Abstract
In the absence of widespread testing, syndromic surveillance approaches may be useful for understanding potential undocumented coronavirus disease 2019 (COVID-19) in the United States. We used publicly available data from the Centers for Disease Control and Prevention FluView Interactive to evaluate its potential for COVID-19 syndromic surveillance. Unlike the prior 3 influenza seasons, we found a 76% decrease in influenza positive tests and a 27% increase in influenza like illness during the weeks since COVID-19 outbreaks began in the United States, which suggests FluView's potential utility for COVID-19 syndromic surveillance.
Key Words: SARS-CoV-2, Coronavirus, Syndrome, Influenza, Contact tracing
Coronavirus disease 2019 (COVID-19) is a novel illness caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2).1 Originating in Wuhan, China, it has spread internationally, resulting in the World Health Organization (WHO) declaring it a pandemic on March 11, 2020.2 In the United States, difficulty in developing, distributing, and operationalizing SARS-CoV-2 testing is hindering our prevention efforts, resulting in the potential for widespread, undocumented infection. Since COVID-19 may result in influenza-like illness (ILI),3 syndromic surveillance may assist investigations in the absence of laboratory-confirmed disease. In fact, syndromic surveillance efforts were proven useful during the 2003 outbreak of Severe Acute Respiratory Syndrome (SARS).4 An enhanced understanding of COVID-19 syndromic surveillance can assist in driving evidence-based prevention efforts to lessen the impact of the pandemic. If undocumented cases of COVID-19 are present in the United States and individuals infected display an ILI syndrome, we hypothesized there may be a differential burden of ILI vs reported laboratory-confirmed influenza disease. The objective of this study was to use publicly available influenza surveillance data to investigate their utility for understanding potential undocumented COVID-19 in the United States.
Methods
This was a secondary analysis of publicly available data from the Centers for Disease Control and Prevention FluView Interactive.5 Data utilized included the weighted percentage of ILI, the total number of influenza diagnostics obtained, as well as confirmed influenza diagnostics from clinical and public health laboratories. Although the 2019/2020 influenza season was the focus of analysis, data were gathered from week 40, 2017 through week 12, 2020 to allow historical comparison during similar periods. Line charts were constructed to compare the burden of ILI vs positive influenza tests for each of the 3 seasons. R v3.6.2 (R Foundation for Statistical Computing, Vienna, Austria) was used for all analyses. To ensure consistent updating of weeks after analyses ended, we created a free web application which scrapes the most recent influenza season data from FluView and re-creates the figures, on-demand with the most updated data. This application can be found at https://surveillance.shinyapps.io/covidsyndromic/.
Results
Through week 12, 2020 of the 2019/2020 influenza season, there have been 1,144,339 influenza tests submitted from 33,302,530 patients. Of these, 262,196 were positive for an influenza virus. During this same period, there were 1,429,499 ILI reported.
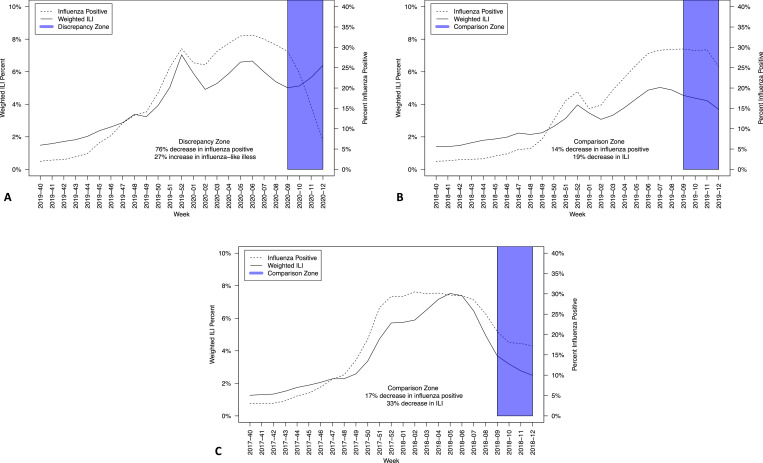
Evaluation of ILI vs positive influenza tests suggests an increasing burden of ILI with decreasing laboratory-confirmed influenza disease. Figure 1 , panel A depicts this anomalous data in week 09 to week 12, 2020, indicating a 76% decrease in influenza-positive diagnostics. During this same period, there was an increase (27%) in the weighted percentage of ILI. Similar trends were not identified in weeks 09-12, 2019 (Fig 1, Panel B) or weeks 09-12, 2018 (Fig 1, Panel C).
Fig 1.
