Summary
Background
Effective antiviral therapy is important for tackling the coronavirus disease 2019 (COVID-19) pandemic. We assessed the efficacy and safety of combined interferon beta-1b, lopinavir–ritonavir, and ribavirin for treating patients with COVID-19.
Methods
This was a multicentre, prospective, open-label, randomised, phase 2 trial in adults with COVID-19 who were admitted to six hospitals in Hong Kong. Patients were randomly assigned (2:1) to a 14-day combination of lopinavir 400 mg and ritonavir 100 mg every 12 h, ribavirin 400 mg every 12 h, and three doses of 8 million international units of interferon beta-1b on alternate days (combination group) or to 14 days of lopinavir 400 mg and ritonavir 100 mg every 12 h (control group). The primary endpoint was the time to providing a nasopharyngeal swab negative for severe acute respiratory syndrome coronavirus 2 RT-PCR, and was done in the intention-to-treat population. The study is registered with ClinicalTrials.gov, NCT04276688.
Findings
Between Feb 10 and March 20, 2020, 127 patients were recruited; 86 were randomly assigned to the combination group and 41 were assigned to the control group. The median number of days from symptom onset to start of study treatment was 5 days (IQR 3–7). The combination group had a significantly shorter median time from start of study treatment to negative nasopharyngeal swab (7 days [IQR 5–11]) than the control group (12 days [8–15]; hazard ratio 4·37 [95% CI 1·86–10·24], p=0·0010). Adverse events included self-limited nausea and diarrhoea with no difference between the two groups. One patient in the control group discontinued lopinavir–ritonavir because of biochemical hepatitis. No patients died during the study.
Interpretation
Early triple antiviral therapy was safe and superior to lopinavir–ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19. Future clinical study of a double antiviral therapy with interferon beta-1b as a backbone is warranted.
Funding
The Shaw-Foundation, Richard and Carol Yu, May Tam Mak Mei Yin, and Sanming Project of Medicine.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected more than 3 million patients with more than 200 000 deaths in more than 230 countries.1 COVID-19 spreads quickly from person to person,2 and is primarily an acute viral pneumonia leading to respiratory failure as reported in autopsy studies and animal models,3, 4 although cytokine storm and extrapulmonary involvements have been occasionally reported.3, 4 Besides respiratory and intensive care support to the extent of extracorporeal membrane oxygenation, no specific antiviral treatment has been recommended because of insufficient evidence from randomised trials. Many repurposed drugs have been shown to have in-vitro activity against the close relatives of SARS-CoV-2, which are all beta-coronaviruses. Lopinavir and many interferons, particularly interferon beta, have been shown to have modest activity in vitro against SARS-CoV and Middle East respiratory syndrome (MERS)-CoV, and can be used synergistically with ribavirin.5, 6 In 2003, we did an open-label trial using historical controls, and showed that a combination of lopinavir–ritonavir with ribavirin reduced the mortality and need for intensive respiratory support of patients with SARS who had been admitted to hospital.7 Moreover, lopinavir–ritonavir or interferon beta-1b has been shown to reduce viral load and improve lung pathology in a common marmoset model.8 However the viral load of SARS and MERS peaks at around day 7–10 after symptom onset, whereas the viral load of COVID-19 peaks at the time of presentation, similar to influenza.9, 10 Experience from the treatment of patients with influenza who are admitted to hospital suggested that a combination of multiple antiviral drugs is more effective than single drug treatments in this setting of patients with a high viral load at presentation.11, 12 Therefore, we did this phase 2 randomised trial to establish whether a combination of three modestly active drugs against SARS-CoV-2 can improve the viral load profile and clinical parameters in adults with COVID-19 requiring hospital admission.
Research in context.
Evidence before this study
We searched PubMed on March 30, 2020, using the terms “Covid-19”, “interferon beta 1b”, “lopinavir/ ritonavir”, “treatment”, “hospitalized”, “patients”, “phase 2”, and “trial” for articles in English published up to the date of the search. Our search did not show any randomised controlled trials assessing a combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients with coronavirus disease 2019 (COVID-19).
Added value of this study
This is the first randomised controlled trial on the triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin, compared with single-drug lopinavir–ritonavir in the treatment of patients admitted to hospital with COVID-19. Treatment with the triple combination effectively suppressed viral load in all clinical specimens, including the nasopharyngeal swab, throat saliva, posterior oropharyngeal saliva, and stool in most patients 8 days from treatment commencement, which was significantly shorter than the time taken in the control group, treated with lopinavir–ritonavir alone. The triple combination also alleviated symptoms completely within 4 days—a significantly shorter time than the control. The triple combination also suppressed IL-6 levels. The clinical and virological efficacy resulted in shorter hospital stays and facilitated infection control. This treatment regimen was also shown to be safe, with minor and self-limiting gastrointestinal adverse events of diarrhoea and vomiting. Increased liver enzymes were uncommon, and resolved upon stopping the medications.
Implications of all the available evidence
This study showed that early treatment with the triple combination of antiviral therapy with interferon beta-1b, lopinavir–ritonavir, and ribavirin is safe and highly effective in shortening the duration of virus shedding, decreasing cytokine responses, alleviating symptoms, and facilitating the discharge of patients with mild to moderate COVID-19. Furthermore, the triple antiviral therapy rapidly rendered viral load negative in all specimens, thereby reducing infectiousness of the patient.
Methods
Study design and patients
This was a phase 2, multicentre, open-label, randomised trial. Adult patients aged at least 18 years admitted to hospital from Feb 10, 2020, for virologically confirmed COVID-19, were recruited from the Queen Mary Hospital, Pamela Youde Nethersole Hospital, Ruttonjee Hospital, United Christian Hospital, Queen Elizabeth Hospital, and Tuen Mun Hospital in Hong Kong. These six major public hospitals are positioned across five of the seven hospital clusters, and serve 75% of the 7·5 million population. Public health ordinance in Hong Kong required all patients tested positive for COVID-19 be admitted to hospital. Eligibility criteria for the study were age at least 18 years, a national early warning score 2 (NEWS2) of at least 1, and symptom duration of 14 days or less upon recruitment (appendix pp 9–10). The institutional review board of the University of Hong Kong Hospital Authority approved this study (UW20–074). All patients gave written consent for participation in the study.
Randomisation and masking
Patients were randomly assigned to either the triple combination lopinavir–ritonavir, ribavirin, and interferon beta-1b group or the control group (lopinavir–ritonavir only), in the ratio of 2:1, by simple randomisation with no stratification. Randomised treatment was open-label. Patients were assigned to a serial number by the study coordinator. Each serial number was linked to a computer-generated randomisation list assigning the antiviral treatment regimens. The study medications were dispensed by the hospital pharmacy and then to the patients by the medical ward nurses.
