Abstract
Waterborne enteric viruses are an emerging cause of disease outbreaks and represent a major threat to global public health. Enteric viruses may originate from human wastewater and can undergo rapid transport through aquatic environments with minimal decay. Surveillance and source apportionment of enteric viruses in environmental waters is therefore essential for accurate risk management. However, individual monitoring of the >100 enteric viral strains that have been identified as aquatic contaminants is unfeasible. Instead, viral indicators are often used for quantitative assessments of wastewater contamination, viral decay and transport in water. An ideal indicator for tracking wastewater contamination should be (i) easy to detect and quantify, (ii) source-specific, (iii) resistant to wastewater treatment processes, and (iv) persistent in the aquatic environment, with similar behaviour to viral pathogens. Here, we conducted a comprehensive review of 127 peer-reviewed publications, to critically evaluate the effectiveness of several viral indicators of wastewater pollution, including common enteric viruses (mastadenoviruses, polyomaviruses, and Aichi viruses), the pepper mild mottle virus (PMMoV), and gut-associated bacteriophages (Type II/III FRNA phages and phages infecting human Bacteroides species, including crAssphage). Our analysis suggests that overall, human mastadenoviruses have the greatest potential to indicate contamination by domestic wastewater due to their easy detection, culturability, and high prevalence in wastewater and in the polluted environment. Aichi virus, crAssphage and PMMoV are also widely detected in wastewater and in the environment, and may be used as molecular markers for human-derived contamination. We conclude that viral indicators are suitable for the long-term monitoring of viral contamination in freshwater and marine environments and that these should be implemented within monitoring programmes to provide a holistic assessment of microbiological water quality and wastewater-based epidemiology, improve current risk management strategies and protect global human health.
Keywords: Gastroenteric viruses, Environmental sampling, Viral indicators, Sewage contamination, Risk assessment
Graphical abstract
Highlights
-
•
Human mastadenoviruses are robust indicators for human-associated pollution in water.
-
•
Bacteroides-associated phages and crAssphage are promising indicators.
-
•
Multiple indicators should be used to assess wastewater treatment efficiency.
-
•
Survival and abundance of indicator viruses should be further assessed.
1. Introduction
1.1. Waterborne enteric viruses
Waterborne diarrheal diseases account for approximately 4 billion cases annually, resulting in 2 million deaths, most of which occur in children under five (WHO, 2010). A significant proportion of these illnesses are caused by enteric viral infections (Ramani and Kang, 2009). Enteric viruses are transmitted via the faecal-oral route and the most important route of transmission is direct contact with infected individuals (Katayama and Vinje, 2017). Nonetheless, most enteric viruses are persistent in environments affected by domestic wastewater discharge and are often associated with waterborne outbreaks (Gibson, 2014; Kauppinen et al., 2018; Sekwadi et al., 2018). Wastewater often receives treatment prior to release into the environment, although traditional wastewater treatment methods can be relatively ineffective at removing enteric viruses (Kitajima et al., 2014; Qiu et al., 2015; Sidhu et al., 2017b). In developing countries, many areas lack adequate sanitary infrastructure and wastewater treatment facilities and hence faecal matter contaminates the environment and drinking water sources (Bain et al., 2014). Furthermore, large volumes of untreated wastewater may also be discharged via combined sewer overflows (CSOs) during heavy rainfall events and via dry water overflows for example during snowmelt, tidal infiltration or system failures and blockages (Ahmed et al., 2020). These events enable the direct entry of enteric pathogens into the environment (Fong et al., 2010), where people in direct or indirect contact with contaminated waters may be at risk of acquiring viral infections (Sinclair et al., 2009). Enteric viruses are readily transported in environmental waters and can adsorb to solid matter present in the water column or accumulate in sediment (Hassard et al., 2016). Subsequently, they may also be taken up by filter feeding aquatic animals such as bivalve shellfish that are harvested for human consumption (Landry et al., 1983; Lowther et al., 2012). Furthermore, wastewater is often used for irrigation in countries experiencing freshwater shortage, and hence, enteric viruses may directly contaminate fruit and salad vegetables and result in foodborne outbreaks (Bosch et al., 2016; Chatziprodromidou et al., 2018; Jasim et al., 2016).
Enteric viruses usually cause gastroenteritis lasting for 2–5 days. In some cases, the infection results in respiratory, neural or epidermal symptoms, or remains asymptomatic (Table 1 ). The viruses most often associated with gastroenteritis are members of the Picornaviridae, Caliciviridae, Reoviridae and Adenoviridae families (Table 1). For example, noroviruses (family Caliciviridae) are responsible for a high proportion of gastroenteritis infections globally, with 685 million cases and approximately 200,000 deaths (CDC, 2016; Katayama and Vinje, 2017), resulting in a total direct cost of US$4.2 billion to the healthcare system and US$60.3 billion in associated societal costs per year (Bartsch et al., 2016). Rotaviruses (family Reoviridae) and group F mastadenoviruses (AdVs) (family Adenoviridae) are the main causative agents of gastroenteritis amongst infants and young children (Desselberger and Gray, 2009; Jiang, 2006). Noroviruses, hepatitis A virus (family Picornaviridae) and AdVs are the most common viral pathogens associated with waterborne and water-associated foodborne outbreaks and infection may result in serious illness, e.g. acute hepatitis (Bellou et al., 2013; Harris et al., 2006; Jiang, 2006; Parshionikar et al., 2003; Sinclair et al., 2009).
Table 1.
Human pathogenic viruses detected in the aquatic environment.
| Family | Genus | Virus types found in water | Structure |
Symptoms | Zoonotic | Reference | ||
|---|---|---|---|---|---|---|---|---|
| Capsid | Genome | Size | ||||||
| Adenoviridae | Mastadenovirus | Mastadenovirus A-F | Icosahedral | dsDNA | 70–90 nm | Gastroenteritisa, respiratory illness, ear infection, conjunctivitis | No | King et al. (2009) |
| Anelloviridae | Alphatorquevirus | Torque teno virus | Icosahedral | ssDNA | 30 nm | Unknown, hepatitisa | Yes | King et al. (2009) |
| Astroviridae | Mamastrovirus | Astrovirus | Icosahedral | ssRNA+ | 28–30 nm | Gastroenteritis | Potentially | De Benedictis et al. (2011); King et al. (2009) |
| Caliciviridae | Norovirus | Norovirus GI, GII | Icosahedral | ssRNA+ | 35–40 nm | Gastroenteritis | No | King et al. (2009) |
| Sapovirus | Sapovirus GI, GII | Gastroenteritis | No | King et al. (2009) | ||||
| Circoviridae | Circovirus | Human-associated circovirus | Icosahedral | ssDNA | 15–25 nm | Unknowna | No | Breitbart et al. (2017) |
| Hepeviridae | Orthohepevirus | Hepatitis E virus type 1-4 | Icosahedral | ssRNA+ | 27–34 nm | Acute hepatitisa | Yes | Purdy et al. (2017) |
| Papillomaviridae | various | assorted papillomaviruses | Icosahedral | dsDNA | 55 nm | Genital tract infection, cancera | No | Van Doorslaer et al. (2018) |
| Parvoviridae | Bocavirus | Human bocavirus type 1-4 | Icosahedral | ssDNA | 22 nm | Gastroenteritis and respiratory disease | No | King et al. (2009) |
| Picornaviridae | Kobuvirus | Aichivirus A-B | Icosahedral | ssRNA+ | 30–32 nm | Gastroenteritisa | No | Zell et al. (2017) |
| Cosavirus | Cosavirus A | Gastroenteritisa | No | |||||
| Enterovirus | Coxsackievirus B Enterovirus A-D Poliovirus type 1-3 |
Gastroenteritis, mild meningitis, encephalitis, myelitis, myocarditis, conjunctivitisa | No | |||||
| Hepatovirus | Hepatitis A virus | Gastroenteritis, hepatitis | No | |||||
| Polyomaviridae | Alpha-polyomavirus | MC polyomavirus | Icosahedral | dsDNA | 40–45 nm | Cancera | No | Moens et al. (2017) |
| Beta-polyomavirus | BK polyomavirus JC polyomavirus |
Icosahedral | dsDNA | 40–45 nm | Respiratory, urinary tract and skin infection, cancera | No | Moens et al. (2017) | |
| Reoviridae | Reovirus | Rotavirus A | Icosahedral | dsRNA | 60–80 nm | Gastroenteritis | Potentially | Cook et al. (2004); King et al. (2009) |
May be asymptomatic in otherwise healthy individuals.
Rotaviruses, enteroviruses, sapoviruses, astroviruses, Aichi virus (AiV) and hepatitis E virus are also often shown to be associated with wastewater contamination. For example, in Maharashtra state, India in 2017, a rotavirus B outbreak with a 22.8% attack rate (i.e. new cases/number of people) was sourced from contaminated wells used for drinking water (Joshi et al., 2019). In addition, several viral gastroenteritis outbreaks linked to sewage-contaminated drinking water containing AdV, noro-sapo-, astro-, rota-, and enteroviruses have been reported (Kauppinen et al., 2019; Maunula et al., 2009; Rasanen et al., 2010). The largest viral waterborne outbreak affecting approximately 80,000 people in Kanpur, India was associated with hepatitis E virus (Naik et al., 1992). The surveillance of enteric viral illnesses can be challenging, as many of the enteric viral outbreaks are unreported as the symptoms are often subclinical (Cortez et al., 2017; Koff, 1992; Li et al., 2017; Matson et al., 1993; Sakai et al., 2001; Zaoutis and Klein, 1998).
Over the last decade, both newly discovered viruses and known viruses that had previously not been associated with wastewater have been found in environmental waters (Table 1). Human polyomaviruses (PyVs) and papillomaviruses were first discovered in the 1970s and 1950s, respectively, however, they have only recently been found in the faeces and urine of infected individuals (Knowles, 2006; Rachmadi et al., 2016). Some PyVs, including BKPyV, WUPyV, KIPyV, MCPyV and JCPyV have been detected at high concentrations (up to 108 genome copies (gc)/l) in wastewater, river and seawater and sediment, in swimming pools and in tap water (Di Bonito et al., 2017; Dias et al., 2018a, Dias et al., 2018b; Farkas et al., 2018a; Fratini et al., 2014; Hamza and Hamza, 2018; Rachmadi et al., 2016). As these viruses are commonly asymptomatic in healthy individuals, the route of transmission is not yet clear, however, waterborne infections are likely (Fratini et al., 2014).
Bocaviruses (family Parvoviridae), causing respiratory tract infections and gastroenteritis, were first described in 2005 (Allander et al., 2005). They have since been found in untreated and treated wastewater at concentrations of 103-105 genome copies (gc)/l (Hamza et al., 2017; Iaconelli et al., 2016; Myrmel et al., 2015), however, their prevalence in environmental water has not been explored. The torque teno virus (family Anelloviridae), which causes gastroenteritis has also been found in wastewater and in polluted river waters. Similar to bocaviruses, torque teno virus concentrations are considerably lower (up to 106 gc/l) than the concentrations of other, more common enteric viruses (104-109 gc/l) (Hamza et al., 2011; Haramoto et al., 2008). Human picobirnaviruses (family Picobirnaviridae) have also been detected in wastewater (concentration range: 103–106 gc/l) and in contaminated rivers with variable prevalence (Adriaenssens et al., 2018; Hamza et al., 2011; Symonds et al., 2009). In addition, recent comparative genomics analysis has suggested that picobirnaviruses are bacteriophages, likely associated with mammalian gut bacteria (Krishnamurthy and Wang, 2018). Genomes or partial genomes of circoviruses (family Circoviridae) and cardioviruses (family Picornaviridae) along with enveloped viruses (coronaviruses, influenza virus) have also been found in wastewater (Bibby and Peccia, 2013; Blinkova et al., 2009; Ng et al., 2012). Enveloped viruses degrade in water rapidly (Gundy et al., 2009; Lebarbenchon et al., 2011), hence, human infections from waterborne corona- and influenza viruses (e.g. SARS-CoV-2) are unlikely.
1.2. Viral indicators for wastewater contamination
Over 100 types of human enteric viruses are known to be common water pollutants (Melnick, 1984) and with novel and emerging strains, the number is increasing. Due to the diversity of human pathogenic viruses in the environment, surrogates and indicators are often used to investigate the fate and transport of pathogenic strains in the environment. An indicator may be suitable for a broad assessment of wastewater and drinking water treatment efficiency and for studying pathogen abundance, persistence, adsorption and transport in the aquatic environment. Furthermore, quantitative monitoring of viral indicators can provide useful data for microbial source tracking, transport modelling and risk assessment. Traditionally, faecal indicator bacteria (FIB; including coliform bacteria, Escherichia coli, Enterococcus and Streptococcus spp.) have been used to determine levels of faecal contamination in water. However, it has been shown that bacteria are significantly less resistant to wastewater treatment and less persistent in the environment than enteric viruses (Fong et al., 2005; Kim et al., 2009; Lin and Ganesh, 2013; Prez et al., 2015; Sidhu et al., 2017a; Staley et al., 2012). Consequently, FIB are poor indicators of viral infection risk and this suggests that current water quality monitoring programmes based solely on FIB are inadequate.
Ideally, a good viral indicator for wastewater-contamination assessment should have similar inactivation and retention to the target pathogens and should be present in wastewater and in wastewater-contaminated environments throughout the year. That would enable continuous monitoring and inform on the level of contamination and the probability of pathogen presence. Furthermore, an indicator with constant levels in wastewater may serve as a proxy for population size when wastewater-based epidemiology is used to estimate the proportion of infected people during a viral outbreak or pandemic, e.g. COVID-19 (Xagoraraki and O’Brien, 2020). Additionally, it should be source-specific to distinguish between animal- and human-derived pollution (Scott et al., 2002). Some enteric viruses associated with wastewater (as listed in Table 1) have potential to be used as indicators, however, not all of those viruses fulfil these requirements. Influenza viruses, coronaviruses, circoviruses and papillomaviruses have been detected at high concentrations in wastewater but not in polluted environments, which may be due to their rapid decay in water. Furthermore, some enteric viruses (e.g. astrovirus, rotavirus, torque teno virus and hepatitis E virus; Table 1) may be zoonotic, hence their presence in the environment may be a result of e.g. agricultural activities instead of human waste. Hepatitis A and E viruses are abundant in less economically developed countries, however, they are only responsible for sporadic outbreaks in more developed regions (Bosch et al., 2016). Further, enteroviruses, noroviruses and sapoviruses show clear seasonality with peaks either in the summer (enteroviruses) or during the winter (noroviruses and sapoviruses) in temperate climates. Hence, these viruses are not found in wastewater and in the contaminated environment at all times of the year (Farkas et al., 2018a; Nino Khetsuriani et al., 2006; Pons-salort et al., 2018; Prevost et al., 2015). Human AdVs, PyVs and AiVs are frequently found in wastewater and in polluted environments without any distinct seasonality, hence their utility as effective faecal indicators have been suggested (Kitajima and Gerba, 2015; Rachmadi et al., 2016; Rames et al., 2016).
Bacteriophages infecting bacteria associated with the human gut are also common in wastewater. Somatic coliphages (phages infecting E. coli) and F-specific RNA bacteriophages (FRNAP; phages infecting bacteria through the F-pili) are commonly used to assess wastewater contamination. However, as not all strains exclusively associate with human bacteria, they should be used with caution. Bacteriophages infecting Bacteroides spp. also have the potential to indicate wastewater contamination. Amongst these phages are a newly discovered group of viruses called crass-like phages. The type genome, crAssphage sensu stricto (metagenome-assembled genome), belongs to the normal gut virome, having co-evolved with humans (Dutilh et al., 2014; Edwards et al., 2019). Since the discovery of the first crAssphage genome, more crass-like sequences have been found and one phage has been isolated. However, their genomic diversity is large and the crAssphage sensu stricto and the isolated crass-like phage do not belong to the same genus (Shkoporov et al., 2018). As the taxonomy of crass-like phages remains to be established, we refer to crAssphage as a group of viruses with nucleotide similarity to the crAssphage sensu stricto described by Dutilh et al. (2014) and quantified by Stachler et al. (2017).
Interestingly, a plant virus, the pepper mild mottle virus (PMMoV; family Virgaviridae), has also been shown to be associated with human wastewater and found in polluted surface and groundwater and in drinking water (Symonds et al., 2018). The primary source of PMMoV in human excreta is through consumption of peppers (Capsicum spp.) and food products containing peppers that are contaminated with the virus (Zhang et al., 2005). PMMoV is suggested to be a useful indicator for wastewater contamination (Kitajima et al., 2018b; Symonds et al., 2018), however, its shape and size (17 × 300 nm rod-shaped capsid) differs from other pathogenic viruses with icosahedral capsids and hence its fate and behaviour in the environment may be different.
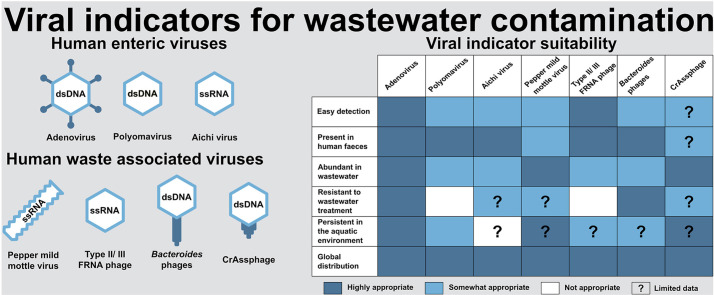
In this review, we evaluated the practicality of a set of human-waste associated viruses as indicators for wastewater contamination of the aquatic environment (Table 2 ). We have extracted data from 127 individual studies to assess the usefulness of viral indicators by addressing specific aspects. The data collected on viral concentrations in wastewater and environmental receiving waters are presented in Tables S1–6, while the corresponding wastewater treatment log removal rates for each virus are presented in Table S7. Together, we used this information to assess the ranges of virus abundance and distribution in global aquatic systems. We included human wastewater-associated viruses, which are often present in wastewater at high concentration without seasonality. We considered enteric viruses, (human AdVs, PyVs and AiVs), PMMoV and human gut bacteria-associated bacteriophages, including FRNAP infecting E. coli (specifically genogroups II and III), and bacteriophages of human gut commensal Bacteroides spp. (including crAssphage). For evaluation, we used the following criteria:
-
1.
Ease of detection and quantification
-
2.
Human waste association
-
3.
Presence in wastewater at high concentrations
-
4.
Resistance to wastewater treatment
-
5.
Persistence in the aquatic environment
-
6.
Global distribution and temporal stability
Table 2.
