Abstract
In April 2019, a suspect cluster of enterovirus cases was reported in a neonatology department in Guangdong, China, resulting in five deaths. We aimed to investigate the pathogen profiles in fatal cases, the circulation and transmission pattern of the viruses by combining metatranscriptomic, phylogenetic, and epidemiological analyses. Metatranscriptomic sequencing was used to characterize the enteroviruses. Clinical and environmental surveillance in the local population was performed to understand the prevalence and genetic diversity of the viruses in the local population. The possible source(s), evolution, transmission, and recombination of the viruses were investigated by incorporating genomes from the current outbreak, from local retrospective surveillance, and from public databases. Metatranscriptomic analysis identified Echovirus 11 (E11) in three fatal cases. Seroprevalence of neutralization antibody to E11 was 35 to 44 per cent in 3–15 age groups of general population, and the viruses were associated with various clinical symptoms. From the viral phylogeny, nosocomial transmissions were identified and all E11 2019 outbreak strains were closely related with E11 strains circulating in local population 2017–19. Frequent recombination occurred among the 2019 Guangdong E11 outbreak strains and various genotypes in enterovirus B species. This study provides an example of combining advanced genetic technology and epidemiological surveillance in pathogen diagnosis, source(s), and transmission tracing during an infectious disease outbreak. The result highlights the hidden E11 circulation and the risk of viral transmission and infection in the young age population in China. Frequent recombination between Guangdong-like strains and other enterovirus genotypes also implies the prevalence of these emerging E11 strains.
Keywords: metatranscriptomic, Echovirus 11, viral transmission, recombination, precision epidemiology
1. Introduction
Enteroviruses belong to the genus Enterovirus, family Picornaviridae and are some of the most common viral pathogens in young children (Zell et al. 2017). According to the International Committee on Taxonomy of Viruses classification, human enteroviruses can be classified into four species and include 103 serotypes: EV-A (16), EV-B (59), EV-C (23), and EV-D (5) (http://www.picornaviridae.com/enterovirus/enterovirus.htm). A broad spectrum of clinical illnesses can be caused by enterovirus infection, ranging from minor symptoms (e.g. febrile rash, mild hand, foot, and mouth diseases (HFMD)) to severe, potentially fatal conditions (e.g. aseptic meningitis, encephalitis, acute flaccid paralysis (AFP), myocarditis, and neonatal enteroviral sepsis), complicating the clinical diagnosis of enterovirus infections (Rotbart, 1995).
A wide range of enteroviruses may circulate in a population without apparent increases in illness reports (Zheng et al. 2013). However, occasionally a previously rarely reported enterovirus genotype may cause large-scale outbreaks or continuous epidemics. For instance, HFMD associated enteroviruses were sporadically identified in mainland China before 2007 but in 2007–08, EVA71 C4a genotype emerged and caused an HFMD outbreak in eastern China (Zhang et al. 2010), followed by a nation-wide epidemic and millions of HFMD cases in mainland China (Xing et al. 2014). In 2014, a rarely reported enterovirus EVD68 caused an endemic outbreak of severe respiratory illness in the USA, with more than 1,150 laboratory-confirmed cases (Greninger et al. 2015). EV-D68 again constituted a large percentage (40.9%) of reported enterovirus types in 2016. In addition, novel human enterovirus genotypes are frequently reported, sometimes regionally, potentially reflecting large enterovirus outbreaks (Han et al. 2018; Huang et al. 2018). Some genotypes like echoviruses 30, 9, and 6 cause large epidemics every 5–6 years in the EU and European Economic Area region and a rapid genogroup switch within EVA71 genotype likely took place in southeast of Asia (Geoghegan et al. 2015; Bubba et al. 2020). Therefore, a comprehensive surveillance and precise investigation of viral prevalence and transmission is critical for enterovirus disease control and intervention.
In April 2019, a potential nosocomial infection was reported from a neonatal department in Guangdong, China (http://en.nhc.gov.cn/). The infection cluster was associated with five deaths, which tested positive for enteroviruses by Multiplex real-time reverse transcription-polymerase chain reaction (RT-PCR). In this study, we used next-generation sequencing (NGS) to characterize the enteroviruses and investigate the other potential pathogen(s) in fatal cases. Echovirus 11 (E11) was identified as the causative viral pathogen. Clinical and environmental surveillances were performed to understand the hidden circulation and genetic diversity of E11 viruses in the local population. Finally, phylogenetic analysis was used to trace the possible source(s) of the virus, to identify transmission chains, and to determine recombination events of the viruses by incorporating sequences from the current outbreaks, from retrospective surveillance, and from public databases.
2. Results
2.1 Epidemiology of a nosocomial outbreak
A possible cluster of cases occurred in the neonatal intensive care units (NICUs) of hospital 1 (H1), Guangdong, China with the initial case identified on 1 April 2019. Between 1 and 20 April, a total of 120 neonates were admitted to the NICU of H1 and the cluster was associated with 5 deaths and 27 fever cases. Thirty-seven suspected cases, including the three fatal cases from H1, were transferred to four neighboring hospitals (H2–H4) during the outbreak without any notification of their possible infectious status. The fatal cases and hospital transfers caused public concern on the severity and the spread of the disease. The following fundamental questions, which were critical for public health response needed to be addressed: 1, what is the causative pathogen during the outbreak especially in the fatal cases? 2, Are all cases part of a single or multiple nosocomial transmission chain? 3, Did the nosocomial outbreak result a wider circulation of the pathogen in the population?
