Significance
Desiccation tolerance likely evolved independently several times in grasses, providing an ideal comparative system to identify genetic elements controlling this trait. Using comparative genomics, we identified genomic and expression changes distinguishing the desiccation-tolerant grass Eragrostis nindensis from its desiccation-sensitive crop relative Eragrostis tef. We expanded these analyses to include several cereals to identify broadly conserved and divergent patterns during water stress. We found that the distinction between drought and desiccation in grasses is subtle, where genes with essential roles in seed development are broadly expressed under water stress. Thus we propose that seeds and leaves share common sets of coregulated genes of likely ancient origin, with only a few genes uniquely expressed for desiccation tolerance.
Keywords: drought, grasses, evolution, desiccation tolerance, rewiring
Abstract
Grasses are among the most resilient plants, and some can survive prolonged desiccation in semiarid regions with seasonal rainfall. However, the genetic elements that distinguish grasses that are sensitive versus tolerant to extreme drying are largely unknown. Here, we leveraged comparative genomic approaches with the desiccation-tolerant grass Eragrostis nindensis and the related desiccation-sensitive cereal Eragrostis tef to identify changes underlying desiccation tolerance. These analyses were extended across C4 grasses and cereals to identify broader evolutionary conservation and divergence. Across diverse genomic datasets, we identified changes in chromatin architecture, methylation, gene duplications, and expression dynamics related to desiccation in E. nindensis. It was previously hypothesized that transcriptional rewiring of seed desiccation pathways confers vegetative desiccation tolerance. Here, we demonstrate that the majority of seed-dehydration–related genes showed similar expression patterns in leaves of both desiccation-tolerant and -sensitive species. However, we identified a small set of seed-related orthologs with expression specific to desiccation-tolerant species. This supports a broad role for seed-related genes, where many are involved in typical drought responses, with only a small subset of crucial genes specifically induced in desiccation-tolerant plants.
Approximately 470 million years ago charophyte green algae emerged from their watery habitat to colonize land (1). Exposure to a harsh, dry atmosphere was the main biophysical constraint facing early land plants, resulting in strong selective pressure favoring adaptive mechanisms to prevent dehydration (2). These early protective mechanisms likely served as a foundation for evolving desiccation-tolerant seeds and pollen, which was critical to the success of seed plants (3). Although most plants have desiccation-tolerant seeds and pollen, comparatively few can withstand drying of vegetative tissues. Vegetative desiccation tolerance is rare in flowering plants, but it is widespread among other plant lineages (4). The appearance of vegetative desiccation tolerance in phylogenetically distant lineages suggests multiple independent evolutionary origins. In the ecologically and economically important plant family Poaceae, vegetative desiccation tolerance is found within nine separate genera across five different tribes (SI Appendix, Table S1), suggesting that it evolved independently multiple times (5, 6). The current consensus hypothesis is that vegetative desiccation tolerance in angiosperms arose convergently through rewiring of common seed desiccation pathways (7, 8).
Transcriptomic studies on desiccation-tolerant angiosperms consistently show activation of seed-related genes during water-deficit stress (7–13). However, many of these genes are also highly expressed during water-deficit stress responses in desiccation-sensitive species. The phytohormone abscisic acid (ABA) is critical for seed maturation and drought tolerance where it is hypothesized to play a major role in regulating desiccation tolerance (5, 14, 15) and drought-responsive pathways, respectively (16). Thus, many of the downstream genes that are activated via ABA-dependent mechanisms are expressed broadly during seed development and in leaf tissues under mild and severe water deficit (desiccation). Accumulation of osmoprotectants and activation of reactive oxygen species quenching mechanisms are also shared responses between these conditions. Thus, it is important to distinguish desiccation-tolerance responses from broader water-deficit stress responses.
While numerous transcriptomic studies of desiccation-tolerant plants have been published, few previous studies have compared the responses of desiccation-sensitive and desiccation-tolerant plants with a close phylogenetic relationship. Previous work comparing the eudicot species Lindernia brevidens (desiccation tolerant) and Lindernia subracemosa (desiccation sensitive) provided some insight into genes that are involved in desiccation tolerance and not drought (13). However, the Linderniaceae family is of little economic importance and is only distantly related to any crop plants, making it difficult to translate these discoveries. Cereals from the grass family (Poaceae) are the most important crops for global food security, and our current study with desiccation-tolerant Poaceae species is likely more readily translatable. The Chloridoideae subfamily of grasses contains the majority of desiccation-tolerant species with multiple independent phylogenetic origins. Chloridoideae also contains the cereals finger millet (Eleusine coracana) and tef (Eragrostis tef), which are widely consumed in semiarid regions of Eastern Africa and Asia. To our knowledge, Eragrostis is the only genus with both desiccation-tolerant and cereal crop species. Thus, Eragrostis and, more broadly, the Chloridoideae subfamily, are ideal systems in which to identify genes involved in desiccation tolerance that are potential targets for improving drought resilience in crops.
Chromosome-scale genome assemblies of the Chloridoideae grasses E. tef (desiccation sensitive) and Oropetium thomaeum (desiccation tolerant) were recently completed (17, 18). Here, we sequenced the desiccation-tolerant grass Eragrostis nindensis and performed detailed comparative genomics within Chloridoideae and across the grass family to search for patterns of convergence in the evolution of desiccation tolerance. We conducted parallel dehydration experiments with E. nindensis and E. tef using matched physiological sampling points to identify signatures that distinguish water stress and desiccation responses. We leveraged seed expression data of E. tef and other grass species to test if desiccation tolerance in grasses arose through co-option of seed dehydration pathways. Together, our results identify similar signatures of water-deficit (drought) stress and desiccation responses with a smaller number of genomic features and expression changes unique to desiccation-tolerant grasses. We propose a model where seeds and leaves share common sets of coregulated genes under water deficit with only a few genes uniquely involved in desiccation responses.
Results
Genome Evolution of Chloridoid Grasses.
Comparative systems with phylogenetically similar desiccation-sensitive and -tolerant species are a powerful tool to elucidate the genetic basis of desiccation tolerance. Only one previous study has conducted genome-wide comparisons between a desiccation-sensitive and -tolerant angiosperm (13), and no such systems are currently available for the grasses. We assembled a draft genome of the desiccation-tolerant grass E. nindensis and compared it to the recently sequenced E. tef genome (18) to distinguish genetic elements associated with desiccation tolerance from those more broadly linked with drought response. Similar to most (∼90%) of chloridoid grasses, E. nindensis and E. tef are polyploid, and they have the same karotype (2n = 4× = 40) (SI Appendix, Fig. S1) (19, 20). We utilized a single-molecule real-time sequencing approach to overcome assembly issues related to tetraploidy and heterozygosity in E. nindensis. In total, we generated 64 Gb of PacBio data representing 63× coverage of the 1.0-Gb E. nindensis genome. We used Canu with parameters optimized to assemble all haplotypes yielding an initial assembly of roughly twice the haploid genome size (SI Appendix, Table S2). We then applied the pseudohaploid algorithm to filter out redundant haplotypes from the assembly (see Materials and Methods for details) (21). This filtering approach produced a total haploid assembly of 986 Mb across 4,368 contigs with an N50 of 520 kb, hereafter referred to as E. nindensis V2.1. We used the MAKER-P pipeline (22) to annotate the E. nindensis genome, and after filtering the annotation based on orthology, pfam domains, and expression, we identified a set of 107,683 high-confidence genes (Materials and Methods).
