Abstract
The Arctic is warming at an unprecedented rate, with unknown consequences for endemic fauna. However, Earth has experienced severe climatic oscillations in the past, and understanding how species responded to them might provide insight into their resilience to near-future climatic predictions. Little is known about the responses of Arctic marine mammals to past climatic shifts, but narwhals (Monodon monoceros) are considered one of the endemic Arctic species most vulnerable to environmental change. Here, we analyse 121 complete mitochondrial genomes from narwhals sampled across their range and use them in combination with species distribution models to elucidate the influence of past and ongoing climatic shifts on their population structure and demographic history. We find low levels of genetic diversity and limited geographic structuring of genetic clades. We show that narwhals experienced a long-term low effective population size, which increased after the Last Glacial Maximum, when the amount of suitable habitat expanded. Similar post-glacial habitat release has been a key driver of population size expansion of other polar marine predators. Our analyses indicate that habitat availability has been critical to the success of narwhals, raising concerns for their fate in an increasingly warming Arctic.
Keywords: climate change, mitochondrial genomes, demographic history, phylogeography, Arctic
1. Introduction
Ongoing global warming is impacting ecosystems across our planet at an unprecedented rate [1]. The Arctic is warming faster than any other region on Earth, and Arctic summer sea ice is likely to disappear before mid-century [2]. There is evidence of rapid biophysical changes in Arctic ecosystems, which is impacting the range, abundance, phenology, growth and condition of native fauna [3,4]. Arctic endemics are at risk from climate warming due to habitat fragmentation and loss, and increasing anthropogenic activities, as previously impenetrable areas become available for human exploitation [5]. Climatic perturbations are further affecting the access of Arctic marine mammals to mating, nursing and foraging areas, which is key to population viability [6]. Arctic food webs are dependent on sea ice associated algae and other sympagic organisms, and on the ocean's vertical structure. Biophysical changes in sea ice dynamics or ocean circulation can therefore cascade up through the trophic layers, with major impacts on top predators [7,8].
Investigations of the impact of past climatic shifts on populations might provide insight into faunal resilience to near-future predictions of global warming. Earth has experienced severe and rapid climatic oscillations in the past, including the interstadial events of the last glacial period 110 to 11.65 thousand years ago (ka), and the Pleistocene/Holocene transition (11.65 ka) [9]. During the Last Glacial Maximum (LGM; 26.5 to 19 ka), sea ice coverage extended considerably towards lower latitudes and then rapidly contracted in the early Holocene [10].
Studies reconstructing the impact of past climatic perturbations on Arctic mammalian faunas have mainly focused on terrestrial populations; investigations of the responses of Arctic marine mammals to past long-term environmental changes are largely lacking. During the warm interstadials of the last glacial period and after the LGM, decreases in suitable habitat contributed to the decline of Holarctic terrestrial mammals, which led to the extirpation and in some cases the extinction of taxa [11–14]. By contrast, several populations of cold-adapted marine predators in the southern hemisphere, both in the sub Antarctic and the Antarctic, including southern elephant seals (Mirounga leonina) and various penguin species, increased after the LGM. These expansions reflect access to new island breeding sites in areas previously covered by ice and/or access to productive foraging grounds proximate to breeding locations [15–17].
Three cetacean species are endemic to the Arctic: narwhals (Monodon monoceros), belugas (Delphinapterus leucas, also called white whales) and bowhead whales (Balaena mysticetus). Each of these whale species is uniquely adapted to the extreme conditions of an Arctic existence, and their evolution and ecology are intrinsically tied to the past and present sea ice dynamics of the region. Mitochondrial control region sequences retrieved from Holocene subfossil remains have shown that North Atlantic bowhead whale populations increased after the LGM [18], suggesting the species responded differently than its terrestrial counterparts to past climatic shifts. However, it is not known whether the pattern observed in bowheads is mirrored in narwhals and belugas.
Eleven marine mammal species are found in the Arctic; in addition to the three cetacean species, these include six pinniped species and polar bears. Narwhals are considered one of the most sensitive of these Arctic marine mammals to sea ice loss and associated trophic cascades, and also to increases in human activities as sea ice disappears [5,19]. Narwhals are distributed across the Atlantic region of the High Arctic, from eastern Arctic Canada to Franz Josef Land in Russia [20] (figure 1a; electronic supplementary material, figure S1). Most narwhal populations are seasonally migratory and follow traditional routes of up to 1700 km along the sea ice edge in autumn and spring. They winter in deep and densely ice-covered offshore habitats, where they forage heavily, and summers are spent in ice-free coastal areas and fjords [22–24]. Satellite-tracking studies since the early 1990s indicate that different groups of individuals (termed stocks) show fidelity to different summering grounds, such as fjords, bays or sounds [23,24]. Narwhals in the Barents Sea differ; they rarely come into coastal shelf areas, but rather remain in deep, ice-covered areas year-round [21,25].
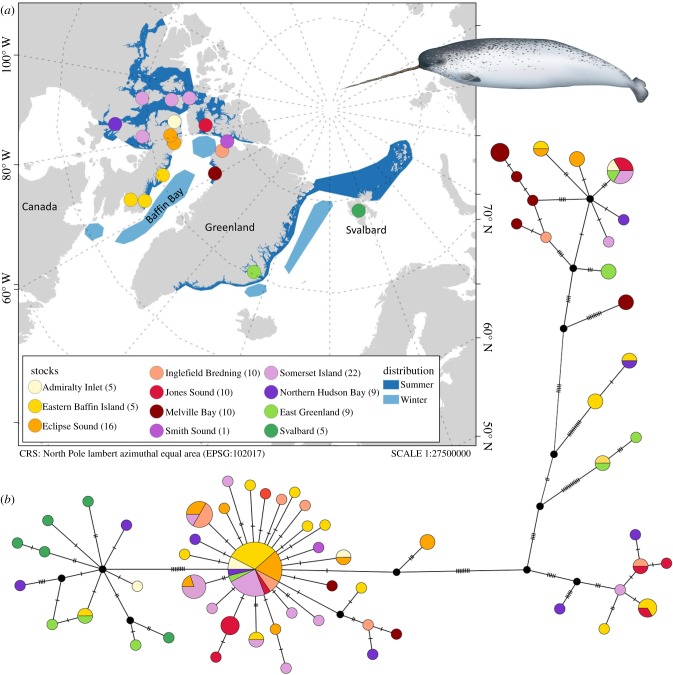
Figure 1.
