Significance
A user-friendly workflow, termed Tomahto, enables real-time targeted pathway proteomics assays using two-dimensional multiplexing technology. Hundreds of proteins of interest from a multitude of samples can be targeted and accurately quantified in a remarkably sensitive fashion. We highlight Tomahto’s ease of use, sensitivity, and accuracy and present proof-of-principle utility by targeting 260 metabolism- and inflammation-related proteins across 90 samples (nine tissues from five old and five young mice). Tissue-specific aging effects are presented with specific interest in metabolic and inflammatory pathways of white adipose tissue. We validated our approach through comparison with a global proteome survey across the tissues, work that we also provide as a general resource for the community.
Keywords: targeted pathway proteomics, tissue-specific aging, isobaric labeling, real-time instrument control, Tomahto
Abstract
Pathway proteomics strategies measure protein expression changes in specific cellular processes that carry out related functions. Using targeted tandem mass tags-based sample multiplexing, hundreds of proteins can be quantified across 10 or more samples simultaneously. To facilitate these highly complex experiments, we introduce a strategy that provides complete control over targeted sample multiplexing experiments, termed Tomahto, and present its implementation on the Orbitrap Tribrid mass spectrometer platform. Importantly, this software monitors via the external desktop computer to the data stream and inserts optimized MS2 and MS3 scans in real time based on an application programming interface with the mass spectrometer. Hundreds of proteins of interest from diverse biological samples can be targeted and accurately quantified in a sensitive and high-throughput fashion. It achieves sensitivity comparable to, if not better than, deep fractionation and requires minimal total sample input (∼10 µg). As a proof-of-principle experiment, we selected four pathways important in metabolism- and inflammation-related processes (260 proteins/520 peptides) and measured their abundance across 90 samples (nine tissues from five old and five young mice) to explore effects of aging. Tissue-specific aging is presented here and we highlight the role of inflammation- and metabolism-related processes in white adipose tissue. We validated our approach through comparison with a global proteome survey across the tissues, work that we also provide as a general resource for the community.
In proteomics, both targeted and untargeted approaches are employed depending on the experiment’s goal (1). While the untargeted approach is widely used in a discovery phase to identify potential protein signatures that underlie cellular processes (2), it often requires substantially more starting material, extensive sample fractionation, and more instrument time relative to targeted approaches (3, 4). It also can suffer from lower sampling reproducibility due to its stochastic nature. In contrast, targeted approaches are favored if a preselected set of proteins needs to be consistently measured across a large cohort of samples, such as in clinical settings, or when sample amount is limited (5). To further improve throughput and reduce quantitative variability, two-dimensional (2D) multiplexing (i.e., multiplexing peptide targets and multiplexing samples) can be achieved via isobaric labeling strategies, such as using tandem mass tags (TMT) (6). Although the quantitative accuracy using isobaric tags can be distorted by cofragmenting peptide species within the isolation window (7, 8), it can be readily restored by using synchronous precursor selection and MS3 scans (SPS-MS3) (9). Previously, our laboratory introduced a 2D multiplexing method (triggered by offset, multiplexed, accurate-mass, high-resolution, and absolute quantification—TOMAHAQ), which incorporates TMT and SPS-MS3 with targeted proteomics. The method exploits the use of synthetic trigger peptides, which completely obviates the need for peptide detection prior to quantification. However, the method requires a very cumbersome method structure, limiting its application (10, 11).
High-throughput and precise quantitation of protein pathways is essential for systematic understanding the molecular basis for complex physiological adaptation. This is perhaps best exemplified in the case of chronic physiological changes that occur with age. Tissue aging is characterized by a homeostatic decline in a variety of physiological and psychological functions and is a major risk factor in many diseases. There has been a continuous effort to understand the underlying cellular and molecular causes, and various hypotheses exist (12). Changes in the genome, epigenome, and transcriptome have all been observed and, as a result, genome instability, gene methylation levels, and transcriptional changes have all been implicated in aging (13–15). In addition, metabolism remodeling, inflammatory response, and the homeostasis of multiple organelles were frequently implicated (16, 17). However, little work has been done to investigate proteome changes (18, 19) and, surprisingly, only a minimal disturbance was reported in the tissues studied (20). In addition to a lack of proteomic characterization in aging organisms, current knowledge suggests different tissues in an organism share common altered pathways due to aging but also have their own unique responses which necessitate tissue-specific investigation (21, 22). For example, Christensen et al. found strikingly different gene methylation profiles in multiple human tissues (21) and Hudgins et al. reported highly variable expression of senescence biomarkers in age- and tissue-specific manners (23).
In this paper we present an application programming interface (API)-based algorithm, termed Tomahto, which enables real-time instrument control and decision making. Tomahto provides an array of functionalities including MS1 peak detection, MS2 real-time peak matching (RTPM), MS2 fragmentation pattern match, SPS ion purity filter, MS3 automatic gain control, MS3 quant scan insertion, and target peptide closeout. Remarkably, the only required input from a user is a list of peptide targets, greatly simplifying the experimental setup. We first benchmarked the technology with three well-established human cell lines by targeting 154 peptides corresponding to 77 kinases. Next, to enable targeted protein profiling of age-related metabolic and inflammatory changes, we created a large Tomahto assay (520 peptides corresponding to 260 proteins). The assay can be readily adopted by any TMT-based targeted analysis in murine systems. We demonstrate that white adipose tissue (WAT) was by far the most affected tissue by aging in terms of protein expression, showing alterations in lipid metabolism, central carbon metabolism, electron transport chain complexes, and inflammation. Finally, we went on to collect proteome-wide measurements on these same tissues validating the Tomahto findings and providing a resource for understanding aging in mice.
Results
Overview of the Real-Time Monitoring Software, Tomahto.