2.2 NGS analysis
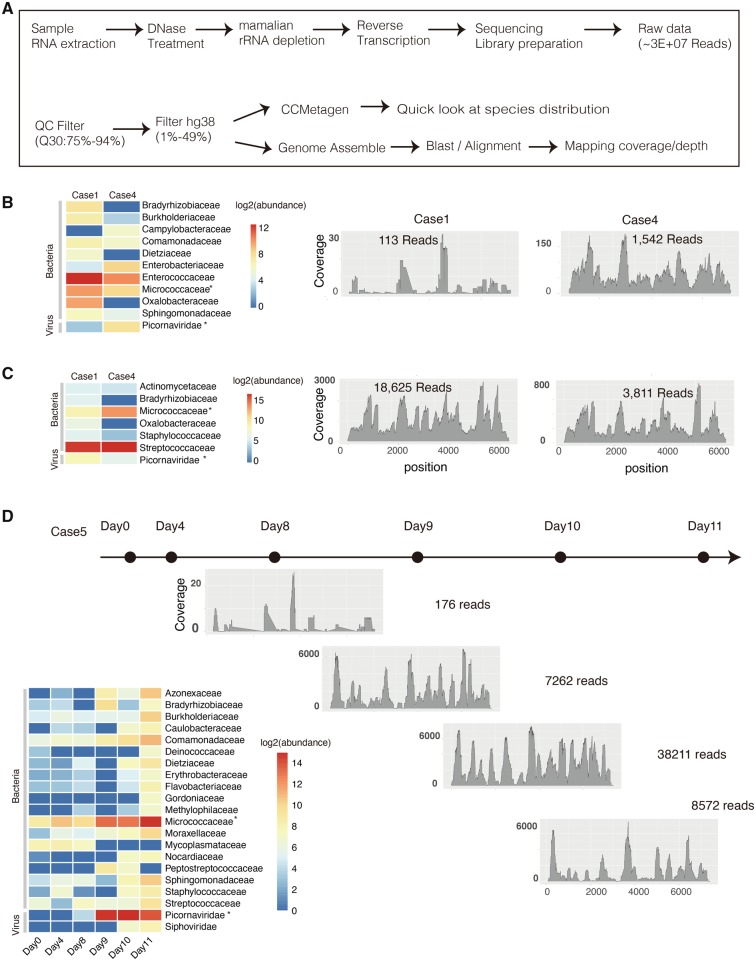
The suspected cluster of cases in H1 was first reported to Guangdong Provincial Center for Diseases Control and Prevention (GDCDC) on 25 April. Throat swabs, anal swabs and blood samples from twenty-five cases were sent to Guangdong provincial CDC for molecular diagnosis. Due to the respiratory disease-related symptoms like progressive dyspnea and pneumonia presenting in multiple cases, a multiplex RT-PCR Respiratory Pathogen Panel was performed and nine of fifteen (60%) throat swab samples from H1 were positive for enterovirus. To further genotype the enterovirus and to identify other possible pathogens in fatal cases, metatranscriptomic NGS was performed on oropharyngeal and anal swabs from fatal Cases 1 to 4, and on serial blood samples from fatal Case 5. The workflow of the metatranscriptomic sequencing is illustrated in Fig. 1A. In total, around 30 million reads (25–49 million reads) were generated per sample, of which 82 to 94 per cent reads had minimum base quality score Q30 (Table 1). After the depletion of reads mapping to the Homo sapiens genome, 2.4 to 49.2 per cent of the sequence reads remained. Notably, the number of reads drastically reduced in the anal swab and blood datasets (86.9–97.5% of reads filtered) but to a lesser extent for oropharyngeal datasets (50.8–68.2% of reads filtered). After the host genome depletion, CCMetagen was used to rapidly identify known microbial species by screening with the NCBI nucleotide (nt) database. The abundances of transcriptionally active microbes were illustrated by investigating the level of their specific transcriptomes. Specific distributions of microbe transcriptomes were found on different types of clinical samples (Fig. 1B–D). As expected, enterococci and streptococci were found as predominant microbes in anal swabs and oral swabs, respectively. Micrococci were commonly identified in all sample types with growth of abundance in time-series samples of Case 5. For viral pathogens, Picornavirius sequences were found in all sample types from the three fatal cases, and they were the most abundant in blood samples collected at the last 3 days of Case 5 (Fig. 1D).
Figure 1.
Metatranscriptomics analysis on different samples types from the three fatal cases. (A) The workflow for metatranscriptomics NGS sequencing and sequencing data analysis. Anal (B) and oral swabs (C) were sampled from Cases 1 and 3 in autopsy diagnosis. The transcriptomes abundance of different microbe families in Cases 1 and 3 was investigated by CCMetagen. The relative coverage and depth at each position of E11 genome is shown by aligning sequencing data from Cases 1 and 3 to the E11 reference genome (KY981558); (D) Serial blood samples were collected from the fatal Case 5 at different time points (marked with black dots). The dynamic change of transcriptomes abundance of different microbe families was shown in left panel. The relative coverage and depth mapping to E11 genome were shown from Day 8. Micrococci and Picornavirus which were commonly identified in all types of samples with a relative high abundance were marked with asterisk.
Table 1.
Number of reads after QC and host genome depletion as described in the workflow in Fig. 2.
| Sample | QC | Host genome depletion | Echo 11 reads | Coverage | Depth (X) | E11 RT-PCR (CT value) | |
|---|---|---|---|---|---|---|---|
| Case1 | Anal Swab | 3.5E+07 | 8.5E+05 (2.4%) | 113 | 3734 (50.2%) | 6.52 | 28 |
| Oral Swab | 3.7E+07 | 1.2E+07 (31.8%) | 18625 | 7420 (99.9%) | 1080 | 24 | |
| Case4 | Anal Swab | 4.4E+07 | 1.1E+06 (2.5%) | 1542 | 7386 (99.4%) | 62.5 | 28 |
| Oral Swab | 2.7E+07 | 1.3E+07 (49.2%) | 3811 | 7385 (99.4%) | 246.2 | 24 | |
| Case5 | Day0_Blood | 4.2E+07 | 8.5E+05 (2.0%) | 6 | 775 (10.4%) | 0.96 | NT |
| Day4_Blood | 5.8E+07 | 1.3E+06 (2.2%) | 2 | 61 (0.8%) | 0.96 | NT | |
| Day8_Blood | 2.3E+07 | 6.4E+05 (2.7%) | 176 | 2309 (31.1%) | 1.59 | 24 | |
| Day9_Blood | 3.0E+07 | 2.5E+06 (8.2%) | 7262 | 7130 (95.9%) | 1769 | 18 | |
| Day10_Blood | 2.6E+07 | 6.7E+05 (2.5%) | 38211 | 7386 (99.4%) | 2702 | 18 | |
| Day11_Blood | 2.1E+07 | 2.8E+06 (13.1%) | 8572 | 7225 (97.2%) | 1252 | 21 |
The number of reads and the coverage depth of Echo 11 virus were calculated.