Number of reviewed studies for each indicator at each region.
| North America | South America | Africa | Europe | Asia | Oceania | Global detection rate | ||
|---|---|---|---|---|---|---|---|---|
| AdV | Raw wastewater | 7 | 2 | 2 | 13 | 3 | 5 | 94% (772/823) |
| Treated wastewater | 10 | 7 | 2 | 13 | 3 | 3 | 86% (1223/1436) | |
| Surface freshwater | 4 | 5 | 4 | 7 | 4 | 0 | 65% (835/1283) | |
| Groundwater | 1 | 2 | 0 | 1 | 1 | 0 | 65% (40/62) | |
| Seawater | 2 | 6 | 0 | 4 | 0 | 0 | 60% (229/381) | |
| Total | 13 | 16 | 5 | 20 | 5 | 4 | 76% (3099/3985) | |
| PyV | Raw wastewater | 5 | 5 | 1 | 7 | 2 | 3 | 93% (542/581) |
| Treated wastewater | 6 | 5 | 1 | 6 | 2 | 2 | 68% (608/892) | |
| Surface freshwater | 1 | 2 | 0 | 6 | 2 | 0 | 52% (326/631) | |
| Groundwater | 0 | 0 | 0 | 1 | 0 | 0 | 48% (10/21) | |
| Seawater | 4 | 0 | 0 | 2 | 2 | 1 | 24% (83/350) | |
| Total | 9 | 7 | 1 | 10 | 4 | 3 | 63% (1569/2475) | |
| AiV | Raw wastewater | 5 | 0 | 0 | 0 | 4 | 0 | 91% (92/101) |
| Treated wastewater | 5 | 0 | 0 | 1 | 3 | 0 | 74% (184/250) | |
| Surface freshwater | 2 | 0 | 0 | 1 | 3 | 0 | 33% (77/236) | |
| Groundwater | 1 | 0 | 0 | 0 | 0 | 0 | 55% (26/47) | |
| Seawater | 0 | 0 | 0 | 0 | 0 | 0 | NA | |
| Total | 6 | 0 | 0 | 1 | 6 | 0 | 60% (379/634) | |
| PMMoV | Raw wastewater | 6 | 0 | 1 | 1 | 2 | 1 | 100% (110/110) |
| Treated wastewater | 6 | 0 | 1 | 1 | 2 | 0 | 99% (135/137) | |
| Surface freshwater | 2 | 0 | 0 | 1 | 4 | 0 | 87% (278/319) | |
| Groundwater | 1 | 0 | 0 | 0 | 0 | 0 | 72% (18/25) | |
| Seawater | 1 | 0 | 0 | 0 | 0 | 1 | 55% (45/82) | |
| Total | 7 | 0 | 1 | 1 | 5 | 1 | 87% (586/673) | |
| Bacteroides phages | Raw wastewater | 2 | 2 | 0 | 14 | 2 | 0 | 97% (531/549) |
| Treated wastewater | 0 | 1 | 0 | 8 | 1 | 0 | 75% (911/1216) | |
| Surface freshwater | 2 | 1 | 0 | 4 | 1 | 0 | 66% (280/427) | |
| Groundwater | 0 | 0 | 0 | 3 | 0 | 0 | 38% (48/127) | |
| Seawater | 0 | 0 | 0 | 3 | 0 | 0 | 42% (43/102) | |
| Total | 3 | 2 | 0 | 19 | 2 | 0 | 72% (1741/2421) | |
| FRNAP (II/III) | Raw wastewater | 0 | 0 | 0 | 1 | 3 | 0 | 73% (96/132) |
| Treated wastewater | 0 | 0 | 0 | 1 | 4 | 0 | 81% (219/270) | |
| Surface freshwater | 0 | 0 | 0 | 2 | 5 | 0 | 59% (375/634) | |
| Groundwater | 0 | 0 | 0 | 0 | 1 | 0 | 0% (0/10) | |
| Seawater | 0 | 0 | 0 | 0 | 0 | 0 | NA | |
| Total | 0 | 0 | 0 | 3 | 8 | 0 | 66% (690/1046) | |
2. Data collection
We collected viral concentration data published in peer-reviewed journal articles since 2005 (Tables S1–7; Fig. 1 ). Articles were identified via Google Scholar in September 2018–October 2019 using the following keywords: ‘wastewater adenovirus’, ‘wastewater polyomavirus’, ‘wastewater Aichi virus’, ‘crAssphage’, ‘wastewater pepper mild mottle virus’, ‘wastewater AND (“F-specific RNA” OR F + OR “FRNA” OR “male specific”) AND ∗phage AND genogroup’ and ‘wastewater Bacteroides bacteriophage’‘. The Google Scholar search included these terms or part of them in the whole text, hence enabling the identification of studies on the aquatic environment where wastewater contamination was assessed using the target viruses. The studies were screened based on the title and abstract and initially 243 papers were selected.
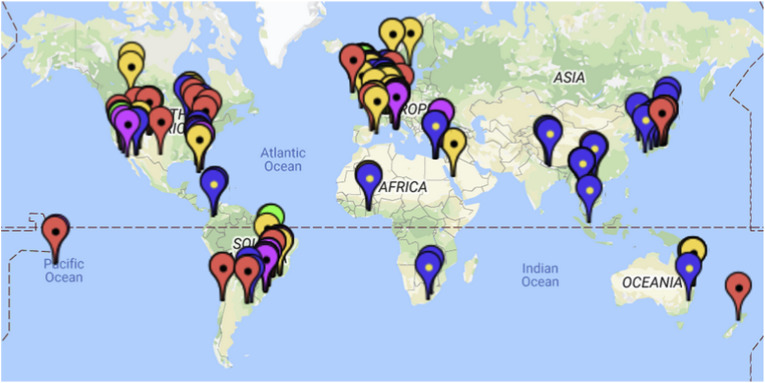
Fig. 1.
Map illustrating the sampling sites where viral indicators have been detected in untreated wastewater (red), treated wastewater (yellow) surface freshwater (blue), groundwater (green), seawater (purple). To zoom in to a particular region visit https://j.mp/2VdQVpY. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The assessment of enteric viruses, PMMoV and crAssphage concentrations usually involved the concentration of large volumes of water (1–10 l), and the efficiency of those procedures may therefore affect the outcomes. Hence, studies where viral concentrations were not determined or sample process recovery efficiency and/or quantitative PCR (qPCR) performance was not addressed were excluded from the study. Studies where sample process recovery was <10% were also excluded. After the quality screen, 127 peer-reviewed research papers were included in the review (Table 2). Viral concentration data were classified by water type (untreated and treated wastewater, surface freshwater, groundwater and seawater) and the detection rates (i.e. positive samples/all samples x 100%), mean/median concentrations and/or minimum-maximum concentrations were extracted (Fig. 2 , Tables S1–S6). In most studies only the mean/median and/or minimum-maximum concentrations were reported, hence further meta-analysis was not performed. Virus removal rates reported during wastewater treatment processes were also retrieved (Fig. 3 ; Table S7). Additionally, the primers and probes used for the qPCR detection and quantification of viruses have also been summarised (Table S8).
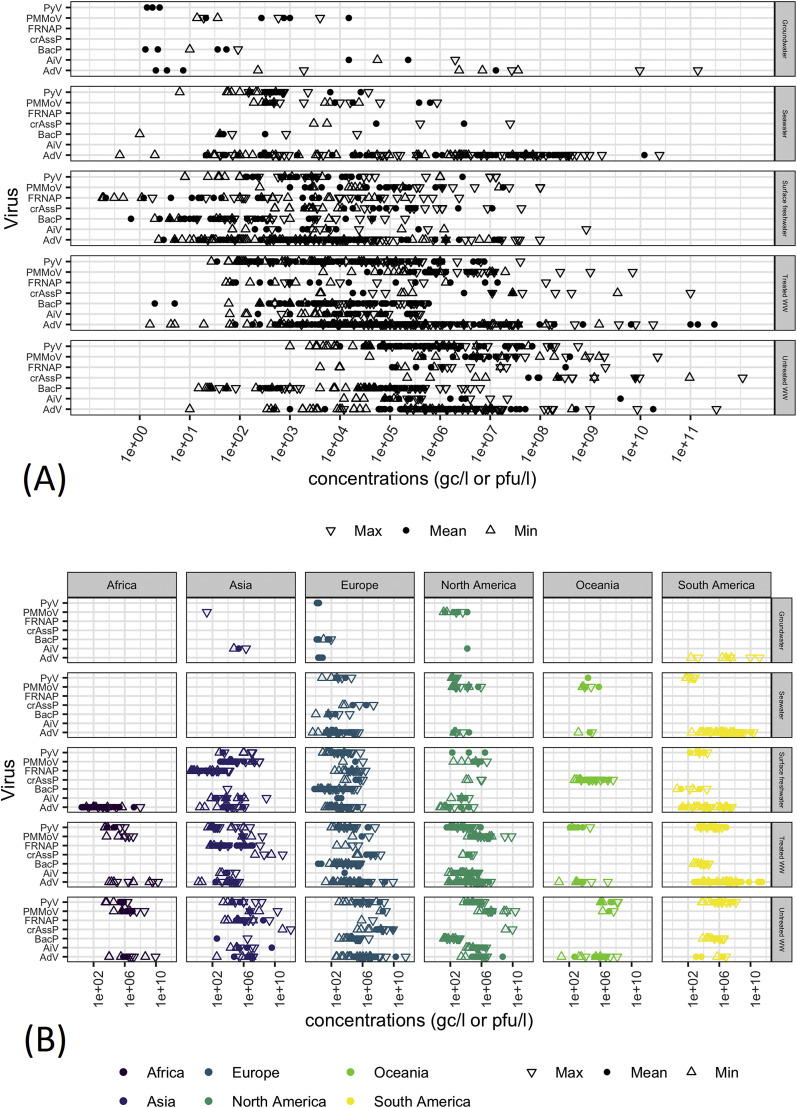
Fig. 2.
Viral concentrations (mean, minimum and maximum values in genome copies (gc) or plaque-forming units (pfu) per litre) extracted from the reviewed studies. (A) All data; (B) Distribution of the data in A grouped by continent. AdV: human mastadenoviruses; PyV: human polyomavirus JC, BK and MC; AiV: human Aichi viruses; PMMoV: pepper mild mottle virus; crAssP: crAssphage; BacP: culturable phages infection Bacteriodes spp.; FRNAP: FRNA phages II and III; WW: wastewater.
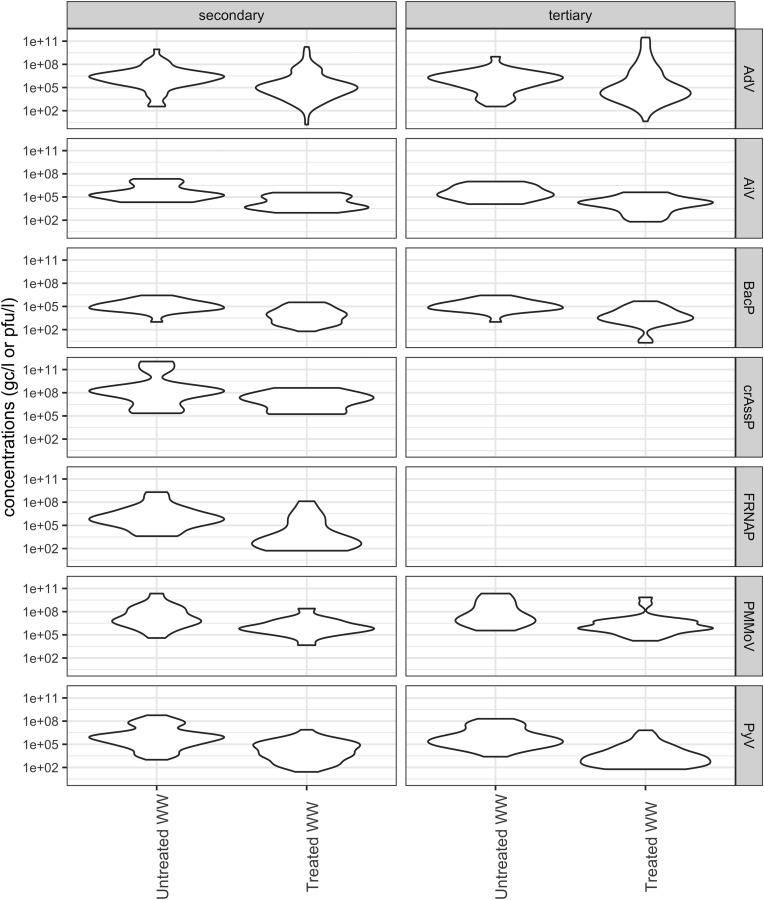
Fig. 3.
Violin plots of viral concentrations observed before and after secondary and tertiary wastewater treatment processes. Data are composite observations of mean and range values extracted from the analysed studies.
3. Evaluation of viral indicators
3.1. Criterion 1: Ease of detection and quantification
For the accurate detection of low viral titres, environmental samples are often concentrated prior to virus detection. Ultracentrifugation, ultrafiltration, adsorption/elution and flocculation are often used for the concentration of water samples, and their effectiveness and limitations have been reviewed previously (Barardi et al., 2012; Bofill-Mas and Rusiñol, 2020; Cashdollar and Wymer, 2013; Haramoto et al., 2018; Ikner et al., 2012). The efficiency of viral recovery depends on the type of concentration method used, the sample type and the virus type. Hence, viruses that can be easily and reproducibly recovered using simple concentration methods should be used as indicator viruses.
Many approaches are available for the detection and quantification of viruses in environmental samples, including PCR and isothermal amplification of target genes, microfluidics, metagenomics, biosensors, microarrays and culturing-based techniques, as reviewed recently (Bonadonna et al., 2019; Farkas et al., 2020; Hamza and Bibby, 2019). Many of the emerging approaches show great potential to detect low concentrations of viruses in difficult matrices (Dhar and Lee, 2018; Farkas et al., 2020; Gyawali et al., 2019b), however, to date they have not been implemented in the monitoring of viral contamination in the aquatic environment.
In most studies, enteric viruses and proposed indicators were detected and quantified using real-time quantitative PCR (qPCR)-based approaches (Girones et al., 2010; Haramoto et al., 2018), which are rapid, easy, and cheap methods enabling strain-level detection. For instance, targeting different regions of the hexon capsid protein gene, all human AdVs or only enteric AdVs (AdV genogroup F) can be quantified (Tables S1 and S8). qPCR can be easily multiplexed, enabling the simultaneous detection of 2–5 viral targets (Ahmed et al., 2019a; Farkas et al., 2017b; Jiang et al., 2014; Lee et al., 2016; Montazeri et al., 2015). Hence, it is widely used for the analysis of the level and spread of viral contamination in the aquatic environment (Staggemeier et al., 2017). More recently, digital PCR approaches, enabling absolute quantification without relying on standards, have also been used for the estimation of viral counts in wastewater and in environmental waters (Ishii et al., 2014; Jumat et al., 2017; Kishida et al., 2014; Sedji et al., 2018). These methods are also efficient, sensitive and often provide more accurate results that qPCR (Ishii et al., 2014; Kishida et al., 2014).
The primers and probes repeatedly used in environmental studies for the detection and quantification of the potential indicator viruses with qPCR, reverse transcription (RT-)qPCR, and dPCR, are listed in Table S8. In general, hydrolysis probe-based assays were predominantly used for viral detection. The specificity and sensitivity of the primer and probe sets had been assessed (empirically or in silico) using a set of target and non-target sequences and dilution series and shown to be adequate for the quantification of the target sequences (Barrios et al., 2018; Chehadeh and Nampoory, 2013; Dumonceaux et al., 2008; Goh et al., 2009; Gröndahl et al., 1999; Heim et al., 2003; Hernroth et al., 2002; Jothikumar et al., 2005; Kitajima et al., 2013; Ko et al., 2005; McQuaig et al., 2009; Ogorzaly and Gantzer, 2006; Pal et al., 2006; Pang et al., 2012; Prevost et al., 2015; Rusiñol et al., 2015; Stachler et al., 2017; van Maarseveen et al., 2010; Wolf et al., 2010, 2008; Xagoraraki et al., 2007). The high detection rates (Fig. 2; Table 2) also suggest that the primer and probe sets were suitable for the sensitive detection of the target viruses. Nonetheless, the specificity of the sets should be revised frequently in order to assure that novel strains are detected.
PCR-based approaches however have some disadvantages. qPCR and especially RT-qPCR are often inhibited by organic substances, e.g. polyphenolic compounds, found in environmental samples (Ahmed et al., 2015; Farkas et al., 2017a; Girones et al., 2010; Matheson et al., 2014). Therefore, the use of DNA viruses as indicators (e.g. AdV, PyV, crAssphage) for wastewater-derived viral contamination may be more feasible than the use of RNA viruses (AiV, PMMoV, FRNAP) due to the more robust molecular detection of DNA targets (Farkas et al., 2017a; Hata et al., 2011). A major disadvantage of all PCR-based viral detection approaches is that they do not give any indication on the infectivity of the target, and hence often overestimate viral concentration and human health risks (Knight et al., 2013). Detecting segments of indicator genes is helpful for the evaluation of the magnitude of faecal contamination and source tracking, nonetheless, an ideal indicator should be capable of being cultured in vitro to enable the direct assessment of viral infectivity and decay in wastewater and in the aquatic environment.
In the studies subject to this review, enteric viruses, crAssphage and PMMoV were predominantly detected and quantified using qPCR-based assays. However, in a few studies plaque assay or integrated cell culture-qPCR (ICC-qPCR) were used for AdV detection (Table S1). The combination of cell culture and qPCR detection of viral replication enabled the detection of infectious viruses, which grew slowly and/or failed to produce cytopathic effects. Using this approach, the time required for infectivity analysis has been reduced from one week to two days, enabling rapid detection. Overall, qPCR/dPCR gave 1–5 log10 higher AdV concentrations than plaque assay and ICC-qPCR due to the presence of damaged virus particles and free viral DNA derived from degraded viruses in the environmental samples (Fongaro et al., 2015, 2013; Hamza et al., 2011; Hewitt et al., 2011; Rigotto et al., 2010; Rodríguez et al., 2013; Sassoubre et al., 2012; Sedji et al., 2018). The higher concentrations detected using ICC-qPCR compared to the traditional culturing assays suggest that using qPCR-based quantification of cultured viruses is more sensitive and hence more reliable in environmental settings (Fongaro et al., 2013; Sedji et al., 2018). PyVs and AiV are also culturable, however, the propagation process is time consuming (2–4 and 4–6 weeks, respectively) and often inconclusive (Reuter et al., 2011; Seehafer et al., 1978), hence, these approaches have not been adapted to environmental studies. ICC-qPCR-based approaches may be suitable for assessing the infectivity of these viruses in the environment, however, to date no ICC-qPCR assays have been developed for these targets. The disadvantage of any culturing-based assay for enteric virus detection is the need for specific equipment (e.g. CO2 incubator), environment (BSL2 or BSL3) and staff for the maintenance of specific cell lines, which may not be available in routine monitoring laboratories.