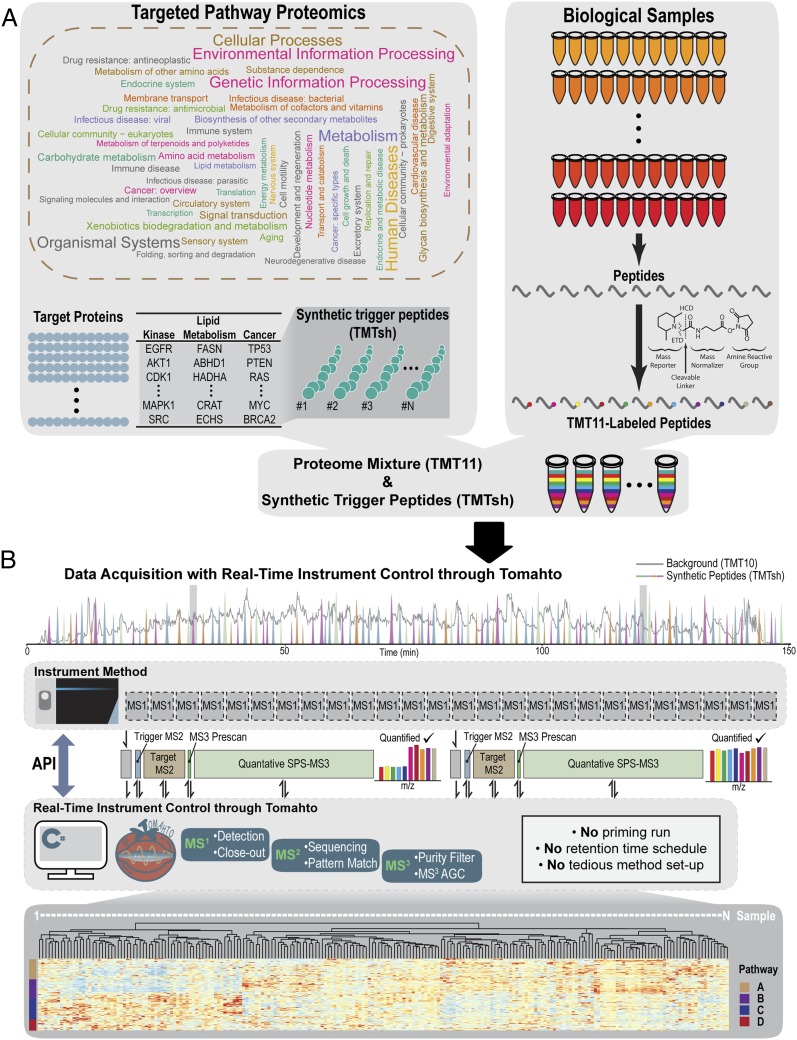
Tomahto was written in C# in the .NET Framework (v4.6.2). As originally reported, spiked-in trigger peptides are used as an alternative to traditional retention time scheduling to ensure correct sampling of target peptides (10). The trigger peptides are labeled with TMT superheavy (TMTsh) reagent to create a mass offset relative to their standard TMT10-labeled endogenous counterparts. In contrast to requiring priming runs with pure trigger peptides and manually curating cumbersome methods (10), Tomahto takes a list of peptides and autopopulates all of the information. During a TOMAHAQ liquid chromatography–mass spectrometry (LC-MS) experiment, Tomahto monitors MS1 scans on the fly and detects precursors that are potentially trigger peptides (Fig. 1B and SI Appendix, Fig. S1). Upon detection, Tomahto may insert as many as four new scans to the instrument. 1) A custom MS2 scan is inserted and used to confirm the trigger peptide’s sequence by RTPM. The presence of a trigger peptide signals the concomitant coelution of all 10 target endogenous peptides regardless of the detection of their precursor peaks (Fig. 1 B and C). 2) Tomahto then triggers an MS2 scan at an offset m/z value to verify that the target peptide is present and detectable. This scan is measured in the Orbitrap at high resolution. Peaks are matched within a tight mass accuracy tolerance, and the fragmentation pattern must match the trigger peptide’s pattern. Interference-free SPS precursor ions are selected from this scan. 3) Once verified, a quick MS3 prescan (collected in the ion trap) ensures accurate injection time scaling for the time-consuming MS3 Orbitrap quantification scan (SI Appendix, Figs. S1 and S3). 4) A highly optimized SPS-MS3 scan is inserted to quantify the abundance of the peptide across the 10 samples with selected SPS ions. This MS3 scan is remarkably sensitive since it can accumulate ions for as long as 5 s, if needed, to improve TMT reporter ion signal. This entire four-scan process can be repeated such that many MS3 scans are inserted as the peak elutes until a threshold is met, as can be specified by the close-out option (SI Appendix, Fig. S1). Once the threshold is met (i.e., number of quantified scans, total summed signal-to-noise ratio), the peptide is excluded from triggering any further scan events.
Fig. 1.
Multiplexed targeted pathway proteomics assisted by Tomahto with real-time instrument control. (A) TOMAHAQ enables 2D multiplexed targeted pathway proteomics. Constructing an assay starts with selecting protein targets and generating synthetic peptides for pathways of interest. These synthetic trigger peptides are labeled with TMTsh reagent, whereas peptides from biological samples are labeled with regular TMT reagents (e.g., TMT10 or TMT11), creating a mass difference of 6 Da. (B) A fully automated API-based algorithm, termed Tomahto, is presented in this work. Tomahto eliminates the need for a priming run and constructing elaborate method files while also enabling implementation of several advanced data acquisition options. The sophisticated data acquisition is handled by Tomahto such that the instrument method contains only MS1 scans. Tomahto monitors the elution of synthetic trigger peptides throughout an LC-MS run. When one is detected, Tomahto prompts the insertion of up to four custom scans in order to 1) sequence verify the trigger peptide, 2) sequence verify the target peptide, 3) optimize the quantification scan, and 4) collect the optimized quantification MS3 scan using only b- and y-type ions that exceed a purity filter. MS3 quantification scans contain the released TMT reporter ions. The final result is one or more pathways profiled for protein expression differences across all samples.
Benchmarking the Tomahto-Based Assay in Common Human Cell Lines.