Next, de novo assembly was performed. The longest assembled sequences recovered from three cases were E11 viruses which belong to enterovirus B genogroup of the Picornaviridae family. The full-genome of E11 could be recovered from the oropharyngeal sample of Case 1 (coverage 99.5%) and Case 4 (coverage 99.4%), when aligning contigs to the closely related E11 reference genome KY981558 (Fig. 2B). Consistent with the species abundance in short reads, a growth dynamic of E11 was observed in blood samples of Case 5 (Fig. 1D). Few E11 reads (Table 1) were detected in samples collected on Days 0 and 4 of Case 5. However, a sharp increase of E11 read number was observed since Day 8 (176 reads) and the abundance of blood viral load reach peak at Day 10 (38,211 reads) since the sequenced reads number was similar for these samples. The dynamic of E11 viral load in serial blood samples were also confirmed by RT-PCR method (Table 1).
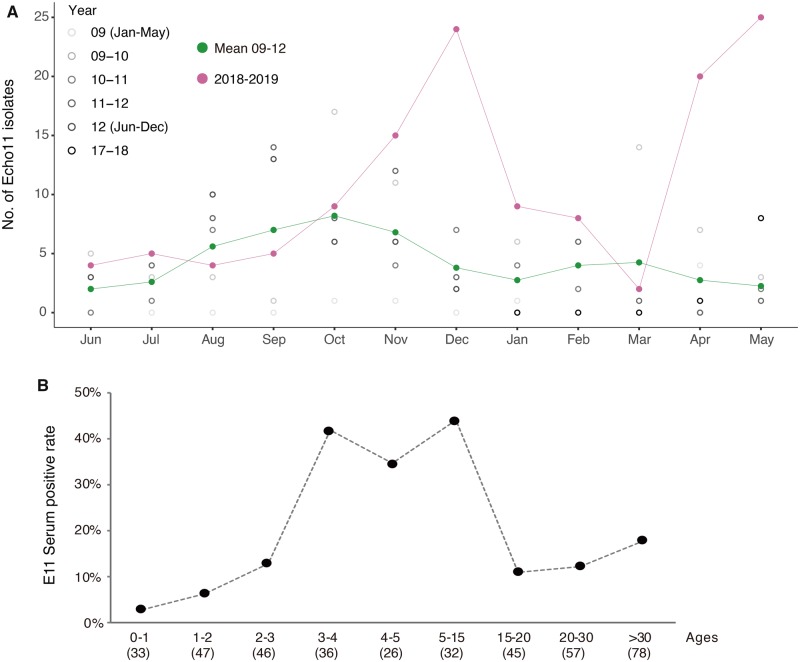
Figure 2.
Environmental surveillance and seroprevalence of E11. (A) The number of E11 isolates detected each month by cell-culture isolation from 2009 to 2012 and June 2018 to May 2019. To clearly illustrate the seasonal pattern and the increase in E11 circulation between 2009 and 2012, the number of E11 isolates from 2009 to 2012 were marked with black dots of a gradient color and presented between May to next June for each season. The mean number of E11 isolates from 2009 to 2012 and number of E11 isolates from 2018 to 2019 were marked with green line and magenta line, respectively. (B) Seroprevalence of neutralizing antibodies to E11 in different age groups of population in Guangdong, China. The number of serums tested for each group was shown in brackets.
2.3 Clinical and environmental surveillance
After the identification of echovirus E11 as causative agent, we next screened seventy samples collected from twenty-five suspicious cases from H1 (25) by using a specific E11 RT-PCR. In total, twelve of twenty-five cases from H1 were positive for E11 during the outbreak. This suggests that E11 virus was the primary causative agent for the outbreak and fatal cases, given the prevalence of the virus in all samples tested. We also enhanced the surveillance in H2–H4 where the H1 E11 patients were transferred in order to trace the possible viral transmission among hospitals. Between April and May 2019, 5 E11 infections were identified in 149 cases admitted into H2–H4 after the outbreak in H1.
Since E11 was rarely reported from clinical cases before in mainland China, the local prevalence of E11 was largely unknown based on the current national HFMD surveillance system. To address this question, we retrospectively screened E11 in ARI (acute respiratory infection), HFMDs, and AFP clinical samples collected from Guangdong provincial surveillance system 2017–18. These samples were previously detected as enterovirus positive but were not genotyped. E11 infection was found in 13 of 792 HFMD cases, three of fifty-nine AFP cases and during an ARI outbreak. The ARI outbreak occurred in a kindergarten in 2018 with ten cases developed a fever and presented respiratory infection symptoms like acute pharyngitis. E11 viruses were detected from six to ten cases in their throat swabs including a young teacher (22 years old) and five children whose age ranging from 2 to 5 years old.