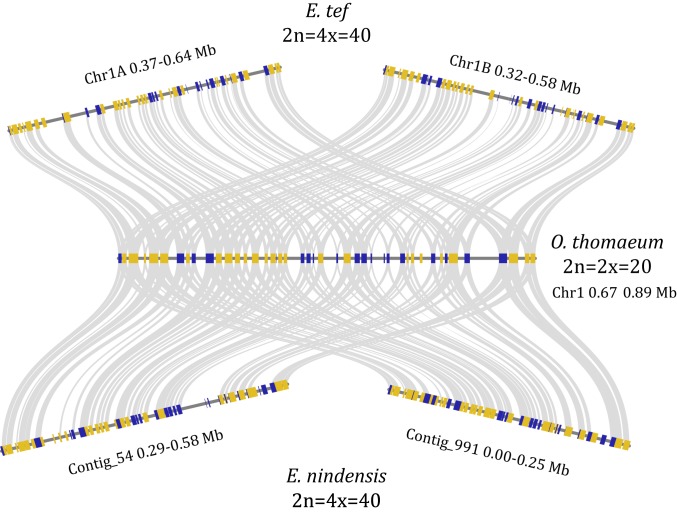
The three chrloridoid grass genomes of E. nindensis, E. tef, and O. thomeaum have largely conserved gene content and order (synteny) (Fig. 1). A high proportion of the E. tef and E. nindensis genomes matched the expected 2:2 ratio of syntenic gene blocks given their tetraploidy, but a substantial portion of the E. nindensis genome has three or four blocks for each homeologous region of E. tef (SI Appendix, Fig. S2B). E. nindensis and the diploid O. thomaeum have 2:1 synteny with similarly duplicated blocks of three or more in some regions (SI Appendix, Fig. S2A). This syntenic pattern is likely a result of assembling multiple haplotypes for each homeologous region in E. nindensis and a single haplotype for E. tef and O. thomaeum. This is supported by the distribution of synonymous substitutions (Ks) between orthologous genes in E. nindensis (SI Appendix, Fig. S3). We observed a strong peak of Ks corresponding to haplotype sequences and homeologous gene pairs. The high degree of collinearity and shared gene content between the three sequenced Chloridoideae grasses allowed us to identify shared and unique genomic signatures of desiccation tolerance.
Fig. 1.
Collinearity of Chloridoideae grasses. Microsynteny of collinear regions between allotetraploid E. nindensis and E. tef and diploid O. thomaeum is shown. Genes in the forward and reverse orientation are shown in gold and blue, respectively, and syntenic gene pairs are connected by gray lines.
Comparative Water-Deficit Responses between E. nindensis and E. tef.
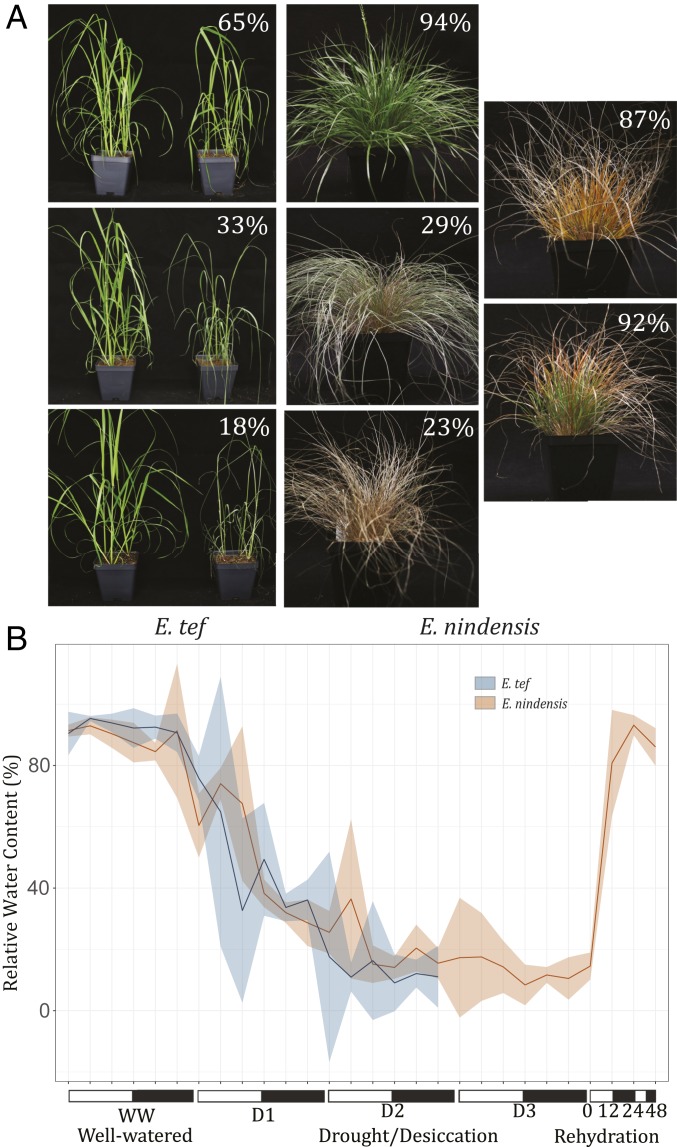
We conducted parallel dehydration time courses for E. nindensis and E. tef under comparable conditions to distinguish drought- and desiccation-associated responses (Fig. 2 and Dataset S1). Water-deficit treatment group plants were dried in a controlled manner until they reached mild water-deficit stress [75.8 and 60.4% relative water content (RWC), respectively]. We then sampled during moderate and severe dehydration stress for both species, with only E. nindensis being sampled after prolonged desiccation, and on recovery, since E. tef does not survive below ∼30% RWC (23). We collected tissue for leaf RNA-Sequencing (RNA-Seq) data in parallel with physiological data for eight time points in E. nindensis and three for E. tef. In E. nindensis, these corresponded to the fully hydrated control (well watered [WW], RWC 90.36%) moderate dehydration (D1, RWC 67.57%), severe dehydration (D2, 15.18% RWC) and desiccated (D3, 14.28% RWC) and on rehydration for 0 h (14.56% RWC), 12 h (80.82% RWC), 24 h (93.16% RWC), and 48 h (86.05% RWC). In E. tef, these included the WW control (93.89% RWC), D1 (32.68% RWC), and D2 (16.32% RWC). We collected E. tef samples below the minimum RWC from which they can recover in order to capture their response to a lethal water-deficit stress; however, we did not collect samples from E. tef plants maintained at this low RWC level for a longer period of time since the leaves would already have senesced. Nocturnal upticks in RWC across the drying time courses are associated with nighttime hydraulic redistribution (24).
Fig. 2.
Comparative parallel dehydration experiments in Eragrostis grasses. (A) Parallel drought/desiccation time course in E. tef (Left) and E. nindensis (Right). E. nindensis plants at 24 and 48 h post rehydration are also shown. E. tef droughted plants are shown next to a well-watered plant in each image. Each pot contains individual E. tef and E. nindensis, and these plants were collected separatley from the timecourses depicted in B. (B) Relative water content changes across the desiccation and rehydration experiments for E. tef (blue) and E. nindensis (orange) with 95% confidence intervals plotted for each time point. The sampling points for RNA-seq are labeled WW (well watered), D1, D2, and D3 (first, second, and third drought-sampling point, respectively), and 0, 12, 24, and 48 h post rehydration).