FRNAP are easy to culture and readily form plaques on a lawn of the host bacterium, which is usually the WG49 strain of Salmonella typhimurium or Escherichia coli HS(pFamp)R (USEPA, 2001). Higher volumes (up to 100 ml) of samples or concentrates are typically used for culture-based assays than for qPCR (a few microliters of nucleic acid extract) and so culturing can be more sensitive than direct qPCR. However, this method will produce plaques of a range of different strains that cannot be differentiated based on morphology. Therefore, to identify and quantify specific FRNAP genogroups, it is necessary to use genogroup-specific molecular detection methods. Such methods include RT-PCR analysis of plaques (Haramoto et al., 2015, 2012) and 1-day ICC-RTqPCR (Hartard et al., 2017), most-probable number assays (Hata et al., 2016) or in-situ plaque membrane hybridisation techniques (Flannery et al., 2013).
Many Bacteroides-associated phages are also culturable using appropriate hosts, including Bacteroides strains GB-124, RYC 2056, GA17 and ARABA-84, with double-layer agar method to quantify the number of plaque forming units (pfu). However, the assay is more challenging than the plaque assay for FRNAP as culturing Bacteroides spp. require anaerobic conditions, which may not be available in most laboratories. The recent isolation and in vitro maintenance of phage ΦCrAss001 infecting Bacteroides sp. indicates that plaque assays for this type of phage may be used in future environmental studies (Shkoporov et al., 2018). It is important to mention, though, that the crAssphage qPCR assay (Table S7) does not detect ΦCrAss001, as this phage is reported to belong to a different genus.
In order to estimate viral decay where no in vitro infectivity assay is routinely available (e.g. hepatitis E virus, noro- and sapovirus), capsid integrity assays can be performed based on the assumption that an intact virus particle is infectious. Capsid integrity can be assessed by the elimination of free viral nucleic acids using enzymatic treatment, such as DNase, RNase (Fongaro et al., 2013) and intercalating dye pre-treatment (Prevost et al., 2016) or by capturing only the intact virus particles using immunomagnetic separation (IMS) (Haramoto et al., 2010). PCR-based enumeration following integrity assays show lower viral concentrations than direct qPCR, as the free viral nucleic acids are eliminated. However, as the intact virus particles may be damaged and hence non-infectious, these approaches may still overestimate viral counts (Fongaro et al., 2013; Walker et al., 2019). Nonetheless, integrity assays are valuable tools for estimating the number of viral particles in environmental samples and their use may improve viral risk assessment.
3.2. Criterion 2: Human waste association
Viruses, such as AdV, PyV and AiV strains (Tables S1–S3), which specifically infect humans, are logical choices for indicators for human faecal contamination. Using these viruses and their corresponding animal associated strains, the source of contamination (e.g. human vs wildlife, livestock, etc.) can be assessed. For example, Staggemeier et al. (2015) used SYBR Green qPCR for the detection and quantification of AdVs in water and sediment samples by distinguishing human, bovine, porcine, canine and avian AdV genome sequences based on their melting temperature. Human and porcine AdVs, bovine PyV and porcine circovirus have also been used to assess the level of agriculture-related and human sewage-associated contamination in recreational, groundwater and drinking water (Fongaro et al., 2015; Garcia et al., 2012; Rusiñol et al., 2014). In the studies evaluated in this review, the AdV qPCR assays targeted all human AdV groups (A-G), the most common groups (A-F) or specific groups (C and F; Table S1). All of these groups are human-specific, demonstrating that waterborne infections of AdV F (predominantly type 41) are the most prevalent in wastewater and in the aquatic environment (Bofill-Mas et al., 2010; Chigor and Okoh, 2012; Fong et al., 2010; Fumian et al., 2013; Haramoto et al., 2007; Hewitt et al., 2011; Iaconelli et al., 2017; Ibrahim et al., 2018; Lun et al., 2019; Myrmel et al., 2015; Ogorzaly et al., 2015; Shih et al., 2017). The most common PyVs associated with wastewater are the JC and BK strains (Table S2), however, MC PyV is also found in wastewater and in wastewater-contaminated water (Di Bonito et al., 2014; Rusiñol et al., 2015). All known human AiV (group A and B) have been found in human wastewater (Table S3). These viruses are highly human-specific and have not been found to associate with animal diseases. The JC and BK strain have not been found in animal waste (McQuaig et al., 2009), whereas to date, animal waste samples have not been tested for human AiV.
PMMoV has been found at high concentrations in domestic wastewater (raw and treated) and in wastewater-polluted environments and shown to correlate well with other human markers (Bacteriodes HF183, PyV) (Kitajima et al., 2018b; Symonds et al., 2018, 2016) implying it associates with human waste. Nonetheless, the primary source of the virus are bell and chilli peppers, with the suggestion that it should not be used as a faecal indicator nearby to commercial pepper plant production areas. qPCR assays targeting PMMoV show high sensitivity, however, the viruses were also detected in avian, bovine and dog faeces at low concentrations (Gyawali et al., 2019a; Hamza et al., 2011; Rosario et al., 2009) suggesting that animals may also access pepper as a food source. Furthermore, it has been suggested that PMMoV is more abundant in faeces and in wastewater where more pepper products are consumed (Symonds et al., 2018), therefore the prevalence of PMMoV should be further investigated.
Coliphages are commonly used as indicators for faecal viral contamination in water (McMinn et al., 2017). FRNAP genogroups II and III (FRNAP-II and FRNAP-III) have been shown to be associated with human sources, while genogroups I and IV (FRNAP-I and FRNAP-IV) are generally associated with non-human sources (Lee et al., 2011; Stewart-Pullaro et al., 2006). For this reason, several studies have used methods (described in Section 3.1) to distinguish between FRNAP genogroups to determine faecal sources. However, while there does appear to be a general association between genogroups and faecal sources, the bacterial host (E. coli expressing F-pili) is not source-specific and there is often overlap between source types for each genogroup (Cole et al., 2003; Harwood et al., 2013).
Bacteriophages infecting Bacteroides, common human gut bacteria, have also shown potential as indicators for faecal contamination in the environment. The most commonly used strains, which are phages that infect Bacteroides BG-124 (BacBG124P), RYC-2056 (BacRYC 2056P), GA-17 (BacGA17P) and ARABA-84 (BacARABA84P), were shown to be human specific. However, one study detected BacRYC 2056P in wastewater samples derived from abattoirs, suggesting animal association (Wicki et al., 2015). qPCR based assays targeting crAssphage have shown good human specificity. While some cross-reactivity has been shown with dog, gull, poultry, pig and cattle faeces (Ahmed et al., 2018a; García-Aljaro et al., 2017; Stachler et al., 2017), the levels of crAssphage found in these non-human sources were several orders of magnitude lower than that of human sources. The highest crAssphage concentrations in animal sources were found by García-Aljaro et al. (2017) which may be attributed to pooled samples and/or the use of a different qPCR assay. García-Aljaro et al. (2017) also found that by normalising crAssphage levels against a general faecal indicator (E. coli), it was still possible to distinguish between human and non-human sources. Nevertheless, the cross-reactions of the qPCR with animal excreta should be further investigated to assess human specificity.
3.3. Criterion 3: Presence in wastewater at high concentrations
AdVs, PyV, AiV, human gut-associated bacteriophages and PMMoV are all frequently found in raw sewage and untreated wastewater at high concentrations (Fig. 2; Tables S1–6). The highest concentrations among the potential indicators were noted for crAssphage with concentrations of 1010–1012 gc/l in raw sewage detected in samples taken in Japan. CrAssphage concentrations were lower in wastewater samples taken in the US (Florida; 109–1010 gc/l) (Ahmed et al., 2018a) and in the UK (Wales; 105–108 gc/l) (Farkas et al., 2019). CrAssphage is not currently well characterised and while the current primer and probe set do not align to any recently discovered relatives of crAssphage, it is possible that the qPCR-based detection assay is not specific to a single strain.
AdVs were detected at 1011 gc/l concentration in wastewater influent, in Pisa, Italy in 2009–2010 (Carducci and Verani, 2013). In other studies using the same primer and probe set (Hernroth et al., 2002), the concentration of AdV was lower, between 103 and 109 gc/l with the highest concentrations measured in other wastewater treatment plants in Italy (La Rosa et al., 2010), followed by peak concentrations of 108 gc/l in Rome, Italy (Muscillo et al., 2008), Barcelona, Spain (Bofill-Mas et al., 2006), and in Minas Gerais and Rio de Janeiro, Brazil (Assis et al., 2017). Similar peak concentrations (108 gc/l) were observed when AdV groups A-G were targeted in Germany (Hamza et al., 2009a) and in Queensland, Australia (Sidhu et al., 2017b). The highest concentrations of AdVF (108-1010 gc/l) were observed in the US (Michigan) (Simmons et al., 2011) and in Giza, Egypt (Elmahdy et al., 2019) further verifying that this group of AdV is highly prevalent.
PMMoV are also present in wastewater at high concentrations (up to 1010 gc/l). The highest PMMoV concentrations were reported in Florida and other states in the US (Rosario et al., 2009) followed by Germany (107–108 gc/l) (Hamza et al., 2011), New Zealand (107 gc/l) (Gyawali et al., 2019a), Vietnam and the US (Arizona; 106–107 gc/l) (Kitajima et al., 2014; Kuroda et al., 2015; Schmitz et al., 2016). Slightly lower FRNAP concentrations were noted in wastewater with the FRNAP-II appears to be more prevalent (107–109 gc/l) than FRNAP-III (104–107 gc/l) (Fig. 2). However, more studies are needed to further investigate FRNAP-II/III concentrations in wastewater.
Among PyVs, JCV had the highest concentrations (107-108 gc/l) in wastewater collected in Brazil and Chile (Fumian et al., 2010; Levican et al., 2019), however, it was less prevalent (103-106 gc/l) in the US (Arizona), Spain and in the UK (Wales) (Bofill-Mas et al., 2006; Farkas et al., 2018a; Kitajima et al., 2014; Rusiñol et al., 2015; Schmitz et al., 2016). BKV and MCV are probably less abundant in wastewater than JCV with concentration ranges of 103-107 gc/l and 104-105 gc/l, however, limited surveillance has been done on these viruses in wastewater. AiV has only been sought in untreated wastewater in the USA (Arizona), Vietnam and Nepal (Haramoto and Kitajima, 2017; Kitajima et al., 2014; Kuroda et al., 2015; Schmitz et al., 2016) with concentrations between 104 and 106 gc/l, and shown to be less prevalent than the other indicators. The detection rate and concentration of AdV, PyV, AiV, FRNAP-II, crAssphage and PMMoV usually exceeded the concentration of noroviruses, sapovirus, enterovirus, astrovirus, rotavirus and hepatitis E virus (Farkas et al., 2019, 2018a; Farkas et al., 2018; Flannery et al., 2013; Fumian et al., 2013; Grøndahl-Rosado et al., 2014a; Hata et al., 2014; Kitajima et al., 2014, 2013; Masclaux et al., 2013; Prevost et al., 2015; Qiu et al., 2015; Simmons et al., 2011). However, in some cases norovirus and rotavirus showed higher concentrations than AdV and PyV (Kaas et al., 2018; Prado et al., 2019).
The peak concentrations of cultured phages infecting Bacteroides in untreated wastewater was 106 pfu/l, however, this number cannot be directly compared with the concentration of other indicators due to the different methods used for detection. The viral infectivity rate (i.e. gc: infective units) of virus may be as high as 1000:1 as determined for AdV (Hewitt et al., 2011), however, the actual infectivity rates for Bacteroides-associated phages is yet to be determined.
All potential indicator viruses showed high (>90%) detection rates in untreated wastewater, except BacRYC 2056P, which was found only in 82% and 38% of the analysed samples. AdVs and PyVs were frequently detected in wastewater from large and small wastewater treatment plants in the Americas, Europe, Asia and Australia (Fig. 1; Table 2; Tables S1 and S2). Prevalence and concentration information for AiV was only reported for untreated wastewater samples from large wastewater treatment plants in the USA (Arizona) and Vietnam (Kitajima et al., 2018a, 2014; 2013; Kuroda et al., 2015; Schmitz et al., 2016). The PMMoV titre was assessed in wastewater samples derived from the USA, Germany, New Zealand and Vietnam (Hamza et al., 2011; Kitajima et al., 2014; Kuroda et al., 2015; Rosario et al., 2009; Schmitz et al., 2016; Symonds et al., 2016). FRNAP-II prevalence in wastewater was only assessed in Japan and Ireland (Flannery et al., 2013; Haramoto et al., 2015, 2012; Lee et al., 2018), while FRNAP-III prevalence in wastewater was only assessed in Japan (Haramoto et al., 2015, 2012; Lee et al., 2018; Setiyawan et al., 2014, 2013). In wastewater, the concentrations of phages associated with Bacteroides BG-124 were assessed in the USA, Brazil, the UK (England) and Switzerland (E. Dias et al., 2018a; McMinn et al., 2014; Prado et al., 2018; Purnell et al., 2015; Stefanakis et al., 2019; Wicki et al., 2015), BacRYC 2056P were investigated in Colombia, the UK, Spain, France, Cyprus, Sweden, Switzerland and Thailand (Costán-Longares et al., 2008; Gomila et al., 2008; Payan et al., 2005; Venegas et al., 2015; Wangkahad et al., 2017; Wicki et al., 2015, 2011), BacGA17P were found in Colombia and several European countries (Casanovas-Massana et al., 2015; Costán-Longares et al., 2008; Gomila et al., 2008; Mayer et al., 2016; Payan et al., 2005; Venegas et al., 2015; Wicki et al., 2015) and BacARABA84P was identified in Switzerland (Wicki et al., 2015, 2011). CrAssphage concentrations were only determined in wastewater in the UK (Wales), Australia and the USA (Florida) (Ahmed et al., 2019a, 2018b; Farkas et al., 2018a). Due to the limited number of studies (Table 2), further testing is necessary to evaluate the prevalence and distribution of AiV, FRNAP-II, FRNAP-III, culturable phages infecting Bacteroides, crAssphage and PMMoV in untreated wastewater to assess their usefulness as indicators of wastewater pollution. Current data suggest that all assessed viral indicators, are present in untreated wastewater at high concentration and hence they are potentially good indicators for wastewater contamination.
3.4. Criterion 4: Resistance to wastewater treatment
Enteric viruses have been shown to be extremely resistant to traditional wastewater treatment procedures (Fig. 3; Table S7). As the removal efficiency varies amongst sites and the type of treatment process, comparative studies have been performed to study the resistance of enteric viruses and potential indicators during wastewater treatment.
In this study, 24 studies comparing the virus removal efficiency of different wastewater treatment processes were evaluated (Fig. 3). Fifteen of these studies exclusively used qPCR and RT-qPCR for the quantitative analysis of viral concentrations. As discussed in Section 3.1, these molecular techniques give no indication on the infectivity state of the viruses and hence may overestimate infective viral titres in untreated and treated wastewater and other environmental samples. This was demonstrated by Flannery et al. (2013) whose data showed that while infectious FRNAP-II in UV-treated effluent was approximately 2.3 log10 less than influent, only a 0.54 log10 reduction was found when using RT-qPCR alone. Most of this reduction in FRNAP-II infectivity occurred during the secondary treatment stage (1.69 log10 reduction), but the type of secondary treatment used in that study was not specified. In a study by Lee et al. (2018), an activated sludge process (a form of secondary treatment) resulted in 2.1 and 3.1 log10 reductions of infectious FRNAP-II and FRNAP-III, respectively. RT-qPCR analysis of the same samples showed log10 reductions of 1.6 and 2.5 for FRNAP-II and FRNAP-III, respectively. The differences between infectious virus and genome removal were not significant. These studies therefore reached conflicting conclusions with the former showing that infectivity studies are vital and the latter showing that they are unnecessary. It is possible that the activated sludge process used by Lee et al. (2018) resulted in physical removal of viruses, while the process used by Flannery et al. (2013) inactivated the viruses without physically removing them from the treated water. This highlights the importance of including the specific mechanisms used within a sewage treatment process when reporting such data, as it is not clear whether the secondary treatment processes in the two studies shared any mechanistic similarities.
Low removal rates have been reported for BacGB124P and BacGA17P during wastewater treatment. In most studies, the removal of these phages was ≤2 log10, regardless of the treatment method used (Dias et al., 2018a, Dias et al., 2018b; Mayer et al., 2016; Prado et al., 2018; Stefanakis et al., 2019), except for one study showing >5.6 log10 removal of BacGB124P when disk filtration and chlorination was used as tertiary treatment (Prado et al., 2018).
Data obtained from a wide range of qPCR-based viral quantification studies have shown limited removal of AdV, PyV, AiV, crAssphage and PMMoV during wastewater treatment. Activated sludge treatment and biofiltration, without further treatment resulted in 0.6–1.9 and 0.3–3.0 log10 removal of AdV and PyV, respectively (Fig. 3; Table S7). Tertiary treatment processes resulted in an additional 1–3.5 log10 removal of AdV and PyV with membrane bioreactors coupled with additional chlorination, filtration and UV treatment being the most efficient method for viral removal (Qiu et al., 2018; Rusiñol et al., 2015; Simmons et al., 2011). Furthermore, AdVs have been shown to be more resistant to UV treatment than poliovirus, rotavirus, caliciviruses and hepatitis A virus (Hijnen et al., 2006). However, laboratory-scale studies suggest that AdVs are more susceptible to chlorine treatment than enteroviruses and caliciviruses (Cromeans et al., 2010; Kahler et al., 2010; Thurston-Enriquez et al., 2005, 2003). Interestingly, significant differences were found for the removal of PyV strains. MCV was found to be the most resistant to treatment followed by JCV and BKV (Rusiñol et al., 2015).
