Abstract
Machine learning (ML) has the potential to improve the dermatologist’s practice from diagnosis to personalized treatment. Recent advancements in access to large datasets (e.g., electronic medical records, image databases, omics), faster computing, and cheaper data storage have encouraged the development of ML algorithms with human-like intelligence in dermatology. This article is an overview of the basics of ML, current applications of ML, and potential limitations and considerations for further development of ML. We have identified five current areas of applications for ML in dermatology: (1) disease classification using clinical images; (2) disease classification using dermatopathology images; (3) assessment of skin diseases using mobile applications and personal monitoring devices; (4) facilitating large-scale epidemiology research; and (5) precision medicine. The purpose of this review is to provide a guide for dermatologists to help demystify the fundamentals of ML and its wide range of applications in order to better evaluate its potential opportunities and challenges.
Keywords: Artificial intelligence, Convolutional neural network, Deep learning, Dermatology, Image classification, Machine learning, Mobile applications, Personal monitoring devices, Precision medicine
Key Summary Points
| Machine learning (ML) has the potential to improve the dermatologist’s practice from diagnosis to personalized treatment. |
| This review article is a guide for dermatologists to help demystify the fundamentals of ML and its wide range of applications in order to better evaluate its potential opportunities and challenges. |
| We have identified five current areas of applications for ML in dermatology: (1) disease classification using clinical images; (2) disease classification using dermatopathology images; (3) assessment of skin diseases using mobile applications and personal monitoring devices; (4) facilitating large-scale epidemiology research; and (5) precision medicine. |
| While ML models are powerful, dermatologists should be cognizant of the potential limitations of ML (e.g. algorithmic bias and black box nature of ML models) and how to make these technologies inclusive of skin of color. |
| Involving more dermatologists in the development and testing of ML models is imperative for creating useful and clinically relevant technology. |
Introduction
In dermatology and medicine at large, the abundance of data in clinical records, patient demographic information, results from imaging examinations, and data collected from questionnaires represent a wealth of information that has the potential to revolutionize personalized medicine [1]. Translational research in dermatology is already abundant, with data from the genome, epigenome, transcriptome, proteome, and microbiome, areas of research that are often referred to by the shortened term “omics” [2]. Recent advancements in faster processing and cheaper storage have allowed for the development of machine learning (ML) algorithms with human-like intelligence that have numerous applications in dermatology [3–5]. To assess the effectiveness of these emerging technologies, it is imperative that dermatologists have a basic understanding of artificial intelligence and ML. In this review, we first provide an overview of artificial intelligence and ML and how algorithms are developed. Second, we examine the current applications of ML that are relevant to dermatologists. Lastly, we explore potential challenges and limitations for the future development of ML. This review is a guide for dermatologists to help demystify the fundamentals of ML and its wide range of applications in order to better evaluate its potential opportunities and challenges.
Methods
This review is based on a literature search performed in Medline, Embase, and Web of Science databases of articles pertaining to artificial intelligence and ML in dermatology. The search was conducted in December 2019. Articles from 2000 to 2019 were included to focus on emerging methods. Only articles written in English were included, and articles that were repeated were excluded. The following primary keywords were used: “artificial intelligence,” “machine learning,” and “dermatology.” After the preliminary results were reviewed, the Boolean operators “AND” and “OR” were used with the following secondary keywords: “personalized medicine,” “teledermatology,” “smartphone apps,” “skin cancer,” “nonmelanoma skin cancer,” “melanoma,” “psoriasis,” “atopic dermatitis,” and dermatopathology.” Our literature search yielded a total of 899 articles, among which 70 articles were deemed relevant to this review.
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Overview of Artificial Intelligence and Machine Learning
What is the Difference Between Artificial Intelligence and Machine Learning?
Artificial intelligence is a branch of computer science that uses machines and programs to simulate intelligent human behavior. Artificial intelligence dates back to the 1950s to Alan Turing’s question “Can machines think?” [6]. By the 1970s, software engineers had created algorithms with explicit rules for computers on how to process data. However, the heuristics of human decision making in medicine were not easy to program into explicit rules.
ML is a tool comprising a subset of artificial intelligence that enables the goals of artificial intelligence to be achieved (Fig. 1). Recently, ML has piqued attention for its broad range of uses in daily life from personalized online recommendations for videos and news to self-driving cars.
Fig. 1.
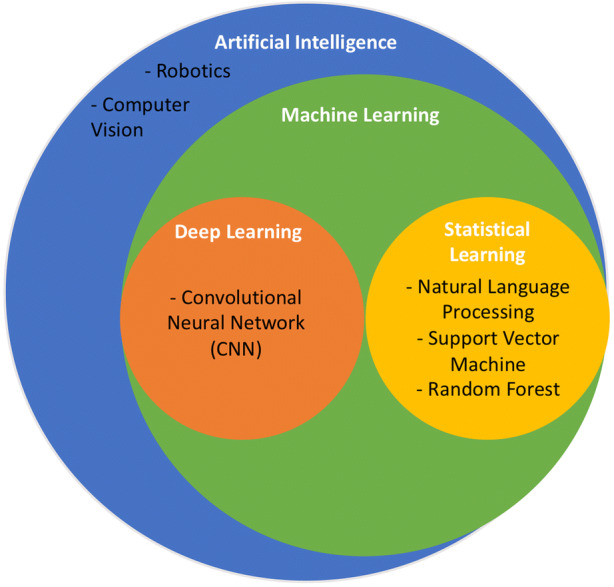
Artificial intelligence and machine learning. Machine learning is a type of artificial intelligence. Some common types of machine learning approaches used in dermatology include convolutional neural network (CNN), natural language processing (NLP), support vector machine, and random forest. Notably, there are many other possible machine learning approaches that are not listed and out of the scope of this review
ML covers a variety of algorithms and statistical methods, including logistic regression, random forest, and deep learning. Although ML can seem enigmatic at first, it can be deeply related to traditional statistical models recognizable to most dermatologists.
Machine Learning Approaches
Machine learning approaches can be divided into three broad categories: supervised learning, unsupervised learning, and reinforcement learning [7]. Supervised learning requires a dataset to be presented as inputs (called features) and outputs (called labels) [7]. For example, in an algorithm that classifies a pigmented lesion as a melanoma or benign, images of pigmented lesions are “features” and the categorical data of whether it is malignant or benign are “labels.” The algorithm is first trained with labeled images of melanoma and benign pigmented lesions and then the computer generalizes this information to a new, unseen set of images of skin. Supervised learning is the most common type of learning used in dermatology. In contrast, unsupervised learning only requires inputs (unlabeled data), and this approach can identify unknown clusters or anomalies in data [7]. Reinforcement learning is a hybrid of both supervised and unsupervised learning which learns by trial and error and input from the environment [7]. An example of reinforcement learning is the algorithm in AlphaGo [8]. Reinforcement learning has yet to be explored in dermatology.
Machine Learning Algorithms
There are a variety of ML algorithms commonly used in dermatology. Most ML algorithms are examples of statistical learning; for example, some of the most common statistical learning methods are linear regression, logistic regression, k-nearest neighbor (k-NN), support vector machine (SVM), random forest (RF), and natural language processing (NLP). k-NN is used for data classification and regression based on the number of k neighbors [9, 10]. SVMs are used to classify data by finding a hyperplane to differentiate between groups [11]. RFs generate a network of random decision trees to find the most common outcome among all the randomly generated decision trees [12]. NLP analyzes large bodies of text in order to identify patterns [13].
Neural Networks and Deep Learning
Deep learning is a subset of ML that uses statistical and mathematical models to mimic how neurons process information. Artificial neural networks (ANNs), or neural networks (NNs), are based on a collection of connected units (e.g., nodes, neurons, or process layers) [14]. ANNs are inspired by the network of neurons in the human brain. The neurons, or nodes, that make up the ANN are organized into linear arrays called layers [14]. Each node receives inputs from other connections that have associated weights [14]. Creating an ANN includes choosing the number of nodes in each layer, the number of layers in the network, and the path of the connections among the nodes [14], and the typical ANN has input layers, output layers, and hidden layers. ANNs are trained to perform specific tasks, such as classification, through a learning process. Learning within ANNs can be supervised or unsupervised; however, supervised learning is more common. In supervised learning, a training set contains examples of input targets and output targets [14]. As the ANN is trained, the weights of the inputs are adjusted to minimize the error between the network output and the correct output [14]. Once the network produces the desired outputs for a series of inputs, the weights are fixed and the NN can be applied to other datasets [14].
Convolutional NNs (CNNs) are a special subclass of ANNs that contain one or more layers called convolutional units (pooling units). CNNs take in two-dimensional or three-dimensional inputs which are passed through multiple hidden layers. An image can be broken down into motifs, or a collection of pixels that form a basic unit of analysis. The first few layers of the CNN compare each part of an input image against some small sub-image [5]. Each node is assigned a certain feature (e.g., color, shape, size, etc.), and the node’s output to the next layer depends on how much a part of the image resembles the feature, a process performed by convolution [5]. After these convolutional layers, pooling layers, which are a standard NN, classify the overall image [5]. CNNs first showed promise for medical image classification at the historic 2012 ImageNet Large Scale Visual Recognition (ILSVRC) conference. A CNN, called AlexNet, was trained to classify 1.2 million images into 1000 different categories with a top-5 error rate of 15.3%, which is the percentage of images for which the correct class was not among the top five predicted classes [15]. This was the first CNN to display such a low error rate. Previous image datasets were relatively small, comprising only of tens of thousands of images [16–18]. By 2016, all methods to classify medical images at the 2016 International Symposium on Biomedical Imaging used CNNs [19].