We tracked electrolyte leakage across the desiccation time course as a proxy for membrane damage and cell death. In both E. nindensis and E. tef, the percentage of total leakage increased significantly during the most severe dehydration and desiccation time points (E. nindensis D2, D3, R0, and E. tef D2) compared to the well-watered control (SI Appendix, Fig. S4). During the rehydration experiment, the electrolyte leakage percentage decreased back to the well-watered level by 12 h post rehydration in E. nindensis (SI Appendix, Fig. S5), suggesting that, while some membrane reorganization or damage might occur during desiccation, the damage is repaired on rehydration. Similar data have been reported previously for both species with the percentage of leakage returning to a predesiccated state in E. nindens, but with levels further increasing on rehydration for E. teff plants that have been dehydrated below 30 to 40% RWC (23).
Across the time courses, 26,275 genes (24.4% of high-confidence genes) in E. nindensis were differentially expressed between well-watered leaves and at least one drought or rehydration time point. Of these, 7,504 increased in transcript abundance (hereafter up-regulated) and 19,506 decreased in transcript abundance (hereafter down-regulated). Down-regulated genes under drought in E. nindensis were significantly enriched in numerous gene ontology (GO) terms related to photosynthesis, while up-regulated genes were significantly enriched in abiotic stress-response–related terms (Dataset S2). We compared GO enrichment between E. nindensis and E. tef by grouping GO terms into categories of related terms (SI Appendix, Supplemental Materials and Methods). In E. nindensis, two seed-related terms were enriched while one seed-related term was enriched in E. tef (SI Appendix, Figs. S6 and S7, and Dataset S3). Interestingly, six light response terms were enriched in E. nindensis but no light response terms were enriched in E. tef. Genes up-regulated during rehydration were significantly enriched in GO terms related to photosynthesis and, specifically, in photoprotection and regulation of photomorphogenesis (Dataset S4).
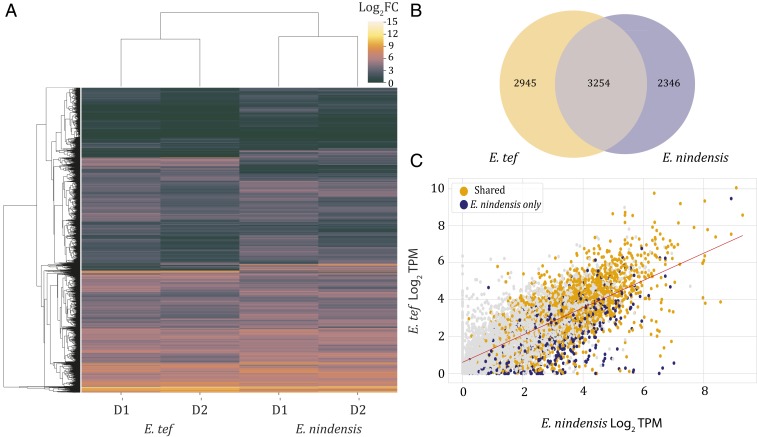
Because of the polyploid nature of E. nindensis and E. tef, we compared expression patterns between the two species using sets of “syntenic orthogroups” (conserved collinear genes) rather than individual gene pairs. Syntenic orthogroups were defined using the MCscan algorithm in blocks of 2:4 corresponding to the allotetraploidy in E. tef and allotetraploidy plus heterozygosity in E. nindensis. Of the conserved syntenic gene groups between E. tef and E. nindensis, 5,600 and 6,199 syntenic groups were up-regulated during the first two water-stress time points (D1, D2) in E. nindensis and E. tef, respectively (Fig. 3A). The majority of these syntenic orthogroups (3,254) were up-regulated in both species (Fig. 3B), supporting a broad conservation of drought responses. To further examine the degree of conservation between E. nindensis and E. tef, we compared the maximum expression in each syntenic group for the well-watered, D1, and D2 time points. Expression was significantly correlated between the two species (F-test, P < 0.001; Pearson’s r2 = 0.54), suggesting that similar pathways are involved in response to water deficit (Fig. 3C). A subset of orthogroups were uniquely expressed in each species, and across the comparable time points, 2,346 syntenic orthogroups were uniquely up-regulated in E. nindensis (Fig. 3B), and 2,945 syntenic orthogroups were uniquely up-regulated in E. tef. Candidate genes that confer vegetative desiccation tolerance are most likely to come from the gene set unique to E. nindensis.
Fig. 3.
Shared and unique desiccation-associated expression changes. (A) Heatmap of shared and unique syntenic orthogroups up-regulated in drought/desiccation time points of E. tef and E. nindensis. D1 and D2 represent the first and second drought time point for E. nindensis and E. tef, respectively. (B) Venn diagram of shared and uniquely up-regulated syntenic orthogroups. (C) Correlation of expression, measured as log2 transcripts per million (TPM), between the syntenic orthogroups of E. tef and E. nindensis where syntenic orthogroups with similar up-regulation are shown in brown, and uniquely up-regulated orthogroups in E. nindensis are shown in blue.
Induction of “Seed-Related Pathways” during Dehydration and Desiccation.
The long-standing hypothesis that vegetative desiccation tolerance evolved from the rewiring of seed development pathways has been supported by several recent genome-scale analyses (5, 7, 8). We tested this hypothesis in E. nindensis by examining whether genes with typically seed-specific expression are induced during vegetative desiccation. We then used a comparative approach to test if these seed- related genes are more broadly expressed during dehydration in E. tef and other cereals. We created a list of “seed-related” genes in grasses through comparing seed and leaf expression datasets from four cereals (E. tef, Oryza sativa, Zea mays, and Sorghum bicolor) and identifying genes with high expression in seeds relative to well-watered leaves (Materials and Methods). To facilitate cross-species comparisons, we identified pairwise syntenic orthologs between each of the six grass species with comparative expression datasets (E. nindensis, E. tef, O. thomaeum, O. sativa, Z. mays, and S. bicolor), herein referred to as syntelog groups (Materials and Methods). In total, we identified 640 syntelog groups with conserved induction in seeds compared to well-watered leaves. We clustered the syntelog groups into 386 orthogroups; of these, we found that 189 seed-related orthogroups were up-regulated in E. nindensis leaves in at least one of the stages of dehydration (SI Appendix, Fig. S6B). Using a permutation test, we determined that this number was significantly more than expected by chance (permutation test: P < 0.001), suggesting that expression of seed-related genes is critical for drought response in E. nindensis leaves (25).