Fewer studies evaluated the removal of AiV, crAssphage and PMMoV, than the removal of AdVs, PyVs and phages. Overall, AiV showed 1–3 log10 reduction during secondary and 1–2 log reduction during tertiary wastewater treatment (Kitajima et al., 2018a, 2014; 2013; Schmitz et al., 2016). The removal of crAssphage was also in the range of 1.0–1.2 log10 during secondary wastewater treatment (Farkas et al., 2019), however, crAssphage removal has not been assessed during tertiary treatment yet. The current data suggests that PMMoV is stable during secondary treatment and chlorination, which results in <2 log reduction (Symonds et al., 2018). Larger PMMoV removal (>4 log) was only observed using electrocoagulation and Bardenpho (aerobic/anaerobic multi-reactor) technologies (Schmitz et al., 2016; Symonds et al., 2015). Further research is needed to evaluate the reduction of PMMoV during UV treatment and other wastewater treatment procedures to evaluate its usefulness as an indicator. The major advantage of crAssphage and PMMoV is that their concentrations are usually high in wastewater, and hence the efficiency of their removal can be easily monitored. Nonetheless, their infectivity and decay have not been investigated due to the current lack of in vitro culturing-based methods.
In studies where the removal of indicator viruses was compared with the removal of common pathogenic enteric viruses, indicator viruses showed similar or less removal than the pathogens (Carducci and Verani, 2013; Farkas et al., 2018a; Kitajima et al., 2014; Prado et al., 2019; Rusiñol et al., 2015; Schmitz et al., 2016). Furthermore, a meta-analysis on the efficiency of secondary wastewater processes showed that activated sludge treatment resulted in 0.20–2.18 log10 reduction of rotavirus, and norovirus GI and GII, whereas biofiltration resulted in higher removal (1.52–4.30 log10) of norovirus GII and enteroviruses (Sano et al., 2016). These removal rates are higher than the removal rates determined for the indicators reviewed here suggesting that the indicators can represent the removal of the most resistant viruses. However, three studies showed higher removal rates of BKV and JCV than norovirus, sapovirus, enterovirus and rotavirus (Farkas et al., 2018a; Fumian et al., 2013; Schmitz et al., 2016) suggesting that PyVs are less resistant than the pathogenic viruses and hence should be used with caution as an indicator. Current data shows that AdV, AiV, FRNAPII/III, crAssphage and PMMoV may be suitable for the assessment of wastewater treatment processes.
As different viruses have varying reactions to wastewater treatment processes, the use of multiple indicators is recommended. For indicators other than bacteriophages reviewed here, the exclusive use of molecular detection and quantitation is a major limitation in understanding enteric virus removal. Hence, combinations of infectivity studies and molecular assays should be performed for viruses that can be cultured in vitro in order to assess viral survival during wastewater treatment.
3.5. Criterion 5: Persistence in the aquatic environment
Many of the studies that have been conducted to estimate viral persistence in natural waters have relied solely on qPCR-based quantification. While the reliance on qPCR data alone may lead to overestimations of infectious viral persistence, its use is nonetheless important especially when considering unculturable enteric viruses. When measuring persistence of viral indicators in the aquatic environment, researchers should therefore be clear whether they are studying the persistence of a viral signal (for example nucleic acids detected by qPCR) or the infectivity of viruses (for example by culture).
3.5.1. Indicators in surface freshwater
Most research has focused on the occurrence and survival of indicator viruses in surface water. When quantified in surface freshwater bodies (lakes, rivers, streams, etc.) by qPCR, these viral indicators (e.g. AiV, AdV, JCV, PMMoV) are typically detected at up to 4 log10 higher concentrations than common enteric pathogenic viruses, e.g. norovirus, enterovirus and rotavirus (Hata et al., 2014; Jurzik et al., 2010; Rusiñol et al., 2015; Sassi et al., 2018). All indicator concentrations in river water correlated with the distance of sampling point from the source of contamination (wastewater treatment plant), with significantly higher concentrations occurring near the wastewater treatment plant than further downstream or upstream (Ebdon et al., 2007; Farkas et al., 2018a; Prevost et al., 2015; Rusiñol et al., 2015; Sassi et al., 2018; Sibanda and Okoh, 2012; Tandukar et al., 2018; Venegas et al., 2015; Wangkahad et al., 2017). Comparative studies showed that PMMoV occurred at higher concentration than AdV, AiV and PyV in surface water bodies in the USA (Arizona: 103-106 gc/l and Colorado: 104-105 gc/l), Germany (104-105 gc/l) and Vietnam (104-106 gc/l) (Betancourt et al., 2014; Hamza et al., 2011; Kuroda et al., 2015; Sassi et al., 2018). The concentration of AdV and AiV (up to 104 gc/l) were similar in river water collected in the USA (Colorado) and Japan (Betancourt et al., 2014; Hata et al., 2014; Sassi et al., 2018), whereas AdV was more prevalent than PyV in river water samples collected in Spain (76% vs 48% detection rates), the UK (Wales: 88% vs 65%), Japan (61% vs 11%) and Germany (79% vs 59%) (Albinana-Gimenez et al., 2009; Farkas et al., 2018a; Haramoto et al., 2010; Jurzik et al., 2010; Rusiñol et al., 2015). However, detection rates and concentrations of AdV and JCV were similar in highly polluted rivers close to the wastewater discharge points near Barcelona, Spain (100%, 103-104 gc/l) and Rio de Janeiro, Brazil (100%, 102-105 gc/l) (Calgua et al., 2013). Taken together, our analysis shows that these indicator viruses are present in wastewater-polluted surface freshwater at high concentrations, which enables the accurate detection of the viruses and the comparative analysis of the rate of pollution (Crank et al., 2019; Zhang et al., 2019).
Culturable bacteriophages were also present in surface freshwater (Fig. 2; Table 2). FRNAP-II were detected most frequently with concentrations up to 106 pfu/l followed by FRNAP-III and phages infecting Bacteroides (up to 105 pfu/l). FRNAP-II concentrations correlated with AdV concentrations in river water in France (Ogorzaly et al., 2009) and with AdV, norovirus, astrovirus and rotavirus in tropical freshwater samples in Singapore (Vergara et al., 2015). To date, no comparative studies have been done to compare enteric viruses and phages infecting Bacteroides spp. in surface freshwater. More research is essential on the prevalence of culturable human gut associated phages to assess their usefulness as indicators.
3.5.2. Indicators in seawater
As for freshwater environments, similar viral indicator trends have been observed in coastal waters where wastewater contamination is present. PMMoV was present at higher concentrations (102-105 gc/l) than PyV (102 gc/l) in coastal water at Miami, Florida (Symonds et al., 2016) and crAssphage was also present at higher concentrations (103-105 gc/l) than AdV (102-104 gc/l) and JCV (102-103 gc/l) in seawater collected at Conwy, Wales (Farkas et al., 2018a). AdV also had higher concentrations than PyV in seawater collected at Rio de Janeiro and Santa Caterina, Brazil (102-105 gc/l vs 101-103 gc/l), Florianopolis, Brazil (103-107 gc/l vs < 10 gc/l), North Wales (102-104 gc/l vs 102-103 gc/l), and Catalonia, Spain (101-105 gc/l vs 100-102 gc/l) (Dias et al., 2018a, Dias et al., 2018b; Farkas et al., 2018; Moresco et al., 2012; Rusiñol et al., 2015). CrAssphage, PMMoV and AdV and PyV are usually present up to 4 log10 higher concentrations in seawater than hepatitis A virus, norovirus and sapovirus (Dias et al., 2018a, Dias et al., 2018b; Farkas et al., 2018; Fongaro et al., 2015; Moresco et al., 2012; Rusiñol et al., 2015; Symonds et al., 2018), however, one study found that the concentration of indicators and norovirus GII, rotavirus and sapovirus were similar, approx. 104 gc/l, in seawater collected at the Tahiti coast (Kaas et al., 2018). BacGB124P, BacRYC 2056P and BacGA17P were also found in seawater at concentrations up to 104 pfu/l (Olalemi et al., 2016), however, these concentrations were not compared with enteric viruses. To date, FRNAP-II/III concentrations have not been measured in seawater samples. Based on the data reviewed here, PMMoV, crAssphage and AdV are suitable markers for wastewater contamination in seawater.
3.5.3. Indicators in groundwater
Very few studies have evaluated the concentration of enteric viruses and viral indicators in groundwater. AdV, JCV, AiV, PMMoV, BacGB124P and BacARABA84P were detected in polluted groundwater in the USA (Arizona and Colorado) and Vietnam at very low concentrations (Albinana-Gimenez et al., 2009; Betancourt et al., 2014; Kuroda et al., 2015), hence the concentrations cannot be compared (Fig. 2; Table 2). Future studies may include the efficient concentration of high volumes (>100 l) of groundwater to accurately determine viral concentrations and the associated risks.
3.5.4. Persistence of indicators in water
Understanding how long pathogenic and indicator viruses survive in the environment is crucial for accurate risk assessment and management. The mechanisms and factors influencing viral decay, such as virus type, temperature, microbial activity, pH, water type/conductivity, UV/sunlight radiation and the presence of solid/organic matter, have been assessed (Jin and Flury, 2002; Rzeżutka and Cook, 2004; Verbyla and Mihelcic, 2015). Many studies have shown that enteric viruses are more stable in the aquatic environment than traditional indicators, such as coliform bacteria and coliphages (El-Senousy et al., 2014; Fattal et al., 1983; Keswick et al., 1982; Muscillo et al., 2008; Ogorzaly et al., 2010; Wait and Sobsey, 2001).
FRNAP are easily cultured, and their persistence in surface waters has been studied in both surface freshwaters and seawater (Hata et al., 2016; Muniesa et al., 2009; Ravva and Sarreal, 2016; Yang and Griffiths, 2013). In general, FRNAP-I has been found to be the most persistent followed by FRNAP-II, FRNAP-III and then FRNAP-IV. Using simulated sunlight, Flannery et al. (2013) studied the effect of solar radiation on the persistence of FRNAP-II and norovirus in seawater. The reductions in RT-qPCR detectable viruses was similar for norovirus and FRNAP-II under both summer and winter sunlight conditions. However, it took between 81% and 88% longer for a 90% reduction in RT-qPCR detectable FRNAP-II than for infectious FRNAP-II. This highlights again the need to consider infectivity when studying viral persistence in the environment. Brion et al. (2002) also studied the survival of different FRNAP genogroups in surface water. Environmental isolates of FRNAP-II had the highest variability in survival between isolates, while FRNAP-III had the lowest variability in survival between isolates. They concluded that FRNAP-III is suited to determining whether there had been recent contamination to a water body by human faeces. In contrast, FRNAP-II was more suited to indicating contamination by distant or sporadic human source contamination.
Due to its easy molecular detection and relatively straightforward in vitro culturing, the survival of AdV has also been well-studied. AdVs have shown 1–2 log10 reduction in infectivity in raw and sterilised groundwater and surface water over 120–180 days (Ogorzaly et al., 2010; Rigotto et al., 2011). In seawater, the decrease of viral infectivity was more rapid than in groundwater, with 1.2–1.4 log10 AdV reductions in 28 days (Enriquez et al., 1995) and sunlight significantly enhancing degradation at a rate of at least 2 log10 reduction per day (Liang et al., 2017). AdVs were more stable in groundwater and surface water than poliovirus, rotavirus and hepatitis A virus (El-Senousy et al., 2014; Enriquez et al., 1995). Ogorzaly et al. (2010) showed that AdV persists for longer in ground water than both MS2 (FRNAP-I) and GA phages (FRNAP-II). This difference in survival and persistence was greatly increased with an increase in temperature from 4 °C to 20 °C. These studies highlight the stability of AdV compared to other viruses, however, these studies were conducted in laboratory experiments and the viral stability may differ in field conditions.
PyV has also been shown to be as resistant to sunlight in seawater as AdV (Ahmed et al., 2019b; Liang et al., 2017), however, the monitoring experiments detailed in Section 3.5.2 suggest that PyV degrades in water more rapidly than AdV. In contrast, crAssphage proved to be as persistent as AdV and PyV in coastal bathing water (Ahmed et al., 2019b). The temporal decay of AiV, PMMoV and culturable bacteriophages infecting Bacteroides is not yet known and should be investigated and compared with AdV decay to determine their usefulness as indicators. The comparison of the mechanisms of decay of PMMoV and the other viral indicators would be especially interesting due to the differences in the structure of the virions (tubular vs. icosahedral).
3.6. Criterion 6: Global distribution and temporal stability
All potential indicator viruses reviewed here have been detected in environmental waters, wastewater or stool samples of individuals in Asia, Europe, Australia, Africa and the Americas, highlighting the global distribution of these viruses (Cinek et al., 2018; Fratini et al., 2014; Friedman et al., 2009; Guido et al., 2016; Jofre et al., 2014; Kitajima and Gerba, 2015; Rames et al., 2016; Schaper et al., 2002; Symonds et al., 2018). In the reviewed studies, the indicator viruses were detected and quantified in 31 countries (including 10 US states), with the majority of studies conducted in the US, Brazil, Western Europe, Japan and Australia (Fig. 1; Table 2). While the available data suggest that these viruses are distributed globally, very limited information on enteric and indicator virus quantities is available from developing countries (e.g. India, Northern Asia and most African countries) (Table 2). To date, none of these viruses have been studied in water from Antarctica, where they could point towards contamination of pristine areas by research scientists, or long-distance dispersal.
During long-term monitoring surveys, in the Katsura River to the west of Kyoto, Japan, FRNAP-II was shown to have very limited seasonality, with similar levels in the winter and summer months (Hata et al., 2016). However, in a tributary of the Uji River to the South of Kyoto, FRNAP-II was detected only during winter. FRNAP-III was also found to be more prevalent during winter at both sites, a trend also observed in effluent from Johkasou effluent by Setiyawan et al. (2013). Culturable phages infecting Bacteroides showed no seasonal changes in their concentration in river water in the UK (Ebdon et al., 2007) and in wastewater collected in seven US states (McMinn et al., 2014) and in Brazil (Prado et al., 2018).
In the studies reviewed, crAssphage, AdV and PyV showed no seasonal changes in concentrations in untreated and treated wastewater, river and seawater samples (Carducci and Verani, 2013; Farkas et al., 2018a, Farkas et al., 2018; Fumian et al., 2013; Iaconelli et al., 2017; Masclaux et al., 2013; Qiu et al., 2015; Rusiñol et al., 2015; Schmitz et al., 2016). AiV and PMMoV also showed stable titres in treated and untreated wastewater over a year (Iaconelli et al., 2016; Kitajima et al., 2014; Myrmel et al., 2015; Schmitz et al., 2016), however, peak concentrations for AiV were noted in wastewater in Japan during winter and spring (Kitajima et al., 2013). PMMoV showed no seasonality in river water either (Haramoto et al., 2013; Rosario et al., 2009). Higher AdV concentrations were observed in treated wastewater collected in Wales during summer than in winter and spring, which was most likely due to dry weather and a transient increase in population due to tourism in the summer months (Farkas et al., 2018). Furthermore, higher AdV concentrations were detected in untreated wastewater in Norway during January–March compared to the concentrations observed during April–December (Myrmel et al., 2015). The prevalence of AdV was also higher during autumn-winter than during the spring-summer in wastewater collected in Egypt (Elmahdy et al., 2019). Similarly, AdVs were detected at low concentrations during the summer and autumn months in river water samples collected in Japan and in Germany, respectively (Hamza et al., 2009b; Kishida et al., 2012), probably due to dry weather conditions. Overall, these findings suggest that the indicators are detectable and quantifiable throughout the year, which enables the continuous evaluation of wastewater contamination. The current data imply that precipitation has more effect on viral loads than temporal changes in the number of infections. Nonetheless, this should be further investigated by comparing epidemiological data, viral loads in wastewater and precipitation over several years.
In terms of fine-scale temporal variability, the effect of rainfall on virus concentrations in surface water is variable. On one hand, a decrease in virus concentrations in surface water has been reported due to dilution of the water body (Grøndahl-Rosado et al., 2014b). In contrast, a number of studies have shown association between precipitation and elevated enteric viral concentrations in water (Ebdon et al., 2007; Wicki et al., 2015). In regions with combined sewers, much of this increase in contamination is likely due to the additional wastewater input via CSOs and storm water drainage.
CSOs discharge largely untreated (screened, partially settled or untreated) wastewater into the environment. This almost certainly results in higher numbers of enteric viruses being discharged into receiving waters than would otherwise be the case from fully treated sewage effluents (Fong et al., 2010; Hata et al., 2014). Furthermore, relatively few CSOs have spill duration monitoring and almost none have microbiological or chemical monitoring requirements. Therefore, in most cases the input of contamination can only be monitored via the surveillance of wastewater-derived contaminants in water bodies.
4. Conclusions and future research
The viruses reviewed here have all been shown to have potential to indicate wastewater-derived pollution in the aquatic environment (Table 3 ). Due to their wide distribution, they may be implemented in water quality risk assessments worldwide. The major advantage of enteric viral indicators (AdV, PyV, AiV) is that they are human specific, hence their use as indicators enables us to track human-derived contamination exclusively. In addition, crAssphages and other phages, which infect commensal bacteria associated with human gut, and PMMoV, which is a plant virus found in the human gut due to the consumption of infected plant-derived food, are also associated primarily with domestic wastewater contamination. A major advantage of phages and PMMoV is that they are not infectious to humans and hence their detection and culturing in the laboratory pose no risk of infection to the operators. FRNAP-II and FRNAP-III have also been shown to be useful in determining human sources of viral contamination due to their prevalence in human waste. However, due to the non-specific nature of their natural E. coli hosts, it is not certain what the reason is for the specific prevalence in human waste relative to FRNAP-I and FRNAP-IV. As such, it is unclear how well these can be applied globally and indeed how stable that relationship is.
Table 3.
