Fig. 3D and table S7); (v) loss of asymmetric anterior-posterior wing patterning (Fig. 3E), a classical developmental phenotype (24); and (vi) a possible signaling defect resembling the Notch phenotype in the wing (Fig. 3F). These data revealed that when the normal expression patterns of these new genes were disrupted, the development of the adult organs was affected. Taken together, knocking down young genes led to stage-specific termination of developmental processes as well as morphological defects. The developmental phenotypes of the lineage-specific genes indicate that different species likely have evolved distinct genetic components for their own development. The young gene HP6 in the D. melanogaster subgroup species is one such example (table S1) (13).
Fig. 3.
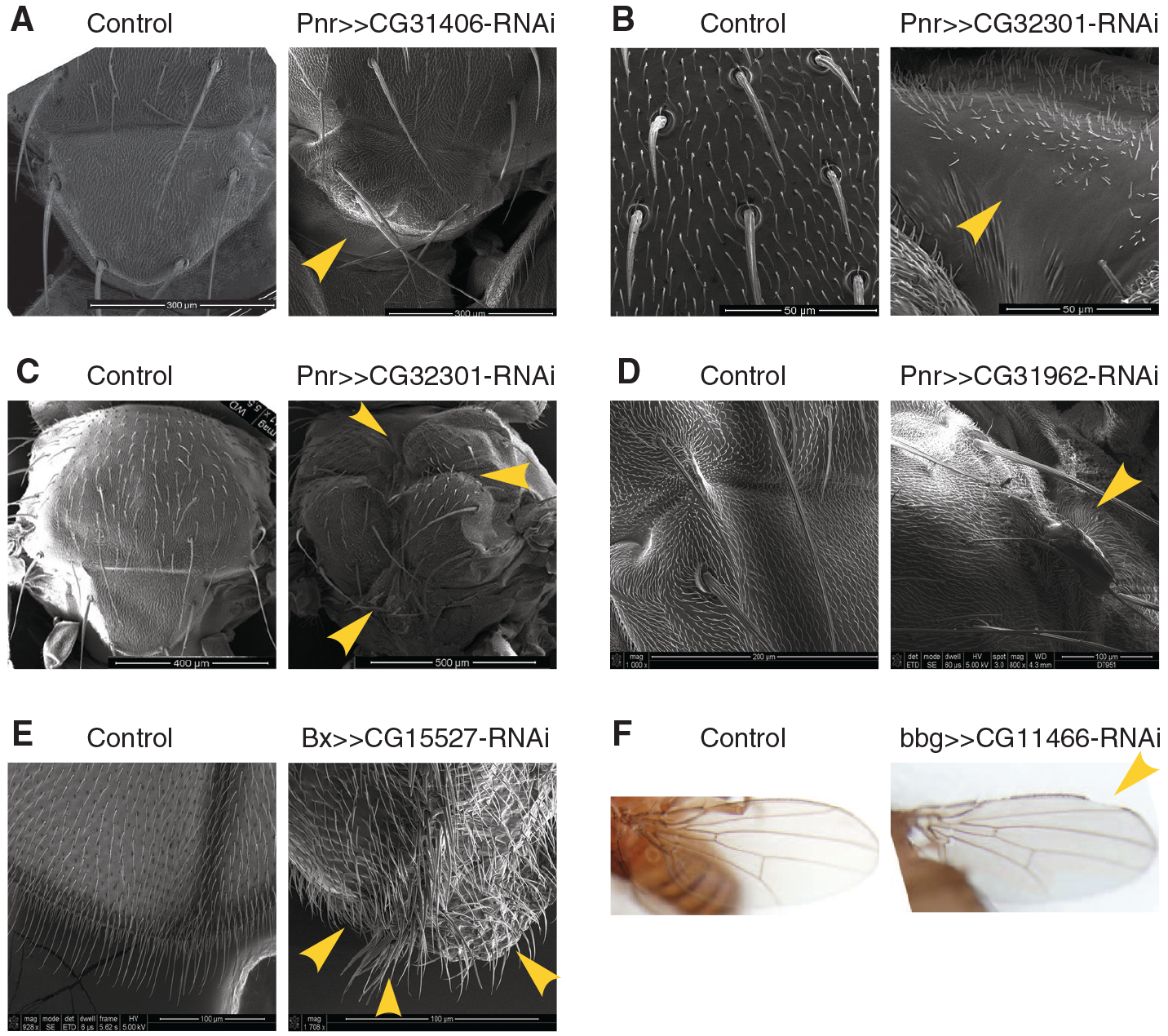
Representative cellular and developmental defects. Representative tissue-specific LOF of young essential genes leading to (A) defects in notum scutellar morphologies, (B) irregular bristle patterning and loss of bristles, (C) necrosis and tissue death at multiple places in the notum, (D) tumor formation at the junction between scutum and scutellum, (E) loss of asymmetric patterning with mirror-like wings and ectopic bristles, and (F) possible signaling defect with wing notches. Genotypes of flies are shown above each image, with scale bars in the lower right corners. Yellow arrowheads point to particular phenotypic defects.