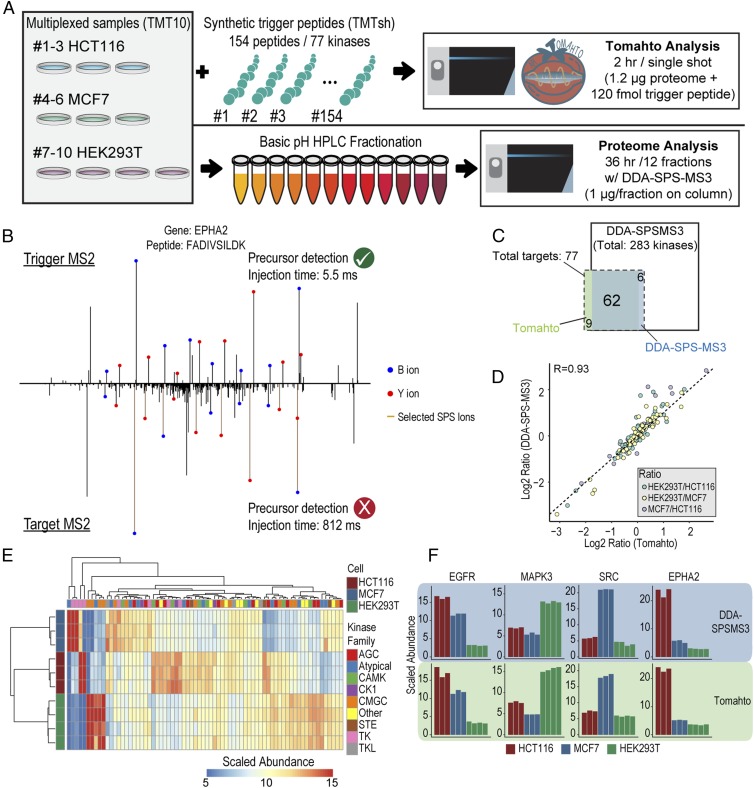
The Tomahto method was initially benchmarked and validated using three human cell line proteomes and a set of peptides to measure kinase levels across these lines (Fig. 2A). A TMT10plex was created consisting of biological triplicates of HCT116 and MCF7 and quadruplicates of HEK293T cells. Synthetic peptides were readily available for 77 kinases (154 peptides; see Dataset S1) that could be used in a Tomahto assay to quantify kinase levels across these three cell lines. The sensitivity of Tomahto was exemplified by peptide FADIVSILDK from EPHA1 (Fig. 2B). The trigger peptide was readily detected and its sequence confirmed in real time by peak matching, whereas the precursor of its endogenous counterpart was undetectable in MS1 scans. However, Tomahto allowed extensive yet specific accumulation of signals (>800 ms) to confirm its presence by an Orbitrap MS2 spectrum which resulted in a subsequent successful quantification event. Seventy-one of the 77 targeted kinases were detected and quantified from a 2-h gradient of the unfractionated mixture (2 h total) (Fig. 2C). The average coefficient of variation for replicates from the three cell lines was 3.7 ± 2.9% when signal-to-noise ratios from individual scans were summed for respective targets.
Fig. 2.
Benchmarking Tomahto by measuring protein levels for 77 kinases across three human cell lines. (A) Lysates from biological triplicates of HCT116 and MCF7 and quadruplicates of HEK293T were processed and labeled with TMT10 reagents. The Tomahto assay was performed with 154 spiked-in trigger peptides corresponding to 77 kinases and a 2-h single-shot method. In addition, shotgun analysis was performed on the same samples after fractionation. Twelve fractions were analyzed with 3-h gradients using the standard DDA-SPS-MS3 method (36 h total). (B) Representative trigger and target MS2s. These are used to confirm the identity of both the trigger and the target precursor peaks. The target MS2 is often of very low abundance and requires long injections times (812 ms here). Note that all labeled fragment masses differ by 6 Da between plots due to the TMTsh labeling on both termini. (C) Overlap for the kinases quantified by both methods. (D) Correlation between standard DDA-SPS-MS3 (fractionated, 36-h analysis) and Tomahto (unfractionated, 2-h analysis). (E) Hierarchical clustering of kinases quantified by Tomahto. Replicates perfectly clustered and many signature up- or down-regulated kinases were identified. (F) Bar plots of example proteins, EGFR, MAPK3, SRC, and EPHA2.
To validate the method’s accuracy orthogonally, we also performed a proteome-wide analysis of the same experiment. This required basic pH peptide separation (24 fractions) and analysis of 12 fractions via a standard data-dependent acquisition (DDA)-SPS-MS3 method (3 × 12 h = 36 h). More than 8,000 proteins were quantified across all 10 samples including 283 kinases, out of which 68 were also targets of Tomahto. Examining the overlap with our targeted set resulted in 62 kinases quantified in both experiments (Fig. 2C). An excellent correlation was found between the two approaches (Pearson R = 0.93; Fig. 2 D and F) even though Tomahto dealt with a much more complex mixture (∼24×), lower sample amounts (∼24×), and a shorter analysis time (∼18×).
Tissue-Specific Metabolic and Inflammatory Alteration Revealed by Tomahto.
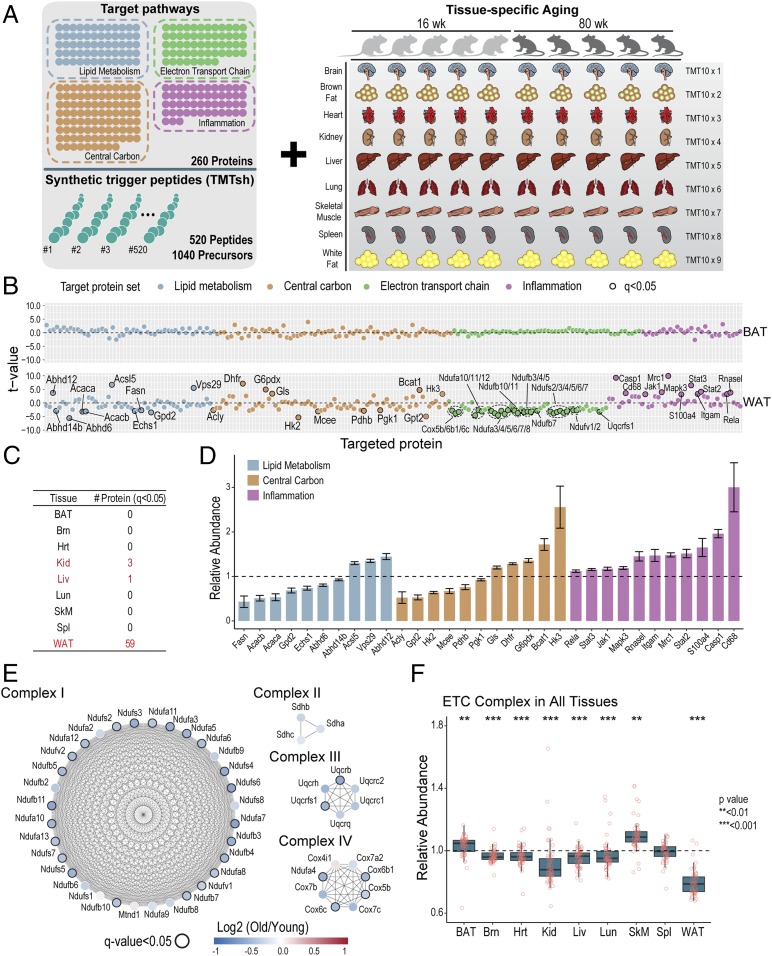
Following benchmarking with human cell lines, we next sought to apply the method to a large-scale analysis across dozens of samples (Fig. 3). Metabolism and inflammation have long been implicated in aging (17, 24). Thus, we synthesized a set of 520 peptides targeting 260 proteins spanning lipid metabolism, central carbon metabolism, electron transport chain, and inflammation (Dataset S2). Ten mice, five “young” (aged 16 wk) and five “old” (aged 80 wk), were killed. Nine tissues from each animal were harvested and processed, including brown adipose tissue (BAT), brain (Brn ), heart (Hrt), kidney (Kid), liver (Liv), lung (Lun), skeletal muscle (SkM), spleen (Spl), and WAT. Resulting peptides were labeled with TMT10 reagents while spike-in trigger peptides were labeled with TMTsh. Including multiple charge states, more than 1,000 precursors were constantly monitored.
