Abstract
To better understand cerebellum-related diseases and functional mapping of the cerebellum, quantitative measurements of cerebellar regions in magnetic resonance (MR) images have been studied in both clinical and neurological studies. Such studies have revealed that different spinocerebellar ataxia (SCA) subtypes have different patterns of cerebellar atrophy and that atrophy of different cerebellar regions is correlated with specific functional losses. Previous methods to automatically parcellate the cerebellum—that is, to identify its sub-regions—have been largely based on multi-atlas segmentation. Recently, deep convolutional neural network (CNN) algorithms have been shown to have high speed and accuracy in cerebral sub-cortical structure segmentation from MR images. In this work, two three-dimensional CNNs were used to parcellate the cerebellum into 28 regions. First, a locating network was used to predict a bounding box around the cerebellum. Second, a parcellating network was used to parcellate the cerebellum using the entire region within the bounding box. A leave-one-out cross validation of fifteen manually delineated images was performed. Compared with a previously reported state-of-the-art algorithm, the proposed algorithm shows superior Dice coefficients. The proposed algorithm was further applied to three MR images of a healthy subject and subjects with SCA6 and SCA8, respectively. A Singularity container of this algorithm is publicly available.
Keywords: Cerebellum, parcellation, convolutional neural network
1. INTRODUCTION
The cerebellum is associated with both motor and cognitive functions.1 To better understand the diseases and functional mappings of the cerebellum, quantitative measurements from magnetic resonance (MR) images have been used in recent studies.2 Since manual labeling is laborious and time-consuming, accurate automated cerebellum parcellation algorithms are required, especially in large-scale studies. Previously reported algorithms are mainly based on multi-atlas segmentation. CGCUTS3 uses multi-atlas registration and label fusion which is refined in a graph cut framework. CERES4 uses a multi-atlas patch-based segmentation and a non-local label fusion algorithm to perform the parcellation.
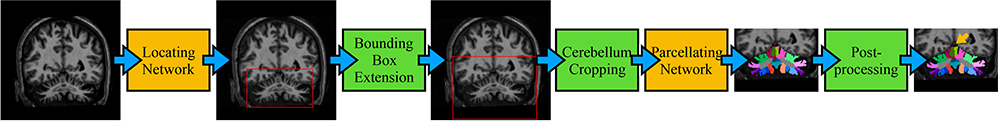
In recent years, convolutional neural networks (CNNs) have been successfully used in cerebral sub-cortical structure segmentation.5 They essentially act by learning a set of non-linear filters, producing feature maps of local to global information to segment the image. It has been shown that CNN-based cerebral sub-cortical structure segmentation can achieve superior results in a shorter execution time compared with conventional methods such as FreeSurfer.5 However, CNNs have not been applied to cerebellum parcellation. Inspired by Mask R-CNN,6 we used a two-step strategy to parcellate the cerebellum in this work. We first used a locating network to predict a bounding box around the cerebellum then a parcellating network to parcellate the cropped-out cerebellum (Fig. 1). The proposed algorithm was compared with a state-of-the-art algorithm, CGCUTS3*, using leave-one-out cross validation and applied to other images to show its broad applicability. A Singularity container of this algorithm is publicly available †.
Figure 1.
Flowchart of the proposed algorithm. Before and after post-processing difference is marked by a yellow arrow.
2. METHODS
2.1. Network Architectures
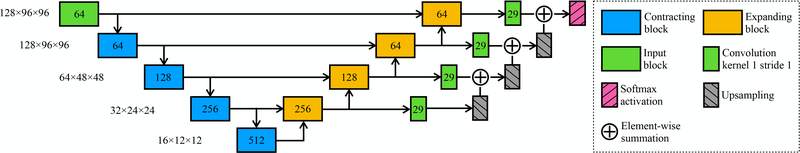
Unlike Mask R-CNN which relies on region proposals, our locating network only computes a single bounding box around the cerebellum since there is one and only one cerebellum presented in the image. The locating network was modified from the pre-activation ResNet,7 taking a 3D image as input and outputting six numbers specifying the extends along x, y, and z axes (Fig. 2, 4). Instead of using “RoIAlign” which interpolates the cropped region into a fixed size as in Mask R-CNN, we chose to symmetrically extend the bounding box (Fig. 1) to a size of 128 × 96 × 96 voxels thus keeping the original resolution. We modified a 3D U-Net8 (Fig. 3) to parcellate the cropped-out image into 28 regions (29 including the background). Note that instance normalization9 was used instead of batch normalization since it is invariant to additive and multiplicative transformations of the image intensities. As a result, image intensity normalization such as white matter peak normalization is not required, which simplifies the pre-processing.