Another subtype of CNNs is called a region-based CNN (R-CNN). R-CNN is a type of CNN that can detect a desired object within an image. In the case of dermatology, it can detect the location of cutaneous lesions by combining region proposal algorithms with CNNs.
Transfer Learning and Ensemble Learning
Transfer learning utilizes the power of a pretrained CNN. These pretrained CNNs are often trained on databases that include millions of images, so they are able to distinguish images with much higher accuracy than a CNN that is only trained on databases of only a few hundred or few thousand images. The last fully connected layer of a pretrained CNN is modified and trained with images for the more specific classification task. Examples of common pretrained CNNs are AlexNet, Google Inception V3, ResNet-50, Xception, VGG-19, and VGG-16. This layered architecture allows researchers to use a pretrained network without its final layer as a fixed feature extractor for other tasks. The learning process of a pretrained CNN can be faster because it relies on previously learned tasks. Using a pretrained CNN that is trained on 1–2 million images is more accurate than a CNN that is trained on a smaller number of images of the more specific classification tasks. Ensemble learning improves ML results by combining several models (meta-algorithms) or the power of multiple of CNNs together [7].
Metrics for Assessment and Validation of Machine Learning Models
Machine learning models are assessed according to variety of metrics based on the number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) from a ML prediction. These metrics include sensitivity [TP/(TP + FN)], specificity [TN/(TN + FP)], positive predictive value [TP/(TP + FP)], negative predictive value [TN/(TN + FN)], and accuracy [(TP + TN)/(TP + TN + FP + FN)]. ML models are also evaluated on the area under the receiver operating characteristic (AUROC or AUC). The receiver operating characteristic (ROC) curve is calculated by plotting the sensitivity versus 1-specificity [20]. The further the ROC curve deviates from the diagonal and the larger the AUROC, the better the algorithm. An AUROC of 1.0 indicates perfect classification, and an AUROC of 0.5 indicates classification that is no better than random chance.
Another important metric is the generalizability of an ML model, or how well a ML model can learn concepts and apply these concepts to examples the model has never seen before [21]. There are two terms to describe the generalizability of an ML model: overfitting and underfitting. Overfitting occurs when a ML model represents the training dataset too well by capturing the noise of the dataset [7]. Algorithms that are trained and validated with the same set of images or with the same dataset risk overfitting to the data. However, current studies are limited by the images available and may be trained and validated on images from the same dataset. Validation or cross-validation to compare the predictive accuracies of datasets can help prevent overfitting. In contrast, underfitting occurs when a ML model cannot represent the training dataset and cannot generalize to a new dataset [7]. Overfitting is more common than underfitting in ML models.
Dermatology and Machine Learning
There are many promising opportunities for ML in the dermatologist’s practice. The classification of images through CNN has garnered the most attention for its potential to increase accessibility of skin cancer screenings and streamline the workflow of dermatologists. CNN has already proven successful in many other fields, such as ophthalmology, pathology, and radiology. In 2018, the US Food and Drug Administration (FDA) approved a CNN in the IDx-DR diagnostic system (IDx Technologies Inc., Coralville, IA, USA) [22] for independent use in diabetic retinopathy screening after being validated in a pivotal prospective clinical trial of over 900 patients. This diagnostic system achieved a sensitivity of 87.2% and specificity of 90.7% and was compared to an independent, high-quality gold standard of imaging protocols [23]. However, CNN for the screening of melanoma or non-melanoma skin cancers has not been validated in prospective clinical trials and still has notable limitations. Beyond the simplistic task of differentiating malignant from benign lesions, ML can also be applied in differential diagnosis, dermatopathology, teledermatology, mobile applications, and personalized medicine. Understanding what type of deep learning methods are currently being used and how these methods are evolving is crucial for regulating and optimizing these technologies for patients.
Classification of Dermatological Diseases Using Clinical Images
The majority of studies implementing ML in dermatology focus on classifying skin lesions for a variety of diseases, including melanoma, non-melanoma skin cancer (NMSC), psoriasis, atopic dermatitis, onychomycosis, and rosacea. These studies primarily rely on CNN for image recognition and classification. Initially, a pretrained CNN (i.e., AlexNet) was only used to extract features, and these features were then classified by a more simplistic ML algorithm, such as k-nearest neighbors or SVMs [24–26]. Currently, most CNNs can both extract features and classify images through end-to-end learning.
Melanoma
Melanoma is the fifth most common invasive cancer in the USA, and its incidence is increasing around the world [27, 28]. Melanomas are also responsible for the vast majority of skin cancer-related mortalities [28]. Skin cancer is screened visually with a total body skin examination. Unfortunately, data from the National Health Interview Survey indicate that screening rates are remarkably low (16% in men and 13% in women) [29]. Consequently, the first deep learning algorithms focused on classifying melanoma to address low screening rates and increase access. One of the first milestone studies to classify malignant melanoma with notable accuracy was that of Esteva et al. in 2017 [30]. This historic study used a CNN called Google Inception V3 that was previously pretrained on 1.28 million images of general objects. Using transfer learning, the authors trained the algorithm using 129,450 dermoscopic and clinical images. This was the first classifier to show comparable accuracy to dermatologists when classifying keratinocyte carcinoma versus seborrheic keratosis, and malignant melanoma versus benign nevi. The CNN achieved 72.1% accuracy while two dermatologists achieved 65.56 and 66% accuracy, respectively. The CNN achieved an overall AUC of > 91%, which was similar to the average output predications of 21 dermatologists. Many studies since then have leveraged transfer learning to classify lesions into a number of skin cancer classes and determine the probability of malignancy; these studies showed comparable accuracy, AUROC, sensitivity, and/or specificity to board-certified dermatologists or dermatologists in training [19, 31–39]. It is also important to note the average dermatologist’s diagnostic accuracy (e.g., sensitivity and specificity) when evaluating ML models for general screening. The authors of a systematic review of prospective studies note that regarding the diagnostic accuracy of melanoma, the sensitivity for dermatologists was 81–100% and that for primary care physicians (PCPs) was 42–100% [40]. While none of the studies in the review reported the specificity for dermatologists, one study did report specificity for PCPs to be 98%. For biopsy or referral accuracy, the sensitivity for dermatologists and PCPs ranged from 82 to 100% and from 70 to 88%, respectively, while the specificity for dermatologists and PCPs ranged from 70 to 89% and from 70 to 87%, respectively [40]. Therefore, ML models demonstrating similar diagnostic and biopsy/referral accuracy may prove useful for general screening of melanoma.
Studies by Brinker and colleagues have demonstrated that CNNs can exhibit superior sensitivity and specificity in melanoma classification as compared to board-certified dermatologists and dermatologists in training [41, 42]. In one of their studies, these researchers used transfer learning on a pretrained ResNet50 CNN and trained it with 4204 biopsy-proven images of melanoma and nevi (1:1). They also asked 144 dermatologists (52 board-certified and 92 junior dermatologists) to evaluate 804 biopsy-proven dermoscopic images for melanoma versus nevi. The trained CNN achieved a higher sensitivity (82.3 vs. 67.2%) and specificity (77.9 vs. 62.2%) than both the board-certified and junior dermatologists. Although these findings are promising, images from the same overall dataset were used for both training and validation. This algorithm has also not been externally validated; therefore, it is unclear whether these results are generalizable to other datasets. This limitation is discussed by the authors who suggest that future research could fine-tune the CNN with a small sample of new images before being applied to new datasets. Nevertheless, further validation in prospective clinical trials in more real-world settings is necessary before claiming superiority of algorithm performance over dermatologists. While most of these studies pitted artificial intelligence algorithms against dermatologists, a recent study by Hekler et al. [43] found that combining human and artificial intelligence accomplishes a better classification of images as compared to only dermatologists or only classification by a CNN [43]. The mean accuracy increased by 1.36% when dermatologists worked together with ML. While the results were not statistically significant, it is an important step toward finding the best way to maximize combining human and artificial intelligence.
A few studies have also integrated clinical metadata, such as patient demographics (i.e. age, gender, etc.), with images of lesions and have shown that a classifier based on both patient metadata and dermoscopic images is more accurate than a classifier based only on dermoscopic images [32, 44]. Other studies have combined dermoscopic images with macroscopic images [45] or with a sonification layer [39] to increase the accuracy of its classifier. When diagnosing unknown skin lesions, clinicians typically do not only visually inspect the skin, but they simultaneously take into account many points of clinical data, including patient demographics, laboratory tests, etc. Accounting for clinical patient data into future CNNs has the potential for creating more accurate algorithms and significant biomarkers.
It is difficult to validate the results of many of these studies since their algorithms are not publicly available. However, the authors of one study made their algorithm easily accessible online [31]. In this study, Han et al. transferred learning with the pretrained CNN, the Microsoft ResNet-152 model, to train 19,398 images from four databases of Asian patients and then validated the model on both Asian and Caucasian patients for 12 diagnoses, including basal cell carcinoma, squamous cell carcinoma, intraepithelial carcinoma, and melanoma. The AUCs for the corresponding diseases were all comparable to those of 16 dermatologists. However, the AUCs for basal cell carcinoma (0.96 for the Asian dataset vs. 0.90 for the Caucasian dataset) and melanoma (0.96 for the Asian dataset vs. 0.88 for the Caucasian dataset) were slightly lower for the Caucasian dataset than for the Asian dataset [31], demonstrating that the accuracy of algorithms can be affected by differences in patient ethnicity. Both clinical presentation and prevalence of diseases can differ between ethnicities. For example, basal cell carcinoma presents with a brown, glossy pigmentation in 75% of basal cell carcinomas in Japanese patients but in only 6% of basal cell carcinomas in Caucasian patients [46]. In addition, melanomas have a low incidence in Asian populations and are more common in Caucasian populations [46]. Asians are more likely to develop a rare subtype melanoma called acral lentiginous melanoma, which accounted for 69.6% of melanomas in the Asian dataset that trained Han et al.’s algorithm [31, 46]. These algorithms need to be trained and validated in more diverse population sets to better reflect the world’s population. Most algorithms are trained on either Caucasian or Asian patients.