Fig. 3.
A targeted Tomahto assay to profile proteome changes in metabolism and inflammation applied to aging mouse tissues. (A) Five “young” (16-wk-old) and five “old” (80-wk-old) mice were killed, and nine tissues from each animal were obtained and processed. Resulting tryptic peptides were labeled with TMT10 reagents. For the targeted assay, synthetic trigger peptides targeting four different panels of proteins (i.e., lipid metabolism, electron transport chain, central carbon metabolism, and inflammation), totaling 260 proteins and 520 peptides, were labeled with TMTsh reagent and spiked into multiplexed endogenous proteomes. (B) Examples of protein expression differences from the TOMAHAQ pathway analysis in brown compared to WAT (BAT vs. WAT). t tests (n = 5; old vs. young mice) were performed for each target protein with FDR correction. t values for each quantified protein are plotted and proteins passing a q value of 0.05 are labeled. BAT did not exhibit any change with q < 0.05, whereas WAT was the most affected tissue by aging with 59 proteins changed significantly by the same standard. See also SI Appendix, Fig. S4A. (C) Number of significantly changed (q < 0.05) protein targets in nine tissues. (D) Relative abundance of significantly changed (q < 0.05) proteins from WAT. Proteins are colored according to primary pathway. Bars represent mean ± SEM. (E) Quantification of targeted ETC complex proteins from WAT. Many ETC complex subunits (24) in WAT exhibited statistically significant (q < 0.05) decreases in old mice and most of the other complex members exhibited a decreasing trend. Color scale represents the log2-transformed ratio of protein abundance relative to young mice. Nodes with q value < 0.05 are circled with a black border. (F) Distribution of mean relative abundance of quantified ETC complex subunits in nine tissues. Relative abundance in old mice was normalized to mean young mice values (n = 5). Each point represents mean relative abundance of a quantified protein. t tests were used to detect differences between young and old mice using the mean relative abundances of quantified ETC proteins. In addition to WAT, ETC complexes as a group (but not alone) showed significant decreases in five tissues (P < 0.05). See also SI Appendix, Fig. S4B.
Overall, we profiled protein expression levels for 260 proteins across nine tissues from 10 animals in ∼1 d with single shots. More than half (54%) of the resulting MS3 scans met the ion injection time maximum of 5 s, suggesting they were present at low levels (SI Appendix, Fig. S3F). Student’s t tests on each quantified protein were corrected for multiple hypothesis testing, and we observed tissue-specific responses to age even in different fat tissues (BAT and WAT) (Fig. 3B). Although most tissues presented minimal changes at the individual protein level (q < 0.05) (SI Appendix, Fig. S4), which corroborates previous reports (20), WAT had 59 proteins that were significantly changed (q < 0.05) (Fig. 3C). Although traditionally thought as a specialized tissue for energy storage, the appreciation for WAT as a highly influential driver in the regulation of systemic metabolic homeostasis has expanded greatly in recent years (25, 26). Dysfunctional WAT is linked to insulin resistance, inflammation, and other metabolic disorders (26), all of which are, interestingly, also implicated in aging (17, 27). Therefore, it prompted us to think that WAT could be critical in an organism’s adaptation and response to aging. In WAT, we found that proteins involved in fatty acid/lipid biogenesis (e.g., Fasn, Acaca, and Acacb), lipid catabolism (e.g., Abhd6, Abhd12, and Abhd14b), and fatty acid beta-oxidation (e.g., Echs1) were significantly altered (Fig. 3D). Similarly, proteins involved in glycolysis (e.g., Hk2, Hk3, Pdhb, and Pgk1), amino acid metabolism (e.g., Bcat1, Dhfr, Gls, and Gpt2) were altered to various extents (Fig. 3D). Interestingly, consistent up-regulation of inflammatory proteins was observed, reflecting a general chronic inflammation, which is one of the hallmarks of aging (17) (Fig. 3D).
In contrast to higher expression levels of inflammatory proteins, subunits of electron transfer chain (ETC) complexes presented a universal decrease in WAT (Fig. 3E). Accumulating evidence has suggested a causative link between mitochondrial dysfunctional deterioration and major phenotypes associated with aging (28). Although not statistically significant (q < 0.05) as individual proteins after correction for multiple hypothesis testing, ETC subunits, as a group, also presented a significant and similar underrepresentation in five other tissues explored (Brn, Hrt, Kid, Liv, and Lun) (Fig. 3F), implying that ETC functions were impacted to various extents with age.
Comparing Tomahto to the Proteome-Wide, SPS-MS3 Method.