(a) Map of sampled narwhal stocks. Three stocks comprise several sample localities: Eastern Baffin Island, Eclipse Sound and Somerset Island [20]. The summer and winter distributions have been re-drawn from the latest NAMMCO report [20]. Note that a recent study indicated that narwhals between North-East Greenland and Svalbard are present year-round [21] (b) Median-joining haplotype network of the 64 haplotypes found within the 121 narwhal mitogenome sequences. Each circle represents a haplotype and is coloured in proportion of individuals from each stock, where the haplotype is found. Circle size is proportional to haplotype frequency. Black circles indicate intermediate haplotypes not found in our samples. Black dashes indicate mutation steps between haplotypes; of note, distances between haplotypes are not to scale. Narwhal illustration by Uko Gorter.
Over recent decades, environmental impacts in the form of shifts in sea ice extent have altered migratory routes, with possible impacts on the diet of narwhals in Arctic Canada [26]. Understanding recent trends, in conjunction with long-term evolutionary responses, are necessary to reliably assess the resilience of narwhals to an increasingly warming Arctic. Little is currently known about the genetic structure and evolutionary trajectory of the species. Previous population genetic work, based on short (287 bp) mitochondrial control region sequences and 12 microsatellites, reported low levels of genetic structuring and diversity across the narwhal range [27–29]. In addition, low levels of diversity have been reported in the nuclear genome of the species [30].
Here, we present complete mitochondrial genomes (mitogenomes) from 121 narwhals sampled across their range (figure 1a); our study focuses on mitogenomes, as they are a useful marker for inferring phylogeographic and evolutionary processes in species with low genetic diversity (e.g. [31,32]). Our analysis elucidates population structure and diversity patterns across the species. We compare diversity levels in narwhals with those of other cetacean species in the Arctic and elsewhere, to evaluate their relative diversity. To address the resilience of narwhal populations to near-future projections, we use species distribution models to reconstruct their demographic history and assess the impact of past and ongoing climatic shifts.
2. Material and methods
(a). Samples
We collected tissue samples from 121 narwhals sampled across the species range (figure 1a; map produced in QGis v. 3.4.2 [33]). Samples were collected from 11 of the 12 narwhal stocks recognized by the North Atlantic Marine Mammal Commission (NAMMCO) [20]; at the time of this study, no material was available from the North-East Greenland stock. Stocks correspond to summer aggregations and constitute management units based on distribution, telemetry, ecological and, in some cases, genetic data [20]. Three stocks included several sample localities (figure 1a). Samples were collected during summer between 1982 and 2012, except from one sample which is from 1905 and came from subsistence hunts or from animals biopsied during satellite-tagging studies (electronic supplementary material, table S1). Samples were shipped to Denmark under CITES exemption number DK003.
(b). DNA extraction, amplification and sequencing
In short, we extracted DNA from tissue samples using the Qiagen Blood and Tissue Kit, with minor modifications, and fragmented the DNA to approximately 350–550 bp. Libraries were prepared and sequenced according to two different protocols. For 84 samples, libraries were built using the Illumina NeoPrep and sequenced on an Illumina HiSeq 2500 with the 80 bp SE technology. For 37 samples, libraries were built using the BEST protocol (i.e. blunt-end single-tube library building for modern and ancient DNA) [34] and sequenced on an Illumina HiSeq Xten with the 150 bp PE technology. Full details on DNA extraction, library preparation and sequencing are included in the electronic supplementary material.
(c). Bioinformatics
After demultiplexing, sequencing reads were processed using PALEOMIX v. 1.2.13.1 [35] (see electronic supplementary material, text for details). Briefly, the PALEOMIX pipeline (i) trimmed read ends for residual adapters and low-quality stretches, (ii) mapped reads to the published narwhal mitogenome reference (GenBank accession number: NC_005279) [36] using bwa v. 0.7.15 [37], requiring a minimum mapping quality of 30, (iii) removed duplicates and (iv) re-aligned indels. We built consensus sequences using the FastaAlternateReferenceMaker function in GATK v. 3.8.1 [38]. We aligned sequences using the ClustaIW algorithm in MEGA X [39]. Our QC procedures are described in electronic supplementary material, text.
(d). Haplotype network
We used PopArt v. 1. 7 [40] to generate a mitogenome haplotype median-joining network.
(e). Diversity statistics––narwhals
We calculated two measures of genetic diversity, haplotype diversity (h) and nucleotide diversity (π) for each of the ten stocks in DnaSP v. 6.12.03 [41]; Smith Sound (n = 1) was omitted due to small sample size. We tested differences in haplotype diversity and nucleotide diversity among the 10 stocks using genetic_diversity_diffs v. 1.0.3 [42].
(f). Diversity statistics––cetaceans
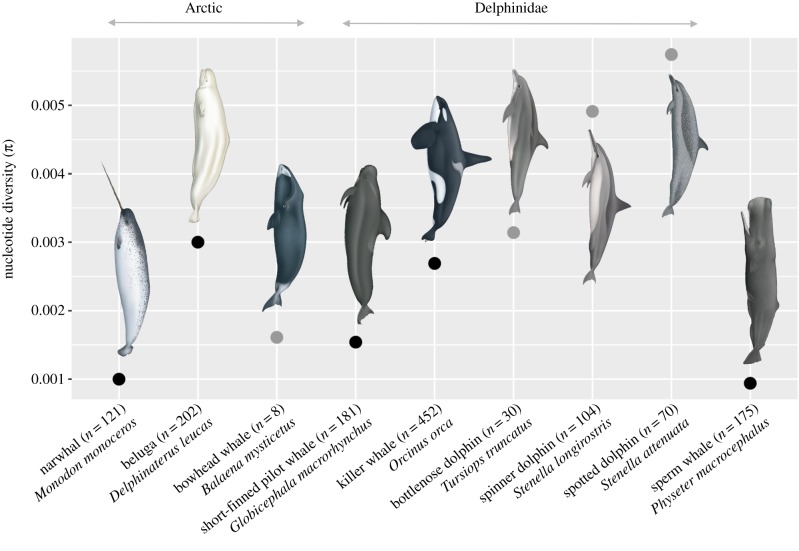
To evaluate the relative level of mitochondrial diversity in narwhals, we compared the range-wide level of nucleotide diversity (π) with available population-scale mitogenome data of cetaceans in the Arctic and elsewhere, from the published literature. Studies were found for eight species, using the keywords ‘mitogenome’ and ‘bowhead whale’, ‘dolphin’ or ‘whale’. Data were obtained from GenBank or from the authors of the studies. These included range-wide data, as for narwhals, and local data for species where range-wide data were unavailable (see electronic supplementary material text for details).