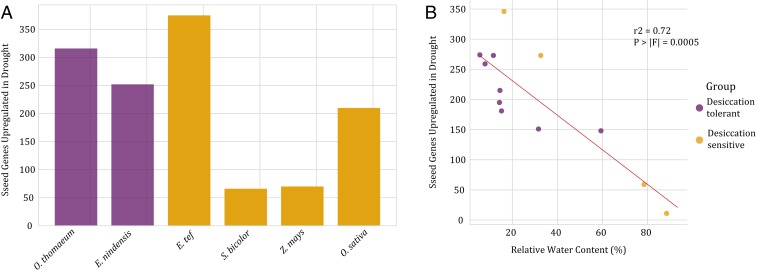
We then compared the E. nindensis expression data with reanalyzed well-watered and drought-stressed leaf expression data from the same four grass species to determine whether the observed enrichment of seed-related genes is unique to desiccation-tolerant plants or if it represents a more broadly shared water-deficit stress response. We counted the number of seed-related syntegroups up-regulated in leaves during water deficit in each of the six species. E. tef had the greatest number of seed-related genes up-regulated in dehydrated leaves, followed by O. thomaeum, E. nindensis, and O. sativa (Fig. 4A). S. bicolor and Z. mays had fewer seed-related genes up-regulated in water-stressed leaves compared to the other four species. The number and severity of water-deficit stress time points differed between species, limiting our power to draw conclusions about differences in expression of seed-related genes during water deficit between the species. However, these data do suggest that the number of such genes up-regulated during water-deficit stress in the desiccation-tolerant grasses E. nindensis and O. thomaeum is similar to the number observed during severe water-deficit conditions in desiccation-sensitive grasses.
Fig. 4.
Induction of seed pathways during drought in grasses. (A) Bar graph of seed-related gene expression in drought-treated leaf tissue of the desiccation-tolerant grasses O. thomaeum and E. nindensis (purple) and desiccation-sensitive cereals E. tef, S. bicolor, Z. mays, and O. sativa (gold). (B) Correlation of leaf RWC vs. induced seed-related pathways for drought-treated desiccation-tolerant (purple) and -sensitive (gold) species.
For the four species for which RWC data were available for the sampled time points (E. nindensis, E. tef, O. thomaeum, and S. bicolor), we compared the number of seed-related syntelog groups up-regulated during each drought time point with the mean RWC of all replicates from that time point (Fig. 4B). We found that RWC was a significant predictor of the number of seed-related syntelogs up-regulated in leaves during drought (F-test, P < 0.001), and RWC also explained a substantial amount of the variation in the number of seed-related syntelogs up-regulated in leaves during drought (Pearson’s, r2 = 0.72). Conversely, whether a sample was derived from a desiccation-tolerant or -sensitive plant did not significantly predict the number of seed-related syntelog groups up-regulated in leaves during drought (F-test, P = 0.53). This model did not explain much of the observed variation (r2 = 0.041). The spread of the model residuals was also much larger for this desiccation tolerance model as compared with the RWC model, indicating a better fit for the RWC model (F-test, P = 0.001) (SI Appendix, Fig. S8). While this analysis is somewhat limited by the paucity of RWC measurements, and diverse time points over which plants were dried out, we can tentatively conclude that more seed-related genes are up-regulated at lower RWC in grasses regardless of whether the species possesses desiccation tolerance.
Unique Components of Desiccation Tolerance in Grasses.
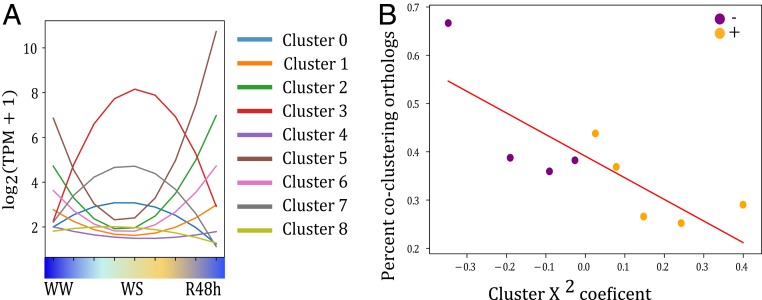
Comparisons of drought-induced expression across grasses suggest that seed pathways are not uniquely induced in resurrection plants but instead represent a conserved response to water deficit. Using this comparative framework, we searched for unique patterns that distinguish tolerant and sensitive species. E. nindensis and O. thomaeum represent separate tribes and likely independent origins of desiccation tolerance within the Chloridoideae subfamily. These species also utilize different photoprotective strategies under desiccation where E. nindensis largely degrades and O. thomaeum retains chlorophyll. We distinguished conserved expression patterns indicative of convergent evolution from species-specific patterns reflecting differences in desiccation strategies. We used k-means clustering to group E. nindensis and O. thomaeum genes based on a curve fit to their expression pattern during separate desiccation and rehydration time series (Fig. 5A). For each resulting cluster, we counted the number of O. thomaeum genes with at least one coclustering E. nindenis syntenic ortholog. Genes in clusters that on average contained genes more strongly up-regulated during desiccation were more likely to cocluster with their E. nindensis syntenic orthologs. In addition, the x2 coefficient for syntenic ortholog pairs in both species were significantly correlated (Pearson’s, r = 0.53; F-test, P < 0.001), suggesting overall conservation of expression patterns between the two species (Fig. 5B). In particular, expression patterns of seed-related genes were significantly more strongly correlated (Pearson’s r = 0.69), t test for sample correlation coefficient (P < 0.001) than either all pairs (Pearson’s r = 0.53) or all syntenic pairs up-regulated in both species (Pearson’s r = 0.55). This pattern of conservation suggests that seed-related genes may have similar roles, and possibly similar regulation, during dehydration and rehydration in both E. nindensis and O. thomaeum.
Fig. 5.
Comparative network dynamics across desiccation-tolerant grasses. (A) K-mean clusters for the E. nindensis and O. thomaeum desiccation and rehydration time courses. (B) Cluster coefficient of expression patterns of syntenic orthogroups.
We identified a set of 239 syntenic orthogroups which were up-regulated under water-deficit stress in both E. nindensis and O. thomaeum but not in any of the four desiccation-sensitive species examined. Based on a permutation test, an overlap of 239 genes is significantly more than expected by random chance (permutation test: P < 0.001) (25). Among the conserved desiccation-associated genes in E. nindensis and O. thomaeum are a wide array of transcription factors (SI Appendix, Table S3) including orthologs of Arabidopsis DOG1 and HY5, which regulate seed dormancy and photomorphogenesis, respectively. The ABA-dependent transcription factor ABI3 is important for controlling seed development and is thought to be critical for vegetative desiccation tolerance (7). However, the set of conserved up-regulated transcription factors does not include orthologs of Arabidopsis ABI3 or any other LAFL transcription factors responsible for regulating seed dormancy. Of the 23 orthogroups containing target genes of the ABI3 regulon in Arabidopsis (26), 16 and 20 contained at least one gene which was up-regulated during desiccation in E. nindensis and O. thomaeum, respectively. However, 16 of the 23 orthogroups were also up-regulated during water-deficit stress in E. tef, and 12 ABI3-regulated orthogroups were up-regulated during water stress in O. sativa. We identified only one ABI3-regulated orthogroup (OG0002708) containing cupin seed storage proteins that is uniquely up-regulated in only the desiccation-tolerant grasses. Furthermore, of the 239 genes with vegetative expression unique to the two desiccation-tolerant species, only 14 were from our list of seed-related genes, which is not more than expected by random chance (permutation test: P = 0.37). Taken together, this suggests that much of the ABI3 regulon, and seed-related genes more broadly, are induced in leaves during both dehydration and desiccation, implying that expression of seed-related genes alone is insufficient to confer desiccation tolerance.