Procedures
In the combination group, patients who were recruited and treated less than 7 days from symptom onset received a triple combination of 14 days of oral lopinavir–ritonavir (lopinavir 400 mg and ritonavir 100 mg) every 12 h (via nasogastric tube to intubated patients), ribavirin 400 mg every 12 h, and subcutaneous injection of one to three doses of interferon beta-1b 1 mL (8 million international units [IU]) on alternate days depending on the day of drug commencement (if commenced on day 1–2 from symptom onset, the patient received all three doses of interferon beta-1b; if commenced on day 3–4, the patient received two doses; if commenced on day 5–6, the patient received one dose). For those recruited and treated between days 7 and 14, interferon beta-1b injection was omitted to avoid its proinflammatory effects. Patients assigned to the control group received only oral lopinavir–ritonavir (lopinavir 400 mg and ritonavir 100 mg) every 12 h for 14 days. For patients who had no history of prolonged QTc syndrome, but were found to have prolonged QTc less than 480 ms, first-degree heart block or bundle branch block, or bradycardia upon ECG examination, and those who developed increased alanine transaminase of three times the upper limit of normal (ULN), the lopinavir–ritonavir treatment was reduced to once per day. Lopinavir–ritonavir would be stopped if alanine transaminase levels exceeded six times the ULN. The randomisation window from symptom onset was extended from 10 to 14 days after trial commencement after knowing that the incubation period could go beyond 14 days. Because a placebo group was generally not accepted in Chinese culture, and our previous study showed that interferon beta-1b and lopinavir–ritonavir are active against SARS-CoV and MERS-CoV, lopinavir–ritonavir was used in the control group whereas interferon beta-1b, lopinavir–ritonavir, and ribavirin were used in the combination group for patients admitted less than 7 days from symptom onset.
The intervention treatment had to be started within 48 h after hospital admission. Standard of care included oxygen, non-invasive and invasive ventilatory support, extracorporeal membrane oxygenation support, dialysis support, and antimicrobial treatment for secondary bacterial infection as indicated clinically. Stress doses of corticosteroid (50 mg hydrocortisone every 8 h intravenously, tapering over 7 days) were given to patients who developed oxygen desaturation and required oxygen support. Non-invasive or invasive ventilatory support beyond day 7 from symptom onset was at the discretion of the consultants.
Clinical and laboratory monitoring
Clinical findings including history and physical examination, and laboratory and radiological investigation results were entered into a predesigned database. Chest radiograph and ECG were taken at baseline and at regular intervals for monitoring of patient progress and to detect early cardiac rhythm changes. Patients with underlying cardiac conditions were put on cardiac monitoring. High-resolution CT was done at the consultants' discretion. All patients were followed up at the infectious disease clinic within 30 days after discharge.
Initial diagnosis of SARS-CoV-2 infection was made upon admission. All recruited patients had to have laboratory confirmed SARS-CoV-2 infection by RT-PCR in the nasopharyngeal swab. Daily nasopharyngeal swab, posterior oropharyngeal saliva, throat swab, stool or rectal swabs, and urine if available, were obtained until discharge, for quantification of viral load and genetic mutation testing (appendix pp 20–23). Complete blood count, liver and renal function tests, lactate dehydrogenase, creatine kinase, C-reactive protein, erythrocyte sedimentation rate, and cytokine profile were regularly checked until discharge (appendix p 21). Blood and urine samples for bacterial culture were taken when clinically indicated. The nasopharyngeal swab upon admission was assessed by BioFire FilmArray Respiratory Panel 2 plus (bioMérieux, Marcy l'Etoile, France). Methods for assays by quantitative RT-PCR, serum cytokine profiling, and nanopore sequencing for nsp5 mutation are in the appendix (pp 20–23).
Outcomes
The primary endpoint was time to achieve a negative RT-PCR result for SARS-CoV-2 in a nasopharyngeal swab sample. Secondary clinical endpoints were time to resolution of symptoms defined as a NEWS2 of 0 maintained for 24 h; daily NEWS2 and sequential organ failure assessment (SOFA) score; length of hospital stay; and 30-day mortality. Other virological endpoints included the time to achieve negative SARS-CoV-2 RT-PCR in all clinical samples, including nasopharyngeal swab, posterior oropharyngeal saliva, throat swab, stool, and urine; daily viral load changes in the first 7 days; and emergence of amino acid mutations in the nsp5 gene encoding a 3C-like protease. The serum cytokine response was also measured. Safety endpoints were the frequencies and duration of adverse events.
Statistical analysis
It is important to note that COVID-19 is a new disease caused by SARS-CoV-2, which is phylogenetically closest to the 2003 SARS-CoV. At the time of study design in mid-January, 2020, there was insufficient information on the mortality of COVID-19. Thus, we based our sample size calculation on our own findings of lopinavir–ritonavir treatment in a trial on the 2003 SARS-CoV.7 The current study was designed on the basis of an estimated difference of 26·4% in the 21-day mortality or acute respiratory distress syndrome rate in patients with severe SARS-CoV-2 infection, when treated with lopinavir–ritonavir (2·4%) versus historical controls without antiviral treatment (28·8%). The necessary sample size had been calculated to be 30 patients per group to detect such a difference at a two-sided α level of 0·05, with 80% power. The protocol proposed recruiting at least 35 patients per group to allow for a 17% dropout rate.
The primary endpoint was assessed in the intention-to-treat population of all randomised patients. Safety was assessed in all patients who received at least one dose of their assigned drug. Categorical variables were compared using the χ2 test and continuous variables were compared using the Mann-Whitney U test, for both intention-to-treat and subgroup analyses. For viral load, specimens with undetectable viral load were assigned a value of 1 log10 copies per mL for the purpose of statistical analysis. Hazard ratios (HRs) with 95% CIs were calculated by Cox proportional hazards model. Factors significant at univariable analysis (p<0·10) were further assessed by means of a multivariable analysis by Cox proportional hazards model to identify the independent factors for negative nasopharyngeal swab RT-PCR on day 7 after treatment. A p value of less than 0·05 was considered statistically significant. Statistical analysis was performed using SPSS, version 26.0 and PRISM, version 8. The study is registered with ClinicalTrials.gov, NCT04276688.