Figure 2.
Architecture of the locating network. The output number of features are marked on each block.
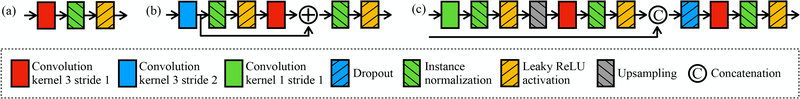
Figure 4.
Architecture of building blocks: (a) input block, (b) contracting block, and (c) expanding block.
Figure 3.
Architecture of the parcellating network. The output number of feature maps are marked on each block. The input spatial shape is marked on the left of each contracting block.
2.2. Data and Pre-processing
Training Data. Fifteen magnetization prepared rapid gradient echo (MPRAGE) images were expertly delineated.10 These images were acquired on a 3.0 T MR Intera scanner (Phillips Medical Systems, Netherlands) with the following parameters: slice thickness = 1.1 mm, flip angle = 8◦, TE = 3.9 ms, TR = 8.43 ms, field of view = 20.5 × 20.5 × 14.5 cm, and image matrix = 256 × 256. The images were resampled to 1 mm isotropic resolution. Testing Data. Three images of a healthy control, a subject with SCA subtype 6, and a subject with SCA subtype 8 were acquired using the same protocol (except that the SCA6 image was acquired with 0.9 mm isotropic resolution). Pre-processing. The images were first rigidly transformed to the 1 mm isotropic ICBMc 2009c template11 in the MNI space then N4 inhomogeneity corrected12 using the ANTs registration suite‡. No skull-stripping or intensity normalization was performed.
2.3. Training Networks
Data Augmentation. Right-left flipping, random translation, and random rotation were used in the training of the locating network. Right-left flipping, random rotation, and random elastic deformation were used in the training of the parcellating network. When flipping, the corresponding right-and-left labels were swapped. The translations were uniformly sampled from 0 to 30 voxles independently for the x, y, and z axes. The rotation angles were uniformly sampled from 0 to 15 degrees independently for the three axes. Elastic deformations were generated by smoothing voxel-wise random translations. Examples of data augmentation are shown in Fig. 5. Training. We used a smooth L1 loss13 to train the locating network and one minus the mean Dice coefficient over all the labels14 (the weight for each label is equal to each other) to train the parcellating network. The Adam optimizer15 with learning rate 0.001, β1 = 0.9, β2 = 0.999, and ϵ = 1 × 10−7 was used. The locating and parcellating networks were trained for 200 and 400 epochs, respectively, with a batch size of one.
Figure 5.
Data augmentation: (a) the original image, (b) the flipped image, (c) the translated image, (d) the rotated image, and (e) the deformed image, overlaid with their corresponding parcellation.
2.4. Post-processing
Since CNNs perform per-pixel labeling, isolated mislabeling can occur (see Fig. 1). To correct this, we first calculate the connected components of each label. Using a threshold for the fraction of the volume of the largest connected components, these components were divided into two categories: the large size or the small size. A large-size component keeps its original label, while a small-size component changes its label depending on its adjacency to other labels. If a small-size component is not adjacent to any non-background large-size components, it is discarded; otherwise, it is assigned with the label of the region with which it shares most of its boundary. Finally, we fill the holes of each label using binary-image morphological operations.