Late embryogenesis abundant (LEA) proteins are believed to play an important role in protecting cellular components from damage during desiccation (27). It is likely that certain LEA proteins (or groups of LEAs) play an essential and conserved role in vegetative desiccation tolerance, but some are also more broadly involved in drought response pathways among desiccation-sensitive plants (28). We identified LEA genes belonging to the eight LEA subfamilies in the genomes of E. nindensis, E. tef, and O. thomaeum using PFAM domains (SI Appendix, Fig. S9). We found no evidence of expansion of any LEA subfamilies in E. nindensis relative to E. tef (SI Appendix, Figs. S9 and S10). The LEA5 and LEA6 (also referred to as LEA18) subfamilies were uniquely induced during desiccation and rehydration in E. nindensis and O. thomaeum, but neither subfamily showed increased expression during water stress in E. tef. The remaining LEA subfamilies showed similar patterns across all three species (SI Appendix, Fig. S10).
All previously sequenced resurrection plant genomes have massive tandem arrays of early light-induced proteins (ELIPs) to protect against photooxidative damage during prolonged desiccation (29). Consistent with this pattern, the E. nindensis genome has 27 ELIPs, and most are found in large tandem arrays (SI Appendix, Fig. S11A). The ELIPs in E. nindensis are nonsyntenic to the 22 orthologs in O. thomaeum and the 5 orthologs in E. tef, suggesting that they translocated and duplicated after the divergence of these grasses. The nonsyntenic nature of the ELIP tandem arrays in E. nindensis and O. thomaeum supports an independent origin of desiccation tolerance, as distinct ELIPs were duplicated in each species. ELIPs are induced under water stress with 23 of the 27 up-regulated during desiccation or rehydration in E. nindensis (SI Appendix, Fig. S11B). ELIPs are most highly expressed 12 and 24 h post rehydration, contrasting with most other species where ELIPs are highest in desiccated tissues (29). Unlike O. thomaeum, E. nindensis largely degrades its chlorophyll and dismantles thyllakoids (a strategy termed poikilochlorophylly) during desiccation, with the thyllakoids and chlorophyll being reconstituted upon rehydration. In turn, this results in a comparatively slow post-rehydration recovery.
Chlorophyll degradation is catalyzed by chlorophyllase enzymes, and we observed different expression patterns in E. nindensis and O. thomaeum, reflecting alternate strategies of chlorophyll degradation or retention during desiccation (SI Appendix, Fig. S12). An E. nindensis chlorophyllase (En_0076685) was up-regulated 3.5-fold under desiccation but had no change in expression during any other time point. The syntenic ortholog in O. thomaeum (Ot_Chr2_06331) was not up-regulated during any desiccation time point, but was slightly up-regulated 48 h post rehydration. The two E. tef chlorophyllases (Et_2A_015704 and Et_2B_019834) are up-regulated in seeds but not in leaves, suggesting that desiccation-associated expression of chlorophyllase may be specific to chlorophyll-degrading resurrection plants. Senescence-related chlorophyll degradation is catalyzed by pheophytinase (30, 31) which is up-regulated in all three species, suggesting that pheophytinase activity is a more general drought response (SI Appendix, Fig. S12).
Chromatin Dynamics and Epigenetic Changes during Desiccation.
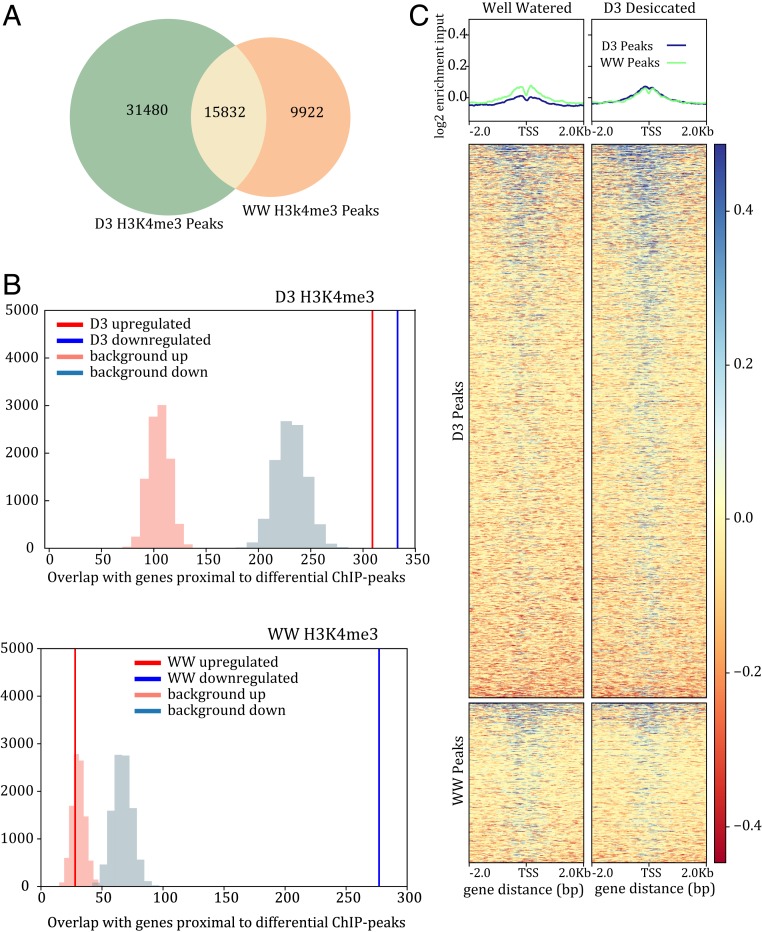
Alterations of histone modifications and DNA methylation are correlated with stress-induced gene expression, and these processes are integral for many stress responses (32). Histone modifications are also important for regulating seed dormancy possibly through a thermal-sensing role (33). It was previously suggested that chromatin modifications may be partly responsible for gene regulatory changes required for desiccation tolerance (34, 35). We surveyed methylation changes and chromatin dynamics in well-watered and desiccated E. nindensis leaves using Bisulfite-seq and ChIP-seq with a histone modification associated with open chromatin (H3K4me3). H3K4me3 is correlated with active transcription, and these histone marks accumulate immediately upstream of the transcriptional start site of actively transcribed genes (36). We identified regions with differential binding of H3K4me3 antibody between well-watered E. nindensis leaves and desiccated leaves from the D3 time point. Across the three replicates of well-watered leaves, we identified 25,754 H3K4me3 peaks with significant enriched coverage (Wald test q < 0.05) over the input control (Fig. 6A). The D3 samples contained 47,312 peaks, and 15,832 peaks overlapped by at least one base with the well-watered peaks. Despite the large number of unique peaks in each condition, only 3,757 peaks had significantly greater binding in D3 compared to WW (Wald test q < 0.05), and 949 peaks had significantly more binding in WW compared to D3 (Fig. 6C). We identified the closest gene to each of these differentially bound peaks and tested for enrichment of genes with up- or down-regulated expression in D3 compared with WW. We found significant enrichment of both up- and down-regulated genes among genes proximal to the peaks with increased binding in D3 (Fig. 6B). Only genes down-regulated in D3 compared to WW were enriched for H3K4me3 peaks with increased binding in the WW samples. The significant number of altered H3K4me3 histone modifications and overlap with differentially expressed genes suggests that chromatin dynamics play a central role in desiccation tolerance.
Fig. 6.
Changes in histone modifications associated with desiccation in E. nindensis. (A) Venn diagram of overlapping H3K4me3 peaks between well-watered and desiccated samples. (B) Enrichment of H3K4me3 peaks and up-regulated genes during desiccation. The expected background distribution is shown in light blue or light red, and the observed is shown in red and blue for up-regulated and down-regulated genes, respectively. (C) Heatmap of fold enrichment of peaks at the transcriptional start site of genes. The color scale represents log2 enrichment values over the input.