The algorithm published by Han et al. [31] is one of the few that are publicly available and, consequently, Navarrete-Dechent et al. were able to evaluate its generalizability [47]. These authors tested the algorithm with 100 biopsy-proven, high-quality images from the International Skin Imaging Collaboration Archive; all 100 images were lesions from Caucasians in the USA. They found that the top-1 error rate was 71% and the top-5 error rate was 42%. Although the sample size of the images tested by Navarrete-Dechent et al. [47] was small, these findings suggest that the sensitivity of the algorithm was significantly lower than had been presented in the original publication [31]. It is known that published algorithms may underperform in less than ideal conditions or when validated through external testing. Therefore, just as with new drug treatments and other biomedical technologies, it is crucial to require more external testing and validation of these algorithms.
There has been only one prospective trial examining the accuracy of a deep learning algorithm for the accurate diagnosis of melanoma [48]. This study used the artificial intelligence algorithm Deep Ensemble for Recognition of Malignancy, developed by Skin Analytics Ltd. (London, UK), to identify melanoma in dermoscopic images of lesions taken with a smartphone and digital single-lens reflex (DSLR) camera [48]. This project trained the algorithm on dermoscopic images of biopsy-proven and control lesions from 514 patients. The inclusion of controls, or lesions believed to be benign, was useful because the algorithm maintained a high specificity. Most studies have only been trained on datasets of lesions already considered to be suspicious of melanoma, which creates a biased dataset. The algorithm achieved comparable AUROCs for biopsied lesions and all lesions captured with Apple iPhone 6 s images (90.1% for biopsied lesions and 95.8% for all lesions), Samsung Galaxy S6 images (85.8% for biopsied lesions and 86.9% for all lesions), and DSLR camera images (86.9% for biopsied lesions and 91.8% for all lesions). Specialists achieved an AUROC of 77.8% for all lesions; however, the type of specialist and the credentials of whether the specialist was board-certified in dermatology were not specified. Previous studies have found significant differences in the accuracy of classification of malignant versus benign melanoma between PCPs and dermatologists.
It is also important to be aware of potential diagnostic confounders before implementing these CNNs on a large scale. Artifacts, such as air bubbles, skin hairs, or ruler markers, have been noted to disrupt automated melanoma detection [49]. Suspicious lesions are routinely marked with gentian violet surgical skin markers. Ink markings have been found to be more prevalent among malignant lesions than among benign lesions in test image datasets [47], and it has been reported that ink markings can significantly interfere with the CNN’s correct diagnosis of nevi by increasing both the likelihood of the lesion being a melanoma and the false-positive rate of the classifier [50]. Therefore, it is recommended to avoid ink markings in dermoscopic images analyzed by a CNN. Such confounders are difficult to identify because ML algorithms cannot necessarily explain their outputs.
Non-Melanoma Skin Cancer
New studies have focused on classifying NMSCs or skin cancers that occur on specific regions (i.e., lips or face). NMSCs, such as basal cell carcinoma and squamous cell carcinoma, are the most common cancers in Caucasians [51]. Diagnosing NMSC is a complex classification problem because the differential diagnosis for NMSC includes benign and malignant neoplasms, cysts, and inflammatory diseases. In contrast, determining the diagnosis of melanoma versus benign pigmented nevus is a simpler binary classification problem. In 2019, Tschandl et al. [52] demonstrated that a combined CNN (cCNN) can classify dermoscopic and clinical images of nonpigmented lesions on par with experts, namely 95 human raters of whom 62 were board-certified dermatologists. The authors combined the outputs of two CNNs, one trained with dermoscopic images and the other with clinical images. While the AUROC of the trained cCNN was higher than that of the human raters (0.742 vs. 0.695), the cCNN did not achieve a higher percentage of correct specific diagnoses when compared with experts (37.3 vs. 40%). The classifier exceeded the accuracy of human raters for common nonpigmented skin cancers such as basal cell carcinoma, actinic keratoses, squamous cell carcinoma, and keratoacanthoma, but the classifier was not as accurate as human raters for rare malignant nonpigmented lesions such as amelanotic melanoma and benign nonpigmented lesions. This study is an important example of how the data input into a CNN can determine the accuracy of the CNN’s outputs, as the CNN used in this study was trained on very few images of the rare nonpigmented lesions. Tschandl and colleagues admit that this algorithm is not ready to be implemented in the clinic, but the results of the study do demonstrate that CNNs are capable of more complex diagnoses and call for the collection of more dermoscopic and clinical images of rare malignant lesions. Marka et al. [3] carried out a systematic review of 39 studies on the automated detection of NMSC and found that most studies report model performance greater than or equal to the reported diagnostic accuracy of the average dermatologist, but that relatively few studies have presented a high level of evidence.
Some skin disorders occur more frequently or exclusively on particular areas of the skin. For example, many skin cancers occur on sun-exposed areas such as the face or neck. Early screening and detection of carcinomas that occur on the face is crucial considering the significant impact on quality of life and cosmetic disfiguration if diagnosis is delayed. The targeting of specific regions where skin cancers occur more frequently has been explored in two very recent studies [53, 54]. In one of these studies, Cho et al. [53] focused on classifying lip diseases at a similar level to dermatologists [53]. The CNN performed on par with dermatologists and outperformed non-dermatologists in classifying malignant lip diseases. In the other study, a more recent study by Han et al. [54], R-CNNs were used to detect keratinocyte cancer on the face [54], such that an R-CNN was used to detect the location of the lesion and a traditional CNN was used to classify the lesion. The R-CNN generated 924,538 possible lesions from 182,348 clinical photographs. The CNN was trained on 1,106,886 image crops of the possible lesions. The authors reported an AUC of 0.910, sensitivity of 76.8%, and specificity of 90.6%. Again, the combined performance was on par with dermatologists and outperformed non-dermatologists. R-CNNs have been used in fracture detection in radiology [55] and the detection of the nail plate in onychomycosis [56]. Algorithms could potentially detect and diagnosis skin cancer without any preselection of suspicious lesions by dermatologists.
Other Dermatological Diseases
Deep learning algorithms have also been implicated in classifying other important dermatological diseases. Similar to the accuracy of CNNs used to classify melanomas, a study using a CNN for psoriasis achieved an AUC of 0.981 and outperformed 25 Chinese dermatologists. However, this model was limited to classifying psoriasis located on large areas of exposed skin due to the low quality and lack of scalp and nail psoriasis images. This is a notable limitation considering that the incidence of scalp psoriasis is 45–56% and nail psoriasis is 23–27% among psoriatic patients [57]. CNNs have been created for other diseases, such as atopic dermatitis [58], onychomycosis [56], and rosacea [59]. To classify onychomycosis, Han et al. [56] used a R-CNN to generate a training datasets of 49,567 images of nails and found that a combination of their datasets performed better than dermatologists.
When compared to the number of studies on skin cancer, there is still a significant shortage of research conducted on classifying other types of cutaneous diseases. These diseases may be harder to classify because of greater clinical heterogeneity (i.e., atopic dermatitis) [60], the numerous subtypes (i.e., psoriasis), or more variance in severity. Therefore, some studies have focused on assessing specific features that contribute to the severity of a disease. For example, for psoriasis, dermatologists use the Psoriasis Area and Severity Index (PASI) as the gold standard to assess severity based on body surface area involved, erythema, induration, and scaling. ML models have been developed to singly assess body surface area [61, 62], scaling [63], induration/color [64–67], and erythema only [68] in patients with psoriasis.
Dermatopathology
Deep learning algorithms are useful for classifying images of lesions and histopathological images. Advancements in digital pathology, such as greater computing power and cheaper data storage, have facilitated whole-slide imaging. Whole-slide images allow entire high-resolution slides to be stored permanently in a digital format, making it easier to classify these images using an algorithm. The complexity of examining histopathology was first captured in one study that developed a framework for an unsupervised model to identify learned features of basal cell carcinoma histopathology [69]. Unsupervised models are not yet frequently used in medicine and do not require the input of a clinician to label images for training the model. The unsupervised learning model performed with an AUROC of 98.1% [69]. However, with this approach, explanations for why certain patterns and features chosen by the algorithm that discriminate between cancer and healthy tissue are not always apparent. In some cases, patterns discovered that are thought to be specific for cancer actually identify cell proliferation patterns seen in healthy tissue [69]. Therefore, more recent studies have relied on supervised learning, in which a dermatopathologist labels images to help train the model [70, 71]. These studies highlight the importance of the dermatopathologist in creating models to classify melanocytic lesions. Models trained with image curation done by a dermatopathologist were found to be 50% more accurate and to take significantly less time to train [70]. While most studies have focused on using CNNs to classify whole-slide images, one study has reported that basal cell carcinoma can also be identified by using microscopic ocular images (MOIs) of histopathological tissue sections [72]. MOIs are images taken on a smartphone equipped with a microscope eyepiece. In that study, using transfer learning onto a CNN, the authors study achieved an AUC comparable to classification using whole-slide images [72].