Summary on how the reviewed viruses meet the criteria for wastewater indicator.
| Criterion | AdV | PyV | AiV | PMMoV | FRNAP (II/III) | Culturable Bacteroides phages | CrAssphage |
|---|---|---|---|---|---|---|---|
| Methods used for detection in environmental samples | qPCR; ICC-qPCR; culturing |
qPCR | qRT-PCR | qRT-PCR; plant infectivity assay |
qRT-PCR; culturing |
culturing | qPCR |
| Human association | Human-specific | Human-specific | Human-specific | Human waste and agricultural sites | Primarily human gut-associated | Primarily human gut-associated, have been found in animal faeces at low titres | Primarily human gut-associated, have been found in animal faeces at low titres |
| Concentration in wastewater (gc/l) | 1 × 101 – 3 × 1011 | 1 × 103 – 6 × 108 | 1 × 104 – 4 × 106 | 3 × 105 – 2 × 1010 | 4 × 103-2x109 | 1 × 101 – 6 × 106 | 2 × 105 – 1 × 1012 |
| Log10 removal during wastewater treatment | 0.2–5.5 (n = 500) | 0.3–4.2 (n = 407) | 0.8–2.7 (n = 72) | 0–2.7 (n = 106) | 0.1–3.1 (n = 172) | 0.5–5.6 (n = 304) | 1–1.2 (n = 39) |
| Concentration in the aquatic environment (gc/l) | 4–2 × 1010 | 1–1 × 107 | 7 × 101 – 8 × 108 | 1 × 101 – 8 × 108 | 0.2–2x106 | 1–2 × 105 | 1 × 103 – 3 × 107 |
| Global distribution and temporal stability | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations | Detected in clinical samples globally; limited seasonal variations |
The viruses reviewed here can be easily detected by qPCR-based methods, however, no such assay has been developed yet for culturable phages infecting Bacteroides spp. When using molecular methods, DNA viruses (AdV, PyV and crAssphage) may be easier and more affordable to monitor than RNA viruses (AiV, PMMoV, FRNAP). The infectivity of FRNAP can be easily studied using a simple and rapid plaque assay. Furthermore, the infectivity state of AdV can also be monitored using ICC-qPCR. Infectivity assays are also available for PyV, AiV and crAssphage, however, the usefulness of those assays in environmental setting have not been critically evaluated. Furthermore, there are emerging technologies, such as isothermal amplification, biosensors and microfluidics approaches, which may be useful for the routine monitoring of viruses in the environment (Farkas et al., 2020). In some cases, these may offer the potential for near real-time reporting of viral concentrations in water, however, their applicability still needs to be critically evaluated from a scientific, practical and economic perspective. This is particularly the case for in situ devices where biofouling, cross-reactivity and sensor drift represent major problems when translating technologies developed in the laboratory to the field (Lin and Li, 2020).
Here, the review of global studies suggests that AdVs, AiV, FRNAP-II, FRNAP-III, crAssphage and PMMoV are detected more frequently and at high concentrations in wastewater and within polluted water bodies than the other indicators reviewed. PyVs are also present in wastewater at high concentrations, however, they are less prevalent in the environment than AdV (Albinana-Gimenez et al., 2009; Bortagaray et al., 2019; Dias et al., 2018a, Dias et al., 2018b; Haramoto et al., 2010; Moresco et al., 2012), suggesting rapid degradation. Similarly, while FRNAP-II and FRNAP-III are initially found at high concentrations in wastewater, it is likely that they degrade more rapidly in the environment than AdV. The concentration of BacGB124P, BacGA17P and BacARABA84P was also high in wastewater and have been found in wastewater polluted environments, however, only a limited number of studies have been conducted to date on viral decay prompting the need for more research in this area.
Based on their ease of detection, high concentrations in wastewater and environmental persistence, our review suggests that AdVs are the most useful viral indicators of wastewater contamination. However, AiV, crAssphage and PMMoV also show potential. More research is essential to evaluate the usefulness of these viruses and indicators. Future research should therefore focus on:
-
(i)
Careful monitoring of the association of crAssphage and PMMoV with non-human contamination.
-
(ii)
Monitoring the concentration and persistence of AiV, crAssphage and PMMoV in the aquatic environment, especially in groundwater and in seawater. The effect of extreme weather events on viral concentrations should also be investigated.
-
(iii)
Development of a simple and rapid standard operating procedure for concentrating and detecting viruses from water to facilitate the accurate detection of selected indicator virus (es).
-
(iv)
The development of multiplex qPCR assays to simultaneously detect a panel of the best markers, potentially tailored to differences in geographical diversity (particularly for PMMoV).
-
(v)
Critical evaluation and application of new and emerging rapid approaches for viral surveillance.
-
(vi)
Survival and maintenance of infectivity monitoring of AiV, crAssphage and PMMoV in wastewater and in the water environment. For that, the usefulness of infectivity assays for these viruses should be developed and evaluated.
-
(vii)
Undertake comprehensive field campaigns in areas where data is not available (e.g. Africa, Asia, Oceania) to validate the use of viral indicators as an effective way to monitor wastewater pollution.
-
(viii)
Use these viral indicators to validate current mathematical models which predict viral dispersal and which are used for risk assessment purposes.
-
(ix)
Better establish the relationship between viral indicators and wastewater pollution to enable the development of legislative standards for viral contamination of waterbodies.
A greater understanding of the fate and behaviour of these viruses will allow them to be routinely implemented for water quality monitoring and for viral risk assessment. With a standardised protocol for the detection and quantification of proposed indicators, viral contamination can be efficiently addressed by regulators and hence the number of waterborne and foodborne viral diseases can be reduced, ultimately enhancing global human health.
Contributors
KF and DIW conducted the literature search and the collection of data on viral concentrations. All authors contributed to structuring and writing the article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the UK Natural Environment Research Council (NERC) and the Food Standards Agency (FSA) under the Environmental Microbiology and Human Health (EMHH) Programme (NE/M010996/1) and by the Shellfish Centre RD&I operation, part-funded by the EU’s West Wales and the Valleys European Regional Development Fund (ERDF) Operational Programme through the Welsh Government. EMA was funded by the Biotechnology and Biological Sciences Research Council (BBSRC); this research was funded by the BBSRC Institute Strategic Programme Gut Microbes and Health BB/R012490/1 and its constituent projects BBS/E/F/000PR10353 and BBS/E/F/000PR10356. LSH was supported by a Soils Training and Research Studentship (STARS) grant from the Biotechnology and Biological Sciences Research Council (BBSRC) and Natural Environment Research Council (NE/M009106/1). We thank I. Bertrand (University of Lorraine, France), G. Fongaro, (Federal University of Santa Caterina, Brazil), I. Hamza (National Research Centre, Egypt), A. Hata (University of Tokyo, Japan), M.R. Jumat (King Abdullah University of Science and Technology, Saudi Arabia), M. Rusiñol (University of Barcelona, Spain), T. Sibanda (University of South Africa, South Africa), R. Staggemeier (Feevale University, Brazil), E. Symonds (University of Florida, USA), M. Verani (University of Pisa, Italy), S. Wurtzer (University of Paris, France) for providing data for Tables S1–6.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.watres.2020.115926.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adriaenssens E.M., Farkas K., Harrison C., Jones D.L., Allison H.E., McCarthy A.J., Jones D.L., Adriaenssens E.M., Harrison C., McCarthy A.J. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. mSystems. 2018;3 doi: 10.1128/mSystems.00025-18. e00025-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P.S.P.S., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81:2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Lobos A., Senkbeil J., Peraud J., Gallard J., Harwood V.J. Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida. Water Res. 2018;131:142–150. doi: 10.1016/j.watres.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Besley C., Power K. Novel crAssphage marker genes ascertain sewage pollution in a recreational lake receiving urban stormwater runoff. Water Res. 2018;145:769–778. doi: 10.1016/j.watres.2018.08.049. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Besley C. A duplex PCR assay for the simultaneous quantification of Bacteroides HF183 and crAssphage CPQ_056 marker genes in untreated sewage and stormwater. Environ. Int. 2019;126:252–259. doi: 10.1016/j.envint.2019.01.035. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Zhang Q., Kozak S., Beale D., Gyawali P., Sadowsky M.J., Simpson S. Comparative decay of sewage-associated marker genes in beach water and sediment in a subtropical region. Water Res. 2019;149:511–521. doi: 10.1016/j.watres.2018.10.088. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Payyappat S., Cassidy M., Harrison N., Besley C. Sewage-associated marker genes illustrate the impact of wet weather overflows and dry weather leakage in urban estuarine waters of Sydney, Australia. Sci. Total Environ. 2020;705:135390. doi: 10.1016/j.scitotenv.2019.135390. [DOI] [PubMed] [Google Scholar]
- Albinana-Gimenez N., Miagostovich M.P., Calgua B., Huguet J.M., Matia L., Girones R. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009;43 doi: 10.1016/j.watres.2009.01.025. 2011–2019. [DOI] [PubMed] [Google Scholar]
- Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U.S.A. 2005;102 doi: 10.1073/pnas.0504666102. 12891 LP – 12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis A.S.F., Otenio M.H., Drumond B.P., Fumian T.M., Miagostovich M.P., da Rosa e Silva M.L. Optimization of the skimmed-milk flocculation method for recovery of adenovirus from sludge. Sci. Total Environ. 2017;583:163–168. doi: 10.1016/j.scitotenv.2017.01.045. [DOI] [PubMed] [Google Scholar]
- Bain R., Cronk R., Hossain R., Bonjour S., Onda K., Wright J., Yang H., Slaymaker T., Hunter P., Prüss-Ustün A., Bartram J. Global assessment of exposure to faecal contamination through drinking water based on a systematic review. Trop. Med. Int. Health. 2014;19:917–927. doi: 10.1111/tmi.12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barardi C.R., Viancelli A., Rigotto C., Correa A.A., Moresco V., Souza D.S., Elmahdy M.E., Fongaro G., Pilotto M.R., Nascimento M.A. Monitoring viruses in environmental samples. Int. J. Environ. Sci. Eng. Res. 2012;3:62–79. [Google Scholar]
- Barrios M.E., Blanco Fernández M.D., Cammarata R.V., Torres C., Mbayed V.A. Viral tools for detection of fecal contamination and microbial source tracking in wastewater from food industries and domestic sewage. J. Virol. Methods. 2018;262:79–88. doi: 10.1016/j.jviromet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Bartsch S.M., Lopman B.A., Ozawa S., Hall A.J., Lee B.Y. Global economic burden of norovirus gastroenteritis. PloS One. 2016;11:1–16. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellou M., Kokkinos P., Vantarakis A. Shellfish-borne viral outbreaks: a systematic review. Food Environ. Virol. 2013;5:13–23. doi: 10.1007/s12560-012-9097-6. [DOI] [PubMed] [Google Scholar]
- Betancourt W.Q., Kitajima M., Wing A.D., Regnery J., Drewes J.E., Pepper I.L., Gerba C.P. Assessment of virus removal by managed aquifer recharge at three full-scale operations. J. Environ. Sci. Heal. - Part A Toxic/Hazardous Subst. Environ. Eng. 2014;49:1685–1692. doi: 10.1080/10934529.2014.951233. [DOI] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinkova O., Rosario K., Li L., Kapoor A., Slikas B., Bernardin F., Breitbart M., Delwart E. Frequent detection of highly diverse variants of Cardiovirus, Cosavirus, Bocavirus, and Circovirus in sewage samples collected in the United States. J. Clin. Microbiol. 2009;47:3507–3513. doi: 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr. Opin. Environ. Sci. Heal. 2020;16:7–13. doi: 10.1016/j.coesh.2020.01.006. [DOI] [Google Scholar]
- Bofill-Mas S., Albinana-Gimenez N., Clemente-Casares P., Hundesa A., Rodriguez-Manzano J., Allard A., Calvo M., Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006;72:7894–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Calgua B., Clemente-Casares P., la Rosa G., Iaconelli M., Muscillo M., Rutjes S., de Roda Husman A.M., Grunert A., Gräber I., Verani M., Carducci A., Calvo M., Wyn-Jones P., Girones R. Quantification of human adenoviruses in European recreational waters. Food Environ. Virol. 2010;2:101–109. doi: 10.1007/s12560-010-9035-4. [DOI] [Google Scholar]
- Bonadonna L., Briancesco R., La Rosa G. Innovative analytical methods for monitoring microbiological and virological water quality. Microchem. J. 2019;150:104160. doi: 10.1016/j.microc.2019.104160. [DOI] [Google Scholar]
- Bortagaray V., Lizasoain A., Piccini C., Gillman L., Berois M., Pou S., Díaz M. del P., Tort F.L., Colina R., Victoria M. Microbial source tracking analysis using viral indicators in Santa lucía and Uruguay rivers, Uruguay. Food Environ. 2019;11:259–267. doi: 10.1007/s12560-019-09384-2. Virol. [DOI] [PubMed] [Google Scholar]
- Bosch A., Pinto R.M., Guix S. Foodborne viruses. Curr. Opin. Food Sci. 2016;8:110–119. doi: 10.1016/j.cofs.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M., Delwart E., Rosario K., Segalés J., Varsani A., Consortium I.R. ICTV virus taxonomy: Circoviridae. J. Gen. Virol. 2017;98:1997–1998. doi: 10.1099/jgv.0.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion G.M., Meschke J.S., Sobsey M.D. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 2002;36:2419–2425. doi: 10.1016/s0043-1354(01)00547-4. [DOI] [PubMed] [Google Scholar]
- Calgua B., Fumian T., Rusiñol M., Rodriguez-Manzano J., Mbayed V.A., Bofill-Mas S., Miagostovich M., Girones R. Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res. 2013;47:2797–2810. doi: 10.1016/j.watres.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Carducci A., Verani M. Effects of bacterial, chemical, physical and meteorological variables on virus removal by a wastewater treatment plant. Food Environ. Virol. 2013;5:69–76. doi: 10.1007/s12560-013-9105-5. [DOI] [PubMed] [Google Scholar]
- Casanovas-Massana A., Gómez-Doñate M., Sánchez D., Belanche-Muñoz L.A., Muniesa M., Blanch A.R. Predicting fecal sources in waters with diverse pollution loads using general and molecular host-specific indicators and applying machine learning methods. J. Environ. Manag. 2015;151:317–325. doi: 10.1016/j.jenvman.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Cashdollar J.L., Wymer L. Methods for primary concentration of viruses from water samples: a review and meta-analysis of recent studies. J. Appl. Microbiol. 2013;115:1–11. doi: 10.1111/jam.12143. [DOI] [PubMed] [Google Scholar]
- Cvdc Norovirus worldwide [WWW document] 2016. https://www.cdc.gov/norovirus/worldwide.html URL. accessed 5.29.19.