We estimated π in DnaSP using range-wide data for belugas (Skovrind et al. 2019, unpublished data) and local-scale data for bowhead whales, as data are only available from Spitsbergen for the latter [43]. For belugas, which have a circumpolar distribution and for which range-wide data were available, we also estimated π for the Atlantic Arctic, to enable direct comparison with the distribution of narwhals. In addition, we estimated π for several Delphinidae species using range-wide data for killer whales (Orcinus orca), short-finned pilot whales (Globicephala macrorhynchus), local-scale data for bottlenose dolphins (Tursiops truncatus), spinner dolphins (Stenella longirostris) and spotted dolphins (Stenella attenuata), as well as for the physeterid sperm whales (Physeter macrocephalus) using range-wide data [31,32,44–46].
(g). Fixation statistics
To address levels of genetic differentiation among summer localities, we estimated ФST and FST using Arlequin [47]. We chose the Tamura & Nei [48] model of substitution, which was selected with PartitionFinder v. 2.1.1 [49].
(h). Phylogenetic analysis
To estimate divergence times among narwhal clades, we used a two-phase approach following the procedure outlined previously [31,32,44]. First, we built an odontocete fossil-calibrated phylogenetic tree with one sequence from each of 14 species of toothed whales, and four narwhal sequences from the most divergent groups identified in the haplotype network. The objective was to determine a root calibration (time to the most recent common ancestor, TMRCA) for use in the narwhal phylogeny. The odontocete phylogenetic tree construction is described in electronic supplementary material, text. The two phylogenetic analyses were run in Beast v. 2.5.1 [50].
The phylogenetic analysis of narwhals included the 64 unique mitogenome haplotypes identified. We combined the data into six subsets that were (i) first, (ii) second and (iii) third codon positions of all protein coding regions, (iv) tRNAs, (v) rRNAs and (vi) control region. Optimal substitution models for each of the six subsets were identified in PartitionFinder (electronic supplementary material, table S3b). We also ran the analysis on the 43 unique haplotypes of the third codon positions of the protein coding regions only, as the first and second positions may be impacted by purifying selection, leading to an overestimation of substitution rates at recent timescales [51]. We used a constant size coalescent tree model because the sequences were from a single species, and applied a strict clock, because we expect little heterogeneity in clock rate within narwhals. We applied a calibration to the root age of the tree (TMRCA), which is the mean and 95% highest posterior density (HPD) of the oldest divergence within narwhals, as estimated in the odontocete phylogeny. For further details on the analysis, see electronic supplementary material, text. Clades which lack support (posterior values < 0.9) were collapsed.
(i). Bayesian skyline analysis
To reconstruct the demographic history of narwhals, we ran a coalescent Bayesian skyline analysis in BEAST [50], using both the fully partitioned mitogenome sequences and only the third codon positions of the protein coding regions of all 121 sequences (see electronic supplementary material, text for the latter result). Our sampling across the range of the species (11 of 12 recognized stocks were included in our analysis) has been shown to be the most reliable for inferring demographic changes [52]. We calibrated the age of the root using the mean and 95% HPD estimate from the phylogenetic tree of phase 2 (details on the analysis are provided in the electronic supplementary material). We used a generation time of 30 years to scale the population size estimates [53].
(j). Species distribution models
We used AquaMaps to predict suitable habitat for narwhals for three climatic periods: the LGM, representing a glacial period, the present, representing an interglacial period, and year 2100; [54–56] (see details in electronic supplementary material, text). Narwhal summer, winter and year-round species envelopes were generated using distribution maps published in the Global Review of Monodontids report (figure 1a; electronic supplementary material, figure S1) [20]. Ideally, raw sighting data points should have been used, but these data do not exist for all regions.
The final envelopes included three parameters that are known to drive the distribution of the species: sea ice concentration, depth and sea surface temperature [57,58]. Models including more environmental parameters were also run and are detailed in the electronic supplementary material, text. Using the environmental envelopes, we computed predictions of habitat suitability in the Atlantic Arctic. Predictions were plotted using QGIS, considering a probability threshold of 0.6, which represents habitats of high suitability [54]. We restricted the predictions to the Atlantic as narwhals are not distributed in the Pacific, and we calculated and compared the size and mean latitude of suitable habitat between the three periods. We note that densities are not considered, and hence predictions do not reflect fine habitat preferences. We used mean annual average values for the parameters for the three periods, despite the availability of higher-resolution data for the present. This was done to enable direct comparison of suitable habitat estimates across time bins.
3. Results
Mean read coverage of the complete mitogenome sequences from 121 narwhals spanning the geographic range of the species (figure 1a) was 439x (26x–9400x). QC statistics are presented in the electronic supplementary material.
(a). Haplotype network
The 121 sequences included 164 variable sites identifying 64 unique haplotypes. The most divergent haplotypes were separated by 55 mutations. We identified several major clades in the haplotype network, although these were not geographically structured (figure 1b). Each clade had a star-like topology, indicating population expansion. Svalbard (n = 5) was the only location where all individuals shared haplotypes that were closely related, separated by eight mutations at most.
(b). Diversity statistics
Mean haplotype diversity was 0.95 (s.d. = 0.02) and mean nucleotide diversity was 0.001 (s.d. = 0.0001). Nucleotide diversity was the highest in East Greenland and the lowest in Svalbard (electronic supplementary material, figure S2a). We present the values per summer locality in electronic supplementary material, figures S2a,b and the test for significant differences in diversity between stocks in electronic supplementary material, table S4 and text.
The level of mitogenome nucleotide diversity across narwhals (0.001) was among the lowest recorded among cetaceans for which population-level mitogenome data are publicly available (figure 2). The value estimated in narwhals was lower than in belugas and bowhead whales, and in any other species of Delphinidae studied at their range-wide or local scale. For belugas, we also estimated π for the Atlantic Arctic (n = 113) to reflect the geographic range of narwhals and obtained the same value of 0.003 as in the circumpolar beluga data. The value estimated in narwhals was similar to the range-wide estimate of another toothed whale, the sperm whale, which has previously been reported to have extremely low levels of mitogenomic diversity [32].
Figure 2.
Mean nucleotide diversity (π) from published, population-level cetacean mitogenome studies from the Arctic and elsewhere. Dark dots indicate range-wide data and grey dots local data. The number of samples comprising each dataset is shown. Cetacean illustrations by Uko Gorter.