Role of the funding source
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Between Feb 10 and March 20, 2020, 144 patients were screened, and 127 patients were recruited (figure 1 ). The number of patients screened accounted for 80% of the confirmed COVID-19 cases in Hong Kong during this period. Nine patients did not fulfil the inclusion criteria (four with second-degree and third-degree cardiac arrhythmia, two with severe depression, and three because of pregnancy) and eight patients declined the treatment regimen. One patient in the control group required discontinuation of lopinavir–ritonavir because of alanine transaminase six times greater than the ULN after 1 week of treatment. The median age was 52 years (IQR 32–62); 68 (54%) patients were men versus 59 (46%) women (table 1 ). 51 (40%) patients had underlying diseases. The median time to hospital admission from symptom onset was 5 days (IQR 3–7).
Figure 1.
Trial profile
Table 1.
Baseline demographics of the study population
| Combination group (n=86) | Control group (n=41) | |||
|---|---|---|---|---|
| Age | 51·0 (31·0–61·3) | 52·0 (33·5–62·5) | ||
| Sex | ||||
| Men | 45 (52%) | 23 (56%) | ||
| Women | 41 (48%) | 18 (44%) | ||
| Time from symptoms onset to start of treatment, days | 5 (4–7) | 4 (3–8) | ||
| Underlying diseases | ||||
| Diabetes | 11 (13%) | 6 (15%) | ||
| Hypertension | 23 (27%) | 13 (32%) | ||
| Coronary artery disease | 5 (6%) | 5 (12%) | ||
| Cerebrovascular disease | 1 (1%) | 1 (2%) | ||
| Hyperlipidaemia | 18 (21%) | 11 (27%) | ||
| Thyroid disease | 3 (3%) | 1 (2%) | ||
| Obstructive sleep apnoea | 1 (1%) | 1 (2%) | ||
| Crohn's disease | 1 (1%) | 0 | ||
| Epilepsy | 1 (1%) | 0 | ||
| Tuberculosis | 2 (2%) | 0 | ||
| Chronic hepatitis B | 2 (2%) | 1 (2%) | ||
| Chronic hepatitis C | 0 | 1 (2%) | ||
| Malignancy | 1 (1%) | 1 (2%) | ||
| Smoker | 6 (7%) | 1 (2%) | ||
| Symptoms and signs | ||||
| Fever | 70 (81%) | 32 (78%) | ||
| Chills | 13 (15%) | 6 (15%) | ||
| Cough | 45 (52%) | 23 (56%) | ||
| Sputum | 29 (34%) | 13 (32%) | ||
| Shortness of breath | 7 (8%) | 7 (17%) | ||
| Sore throat | 16 (19%) | 10 (24%) | ||
| Myalgia | 10 (12%) | 8 (20%) | ||
| Malaise | 19 (22%) | 5 (12%) | ||
| Nausea or vomiting | 1 (1%) | 0 | ||
| Diarrhoea | 17 (20%) | 7 (17%) | ||
| Rhinorrhoea | 14 (16%) | 10 (24%) | ||
| Anosmia | 4 (5%) | 1 (2%) | ||
| Headache | 3 (3%) | 3 (7%) | ||
| Chest tightness | 2 (2%) | 0 | ||
| Anorexia | 1 (1%) | 0 | ||
| Baseline laboratory findings (normal range) | ||||
| Haemoglobin (11·5–14·8 g/dL) | 13·4 (12·7–14·9) | 13·5 (12·7–14·8) | ||
| White cell count (3·89–9·93 × 109 per L) | 4·9 (3·7–6·2) | 5·4 (4·6–6·4) | ||
| Neutrophils (2·01–7·42 × 109 per L) | 3·4 (2·4–4·3) | 3·5 (2·9–4·5) | ||
| Lymphocytes (1·06–3·61 × 109 per L) | 1·0 (0·8–1·5) | 1·3 (0·9–1·6) | ||
| Platelets (154–371 × 109 per L) | 195·0 (171·8–260·0) | 192·0 (160·5–244·5) | ||
| Alanine aminotransferase (8–45 U/L) | 23·0 (15·0–33·3) | 26·0 (14·5–43·0) | ||
| Alkaline phosphatase (42–110 U/L) | 58·0 (48·0–75·0) | 65·0 (52·5–75·0) | ||
| Lactate dehydrogenase (143–280 U/L) | 194·0 (159·8–249·0) | 167·5 (142·0–200·0) | ||
| Bilirubin (4–23 μmol/L) | 7·9 (5·5–9·0) | 7·5 (6·0–10·8) | ||
| Creatinine (49–82 μmol/L) | 75·5 (65·0–92·0) | 76·0 (62·5–96·0) | ||
| Urea (2·9–8·0 mmol/L) | 4·0 (2·9–4·8) | 3·7 (2·7–4·6) | ||
| Creatine kinase (22–198 U/L) | 79·0 (50·0–151·0) | 90·5 (54·5–141·5) | ||
| C-reactive protein (<0·76 mg/dL) | 3·0 (2·0–9·2) | 3·0 (1·5–7·2) | ||
| Erythrocyte sedimentation rate (<12 mm/h) | 19·0 (11·0–48·0) | 19·0 (9·8–37·8) | ||
| Baseline radiological findings (%) | ||||
| Abnormal chest x-ray | 64 (74%) | 32 (78%) | ||
| Right upper zone infiltrate | 0 | 0 (0%) | ||
| Right middle zone infiltrate | 4 (5%) | 6 (15%) | ||
| Right lower zone infiltrate | 38 (44%) | 18 (44%) | ||
| Left upper zone infiltrate | 1 (1%) | 0 | ||
| Left middle zone infiltrate | 7 (8%) | 7 (17%) | ||
| Left lower zone infiltrate | 27 (31%) | 10 (24%) | ||
| High-resolution CT (performed in 22 patients) | 14 (16%) | 6 (15%) | ||
Data are n (%) or median (IQR). In the combination group 52 patients were treated with triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin and 34 patients were treated with lopinavir–ritonavir and ribavirin; in the control group, 41 patients were treated with lopinavir–ritonavir. U/L=units per L.
Among the 127 patients, 86 were randomly assigned to the combination group and 41 patients were assigned to the control group. Within the combination group, 52 patients were admitted to hospital less than 7 days from symptom onset and received the lopinavir–ritonavir, ribavirin, and interferon beta-1b regimen, and 34 patients who were admitted 7 days or more after symptom onset received the lopinavir–ritonavir and ribavirin only regimen. The median number of doses of interferon beta-1b received was two. Median time from symptom onset to start of treatment was 5 days (4–7) for the combination group and 4 days (3–8) for the control group (table 1). The age, sex, and baseline demographics in each group were similar. Fever and unproductive cough were the most common presenting signs and symptoms. Both diarrhoea and anosmia were infrequent. Most patients had lymphopenia and increased C-reactive protein and erythrocyte sedimentation rate upon presentation. One patient in the combination group had concomitant rhinovirus infection upon presentation. Disease severity upon presentation was mild based on NEWS2 and SOFA scores (table 2 ).