Urban sewage can serve as an indicator of enterovirus prevalence in a given population (Hellmér et al. 2014; Lu et al. 2015). We analyzed sewage samples collected monthly from the treatment plant of Guangzhou city which was 50 km away from H1. Twenty-seven E11 viruses were isolated and sequenced from sewage samples collected between January 2018 and April 2019 (See detail in Section 4). As previously described, the relative viral abundance was estimated by the number of viral isolates recovered from concentrated sewage (Zheng et al. 2013). E11 typically increased from February and generally peaked in October (Fig. 2). When compared with the average number of E11 isolates between 2009 and 2012, more than five fold of E11 isolate numbers were observed in the winter peak of 2018 (December) and the summer peak of 2019 (May) suggesting a possible increase of viral prevalence in the local population (Fig. 2A).
To understand the E11 circulations among different age groups of the population, the seroprevalence of E11 neutralizing antibody (NA) was investigated from 400 healthy individuals’ serums obtained between August and October 2019 in Guangzhou city of Guangdong Province. The overall positive rate of E11 NA was 16.4 per cent (38 in 232) for male and 21.4 per cent for female (36 in 168). Notably, there was a significant increase of E11 seroprevalence with age from 0 to 4 years old (Fig. 2B). When compare with those 0 to 2 years old, the proportion of E11 positive individuals was much higher in the 3–4 age group (41.7%). The seroprevalence of E11 stayed in a relative high level in 3–15 years old individuals (34.6–43.8%) and decreased in those age over 15.
2.4 Source(s) and transmission of the viruses
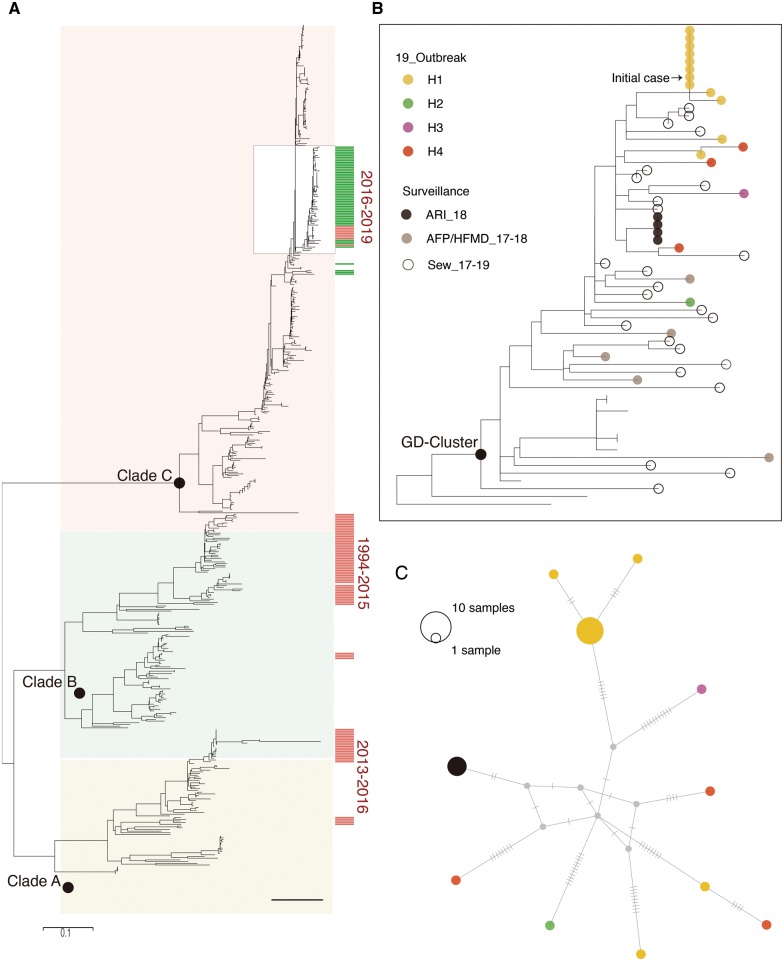
In total, fifty-four full E11 VP1 genes were sequenced from the outbreak cases in H1 (twelve cases), cases from H1 neighboring hospitals after the outbreak (five cases), and the retrospective clinical and environmental surveillance (thirty-seven sequences, 2017–18). The analysis was performed by using newly generated VP1 sequences and all published VP1 sequences from 1978 to 2016 (Fig. 3A). The phylogeny of VP1 showed E11 strains had diverged into three clades (Fig. 3A) in which each phylogenetic clade had pairwise nt identities above 75 per cent (76.9%, 76.4%, 76.4% for Clade A, Clade B, and Clade C strains, respectively). Majority (fifty of fifty-four) of E11 Guangdong strains clustered together into Clade C (Fig. 3A and B), with a common ancestor about 3 years ago, estimated by molecular clock analysis (TMRCA May 2014, 95%HPD December 2013–January 2015, Supplementary Fig. S1). Notably, all E11 sequences from the clinical cases 2017–19 were closely related to E11 viruses serially sampled from local sewage 2018–19 (Fig. 3B) reflecting these E11 clinical infections were most likely caused by the hidden circulation of E11 in Guangdong local population.
Figure 3.
Phylogenetic analysis of E11 VP1 genes. (A) ML tree of VP1 genes was constructed including the sixty-six new E11 VP1 gene sequences identified in this study (marked in orange bars on the left). To provide a background of the diversity of E11 lineages in China, the E11 sequences collected from other regions of China (1994–2016) were highlighted with a green bar. Black circles indicate bootstrap support >0.9 at the root node of selected clade. The Guangdong subclade of E11 including the outbreaks strains and retrospectively detected strains between 2017 and 2019 were enlarged in the box (B) twenty-seven clinical cases from five different hospitals (H1–H4) were highlighted with different color dots according to their administration hospitals. The E11 strains sampled from sewage each month from January 2018 to May 2019 are marked with grey dots. (C) Median-joining haplotype network constructed with clinical sequences from H1 to H4 and the ARI outbreak with the same color scheme in (B).