Hekler and colleagues claimed that a CNN which they tested outperformed 11 pathologists in the classification of histopathological melanoma images [73]. In this study, 695 lesions were classified by one expert histopathologist using tissue slides stained with hematoxylin and eosin. These slides were randomly cropped, producing 595 images to train the algorithm and 100 images to validate the algorithm. A questionnaire with 100 randomly cropped images was sent out to dermatologists, with the results showing that 157 dermatologists achieved a mean sensitivity of 74.1% and specificity of 60%. At a mean sensitivity of 74.1%, the CNN achieved a mean specificity of 86.5%. The 157 dermatologists ranged from junior to chief physicians. Chief physicians had the highest mean specificity of 69.2% and a mean sensitivity of 73.3%. At the same mean specificity of 69.2%, the CNN had a mean sensitivity of 84.5% [73].
However, in a reply, Géraud et al. [74] caution us to evaluate these studies with the same metrics that we would use on other double-blind peer-reviewed clinical trials or statistical analyses [74]. These authors point out three problematic limitations to the methods of Hekler and colleagues’s paper. The first limitation was that dermatopathologists were only given 20 min to assess 100 images (12 s per image), which is unrealistic in a clinical setting. In a survey of dermatopathologists, more than 70% of dermatopathologists noted that the quality and clarity of clinical information has a ‘large’ impact on their diagnostic confidence and diagnostic accuracy, and some 44.7% of respondents spent 30 min or more searching for clinical information to assist with their histopathological interpretation [75]. Future research could use natural language processing to comply relevant clinical information for each set of histopathological images to reduce the time spent searching for clinical information.
A second major limitation was that only one histopathologist labeled the images to train the ML model. Between histopathologists there can be 25–26% discordance between diagnoses for cutaneous melanoma and benign lesions [76]. Given this possibility of a significant amount of disagreement between histopathologists, having only one histopathologist label the images used to train the model will magnify both the accuracy and mistakes of that one histopathologist. Géraud et al. [74] suggest that studies should include a larger cohort of images that are labeled by at least three dermatopathologists.
Finally, a major limitation was that cropped images only allow a small, random portion of the lesion to be analyzed. Dermatopathologists make diagnoses by examining the entire lesion—not by examining only a small part of the lesion. Géraud et al. [74] found that 15% of the images used in the trial had no recognizable melanocytic lesion whatsoever, which indicates that these images only contained perilesional normal skin tissue. Hekler et al. [77] also released a similar study a few months earlier with the same methods and model, claiming that their models achieved pathologist-level classification of histopathological melanoma images, but this study still has the same pitfalls.
Most ML methods used to classify histopathology have focused on skin cancer, but there are early studies starting to work on classifying other diseases. CNN can differentiate dermis from epidermis in the histopathology of psoriasis lesions [78]. This is the first step toward developing a ML solution for automatic segmentation and diagnosis of psoriasis pathology. Further research will need to go beyond this basic segmentation and find more specific features, such as detecting changes in the epidermis and the presence of immune and nucleated cells.
Mobile Applications and Personal Monitoring Devices
Mobile applications and personal monitoring combined with ML algorithms hold great potential due to their portability and convenience. Melanoma screening through a mobile application could ultimately increase the accessibility of screening, especially in rural areas with limited availability to dermatologists. There are currently two types of mobile applications for melanoma screening: (1) store-and-forward teledermatology and (2) automated smartphone apps. The store-and-forward teledermatology applications send pictures taken by the patient to a remote dermatologist for evaluation. In contrast, automated smartphone apps use a ML algorithm to determine the probability of malignancy on the spot without consultation from a dermatologist. In one survey, 70% of the patients using a store-and-forward teledermatology program stated they would not have seen a dermatologist without the teledermatology program, indicating that these applications can significantly impact outreach [79]. It is important to note that these applications are not a replacement for a face-to-face consultation, especially since patients may miss key lesions without a full body examination. The authors of a 2018 systematic review report that the sensitivity of these automated applications can range from 7 to 87% and that there is an overall lack of evidence regarding the safety for using these automated smartphone applications [80]. As of 2018, none of the automated smartphone applications for melanoma screening had been approved by the US FDA. Since the publication of the last systematic review of smartphone applications, a study on a smartphone application called SkinVision reported improved results for an algorithm trained on more than 130,000 images by more than 30,000 users [81]. This algorithm achieved a higher sensitivity (95.1 vs. 80.0%) and similar specificity (78.3 vs. 78%) than previously reported [81, 82]. However, these results should still be taken with caution given the lower specificity compared to other experimental deep learning melanoma classifications. The lack of regulation of these applications and potential for false negative/positives makes these applications not adequate for patient use at the present time. In the future, use of these applications under careful physician consultation could allow for patients to better communicate with their healthcare professionals about their skin concerns.
Data from personal monitoring devices can also be useful in dermatology for quantifying pruritus [83] or tracking skin over time [84]. One scientific method for assessing pruritus is through video recording of patients, which is tedious and not practical outside an experimental setting [85]. Using a ML algorithm to analyze data from a wrist actigraphy device to quantify nocturnal scratching could lead to the creation of novel therapies for atopic dermatitis and other pruritic disorders [83].
Facilitating Large-Scale Epidemiology Research
“Big data” is defined by immense and complex datasets for which traditional data processing methods may be inadequate. These large datasets can derive from electronic medical records (EMR), insurance claims, the internet, mobile applications, personal monitoring devices, and omics databases. Big data in dermatology presents a promising hypothesis-generating framework to conduct research by identifying unseen patterns within the data [1]. ML is the perfect tool to analyze and harness the power of this enormous amount of information. This section will evaluate the use of ML in large datasets for epidemiology applications and for understanding patient experiences.
Using EMR data, ML has been used to explore electronic health record-phenotyping [86–88], to conduct population-based analysis [89], and to evaluate patient experiences [90]. NLP has been used to analyze EMR with the aim to identify atopic dermatitis. This method is useful for phenotyping patients for a genome-wide association study. Previous atopic dermatitis phenotyping done by human reviewers of EMR had very high positive predictive value (95.2%) but low sensitivity (8.7%), which limited the number of patients included [91]. Using the ML algorithm for atopic dermatitis phenotyping, they achieved a similar positive predictive value of (84.0%) with a much higher sensitivity (75.0%), confirming that this approach may be effective for developing phenotype algorithms. ML methods can also be used to phenotype rare diseases, such as systemic sclerosis in EMR data. One study found that the highest performing ML methods to phenotype systemic sclerosis incorporated clinical data with billing codes [88]. Another study used NLP to develop the first population-based estimates of melanocytic lesions from EMR pathology reports [89]. Further research can use NLP to explore the epidemiology of other cutaneous diseases using EMR. NLP can be used on other sets of unstructured data. In one study, social media data on Reddit were analyzed using NLP to evaluate dermatology patient experiences and therapeutics [90]. An examination of 176,000 comments suggested the utility of social media data for dermatology research and engagement with the public.
Machine Learning and Precision Medicine
The convergence of ML and stores of big data has fueled the acceleration of the possibilities for precision (also known as personalized or individualized) medicine. The aim of precision medicine is to develop targeted treatments based on data from multi-omics platforms or other phenotypic or psychosocial characteristics to improve clinical outcomes and reduce unnecessary side effects for those less likely to respond to a certain treatment [92]. Studies not using ML have taken the first steps toward precision medicine in dermatology by identifying new genetic biomarkers and showing differential response to therapy based on these biomarkers. However, by harnessing the power of ML, millions of datapoints can be analyzed, thereby expanding the power of its predictive results. Furthermore, genetic biomarkers can be discovered faster than previously possible. Here we discuss in more detail research being conducted on the following cutaneous diseases: psoriasis, psoriatic arthritis, and skin cancer.
Psoriasis
Despite the large amount of clinical trial data on the efficacy of 11 FDA-approved biologics, choosing a biologic for a patient is still based on trial and error. It often takes 12–16 weeks for a clinical response to be meaningful, and the efficacy of the drug can range between a 30 and 80% success rate [93]. This creates an “assessment gap” between a patient’s response to a treatment that may in part be biologically determined and when a response can be clinically determined. Changes in biopsy or gene expression profiles can show an improvement and response to treatment faster than clinical improvement. Therefore, ML can address this assessment gap by predicting the long-term outcomes of biologics in psoriasis patients. Several studies have created ML prediction models to determine the long-term treatment response to biologics [93–96]. The first study to assess this gap created two ML models to examine gene expression data from skin biopsies [93]. The models predicted the PASI 75 response (i.e., a ≥ 75% improvement in PASI score from baseline) after 12 weeks of treatment by evaluating the molecular profile of the short-term (2–4 weeks) treatment. Both of these models predicted the PASI 75 response with high accuracy (AUC > 0.80) and decreased the psoriasis assessment gap by 2 months. Assessment of baseline samples of gene expression from skin biopsies may also be able to predict a patient’s response to biologic therapies, a strategy which was previously thought of as unrealiable [94]. Another study used multi-omics to examine patients with severe psoriasis on etanercept and found indications of treatment response in genes and pathways associated with tumor necrosis factor (TNF) signaling and the major histocompatibility complex [94]. Although the assessment of gene expression data from skin biopsies is a promising method to predict treatment response, skin biopsies are still invasive and expensive. To address this issue, Tomalin et al. [95] created a ML predictive model from blood biochemical measurements rather than skin biopsies. These authors measured the longitudinal profiles for 92 inflammatory and 65 cardiovascular disease proteins at baseline and 4 weeks following the respective treatment. The accuracy of the prediction of the 12-week efficacy endpoint following treatment with tofacitinib or etanercept was AUROC 78% and AUROC 71%, respectively. Interestingly, simple models based on PASI scores performed better than the blood predictive model, which indicates that future studies may need to measure more proteins. A very recent study used six different ML models based on basic health information (i.e., drug discontinuations, adverse events, etc.) to predict long-term response to treatment [96]. The best model of these authors predicted treatment outcomes with 18% classification error, demonstrating the utility of basic clinical information. In addition to identifying the treatment response for psoriasis, ML models can also discover potential off-label treatments for psoriasis, atopic dermatitis, and alopecia areata [97]. This model uses a combination of an unsupervised word embedding model summarized drug information from over 20 million articles and application of classification of disease ML models to identify potential drugs for immune-mediated cutaneous diseases.