Table 2.
Clinical, viral load, and cytokine profile and concomitant treatments
| Combination group (n=86) | Control group (n=41) | p value | ||
|---|---|---|---|---|
| NEWS2 | ||||
| Baseline | 2 (1–2) | 2 (2–2) | 0·52 | |
| Day 1 | 1 (1–2) | 2 (2–2) | <0·0001 | |
| Day 2 | 1·0 (0·0–2·0) | 2·0 (1·5–3·0) | <0·0001 | |
| Day 3 | 0 (0–1) | 2 (1–3) | <0·0001 | |
| Day 4 | 0 (0–1) | 2 (1–2) | <0·0001 | |
| Day 5 | 0 (0–1) | 2 (1–2) | <0·0001 | |
| Day 6 | 0·0 (0·0–1·0) | 1·5 (1·0–2·0) | <0·0001 | |
| Day 7 | 0·0 (0·0–1·0) | 1·0 (0·8–2·0) | 0·0010 | |
| Time to NEWS2 of 0, days | 4 (3–8) | 8 (7–9) | <0·0001 | |
| SOFA score | ||||
| Baseline | 0 (0–1) | 0 (0–1) | 0·38 | |
| Day 1 | 0 (0–1) | 0 (1–1) | 0·21 | |
| Day 2 | 0 (0–2) | 1 (0–2) | 0·025 | |
| Day 3 | 0 (0–2) | 1 (0–2) | 0·010 | |
| Day 4 | 0·0 (0·0–1·3) | 1·0 (0·0–2·0) | 0·012 | |
| Day 5 | 0 (0–1) | 1 (0–2) | 0·010 | |
| Day 6 | 0 (0–1) | 1 (0–2) | 0·035 | |
| Day 7 | 0 (0–1) | 1 (0–2) | 0·028 | |
| Time to SOFA score of 0, days | 3·0 (1·0–8·0) | 8·0 (6·5–9·0) | 0·041 | |
| Duration of hospital stay, days | 9·0 (7·0–13·0) | 14·5 (9·3–16·0) | 0·016 | |
| 30-day mortality | 0 (0) | 0 (0) | 1·00 | |
| Time to negative viral load, days | ||||
| Nasopharyngeal swab | 7 (5–11) | 12 (8–15) | 0·0010 | |
| Posterior oropharyngeal saliva | 6·0 (3·0–8·0) | 8·0 (5·3–10·8) | 0·044 | |
| Throat swab | 4·5 (1·3–6·8) | 7·0 (3·0–12·0) | 0·039 | |
| Stool | 5 (2–5) | 7 (4–8) | 0·030 | |
| All specimens | 8 (6–12) | 13 (8–15) | 0·0010 | |
| Virological findings (RT-PCR), log10 copies per mL | ||||
| Nasopharyngeal swab (baseline) | 6·4 (4·5–8·0) | 6·4 (3·9–7·7) | 0·70 | |
| Posterior oropharyngeal saliva (baseline)* | 5·2 (3·8–7·0) | 5·3 (4·3–7·1) | 0·54 | |
| Throat swab (baseline) | 4·6 (2·9–6·1) | 4·5 (3·7–5·7) | 0·85 | |
| Stool (baseline) | 3·3 (2·7–5·3) | 3·8 (2·6–7·3) | 0·53 | |
| Cytokine concentration, log10 pg/mL | ||||
| IL-6 (baseline) | 1·4 (1·0–1·4) | 1·4 (1·0–1·6) | 0·43 | |
| TNFα (baseline) | 1 (1–1) | 1 (1–1) | 1·00 | |
| Concomitant treatments | ||||
| Oxygen therapy | 12 (14%) | 5 (12%) | 0·72 | |
| Non-invasive ventilator support | 3 (3%) | 2 (5%) | 0·75 | |
| Ventilator support | 0 | 1 (2%) | 0·15 | |
| Antibiotics | 44 (51%) | 25 (61%) | 0·33 | |
| Amoxicillin–clavulanate | 29 (34%) | 21 (51%) | 0·080 | |
| Azithromycin | 7 (8%) | 4 (10%) | 0·76 | |
| Ceftriaxone | 12 (14%) | 8 (20%) | 0·42 | |
| Doxycycline | 13 (15%) | 8 (20%) | 0·53 | |
| Levofloxacin | 11 (13%) | 3 (7%) | 0·36 | |
| Piperacillin–tazobactam | 5 (6%) | 0 | 0·12 | |
| Corticosteroid (stress dose)* | 6 (7%) | 2 (5%) | 0·65 | |
Data are median (IQR) or n (%). NEWS2=national early warning score 2. SOFA=sequential organ failure assessment.
Stress-dose steroid was hydrocortisone 50 mg every 8 h intravenously, tapered over 5–7 days.
For the primary endpoint of time from start of study treatment to negative nasopharyngeal swab, the combination group had a significantly shorter median time (7 days [IQR 5–11]) than the control group (12 days [8–15]; HR 4·37 [95% CI 1·86–10·24], p=0·0010; table 2).