The molecular phylogeny also provided clues on the transmission chains of viruses. As shown in Fig. 3B, two small clusters were formed by sequences from H1 and an ARI infection in a kindergarten. For the outbreaks in H1, the initial case was the fatal Case 1 sampled on 1 April (Fig. 3B). Nine other E11 cases were sampled between 9 and 25 April, and have identical VP1 gene sequences or 2 and 3 nt synonymous changes from the sequences of the initial patient. Besides the nosocomial outbreak identified in H1, a cluster infection was also observed in cases from an ARI outbreak in October 2018 with sequences from a teacher and three children phylogenetically clustered and identical. From the phylogeny (Fig. 3B and C), we also noticed there were two cases from H1 having the viral sequencing out of the outbreak cluster suggesting these cases were not likely caused by the nosocomial outbreak strain. In other words, twelve E11 infections in H1 were likely caused by three different E11 strains highlighting the complexity of E11 circulation in Guangdong. During the first outbreak, thirty-seven cases from H1 were transferred to H2–H4 and five additional E11 positive cases were identified from these hospitals. This raised a question on the viral transmissions among hospitals. Phylogenetic analysis showed that these five cases were scattered and distinct from the outbreak strains in H1, suggesting the outbreak strains in H1 most likely did not spread to other hospitals following transfer of the E11 positive cases. However, we also noticed a sporadic infection case from H1 that was closely related to a E11 case in H4 indicating a possible transmission (Fig. 3B and C). The relationship was more clearly determined by constructing a median-joining haplotype network, which was constructed with twenty-one VP1 sequences from the clinical cases. From the topology, most of the other sample nodes could only be connected by unsampled intermediate hypothetical haplotypes except for the two infection clusters and two closely related cases from H1 to H4. These results highlighted the transmission risk of E11 viruses in young children evidenced by the nosocomial outbreak in H1 and the ARI cluster infection in kindergarten.
2.5 Recombination between E11 and other enteroviruses
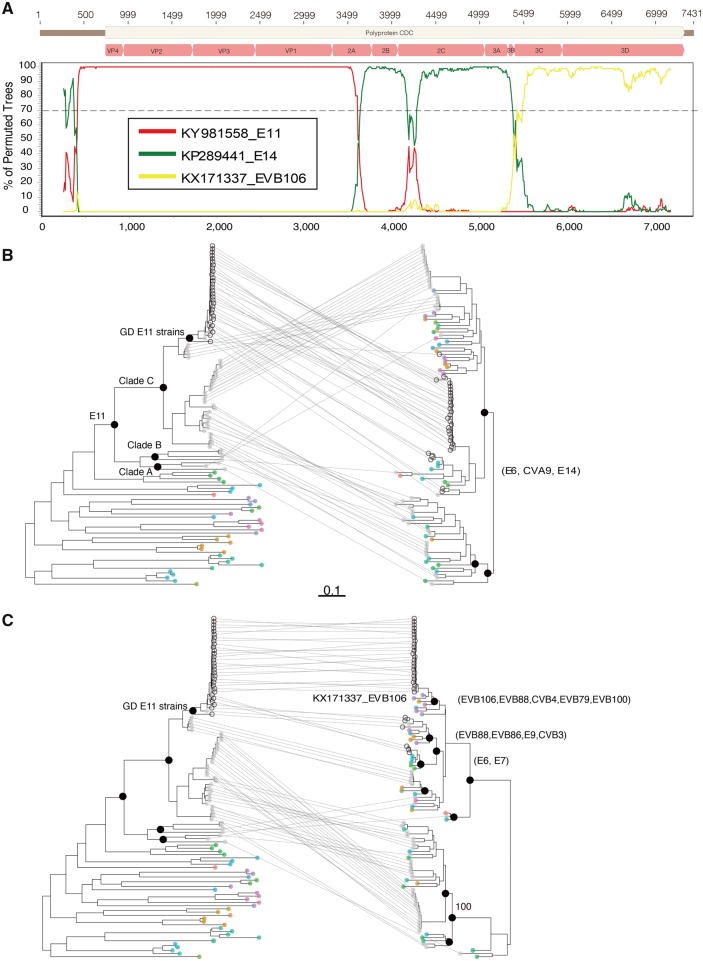
The phylogeny based on VP1 sequences suggest that the E11 strains from Guangdong 2017–19 and some strains from other parts of China 2016 formed an endemic cluster (highlighted in Fig. 3A). The de novo assembly showed some short contigs were mapped to other Enterovirus B genotypes suggesting possible recombination. To characterize these strains at genome level, we performed whole genome sequencing on thirty-two E11 strains from the 2019 outbreak, sporadic infections and sewage samples. Analyses by using Simplot suggested that two possible recombination events occurred with bootstrap values over 70 per cent. The first was identified between Guangdong E11 strains and Echovirus 14 (KP289441) in the region ‘2A-2B’ and EVB106 and the second was observed between Guangdong E11 strains and EVB106 (KX171337) in ‘3B-3’UTR’ (Fig. 4A) To further investigate the phylogenetic relationship among E11 and other enterovirus B genotypes, we constructed three datasets including the ‘5’UTR-2A, 2B-3B, and 3C-3’UTR’ part of E11 genome and other closely related enterovirus sequences (see detail in Section 4). The phylogeny of ‘5’UTR-2A’ (Fig. 4B) containing the structure protein coding sequences were consistent with the phylogeny of VP1 in Fig. 3. First, the Echo 11 genome sequences from this study (n = 30, dark grey dots), together with E11 genome sequences in public database (n = 41, light grey dots) clustered together and distinct from other enterovirus genotypes (other color dots in Fig. 3B). Second, all Guangdong E11 strains isolated in 2017–19 formed an endemic cluster in C1 clade. In contrast, the 2B-3B and 3C-3’UTR sequences of E11 were more diversified according to the phylogeny topology. For 2B-3B, there were two major clades with confident bootstrap value (>70%; Fig. 4B). E11 strains from both Clades A and B of the 5’UTR-2A phylogeny have 2B-3B sequences that fell into two different clades according to the 2B-3B phylogeny. Consistent with the Simplot result, there were E6, E14, and CVA9 strains closely related with Guangdong E11 strains based upon 2B-3B phylogeny (Fig. 2B and Supplementary Fig. S2). For 3C-3’UTR sequences, frequent recombination was observed with confident bootstrap support values among E11 strains and other enterovirus genotypes based upon the phylogeny. In particular, in the E11 Guangdong strains, at least ten different enterovirus B genotypes formed sub-clades with E11 Guangdong strains suggesting potential recombination events (Fig. 4C and Supplementary Fig. S2).