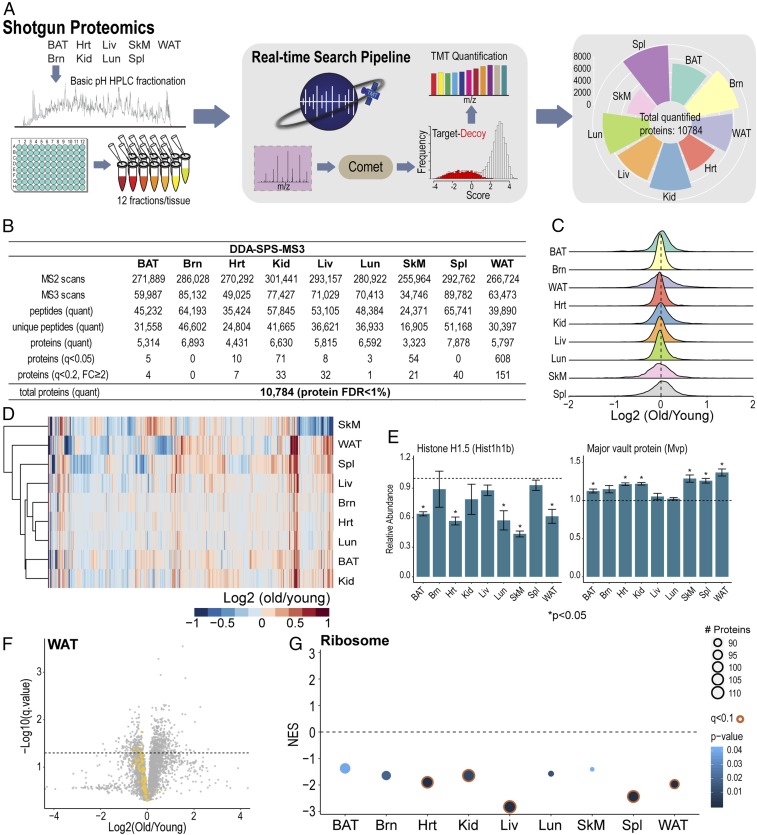
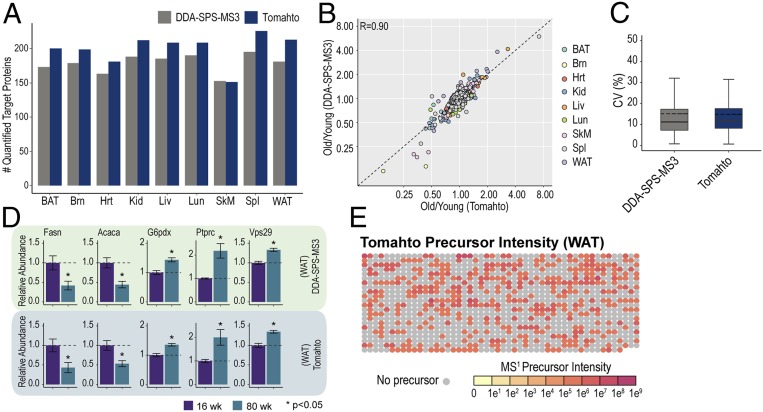
Targeted protein quantification boosts sensitivity and reproducibility while reducing analysis time as well as starting material. Two-dimensional multiplexing (sample and analyte) facilitated by the Tomahto software makes these advantages even more pronounced. For example, 2D multiplexing relies on mixtures of low starting amounts (∼10 µg total material) as opposed to hundreds of micrograms of starting material needed to perform a deep fractionation shotgun experiment. As a reference, we also conducted a shotgun proteomics experiment. For each tissue, TMT10-labeled peptides were fractionated and consolidated into 12 fractions, each of which was analyzed with a 2-h LC-MS method with the DDA-SPS-MS3 method but assisted by our real-time search algorithm termed Orbiter (29) (see Fig. 5A). This strategy improves the data acquisition process (29, 30). Overall, 10,784 proteins were quantified including all tissues with a protein false discovery rate (FDR) <1% (see Fig. 5B). Fig. 4A shows that a greater number of the targeted proteins were consistently quantified using Tomahto (mean 200 ± 22 vs. 179 ± 14). Yet, the quantification results were remarkably well correlated (R = 0.90) with the highly fractionated and carefully optimized DDA-SPS-MS3 method (Fig. 4 B and D). Moreover, the coefficient of variation (percent) among five biological replicates was virtually the same between Tomahto (mean = 14.9) and DDA-RTS (mean = 15.1) (Fig. 4C). Besides comparable accuracy and precision, Tomahto can show remarkable sensitivity due to obviating the need for MS1 detection of peptides. To highlight this, we examined MS1 precursor intensity for each quantified MS3. In one representative 3-h Tomahto experiment, 1,097 MS3 scans were collected and met our quantification thresholds. More than half (668) of these had precursors that were not detected in the preceding MS1 scan (Fig. 4E, gray circles). These would be impossible to quantify if the method relied on detection of precursor peaks.
Fig. 5.
Tissue-specific proteome profiling and common alterations of the proteome in old mice. (A) For proteome-wide quantitative analysis, each of the TMT10-labeled tissue samples was fractionated by basic pH reversed-phase high-performance LC, and 12 fractions were then analyzed with an established analytical pipeline that included a real-time database search (29). Including all nine tissues (216 h of analysis), 10,784 proteins were quantified. (B) Untargeted dataset overview. FC: fold change. (C) Density distributions for log-transformed old-to-young ratios in nine tissues. (D) Unsupervised hierarchical clustering using commonly quantified proteins in nine tissues. (E) Example bar plots of suggested aging marker proteins. Bars represent mean ± SEM. (F) Volcano plot of quantified proteins in WAT. Yellow dots indicate quantified members (n = 94) of cytoplasmic and mitochondrial ribosome complexes. Dotted line indicates q = 0.05. (G) GSEA analysis of ribosome proteins comparing old and young mice for nine tissues. Normalized enrichment scores (NES) are plotted for each tissue.
Fig. 4.
Comparison of TMT-based measurements made using the targeted vs. untargeted approach. (A) Number of quantified target proteins by Tomahto and Standard DDA-SPS-MS3. Tomahto achieved better coverage quantifying target proteins compared to Standard MS3 and consumed much less starting material and instrument time. (B) Pearson correlation was calculated for all target proteins in nine tissues (r = 0.90). (C) The coefficient of variation (CV) among biological replicates for each protein within each tissue, commonly quantified by Standard MS3 and Tomahto, was calculated and assessed for reproducibility. (D) Example bar charts for protein quantifications using both methods. Bars represent mean ± SEM (n = 5). (E) Most peptides are quantified even in the absence of a visible MS1 feature. Array plot of precursor intensity for a representative Tomahto experiment. Each dot represents the MS1 precursor intensity of an inserted MS3 quantification scan. Dot color scale represents MS1 intensity and gray dot indicates absence of any peak within the elution window.
Untargeted Quantitative Characterization of Tissue-Specific Changes between Young and Old Mice.
In addition to characterizing targeted proteins with Tomahto, the shotgun proteomics experiment allowed us to gain a general picture of nine aging tissues (Fig. 5B). For the 10,784 quantified proteins in nine tissues, we performed t tests and adjusted the resulting P values to account for multiple hypothesis testing (31). Strikingly, with the exception of WAT (608 proteins with q < 0.05), we observed few proteins that met statistical significance criteria between the two age groups (Fig. 5B). While this may in part be due to limited power of the n = 5 experiment, we note that the majority of measured proteins had small fold changes (Fig. 5C). For example, among all proteins with P values <0.05, the majority only presented ∼20% change in old mice (SI Appendix, Fig. S5A). When fold changes were considered to find proteins affected by aging, fewer than 1% of all proteins had q < 0.2 and a fold change ≥2 in each tissue except for WAT (2.6%) (Fig. 5B).Thus, in the comparison of the two age groups presented here we did not observe large, consistent rewiring of the murine tissue proteomes. Unsupervised hierarchical clustering with commonly quantified proteins showed no clear pattern of changes across all tissues (Fig. 5D), indicating tissue-specific alterations in aged mice. Among commonly quantified proteins, a few did present consistent changes across tissues, including Hist1h1b and Mvp (Fig. 5E and SI Appendix, Fig. S5B). Other example proteins included Ppt1, Ctsd, which indicated involvement of lysosome, Igkc, Fkbp4, and Abcf1 (SI Appendix, Fig. S5).