The vast majority (56 of 59, 95%) of young essential genes were generated through gene duplication, including DNA-based duplication and RNA-based retroposition (Fig. 1, B to D, and table S2). These new duplicates often show novel chimeric gene structures, including new coding regions and untranslated regions (Fig. 1, C and D, fig. S1, and table S2). The protein sequences of these genes have drastically diverged from those of their parental copies, with a median divergence of 47.3% (table S2). A few (3 of 59) young essential genes originated de novo (Fig. 1, B and E, and table S2). In general, the proportions of new genes that are essential do not differ significantly among the three types of origination mechanisms: 32% (50 of 156) for DNA-based duplication, 26% (6 of 23) for RNA-based retroposition, and 19% (3 of 16) for de novo origination (table S9, P > 0.4).
Young essential genes appeared predominantly autosomal (57 of 59), with only two X-linked (table S2). Only 15% (2 of 13) X-linked genes examined were essential for viability, compared with the ~30 to 35% observed for both young and old autosomal genes (fig. S5), which suggests that X-linked genes are less likely to be essential for viability (two-tailed Fisher’s exact test, P = 0.047).
Sequence evolution (8) shows that young essential genes have higher protein substitution rates (fig. S7A; two-tailed Fisher’s exact test, P = 5 × 10−8) and higher Ka/Ks ratios (ratios of the rate of amino acid substitution to silent substitution) than their parental genes (fig. S7B; Wilcoxon rank test, P = 0.03), likely caused by either relaxation of functional constraint or positive selection. We measured the proportion of substitution under positive selection (α) by comparing between-and within-species variation (8). We found that old essential genes were highly constrained with a highly negative α (−1.48) (fig. S8). The essential genes aged 11 to ~35 My have a slightly negative α (−0.32), significantly higher than the previous group (likelihood ratio test, P < 0.01) (fig. S8). The youngest essential genes (<11 My) have a positive α (+0.25) (fig. S8), significantly higher than the two previous groups and their parental genes (likelihood ratio tests, P < 0.01). These analyses reveal adaptive evolution with young genes and increased purifying selection as genes become older, similar to the pattern of Adh-duplicated new genes (25).
We finally investigated the viability phenotype of the parental genes with available RNAi lines (table S10) and retrieved the phenotypic information of several additional genes from previous studies (10). We summarized the essentiality relationship between parental gene–new gene pairs and found that the parental gene of a young essential gene can be either essential or nonessential, and vice versa (tables S10 and S11). These data suggested that a new essential gene can rise from either an essential or a nonessential parent (given that it represents the ancestral state of essentiality) and that either essential genes or nonessential genes can give rise to each type of gene. These processes appeared to be relatively independent (table S11, Fisher’s Exact test, two-tailed, P = 0.296).
A previous case study of the sterile phenotype of a paternal-effect gene suggested that genes essential for fertility could arise in 10 My (26). Our observation of lethal phenotypes caused by the knockdown of young genes suggested that essential vital genes have been frequently generated in recent evolutionary periods. A new gene might not have become essential immediately after its origination. It, however, can integrate into a vital pathway by interacting with existing genes, and such interaction would be optimized by mutation and selection. This coevolution may lead to the new gene becoming indispensable. This observation is supported by our modeling (8) with large-scale interaction data (27, 28), revealing genome-wide interactions of young essential genes with many previously unrelated genes (fig. S6).