(c). Fixation statistics
Based on ФST, we found that Svalbard and Melville Bay stocks were significantly differentiated from all other stocks; FST values mainly differentiated Melville Bay from the rest (electronic supplementary material, table S5). Narwhals from East Greenland were significantly differentiated from six of the ten other stocks based on ФST. For Melville Bay, closely related individuals could have been sampled, as 6 out of 10 samples were collected on the same day. The six samples include closely related sequences, separated by seven mutations at most across the mitogenome. This could skew the results by increasing differentiation among stocks, if individuals within a group are more related than by chance. The Svalbard and Admiralty Inlet stocks included only five samples each, and results from these stocks should therefore be interpreted with caution.
(d). Phylogenetic analysis
The time-calibrated phylogeny of 14 toothed whale species and four narwhal sequences estimated the divergence of narwhals and their closest relative, belugas, at 4.98 million years ago (Ma; 95% HPD: 6.84–3.10; electronic supplementary material, figure S3). This estimate was within the confidence interval of previous estimates [59,60]. Within narwhals, lineages started to diverge from each other around 130 ka (95% HPD: 216–53 ka).
In our time-calibrated phylogeny of 64 narwhal haplotypes, we found support for several clades (posterior values > 0.9, figure 3a; electronic supplementary material, figure S4). The TMRCA of the narwhal mitochondrial tree was estimated at 105 ka (95% HPD: 175–53 ka). Clades in the narwhal tree were not geographically structured, except for the five Svalbard samples, which grouped in a clade with sequences from four other stocks (East Greenland, Northern Hudson Bay, Admiralty Inlet and Eastern Baffin Island). This clade, situated towards the top of the tree, diverged 22 ka (95% HPD: 43–8 ka).
Figure 3.
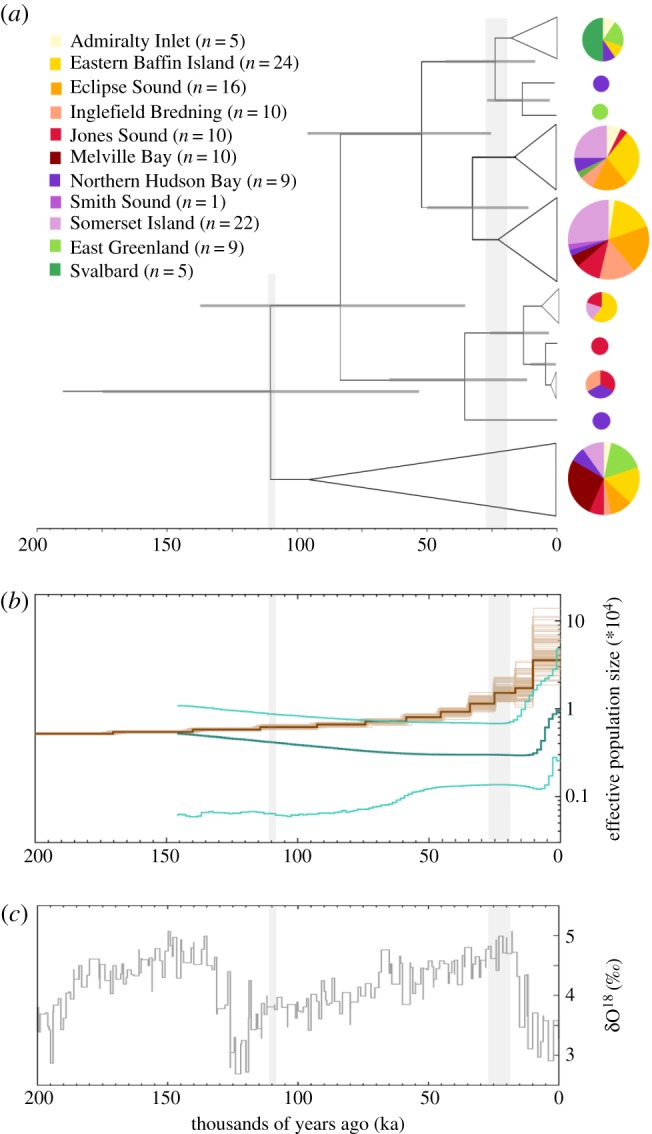
(a) Time-calibrated phylogeny of 64 haplotypes found among the 121 narwhal mitogenome sequences. The 95% HPD of divergence time estimates are represented by horizontal grey bars. Nodes that lack support (posterior values < 0.9) are collapsed. Pie chart colours and relative size indicate which stocks are represented in each clade and relative number of individuals, respectively. (b) Narwhal demographic history based on 121 mitogenomes (green line; lighter green indicates 95% CI) and a nuclear genome (brown line; lighter brown indicates bootstrap values, adapted from Westbury et al. [30]). (c) δ18O levels from Zachos et al. as a proxy for global temperatures [61]. The timing of the onset of the last glacial period (110 ka) and the LGM (26.5 to 19 ka) is indicated in grey shading across the three panels.
When the phylogeny was run with third codon positions of the protein coding regions, we estimated a divergence time of 110 ka (95% HPD: 175–54 ka) for the first node, similar to the analysis using all partitions discussed above (electronic supplementary material, figure S5). Due to a lack of support (posterior values < 0.9), several clades identified in the analysis with all the mitogenome partitions, collapsed. The effect of purifying selection on more recent nodes could therefore not be tested directly. However, all divergence times occurred during the last 110 thousand years, leading to similar conclusions. Convergence results for all analyses run in Beast are presented in electronic supplementary material, tables S6 to S8.
(e). Bayesian skyline analysis
The Bayesian skyline analysis indicated that narwhals have had a low female effective population size (Nef) for at least the past 150 kyr. Nef increased approximately threefold to a median current size of approximately 9500 (95% CI: 2559–47 554) approximately 9 ka (figure 3b).
(f). Species distribution models
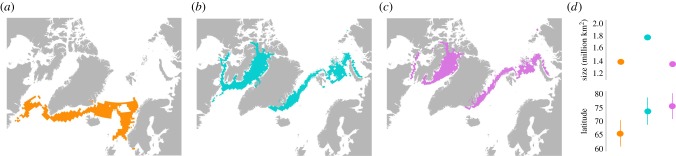
Modelling of suitable habitat indicated an 8° northward shift and an 21% increase in the size of available habitat between the LGM and the present (figure 4a,b,d). This trend was consistent for all runs, regardless of whether they were based on narwhal winter distribution (figure 4a,b,d), summer distribution or year-round distribution (electronic supplementary material, figures S7 and S8). Our models including sea ice concentration, depth and sea surface temperature in the winter were the most conservative and are presented in figure 4a–c. We present the results for the winter in the main text as it is the critical season for narwhals, when most of the annual feeding occurs [22–24]. When we considered the other periods (electronic supplementary material, figures S7 and S8) or include salinity in the models (electronic supplementary material, figures S9–11), the sizes of the habitat increase between the LGM and present were more drastic. The estimate for year 2100 shows a 1.6° northwards shift and decline in suitable habitat size of 25% relative to the present (figure 4b–d).