3. EXPERIMENTS AND RESULTS
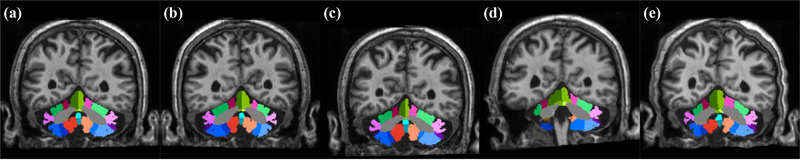
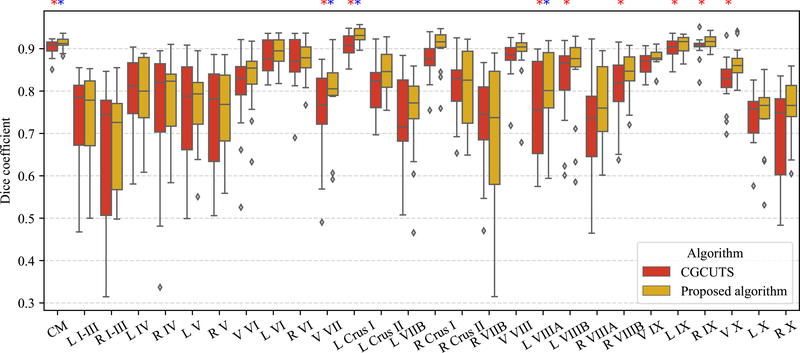
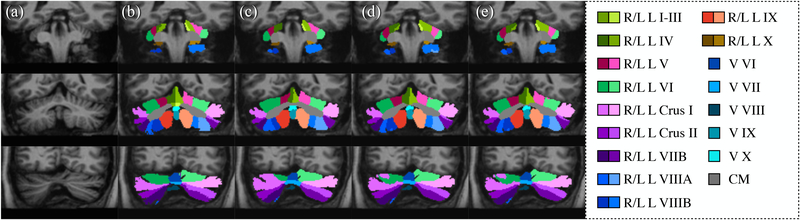
Leave-one-out cross validation was performed using the fifteen manually delineated images and the results were compared with CGCUTS. The Dice coefficient between the prediction and manual delineation per cerebellar region was calculated and shown as box plots in Fig. 6. A two-sided paired Wilcoxon signed-rank test was performed per region with the null hypothesis that the CNNs with post-processing and CGCUTS have the same performance. The p-values were further adjusted by the BH (false discovery rate) method. Nine regions (corpus medullare, vermal lobule VII, left lobule crus I, left lobule VIIIA, left lobule VIIIB, right lobule VIIIB, left lobule IX, right lobule IX, and vermis X) with significant p-values (p < 0.05) are marked by red asterisks. Four regions (corpus medullare, vermal lobule VII, left lobule crus I, and left lobule VIIIA) with significant adjusted p-values (p < 0.05) are marked by blue asterisks (Fig. 6). An example visual comparison is shown in Fig. 7.
Figure 6.
Comparison with CGCUTS. Significantly improved regions are marked by asterisks (p < 0.05); red asterisks for p-values and blue asterisks for adjusted p-values. CM: corpus medullare, L: left, R: right, and V: vermis.
Figure 7.
Cerebellum parcellation by the manual rater and three different algorithms on three coronal slices: (a) image, (b) manual rater, (c) CGCUTS, (d) CNNs without post-processing, and (e) CNNs with post-processing. CM: corpus medullare, L: left, R: right, and V: vermis.
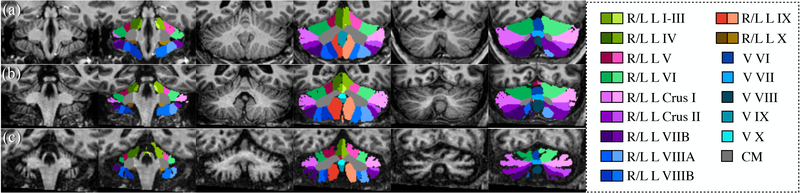
Furthermore, the algorithm was also applied to three images of a healthy control, a subject with SCA6, and a subject with SCA8, respectively, using the networks trained from all fifteen manually delineated images, and the results are shown in Fig. 8.
Figure 8.
Parcellation results on three coronal slices: (a) healthy control, (b) subject with SCA6, and (c) subject with SCA8. CM: corpus medullare, L: left, R: right, and V: vermis.
The two CNNs take approximately 50 seconds in total to run using a Tesla K40c GPU (NVIDIA Corporation, USA). The post-processing takes approximately 74 seconds using an Xeon E5–2623 v3 CPU (Intel Corporation, USA) without parallel computing. As a comparison, CERES takes 212 seconds4 without pre-processing and CGCUTS takes approximately 7 hours.3
4. DISCUSSION AND CONCLUSIONS
The proposed algorithm to parcellate the cerebellum uses locating and parcellating networks instead of multi-atlas segmentation. The paired Wilcoxon signed-rank tests show that the proposed algorithm provides superior results compared with a state-of-the-art algorithm, CGCUTS. A Singularity container of the proposed algorithm is provided.
However, the proposed methods have several limitations. First, different from fully convolutional neural networks, the proposed locating network is not inherently translation-invariant. Therefore, the locating network usually cannot successfully capture the cerebellum if the MNI alignment fails. Although this can be dealt to some extent by the translation data augmentation, a better approach could be appending a coordinate map as an additional channel for the input to this network. Another approach could be using a coarse fully convolutional neural network (the U-Net used as the parcellating network for example) to locate the cerebellum instead.
