Significance
There is concern for design of a safe vaccine for Zika virus because antibodies (Abs) elicited against Zika may also bind flaviviruses that share a similar envelope protein. If Abs elicited by a Zika vaccine bind, but do not effectively neutralize other flaviviruses, they may enhance virus entry into cells through the process of Ab-dependent enhancement of infection, potentially leading to more severe disease. By directly comparing how mature Zika-neutralizing Abs and their germline precursors bind different flaviviruses, we provide insight into the Ab maturation process and the molecular interactions important for strong, neutralizing binding to Zika versus weak, cross-reactive binding to other flaviviruses.
Keywords: Zika, germline antibody, flavivirus, affinity maturation, antibody-dependent enhancement
Abstract
Recent epidemics demonstrate the global threat of Zika virus (ZIKV), a flavivirus transmitted by mosquitoes. Although infection is usually asymptomatic or mild, newborns of infected mothers can display severe symptoms, including neurodevelopmental abnormalities and microcephaly. Given the large-scale spread, symptom severity, and lack of treatment or prophylaxis, a safe and effective ZIKV vaccine is urgently needed. However, vaccine design is complicated by concern that elicited antibodies (Abs) may cross-react with other flaviviruses that share a similar envelope protein, such as dengue virus, West Nile virus, and yellow fever virus. This cross-reactivity may worsen symptoms of a subsequent infection through Ab-dependent enhancement. To better understand the neutralizing Ab response and risk of Ab-dependent enhancement, further information on germline Ab binding to ZIKV and the maturation process that gives rise to potently neutralizing Abs is needed. Here we use binding and structural studies to compare mature and inferred-germline Ab binding to envelope protein domain III of ZIKV and other flaviviruses. We show that affinity maturation of the light-chain variable domain is important for strong binding of the recurrent VH3-23/VK1-5 neutralizing Abs to ZIKV envelope protein domain III, and identify interacting residues that contribute to weak, cross-reactive binding to West Nile virus. These findings provide insight into the affinity maturation process and potential cross-reactivity of VH3-23/VK1-5 neutralizing Abs, informing precautions for protein-based vaccines designed to elicit germline versions of neutralizing Abs.
Zika virus (ZIKV) is an arthropod-borne flavivirus first discovered in 1947, with the first reported human case in 1964 and large outbreaks in 2007 and 2013 to 2015 (1–8). Although commonly transmitted by mosquitoes, ZIKV can also be transmitted sexually and can persist in infected individuals for up to several months (9–12). Infection is either asymptomatic or causes mild symptoms, including fever, conjunctivitis, headache, rash, and arthralgia in ∼20% of cases, and severe neurologic problems, such as meningoencephalitis or Guillain-Barré syndrome, in rare cases (4, 13–15). There are major concerns about ZIKV infection during pregnancy, which can cause fetal neurodevelopmental abnormalities, such as microcephaly (10, 16–19). Given the large-scale spread, symptom severity, and lack of treatment or prophylaxis, a safe and effective ZIKV vaccine is urgently needed.
The ZIKV envelope (E) protein—containing envelope protein domain (ED)I, EDII, and EDIII—is similar to the E protein of other flaviviruses, including dengue virus serotypes 1 to 4 (DENV1-4), West Nile virus (WNV), and yellow fever virus (YFV) (20–22). EDIII is an important target for neutralizing antibodies (Abs) (23–25). Indeed, many Abs against the ZIKV EDIII domain are strongly neutralizing and are an important component of the response to infection (26–38). A set of recurrent Abs (commonly occurring in multiple individuals, also referred to as “public Abs”) identified from a large cohort of patients in Brazil and Mexico potently neutralize both ZIKV and DENV1 by binding the lateral ridge of the EDIII domain (28). These Abs share the germline variable heavy (VH) gene segment VH3-23 and the germline variable κ (VK) gene segment VK1-5. One of these VH3-23/VK1-5 Abs, Z004, exhibited protection against ZIKV infection in mice, and when used in combination with another Ab, Z021, reduced viremia and prevented the emergence of ZIKV escape mutations in infected macaques (28, 29).
Strongly neutralizing anti-ZIKV Abs that are derived from known germline Ab precursors represent a potential target for a germline-targeting approach to vaccine design. Such approaches rely on the Ab response to an antigen being initiated through antigen binding to a B cell receptor in its germline configuration, triggering B cell activation and subsequent affinity maturation through the process of somatic hypermutation (39). However, a potential concern for vaccine design efforts targeting the epitope for VH3-23/VK1-5 Abs or other flavivirus epitopes is that generation of weakly neutralizing or nonneutralizing Abs against ZIKV that cross-react with different flaviviruses could result in enhanced infection through the process of Ab-dependent enhancement (ADE) (22, 40–44). It is thought that ADE can result when the binding of cross-reactive—but non- or only poorly neutralizing—Abs promote viral entry into Fcγ receptor (FcγR)-expressing cells, thereby providing an alternative route of infection and causing increased virus production and symptom severity (22, 41, 42, 45–48). Therefore, understanding the ability of germline Abs to bind flavivirus envelope proteins and mature into specific, potently neutralizing Abs is important for development of a safe vaccine.