- Chatziprodromidou I.P., Bellou M., Vantarakis G., Vantarakis A. Viral outbreaks linked to fresh produce consumption: a systematic review. J. Appl. Microbiol. 2018;124:932–942. doi: 10.1111/jam.13747. [DOI] [PubMed] [Google Scholar]
- Chehadeh W., Nampoory M.R. Genotypic diversity of polyomaviruses circulating among kidney transplant recipients in Kuwait. J. Med. Virol. 2013;85:1624–1631. doi: 10.1002/jmv.23639. [DOI] [PubMed] [Google Scholar]
- Chigor V.N., Okoh A.I. Quantitative detection and characterization of human adenoviruses in the Buffalo River in the eastern Cape Province of South Africa. Food Environ. Virol. 2012;4:198–208. doi: 10.1007/s12560-012-9090-0. [DOI] [PubMed] [Google Scholar]
- Cinek O., Mazankova K., Kramna L., Odeh R., Alassaf A., Ibekwe M.A.U., Ahmadov G., Mekki H., Abdullah M.A., Elmahi B.M.E., Hyöty H., Rainetova P. Quantitative CrAssphage real-time PCR assay derived from data of multiple geographically distant populations. J. Med. Virol. 2018;90:767–771. doi: 10.1002/jmv.25012. [DOI] [PubMed] [Google Scholar]
- Cole D., Long S.C., Sobsey M.D. Evaluation of F + RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters evaluation of fϩ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Society. 2003;69:6507–6514. doi: 10.1128/AEM.69.11.6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook N., Bridger J., Kendall K., Gomara M.I., El-Attar L., Gray J. The zoonotic potential of rotavirus. J. Infect. 2004;48(4):289–302. doi: 10.1016/j.jinf.2004.01.018. [DOI] [PubMed] [Google Scholar]
- Cortez V., Meliopoulos V.A., Karlsson E.A., Hargest V., Johnson C., Schultz-Cherry S. Astrovirus biology and pathogenesis. Annu. Rev. Virol. 2017;4:327–348. doi: 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- Costán-Longares A., Montemayor M., Payán A., Méndez J., Jofre J., Mujeriego R., Lucena F. Microbial indicators and pathogens: removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 2008;42:4439–4448. doi: 10.1016/j.watres.2008.07.037. [DOI] [PubMed] [Google Scholar]
- Crank K., Petersen S., Bibby K. Quantitative microbial risk assessment of swimming in sewage impacted waters using CrAssphage and pepper mild mottle virus in a customizable model. Environ. Sci. Technol. Lett. 2019;6:571–577. doi: 10.1021/acs.estlett.9b00468. [DOI] [Google Scholar]
- Cromeans T.L., Kahler A.M., Hill V.R. Inactivation of adenoviruses, enteroviruses, and murine norovirus in water by free chlorine and monochloramine. Appl. Environ. Microbiol. 2010;76:1028–1033. doi: 10.1128/AEM.01342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals – molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Gray J. Viral gastroenteritis. Medicine (Baltim.) 2009;37:594–598. doi: 10.1016/j.mpmed.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar B.C., Lee N.Y. Lab-on-a-Chip Technology for environmental monitoring of microorganisms. Biochip J. 2018;12:173–183. doi: 10.1007/s13206-018-2301-5. [DOI] [Google Scholar]
- Di Bonito P., Libera S. Della, Petricca S., Iaconelli M., Accardi L., Muscillo M., La Rosa G. Frequent and abundant Merkel cell polyomavirus detection in urban wastewaters in Italy. Food Environ. Virol. 2014;7:1–6. doi: 10.1007/s12560-014-9168-y. [DOI] [PubMed] [Google Scholar]
- Di Bonito P., Iaconelli M., Gheit T., Tommasino M., Della Libera S., Bonadonna L., La Rosa G. Detection of oncogenic viruses in water environments by a Luminex-based multiplex platform for high throughput screening of infectious agents. Water Res. 2017;123:549–555. doi: 10.1016/j.watres.2017.06.088. [DOI] [PubMed] [Google Scholar]
- Dias E., Ebdon J., Taylor H. The application of bacteriophages as novel indicators of viral pathogens in wastewater treatment systems. Water Res. 2018;129:172–179. doi: 10.1016/j.watres.2017.11.022. [DOI] [PubMed] [Google Scholar]
- Dias J., Pinto R.N., Vieira C.B., de Abreu Corrêa A. Detection and quantification of human adenovirus (HAdV), JC polyomavirus (JCPyV) and hepatitis A virus (HAV) in recreational waters of Niterói, Rio de Janeiro, Brazil. Mar. Pollut. Bull. 2018;133:240–245. doi: 10.1016/j.marpolbul.2018.05.031. [DOI] [PubMed] [Google Scholar]
- Dumonceaux T.J.J., Mesa C., Severini A. Internally controlled triplex quantitative PCR assay for human polyomaviruses JC and BK. J. Clin. Microbiol. 2008;46 doi: 10.1128/JCM.00844-08. 2829 LP – 2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G.Z., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K., Felts B., Dinsdale E.A., Mokili J.L., Edwards R.A. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebdon J., Muniesa M., Taylor H. The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of faecal pollution in a temperate river catchment. Water Res. 2007;41:3683–3690. doi: 10.1016/j.watres.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Edwards R., Vega A., Norman H., Ohaeri M.C., Levi K., Dinsdale E., Cinek O., Aziz R., McNair K., Barr J., Bibby K., Brouns S., Cazares A., de Jonge P.A., Desnues C., Diaz-Munoz S., Fineran P., Kurilshikov A., Lavigne R., Mazankova K., McCarthy D., Nobrega F., Reyes A., Tapia G., Trefault N., Tyakht A., Vinuesa P., Wagemans J., Zhernakova A., Aarestrup F., Ahmadov G., Alassaf A., Anton J., Asangba A., Billings E., Cantu A., Carlton J., Cazares Lopez D., Cho G.-S., Condeff T., Cortes P., Cranfield M., Cuevas D., De la Iglesia R., Decewicz P., Doane M., Dominy N., Dziewit L., Elmahi B., Eren M., Franz C., Fu J., Garcia-Aljaro C., Ghedin E., Gulino K., Haggerty J., Head S., Hendriksen R.S., Hill C., Hyoty H., Ilina E., Irwin M., Jeffries T., Jofre J., Junge R., Kelley S., Kowalewski M., Kumaresan D., Leigh S., Lisitsyna E., Llagostera M., Maritz J.M., Marr L., McCann A., Khan Mirzaei M., Molshanski-Mor S., Monteiro S., Moreira-Grez B., Morris M., Mugisha L., Muniesa M., Neve H., Nguyen N., Nigro O., Nilsson A., O’Connell T., Odeh R., Oliver A., Piuri M., Prussin A., Qimron U., Quan Z.-X., Rainetova P., Ramirez-Rojas A., Raya R., Rice G., Rossi A., Santos R., Shimashita J., Stachler E., Stene L., Strain R., Stumpf R., Torres P., Twaddle A., Ibekwe M.U., Villagra N., Wandro S., White B., Whiteley A., Whiteson K., Wijmenga C., Zambrano M.M., Zschach H., Dutilh B.E. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat. Microbiol. 2019;4:1727–1736. doi: 10.1038/s41564-019-04904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Senousy W.M., Osman G.A., Melegy A.A. Survival of adenovirus, rotavirus, Hepatitis A virus, pathogenic bacteria and bacterial indicators in ground water. World Appl. Sci. J. 2014;29:337–348. doi: 10.5829/idosi.wasj.2014.29.03.13849. [DOI] [Google Scholar]
- Elmahdy E.M., Ahmed N.I., Shaheen M.N.F., Mohamed E.-C.B., Loutfy S.A. Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health. 2019:1–8. doi: 10.2166/wh.2019.303. [DOI] [PubMed] [Google Scholar]
- Enriquez C.E., Hurst C.J., Gerba C.P. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 1995;29:2548–2553. doi: 10.1016/0043-1354(95)00070-2. [DOI] [Google Scholar]
- Farkas K., Marshall M., Cooper D., McDonald J.E., Malham S.K., Peters D.E., Maloney J.D., Jones D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ. Sci. Pollut. Res. 2018;25(33):33391–33401. doi: 10.1007/s11356-018-3261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Hassard F., McDonald J.E., Malham S.K., Jones D.L. Evaluation of molecular methods for the detection and quantification of pathogen-derived nucleic acids in sediment. Front. Microbiol. 2017;8:53. doi: 10.3389/fmicb.2017.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Malham S.K., Peters D.E., de Rougemont A., McDonald J.E., de Rougemont A., Malham S.K., Jones D.L. Evaluation of two triplex one-step qRT-PCR assays for the quantification of human enteric viruses in environmental samples. Food Environ. Virol. 2017;9:343–349. doi: 10.1007/s12560-017-9293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L., Rougemont A. de, Jones D.L., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Farkas K., Adriaenssens E.M., Walker D.I., Mcdonald J.E., Malham S.K., Jones D.L., David ·, Walker I., Mcdonald J.E., Malham S.K., Davey ·, Jones L. Critical evaluation of crAssphage as a molecular marker for human-derived wastewater contamination in the aquatic environment. Food Environ. Virol. 2019;11:113–119. doi: 10.1007/s12560-019-09369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Mannion F., Hillary L.S., Malham S.K., Walker D.I. Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Curr. Opin. Environ. Sci. Heal. 2020;16:1–6. doi: 10.1016/j.coesh.2020.01.007. [DOI] [Google Scholar]
- Fattal B., Vasl R.J., Katzenelson E., Shuval H.I. Survival of bacterial indicator organisms and enteric viruses in the Mediterranean coastal waters off Tel-Aviv. Water Res. 1983;17:397–402. doi: 10.1016/0043-1354(83)90135-5. [DOI] [Google Scholar]
- Flannery J., Keaveney S., Rajko-Nenow P., O’Flaherty V., Doré W. Norovirus and FRNA bacteriophage determined by RT-qPCR and infectious FRNA bacteriophage in wastewater and oysters. Water Res. 2013;47:5222–5231. doi: 10.1016/j.watres.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Fong T.T., Griffin D.W., Lipp E.K. Molecular assays for targeting human and bovine enteric viruses in coastal waters and their application for library-independent source tracking. Appl. Environ. Microbiol. 2005;71:2070–2078. doi: 10.1128/AEM.71.4.2070-2078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Do Nascimento M.A., Rigotto C., Ritterbusch G., da Silva A.D., Esteves P.A., Barardi C.R.M. Evaluation and molecular characterization of human adenovirus in drinking water supplies: viral integrity and viability assays. Virol. J. 2013;10:1. doi: 10.1186/1743-422X-10-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fongaro G., Padilha J., Schissi C.D., Nascimento M.A., Bampi G.B., Viancelli A., Barardi C.R.M. Human and animal enteric virus in groundwater from deep wells, and recreational and network water. Environ. Sci. Pollut. Res. 2015;22:20060–20066. doi: 10.1007/s11356-015-5196-x. [DOI] [PubMed] [Google Scholar]
- Fratini M., Di Bonito P., La Rosa G. Oncogenic Papillomavirus and Polyomavirus in water environments: is there a potential for waterborne transmission? Food Environ. Virol. 2014;6:1–12. doi: 10.1007/s12560-013-9134-0. [DOI] [PubMed] [Google Scholar]
- Friedman S.D., Cooper E.M., Casanova L., Sobsey M.D., Genthner F.J. A reverse transcription-PCR assay to distinguish the four genogroups of. J. Virol. Methods. 2009;159:47–52. doi: 10.1016/j.jviromet.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Guimarães F.R., Vaz B.J.P., Da Silva M.T.T., Muylaert F.F., Bofill-Mas S., Gironés R., Leite J.P.G., Miagostovich M.P. Molecular detection, quantification and characterization of human polyomavirus JC from waste water in Rio de Janeiro, Brazil. J. Water Health. 2010;8:438–445. doi: 10.2166/wh.2010.090. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Vieira C.B., Leite J.P.G., Miagostovich M.P. Assessment of burden of virus agents in an urban sewage treatment plant in Rio de Janeiro, Brazil. J. Water Health. 2013;11:110–119. doi: 10.2166/wh.2012.123. [DOI] [PubMed] [Google Scholar]
- Garcia L.A.T., Viancelli A., Rigotto C., Pilotto M.R., Esteves P.A., Kunz A., Barardi C.R.M. Surveillance of human and swine adenovirus, human norovirus and swine circovirus in water samples in Santa Catarina, Brazil. J. Water Health. 2012;10:445–452. doi: 10.2166/wh.2012.190. [DOI] [PubMed] [Google Scholar]
- García-Aljaro C., Ballesté E., Muniesa M., Jofre J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017;10:1775–1780. doi: 10.1111/1751-7915.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.E. Viral pathogens in water: occurrence, public health impact, and available control strategies. Curr. Opin. Virol. 2014;4:50–57. doi: 10.1016/j.coviro.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girones R., Ferrus M.A., Alonso J.L., Rodriguez-Manzano J., Calgua B., de Abreu Corrêa A., Hundesa A., Carratala A., Bofill-Mas S. Molecular detection of pathogens in water–the pros and cons of molecular techniques. Water Res. 2010;44:4325–4339. doi: 10.1016/j.watres.2010.06.030. [DOI] [PubMed] [Google Scholar]
- Goh S., Lindau C., Tiveljung-Lindell A., Allander T. Merkel cell polyomavirus in respiratory tract secretions. Emerg. Infect. Dis. 2009;15:489–491. doi: 10.3201/eid1503.081206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila M., Solis J.J., David Z., Ramon C., Lalucat J. Comparative reductions of bacterial indicators, bacteriophage-infecting enteric bacteria and enteroviruses in wastewater tertiary treatments by lagooning and UV-radiation. Water Sci. Technol. 2008;58:2223–2233. doi: 10.2166/wst.2008.584. [DOI] [PubMed] [Google Scholar]
- Gröndahl B., Puppe W., Hoppe A., Kühne I., Weigl J.A.I., Schmitt H.-J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 1999;37 doi: 10.1128/jcm.37.1.1-7.1999. 1 LP – 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grøndahl-Rosado R.C., Tryland I., Myrmel M., Aanes K.J., Robertson L.J. Detection of microbial pathogens and indicators in sewage effluent and river water during the temporary interruption of a wastewater treatment plant. Water Qual. Expo. Heal. 2014;6:155–159. doi: 10.1007/s12403-014-0121-y. [DOI] [Google Scholar]
- Grøndahl-Rosado R.C., Yarovitsyna E., Trettenes E., Myrmel M., Robertson L.J. A one year study on the concentrations of norovirus and enteric adenoviruses in wastewater and a surface drinking water source in Norway. Food Environ. Virol. 2014;6:232–245. doi: 10.1007/s12560-014-9161-5. [DOI] [PubMed] [Google Scholar]
- Guido M., Tumolo M.R., Verri T., Romano A., Serio F., De Giorgi M., De Donno A., Bagordo F., Zizza A. Human bocavirus: current knowledge and future challenges. World J. Gastroenterol. 2016;22:8684–8697. doi: 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of Coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Gyawali P., Croucher D., Ahmed W., Devane M., Hewitt J. Evaluation of pepper mild mottle virus as an indicator of human faecal pollution in shellfish and growing waters. Water Res. 2019;154:370–376. doi: 10.1016/j.watres.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Gyawali P., Sanjaya K.C., Beale D.J., Hewitt J. Current and emerging technologies for the detection of norovirus from shellfish. Foods. 2019;8 doi: 10.3390/foods8060187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.A., Bibby K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods. 2019;266:11–24. doi: 10.1016/j.jviromet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Hamza H., Hamza I.A. Oncogenic papillomavirus and polyomavirus in urban sewage in Egypt. Sci. Total Environ. 2018;610–611:1413–1420. doi: 10.1016/j.scitotenv.2017.08.218. [DOI] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Stang A., Sure K., berla K., Wilhelm M. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 2009;43:2657–2668. doi: 10.1016/j.watres.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Wilhelm M., Uberla K., Überla K. Detection and quantification of human bocavirus in river water. J. Gen. Virol. 2009;90:2634–2637. doi: 10.1099/vir.0.013557-0. [DOI] [PubMed] [Google Scholar]
- Hamza I.A., Jurzik L., Überla K., Wilhelm M. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 2011;45:1358–1368. doi: 10.1016/j.watres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Hamza H., Leifels M., Wilhelm M., Hamza I.A. Relative abundance of human bocaviruses in urban sewage in Greater Cairo, Egypt. Food Environ. Virol. 2017;9:304–313. doi: 10.1007/s12560-017-9287-3. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M. Quantification and genotyping of Aichi virus 1 in water samples in the Kathmandu Valley, Nepal. Food Environ. Virol. 2017;9:350–353. doi: 10.1007/s12560-017-9283-7. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Oguma K., Ohgaki S. Quantitative analysis of human enteric adenoviruses in aquatic environments. J. Appl. Microbiol. 2007;103:2153–2159. doi: 10.1111/j.1365-2672.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Katayama H., Ohgaki S. Quantification and genotyping of torque teno virus at a wastewater treatment plant in Japan. Appl. Environ. Microbiol. 2008;74:7434–7436. doi: 10.1128/AEM.01605-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Katayama H., Ohgaki S. Real-time PCR detection of adenoviruses, polyomaviruses, and torque teno viruses in river water in Japan. Water Res. 2010;44:1747–1752. doi: 10.1016/j.watres.2009.11.043. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Otagiri M., Morita H., Kitajima M. Genogroup distribution of F-specific coliphages in wastewater and river water in the Kofu basin in Japan. Lett. Appl. Microbiol. 2012;54:367–373. doi: 10.1111/j.1472-765X.2012.03221.x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79:7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Fujino S., Otagiri M. Distinct behaviors of infectious F-specific RNA coliphage genogroups at a wastewater treatment plant. Sci. Total Environ. 2015;520:32–38. doi: 10.1016/j.scitotenv.2015.03.034. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Harris L.J., Farber J.N., Beuchat L.R., Parish M.E., Suslow T.V., Garrett E.H., Busta F.F. Outbreaks associated with fresh produce: incidence, growth, and survival of pathogens in fresh and fresh-cut produce. Compr. Rev. Food Sci. Food Saf. 2006;2:78–141. doi: 10.1111/j.1541-4337.2003.tb00031.x. [DOI] [Google Scholar]
- Hartard C., Banas S., Rivet R., Boudaud N., Gantzer C. Rapid and sensitive method to assess human viral pollution in shellfish using infectious F-specific RNA bacteriophages: application to marketed products. Food Microbiol. 2017;63:248–254. doi: 10.1016/j.fm.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Harwood V.J., Boehm A.B., Sassoubre L.M., Vijayavel K., Stewart J.R., Fong T., Caprais M., Converse R.R., Diston D., Ebdon J., Fuhrman J.A., Gourmelon M., Gentry-shields J., Griffith J.F., Kashian D.R., Noble R.T., Taylor H., Wicki M. Performance of viruses and bacteriophages for fecal source determination in a multi-laboratory , comparative study. Water Res. 2013;47:6929–6943. doi: 10.1016/j.watres.2013.04.064. [DOI] [PubMed] [Google Scholar]
- Hassard F., Gwyther C.L., Farkas K., Andrews A., Jones V., Cox B., Brett H., Jones D.L., McDonald J.E., Malham S.K. Abundance and distribution of enteric bacteria and viruses in coastal and estuarine sediments-A review. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kitajima M., Visvanathan C., Nol C., Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Appl. Environ. Microbiol. 2011;77 doi: 10.1128/AEM.00077-11. 4336 LP – 4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Katayama H., Kojima K., Sano S., Kasuga I., Kitajima M., Furumai H. Effects of rainfall events on the occurrence and detection efficiency of viruses in river water impacted by combined sewer overflows. Sci. Total Environ. 2014;468–469:757–763. doi: 10.1016/j.scitotenv.2013.08.093. [DOI] [PubMed] [Google Scholar]