One step toward the effective treatment for psoriasis is identifying the predictors of disease and its co-morbidities. Psoriasis is associated with an elevated risk of cardiovascular disease. While coronary plaques can be characterized through coronary computed tomography angiograph, identifying potential risk factors is important for predicting prospective cardiac events. A recent study used ML to identify top predictors of non-calcified coronary burden in psoriasis [98]. These authors identified that obesity, dyslipidemia, and inflammation are important comorbidities/risk factors in atherosclerosis.
Psoriatic Arthritis
Approximately 25% of patients with psoriasis also develop chronic inflammatory arthritis called psoriatic arthritis [99]. There is currently no method to predict the development of psoriatic arthritis in a patient with only psoriasis before symptoms appear. ML could be useful in developing a quantitative assessment for psoriatic arthritis risk among psoriasis patients based on underlying genetic differences. Using data from over 7,000 genotyped psoriatic arthritis and psoriasis patients, Patrick et al. 2018 identified 9 new loci for psoriasis or its subtypes [100]. They used ML methods to differentiate psoriatic arthritis from psoriasis based on 200 genetic markers and achieved an AUROC of 0.82 [100]. This is the first study to show a robust prediction of psoriatic arthritis using only genetic data and presents the first step toward a personalized approach to psoriatic arthritis management.
Skin Cancer
Machine learning has been employed to develop cancer risk models for melanoma [101] and NMSC [102]. The dataset used to create the risk model of melanoma was unique because it encompassed over four million dermatology patients in the USA from a cloud-based dermatology-specific EMR called Modernizing Analytics for Melanoma [101]. Given the vast size of the data, the authors used a hybrid method of distributed computing (using multiple computers to maximize efficiency) and nondistributed computing (using one computer) to analyze the data. The distributed computing method was used for collecting and formatting the data and nondistributed computing was used for ML. A good example of how ML and big data can be used to examine a novel hypothesis is a study done by Roffman et al. in 2018 [102]. While ultraviolet radiation exposure and family history are major associated risk factors for NMSC, these authors aimed to create an ANN to predict personal NMSC risk solely based on 13 parameters of personal health data: gender, age, basal metabolic index, diabetic status, smoking status, emphysema, asthma, Hispanic ethnicity, hypertension, heart diseases, vigorous exercise habits, and history of stroke. Given that the model performed with an AUROC of 0.81 without any evaluation of the major associated risk factors or images, it has the potential for improving the diagnosis and management of NMSC. Another study predicted the likelihood of the development of NMSC by analyzing two million randomly sampled patients from the Taiwan National Health Insurance Research Database [103]. A CNN analyzed 3 years of clinical diagnostic information [i.e., International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnosis and procedure codes and prescriptions; https://www.cdc.gov/nchs/icd/icd9cm.htm] and temporal-sequential information (i.e., dates of clinical visits and days of prescriptions) to predict the development of NMSC of a given patient within the next year and achieved an AUROC of 0.89 [103].
Discussion
In this review, we have summarized the basic principles of ML and its current applications in dermatology. We have reviewed that ML has numerous potential applications in the dermatologist’s workflow from diagnosis to treatment. Based on the literature, we have identified five areas of applications of ML in dermatology (Fig. 2).
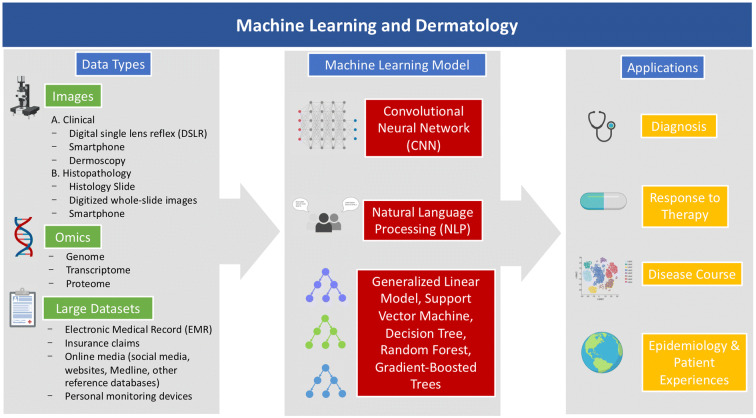
Fig. 2.
Applications of machine learning in dermatology. Flowchart demonstrating the various sources of data in dermatology, machine learning models, and potential applications. Icons were created with the web-based program BioRender (https://biorender.com)
All five areas have benefited from the advent of powerful deep learning algorithms that can analyze large datasets. In the first area, deep learning algorithms are helpful for identifying melanoma, NMSC, and other dermatological diseases from dermoscopic, DSLR, and smartphone images. Classification algorithms for melanoma and NMSC are the most well-studied ML algorithms in dermatology and can potentially address low skin cancer screening rates by increasing access. Classification of other diseases is still in its nascency and will likely involve more complex algorithms to grade disease severity and produce accurate differential diagnoses. In the second area, deep learning to classify histopathology is also still in its early stages and has underscored the importance of involving dermatologists and dermatopathologists in the development of ML studies, as the models perform better with image curation done by dermatopathologists. Most of these studies were published without a dermatologist listed as an author, which suggests that more dermatologists should be involved in the development of these ML models. In the third area, mobile applications for classifying lesions and skin cancer are unregulated and are not currently not sufficiently accurate or sensitive to serve as useful screening tools. Once classification algorithms are more accurate and have been properly clinically validated, mobile applications can disseminate these screening tools to populations in need. In the fourth area, ML can analyze large datasets, such as EMR data and insurance claims, that are useful for epidemiological studies. Lastly, ML can be useful as a tool to predict treatment response and supplement diagnosis for patients with psoriasis, psoriatic arthritis, and skin cancer.
Machine Learning: Limitations and Considerations
Before any of these technologies are implemented in a real-life setting, it is critical to discuss considerations and limitations for the development of these technologies (Fig. 3). These algorithms are useful for making specialty diagnoses more accessible in areas where there is a paucity of dermatologists. However, the accuracy of these algorithms is hard to determine when they are used without any physician input. A major limitation of ML is that it is hard to explain how these algorithms come to their conclusions. A ML algorithm can be compared to a black box that takes in inputs and produces outputs with no explanation of how it produced the outputs. If an algorithm misdiagnoses a malignant lesion, the algorithm cannot explain why it chooses a certain diagnosis. While the outputs can be helpful, if the model is unable to explain to a patient why it diagnosed a lesion as malignant versus benign or how it chose a particular therapy, it is potentially dangerous and problematic for the patient. Physician interpretation is necessary to explain why a diagnosis or treatment should be chosen. In addition to the black box nature of these algorithms, ML is also prone to the maxim “garbage in, garbage out.” This maxim indicates that the quality of the dataset input determines the quality of the output. Therefore, if these images’ inputs are poorly labeled, then the algorithm’s outputs will reflect these inaccuracies.
Fig. 3.
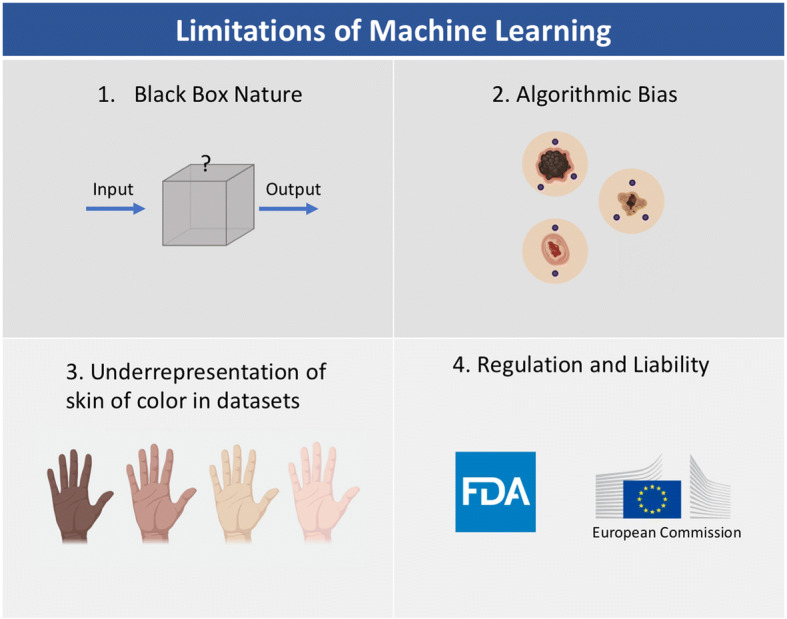
Limitations of machine learning. Icons were created with the web-based program BioRender (https://biorender.com)
Regarding the impact of bias on clinical decisions for patient care, it is also crucial to consider how bias in algorithms can potentially impact millions of patients. As noted earlier, artifacts, such as pen markings, air bubbles, or hairs, can interfere with the algorithm’s correct diagnosis by falsely associating these markings with the prevalence of a disorder [48]; this limitation reinforces the black box nature of ML. The outcomes decided by the algorithm are not always based on clinical evidence. Therefore, ML appears to be a useful tool to supplement physician diagnosis and treatment, but it should not replace decisions made by physicians based on clinical evidence.