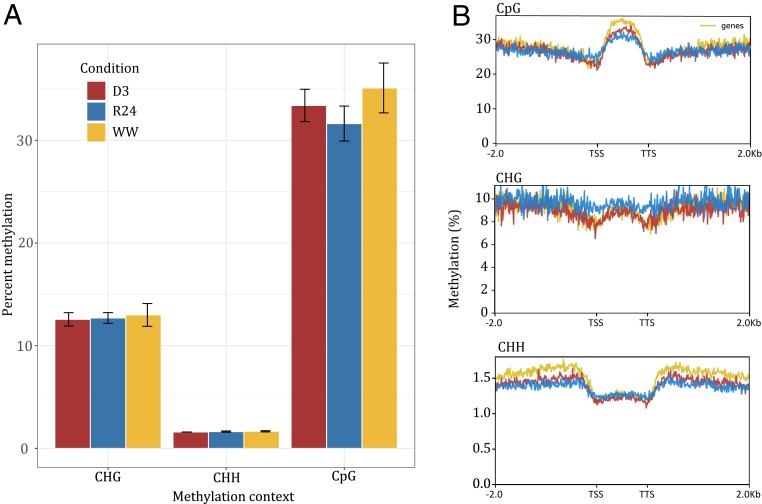
We surveyed changes in DNA methylation in desiccated (D3), rehydrated, and well-watered leaf tissue. Similar to other plants, E. nindensis has low levels of CHH methylation across the genome, and moderate levels of CpG and CHG methylation (Fig. 7A). There was no global difference in methylation levels across the surveyed drought and rehydration time points. However, methylation levels upstream, downstream, and within the gene body varied across the three time points (Fig. 7B). CpG and CHH gene body methylation was lower in desiccated and rehydrated leaf tissue compared to well-watered leaf tissue. Interestingly, CHG gene body methylation was higher in rehydrated leaf tissue compared to well-watered and desiccated leaf tissue, with no reduction in methylation level around the upstream transcriptional start site or downstream transcriptional termination site (Fig. 7B). This general pattern is consistent with stress-induced hypomethylation and transcriptional reprogramming (37).
Fig. 7.
Desiccation-induced changes in DNA methylation in E. nindensis. (A) Global patterns of CHH, CpG, and CHG methylation across the genome for WW, desiccated (D3), and 24-h postrehydration (R24) leaf samples. (B) Gene body methylation for the three methylation contexts in the surveyed samples. Methylation is plotted in a rolling window for all genes in the upstream (transcriptional start site: TSS), downstream (transcriptional termination site: TTS), and body of genes.
Discussion
Desiccation tolerance has evolved recurrently across plants, animals, and microbes as a common adaptation to water-limited environments. Surviving extreme drying requires the coordinated deployment of complex processes to prevent oxidative damage and protect the macromolecules and membranes of cells. Some core elements of desiccation responses are shared across diverse eukaryotes (38), and the potential to evolve tolerance is widespread. Desiccation-tolerant plants have the added challenge of withstanding excess light during prolonged desiccation and have evolved unique photoprotective mechanisms such as early light-induced protein gene family expansion in response (29, 39). The origin of desiccation tolerance in plants is likely linked to the colonization of land, where the algal ancestors of plants would experience rapid and prolonged drying. It was previously hypothesized that this tolerance was subsequently lost in vegetative tissues of most land plants, but retained in seeds, spores, and pollen. Vegetative desiccation tolerance was likely regained independently multiple times in angiosperms (5). The alternative explanation for the distribution of vegetative desiccation tolerance among angiosperms is multiple reversions in desiccation-sensitive lineages similar to the pattern previously described for the evolution of nitrogen-fixing symbiosis (40). However, multiple independent gains of vegetative desiccation tolerance are more likely due to the large number of reversions required to explain the trait’s distribution. In addition, genomic evidence of independent gains of involved components, including the apparent independent expansion of early light-induced proteins in desiccation-tolerant lineages, suggests multiple independent gains of the desiccation-tolerant phenotype (29). The recurrent evolution of vegetative desiccation tolerance across diverse flowering plants has previously been attributed to rewiring of seed and pollen desiccation pathways (4). Studies examining gene expression during desiccation have repeatedly supported this claim by identifying increased expression of seed-related genes in vegetative tissues during drying (7, 8). However, few studies have conducted genome-wide comparisons of tolerant and sensitive species to test whether these seed-related genes are truly unique to desiccation-tolerant plants. Another possibility is that these broad seed-related pathways were never lost in vegetative tissues, but were instead repurposed for roles in typical drought responses.
Using a comparative approach, we observed a similar pattern of seed-related pathway expression under water deficit in desiccation-tolerant and -sensitive grasses. This is not totally surprising as some seed-dehydration–associated pathways have well-characterized overlap with drought responses such as accumulation of osmoprotectants, LEA proteins, and ROS scavengers (41–43). For example, 22 of the 57 LEA genes in Arabidopsis have drought-induced expression but only 10 LEA genes have overlapping expression in drying seeds (44). Across desiccation-tolerant and -sensitive grasses, we observed a substantial overlap between LEA expression under water deficit in leaves and developing seeds. More broadly, ABA is a central signaling molecule for environmental stress and seed development, and ABA-based regulatory networks have some overlap between these processes (45). The ABA-responsive transcription factor ABI3 is a well-characterized regulator of seed maturation drying pathways (46, 47). In the desiccation-tolerant monocot Xerophyta viscosa, orthologs to the majority of ABI3-regulated genes are expressed in leaves during dehydration (7). Similarly, we found that most of the 23 orthogroups containing ABI3-responsive genes were up-regulated during desiccation in both E. nindensis and O. thomaeum. However, many of these orthogroups were also up-regulated during water-deficit stress in the desiccation-sensitive grasses E. tef and O. sativa. Although most ABI3-responsive genes are induced under water deficit, expression of ABI3 orthologs was low in both E. nindensis and O. thomaeum. This is consistent with recent findings from Xerophyta humilis, where there was no evidence that ABI3 or the canonical LAFL seed maturation regulatory network was responsible for desiccation tolerance (48). Taken together, this suggests that many “seed-related” genes are expressed as a universal response to water deficit, but the regulation of these processes is likely distinct from seed regulatory networks.
We identified a strong correlation between the induction of seed-related genes and the severity of the drought treatment in grasses. Comparatively few seed-related genes were expressed under mild drought in any grass, but the number rose dramatically as relative water content decreased. This trend may have been overlooked in previous studies as most water-deficit experiments are comparatively mild, and few survey lethal or sublethal stresses. These seed-related pathways may be induced as a last ditch effort under severe conditions, but are insufficient or too late to prevent fatal damage. In developing seeds, water content decreases as the accumulation of food reserves drives out cellular water (49). Seeds begin to acquire desiccation tolerance starting at ∼50% RWC, and 50% of seeds were tolerant at ∼30% RWC (50). This indicates that a generic water-deficit response occurs in seeds prior to a desiccation response. This pattern is very similar to what is observed in dehydrating resurrection species and suggestive of a common ancestral mechanism related to water content or water potential. This hypothesis warrants further consideration, as only two datasets from desiccation-sensitive grasses with coupled expression and drought physiology data have been collected. Further work comparing the expression of otherwise seed-specific genes during severe drought, and, importantly, captured at low relative water contents, is needed.