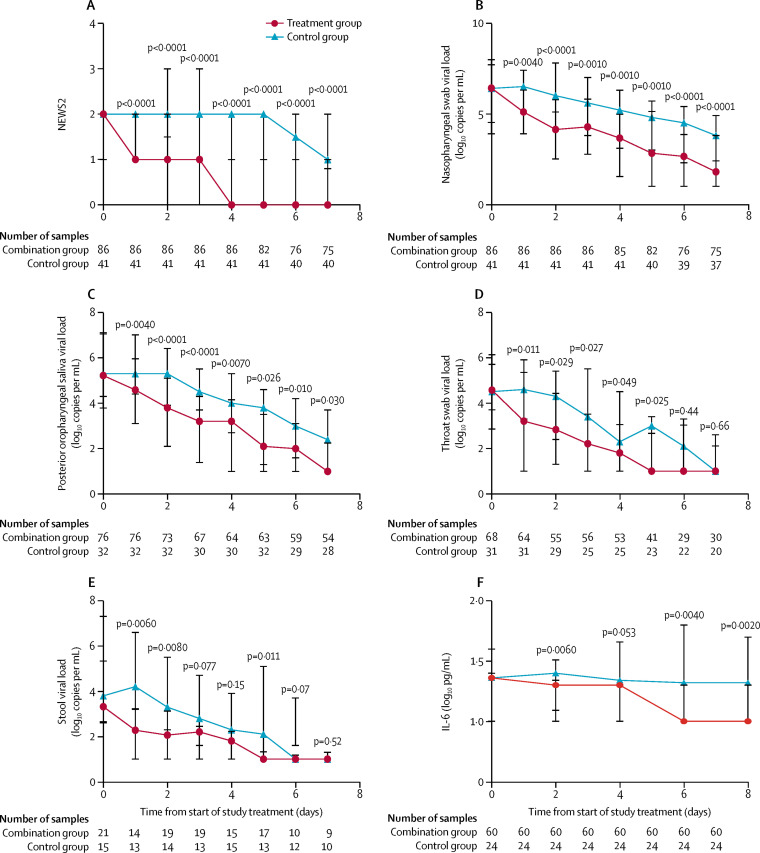
Clinical improvement was significantly better in the combination group, with a significantly shorter time to complete alleviation of symptoms, defined as a NEWS2 of 0 (4 days [IQR 3–8] in the combination group vs 8 days [7–9] in the control group; HR 3·92 [95% CI 1·66–9·23], p<0·0001) and SOFA score of 0 (3·0 days [1·0–8·0] vs 8·0 days [6·5–9·0]; HR 1·89 [1·03–3·49], p=0·041; table 2). A similar pattern was observed on the daily NEWS2 (all p<0·0001; figure 2A ) and daily SOFA score after treatment (all p<0·05 except day 1 [p=0·21]; table 2). The significantly better clinical and virological response is also reflected in the shorter median hospital stay in the combination group than in the control group (9·0 days [7·0–13·0] vs 14·5 days [9·3–16·0]; HR 2·72 [1·2–6·13], p=0·016).
Figure 2.
Outcomes over time
(A) National early warning score 2; (B) nasopharyngeal swab viral load; (C) posterior oropharyngeal saliva viral load; (D) throat swab viral load; (E) stool viral load; and (F) serum IL-6 cytokine concentration (first 60 recruited patients). Data points are medians and error bars are IQRs.
For the virological outcome, the combination treatment was associated with significantly shorter time to negative viral load in all specimens when assessed individually (nasopharyngeal swab, posterior oropharyngeal saliva, throat swab, and stool samples) as well as in all specimens combined (table 2). All urine samples tested negative for viral load.
All patients had a SARS-CoV-2 positive baseline nasopharyngeal swab. With regards to the other clinical samples, 108 (85%) patients provided posterior oropharyngeal saliva samples, 99 (78%) provided throat swabs, 36 (28%) provided stool samples, and 83 (65%) provided urine samples. The baseline viral loads for all specimens were similar between the combination group and control group (table 2). The nasopharyngeal swab viral load was significantly lower in the combination group than in the control group from day 1 to day 7 after treatment (figure 2B). Similar results were found in the posterior oropharyngeal saliva, throat swab, and stool specimens after treatment (figure 2C–E).
Post-hoc subgroup comparison of the 76 patients who started treatment less than 7 days after onset of symptoms showed better clinical and virological outcomes in the combination group (52 patients, receiving lopinavir–ritonavir, ribavirin, and interferon beta-1b) than in the control group (24 patients; table 3 ) across all measured variables except stool samples. However, no significant differences between the treatment groups were measured in these outcomes in the 51 patients who were treated 7 days or more after symptom onset (34 in the combination group [receiving lopinavir–ritonavir and ribavirin only] and 17 in the control group; appendix p 31).
Table 3.
Subgroup analysis of clinical, viral load, and cytokine profile
|
Started treatment <7 days from symptom onset |
Started treatment ≥7 days from symptom onset |
||||||
|---|---|---|---|---|---|---|---|
| Combination group (with interferon beta-1b; n=52) | Control group (n=24) | p value | Combination group (without interferon beta-1b; n=34) | Control group (n=17) | p value | ||
| NEWS2 | |||||||
| Baseline | 2 (1–2) | 2 (2–2) | 0·11 | 2 (1–2) | 2 (1–2) | 0·49 | |
| Day 1 | 1 (1–1) | 2 (2–2) | <0·0001 | 2 (1–2) | 2 (1–2) | 0·71 | |
| Day 2 | 1·0 (0·0–1·0) | 2·0 (1·5–3·0) | <0·0001 | 1·5 (1·0–2·0) | 2·0 (1·0–2·8) | 0·41 | |
| Day 3 | 0·0 (0·0–1·0) | 2·0 (1·0–3·0) | <0·0001 | 1·0 (1·0–2·0) | 2·0 (0·3–2·8) | 0·16 | |
| Day 4 | 0·0 (0·0–0·0) | 2·0 (1·0–2·5) | <0·0001 | 1·0 (1·0–2·0) | 2·0 (0·3–2·0) | 0·37 | |
| Day 5 | 0 (0–0·5) | 2 (1–2) | <0·0001 | 1 (0–1) | 2 (0–2) | 0·040 | |
| Day 6 | 0 (0–0·3) | 1 (1–2) | <0·0001 | 1 (0–1) | 1 (0–2) | 0·14 | |
| Day 7 | 0 (0–0) | 1 (0–2) | <0·0001 | 1 (0–1) | 1 (0–1) | 0·68 | |
| Time to NEWS2 of 0, days | 4·0 (3·0–5·0) | 8·0 (6·5–9·0) | <0·0001 | 6·0 (5·0–10·8) | 8·0 (5·5–8·0) | 0·90 | |
| SOFA score | |||||||
| Baseline | 0 (0–1) | 0 (0–1) | 0·99 | 1 (0–1) | 0 (0–1) | 0·17 | |
| Day 1 | 0·0 (0·0–1·0) | 1·0 (0·0–1·0) | 0·030 | 1·0 (0·0–2·0) | 1·0 (0·0–1·5) | 0·67 | |
| Day 2 | 0 (0–1) | 1 (0–2) | 0·0060 | 1 (0–2) | 1 (0–2) | 0·72 | |
| Day 3 | 0 (0–1) | 1 (0–2) | 0·0050 | 1 (0–2) | 1 (0–3) | 0·49 | |
| Day 4 | 0 (0–1) | 1 (0–2) | 0·0060 | 1 (0–2) | 1 (0–3) | 0·48 | |
| Day 5 | 0·0 (0·0–0·8) | 1·0 (0·0–2·0) | 0·0030 | 1·0 (0·0–2·0) | 1·0 (0·0–3·0) | 0·55 | |
| Day 6 | 0·0 (0·0–0·0) | 0·5 (0·0–2·0) | 0·0010 | 1·0 (0·0–2·0) | 1·0 (0·0–2·0) | 0·88 | |
| Day 7 | 0·0 (0·0–0·0) | 0·5 (0·0–2·0) | <0·0001 | 1·0 (0·0–2·0) | 1·0 (0·0–2·0) | 0·88 | |
| Time to SOFA score of 0, days | 3 (1–5) | 7 (1–9) | 0·0010 | 8 (1–8) | 8 (1–9) | 0·23 | |
| Duration of hospital stay, days | 8 (6–12·5) | 15 (9–16) | 0·0030 | 13 (8–15) | 13·5 (12·3–21·8) | 0·090 | |
| 30-day mortality | 0 (0) | 0 (0) | 1·00 | 0 (0) | 0 (0) | 1·00 | |
| Time to negative viral load, days | |||||||