The Ab affinity maturation process for EDIII recognition can be investigated by structural comparisons of germline and mature Ab recognition of antigen. This approach provided insights into the affinity maturation of Abs against other viruses, including an increased understanding of modes of binding and somatic hypermutation in broadly neutralizing Abs against HIV-1 (49–53). In the case of ZIKV, knowledge of how both germline and mature versions of potently neutralizing Abs bind flaviviruses may enhance our understanding of the interactions that give rise to potent neutralization versus weak cross-reactivity that could contribute to risk for ADE.
Here we report binding and structural studies to gain insight into affinity maturation and cross-reactivity of the VH3-23/VK1-5 class of anti-ZIKV Abs. Through sequence alignments, surface plasmon resonance (SPR), neutralization assays, ADE assays, and structural studies, we compared mature and inferred germline (iGL) Ab binding to flavivirus EDIII domains from ZIKV, DENV1-4, WNV, and YFV. As part of this analysis, we compared two crystal structures, an iGL Ab bound to ZIKV EDIII and a cross-reactive mature Ab bound to WNV EDIII, with two previously determined crystal structures of potently neutralizing mature Abs bound to ZIKV and DENV1 EDIII (28). These findings revealed components of germline maturation, including contributions of somatic hypermutation in the variable domain (VL) of the Fab, important for development of VH3-23/VK1-5 Abs that potently neutralize ZIKV and inform cross-reactivity precautions for flavivirus vaccine design and passive delivery of ZIKV Abs.
Results
Selection of Anti-ZIKV Abs for Binding and Structural Studies.
Potent neutralizing and recurring VH3-23/VK1-5 Abs against ZIKV and DENV1 were previously identified in multiple donors exposed to ZIKV in Mexico and Brazil (28). To investigate germline Ab maturation and cross-reactivity of VH3-23/VK1-5 Abs, we selected a set of seven Abs identified from the memory B cells of three of the donors: Z004mature from donor MEX 18; Z006mature from donor MEX 105; and Z031mature, Z032mature, Z034mature, Z035mature, and Z036mature from donor BRA 112 (SI Appendix, Figs. S1–S3). Since crystal structures were previously determined for Z004mature and Z006mature Fabs complexed with DENV1 EDIII and ZIKV EDIII, respectively (PDB ID codes 5VIC and 5VIG), these Abs were of particular interest for comparison to germline versions (28). Additionally, Z031mature, Z032mature, Z034mature, Z035mature, and Z036mature—which we term the Z03Xmature series—were selected since VH3-23/VK1-5 Abs from patient BRA 112 were previously shown to neutralize ZIKV, but no structural information on EDIII recognition was known (28).