Second, the proposed methods should be preferentially applied to MPRAGE images with similar tissue contrast as the training images. We noted that the parcellation was less accurate on images acquired with the spoiled gradient recalled echo (SPGR) sequence. Even for the MPRAGE sequence, the algorithm may also be less accurate if the sequence parameters are too different. This could potentially be dealt with intensity augmentation during training or image contrast harmonization.16
Third, the largest-connected-component-based post-processing used in this work is not highly accurate. It failed to remove the mislabeled voxels if these voxels were directly connected to the desired region. Additionally, this method can wrongly change the voxel label if the threshold for the connected-component size is chosen inappropriately. More sophisticated post-processing methods such as conditional random fields17 could be used to enforce the topological constraint and reduce mislabeling.
5. ACKNOWLEDGMENTS
The author Shuo Han is in part supported by the Intramural Research Program, National Institute on Aging, NIH.
Footnotes
Downloaded from: http://iacl.jhu.edu/index.php/Resources with minor improvement.
REFERENCES
- [1].Buckner RL, “The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging,” Neuron 80(3), 807–815 (2013). [DOI] [PubMed] [Google Scholar]
- [2].Kansal K, Yang Z, Fishman AM, Sair HI, Ying SH, Jedynak BM, Prince JL, and Onyike CU, “Structural cerebellar correlates of cognitive and motor dysfunctions in cerebellar degeneration,” Brain 140(3), 707–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang Z, Ye C, Bogovic JA, Carass A, Jedynak BM, Ying SH, and Prince JL, “Automated cerebellar lobule segmentation with application to cerebellar structural analysis in cerebellar disease,” NeuroImage 127, 435–444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Romero JE, Coupé P, Giraud R, Ta V-T, Fonov V, Park MTM, Chakravarty MM, Voineskos AN, and Manjón JV, “CERES: A new cerebellum lobule segmentation method,” NeuroImage 147, 916–924 (2017). [DOI] [PubMed] [Google Scholar]
- [5].Mehta R and Sivaswamy J, “M-Net: A convolutional neural network for deep brain structure segmentation,” in [Biomedical Imaging (ISBI), 2017 IEEE 14th International Symposium on], 437–440, IEEE (2017). [Google Scholar]
- [6].He K, Gkioxari G, Dollár P, and Girshick R, “Mask R-CNN,” arXiv preprint arXiv:1703.06870 (2017). [DOI] [PubMed] [Google Scholar]
- [7].He K, Zhang X, Ren S, and Sun J, “Identity mappings in deep residual networks,” arXiv preprint arXiv:1603.05027 (2016). [Google Scholar]
- [8].Kayalibay B, Jensen G, and van der Smagt P, “CNN-based segmentation of medical imaging data,” arXiv preprint arXiv:1701.03056 (2017). [Google Scholar]
- [9].Ulyanov D, Vedaldi A, and Lempitsky V, “Improved texture networks: Maximizing quality and diversity in feed-forward stylization and texture synthesis,” in [Computer Vision and Pattern Recognition (CVPR), 2017 IEEE Conference on], 4105–4113, IEEE (2017). [Google Scholar]
- [10].Bogovic JA, Jedynak B, Rigg R, Du A, Landman BA, Prince JL, and Ying SH, “Approaching expert results using a hierarchical cerebellum parcellation protocol for multiple inexpert human raters,” NeuroImage 64, 616–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fonov VS, Evans AC, McKinstry RC, Almli C, and Collins D, “Unbiased nonlinear average age-appropriate brain templates from birth to adulthood,” NeuroImage 47, S102 (2009). [Google Scholar]
- [12].Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, and Gee JC, “N4ITK: improved N3 bias correction,” IEEE Transactions on Medical Imaging 29(6), 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Girshick R, “Fast R-CNN,” arXiv preprint arXiv:1504.08083 (2015). [Google Scholar]
- [14].Sudre CH, Li W, Vercauteren T, Ourselin S, and Cardoso MJ, “Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations,” in [Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support], 240–248, Springer; (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kingma DP and Ba J, “Adam: a method for stochastic optimization,” arXiv preprint arXiv:1412.6980 (2014). [Google Scholar]
- [16].Jog A, Carass A, Roy S, Pham DL, and Prince JL, “MR image synthesis by contrast learning on neighborhood ensembles,” Medical image analysis 24(1), 63–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Arnab A, Zheng S, Jayasumana S, Romera-Paredes B, Larsson M, Kirillov A, Savchynskyy B, Rother C, Kahl F, and Torr PH, “Conditional random fields meet deep neural networks for semantic segmentation: Combining probabilistic graphical models with deep learning for structured prediction,” IEEE Signal Processing Magazine 35(1), 37–52 (2018). [Google Scholar]