As a field, we should work to make this technology more accessible and be aware of other potential biases that could exacerbate current health disparities. Almost all of these studies reported in this review focus on images and data from Caucasian patients from North America and Europe, with only a few studies focusing on images of Korean, Chinese, or Japanese patients in Asia. It was even noted that studies trained on images of Asian patients performed worse on Caucasian patients [31, 47]. Applying algorithms trained on images of fair skin could be inaccurate on skin of color as many cutaneous diseases manifest differently in skin of color; for example, psoriasis may appear more violaceous and less red in individuals with darker skin than in Caucasian patients. Early screening of skin cancer could have a significant benefit for patients with darker skin who currently experience more advanced disease and lower survival rates due to delays in diagnosis [104, 105]. Since ML models are still in their nascency, we have the opportunity to improve these studies by making sure this technology can be inclusive to patients of all ethnic and racial backgrounds.
Regulation and Liability in Machine Learning
Given the myriad of applications of ML in dermatology, it will be important for clinicians to be involved in determining the appropriate regulation for these devices. In the USA, any changes or modifications to medical devices are typically approved by the US FDA through a supplement to premarket approval or as a new 510(k) submission. However, deep learning algorithms update and change in real time as they are exposed to more clinical examples and experiences. Therefore, as the algorithms continuously update, the outputs could differ from what was initially approved by the FDA.
Continuous software iterations in ML devices will require the development of more specific regulatory approval than that used for traditional physical devices. The first ML-based software approved by the FDA was in 2018 for a program that diagnosed diabetic retinopathy without clinician interpretation. The FDA has only authorized “locked” ML devices, meaning that they do not continually learn or adapt the algorithm in real time. In April 2019, the FDA announced that it is working on a new regulatory framework for modifications to ML-based software or “unlocked” ML algorithms [106]. This framework proposes a “predetermined change control plan” that includes types of anticipated modifications and the associated methodologies being used to incorporate those changes while managing risks to patients. This proposal is not the final regulatory expectation but is gathering input from a wide variety of groups and individuals to draft appropriate guidelines. Interestingly, Hwang et al. [107] noted that 11 of the 14 devices approved by the FDA between 2017 and 2018 were through the 510[k] pathway which only requires “substantial equivalence” to an already-marketed device. This means that these devices are deemed as “moderate risk” products and are not required to have clinical testing. A lifecycle-based framework for regulating ML-based software will be important for ensuring the safety and efficacy of these devices. In addition, all devices should be evaluated in prospective clinical trials and made publicly available in peer-reviewed literature.
Another potential area of concern is the uncharted territory of determining the liability of using a ML device for an erroneous decision in patient care. Currently, physicians are protected as long as they follow “standard of care,” but as ML becomes more accurate it may become the “standard of care” over previous practices. According to Price et al. [108], the safest way to use ML is to use it only as a confirmatory tool to support existing decision-making processes and to check with individual malpractice insurers. Physicians are likely to influence how ML is used in practice and when it should be applied in place of human decision.
Conclusions
Machine learning presents a tremendous potential in dermatology, from diagnosis to predicting more effective and safer treatments. As this technology advances, dermatologists will need to gain an understanding of how ML works, along with when and how it should be appropriately used in a clinical setting. While ML methods are powerful, they are still similar to previous clinical tools in that physician interpretation is crucial for implementation in a real-world setting. We should also be cognizant of how potential biases can interfere with the black box nature of these algorithms. It is also important to make these technologies inclusive of skin of color. Further research in ML should be transparent by making algorithms and datasets available to the public for further validation and testing. Before coming to market, rigorous peer-reviewed prospective clinical trials should be conducted. Overall, involving more dermatologists in the development and testing of ML is imperative for creating useful and clinically relevant technology.
Acknowledgements
Funding
No funding or sponsorship was received for the publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Stephanie Chan, Vidhatha Reddy, Bridget Myers, Quinn Thibodeaux, Nicholas Brownstone have nothing to disclose. Wilson Liao is a member of the journal’s Editorial Board.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article go to 10.6084/m9.figshare.12006789.
References
- 1.Wehner MR, Levandoski KA, Kulldorff M, Asgari MM. Research techniques made simple: an introduction to use and analysis of big data in dermatology. J Investig Dermatol. 2017;137:e153–e58. doi: 10.1016/j.jid.2017.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston A, Sarkar MK, Vrana A, Tsoi LC, Gudjonsson JE. The molecular revolution in cutaneous biology: the era of global transcriptional analysis. J Investig Dermatol. 2017;137:e87–91. doi: 10.1016/j.jid.2016.02.817. [DOI] [PubMed] [Google Scholar]
- 3.Marka A, Carter JB, Toto E, Hassanpour S. Automated detection of nonmelanoma skin cancer using digital images: a systematic review. BMC Med Imaging. 2019;19:21. doi: 10.1186/s12880-019-0307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomsen K, Iversen L, Titlestad TL, Winther O. Systematic review of machine learning for diagnosis and prognosis in dermatology. J Dermatol Treat. 2019;31:1–15. doi: 10.1080/09546634.2019.1682500. [DOI] [PubMed] [Google Scholar]
- 5.Nichols JA, Herbert Chan HW, Baker MAB. Machine learning: applications of artificial intelligence to imaging and diagnosis. Biophys Rev. 2018;11:111–118. doi: 10.1007/s12551-018-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turing AM. Computing machinery and intelligence. Mind. Dordrecht: Springer; 1950. [Google Scholar]
- 7.Murphy KP. Machine learning: a probabilistic perspective. Cambridge: MIT Press; 2012. [Google Scholar]
- 8.Silver D, Huang A, Maddison CJ, et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529:484–489. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 9.Coomans D, Massart DL. Alternative k-nearest neighbour rules in supervised pattern recognition: part 1 k-Nearest neighbour classification by using alternative voting rules. Anal Chim Acta. 1982;136:15–27. doi: 10.1016/S0003-2670(01)95359-0. [DOI] [Google Scholar]
- 10.Altman NS. An introduction to kernel and nearest-neighbor nonparametric regression. Am Stat. 1992;46:175–185. doi: 10.1080/00031305.1992.10475879. [DOI] [Google Scholar]
- 11.Hearst MA, Dumais ST, Osuna E, Platt J, Scholkopf B. Support vector machines. IEEE Intell Syst Appl. 1998;13:18–28. doi: 10.1109/5254.708428. [DOI] [Google Scholar]
- 12.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 13.Manning CD, Schütze H. Foundations of statistical natural language processing. Cambridge: MIT Press; 1999. [Google Scholar]
- 14.Zou J, Han Y, So S-S. Overview of artificial neural networks. In: Livingstone DJ, editor. Artificial neural networks: methods and applications. Totowa: Humana Press; 2009. [Google Scholar]
- 15.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 16.Fei-Fei L, Fergus R, Perona P. Learning generative visual models from few training examples: an incremental Bayesian approach tested on 101 object categories. In: 2004 Conference on Computer Vision and Pattern Recognition Workshop. Washington, DC. 2004, p. 178. https://ieeexplore.ieee.org/document/1384978. Accessed 18 Dec 2019.
- 17.Griffin G, Holub A, Perona P. Caltech-256 Object Category Dataset. 2007. https://resolver.caltech.edu/CaltechAUTHORS:CNS-TR-2007-001. Accessed 20 Feb 2020.
- 18.LeCun Y, Fu Jie Huang, Bottou L. Learning methods for generic object recognition with invariance to pose and lighting. In: Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Washington, DC. 2004, p. 2–104. https://ieeexplore.ieee.org/document/1315150. Accessed 18 Dec 2019.
- 19.Marchetti MA, Codella NCF, Dusza SW, et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: Comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(270–277):e1. doi: 10.1016/j.jaad.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Chen P-HC, Krause J, Peng L. How to read articles that use machine learning: users’ guides to the medical literature. JAMA Am Med Assoc. 2019;322:1806–1816. doi: 10.1001/jama.2019.16489. [DOI] [PubMed] [Google Scholar]
- 22.Abràmoff MD, Lou Y, Erginay A, et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Invest Ophthalmol Vis Sci. 2016;57:5200–5206. doi: 10.1167/iovs.16-19964. [DOI] [PubMed] [Google Scholar]
- 23.Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digit Med. 2018;1:1–8. doi: 10.1038/s41746-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Codella N, Cai J, Abedini M, Garnavi R, Halpern A, Smith JR. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In: Zhou L, Wang L, Wang Q, Shi Y, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2015. pp. 118–126. [Google Scholar]
- 25.Pomponiu V, Nejati H, Cheung N-M, et al. Deepmole: Deep neural networks for skin mole lesion classification. In: 2016 IEEE International Conference on Image Processing (ICIP). Phoenix, AZ; 2016. p. 2623–27. https://ieeexplore.ieee.org/abstract/document/7532834. Accessed 18 Dec 2019.