Beyond individual proteins, the overall abundance of ribosome complex was down-regulated in old mouse tissues (Fig. 5 F and G). WAT, as an example, had 94 cytoplasmic and mitochondrial ribosome proteins quantified, out of which 90 decreased in old mice, with 9 reaching statistical significance (q < 0.05) (Fig. 5F). Gene set enrichment analysis (GSEA) identified global decreases in ribosome abundance across tissues (Fig. 5G). With aging, tissues may undertake a decline in ribosome health and translational capacity (19, 32, 33).
Tissue-Specific Changes Revealed by Global Proteomics.
Overall the proteome-wide shotgun approach quantified 10,784 proteins, providing a resource covering nine major tissues in aging mice. We first asked if young and old mice can be distinguished from a global perspective. Principle component analysis (PCA) revealed that some tissues (e.g., WAT, Liv) could be well separated with major components such as PC1 and PC2 (Fig. 6A and SI Appendix, Fig. S6A). Once again, WAT stood out as the most affected tissue by having 608 proteins with q < 0.05 and 151 proteins with q < 0.2 and fold changes ≥2. Considering the set of proteins targeted by our Tomahto assay, very similar results were obtained (Fig. 6B). Further confirmation came from additional proteins in the same pathways which also showed consistent changes.
Fig. 6.
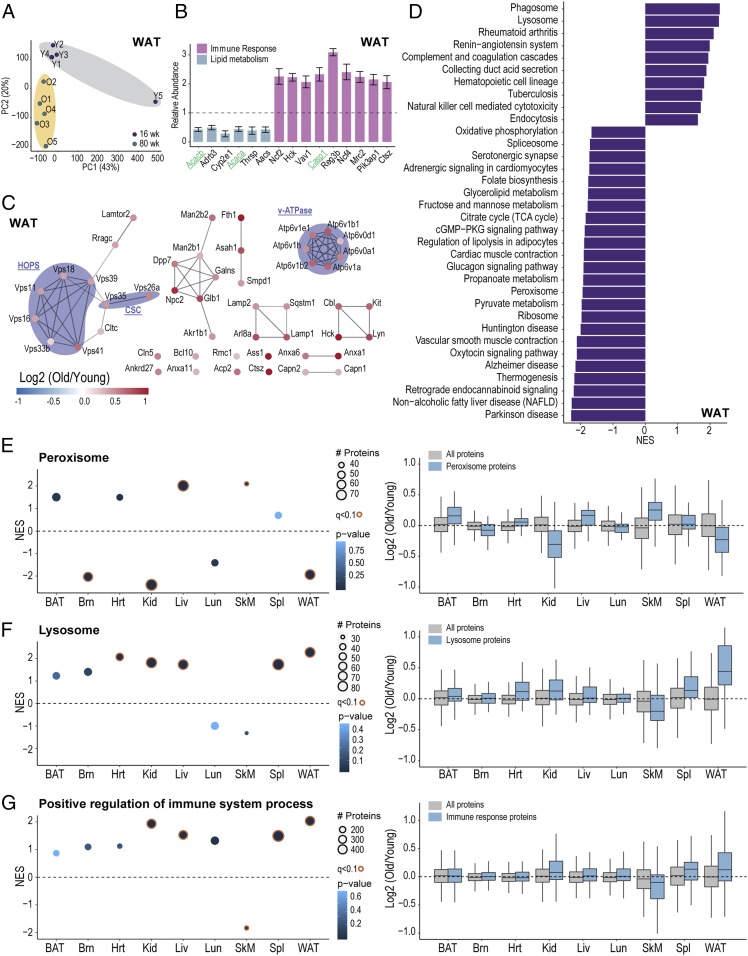
Tissue-specific changes in old mice revealed by proteome-wide profiling. (A) Principal component analysis (PCA) of WAT from 10 mice. (B) Example bar plots of proteins (n = 5, q < 0.05) that also have a minimum 2-fold change in WAT. Bars are colored according to primary pathway. Genes in green were also quantified by Tomahto. Bars represent mean ± SEM. (C) Network of significantly changed (n = 5, q < 0.05) lysosomal proteins in WAT. All genes were up-regulated in old mice. (D) Significantly enriched terms (q < 0.05) from GSEA analysis using all quantified WAT proteins (n = 5,797). (E) GSEA analysis for peroxisome gene set. NES and log2 ratio distribution are plotted for each tissue. (F) GSEA analysis result for the lysosome. NES and log2 ratio distribution are plotted for each tissue. (G) GSEA analysis result of positive regulation of immune system process. NES and log2 ratio distribution are plotted for each tissue.
Gene set enrichment analysis (GSEA) was performed to highlight differentially changed pathways in WAT. After FDR filtering, 34 gene sets remained. For example, lysosome, phagosome, and several inflammation-related pathways were up-regulated in old mice, whereas oxidative phosphorylation, TCA cycle, spliceosome, ribosome, and lipolysis were down-regulated (34, 35). Strikingly, all regulated lysosomal and phagosomal proteins (q < 0.05) (Fig. 6C and SI Appendix, Fig. S6B) showed overexpression. Functionally associating lysosomal proteins revealed tightly connected protein complexes, examples including homotypic fusion and vacuole protein sorting complex (HOPS), cargo selection complex (CSC), and V-ATPase complex (Fig. 6C). Similarly, phagosomal proteins included members of major histocompatibility complex (MHC) and transporter associated with antigen processing protein (TAP) complex (SI Appendix, Fig. S6B), suggesting involvement of immune response.
In addition to WAT, all other tissues were investigated with GSEA, resulting in some with differential regulation. For example, the categories for “peroxisome,” “lysosome,” and “positive regulation of immune system process” are shown in Fig. 6 E and F. It is clear that while changes on a per-protein basis did not often reach significance many gene sets as a whole were differentially regulated.
Discussion
Sample multiplexing represents a powerful approach to both increase sample throughput and to introduce stable isotope labeling for accurate quantification. Recently, the plexing scale for TMT proteomcs was increased to support up to 16 different samples (36). Combining sample multiplexing with targeted analysis of hundreds of samples in a single run increases the throughput even more. To enable the full repertoire of possibilities, software controlling the instrument from the external desktop computer was created (termed Tomahto). Tomahto realizes real-time instrument control based on an API. Importantly, the only scan performed by the instrument method is the MS1 scan. Based on monitoring the elution of TMTsh-labeled trigger peptides in these MS1 scans, Tomahto inserts up to four new scans, including a highly optimized MS3 scan that measures differences in reporter ions across all samples. In the current study, the method was first benchmarked for sensitivity, precision, and accuracy against a fully fractionated, proteome-wide dataset (Fig. 2). The software is freely available (Materials and Methods) and requires an API license from Thermo Fisher Scientific. The same API can be used on any tribrid instrument, including the Fusion, Fusion Lumos, and Eclipse platforms.