- Hata A., Hanamoto S., Shirasaka Y., Yamashita N., Tanaka H. Quantitative distribution of infectious F-Specific RNA phage genotypes in surface waters. Appl. Environ. Microbiol. 2016;82:4244–4252. doi: 10.1128/aem.00621-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A., Ebnet C., Harste G., Pring-Åkerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003;70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]
- Hernroth B.E., Conden-Hansson A.-C., Rehnstam-Holm A.-S., Girones R., Allard A.K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, (Mytilus edulis): the first Scandinavian report. Appl. Environ. Microbiol. 2002;68 doi: 10.1128/AEM.68.9.4523-4533.2002. 4523 LP – 4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J., Leonard M., Greening G.E., Lewis G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011;45:6267–6276. doi: 10.1016/j.watres.2011.09.029. [DOI] [PubMed] [Google Scholar]
- Hijnen W.A.M., Beerendonk E.F., Medema G.J. Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: a review. Water Res. 2006;40:3–22. doi: 10.1016/j.watres.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Iaconelli M., Divizia M., Della Libera S., Di Bonito P., La Rosa G. Frequent detection and genetic diversity of human bocavirus in urban sewage samples. Food Environ. Virol. 2016;8:289–295. doi: 10.1007/s12560-016-9251-7. [DOI] [PubMed] [Google Scholar]
- Iaconelli M., Muscillo M., Della Libera S., Fratini M., Meucci L., De Ceglia M., Giacosa D., La Rosa G. One-year surveillance of human enteric viruses in raw and treated wastewaters, downstream river waters, and drinking waters. Food Environ. Virol. 2017;9:79–88. doi: 10.1007/s12560-016-9263-3. [DOI] [PubMed] [Google Scholar]
- Ibrahim C., Hassen A., Pothier P., Mejri S., Hammami S. Molecular detection and genotypic characterization of enteric adenoviruses in a hospital wastewater. Environ. Sci. Pollut. Res. 2018:1–11. doi: 10.1007/s11356-018-1399-2. [DOI] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4:41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Ishii S., Kitamura G., Segawa T., Kobayashi A., Miura T., Sano D., Okabe S. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl. Environ. Microbiol. 2014;80 doi: 10.1128/AEM.02578-14. 7505 LP – 7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasim S.Y., Saththasivam J., Loganathan K., Ogunbiyi O.O., Sarp S. Reuse of treated sewage effluent (TSE) in Qatar. J. Water Process Eng. 2016;11:174–182. doi: 10.1016/j.jwpe.2016.05.003. [DOI] [Google Scholar]
- Jiang S.C. Human adenoviruses in water: occurrence and health implications: a critical review. Environ. Sci. Technol. 2006;40:7132–7140. doi: 10.1021/es060892o. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Fang L., Shi X., Zhang H., Li Y., Lin Y., Qiu Y., Chen Q., Li H., Zhou L., Hu Q. Simultaneous detection of five enteric viruses associated with gastroenteritis by use of a PCR assay: a single real-time multiplex reaction and its clinical application. J. Clin. Microbiol. 2014;52:1266–1268. doi: 10.1128/JCM.00245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Flury M. Fate and transport of viruses in porous media. Adv. Agron. 2002;77:39–102. doi: 10.1016/S0065-2113(02)77013-2. [DOI] [Google Scholar]
- Jofre J., Blanch A.R., Lucena F., Muniesa M. Bacteriophages infecting Bacteroides as a marker for microbial source tracking. Water Res. 2014;55:1–11. doi: 10.1016/j.watres.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Joshi M.S., Lole K.S., Barve U.S., Salve D.S., Ganorkar N.N., Chavan N.A., Shinde M.S., Gopalkrishna V. Investigation of a large waterborne acute gastroenteritis outbreak caused by group B rotavirus in Maharashtra state, India. J. Med. Virol. 2019;91:1877–1881. doi: 10.1002/jmv.25523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T.L.L., Hill V.R.R., Lu X., Sobsey M.D.D., Erdman D.D.D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005;71 doi: 10.1128/AEM.71.6.3131-3136.2005. 3131 LP – 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumat M.R., Hasan N.A., Subramanian P., Heberling C., Colwell R.R., Hong P.-Y.Y. Membrane bioreactor-based wastewater treatment plant in Saudi Arabia: reduction of viral diversity, load, and infectious capacity. Water (Switzerland) 2017;9:534. doi: 10.3390/w9070534. [DOI] [Google Scholar]
- Jurzik L., Hamza I.A., Puchert W., Überla K., Wilhelm M. Chemical and microbiological parameters as possible indicators for human enteric viruses in surface water. Int. J. Hyg Environ. Health. 2010;213:210–216. doi: 10.1016/j.ijheh.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Kaas L., Ogorzaly L., Lecellier G., Berteaux-Lecellier V., Cauchie H.-M., Langlet J. Detection of human enteric viruses in French Polynesian wastewaters, environmental waters and giant clams. Food Environ. Virol. 2018;11(1):52–64. doi: 10.1007/s12560-018-9358-0. 0, 0. [DOI] [PubMed] [Google Scholar]
- Kahler A.M., Cromeans T.L., Roberts J.M., Hill V.R. Effects of source water quality on chlorine inactivation of adenovirus, coxsackievirus, echovirus, and murine norovirus. Appl. Environ. Microbiol. 2010;76:5159–5164. doi: 10.1128/AEM.00869-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Vinje J. Norovirus and other caliciviruses. In: Meschke J.S., Gironés R., editors. Global Water Pathogen Project. Michigan State University; E. Lansing, MI: 2017. (UNESCO) [Google Scholar]
- Kauppinen A., Pitkänen T., Miettinen I.T. Persistent norovirus contamination of groundwater supplies in two waterborne outbreaks. Food Environ. Virol. 2018;10:39–50. doi: 10.1007/s12560-017-9320-6. [DOI] [PubMed] [Google Scholar]
- Kauppinen A., Pitkänen T., Al-Hello H., Maunula L., Hokajärvi A.-M., Rimhanen-Finne R., Miettinen I.T. Two drinking water outbreaks caused by wastewater intrusion including Sapovirus in Finland. Int. J. Environ. Res. Public Health. 2019;16:4376. doi: 10.3390/ijerph16224376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswick B.H., Gerba C.P., Secor S.L., Cech I. Survival of enteric viruses and indicator bacteria in groundwater. J. Environ. Sci. Heal. . Part A Environ. Sci. Eng. 1982;17:903–912. doi: 10.1080/10934528209375085. [DOI] [Google Scholar]
- Khetsuriani Nino, LaMonte-Fowlkes A., Oberste M.S., Pallansch M.A. Enterovirus surveillance --- United States, 1970--2005. Morb. Mortal. Wkly. Rep. 2006;55:1–20. [PubMed] [Google Scholar]
- Kim W.J., Managaki S., Furumai H., Nakajima F. Diurnal fluctuation of indicator microorganisms and intestinal viruses in combined sewer system. Water Sci. Technol. 2009;60:2791–2801. doi: 10.2166/wst.2009.732. [DOI] [PubMed] [Google Scholar]
- King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; 2009. Virus taxinomy: classification and nomenclature of viruses. [Google Scholar]
- Kishida N., Morita H., Haramoto E., Asami M., Akiba M. One-year weekly survey of noroviruses and enteric adenoviruses in the Tone River water in Tokyo metropolitan area. Japan. Water Res. 2012;46 doi: 10.1016/j.watres.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Kishida N., Noda N., Haramoto E., Kawaharasaki M., Akiba M., Sekiguchi Y. Quantitative detection of human enteric adenoviruses in river water by microfluidic digital polymerase chain reaction. Water Sci. Technol. 2014;70 doi: 10.2166/wst.2014.262. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Gerba C. Aichi virus 1: environmental occurrence and behavior. Pathogens. 2015;4:256–268. doi: 10.3390/pathogens4020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Hata A., Yamashita T., Haramoto E., Minagawa H., Katayama H. Development of a reverse transcription-quantitative PCR system for detection and genotyping of Aichi viruses in clinical and environmental samples. Appl. Environ. Microbiol. 2013;79 doi: 10.1128/AEM.00820-13. 3952 LP – 3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci. Total Environ. 2014;488:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Gerba C.P. Temporal variations in genotype distribution of human sapoviruses and Aichi virus 1 in wastewater in Southern Arizona, United States. J. Appl. Microbiol. 2018;124:1324–1332. doi: 10.1111/jam.13712. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018;1:19. doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- Knight A., Li D., Uyttendaele M., Jaykus L.-A. A critical review of methods for detecting human noroviruses and predicting their infectivity. Crit. Rev. Microbiol. 2013;39:295–309. doi: 10.3109/1040841X.2012.709820. [DOI] [PubMed] [Google Scholar]
- Knowles W.A. In: Discovery and Epidemiology of the Human Polyomaviruses BK Virus (BKV) and JC Virus (JCV) BT - Polyomaviruses and Human Diseases. Ahsan N., editor. Springer New York; New York, NY: 2006. pp. 19–45. [DOI] [PubMed] [Google Scholar]
- Ko G., Jothikumar N., Hill V.R.R., Sobsey M.D.D. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods. 2005;127:148–153. doi: 10.1016/j.jviromet.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Koff R.S. Clinical manifestations and diagnosis of hepatitis A virus infection. Vaccine. 1992;10:S15–S17. doi: 10.1016/0264-410X(92)90533-P. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S.R., Wang D. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology. 2018;516:108–114. doi: 10.1016/j.virol.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Nakada N., Hanamoto S., Inaba M., Katayama H., Do A.T., Nga T.T.V., Oguma K., Hayashi T., Takizawa S. Pepper mild mottle virus as an indicator and a tracer of fecal pollution in water environments: comparative evaluation with wastewater-tracer pharmaceuticals in Hanoi. Vietnam. Sci. Total Environ. 2015:506–507. doi: 10.1016/j.scitotenv.2014.11.021. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Pourshaban M., Iaconelli M., Muscillo M. Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Ann. Ist. Super Sanita. 2010;46:266–273. doi: 10.4415/ANN_10_03_07. [DOI] [PubMed] [Google Scholar]
- Landry E.F., Vaughn J.M., Vicale T.J., Mann R. Accumulation of sediment-associated viruses in shellfish. Appl. Environ. Microbiol. 1983;45:238–247. doi: 10.1128/aem.45.1.238-247.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebarbenchon C., Yang M., Keeler S.P., Ramakrishnan M.A., Brown J.D., Stallknecht D.E., Sreevatsan S. Viral replication, persistence in water and genetic characterization of two influenza a viruses isolated from surface lake water. PloS One. 2011;6 doi: 10.1371/journal.pone.0026566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Lee H., Cho Y., Hur H., Ko G. F + RNA coliphage-based microbial source tracking in water resources of South Korea. Sci. Total Environ. 2011;412–413:127–131. doi: 10.1016/j.scitotenv.2011.09.061. [DOI] [PubMed] [Google Scholar]
- Lee D.Y., Leung K.T., Lee H., Habash M.B. Simultaneous detection of selected enteric viruses in water samples by multiplex quantitative PCR. Water Air Soil Pollut. 2016;227:107. doi: 10.1007/s11270-016-2811-5. [DOI] [Google Scholar]
- Lee S., Suwa M., Shigemura H. Occurrence and reduction of F-specific RNA bacteriophage genotypes as indicators of human norovirus at a wastewater treatment plant. J. Water Health. 2018;17:50–62. doi: 10.2166/wh.2018.367. [DOI] [PubMed] [Google Scholar]
- Levican J., Levican A., Ampuero M., Gaggero A. JC polyomavirus circulation in one-year surveillance in wastewater in Santiago, Chile. Infect. Genet. Evol. 2019;71:151–158. doi: 10.1016/j.meegid.2019.03.017. [DOI] [PubMed] [Google Scholar]
- Li L., Liu N., Yu J., Ao Y., Li S., Stine O.C., Duan Z. Analysis of Aichi virus and Saffold virus association with pediatric acute gastroenteritis. J. Clin. Virol. 2017;87:37–42. doi: 10.1016/j.jcv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Liang L., Goh S.G., Gin K.Y.H. Decay kinetics of microbial source tracking (MST) markers and human adenovirus under the effects of sunlight and salinity. Sci. Total Environ. 2017;574:165–175. doi: 10.1016/j.scitotenv.2016.09.031. [DOI] [PubMed] [Google Scholar]
- Lin J., Ganesh A. Water quality indicators: bacteria, coliphages, enteric viruses. Int. J. Environ. Health Res. 2013;23(6):484–506. doi: 10.1080/09603123.2013.769201. [DOI] [PubMed] [Google Scholar]
- Lin P.-H., Li B.-R. Antifouling strategies in advanced electrochemical sensors and biosensors. Analyst. 2020;145:1110–1120. doi: 10.1039/C9AN02017A. [DOI] [PubMed] [Google Scholar]
- Lowther J.A., Gustar N.E., Powell A.L., Hartnell R.E., Lees D.N. Two-year systematic study to assess norovirus contamination in oysters from commercial harvesting areas in the United Kingdom. Appl. Environ. Microbiol. 2012;78:5812–5817. doi: 10.1128/AEM.01046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun J.H., Crosbie N.D., White P.A. Genetic diversity and quantification of human mastadenoviruses in wastewater from Sydney and Melbourne, Australia. Sci. Total Environ. 2019;675:305–312. doi: 10.1016/j.scitotenv.2019.04.162. [DOI] [PubMed] [Google Scholar]
- Masclaux F.G., Hotz P., Friedli D., Savova-Bianchi D., Oppliger A. High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Res. 2013;47:5101–5109. doi: 10.1016/j.watres.2013.05.050. [DOI] [PubMed] [Google Scholar]
- Matheson C.D., Gurney C., Esau N., Lehto R. Assessing PCR inhibition from humic substances. Open Enzym. Inhib. J. 2014;3:38–45. doi: 10.2174/1874940201003010046. [DOI] [Google Scholar]
- Matson D.O., O’Ryan M.L., Herrera I., Pickering L.K., Estes M.K. Fecal antibody responses to symptomatic and asymptomatic rotavirus infections. J. Infect. Dis. 1993;167:577–583. doi: 10.1093/infdis/167.3.577. [DOI] [PubMed] [Google Scholar]
- Maunula L., Klemola P., Kauppinen A., Söderberg K., Nguyen T., Pitkänen T., Kaijalainen S., Simonen M.L., Miettinen I.T., Lappalainen M., Laine J., Vuento R., Kuusi M., Roivainen M. Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ. Virol. 2009;1:31–36. doi: 10.1007/s12560-008-9004-3. [DOI] [Google Scholar]
- Mayer R.E., Bofill-Mas S., Egle L., Reischer G.H., Schade M., Fernandez-Cassi X., Fuchs W., Mach R.L., Lindner G., Kirschner A., Gaisbauer M., Piringer H., Blaschke A.P., Girones R., Zessner M., Sommer R., Farnleitner A.H. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 2016;90:265–276. doi: 10.1016/j.watres.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMinn B.R., Korajkic A., Ashbolt N.J. Evaluation of Bacteroides fragilis GB-124 bacteriophages as novel human-associated faecal indicators in the United States. Lett. Appl. Microbiol. 2014;59:115–121. doi: 10.1111/lam.12252. [DOI] [PubMed] [Google Scholar]
- McMinn B.R., Ashbolt N.J., Korajkic A. Bacteriophages as indicators of faecal pollution and enteric virus removal. Lett. Appl. Microbiol. 2017;65:11–26. doi: 10.1111/lam.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S.M., Scott T.M., Lukasik J.O., Paul J.H., Harwood V.J. Quantification of human polyomaviruses JC Virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 2009;75 doi: 10.1128/AEM.02302-08. 3379 LP – 3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick J.L. Etiologic agents and their potential for causing waterborne virus diseases. In: Melnick J.L., editor. Enteric Viruses in Water. Kragel; Basel, Switherland: 1984. pp. 1–16. [Google Scholar]
- Moens U., Calvignac-Spencer S., Lauber C., Ramqvist T., Feltkamp M.C.W., Daugherty M.D., Verschoor E.J., Ehlers B., Consortium I.R. ICTV virus taxonomy profile: polyomaviridae. J. Gen. Virol. 2017;98:1159–1160. doi: 10.1099/jgv.0.000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazeri N., Goettert D., Achberger E.C., Johnson C.N., Prinyawiwatkul W., Janes M.E. Pathogenic enteric viruses and microbial indicators during secondary treatment of municipal wastewater. Appl. Environ. Microbiol. 2015;81:6436–6445. doi: 10.1128/AEM.01218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresco V., Viancelli A., Nascimento M.A., Souza D.S.M., Ramos A.P.D., Garcia L.A.T., Simões C.M.O., Barardi C.R.M. Microbiological and physicochemical analysis of the coastal waters of southern Brazil. Mar. Pollut. Bull. 2012;64:40–48. doi: 10.1016/j.marpolbul.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Muniesa M., Payan A., Moce-llivina L., Blanch A.R., Jofre J. Differential persistence of F-specific RNA phage subgroups hinders their use as single tracers for faecal source tracking in surface water. Water Res. 2009;43:1559–1564. doi: 10.1016/j.watres.2008.12.038. [DOI] [PubMed] [Google Scholar]
- Muscillo M., Pourshaban M., Iaconelli M., Fontana S., Di Grazia A., Manzara S., Fadda G., Santangelo R., La Rosa G. Detection and quantification of human adenoviruses in surface waters by nested PCR, TaqMan real-time PCR and cell culture assays. Water Air Soil Pollut. 2008;191:83–93. doi: 10.1007/s11270-007-9608-5. [DOI] [Google Scholar]
- Myrmel M., Lange H., Rimstad E. A 1-Year quantitative survey of Noro-, Adeno-, human Boca-, and Hepatitis E Viruses in raw and secondarily treated sewage from two plants in Norway. Food Environ. Virol. 2015;7:213–223. doi: 10.1007/s12560-015-9200-x. [DOI] [PubMed] [Google Scholar]
- Naik S.R., Aggarwal R., Salunke P.N., Mehrotra N.N. A large waterborne viral hepatitis E epidemic in Kanpur, India. Bull. World Health Organ. 1992;70:597–604. [PMC free article] [PubMed] [Google Scholar]
- Ng T.F.F., Marine R., Wang C., Simmonds P., Kapusinszky B., Bodhidatta L., Oderinde B.S., Wommack K.E., Delwart E. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorzaly L., Gantzer C. Development of real-time RT-PCR methods for specific detection of F-specific RNA bacteriophage genogroups: application to urban raw wastewater. J. Virol. Methods. 2006;138:131–139. doi: 10.1016/j.jviromet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Ogorzaly L., Tissier A., Bertrand I., Maul A., Gantzer C. Relationship between F-specific RNA phage genogroups, faecal pollution indicators and human adenoviruses in river water. Water Res. 2009;43:1257–1264. doi: 10.1016/j.watres.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Ogorzaly L., Bertrand I., Paris M., Maul A., Gantzer C. Occurrence, survival, and persistence of human adenoviruses and F-specific RNA phages in raw groundwater. Appl. Environ. Microbiol. 2010;76:8019–8025. doi: 10.1128/AEM.00917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorzaly L., Walczak C., Galloux M., Etienne S., Gassilloud B., Cauchie H.-M. Human adenovirus diversity in water samples using a next-generation amplicon sequencing approach. Food Environ. Virol. 2015;7:112–121. doi: 10.1007/s12560-015-9194-4. [DOI] [PubMed] [Google Scholar]
- Olalemi A., Purnell S., Caplin J., Ebdon J., Taylor H. The application of phage-based faecal pollution markers to predict the concentration of adenoviruses in mussels (Mytilus edulis) and their overlying waters. J. Appl. Microbiol. 2016;121:1152–1162. doi: 10.1111/jam.13222. [DOI] [PubMed] [Google Scholar]