- 26.Kawahara J, BenTaieb A, Hamarneh G. Deep features to classify skin lesions. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). Prague, Czech Republic; 2016, p. 1397–400. https://ieeexplore.ieee.org/document/7493528. Accessed 18 Dec 2019.
- 27.Arnold M, Holterhues C, Hollestein LM, et al. Trends in incidence and predictions of cutaneous melanoma across Europe up to 2015. J Eur Acad Dermatol Venereol. 2014;28:1170–1178. doi: 10.1111/jdv.12236. [DOI] [PubMed] [Google Scholar]
- 28.Johnson MM, Leachman SA, Aspinwall LG, et al. Skin cancer screening: recommendations for data-driven screening guidelines and a review of the US Preventive Services Task Force controversy. Melanoma Manag. 2017;4:13–37. doi: 10.2217/mmt-2016-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coups EJ, Geller AC, Weinstock MA, Heckman CJ, Manne SL. Prevalence and correlates of skin cancer screening among middle-aged and older white adults in the United States. Am J Med. 2010;123:439–445. doi: 10.1016/j.amjmed.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. 2018;138:1529–1538. doi: 10.1016/j.jid.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 32.Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29:1836–1842. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 33.Kawahara J, Hamarneh G. Multi-resolution-tract CNN with hybrid pretrained and skin-lesion trained layers. In: Wang L, Adeli E, Wang Q, Shi Y, Suk H-I, editors. Machine learning in medical imaging. Cham: Springer International Publishing; 2016. pp. 164–171. [Google Scholar]
- 34.Sun X, Yang J, Sun M, Wang K. A benchmark for automatic visual classification of clinical skin disease images. In: Leibe B, Matas J, Sebe N, Welling M, editors. Computer vision ECCV 2016. Cham: Springer International Publishing; 2016. pp. 206–222. [Google Scholar]
- 35.Romero-Lopez A, Giro-i-Nieto X, Burdick J, Marques O. Skin lesion classification from dermoscopic images using deep learning techniques. Calgary: ACTA Press. 2017. https://www.actapress.com/PaperInfo.aspx?paperId=456417. Accessed 18 Dec 2019.
- 36.Brinker TJ, Hekler A, Enk AH, et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer. 2019;111:148–154. doi: 10.1016/j.ejca.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Cui X, Wei R, Gong L, et al. Assessing the effectiveness of artificial intelligence methods for melanoma: a retrospective review. J Am Acad Dermatol. 2019;81:1176–1180. doi: 10.1016/j.jaad.2019.06.042. [DOI] [PubMed] [Google Scholar]
- 38.Tschandl P, Akay BN, Argenziano G. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 2019;20:938–947. doi: 10.1016/S1470-2045(19)30333-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dascalu A, David EO. Skin cancer detection by deep learning and sound analysis algorithms: a prospective clinical study of an elementary dermoscope. EBioMedicine. 2019;43:107–113. doi: 10.1016/j.ebiom.2019.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen SC, Bravata DM, Weil E, Olkin I. A comparison of dermatologists’ and primary care physicians’ accuracy in diagnosing melanoma: a systematic review. Arch Dermatol. 2001;137:1627–1634. doi: 10.1001/archderm.137.12.1627. [DOI] [PubMed] [Google Scholar]
- 41.Brinker TJ, Hekler A, Enk AH, et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur J Cancer. 2019;113:47–54. doi: 10.1016/j.ejca.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Brinker TJ, Hekler A, Enk AH, et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer. 2019;119:11–17. doi: 10.1016/j.ejca.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 43.Hekler A, Utikal JS, Enk AH, et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur J Cancer. 2019;120:114–121. doi: 10.1016/j.ejca.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Kharazmi P, Kalia S, Lui H, Wang ZJ, Lee TK. A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile. Skin Res Technol. 2018;24:256–264. doi: 10.1111/srt.12422. [DOI] [PubMed] [Google Scholar]
- 45.Yap J, Yolland W, Tschandl P. Multimodal skin lesion classification using deep learning. Exp Dermatol. 2018;27:1261–1267. doi: 10.1111/exd.13777. [DOI] [PubMed] [Google Scholar]
- 46.Kim GK, Del Rosso JQ, Bellew S. Skin cancer in Asians: part 1: nonmelanoma skin cancer. J Clin Aesthet Dermatol. 2009;2:39–42. [PMC free article] [PubMed] [Google Scholar]
- 47.Navarrete-Dechent C, Dusza SW, Liopyris K, Marghoob AA, Halpern AC, Marchetti MA. Automated dermatological diagnosis: hype or reality? J Invest Dermatol. 2018;138:2277–2279. doi: 10.1016/j.jid.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips M, Marsden H, Jaffe W, et al. Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw Open. 2019;2:e1913436. doi: 10.1001/jamanetworkopen.2019.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okur E, Turkan M. A survey on automated melanoma detection. Eng Appl Artif Intell. 2018;73:50–67. doi: 10.1016/j.engappai.2018.04.028. [DOI] [Google Scholar]
- 50.Winkler JK, Fink C, Toberer F, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. 2019 doi: 10.1001/jamadermatol.2019.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol. 2014;810:120–140. doi: 10.1007/978-1-4939-0437-2_7. [DOI] [PubMed] [Google Scholar]
- 52.Tschandl P, Rosendahl C, Akay BN, et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. 2019;155:58–65. doi: 10.1001/jamadermatol.2018.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho SI, Sun S, Mun J-H, et al. Dermatologist-level classification of malignant lip diseases using a deep convolutional neural network. Br J Dermatol. 2019 doi: 10.1111/bjd.18459. [DOI] [PubMed] [Google Scholar]
- 54.Han SS, Moon IJ, Lim W, et al. Keratinocytic skin cancer detection on the face using region-based convolutional neural network. JAMA Dermatol. 2019;156:29–37. doi: 10.1001/jamadermatol.2019.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thian YL, Li Y, Jagmohan P, Sia D, Chan VEY, Tan RT. Convolutional neural networks for automated fracture detection and localization on wrist radiographs. Radiology. 2019;1:e180001. doi: 10.1148/ryai.2019180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han SS, Park GH, Lim W, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS ONE. 2018;13:e0191493. doi: 10.1371/journal.pone.0191493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dopytalska K, Sobolewski P, Błaszczak A, Szymańska E, Walecka I. Psoriasis in special localizations. Reumatologia. 2018;56:392–398. doi: 10.5114/reum.2018.80718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Guzman LC, Maglaque RPC, Torres VMB, Zapido SPA, Cordel MO. Design and evaluation of a multi-model, multi-level artificial neural network for eczema skin lesion detection. In: 2015 3rd International Conference on Artificial Intelligence, Modelling and Simulation (AIMS). Kota Kinabalu, Malaysia; 2015, p. 42–7. https://ieeexplore.ieee.org/document/7604549. Accessed 18 Dec 2019.