The mechanism for the evolution of essentiality would change with the types of new genes. A de novo gene has to evolve essentiality through neofunctionalization because it has no ancestral template. A duplicated gene, generated from an ancestral copy of its parental gene, could become essential from the loss of parents, or from the switch of essentiality from paralogs, or through subfunctionalization (29). However, in our data set, the vast majority of the young essential genes have detectable older and conserved paralogs (table S2) and experienced rapid sequence evolution (table S2 and Fig. S7). The prevalent gene structure renovation (table S2), together with the independence between parental gene essentiality and new gene essentiality (table S11), support the neofunctionalization origin of essentiality for most new protein-coding genes, many of which may contribute to the lineage-specific developmental program.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/330/6011/1682/DC1
Materials and Methods
References
References and Notes
- 1.Krebs JE, Goldstein ES, Kilpatrick ST, Lewin B, Lewin’s Essential Genes (Jones and Bartlett Publishers, Sudbury, Mass, ed. 2nd, 2009) [Google Scholar]
- 2.Miklos GL, Rubin GM, Cell 86, 521 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Wilson AC, Carlson SS, White TJ, Annu. Rev. Biochem 46, 573 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Krylov DM, Wolf YI, Rogozin IB, Koonin EV, Genome Res. 13, 2229 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaessmann H, Vinckenbosch N, Long M, Nat. Rev. Genet 10, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long M, Betrán E, Thornton K, Wang W, Nat. Rev. Genet 4, 865 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Clark AG et al. ; Drosophila 12 Genomes Consortium, Nature 450, 203 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Materials and methods are available as supporting material on Science Online. [Google Scholar]
- 9.Russo CA, Takezaki N, Nei M, Mol. Biol. Evol 12, 391 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Dietzl G et al. , Nature 448, 151 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Keleman K, Micheler T, VDRC project members, Personal communication to FlyBase, FBrf0208510: RNAi-phiC31 construct and insertion data submitted by the Vienna Drosophila RNAi Center (2009); http://fb2010_07.flybase.org/reports/FBrf0208510.html. [Google Scholar]
- 12.Parks AL et al. , Nat. Genet 36, 288 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Joppich C, Scholz S, Korge G, Schwendemann A, Chromosome Res. 17, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ida H et al. , Nucleic Acids Res. 37, 1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller JL, Page JL, Wolfner MF, Genetics 175, 777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrimon N, Lanjuin A, Arnold C, Noll E, Genetics 144, 1681 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh JB, Genetics 139, 421 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreitman M, Comeron JM, Curr. Opin. Genet. Dev 9, 637 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Liao BY, Zhang J, Trends Genet. 23, 378 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Makino T, Hokamp K, McLysaght A, Trends Genet. 25, 152 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Su Z, Gu X, J. Mol. Evol 67, 705 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Liang H, Li WH, Trends Genet. 23, 375 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Gauhar Z et al. , Personal communication to FlyBase, FBrf0205914: Drosophila melanogaster life-cycle gene expression dataset and microarray normalisation protocols (2008); http://flybase.org/reports/FBrf0205914.html. [Google Scholar]
- 24.Williams JA, Paddock SW, Carroll SB, Development 117, 571 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Jones CD, Begun DJ, Proc. Natl. Acad. Sci. U.S.A 102, 11373 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL, Curr. Biol 15, 87 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Giot L et al. , Science 302, 1727 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Methods Mol. Biol 342, 129 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Lynch M, O’Hely M, Walsh B, Force A, Genetics 159, 1789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. We thank C. H. Langley and D. Begun for providing polymorphism data; W. Du, J. Gavin-Smyth, Q. Guo, and M. Guffey for technical assistance and discussion; J. Coyne, M. Kreitman, and X. Ni for critically reading and/or revising the manuscript; and the members of the Manyuan Long laboratory, C. Ferguson, R. Hudson, C. I. Wu, and T. Nagylaki, for valuable discussion. S.C. was supported by University of Chicago Biological Sciences Division Fellowships. This research was supported by National Institutes of Health (R01GM065429–01A1 and R01GM078070–01A1) and National Science Foundation (CAREER Award MCB 0238168) to M.L. Y.E.Z. was also supported by the Searle Funds from Chicago Biomedical Consortium (2009, Spark).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