Comparison of documented Influenza-Like-Illness (ILI) and influenza positive public health and clinical laboratory Reports, United States, Panel A: Week 40, 2019 through Week 12, 2020; Panel B: Week 40, 2018 through Week 12, 2019; Panel C: Week 40, 2017 through Week 12, 2018.
The anomalous data from week 09 to week 12, 2020 was also linked with a 30% decreased frequency of reported patients evaluated for ILI (1,446,678 evaluated in week 09, 2020 and 1,015,321 evaluated in week 12, 2020). During this same period, there was also 17% decrease in the number of reported patients tested for influenza in clinical and public health laboratories (64,745 tested in week 09, 2020 and 53,709 tested in week 12, 2020). This was not consistent with the 2 prior years when there were stable to slight increases in both those tested and evaluated (data not shown).
Discussion
Here, we identify a decrease in positive influenza diagnostics with a concurrent increased burden of ILI. This is concerning as it could be explained by undocumented COVID-19 disease in the United States, particularly given the limited large-scale testing for SARS-CoV-2. These findings highlight how syndromic surveillance using proxy measures to triangulate unavailable data may be possible and provide valuable direction for implementing prevention efforts. Utilization of syndromic surveillance such as this is particularly valuable for public health practitioners working to understand the trajectory of disease. Given the continued burden of COVID-19 infection and mortality around the world, integrating as many approaches to understanding this disease is of utmost necessity.
The WHO suggests using existing influenza surveillance systems to assist in COVID-19 surveillance,6 similar to what we have proposed. However, the WHO guidance relies heavily on SARS-CoV-2 testing, something that continues to be difficult in many countries including the United States. Even when testing kits are available, reagents and genome extraction kits continue to be in limited supply. Due to these issues, a syndromic approach may be more feasible at this time for surveillance practitioners.
A potential limitation of this analysis is that FluView shares public health and clinical laboratory data but may be limited in the number of influenza diagnostic tests conducted as influenza disease is not a reportable illness, therefore misclassification is possible. Further, reporting may be delayed, or influenza testing may be limited due to supply constraints or other burdens of the COVID-19 pandemic. Here, we found a decrease in the number tested for influenza and evaluated for influenza like illness in the current year but not prior years. This could explain some of the discrepancy. This is also the very beginning of the outbreak in the United States, with only a limited number of weeks of potential transmission evaluated. The outbreak is too far in its infancy in the United States to validate our results. Further, if transmission were to be occurring in peak influenza season, it could be difficult to discriminate between influenza and noninfluenza respiratory illness such as COVID-19, potentially impacting the utility of this syndromic approach. These data must all be taken under the consideration that we have never experienced a pandemic of this magnitude during a time when computational and data resources were as available as they are today in a highly globalized environment. Much like predictive models for COVID-19, all analyses may change rapidly due to the constant influx of new information.
Information regarding potential undocumented secondary community transmission for COVID-19 is critical to institute appropriate, targeted prevention interventions, as noted in the current COVID-19 outbreak in Italy.7 Attempting to understand transmission using syndromic surveillance efforts may allow for specific, data-driven allocation of needed resources to communities more vulnerable to severe COVID-19 or COVID-19-related mortality, particularly when laboratory confirmation is difficult or impossible. Organizations with available data should evaluate or share geolocation of testing to facilitate the identification of potential disease clusters. Those locations and community members may be experiencing concentrated COVID-19-related morbidity. The rapid escalation of testing for SARS-CoV-2 must be prioritized to institute the most appropriate prevention efforts.
Footnotes
Conflicts of interest: None to report.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Virtual press conference on COVID–19 - March 11, 2020. Available at:https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-press-conference-full-and-final-11mar2020.pdf?sfvrsn=cb432bb3_2. Accessed May 17, 2020.
- 3.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilder-Smith A, Chiew CJ, Lee VJ. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. 2020;20:e102–e107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Influenza (Flu) weekly U.S. influenza surveillance report (FluView) [updated February 7, 2020]. Available at:https://www.cdc.gov/flu/weekly/index.htm. Accessed May 17, 2020.
- 6.World Health Organization. Operational considerations for COVID-19 surveillance using GISRS. 2020 March 26, 2020. Available at:https://apps.who.int/iris/bitstream/handle/10665/331589/WHO-2019-nCoV-Leveraging_GISRS-2020.1-eng.pdf. Accessed May 17, 2020.
- 7.Grasselli G, Pesenti A, Cecconi M. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response [e-pub ahead of print] JAMA. 2020 doi: 10.1001/jama.2020.4031. [DOI] [PubMed] [Google Scholar]