Our results suggest a strong overlap between drought and desiccation responses, but what elements are unique to desiccation? It was previously shown that expansion of ELIP genes is conserved among all sequenced desiccation-tolerant plants (29). Similar to other desiccation-tolerant plants, E. nindensis contains an expansion of ELIPs, and the majority are highly expressed during dehydration. Interestingly, ELIPs are most highly expressed in E. nindensis during rehydration, in contrast to patterns observed in chlorophyll-retaining species. High ELIP expression was also observed during rehydration in the chlorophyll-degrading species Xerophyta viscosa (7). ELIPs may function to protect leaves during the slow postrehydration recovery in chlorophyll-degrading species, mirroring their role in germinating seeds (51). Consistent with adaptation to light stress, E. nindensis expresses genes for anthocyanin and carotenoid biosynthesis at much higher levels than E. tef. Furthermore, orthologs of the hub regulator for photomorphogenesis HY5 are expressed during drought exclusively in the desiccation-tolerant species. Thus, we infer that mechanisms to protect against photooxidative damage are critical for the desiccation-tolerant phenotype even in chlorophyll-degrading species.
Desiccation tolerance is found in five tribes of chloridoid grasses across nine genera within four distinct clades among numerous desiccation-sensitive species (6). Many chloridoid grasses are drought and heat tolerant, and this preadaptation may have facilitated the recurrent evolution of desiccation tolerance within this subfamily. It is possible that some desiccation tolerance mechanisms are shared with desiccation-sensitive but still highly resilient members of this subfamily. This could explain the strong overlap in expression patterns between E. nindensis and E. tef and the induction of similar seed-related genes. Species like E. tef may represent evolutionary intermediates with induction of some desiccation-related pathways that are not observed in less tolerant plants. Desiccation tolerance strategies vary within grasses, and E. nindensis and O. thomaeum utilize different photoprotective strategies to cope with anhydrobiosis. E. nindensis largely degrades its chlorophyll prior to desiccation, and O. thomaeum retains and protects its chlorophyll and photosystem II complexes. Although many desiccation-specific orthogroups are similarly expressed in both species, we observed unique expression patterns that may reflect differing photoprotective strategies.
Chromatin remodeling plays an important role in seed development, and it was previously proposed that chromatin dynamics could have a similar role in desiccation tolerance (35, 52, 53). We found no global changes in methylation during desiccation; however, we did observe an intriguing pattern of enrichment of H3K4me3 histone modifications upstream of genes down-regulated during desiccation. Typically, H3K4me3 histone marks accumulate upstream of actively transcribed genes, a pattern we also observed in both well-watered and desiccated samples. However, the observed enrichment near down-regulated genes is unusual. In seeds, chromatin remodeling is important for genome compaction as well as for preparing the genome for transcription upon germination (54). We hypothesize that the observed enrichment of H3K4me3 histone marks upstream of down-regulated genes may be a sign of chromatin remodeling in preparation for transcription upon rehydration.
Here, we propose that seed dehydration pathways are important components of both drought and vegetative desiccation responses. Numerous previous studies have shown that seed dehydration genes are important in protecting leaves of desiccation-tolerant species. We identified a similar pattern in E. nindensis; however, comparisons to E. tef and other grasses reveal that seed pathways are also important in leaves of desiccation-sensitive species during drought. The importance of these pathways for general drought response has been understated in the previous literature. Nevertheless, some aspects of seed dehydration pathways do appear to be specific to seeds and leaves of desiccation-tolerant plants. Photoprotective pathways that are important in germinating seeds are also active in desiccated and rehydrating leaf tissue of resurrection plants. The timely, coordinated, and orderly induction of these desiccation-responsive pathways may be essential for engineering improved stress resilience in crop plants.
Materials and Methods
Accessions of E. nindensis (PI 410063) and E. tef (PI 524434) were obtained from the US Department of Agriculture Germplasm Resources Information Network (http://www.ars-grin.gov/). Methodological details of plant growth conditions, water-deficit treatments, nucleic acid isolation, genome assembly, comparative genomics, expression analysis, Bisulfite-Seq, and ChIP-seq are described in SI Appendix, Supplemental Materials and Methods.
Data Availability.
The raw PacBio data, Illumina DNA-seq, RNA-seq data, Bisulfite-seq, and ChIP-seq are available from the National Center for Biotechnology Information (NCBI) Short Read Archive. E. nindensis RNAseq data can be found under BioProject accession no. PRJNA548129 and PRJNA548367, and E. tef RNAseq data can be found under BioProject accession no. PRJNA548000. Bisulfite-seq and ChIP-seq for E. nindensis can be found under BioProject acession no. PRJNA548367. The E. nindensis V2.1 genome can be downloaded from NCBI BioProject accession no. PRJNA622516 and CoGe (ID 54689). Code used to analyze the expression data is available on GitHub (https://github.com/pardojer23/VanBuren_Lab_Genomics_Tools).
Supplementary Material
Acknowledgments
We thank Yao Cao for help with DAPI staining and karyotyping, Alan Yocca for assistance with his Ka/Ks pipeline, and Scott Pardo for reviewing the reporting of statistical results. This work is supported by NSF Grant MCB‐1817347 (to R.V.) and by predoctoral training award T32-GM110523 from the National Institute of General Medical Sciences of the NIH (to J.P.). Hannah Chay was supported by the High School Honors Science, Math, and Engineering Program at Michigan State University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. E.G. is a guest editor invited by the Editorial Board.
Data deposition: The raw PacBio data, Illumina DNA-seq, RNA-seq data, Bisulfite-seq, and ChIP-seq data are available from the National Center for Biotechnology Information (NCBI) Short Read Archive. Eragrostis nindensis data can be found under BioProject accession no. PRJNA548129 and PRJNA548367. Eragrostis tef data can be found under BioProject accession no. PRJNA548000. Oropetium thomaeum RNAseq data can be found under BioProject PRJNA548527. The E. nindensis V2.1 genome can be downloaded from NCBI PRJNA622516 and CoGe databases (ID 54689).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001928117/-/DCSupplemental.