Figure 4.
Recombination analysis of E11 genomes. (A) Bootscanning recombination analysis of E11 strains and other closely related enterovirus B genotypes. The outbreak representative strain (Accession No. MN597939) was used as query sequence. The dashed line indicates >70% bootstrap support. The genome structure of the E11 was annotated according to the E11 reference strain KY981558. ML trees were construct separately for the 5’UTR-3A, 2B-3B (B) and 5’UTR-3A, 3C-3’UTR (C) parts of the E11 genome and genomes from other closely related enterovirus genotypes. The E11 genomes generated in this study (n = 32) and collected from public database (n = 41) were highlighted with black circles and light grey dots, respectively. The different parts of the genome from the same virus were connected with a line. In the phylogeny of 2B-3B and 3C-3’UTR, other enterovirus genotypes clustered with E11 suggesting potential recombination, and name of these enterovirus genotypes were noted on the right.
3. Discussion
‘Precision epidemiology’ recently proposed by Kristian et al. refers to a more targeted approach to infectious disease control at both the individual and population-level via genomic technologies, aided by using NGS and phylogenetic analyses (Grubaugh et al. 2019; Ladner et al. 2019). With the advances of NGS, clinical metatranscriptomics and metatranscriptomics, the comprehensive analysis of microbial and host genetic material (DNA and RNA) in clinical samples, is rapidly changing infectious disease diagnosis and treatment (Grubaugh et al. 2019). It allows us to determine the causative pathogen of infectious diseases and even to characterize entire viral and bacterial genomes from infected individuals nearly real-time without the need for a priori knowledge of the potential causative agent. Incorporation of the genetic sequences and epidemiological information, phylogenetic analysis can be used to trace the source, transmission, and spread of diseases at both time and spatial levels (Dudas et al. 2017; Faria et al. 2018). In response to the emergence of virus outbreaks, the viral genomic sequences are more informative due to the rapid evolution of viruses. Here, we show that using ‘precision epidemiology’ in combination with metatranscriptomics, phylogenetics, and epidemiology surveillance data can be used to untangle critical public health questions related to E11 outbreaks in Guangdong, China 2019.
By using metatranscriptomics, we confirm E11 as the genotype causing the nosocomial outbreaks and the three fatal cases sampled in this study. The retrospective surveillance in Guangdong also identifies E11 infections are associated with a wide range of symptoms including ARI, AFP, and HFMD cases. Previous surveillance data from the USA indicates that E11 infection causes fatal infections in 4.6 per cent of the cases, which is higher than infections with most of the other enterovirus genotypes (Khetsuriani et al. 2006). The most serious, potentially fatal clinical presentation caused by E11 is neonatal systemic illness, presenting like a hepatitis-hemorrhage syndrome (Nagington 1982; Khetsuriani et al. 2006). In this study, we found that pneumonia or acute respiratory diseases were observed in fatal cases at the onset of illness. In addition to the viral reads, RNA metatranscriptomics provided rRNA and mRNA profiles (Bashiardes, Zilberman-Schapira, and Elinav 2016). Micrococcus luteus was the commonly detected and dominant bacteria in oropharyngeal, anal, and blood samples from all three fatal cases (Fig. 1B–D). Similar with the dynamic of E11, the increasing level of M. luteus transcriptomes is also observed accompany with the illness progression of Case 5 (Fig. 1D, left panel). Micrococcus luteus is a constituent of the normal human bacterial flora that inhabit or contaminate the skin, mucosa, and the oropharynx, but has been reported as opportunistic pathogen for immunocompromised patients, associated with bacteremia (Peces et al. 1997), pneumonia (Dürst et al. 1991), septic arthritis (Albertson 1978), endocarditis, and meningitis (Fosse et al. 1985). The co-occurrence of M. luteus and E11 virus is associated with these severe cases is worth further investigation.
E11 has rarely been reported from clinics before in mainland China, and its prevalence and genetic distribution in most parts of the world are largely unknown. However, epidemic activity of E11 has been observed in the USA between 1970 and 2005 (Khetsuriani et al. 2006). We found evidence of widespread circulation and increasing prevalence in local sewage was observed in the years 2018–19. Moreover, the seroprevalence of E11 in young children (3–5 years old) is comparable with that of known epidemic enterovirus genotypes EVA71 and CVA16 in the local population (Li et al. 2013a). These data highlight that the infection risk of E11 may be underestimated based on the current HFMD surveillance system in China. According to the phylogenetic result, E11 strains from China (1996–2019) display a great genetic diversity. Notably, Clade C viruses were rarely detected in China before 2016 but became the dominant subgenotype of E11 in Guangdong, China 2016–19. Whether the emerging Guangdong E11-like strains are also dominant elsewhere is still elusive due to the lack of E11 sequences from public database after 2016. Frequent recombination evens are observed between E11 Guangdong strains and other enterovirus B genotypes through the 3C-3’UTR phylogeny (Fig. 4C). Based on the current public data, it is more likely that these enterovirus genotypes adapt gene fragments from E11 since majority strains in the clade are Guangdong E11 viruses. The recombination result also highlights the prevalence of Guangdong E11 strains.
Changes in prevalence in circulating enterovirus genotypes have been seen observed previously, as evidenced by EVA71 and EVD68 in the past decades. Therefore, closely monitoring the E11 prevalence and evolution in China, as well as in other parts of the world, are essential for future surveillance.