Figure 4.
Extent of suitable habitat for narwhals in winter estimated using Aquamaps with a probability of greater than 0.6 (a) during the LGM (approx. 20 ka, orange), (b) at present (cyan) and (c) for year 2100 (purple). (d) Change in size (million km2) and mean latitude (SD is indicated) of suitable habitat between the LGM, present and year 2100.
4. Discussion
Narwhal populations are particularly vulnerable to rapid climate change due to their relatively restricted range within the Atlantic Arctic, fidelity to migratory routes, low genetic diversity, a specialized diet and sensitivity to human activities [5,19]. Here, we present the first study of their population structure and demographic history based on complete mitochondrial genomes. Our results indicate that climatic shifts since the onset of the last glacial period have impacted narwhals in major ways, shaping their population structure, diversity and demographic history.
(a). Phylogeography
Our analyses show a lack of marked geographic structuring in the haplotype network and phylogeny, and we find low levels of genetic structuring among narwhal stocks (figures 1b and 3a; electronic supplementary material, table S5). This absence of a strong geographic pattern may reflect past environmental perturbations that have driven fluctuations in the extent of Arctic sea ice cover since the onset of the Pleistocene. Our phylogenetic analysis indicates that all clades diverged during the last glacial period (figure 3a). The timing is consistent with the demographic trajectory of a nuclear genome sequenced from a narwhal from West Greenland, which indicated changes in coalescent rates, and therefore potentially alterations in gene flow, around 100 ka (figure 3b) [30]. We suggest the genetic pattern observed in narwhals reflects secondary contact of allopatric lineages that diverged during the last glacial period. Our species distribution models suggest narwhals extended their range northwards as sea ice retreated after the LGM (figure 4), which may have facilitated the admixture of divergent lineages, as isolated populations came into secondary contact.
Alternatively, the lack of any significant geographic structuring in narwhals could be explained by contemporary behavioural processes. Narwhals show fidelity to summering grounds and migratory routes in autumn and spring, but several stocks use the same offshore, densely ice-covered areas during winter, for example in Baffin Bay, which could lead to mixing [23]. However, the estimates of the timing of mating in narwhals spans from late winter [62] to late spring, when narwhals are already separated on their migration routes towards different summering grounds [63]. Another factor that could shape the genetic pattern in narwhals is dispersal. Although narwhals from a specific stock follow similar migratory routes, three males and one female out of 135 tagged animals in Canada and Greenland showed different migratory patterns [23,24], which could eventually lead to gene flow among summering grounds. Long-range natal dispersal, where juveniles disperse after weaning, could also explain the absence of geographic structuring, although no behavioural data exist to test this hypothesis.
Behavioural processes such as site fidelity or fidelity to migratory routes can also drive genetic differentiation [64]. However, for narwhals, these processes are either not strong enough, or have not been in place for a long enough duration, to significantly alter the genetic pattern that has been shaped by longer term climatic processes across their range. The exception is the distinctness of individuals from Svalbard and from East Greenland (electronic supplementary material, table S5). Satellite tracking has not shown any movement of individuals between west and east of Greenland, which supports the fidelity of narwhals from each region to their respective summering grounds and migratory routes [20,23,24]. Mitogenomes are maternally inherited, and the genetic differentiation of Svalbard and East Greenland from animals in other areas could reflect maternally transmitted site fidelity. Furthermore, narwhals are acoustically recorded year-round in the Fram Strait between Svalbard and North-East Greenland, which suggests a more residential behaviour of the Svalbard stock and which may explain its genetic distinctness [21]. Narwhals from Melville Bay are also significantly differentiated from all other stocks, but we cannot exclude this may reflect the sampling of related individuals.
(b). Demographic history
Our analysis of the demographic trajectory of narwhals supports past environmental perturbations as a major driver of population-scale change. Narwhal female effective population size was low (approx. 3000) and relatively stable from approximately 150 ka until after the LGM, when it increased threefold approximately 9 ka (figure 3b). Although Bayesian skyline models are based on coalescence rates, and an increase in Nef could therefore reflect changes in population structure, a population expansion is supported by the star-like topology of most groups in our haplotype network (figure 1b). The timing of the expansion is coincident with temperature shifts associated with the onset of the Holocene (as shown by δ18O levels; figure 3c), and an increase and northwards shift in the amount of available suitable habitat between the LGM and the present (figure 4). The limited size of available habitat may therefore have been a constraining factor on narwhal population size during the last glacial period. Environmentally driven, long-term low population size is indicated by the extremely low genome-wide diversity reported in the narwhal nuclear genome [30], and the low level of genetic diversity observed across our 121 mitogenomes, which is among the lowest observed among the surveyed cetacean species (figure 2).
The post-glacial northwards range shift in narwhals (figure 4) follows the same pattern as has been reported in many terrestrial mammals in the Arctic and also in mammals from northern hemisphere temperate regions [14]. However, our combined findings of a post-glacial increase in both suitable habitat and population size contrasts with the trajectory of many cold-adapted terrestrial mammals during this time; many declined or went extinct during rapid transitions to warmer periods, such as warm interstadials or the Pleistocene/Holocene transition, probably due to habitat loss [11–13].
The impact of past climatic perturbations on Arctic marine mammals has also been reported in bowhead whales. Similar to narwhals, population sizes expanded after the LGM [18]. Both narwhals and bowheads probably benefited from deglaciation, as the amount of available habitat increased [18,30]. The pattern observed in the two species mirrors that reported in several polar marine predators in the Southern Ocean (e.g. southern elephant seals and several penguin species), population sizes of which are also limited by cold conditions. For those species, access to novel, ice-free breeding sites and proximate productive foraging grounds after sea ice retreat likely explains their population size increases [15,16].
Holocene population expansion has also been inferred from around 8 ka for several baleen whale species both in the North Atlantic and the Southern Ocean [65]. These expansions are correlated with the simultaneous increase in North Atlantic baleen whale prey species, such as capelin (Mallotus villosus) and herring (Clupea harengus), 8–6 ka. Capelin is also a potential prey of narwhals, which has a specialized diet composed of few prey species [26,66]. Therefore, together with an increase of available habitat, an increase in prey may have led to the increase in abundance of both North Atlantic baleen whales and narwhals. The mobility of marine predators, few barriers to movements, and possibly low costs of movement in the aquatic environment [67], may have facilitated habitat tracking by the Arctic whales, in contrast with terrestrial mammals.