| Nasopharyngeal swab | 6·5 (4·0–8·0) | 12·5 (8·0–14·8) | <0·0001 | 10·5 (8·0–12·3) | 12·0 (8·0–17·0) | 0·10 | |
| Posterior oropharyngeal saliva | 6·0 (2·0–7·0) | 8·5 (5·3–11·8) | <0·0001 | 8·0 (6·0–9·0) | 8·0 (5·3–9·0) | 0·79 | |
| Throat swab | 4·0 (1·0–6·0) | 8·0 (3·3–12·8) | 0·0010 | 5·0 (1·5–8·0) | 4·5 (2·0–9·0) | 0·52 | |
| Stool | 4·5 (2·0–5·0) | 6·0 (3·0–7·0) | 0·070 | 5·0 (2·0–10·0) | 7·0 (5·5–8·5) | 0·14 | |
| All specimens | 7·0 (4·0–9·0) | 13·0 (8·0–14·0) | <0·0001 | 12·0 (7·8–14·0) | 12·0 (12·0–19·0) | 0·080 | |
| Virological findings (RT-PCR), log10 copies per mL | |||||||
| Nasopharyngeal swab (baseline) | 7 (5·2–8·4) | 6·1 (4·3–7·7) | 0·29 | 5·5 (3·8–7·3) | 6·6 (3·8–8) | 0·65 | |
| Posterior oropharyngeal saliva (baseline) | 5·4 (3·9–7·3) | 5·3 (3·9–7·5) | 0·86 | 4·8 (3·8–6·2) | 5·4 (4·9–6·8) | 0·30 | |
| Throat swab (baseline) | 4·8 (3·2–6·9) | 4·4 (3·5–6·1) | 0·81 | 4·5 (1·0–5·6) | 5·0 (4·0–5·5) | 0·52 | |
| Stool (baseline) | 3·2 (1·9–6·2) | 3·2 (2·9–5·6) | 0·85 | 3·3 (2·8–3·9) | 5·6 (1·9–7·4) | 0·48 | |
| Cytokine concentration, log10 pg/mL | |||||||
| IL-6 (baseline) | 1·4 (1–1·5) | 1·4 (1·4–1·6) | 0·13 | 1·4 (1–1·4) | 1 (1–1·6) | 0·45 | |
| TNFα (baseline) | 1 (1–1) | 1 (1–1) | 0·87 | 1 (1–1) | 1 (1–1) | 0·82 | |
Data are median (IQR). NEWS2=national early warning score 2. SOFA=sequential organ failure assessment.
17 (13%) of 127 patients developed oxygen desaturation and required oxygen treatment (table 2). Six (5%) patients were admitted to the intensive care unit, of whom five required non-invasive ventilator support and one 96-year-old female patient with a past medical history of coronary artery disease required intubation and ventilator support. She was in the control group and was successfully extubated after 10 days of intensive care. 69 (54%) patients received concomitant antibiotics. Eight (6%) patients were given stress doses of corticosteroids in the second week from symptom onset.
The serum cytokine profile was analysed in the first 84 recruited patients. The IL-6 concentration in the combination group was significantly lower than in the control group on days 2, 6, and 8 (figure 2F). TNFα concentrations and IL-10 concentrations were not significantly different between the groups. No significant nsp5 mutations were identified in serial nasopharyngeal swab samples.
Multivariable analysis showed that the combination group and having a normal baseline chest x-ray were independently associated with day 7 negative nasopharyngeal swab viral load. Of the two, the combination group was the most significant independent factor (HR 4·27 [95% CI 1·82–10·02], p=0·0010; appendix p 30).
Adverse events were reported by 41 (48%) of 86 patients in the combination group and 20 (49%) of 41 patients in the control group. The most common adverse events were diarrhoea (52 [41%] of 127 patients), fever (48 [38%] patients), nausea (43 [34%]) and raised alanine transaminase level (18 [14%]; table 4 ). These side-effects mostly resolved within 3 days after drug initiation. Sinus bradycardia was reported by four (3%) patients. There were no differences between incidence of any of the adverse events or durations of nausea or diarrhoea between the treatment groups. The peak median alanine transaminase concentration was 38·0 units per L (24·5–62·5) and peak median bilirubin was 22·0 μmol/L (17·0–32·5), in all patients. No serious adverse events were reported in the combination group. One patient in the control group had a serious adverse event of impaired hepatic enzymes requiring discontinuation of treatment. No patients died during the study.
Table 4.
Adverse events in the study population
| Combination group (n=86) | Control group (n=41) | p value | ||
|---|---|---|---|---|
| Adverse events | ||||
| Nausea | 30 (35%) | 13 (32%) | 0·87 | |
| Diarrhoea | 34 (40%) | 18 (44%) | 0·54 | |
| Increased alanine aminotransferase | 11 (13%) | 7 (17%) | 0·32 | |
| Hyperbilirubinaemia | 4 (5%) | 3 (7%) | 0·54 | |
| Sinus bradycardia | 3 (4%) | 1 (2%) | 0·77 | |
| Fever | 32 (37%) | 16 (39%) | 0·73 | |
| Serious adverse events | 0 | 1 (2%) | 0·15 | |
| Duration of nausea, days | 2 (1–2) | 2 (1–2) | 0·80 | |
| Duration of diarrhoea, days | 3 (3–3) | 3 (3–3) | 0·88 | |
Discussion
In this multicentre randomised open-label phase 2 trial in patients with COVID-19, we showed that a triple combination of an injectable interferon (interferon beta-1b), oral protease inhibitor (lopinavir–ritonavir), and an oral nucleoside analogue (ribavirin), when given within 7 days of symptom onset, is effective in suppressing the shedding of SARS-CoV-2, not just in a nasopharyngeal swab, but in all clinical specimens, compared with lopinavir–ritonavir alone. Furthermore, the significant reductions in duration of RT-PCR positivity and viral load were associated with clinical improvement as shown by the significant reduction in NEWS2 and duration of hospital stay. Most patients treated with the triple combination were RT-PCR negative in all specimens by day 8. The side-effects were generally mild and self-limiting.