In the in-depth pathway proteomics study of aging, we monitored 520 peptides and 1,040 precursors, which was four times the number of peptide targets compared to our original publication (10), and we have tested a higher number by targeting 1,000 peptides (2,000 precursors) from JPT Peptide Technology (catalog no. SPT-ABRF-POOL-L) (SI Appendix, Fig. S8). This was by no means a hard limit. It is possible that future work could target a higher number. For example, the current implementation simply requires that the trigger peptide be eluting but makes no attempt to catch the peak at its apex. Since the target and trigger peaks perfectly coelute, this could further increase the sensitivity while also increasing the number of potential targets in a single analysis.
Aging is associated with a wide spectrum of diseases. Yet, several reports described only minimal perturbation at the proteome level (19, 20). Studies on the genome and transcriptome levels also indicated that each tissue should be analyzed separately as they may respond to aging via distinct mechanisms (21, 22). Tomahto revealed WAT to be the most affected tissue with almost a quarter of all targets significantly (q < 0.05) regulated, including down-regulation of fatty acid synthesis-related proteins and up-regulation of inflammatory proteins, whereas other tissues, as expected, showed minimal change of individual targeted proteins. Our investigation generated a comprehensive resource covering proteome modulations in nine major tissues from aged mice and underscores that each tissue may possess its distinct mechanism. One limitation of the current study lies in the limited resolution within each tissue, given that certain reports have claimed particular regions (37) or cell subpopulations (18) may contribute distinctively to tissue aging or aging-related diseases. In addition, we believe adding more age groups could be of future interest and potentially improve temporal resolution and be beneficial in investigating correlations between transcription and translation, given the corresponding transcriptome study has just become available (38).
In the aging experiments, only a few proteins exhibited tissue-wide changes between the two investigated age groups. For example, the linker histone H1 binds to the DNA entering and exiting the nucleosomal core particle and has an important role in establishing and maintaining higher-order chromatin structures as well as regulating transcription activity. Previous studies reported translational down-regulation of Hist1h1b in microglia (39), SkM (40), and transcriptional down-regulation in mouse retinae (41) due to aging. Our measurement confirmed that Hist1h1b presented a decreasing trend in nine tissues with SkM having the most dramatic change (more than twofold) (Fig. 4E). Another example was major vault protein (Mvp). Mvp is the main component contributing most of the particle mass for the cellular ribonucleoprotein particle known as the vault, which also consists of vault poly(ADP ribose) polymerase (VPARP) and telomerase-associated protein-1 (TEP1) and small untranslated RNA (42). The vault is implicated in multiple cellular processes, including nucleocytoplasmic transport, signaling transduction, cellular differentiation, cell survival, and immune responses (42–44). Ryu et al. (44) demonstrated increased expression of Mvp in aged human diploid fibroblasts and multiple mouse organs and connected its expression level with cellular sensitivity to apoptosis. Similarly, our data suggested Mvp amount increased in all nine tissues. Mvp’s suppressive role in low-grade chronic inflammation ameliorates high-fat-diet–induced obesity, insulin resistance, hepatic steatosis and atherosclerosis in mice (43). The highest up-regulation levels for Mvp observed in WAT were correlated with significantly overexpressed inflammatory proteins as measured by Tomahto.
While outside of WAT few proteins were observed to have substantially altered levels in old vs. young mice, many pathways—when taken as a whole—were detected as altered in our shotgun proteomics experiments. Peroxisomal proteins, as just one example, were significantly up-regulated in Liv and SkM, whereas the opposite was observed in Brn, Kid, and WAT. Accumulating evidence suggests that peroxisomal function declines with aging, linking oxidative stress and dysregulated lipid metabolism to diseases including Alzheimer’s disease, diabetes, and cancer (45). Specifically, peroxisomal biogenesis factors (Pex) are essential for the formation of functional peroxisomes and transport of peroxisomal matrix proteins after synthesis in cytosol (46). We observed Pex3, Pex10, and Pex11a down-regulation in WAT and up-regulation in Liv (SI Appendix, Fig. S6C). Moreover, ATP-binding cassette subfamily d (Abcd) transporters are responsible for transporting different subtypes of acyl-CoA and therefore critical in lipid and fatty acid metabolism (47). Abcd2 and Abcd3 increased in BAT and Liv and decreased in Brn and WAT, in accordance with overall peroxisomal protein changes revealed by GSEA (SI Appendix, Fig. S6C).
In conclusion, we report a strategy which supports extensive multiplexing at both the sample and the analyte levels. More than 500 peptides were targeted in a TMT 10plex environment using the free Tomahto software with instrument control via an API. Future improvements will include 1) adaptation of this method to the new 16plex TMTPro reagents (36), 2) apex triggering to increase sensitivity, and 3) combining the method with high-field asymmetric waveform (48, 49) ion mobility spectrometry to separate coeluting peptides.
Materials and Methods
Tomahto was written in C# in the .NET Framework (v4.6.2). It has three modules, namely data acquisition, real-time data visualization, and data analysis. Kinase peptides for cellular experiments were obtained from Thermo Fisher Scientific and peptides for experiments in aging mouse tissues were synthesized by Cell Signaling Technologies. Samples were analyzed with an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Descriptions of additional experimental procedures can be found in SI Appendix.
All animal-related experiments were approved by Institutional Animal Care and Use Committee of the Beth Israel Deaconess Medical Center.
Data Availability.
All MS raw files haven been deposited to the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with the identifier PXD017385. The software is freely available at https://gygi.med.harvard.edu/software (also smarttmt.org). The use of Tomahto requires an API license (https://github.com/thermofisherlsms/iapi) from Thermo Fisher Scientific.
Supplementary Material
Acknowledgments
We thank members of the S.P.G. laboratory for helpful discussions and Derek Bailey, Philip Remes, Graeme McAlister, Jesse Canterbury, and Shannon Eliuk at Thermo Scientific for technical assistance and advice. This work was funded in part by NIH grant GM67945 (S.P.G.), the Claudia Adams Barr Program (E.T.C. and M.P.J.), the National Cancer Center (H.X.), and Mexican Council for Science and Technology grant 289937 (J.N.-P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.B. is a guest editor invited by the Editorial Board.
Data deposition: All MS raw files haven been deposited to the PRIDE archive with the identifier PXD017385.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919410117/-/DCSupplemental.