5. Conclusion
In summary, we find that past climate change has been a major driver of mitogenomic patterns of structuring and diversity in narwhals. Site fidelity may either not be strong enough, or the time frame of separation of stocks relative to these long-term forces may be too short, to have any strong effect. Environmental shifts further impacted the demographic trajectory of narwhals through a combination of habitat and perhaps also prey resource availability. Our results highlight the value of combining genetic data with ecological models to contextualize evolutionary processes. We find that narwhal populations responded positively to post-LGM warming, similar to several other marine predators from both polar regions [15,16,18,30]. However, at present, many polar marine predators are being negatively affected by global warming, which is decreasing the availability of habitat and prey [6,16,68,69]. Although the range and effective population size of narwhals increased post-LGM, their future in a rapidly changing Arctic is uncertain. Although associated with a large degree of uncertainty, our habitat suitability estimates for year 2100 indicate a 25% decline and 1.6° northwards shift in habitat availability, suggesting narwhal habitat is likely to decrease in size as sea temperatures rise and sea ice continues to decline [2]. Narwhal distribution will be further affected in the near future, as the species also faces increased human encroachment [5], changes in prey availability [68], new competitors and increased predation rate by killer whales [70].
Supplementary Material
Supplementary Material
Acknowledgements
We thank Tony Martin, Ian Gjertz, Hans Lund, Outi Dove and Charmain Hamilton for helping with sample collection. Collection of samples from Greenland, Svalbard and Canada was conducted with support from the Greenland Institute of Natural Resources, the Norwegian Polar Institute and Fisheries and Oceans, Canada. Hunters in Greenland and Canada assisted with the sampling. We thank René Swift for preparing the map for figure 1, Uko Gorter for the illustrations in figures 1 and 2, and Mick Westbury for providing the PSMC curves shown in figure 3. We acknowledge the Danish National High-Throughput DNA Sequencing Centre for assistance in generating the Illumina data. We thank Lars Boehme for discussions about the habitat suitability modelling. We thank Milaja Nykanen, Andy Foote and Phil Morin for providing their published cetacean mitogenome data. We also would like to thank two anonymous reviewers for their input.
Data accessibility
Mitochondrial genome sequences are accessible on GenBank at accession numbers MT251225–MT251289. The fasta alignment used for the analyses is available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4qrfj6q72 [71].
Authors' contributions
E.D.L. and E.W. conceived the project. C.L., K.M.K., E.G., M.P.H.-J., L.P. and S.H.F. provided samples. ML generated the data with the help of M.S. and J.H. M.L., M.S. and J.A.S.C. analysed the data with input from S.G. C.G. and K.K. generated the habitat models. M.L. and E.D.L. wrote the manuscript, with input from all co-authors.
Competing interests
We declare we have no competing interests.
Funding
The work was funded by the Carlsberg Foundation Distinguished Associate Professor Fellowship, grant no CF16-0202, and the Villum Fonden Young Investigator Programme, grant no. 13151, to E.D.L.
References
- 1.Scheffers BR, et al. 2016. The broad footprint of climate change from genes to biomes to people. Science 354, aaf7671 ( 10.1126/science.aaf7671) [DOI] [PubMed] [Google Scholar]
- 2.IPCC. 2013. Climate change 2013: The physical science basis. Contribution of Working Group I to the fifth assessment report of the Intergovernmental Panel on Climate Change (eds Stocker TF. et al. ) Cambridge, UK: Cambridge University Press. [Google Scholar]
- 3.Wassmann P, Duarte CM, Agustí S, Sejr MK. 2011. Footprints of climate change in the Arctic marine ecosystem. Glob. Chang. Biol. 17, 1235–1249. ( 10.1111/j.1365-2486.2010.02311.x) [DOI] [Google Scholar]
- 4.Descamps S, Aars J, Fuglei E, Kovacs KM, Lydersen C, Pavlova O, Pedersen ÅØ, Ravolainen V, Strøm H. 2017. Climate change impacts on wildlife in a High Arctic archipelago - Svalbard, Norway. Glob. Change Biol. 23, 490–502. ( 10.1111/gcb.13381) [DOI] [PubMed] [Google Scholar]
- 5.Hauser DDW, Laidre KL, Stern HL. 2018. Vulnerability of Arctic marine mammals to vessel traffic in the increasingly ice-free Northwest Passage and Northern Sea Route. Proc. Natl Acad. Sci. USA 115, 7617–7622. ( 10.1073/pnas.1803543115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovacs KM, Lydersen C, Overland JE, Moore SE. 2011. Impacts of changing sea-ice conditions on Arctic marine mammals. Mar. Biodivers. 41, 181–194. ( 10.1007/s12526-010-0061-0) [DOI] [Google Scholar]
- 7.Brown TA, Galicia MP, Thiemann GW, Belt ST, Yurkowski DJ, Dyck MG. 2018. High contributions of sea ice derived carbon in polar bear (Ursus maritimus) tissue. PLoS ONE 13, e0191631 ( 10.1371/journal.pone.0191631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grebmeier JM. 2012. Shifting patterns of life in the Pacific Arctic and sub-Arctic seas. Ann. Rev. Mar. Sci. 4, 63–78. ( 10.1146/annurev-marine-120710-100926) [DOI] [PubMed] [Google Scholar]
- 9.Wolff EW, Chappellaz J, Blunier T, Rasmussen SO, Svensson A. 2010. Millennial-scale variability during the last glacial: the ice core record. Quat. Sci. Rev. 29, 2828–2838. ( 10.1016/j.quascirev.2009.10.013) [DOI] [Google Scholar]
- 10.Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The Last Glacial Maximum. Science 325, 710–714. ( 10.1126/science.1172873) [DOI] [PubMed] [Google Scholar]
- 11.Cooper A, Turney C, Hughen KA, Brook BW, McDonald HG, Bradshaw CJA. 2015. Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science 349, 602–606. ( 10.1126/science.aac4315) [DOI] [PubMed] [Google Scholar]
- 12.Prost S, Smirnov N, Fedorov VB, Sommer RS, Stiller M, Nagel D, Knapp M, Hofreiter M. 2010. Influence of climate warming on arctic mammals? New insights from ancient DNA studies of the collared lemming Dicrostonyx torquatus. PLoS ONE 5, e10447 ( 10.1371/journal.pone.0010447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenzen ED, et al. 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359–364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 15.Younger JL, Emmerson LM, Miller KJ. 2016. The influence of historical climate changes on Southern Ocean marine predator populations: a comparative analysis. Glob. Chang. Biol. 22, 474–493. ( 10.1111/gcb.13104) [DOI] [PubMed] [Google Scholar]
- 16.Cristofari R, et al. 2018. Climate-driven range shifts of the king penguin in a fragmented ecosystem. Nat. Clim. Change. 8, 245–251. ( 10.1038/s41558-018-0084-2) [DOI] [Google Scholar]
- 17.Cole TL, et al. 2019. Receding ice drove parallel expansions in Southern Ocean penguins. Proc. Natl Acad. Sci. USA 116, 26 690–26 696. ( 10.1073/pnas.1904048116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foote AD, et al. 2013. Ancient DNA reveals that bowhead whale lineages survived Late Pleistocene climate change and habitat shifts. Nat. Commun. 4, 1677 ( 10.1038/ncomms2714) [DOI] [PubMed] [Google Scholar]
- 19.Laidre KL, Stirling I, Lowry LF, Wiig O, Heide-Jørgensen MP, Ferguson SH. 2008. Quantifying the sensitivity of Arctic marine mammals to climate-induced habitat change. Ecol. Appl. 18, S97–125. ( 10.1890/06-0546.1) [DOI] [PubMed] [Google Scholar]
- 20.NAMMCO. 2018. Report of the Global Review of Monodontids, 13–16 March 2017. Hillerød, Denmark: NAMMCO.