4. Materials and methods
4.1 Clinical surveillance
On 25 March 2019, an outbreak of E11 was reported in a neonatal department of a hospital (H1) in Guangdong, China. Following the pathogen confirmation, at the end of April 2019, the Guangdong Provincial CDC launched an E11 molecular surveillance in H1 and H2–H4 where H1 patients were transferred during the outbreak. The seroprevalence of E11 was evaluated by using serum samples from health population in Guangzhou city of Guangdong province. The population were divided into 0–2, 3–5, 6–25, 26–60 age groups and one hundred serum samples were randomly selected for NA testing from each group. The Ethics Review Committee of the Guangdong CDC confirmed that all methods were performed in accordance with the relevant guidelines and regulations and molecular surveillance and metatranscriptomic analyses were performed for anonymized patient samples. All patients or their guardian(s) were informed about the surveillance before providing written consent.
4.2 NA assay
The NA tests were performed as previously described (Li et al. 2013b). To define the E11 NA, the outbreak strain (Accession No. MN597939) was used. All NA assays were run in 96-well microplates. Serum samples were diluted two fold from 1:8 to 1:1,024. The antibody titer of the sample was defined as the highest dilution that could inhibit CPE development in 50 per cent of the virus-infected wells. A titer ≥ 8 was considered as seropositive.
4.3 Multiplex RT-PCR and metatranscriptomics sequencing
Multiplex RT-PCR (NxTAG Respiratory Pathogen Panel Plate, Luminex, Cat. No. I051C0447) was applied on fifteen throat swab samples from H1 with similar respiratory disease symptoms. For enterovirus positive samples metatranscriptomic analysis was performed to determine the enterovirus genotype and to recovering the enterovirus genome from three fatal cases. E11-specific RT-PCR was used to screen other suspicious samples and for the retrospective surveillance. For metatranscriptomics, total RNAs were extracted from different types of samples by using QIAamp Viral RNA Mini Kit (Qiagen, Cat. No. 52904), followed by DNase treatment and purification with TURBO DNase (Invitrogen, Cat. No. AM2239) and Agencourt RNAClean XP beads (Beckman Cat. No. A63987). Both the concentration and the quality of all isolated RNA samples were measured and checked with the Agilent Bioanalyzer 2100 and Qubit. Libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit v2 (Clontech, Cat. No. 634412) according to the manufacturer’s protocol starting with 10 ng total RNA. Briefly, purified RNA was first fragmented and converted to cDNA using reverse transcriptase. The ribosome CDNA was depleted by using ZapRv2 (mammalian-specific). The resulting cDNA was converted to double-stranded DNA and subjected to end-repair, A-tailing, and adapter ligation. The constructed libraries were amplified using nine to sixteen cycles of PCR. Sequencing of metatranscriptome libraries was conducted using Illumina Novaseq 6000 (150 bp paired-end sequencing of ∼250 bp inserts).
The datasets were quality controlled (QC) using fastp (Chen et al. 2018) to trim artificial sequences (adapters), to cut low-quality bases (quality scores < 30), and to remove short reads (< 50 bp). Reads were then mapped to the human genome (hg38) using Bowtie2 (Langmead and Salzberg 2012) to remove human-related RNA reads. General taxonomic profiles of the microbe were generated using CCMetagen with ncbi_nt database (Marcelino et al. 2019), and all best mappings reach the alignment score threshold (0.5) and P-value threshold (0.5) were printed. The abundance of microbes transcriptomes identified in oral swabs, anal swabs, and serial blood samples were calculated and normalized with number of raw reads using R scrips. De novo assembling was performed using Trinity for each dataset after human RNA depletion. Assembled contigs were annotated using blastn (Camacho et al. 2009) using the complete nt database. The depth and coverage of E11-specific reads were calculated by using samtools (Li et al., 2009). Whole genome of E11 viruses were sequenced by using the Enterovirus Genome Sequencing Kit (Gene Denovo, China). All sequences generated in this study have been submitted to GenBank under the accession numbers MN597923–MN597954, MN121645–MN121661, MN579559–MN579569, and MN198979–MN199003. The metatranscriptomic raw data after filtering with host genome were submitted to National Genomics Data Center (https://bigd.big.ac.cn/) with submission number (subSAM090965).
4.3 Phylogenetic analysis
For VP1 phylogeny, the E11 VP1 sequences generated in this study were combined with all publicly available VP1 genes sequences in GenBank with known sampling dates (Supplementary Table S2). In total, 502 VP1 gene sequences were included in the phylogenetic analysis, including the 54 sequences generated in this study. Maximum-likelihood (ML) trees were estimated for VP1 genes in RaxML (Stamatakis 2014). Median-joining haplotype network of clinical cases was constructed by using Popart (Bandelt, Forster, and Rohl 1999). Potential intertypic and intratypic recombination was analyzed using the SimPlot 3.5.1 program (Lole et al. 1999) with a 500-nt window moving in 40-nt steps and Jukes–Cantor correction. To investigate the recombination relationship among E11 and other enterovirus genotypes, the E11 genome was spliced at the suggested recombination break points. Blast searches were performed by using three different parts of E11 genome sequences (5’UTR-2A, 2A-2B, 3B-3’UTR), separately. The closely related enterovirus genome sequences were selected and three datasets were built for different parts of genome. ML trees were estimated for two datasets separately in RaxML (Stamatakis, 2014). Phylogenetic trees were annotated and visualized with ggtree (Yu et al. 2017).
Data availability
Data are available through Dryad.
Supplementary Material
Acknowledgments
We gratefully acknowledge the efforts of local CDCs in the investigation and sample collection.