- 59.Binol H, Plotner A, Sopkovich J, Kaffenberger B, Niazi MKK, Gurcan MN. Ros-NET: a deep convolutional neural network for automatic identification of rosacea lesions. Skin Res Technol. 2019 doi: 10.1111/srt.12817. [DOI] [PubMed] [Google Scholar]
- 60.Deleuran M, Vestergaard C. Clinical heterogeneity and differential diagnosis of atopic dermatitis. Br J Dermatol. 2014;170(Suppl 1):2–6. doi: 10.1111/bjd.12933. [DOI] [PubMed] [Google Scholar]
- 61.Meienberger N, Anzengruber F, Amruthalingam L, et al. Observer-independent assessment of psoriasis affected area using machine learning. J Eur Acad Dermatol Venereol. 2019 doi: 10.1111/jdv.16002. [DOI] [PubMed] [Google Scholar]
- 62.Fadzil MHA, Ihtatho D, Affandi AM, Hussein SH. Area assessment of psoriasis lesions for PASI scoring. J Med Eng Technol. 2009;33:426–436. doi: 10.1080/07434610902744066. [DOI] [PubMed] [Google Scholar]
- 63.Lu J, Kazmierczak E, Manton JH, Sinclair R. Automatic segmentation of scaling in 2-D psoriasis skin images. IEEE Trans Med Imaging. 2013;32:719–730. doi: 10.1109/TMI.2012.2236349. [DOI] [PubMed] [Google Scholar]
- 64.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Exploring the color feature power for psoriasis risk stratification and classification: a data mining paradigm. Comput Biol Med. 2015;65:54–68. doi: 10.1016/j.compbiomed.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 65.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput Method Programs Biomed. 2016;126:98–109. doi: 10.1016/j.cmpb.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 66.Shrivastava VK, Londhe ND, Sonawane RS, Suri JS. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput Methods Programs Biomed. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 67.George YM, Aldeen M, Garnavi R. Automatic scale severity assessment method in psoriasis psoriasis skin images using local descriptors. IEEE J Biomed Health Inform. 2019;24:577–585. doi: 10.1109/JBHI.2019.2910883. [DOI] [PubMed] [Google Scholar]
- 68.George Y, Aldeen M, Garnavi R. Psoriasis image representation using patch-based dictionary learning for erythema severity scoring. Comput Med Imaging Graph. 2018;66:44–55. doi: 10.1016/j.compmedimag.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 69.Arevalo J, Cruz-Roa A, Arias V, Romero E, González FA. An unsupervised feature learning framework for basal cell carcinoma image analysis. Artif Intell Med. 2015;64:131–145. doi: 10.1016/j.artmed.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Hart SN, Flotte W, Norgan AP, et al. Classification of melanocytic lesions in selected and whole-slide images via convolutional neural networks. J Pathol Inform. 2019;10:5. doi: 10.4103/jpi.jpi_32_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsen TG, Feeser TA, Kent MN. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. 2018;9:32. doi: 10.4103/jpi.jpi_31_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang YQ, Xiong JH, Li HY, et al. Recognizing basal cell carcinoma on smartphone-captured digital histopathology images with a deep neural network. Br J Dermatol. 2019;182:754–762. doi: 10.1111/bjd.18026. [DOI] [PubMed] [Google Scholar]
- 73.Hekler A, Utikal JS, Enk AH, et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur J Cancer. 2019;118:91–96. doi: 10.1016/j.ejca.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 74.Géraud C, Griewank KG. Re: Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur J Cancer. 2019. https://www.sciencedirect.com/science/article/pii/S0959804919307415 [DOI] [PubMed]
- 75.Comfere NI, Peters MS, Jenkins S, Lackore K, Yost K, Tilburt J. Dermatopathologists’ concerns and challenges with clinical information in the skin biopsy requisition form: a mixed methods study. J Cutan Pathol. 2015;42:333–345. doi: 10.1111/cup.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lodha S, Saggar S, Celebi JT, Silvers DN. Discordance in the histopathologic diagnosis of difficult melanocytic neoplasms in the clinical setting. J Cutan Pathol. 2008;35:349–352. doi: 10.1111/j.1600-0560.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 77.Hekler A, Utikal JS, Enk AH, et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur J Cancer. 2019;115:79–83. doi: 10.1016/j.ejca.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 78.Pal A, Garain U, Chandra A, Chatterjee R, Senapati S. Psoriasis skin biopsy image segmentation using Deep Convolutional Neural Network. Comput Methods Programs Biomed. 2018;159:59–69. doi: 10.1016/j.cmpb.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 79.Cazzaniga S, Castelli E, Di Landro A, et al. Mobile teledermatology for melanoma detection: assessment of the validity in the framework of a population-based skin cancer awareness campaign in northern Italy. J Am Acad Dermatol. 2019;81:257–260. doi: 10.1016/j.jaad.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 80.Rat C, Hild S, Rault Sérandour J, et al. Use of smartphones for early detection of melanoma: systematic review. J Med Internet Res. 2018;20:e135. doi: 10.2196/jmir.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Udrea A, Mitra GD, Costea D, et al. Accuracy of a smartphone application for triage of skin lesions based on machine learning algorithms. J Eur Acad Dermatol Venereol. 2019;34:648–655. doi: 10.1111/jdv.15935. [DOI] [PubMed] [Google Scholar]
- 82.Thissen M, Udrea A, Hacking M, von Braunmuehl T, Ruzicka T. mHealth App for risk assessment of pigmented and nonpigmented skin lesions—-a study on sensitivity and specificity in detecting malignancy. Telemed J E Health. 2017;23:948–954. doi: 10.1089/tmj.2016.0259. [DOI] [PubMed] [Google Scholar]
- 83.Moreau A, Anderer P, Ross M, Cerny A, Almazan TH, Peterson B. Detection of nocturnal scratching movements in patients with atopic dermatitis using accelerometers and recurrent neural networks. IEEE J Biomed Health Inform. 2018;22:1011–1018. doi: 10.1109/JBHI.2017.2710798. [DOI] [PubMed] [Google Scholar]
- 84.Li Y, Esteva A, Kuprel B, Novoa R, Ko J, Thrun S. Skin cancer detection and tracking using data synthesis and deep learning. In: Workshops at the Thirty-First AAAI Conference on Artificial Intelligence. San Francisco, CA; 2017. https://www.aaai.org/ocs/index.php/WS/AAAIW17/paper/view/15087. Accessed 26 Dec 2019.
- 85.Ebata T, Aizawa H, Kamide R, Niimura M. The characteristics of nocturnal scratching in adults with atopic dermatitis. Br J Dermatol. 1999;141:82–86. doi: 10.1046/j.1365-2133.1999.02924.x. [DOI] [PubMed] [Google Scholar]
- 86.Gustafson E, Pacheco J, Wehbe F, Silverberg J, Thompson W. A machine learning algorithm for identifying atopic dermatitis in adults from electronic health records. IEEE Int Conf Healthc Inform. 2017;2017:83–90. doi: 10.1109/ICHI.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eide MJ, Tuthill JM, Krajenta RJ, Jacobsen GR, Levine M, Johnson CC. Validation of claims data algorithms to identify nonmelanoma skin cancer. J Invest Dermatol. 2012;132:2005–2009. doi: 10.1038/jid.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jamian L, Wheless L, Crofford LJ, Barnado A. Rule-based and machine learning algorithms identify patients with systemic sclerosis accurately in the electronic health record. Arthritis Res Ther. 2019;21:305. doi: 10.1186/s13075-019-2092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lott JP, Boudreau DM, Barnhill RL, et al. Population-based analysis of histologically confirmed melanocytic proliferations using natural language processing. JAMA Dermatol. 2018;154:24. doi: 10.1001/jamadermatol.2017.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okon E, Rachakonda V, Hong HJ, Callison-Burch C, Lipoff J. Natural language processing of reddit data to evaluate dermatology patient experiences and therapeutics. J Am Acad Dermatol. 2019 doi: 10.1016/j.jaad.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 91.Hsu DY, Dalal P, Sable KA, et al. Validation of international classification of disease ninth revision codes for atopic dermatitis. Allergy. 2017;72:1091–1095. doi: 10.1111/all.13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jameson JL, Longo DL. Precision medicine: personalized, problematic, and promising. N Engl J Med. 2015;372:2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 93.Correa da Rosa J, Kim J, Tian S, Tomalin LE, Krueger JG, Suárez-Fariñas M. Shrinking the psoriasis assessment gap: early gene-expression profiling accurately predicts response to long-term treatment. J Invest Dermatol. 2017;137:305–312. doi: 10.1016/j.jid.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 94.Foulkes AC, Watson DS, Carr DF, et al. A framework for multi-omic prediction of treatment response to biologic therapy for psoriasis. J Invest Dermatol. 2019;139:100–107. doi: 10.1016/j.jid.2018.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomalin LE, Kim J, Correa da Rosa J, et al. Early quantification of systemic inflammatory-proteins predicts long-term treatment response to Tofacitinib and Etanercept: Psoriasis response predictions using blood. J Invest Dermatol. 2019 doi: 10.1016/j.jid.2019.09.023. [DOI] [PubMed] [Google Scholar]
- 96.Emam S, Du AX, Surmanowicz P, Thomsen SF, Greiner R, Gniadecki R. Predicting the long-term outcomes of biologics in psoriasis. Br J Dermatol. 2019 doi: 10.1111/bjd.18741. [DOI] [PubMed] [Google Scholar]
- 97.Patrick MT, Raja K, Miller K, et al. Drug repurposing prediction for immune-mediated cutaneous diseases using a word-embedding–based machine learning approach. J Invest Dermatol. 2019;139:683–691. doi: 10.1016/j.jid.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Munger E, Choi H, Dey AK, et al. Application of machine learning to determine top predictors of non-calcified coronary burden in psoriasis. J Am Acad Dermatol. 2019 doi: 10.1016/j.jaad.2019.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alinaghi F, Calov M, Kristensen LE, et al. Prevalence of psoriatic arthritis in patients with psoriasis: a systematic review and meta-analysis of observational and clinical studies. J Am Acad Dermatol. 2019;80(251–265):e19. doi: 10.1016/j.jaad.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 100.Patrick MT, Stuart PE, Raja K, et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat Commun. 2018;9:4178. doi: 10.1038/s41467-018-06672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richter AN, Khoshgoftaar TM. Efficient learning from big data for cancer risk modeling: a case study with melanoma. Comput Biol Med. 2019;110:29–39. doi: 10.1016/j.compbiomed.2019.04.039. [DOI] [PubMed] [Google Scholar]
- 102.Roffman D, Hart G, Girardi M, Ko CJ, Deng J. Predicting non-melanoma skin cancer via a multi-parameterized artificial neural network. Sci Rep. 2018;8:1701. doi: 10.1038/s41598-018-19907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H-H, Wang Y-H, Liang C-W, Li Y-C. Assessment of deep learning using nonimaging information and sequential medical records to develop a prediction model for nonmelanoma skin cancer. JAMA Dermatol. 2019 doi: 10.1001/jamadermatol.2019.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cormier JN, Xing Y, Ding M, et al. Ethnic differences among patients with cutaneous melanoma. Arch Intern Med. 2006;166:1907–1914. doi: 10.1001/archinte.166.17.1907. [DOI] [PubMed] [Google Scholar]
- 105.Ward-Peterson M, Acuña JM, Alkhalifah MK, et al. Association between race/ethnicity and survival of melanoma patients in the United States over 3 decades: a secondary analysis of SEER data. Medicine. 2016;95:e3315. doi: 10.1097/MD.0000000000003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.US Food and Drug Administration. Artificial intelligence and machine learning in software as a medical device. 2019. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device. Accessed 17 Jan 2020.
- 107.Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle regulation of artificial intelligence and machine learning-based software devices in medicine. JAMA. 2019;322:2285–2286. doi: 10.1001/jama.2019.16842. [DOI] [PubMed] [Google Scholar]
- 108.Price WN, Gerke S, Cohen IG. potential liability for physicians using artificial intelligence. JAMA. 2019;322:1765–1766. doi: 10.1001/jama.2019.15064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.