Specific highly active antiviral drugs are always needed for any novel emerging infectious disease because the development of a new antiviral takes years before its approval for clinical use. Therefore, drug repurposing by testing existing broad-spectrum antiviral drugs that have been used to treat other viral infections is the most feasible approach in a pandemic. Many drugs have been shown to have some in-vitro activity against betacoronaviruses, including remdesivir, favipiravir, nitazoxanide, camostat mesilate, interferons, lopinavir–ritonavir, ribavirin, chloroquine, hydroxychloroquine, and convalescent plasma containing neutralising antibodies.5, 6, 7, 8, 13, 14, 15, 16, 17, 18, 19, 20, 21 These drugs have known pharmacokinetic and pharmacodynamic properties, side-effects, and dosing regimens. As expected, lopinavir–ritonavir alone was shown to have similar effects to placebo on reducing viral load when treatment was initiated at a median of 13 days after symptom onset, despite some improvement in symptoms.22 Up to now, only two open-label non-randomised trials have been reported. One trial used a combination of oral hydroxychloroquine and azithromycin in 20 patients with COVID-19 showing that this combination might reduce viral load significantly by day 6 after treatment, compared with 16 controls from another hospital, which could be due to chance because this combination was not planned a priori and the addition of azithromycin was at the physician's discretion.23 A small retrospective analysis showed that viral load was negative at day 7 post-treatment in 75% of patients with COVID-19 treated with arbidol and lopinavir–ritonavir (16 patients) versus 35% of patients treated with lopinavir–ritonavir alone (17 patients).24
Under the Hong Kong Special Administrative Region public health ordinance, all patients with COVID-19 must stay in hospital until nasopharyngeal swab viral loads are negative on 2 consecutive days. Thus, most patients were admitted to hospital within 7 days of symptom onset, allowing recruitment into the clinical trial during the early course of COVID-19. With the memory of the 2003 SARS pandemic, most patients with COVID-19 in Hong Kong accepted antiviral treatment, which explained our high recruitment rate. Despite being an open-label study, all patients were enrolled consecutively without bias. Our case demographics and proportion of patients with underlying diseases were similar to other reported cohorts in China. The low crude mortality rate in Hong Kong (four [0·4%] of 1041 cases) could be explained by the highly vigilant infection control measures, efficient contact tracing, and early hospital admission and treatment.
Early treatment with a triple combination of modestly active antivirals is appropriate for the treatment of COVID-19 because the viral load of SARS-CoV-2 peaks at around the time of symptom onset. This is unlike the situation of SARS and MERS when the antiviral treatment has time to suppress the viral load before it peaks at around days 7–10 after symptom onset. The viral load profile of COVID-19 is similar to that of influenza, which has a high viral load at the time of initiation of anti-influenza treatment. The emergence of resistant influenza virus quasispecies during treatment has been well reported with single-drug treatment by amantadine, baloxavir marboxil, and oseltamivir in the setting of severe influenza or diseases caused by H5N1, H7N9, or in immunosuppressed hosts.11, 17, 25 Thus, the antiviral combination was considered a reasonable option to improve the outcome of severe influenza. Indeed, we have previously shown that a combination of naproxen and clarithromycin, with weak anti-influenza virus activity in vitro individually, when combined with oseltamivir can improve the morbidity and mortality and shorten the duration of hospital stay in patients with influenza A/H3N2 pneumonia.12 Furthermore, we have previously shown that a combination of lopinavir–ritonavir and ribavirin significantly reduced mortality and respiratory failure in patients during the 2003 SARS outbreak.7 Thus, we hypothesised that a triple combination of modest antiviral drugs might rapidly suppress the high initial viral load, improve the clinical parameters, and reduce risk of health-care workers by reducing the duration and quantity of virus shedding from these treated patients.
An in-vitro study in cell culture-based assays showed that the 50% effective concentration (EC50) of lopinavir against SARS-CoV is about 17 μM and against MERS-CoV it is about 8 μM, whereas the peak serum lopinavir concentration is about 15 μM with a half-life of 7·4–10·8 h after an oral dose of 400 mg lopinavir and 100 mg ritonavir.26 The EC50 of interferon beta-1b is 0·12 IU/mL against SARS-CoV and 17·6 IU/mL against MERS-CoV,6, 27 whereas its peak serum level is about 20 IU/mL with a half-life of 2–5 h after a single subcutaneous dose of 8 million IU. Notably, the maintenance of high serum interferon beta-1b level is not essential once the antiviral status of exposed cells is induced. The EC50 of ribavirin against SARS-CoV-2 was 109 μM,15 which greatly exceeds the drug's serum concentration with the usual oral dosing. Nevertheless, synergistic activity between interferons and a lower dose of ribavirin have been shown in checkerboard assays. However, combining ribavirin with interferons (alfa-2a, alfa-2b, and beta-1a) did not improve outcomes in critically ill patients with MERS.28 Thus, the repurposing of this triple combination of modestly active lopinavir–ritonavir, interferon beta-1b, and ribavirin for the treatment of this novel pandemic virus should be a reasonable therapeutic approach.
Furthermore, SARS-CoV-2 did not significantly induce types I, II, or III interferons in ex-vivo infected human lung tissues compared with 2003 SARS-CoV.29 Thus, the use of interferon beta-1b treatment to jump-start or improve the antiviral response of patients would be a logical approach. Additionally, interferon beta-1b was shown to decrease virus-induced lung fibrosis in a mouse model, which might improve outcomes of patients with COVID-19 complicated by acute respiratory distress syndrome.30
Despite the concern of major side-effects arising from a combination of three drugs, no significant differences in incidence of adverse events between treatment groups were reported in our cohort of 127 patients. No haemolysis occurred from the short duration of low dose ribavirin. We did not use triple combination for patients who started treatment 7 days or more after symptom onset because of the concerns about the proinflammatory side-effects of interferon beta-1b, despite that at most three doses were used for each patient. Liver dysfunction was observed in about 14% of these patients and it was mild and self-limiting, except in one patient in the control group, in whom the biochemical hepatitis warranted the discontinuation of lopinavir–ritonavir treatment.