Funding
This work was supported by grants from Science and Technology Planning Project of Guangdong [2018B020207006], Science and Technology Planning Project of Guangzhou [201804010030], National Natural Science Foundation of China [81660280], and European Union’s Horizon 2020 grant COMPARE [643476].
Conflict of interest: None declared.
References
- Albertson D. (1978) ‘Septic Shock with Micrococcus luteus’, Archives of Internal Medicine, 138: 487. [PubMed] [Google Scholar]
- Bandelt H. J., Forster P., Rohl A. (1999) ‘Median-Joining Networks for Inferring Intraspecific Phylogenies’, Molecular Biology and Evolution, 16: 37–48. [DOI] [PubMed] [Google Scholar]
- Bashiardes S., Zilberman-Schapira G., Elinav E. (2016) ‘Use of Metatranscriptomics in Microbiome Research’, Bioinformatics and Biology Insights, 10:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubba L. et al. (2020) ‘Circulation of Non-Polio Enteroviruses in 24 EU and EEA Countries between 2015 and 2017: A Retrospective Surveillance Study’, The Lancet Infectious Diseases, 20: 350–61. [DOI] [PubMed] [Google Scholar]
- Camacho C. et al. (2009) ‘BLAST+: Architecture and Applications’, BMC Bioinformatics, 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. et al. (2018) ‘Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor’, Bioinformatics, 34: i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G. et al. (2017) ‘Virus Genomes Reveal Factors That Spread and Sustained the Ebola Epidemic’, Nature, 544: 309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürst U. N. et al. (1991) ‘Micrococcus luteus: A Rare Pathogen of Valve Prosthesis Endocarditis’, Zeitschrift fur Kardiologie, 80: 294–8. [PubMed] [Google Scholar]
- Faria N. R. et al. (2018) ‘Genomic and Epidemiological Monitoring of Yellow Fever Virus Transmission Potential’, Science, 361: 894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosse T. et al. (1985) ‘Meningitis Due to Micrococcus luteus’, Infection, 13: 280–1. [DOI] [PubMed] [Google Scholar]
- Geoghegan J. L. et al. (2015) ‘Phylodynamics of Enterovirus A71-Associated Hand’, Journal of Virology, 89: 8871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger A. L. et al. (2015) ‘A Novel Outbreak Enterovirus D68 Strain Associated with Acute Flaccid Myelitis Cases in the USA (2012–14): A Retrospective Cohort Study’, The Lancet Infectious Diseases, 15: 671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D. et al. (2019) ‘Tracking Virus Outbreaks in the Twenty-First Century’, Nature Microbiology, 4: 10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z. et al. (2018) ‘Genetic Characterization and Molecular Epidemiological Analysis of Novel Enterovirus EV-B80 in China’, Emerging Microbes & Infections , 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M. et al. (2014) ‘Detection of Pathogenic Viruses in Sewage Provided Early Warnings of Hepatitis a Virus and Norovirus Outbreaks’, Applied and Environmental Microbiology, 80: 6771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. et al. (2018) ‘Antigenic Characteristics and Genomic Analysis of Novel EV-A90 Enteroviruses Isolated in Xinjiang’, Scientific Reports, 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetsuriani N., et al. ; Centers for Disease Control and Prevention. (2006) ‘Enterovirus Surveillance–United States, 1970-2005’, Morbidity and Mortality Weekly Report Surveillance Summaries, 55: 1–20. [PubMed] [Google Scholar]
- Ladner J. T. et al. (2019) ‘Precision Epidemiology for Infectious Disease Control’, Nature Medicine, 25: 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2013. a) ‘Seroepidemiology of Human Enterovirus71 and CoxsackievirusA16 Among Children in Guangdong Province’, BMC Infectious Diseases, 13: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2013. b) ‘Seroprevalence of Human Enterovirus 71 and Coxsackievirus A16 in Guangdong, China, in Pre- and Post-2010 HFMD Epidemic Period’, PLoS One, 8: e80515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S. et al. (1999) ‘Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination’, Journal of Virology, 73: 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2015) ‘Elucidation of Echovirus 30’s Origin and Transmission during the 2012 Aseptic Meningitis Outbreak in Guangdong, China, through Continuing Environmental Surveillance’, Applied and Environmental Microbiology, 81: 2311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino V. R. et al. (2019). ‘CCMetagen: Comprehensive and Accurate Identification of Eukaryotes and Prokaryotes in Metagenomic Data’, BioRxiv, 641332. [DOI] [PMC free article] [PubMed]
- Nagington J. (1982) ‘Echovirus 11 Infection and Prophylactic Antiserum’, The Lancet, 319: 446. [DOI] [PubMed] [Google Scholar]
- Peces R. et al. (1997) ‘Relapsing Bacteraemia Due to Micrococcus luteus in a Haemodialysis Patient with a Perm-Cath Catheter’, Nephrology Dialysis Transplantation, 12: 2428–9. [DOI] [PubMed] [Google Scholar]
- Rotbart H. A. (1995) ‘Enteroviral Infections of the Central Nervous System’, Clinical Infectious Diseases, 20: 971–81. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics, 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W. et al. (2014) ‘Hand, Foot, and Mouth Disease in China, 2008–12: An Epidemiological Study’, The Lancet Infectious Diseases, 14: 308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. et al. (2017) ‘ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data’, Methods in Ecology and Evolution, 8: 28–36. [Google Scholar]
- Zell R., ICTV Report Consortium, et al. (2017) ‘ICTV Virus Taxonomy Profile: Picornaviridae’, Journal of General Virology, 98: 2421–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2010) ‘An Emerging Recombinant Human Enterovirus 71 Responsible for the 2008 Outbreak of Hand Foot and Mouth Disease in Fuyang City of China’, Virology Journal, 7: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H. et al. (2013) ‘Prevalence of Nonpolio Enteroviruses in the Sewage of Guangzhou City, China, from 2009 to 2012’, Applied and Environmental Microbiology, 79: 7679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available through Dryad.