- 21.Ahonen H, Stafford KM, Lydersen C, de Steur L, Kovacs KM. 2019. A multi-year study of narwhal occurrence in the western Fram Strait—detected via passive acoustic monitoring. Polar Res. 38 ( 10.33265/polar.v38.3468) [DOI] [Google Scholar]
- 22.Laidre KL, Heide-Jorgensen MP. 2005. Winter feeding intensity of narwhals (Monodon monoceros). Mar. Mamm. Sci. 21, 45–57. ( 10.1111/j.1748-7692.2005.tb01207.x) [DOI] [Google Scholar]
- 23.Heide-Jørgensen MP, Richard PR, Dietz R, Laidre KL. 2013. A metapopulation model for Canadian and West Greenland narwhals. Ani. Conser. 16, 331–343. ( 10.1111/acv.12000) [DOI] [Google Scholar]
- 24.Heide-Jørgensen MP, Nielsen NH, Hansen RG, Schmidt HC, Blackwell SB, Jørgensen OA. 2015. The predictable narwhal: satellite tracking shows behavioural similarities between isolated subpopulations. J. Zool. 297, 54–65. ( 10.1111/jzo.12257) [DOI] [Google Scholar]
- 25.Vacquié-Garcia J, Lydersen C, Marques TA, Aars J, Ahonen H, Skern-Mauritzen M, Øien N, Kovacs KM. 2017. Late summer distribution and abundance of ice-associated whales in the Norwegian High Arctic. Endanger. Species Res. 32, 59–70. ( 10.3354/esr00791) [DOI] [Google Scholar]
- 26.Watt CA, Ferguson SH. 2015. Fatty acids and stable isotopes (δ13C and δ15N) reveal temporal changes in narwhal (Monodon monoceros) diet linked to migration patterns. Mar. Mamm. Sci. 31, 21–44. ( 10.1111/mms.12131) [DOI] [Google Scholar]
- 27.De March BGE, Tenkula DA, Postma LD. 2003. Molecular genetics of narwhal (Monodon monoceros) from Canada and West Greenland (1982–2001). Ottawa, Canada: Canadian Science Advisory Secretariat.
- 28.Petersen SD, Tenkula DA, Ferguson SH. 2011. Population genetic structure of narwhal (Monodon monoceros). Ottawa, Canada: Canadian Science Advisory Secretariat.
- 29.Palsbøll PJ, Heide-Jørgensen MP, Dietz R. 1997. Population structure and seasonal movements of narwhals, Monodon monoceros, determined from mtDNA analysis. Heredity 78, 284–292. ( 10.1038/hdy.1997.43) [DOI] [PubMed] [Google Scholar]
- 30.Westbury MV, Petersen B, Garde E, Heide-Jørgensen MP, Lorenzen ED. 2019. Narwhal genome reveals long-term low genetic diversity despite current large abundance size. iScience 15, 592–599. ( 10.1016/j.isci.2019.03.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin PA, et al. 2015. Geographic and temporal dynamics of a global radiation and diversification in the killer whale. Mol. Ecol. 24, 3964–3979. ( 10.1111/mec.13284) [DOI] [PubMed] [Google Scholar]
- 32.Morin PA, et al. 2018. Demography or selection on linked cultural traits or genes? Investigating the driver of low mtDNA diversity in the sperm whale using complementary mitochondrial and nuclear genome analyses. Mol. Ecol. 27, 2604–2619. ( 10.1111/mec.14698) [DOI] [PubMed] [Google Scholar]
- 33.QGIS Development Team 2019. QGIS Geographic Information System. Open Source Geospatial Foundation Project. See http://qgis.org.