- Pal A., Sirota L., Maudru T., Peden K., Lewis A.M. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods. 2006;135:32–42. doi: 10.1016/j.jviromet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Pang X.L., Lee B.E., Pabbaraju K., Gabos S., Craik S., Payment P., Neumann N. Pre-analytical and analytical procedures for the detection of enteric viruses and enterovirus in water samples. J. Virol. Methods. 2012;184:77–83. doi: 10.1016/j.jviromet.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Parshionikar S.U., Willian-True S., Fout G.S., Robbins D.E., Seys S.A., Cassady J.D., Harris R. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl. Environ. Microbiol. 2003;69:5263–5268. doi: 10.1128/AEM.69.9.5263-5268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payan A., Ebdon J., Taylor H., Gantzer C., Ottoson J., Papageorgiou G.T., Blanch A.R., Lucena F., Jofre J., Muniesa M. Method for isolation of Bacteroides bacteriophage Hhost strains suitable for tracking sources of fecal pollution in water. Appl. Environ. Microbiol. 2005;71:5659–5662. doi: 10.1128/AEM.71.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons-salort M., Oberste M.S., Pallansch M.A., Abedi G.R., Takahashi S., Grenfell B.T. The seasonality of nonpolio enteroviruses in the United States : patterns and drivers. 2018. 115, 3078-3083. [DOI] [PMC free article] [PubMed]
- Prado T., Bruni A. de C., Barbosa M.R.F., Bonanno V.M.S., Garcia S.C., Sato M.I.Z. Distribution of human fecal marker GB-124 bacteriophages in urban sewage and reclaimed water of São Paulo city, Brazil. J. Water Health. 2018;16:289–299. doi: 10.2166/wh.2017.011. [DOI] [PubMed] [Google Scholar]
- Prado T., de Castro Bruni A., Barbosa M.R.F., Garcia S.C., de Jesus Melo A.M., Sato M.I.Z. Performance of wastewater reclamation systems in enteric virus removal. Sci. Total Environ. 2019;678:33–42. doi: 10.1016/j.scitotenv.2019.04.435. [DOI] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Prevost B., Goulet M., Lucas F.S., Joyeux M., Moulin L., Wurtzer S. Viral persistence in surface and drinking water: suitability of PCR pre-treatment with intercalating dyes. Water Res. 2016;91 doi: 10.1016/j.watres.2015.12.049. [DOI] [PubMed] [Google Scholar]
- Prez V.E., Gil P.I., Temprana C.F., Cuadrado P.R., Martínez L.C., Giordano M.O., Masachessi G., Isa M.B., Ré V.E., Paván J.V., Nates S.V., Barril P.A. Quantification of human infection risk caused by rotavirus in surface waters from Córdoba, Argentina. Sci. Total Environ. 2015;538 doi: 10.1016/j.scitotenv.2015.08.041. [DOI] [PubMed] [Google Scholar]
- Purdy M.A., Harrison T.J., Jameel S., Meng X.-J., Okamoto H., Van der Poel W.H.M., Smith D.B., Consortium I.R. ICTV virus taxonomy profile: hepeviridae. J. Gen. Virol. 2017;98:2645–2646. doi: 10.1099/jgv.0.000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell S., Ebdon J., Buck A., Tupper M., Taylor H. Bacteriophage removal in a full-scale membrane bioreactor (MBR) – implications for wastewater reuse. Water Res. 2015;73:109–117. doi: 10.1016/j.watres.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Lee B.E., Neumann N., Ashbolt N., Craik S., Maal-Bared R., Pang X.L. Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. J. Appl. Microbiol. 2015;119:1729–1739. doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Rachmadi A.T., Torrey J.R., Kitajima M. Human polyomavirus: advantages and limitations as a human-specific viral marker in aquatic environments. Water Res. 2016;105:456–469. doi: 10.1016/j.watres.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Ramani S., Kang G. Viruses causing childhood diarrhoea in the developing world. Curr. Opin. Infect. Dis. 2009;22:477–482. doi: 10.1097/QCO.0b013e328330662f. [DOI] [PubMed] [Google Scholar]
- Rames E., Roiko A., Stratton H., Macdonald J. Technical aspects of using human adenovirus as a viral water quality indicator. Water Res. 2016;96:308–326. doi: 10.1016/j.watres.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Rasanen S., Lappalainen S., Kaikkonen S., Hamalainen M., Salminen M., Vesikari T. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 2010;138:1227–1234. doi: 10.1017/S0950268809991671. [DOI] [PubMed] [Google Scholar]
- Ravva S.V., Sarreal C.Z. Persistence of f-specific RNA coliphages in surface waters from a produce production region along the central coast of California. PloS One. 2016;11:1–13. doi: 10.1371/journal.pone.0146623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G., Boros Á., Pankovics P. Kobuviruses – a comprehensive review. Rev. Med. Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- Rigotto C., Victoria M., Moresco V., Kolesnikovas C.K., Corrêa A., Souza D.S.M., Miagostovich M.P., Simões C.M.O., Barardi C.R.M. Assessment of adenovirus, hepatitis A virus and rotavirus presence in environmental samples in Florianopolis, South Brazil. J. Appl. Microbiol. 2010;109 doi: 10.1111/j.1365-2672.2010.04827.x. 1979–1987. [DOI] [PubMed] [Google Scholar]
- Rigotto C., Hanley K., Rochelle P.A., De Leon R., Barardi C.R.M., Yates M.V. Survival of adenovirus types 2 and 41 in surface and ground waters measured by a plaque assay. Environ. Sci. Technol. 2011;45:4145–4150. doi: 10.1021/es103922r. [DOI] [PubMed] [Google Scholar]
- Rodríguez R.A., Polston P.M., Wu M.J., Wu J., Sobsey M.D. An improved infectivity assay combining cell culture with real-time PCR for rapid quantification of human adenoviruses 41 and semi-quantification of human adenovirus in sewage. Water Res. 2013;47:3183–3191. doi: 10.1016/j.watres.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75:7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Fernandez-Cassi X., Hundesa A., Vieira C., Kern A., Eriksson I., Ziros P., Kay D., Miagostovich M., Vargha M., Allard A., Vantarakis A., Wyn-Jones P., Bofill-Mas S., Girones R. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 2014;59:119–129. doi: 10.1016/j.watres.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Rusiñol M., Fernandez-Cassi X., Timoneda N., Carratalà A., Abril J.F., Silvera C., Figueras M.J., Gelati E., Rodó X., Kay D., Wyn-Jones P., Bofill-Mas S., Girones R. Evidence of viral dissemination and seasonality in a Mediterranean river catchment: implications for water pollution management. J. Environ. Manag. 2015;159:58–67. doi: 10.1016/j.jenvman.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Rzeżutka A., Cook N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004;28:441–453. doi: 10.1016/j.femsre.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Nakata S., Honma S., Tatsumi M., Numata-Kinoshita K., Chiba S. Clinical severity of Norwalk virus and Sapporo virus gastroenteritis in children in Hokkaido, Japan. Pediatr. Infect. Dis. J. 2001;20:849–853. doi: 10.1097/00006454-200109000-00005. [DOI] [PubMed] [Google Scholar]
- Sano D., Amarasiri M., Hata A., Watanabe T., Katayama H., Amarasiria M., Hata A., Watanabe T., Katayama H. Environment International. Pergamon; 2016. Risk Management of Viral Infectious Diseases in Wastewater Reclamation and Reuse: Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi H.P., Tuttle K.D., Betancourt W.Q., Kitajima M., Gerba C.P. Persistence of viruses by qPCR downstream of three effluent-dominated rivers in the western United States. Food Environ. Virol. 2018;10:297–304. doi: 10.1007/s12560-018-9343-7. [DOI] [PubMed] [Google Scholar]
- Sassoubre L.M., Love D.C., Silverman A.I., Nelson K.L., Boehm A.B. Comparison of enterovirus and adenovirus concentration and enumeration methods in seawater from Southern California, USA and Baja Malibu, Mexico. J. Water Health. 2012;10:419–430. doi: 10.2166/wh.2012.011. [DOI] [PubMed] [Google Scholar]
- Schaper M., Jofre J., Uys M., Grabow W.O.K. 2002. Distribution of Genotypes of F-specific RNA Bacteriophages in Human and Non-human Sources of Faecal Pollution in South Africa and Spain; pp. 657–667. [DOI] [PubMed] [Google Scholar]
- Schmitz B.W., Kitajima M., Campillo M.E., Gerba C.P., Pepper I.L. Virus reduction during advanced bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- Scott T.M., Rose J.B., Jenkins T.M., Farrah S.R., Lukasik J. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 2002;68 doi: 10.1128/AEM.68.12.5796-5803.2002. 5796 LP – 5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedji M.I., Varbanov M., Meo M., Colin M., Mathieu L., Bertrand I. Quantification of human adenovirus and norovirus in river water in the north-east of France. Environ. Sci. Pollut. Res. 2018:30497–30507. doi: 10.1007/s11356-018-3045-4. [DOI] [PubMed] [Google Scholar]
- Seehafer J.P., Carpenter P., Downer D.N., Colter J.S. Observations on the growth and plaque assay of BK virus in cultured human and monkey cells. J. Gen. Virol. 1978;38:383–387. doi: 10.1099/0022-1317-38-2-383. [DOI] [PubMed] [Google Scholar]
- Sekwadi P.G., Ravhuhali K.G., Mosam A., Essel V., Ntshoe G.M., Shonhiwa A.M., McCarthy K., Mans J., Taylor M.B., Page N.A., Govender N. Waterborne outbreak of gastroenteritis on the KwaZulu-natal coast, South Africa. Epidemiol. Infect. 2018;146:1318–1325. doi: 10.1017/S095026881800122X. December 2016/January 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiyawan A.S., Yamada T., Fajri J.A., Li F., Helard D., Horio A., Huang M., Kawaguchi T. Spatial and temporal variation in concentration of F-specific RNA bacteriophages in an open channel receiving Johkasou effluents. 土木学会論文集 G. 2013;69:667–678. doi: 10.2208/jscejer.69.III_667. [DOI] [Google Scholar]
- Setiyawan A.S., Yamada T., Fajri J.A., Li F. Characteristics of fecal indicators in channels of Johkasou systems. J. Water Environ. Technol. 2014;12:469–480. [Google Scholar]
- Shih Y.-J., Tao C.-W., Tsai H.-C., Huang W.-C., Huang T.-Y., Chen J.-S., Chiu Y.-C., Hsu T.-K., Hsu B.-M. First detection of enteric adenoviruses genotype 41 in recreation spring areas of Taiwan. Environ. Sci. Pollut. Res. 2017;24:18392–18399. doi: 10.1007/s11356-017-9513-4. [DOI] [PubMed] [Google Scholar]
- Shkoporov A.N., Khokhlova E.V., Fitzgerald C.B., Stockdale S.R., Draper L.A., Ross R.P., Hill C. ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 2018;9:4781. doi: 10.1038/s41467-018-07225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda T., Okoh A.I.A.I. Assessment of the incidence of enteric adenovirus species and serotypes in surface waters in the Eastern Cape Province of South Africa: Tyume river as a case study. Sci. World J. 2012 doi: 10.1100/2012/949216. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu J.P.S., Ahmed W., Palmer A., Smith K., Hodgers L., Toze S. Optimization of sampling strategy to determine pathogen removal efficacy of activated sludge treatment plant. Environ. Sci. Pollut. Res. 2017;24:19001–19010. doi: 10.1007/s11356-017-9557-5. [DOI] [PubMed] [Google Scholar]
- Sidhu J.P.S., Sena K., Hodgers L., Palmer A., Toze S. Comparative enteric viruses and coliphage removal during wastewater treatment processes in a sub-tropical environment. Sci. Total Environ. 2017;616:669–677. doi: 10.1016/j.scitotenv.2017.10.265. [DOI] [PubMed] [Google Scholar]
- Simmons F.J., Kuo D.H.W., Xagoraraki I. Removal of human enteric viruses by a full-scale membrane bioreactor during municipal wastewater processing. Water Res. 2011;45:2739–2750. doi: 10.1016/j.watres.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Sinclair R.G., Jones E.L., Gerba C.P. Viruses in recreational water-borne disease outbreaks: a review. J. Appl. Microbiol. 2009;107:1769–1780. doi: 10.1111/j.1365-2672.2009.04367.x. [DOI] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative crAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggemeier R., Bortoluzzi M., da Silva Heck T.M., da Luz R.B., Fabres R.B., Soliman M.C., Rigotto C., Baldasso N.A., Spilki F.R., de Matos Almeida S.E. Animal and human enteric viruses in water and sediment samples from dairy farms. Agric. Water Manag. 2015;152:135–141. doi: 10.1016/j.agwat.2015.01.010. [DOI] [Google Scholar]
- Staggemeier R., Heck T.M.S., Demoliner M., Ritzel R.G.F., Röhnelt N.M.S., Girardi V., Venker C.A.C., Spilki F.R. Enteric viruses and adenovirus diversity in waters from 2016 Olympic venues. Sci. Total Environ. 2017;586:304–312. doi: 10.1016/j.scitotenv.2017.01.223. [DOI] [PubMed] [Google Scholar]
- Staley C., Reckhow K.H., Lukasik J., Harwood V.J. Assessment of sources of human pathogens and fecal contamination in a Florida freshwater lake. Water Res. 2012;46:5799–5812. doi: 10.1016/j.watres.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Stefanakis A.I., Bardiau M., Trajano D., Couceiro F., Williams J.B., Taylor H. Presence of bacteria and bacteriophages in full-scale trickling filters and an aerated constructed wetland. Sci. Total Environ. 2019;659:1135–1145. doi: 10.1016/j.scitotenv.2018.12.415. [DOI] [PubMed] [Google Scholar]
- Stewart-Pullaro J., Daugomah J.W., Chestnut D.E., Graves D.A., Sobsey M.D., Scott G.I. F + RNA coliphage typing for microbial source tracking in surface waters. J. Appl. Microbiol. 2006;101:1015–1026. doi: 10.1111/j.1365-2672.2006.03011.x. [DOI] [PubMed] [Google Scholar]
- Symonds E.M., Griffin D.W., Breitbart M. Eukaryotic viruses in wastewater samples from the United States. Appl. Environ. Microbiol. 2009;75:1402–1409. doi: 10.1128/AEM.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Cook M.M., McQuaig S.M., Ulrich R.M., Schenck R.O., Lukasik J.O., Van Vleet E.S., Breitbart M. Reduction of nutrients, microbes, and personal care products in domestic wastewater by a benchtop electrocoagulation unit. Sci. Rep. 2015;5:9380. doi: 10.1038/srep09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Sinigalliano C., Gidley M., Ahmed W., McQuaig-Ulrich S.M., Breitbart M. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J. Appl. Microbiol. 2016;121:1469–1481. doi: 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- Symonds E.M., Nguyen K.H., Harwood V.J., Breitbart M. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 2018;144:1–12. doi: 10.1016/j.watres.2018.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar S., Sherchand J., Bhandari D., Ghaju Shrestha R., Malla B., Haramoto E., Sherchan S. Presence of human enteric viruses, protozoa, and indicators of pathogens in the Bagmati River, Nepal. Pathogens. 2018;7:38. doi: 10.3390/pathogens7020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston-Enriquez J.A., Haas C.N., Jacangelo J., Gerba C.P. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 2003;69:3979–3985. doi: 10.1128/AEM.69.7.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston-Enriquez J. a, Haas C.N., Gerba C.P., Jacangelo J. Inactivation of enteric Adenovirus and feline Calicivirus by chlorine dioxide. Appl. Environ. Microbiol. 2005;71:3100–3105. doi: 10.1128/AEM.71.6.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usepa . 2001. Method 1602 : Male-specific ( F + ) and Somatic Coliphage in Water by Single Agar Layer ( SAL ) Procedure. EPA Document 821-R-01-029 April. [Google Scholar]
- Van Doorslaer K., Chen Z., Bernard H., Chan P.K., DeSalle R., Dillner J., Forslund O., Haga T., McBride A.A., Villa L.L., Burk R.D., Consortium I.R. ICTV virus taxonomy profile: papillomaviridae. J. Gen. Virol. 2018;99:989–990. doi: 10.1099/jgv.0.001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maarseveen N.M., Wessels E., de Brouwer C.S., Vossen A.C.T.M., Claas E.C.J. Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus, Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 2010;49:205–210. doi: 10.1016/j.jcv.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Venegas C., Diez H., Blanch A.R., Jofre J., Campos C. Microbial source markers assessment in the Bogotá River basin (Colombia) J. Water Health. 2015;13:801–810. doi: 10.2166/wh.2015.240. [DOI] [PubMed] [Google Scholar]
- Verbyla M.E., Mihelcic J.R. A review of virus removal in wastewater treatment pond systems. Water Res. 2015;71:107–124. doi: 10.1016/j.watres.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Vergara G., Goh S.G., Rezaeinejad S., Chang S.Y., Sobsey M.D., Gin K.Y.H. Evaluation of FRNA coliphages as indicators of human enteric viruses in a tropical urban freshwater catchment. Water Res. 2015;79:39–47. doi: 10.1016/j.watres.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Wait D.A., Sobsey M.D. Comparative survival of enteric viruses and bacteria in Atlantic Ocean seawater. Water Sci. Technol. 2001;43:139–142. [PubMed] [Google Scholar]
- Walker D.I., Cross L.J., Stapleton T.A., Jenkins C.L., Lees D.N., Lowther J.A. Assessment of the applicability of capsid - integrity assays for detecting infectious norovirus inactivated by heat or UV irradiation. Food Environ. Virol. 2019;11(3):229–237. doi: 10.1007/s12560-019-09390-4. [DOI] [PubMed] [Google Scholar]
- Wangkahad B., Mongkolsuk S., Sirikanchana K. Integrated multivariate analysis with nondetects for the development of human sewage source-tracking tools using bacteriophages of Enterococcus faecalis. Environ. Sci. Technol. 2017;51:2235–2245. doi: 10.1021/acs.est.6b04714. [DOI] [PubMed] [Google Scholar]
- Who . 2010. WHO/UNICEF Joint Monitoring Programme for Water Supply and Sanitation. 2010 Meeting the MDG Drinkingwater and Sanitation Target: A Mid-term Assessment of Progress. (Geneva) [Google Scholar]
- Wicki M., Auckenthaler A., Felleisen R., Tanner M., Baumgartner A. Novel Bacteroides host strains for detection of human- and animal-specific bacteriophages in water. J. Water Health. 2011;9:159–168. doi: 10.2166/wh.2010.165. [DOI] [PubMed] [Google Scholar]
- Wicki M., Auckenthaler A., Felleisen R., Karabulut F., Niederhauser I., Tanner M., Baumgartner A. Assessment of source tracking methods for application in spring water. J. Water Health. 2015;13:473–488. doi: 10.2166/wh.2014.255. [DOI] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Rivera-Aban M., Greening G.E. Detection and characterization of F+ RNA bacteriophages in water and shellfish: application of a multiplex real-time reverse transcription PCR. J. Virol. Methods. 2008;149:123–128. doi: 10.1016/j.jviromet.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Wolf S., Hewitt J., Greening G.E. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl. Environ. Microbiol. 2010;76 doi: 10.1128/AEM.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. Wastewater-based epidemiology for early detection of viral outbreaks. In: O’Bannon D.J., editor. Women in Water Quality. Michigan State University; E. Lansing, MI: 2020. pp. 75–97. UNESCO, East Lansing. [DOI] [Google Scholar]
- Xagoraraki I., Kuo D.H.-W., Wong K., Wong M., Rose J.B. Occurrence of human adenoviruses at two recreational beaches of the Great Lakes. Appl. Environ. Microbiol. 2007;73 doi: 10.1128/AEM.01239-07. 7874 LP – 7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Griffiths M.W. Comparative persistence of subgroups of F-specific RNA phages in river water. Appl. Environ. Microbiol. 2013;79 doi: 10.1128/AEM.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoutis T., Klein J.D. Enterovirus infections. Pediatr. Rev. 1998;19:183–191. doi: 10.1542/pir.19-6-183. [DOI] [PubMed] [Google Scholar]
- Zell R., Delwart E., Gorbalenya A.E., Hovi T., King A.M.Q., Knowles N.J., Lindberg A.M., Pallansch M.A., Palmenberg A.C., Reuter G., Simmonds P., Skern T., Stanway G., Yamashita T., Consortium I.R. ICTV virus taxonomy profile: Picornaviridae. J. Gen. Virol. 2017;98:2421–2422. doi: 10.1099/jgv.0.000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Gallard J., Wu B., Harwood V.J., Sadowsky M.J., Hamilton K.A., Ahmed W. Synergy between quantitative microbial source tracking (qMST) and quantitative microbial risk assessment (QMRA): a review and prospectus. Environ. Int. 2019;130:104703. doi: 10.1016/j.envint.2019.03.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.