References
- 1.Gillet L. C., Leitner A., Aebersold R., Mass spectrometry applied to bottom-up proteomics: Entering the high-throughput Era for hypothesis testing. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 9, 449–472 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Di Stefano B., et al. , Reduced MEK inhibition preserves genomic stability in naive human embryonic stem cells. Nat. Methods 15, 732–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarrete-Perea J., Yu Q., Gygi S. P., Paulo J. A., Streamlined tandem mass tag (SL-TMT) protocol: An efficient strategy for quantitative (Phospho)proteome profiling using tandem mass tag-synchronous precursor selection-MS3. J. Proteome Res. 17, 2226–2236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekker-Jensen D. B., et al. , An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Syst. 4, 587–599.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song E., et al. , Targeted proteomic assays for quantitation of proteins identified by proteogenomic analysis of ovarian cancer. Sci. Data 4, 170091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson A., et al. , Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Ting L., Rad R., Gygi S. P., Haas W., MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenger C. D., et al. , Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat. Methods 8, 933–935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McAlister G. C., et al. , MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson B. K., et al. , A strategy to combine sample multiplexing with targeted proteomics assays for high-throughput protein signature characterization. Mol. Cell 65, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose C. M., et al. , TomahaqCompanion: A tool for the creation and analysis of isobaric label based multiplexed targeted assays. J. Proteome Res. 18, 594–605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenyon C. J., The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Sen P., Shah P. P., Nativio R., Berger S. L., Epigenetic mechanisms of longevity and aging. Cell 166, 822–839 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou S., Meadows S., Sharp L., Jan L. Y., Jan Y. N., Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 97, 13726–13731 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davie K., et al. , A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982–998.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.López-Otín C., Galluzzi L., Freije J. M. P., Madeo F., Kroemer G., Metabolic control of longevity. Cell 166, 802–821 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Ferrucci L., Fabbri E., Inflammageing: Chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 15, 505–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angelidis I., et al. , An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat. Commun. 10, 963 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ori A., et al. , Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 1, 224–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther D. M., Mann M., Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol. Cell. Proteomics 10, M110.004523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen B. C., et al. , Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 5, e1000602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmel J. C., et al. , A murine aging cell atlas reveals cell identity and tissue-specific trajectories of aging. bioRxiv:10.1101/657726 (6 June 2019).
- 23.Hudgins A. D., et al. , Age- and tissue-specific expression of senescence biomarkers in mice. Front. Genet. 9, 59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkel T., The metabolic regulation of aging. Nat. Med. 21, 1416–1423 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Choe S. S., Huh J. Y., Hwang I. J., Kim J. I., Kim J. B., Adipose tissue remodeling: Its role in energy metabolism and metabolic disorders. Front. Endocrinol. (Lausanne) 7, 30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stern J. H., Rutkowski J. M., Scherer P. E., Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metab. 23, 770–784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fink R. I., Kolterman O. G., Griffin J., Olefsky J. M., Mechanisms of insulin resistance in aging. J. Clin. Invest. 71, 1523–1535 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun N., Youle R. J., Finkel T., The mitochondrial basis of aging. Mol. Cell 61, 654–666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweppe D. K., et al. , Full-featured, real-time database searching platform enables fast and accurate multiplexed quantitative proteomics. bioRxiv:10.1101/668533 (12 June 2019). [DOI] [PMC free article] [PubMed]
- 30.Erickson B. K., et al. , Active instrument engagement combined with a real-time database search for improved performance of sample multiplexing workflows. J. Proteome Res. 18, 1299–1306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strimmer K., fdrtool: A versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Kurtz D. I., A decrease in the number of active mouse liver ribosomes during aging. Exp. Gerontol. 13, 397–402 (1978). [DOI] [PubMed] [Google Scholar]
- 33.Cellerino A., Ori A., What have we learned on aging from omics studies? Semin. Cell Dev. Biol. 70, 177–189 (2017). [DOI] [PubMed] [Google Scholar]
- 34.de Magalhães J. P., Curado J., Church G. M., Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmona-Gutierrez D., Hughes A. L., Madeo F., Ruckenstuhl C., The crucial impact of lysosomes in aging and longevity. Ageing Res. Rev. 32, 2–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson A., et al. , TMTpro: Design, synthesis, and initial evaluation of a proline-based isobaric 16-plex tandem mass tag reagent set. Anal. Chem. 91, 15941–15950 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Carlyle B. C., et al. , A multiregional proteomic survey of the postnatal human brain. Nat. Neurosci. 20, 1787–1795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaum N., et al. , The murine transcriptome reveals global aging nodes with organ-specific phase and amplitude. bioRxiv:10.1101/662254 (7 June 2019).
- 39.Flowers A., Bell-Temin H., Jalloh A., Stevens S. M. Jr, Bickford P. C., Proteomic anaysis of aged microglia: Shifts in transcription, bioenergetics, and nutrient response. J. Neuroinflammation 14, 96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cutler A. A., et al. , Biochemical isolation of myonuclei employed to define changes to the myonuclear proteome that occur with aging. Aging Cell 16, 738–749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banday A. R., et al. , Replication-dependent histone genes are actively transcribed in differentiating and aging retinal neurons. Cell Cycle 13, 2526–2541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger W., Steiner E., Grusch M., Elbling L., Micksche M., Vaults and the major vault protein: Novel roles in signal pathway regulation and immunity. Cell. Mol. Life Sci. 66, 43–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben J., et al. , Major vault protein suppresses obesity and atherosclerosis through inhibiting IKK-NF-κB signaling mediated inflammation. Nat. Commun. 10, 1801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryu S. J., et al. , On the role of major vault protein in the resistance of senescent human diploid fibroblasts to apoptosis. Cell Death Differ. 15, 1673–1680 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cipolla C. M., Lodhi I. J., Peroxisomal dysfunction in age-related diseases. Trends Endocrinol. Metab. 28, 297–308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterham H. R., Ebberink M. S., Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta 1822, 1430–1441 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Baker A., et al. , Peroxisomal ABC transporters: Functions and mechanism. Biochem. Soc. Trans. 43, 959–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweppe D. K., et al. , Characterization and optimization of multiplexed quantitative analyses using high-field asymmetric-waveform ion mobility mass spectrometry. Anal. Chem. 91, 4010–4016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hebert A. S., et al. , Comprehensive single-shot proteomics with FAIMS on a hybrid orbitrap mass spectrometer. Anal. Chem. 90, 9529–9537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS raw files haven been deposited to the PRIDE archive (https://www.ebi.ac.uk/pride/archive) with the identifier PXD017385. The software is freely available at https://gygi.med.harvard.edu/software (also smarttmt.org). The use of Tomahto requires an API license (https://github.com/thermofisherlsms/iapi) from Thermo Fisher Scientific.