References
- 1.Delwiche C. F., Cooper E. D., The evolutionary origin of a terrestrial flora. Curr. Biol. 25, R899–R910 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Bateman R. M., et al. , Early evolution of land plants: Phylogeny, physiology, and ecology of the primary terrestrial radiation. Annu. Rev. Ecol. Syst. 29, 263–292 (1998). [Google Scholar]
- 3.Franchi G. G., et al. , Pollen and seed desiccation tolerance in relation to degree of developmental arrest, dispersal, and survival. J. Exp. Bot. 62, 5267–5281 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Oliver M. J., Tuba Z., Mishler B. D., The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 151, 85–100 (2000). [Google Scholar]
- 5.Gaff D. F., Oliver M., The evolution of desiccation tolerance in angiosperm plants: A rare yet common phenomenon. Funct. Plant Biol. 40, 315–328 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Peterson P. M., Romaschenko K., Herrera Arrieta Y., A molecular phylogeny and classification of the Cynodonteae (Poaceae: Chloridoideae) with four new genera: Orthacanthus, Triplasiella, Tripogonella, and Zaqiqah; three new subtribes: Dactylocteniinae, Orininae, and Zaqiqahinae; and a subgeneric classification of Distichlis. Taxon 65, 1263–1287 (2016). [Google Scholar]
- 7.Costa M. D., et al. , A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat. Plants 3, 17038 (2017). [DOI] [PubMed] [Google Scholar]
- 8.VanBuren R., et al. , Seed desiccation mechanisms co-opted for vegetative desiccation in the resurrection grass Oropetium thomaeum. Plant Cell Environ. 40, 2292–2306 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Gechev T. S., et al. , Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 70, 689–709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez M. C. S., et al. , Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 63, 212–228 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Yobi A., et al. , Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol. 17, 67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y., et al. , Global transcriptome analysis reveals acclimation-primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 56, 1429–1441 (2015). [DOI] [PubMed] [Google Scholar]
- 13.VanBuren R., et al. , Desiccation tolerance evolved through gene duplication and network rewiring in Lindernia. Plant Cell 30, 2943–2958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manfre A. J., LaHatte G. A., Climer C. R., Marcotte W. R. Jr, Seed dehydration and the establishment of desiccation tolerance during seed maturation is altered in the Arabidopsis thaliana mutant atem6-1. Plant Cell Physiol. 50, 243–253 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Shinozaki K., Yamaguchi-Shinozaki K., Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Daszkowska-Golec A., “The role of abscisic acid in drought stress: How ABA helps plants to cope with drought stress” in Drought Stress Tolerance in Plants, Vol 2: Molecular and Genetic Perspectives, Hossain M. A., Wani S. H., Bhattacharjee S., Burritt D. J., Tran L.-S. P., Eds. (Springer International Publishing, 2016), pp. 123–151. [Google Scholar]
- 17.VanBuren R., Wai C. M., Keilwagen J., Pardo J., A chromosome-scale assembly of the model desiccation tolerant grass Oropetium thomaeum. Plant Direct 2, e00096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanBuren R., et al. , Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat. Commun. 11, 884 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidse G., Hoshino T., Simon B. K., Chromosome counts of Zimbabwean grasses (Poaceae) and an analysis of polyploidy in the grass flora of Zimbabwe. S. Afr. J. Bot. 52, 521–528 (1986). [Google Scholar]
- 20.Roodt R., Spies J. J., Chromosome studies in the grass subfamily Chloridoideae. II. An analysis of polyploidy. Taxon 52, 736–746 (2003). [Google Scholar]
- 21.Chen L.-Y., et al. , The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 51, 1549–1558 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Campbell M. S., et al. , MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164, 513–524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginbot Z. G., Farrant J. M., Physiological response of selected Eragrostis species to water-deficit stress. Afr. J. Biotechnol. 10, 10405–10417 (2011). [Google Scholar]
- 24.Hinckley T. M., Lassoie J. P., Running S. W., Temporal and spatial variations in the water status of forest trees. For. Sci. 24, a0001–z0001 (1978). [Google Scholar]
- 25.Good P., Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses (Springer, New York,1994). [Google Scholar]
- 26.Magadum S., Banerjee U., Murugan P., Gangapur D., Ravikesavan R., Gene duplication as a major force in evolution. J. Genet. 92, 155–161 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Goyal K., Walton L. J., Tunnacliffe A., LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olvera-Carrillo Y., Luis Reyes J., Covarrubias A. A., Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 6, 586–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.VanBuren R., Pardo J., Man Wai C., Evans S., Bartels D., Massive tandem proliferation of ELIPs supports convergent evolution of desiccation tolerance across land plants. Plant Physiol. 179, 1040–1049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckardt N. A., A new chlorophyll degradation pathway. Plant Cell 21, 700 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk N., et al. , The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett. 581, 5517–5525 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kim J.-M., Sasaki T., Ueda M., Sako K., Seki M., Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6, 114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Footitt S., Müller K., Kermode A. R., Finch-Savage W. E., Seed dormancy cycling in Arabidopsis: Chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 81, 413–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitra J., Xu G., Wang B., Li M., Deng X., Understanding desiccation tolerance using the resurrection plant Boea hygrometrica as a model system. Front. Plant Sci. 4, 446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilhorst H. W. M., Costa M. D., Farrant J. M., A footprint of plant desiccation tolerance. Does it exist? Mol. Plant 11, 1003–1005 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Howe F. S., Fischl H., Murray S. C., Mellor J., Is H3K4me3 instructive for transcription activation? BioEssays 39, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Chinnusamy V., Zhu J.-K., Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berjak P., Unifying perspectives of some mechanisms basic to desiccation tolerance across life forms. Seed Sci. Res. 16, 1–15 (2006). [Google Scholar]
- 39.Challabathula D., Zhang Q., Bartels D., Protection of photosynthesis in desiccation-tolerant resurrection plants. J. Plant Physiol. 227, 84–92 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Griesmann M., et al. , Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361, eaat1743 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Nagaraju M., et al. , Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS One 14, e0209980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cruz de Carvalho M. H., Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 3, 156–165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kido E. A., et al. , Expression dynamics and genome distribution of osmoprotectants in soybean: Identifying important components to face abiotic stress. BMC Bioinformatics 14 (suppl. 1), S7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bies-Ethève N., et al. , Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 67, 107–124 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Song L., et al. , A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parcy F., et al. , Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooms J., Leon-Kloosterziel K. M., Bartels D., Koornneef M., Karssen C. M., Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana (A comparative study using abscisic acid-insensitive abi3 mutants). Plant Physiol. 102, 1185–1191 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyall R., et al. , Vegetative desiccation tolerance in the resurrection plant Xerophyta humilis has not evolved through reactivation of the seed canonical LAFL regulatory network. Plant J., 10.1111/tpj.14596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derek Bewley J., Bradford K., Hilhorst H., Nonogaki H., Seeds: Physiology of Development, Germination and Dormancy (Springer Science & Business Media, 2012). [Google Scholar]
- 50.Verdier J., et al. , A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 163, 757–774 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutin C., et al. , Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. U.S.A. 100, 4921–4926 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baroux C., Pien S., Grossniklaus U., Chromatin modification and remodeling during early seed development. Curr. Opin. Genet. Dev. 17, 473–479 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Wolny E., Braszewska-Zalewska A., Kroczek D., Hasterok R., Histone H3 and H4 acetylation patterns are more dynamic than those of DNA methylation in Brachypodium distachyon embryos during seed maturation and germination. Protoplasma 254, 2045–2052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Zanten M., Carles A., Li Y., Soppe W. J. J., Control and consequences of chromatin compaction during seed maturation in Arabidopsis thaliana. Plant Signal. Behav. 7, 338–341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw PacBio data, Illumina DNA-seq, RNA-seq data, Bisulfite-seq, and ChIP-seq are available from the National Center for Biotechnology Information (NCBI) Short Read Archive. E. nindensis RNAseq data can be found under BioProject accession no. PRJNA548129 and PRJNA548367, and E. tef RNAseq data can be found under BioProject accession no. PRJNA548000. Bisulfite-seq and ChIP-seq for E. nindensis can be found under BioProject acession no. PRJNA548367. The E. nindensis V2.1 genome can be downloaded from NCBI BioProject accession no. PRJNA622516 and CoGe (ID 54689). Code used to analyze the expression data is available on GitHub (https://github.com/pardojer23/VanBuren_Lab_Genomics_Tools).