Our study had several limitations. This trial was open label, without a placebo group, and confounded by a subgroup omitting interferon beta-1b within the combination group, depending on time from symptom onset. A subsequent phase 3 trial with interferon beta-1b as a backbone treatment with a placebo control group should be considered, because subgroup comparison suggested that interferon beta-1b appears to be a key component of our combination treatment. Our absence of critically ill patients did not allow the generalisation of our findings to severe cases.
Triple antiviral therapy with interferon beta-1b, lopinavir–ritonavir, and ribavirin were safe and superior to lopinavir–ritonavir alone in shortening virus shedding, alleviating symptoms, and facilitating discharge of patients with mild to moderate COVID-19.
Data sharing
Qualified researchers can request access to deidentified participant data or anonymised clinical study reports, informed consent forms, and related documents including the study protocol that underlie this Article through submission of a proposal with a valuable research question to the corresponding author, provided that the necessary data protection agency and ethical committee approvals are in compliance with the relevant registration. A contract will also be signed.
Acknowledgments
Acknowledgments
This study was partly supported by Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of Hong Kong; the Theme-Based Research Scheme (T11/707/15) of the Research Grants Council, Hong Kong; Sanming Project of Medicine in Shenzhen, China (no SZSM201911014); the Health and Medical Research Fund, Hong Kong; and the High Level-Hospital Program, Health Commission of Guangdong Province, China; and the donations of Richard Yu and Carol Yu, the Shaw Foundation Hong Kong, Michael Seak-Kan Tong, May Tam Mak Mei Yin, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy, and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, and the Jessie & George Ho Charitable Foundation.
Contributors
IF-NH, K-CL, EY-KT, JC, W-SL, Y-YN, T-CW, and K-YY were responsible for the design, analysing, and writing of the manuscript. IF-NH, K-CL, EY-KT, RL, TW-HC, M-YC, Y-YN, JL, ART, H-PS, VC, AK-LW, K-MS, W-LL, DCL, SS, PY, RLi, KF, AY, T-CW, JW-MC, W-WY, W-MiC, AK-WL, VC-CC, T-LQ, and C-SL were responsible for recruitment and clinical care of the patients. IFN-H, JF-WC, KK-WT, K-YY were responsible for analysing and writing up of the manuscript. CC-YY, RRZ, AY-FF, EY-WY, K-HL, AW-HC, W-MuC, AC-KN, JDI, RLe, KF, DCL, AK-LW, VC-CC, T-LQ, K-HC were responsible for the laboratory analysis. All authors reviewed and approved the final version of the manuscript.
Declaration of interests
We declare no competing interests.
Supplementary Material
References
- 1.WHO Coronavirus disease 2019 (COVID-19) situation report–107. May 6, 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200506covid-19-sitrep-107.pdf?sfvrsn=159c3dc_2
- 2.Chan JF, Yuan S, Kok KH. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao XH, Li TY, He ZC. A pathological report of three COVID-19 cases by minimally invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020;49:E009. doi: 10.3760/cma.j.cn112151-20200312-00193. (in Chinese). [DOI] [PubMed] [Google Scholar]
- 4.Chan JF, Zhang AJ, Yuan S. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in golden syrian hamster model: implications for disease pathogenesis and tansmissibility. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa325. publised online March 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen F, Chan KH, Jiang Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J Clin Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JF, Chan KH, Kao RY. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J Infect. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu CM, Cheng VC, Hung IF. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan JF, Yao Y, Yeung ML. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To KK-W, Tsang OT-Y, Leung W-S. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. published online March 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng VC, Tang BS, Wu AK, Chu CM, Yuen KY. Medical treatment of viral pneumonia including SARS in immunocompetent adult. J Infect. 2004;49:262–273. doi: 10.1016/j.jinf.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunning J, Baillie JK, Cao B, Hayden FG. Antiviral combinations for severe influenza. Lancet Infect Dis. 2014;14:1259–1270. doi: 10.1016/S1473-3099(14)70821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung IFN, To KKW, Chan JFW. Efficacy of clarithromycin-naproxen-oseltamivir combination in the treatment of patients hospitalized for influenza A(H3N2) infection: an open-label randomized, controlled, phase IIb/III trial. Chest. 2017;151:1069–1080. doi: 10.1016/j.chest.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Agostini ML, Andres EL, Sims AC. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonulcease. MBio. 2018;9:e00221–e00318. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Cao R, Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossignol JF. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beigel JH, Nam HH, Adams PL. Advances in respiratory virus therapeutics - a meeting report from the 6th isirv Antiviral Group conference. Antiviral Res. 2019;167:45–67. doi: 10.1016/j.antiviral.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheahan TP, Sims AC, Leist SR. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mair-Jenkins J, Saavedra-Campos M, Baillie JK. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham AC, Goh HP, Koh D. Treatment of COVID-19: old tricks for new challenges. Crit Care. 2020;24:91. doi: 10.1186/s13054-020-2818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 22.Cao B, Wang Y, Wen D. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001282. published online March 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautret P, Lagier JC, Parola P. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. published online March 20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Deng L, Li C, Zeng Q. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: a retrospective cohort study. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.002. published March 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden FG, Sugaya N, Hirotsu N. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med. 2018;379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 26.de Wilde AH, Jochmans D, Posthuma CC. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan EL, Ooi EE, Lin CY. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg Infect Dis. 2004;10:581–586. doi: 10.3201/eid1004.030458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arabi YM, Shalhoub S, Mandourah Y. Ribavirin and interferon therapy for critically ill patients with Middle East Respiratory Syndrome: a multicentre observational study. Clin Infect Dis. 2020;70:1837–1844. doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu H, Chan JF-W, Wang Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa410. published online April 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luckhardt TR, Commes SM, Trujillo G. TLR9-induced interferon β is associated with protection from gammaherpesvirus-induced exacerbation of lung fibrosis. Fibrogenesis Tissue Repair. 2011;4:18. doi: 10.1186/1755-1536-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Qualified researchers can request access to deidentified participant data or anonymised clinical study reports, informed consent forms, and related documents including the study protocol that underlie this Article through submission of a proposal with a valuable research question to the corresponding author, provided that the necessary data protection agency and ethical committee approvals are in compliance with the relevant registration. A contract will also be signed.