- 34.Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP. 2018. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410–419. ( 10.1111/2041-210x.12871) [DOI] [Google Scholar]
- 35.Schubert M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082. ( 10.1038/nprot.2014.063) [DOI] [PubMed] [Google Scholar]
- 36.Arnason U, Gullberg A, Janke A. 2004. Mitogenomic analyses provide new insights into cetacean origin and evolution. Gene 333, 27–34. ( 10.1016/j.gene.2004.02.010) [DOI] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leigh JW, Bryant D. 2015. popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. ( 10.1111/2041-210X.12410) [DOI] [Google Scholar]
- 41.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. ( 10.1093/bioinformatics/btg359) [DOI] [PubMed] [Google Scholar]
- 42.Alexander A, Steel D, Hoekzema K, Mesnick SL, Engelhaupt D, Kerr I, Payne R, Baker CS. 2016. What influences the worldwide genetic structure of sperm whales (Physeter macrocephalus)? Mol. Ecol. 25, 2754–2772. [DOI] [PubMed] [Google Scholar]
- 43.Nyhus ES, Lindqvist C, Kovacs K, Lydersen C, Wiig Ø, Bachmann L. 2016. Mitogenomes of contemporary Spitsbergen stock bowhead whales (Balaena mysticetus). Mitochondrial DNA Part B. 1, 898–900. ( 10.1080/23802359.2016.1258345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nykänen M, et al. 2019. Post-glacial colonization of northern coastal habitat by bottlenose dolphins: a marine leading-edge expansion? J. Hered. 110, 662–674. ( 10.1093/jhered/esz039) [DOI] [PubMed] [Google Scholar]
- 45.Leslie MS, Archer FI, Morin PA. 2019. Mitogenomic differentiation in spinner (Stenella longirostris) and pantropical spotted dolphins (S. attenuata) from the eastern tropical Pacific Ocean. Mar. Mamm. Sci. 35, 522–551. ( 10.1111/mms.12545) [DOI] [Google Scholar]
- 46.Van Cise AM, et al. 2019. Oceanographic barriers, divergence, and admixture:pPhylogeography and taxonomy of two putative subspecies of short-finned pilot whale. Mol. Ecol. 23, 758 ( 10.1093/oxfordjournals.molbev.a040023) [DOI] [PubMed] [Google Scholar]
- 47.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. ( 10.1093/oxfordjournals.molbev.a040023) [DOI] [PubMed] [Google Scholar]
- 49.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. ( 10.1093/molbev/msw260) [DOI] [PubMed] [Google Scholar]
- 50.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ho SYW, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol. Ecol. 20, 3087–3101. ( 10.1111/j.1365-294x.2011.05178.x) [DOI] [PubMed] [Google Scholar]
- 52.Heller R, Chikhi L, Siegismund HR. 2013. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS ONE 8, e62992 ( 10.1371/journal.pone.0062992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garde E, Hansen SH, Ditlevsen S, Tvermosegaard KB, Hansen J, Harding KC, Heide-Jørgensen MP. 2015. Life history parameters of narwhals (Monodon monoceros) from Greenland. J. Mammal. 96, 866–879. ( 10.1093/jmammal/gyv110) [DOI] [Google Scholar]
- 54.Kaschner K, Tittensor DP, Ready J, Gerrodette T, Worm B. 2011. Current and future patterns of global marine mammal biodiversity. PLoS ONE 6, e19653 ( 10.1371/journal.pone.0019653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ready J, Kaschner K, South AB, Eastwood PD, Rees T, Rius J, Agbayani E, Kullander S, Froese R. 2010. Predicting the distributions of marine organisms at the global scale. Ecol. Model. 221, 467–478. ( 10.1016/j.ecolmodel.2009.10.025) [DOI] [Google Scholar]
- 56.Kaschner K, Kesner-Reyes K, Garilao C, Rius-Barile J, Rees T, Froese R.2016. AquaMaps: predicted range maps for aquatic species. Version 08/2016. See www.aquamaps.org .
- 57.Laidre KL, Heide-Jørgensen MP, Logdson ML, Dietz R, Jørgensen OA, Treble MA, Heagerty P, Hobbs RC. 2003. Seasonal narwhal habitat associations in the high Arctic. Mar. Biol. 145, 821–831. ( 10.1007/s00227-004-1371-1) [DOI] [Google Scholar]
- 58.Laidre KL, Heide-Jørgensen MP. 2011. Life in the lead: extreme densities of narwhals Monodon monoceros in the offshore pack ice. Mar. Ecol. Prog. Series. 423, 269–278. ( 10.3354/meps08941) [DOI] [Google Scholar]
- 59.Steeman ME, et al. 2009. Radiation of extant cetaceans driven by restructuring of the oceans. Syst. Biol. 58, 573–585. ( 10.1093/sysbio/syp060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGowen MR, Spaulding M, Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 53, 891–906. ( 10.1016/j.ympev.2009.08.018) [DOI] [PubMed] [Google Scholar]
- 61.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292, 686–693. ( 10.1126/science.1059412) [DOI] [PubMed] [Google Scholar]
- 62.Kelley TC, Stewart REA, Yurkowski DJ, Ryan A, Ferguson SH. 2015. Mating ecology of beluga (Delphinapterus leucas) and narwhal (Monodon monoceros) as estimated by reproductive tract metrics. Mar. Mamm. Sci. 31, 479–500. ( 10.1111/mms.12165) [DOI] [Google Scholar]
- 63.Heide-Jørgensen MP, Garde E. 2011. Fetal growth of narwhals (Monodon monoceros). Mar. Mamm. Sci. 27, 659–664. ( 10.1111/j.1748-7692.2010.00423.x) [DOI] [Google Scholar]
- 64.Musiani M, Leonard JA, Cluff HD, Gates CC, Mariani S, Paquet PC, Vilà C, Wayne RK. 2007. Differentiation of tundra/taiga and boreal coniferous forest wolves: genetics, coat colour and association with migratory caribou. Mol. Ecol. 16, 4149–4170. ( 10.1111/j.1365-294X.2007.03458.x) [DOI] [PubMed] [Google Scholar]
- 65.Cabrera AA, et al. 2018. Strong and lasting impacts of past global warming on baleen whale and prey abundance. bioRxiv 497388 ( 10.1101/497388) [DOI]
- 66.Watt CA, Heide-Jørgensen MP, Ferguson SH. 2013. How adaptable are narwhal? A comparison of foraging patterns among the world's three narwhal populations. Ecosphere. 4, 1–15. ( 10.1890/es13-00137.1) [DOI] [Google Scholar]
- 67.Tucker VA. 1975. The energetic cost of moving about. Am. Sci. 63, 413–419. [PubMed] [Google Scholar]
- 68.Fossheim M, Primicerio R, Johannesen E, Ingvaldsen RB, Aschan MM, Dolgov AV. 2015. Recent warming leads to a rapid borealization of fish communities in the Arctic. Nat. Clim. Change. 5, 673–677. ( 10.1038/nclimate2647) [DOI] [Google Scholar]
- 69.Bost CA, et al. 2015. Large-scale climatic anomalies affect marine predator foraging behaviour and demography. Nat. Commun. 6, 8220 ( 10.1038/ncomms9220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferguson SH, Higdon JW, Westdal KH. 2012. Prey items and predation behavior of killer whales (Orcinus orca) in Nunavut, Canada based on Inuit hunter interviews. Aquat. Biosyst. 8, 3 ( 10.1186/2046-9063-8-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louis M, et al. 2020. Data from: Influence of past climate change on phylogeography and demographic history of narwhals, Monodon monoceros Dryad Digital Repository. 10.5061/dryad.4qrfj6q72. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Louis M, et al. 2020. Data from: Influence of past climate change on phylogeography and demographic history of narwhals, Monodon monoceros Dryad Digital Repository. 10.5061/dryad.4qrfj6q72. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Mitochondrial genome sequences are accessible on GenBank at accession numbers MT251225–MT251289. The fasta alignment used for the analyses is available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.4qrfj6q72 [71].