Design of iGL Versions of Anti-ZIKV Abs.
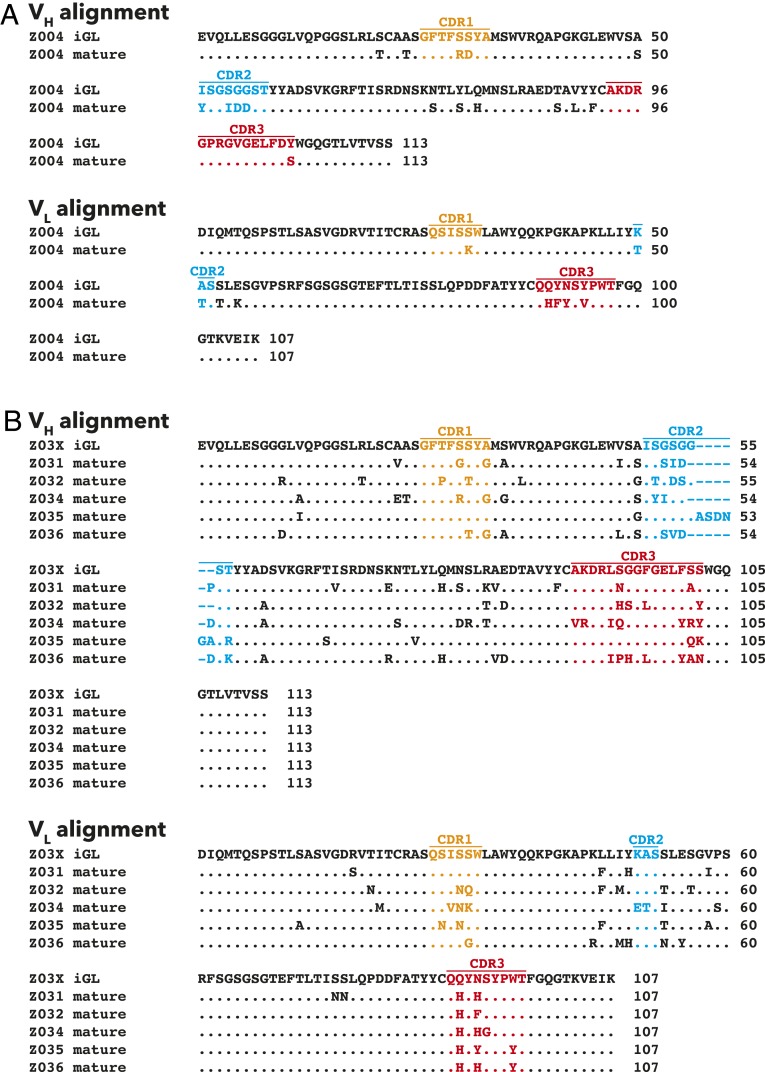
Z004iGL and Z03XiGL were constructed based on the germline gene assignments of mature VH3-23/VK1-5 Abs identified from donors MEX 18 and BRA 112 (SI Appendix, Figs. S1–S3) (28, 54). There are 16 amino acid differences in the VH and 9 differences in the VL of the Z004mature and Z004iGL sequences (Fig. 1), some occurring in complementarity determining regions (CDRs; 3 in CDR1, 6 in CDR2, and 5 in CDR3) (Fig. 1A). Amino acid differences between the Z03Xmature series and the Z03XiGL ranged from 13 to 23 for VH and 8 to 11 for VL (Fig. 1B). The Z03XiGL VH CDR1 (CDRH1) differed from all Z03Xmature sequences except Z035mature, and the VL CDR1 (CDRL1) differed from all Z03Xmature sequences except Z031mature. The Z03XiGL CDRH2 differed from all Z03Xmature sequences by at least three amino acids, and CDRL2 was the same as all Z03Xmature sequences except Z034mature. The CDR3s of the Z03XiGL VH and VL differed by at least two amino acids from all Z03Xmature sequences.
Fig. 1.
Alignments of VH and VL sequences of mature and iGL versions of Abs isolated from patients exposed to ZIKV. (A) Z004 mature and iGL Ab. (B) Mature and iGL Z03X Abs. CDR1, CDR2, and CDR3 are orange, blue, and red, respectively. The Kabat numbering scheme was used.
Assessing Binding of Anti-ZIKV IgGs with Flavivirus EDIIIs.
To investigate whether iGL versions of Abs bind ZIKV EDIIIs and whether any of the mature anti-ZIKV Abs cross-react with EDIIIs of other flaviviruses, we used SPR to determine Ab binding affinities for EDIII domains. To avoid avidity effects, the monomeric EDIII domains were injected over mature and iGL versions of IgGs coupled to biosensor flow cells. The analytes included EDIIIs from ZIKV, DENV1, DENV2, DENV3, DENV4, WNV, and YFV, and the IgG ligands included Z004mature, Z006mature, Z031mature, Z032mature, Z034mature, Z035mature, Z036mature, Z004iGL, and Z03XiGL.
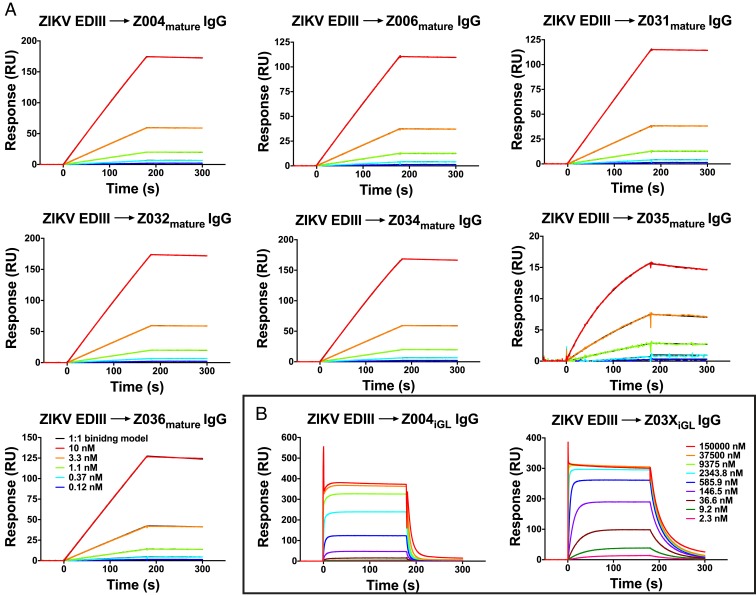
Sensorgrams revealed strong binding of both ZIKV and DENV1 EDIII to all mature IgGs, with low nanomolar or picomolar equilibrium dissociation constants (KDs) (Fig. 2A, Table 1, and SI Appendix, Figs. S4A and S5–S7). The KD values for Z004mature, Z006mature, Z032mature, Z034mature, Z035mature and Z036mature were all ∼2 to 14× lower for ZIKV EDIII than DENV1 EDIII, demonstrating stronger binding to ZIKV EDIII. Z031mature was the only IgG to bind more tightly to DENV1 than ZIKV EDIII. Mature IgGs showed weak binding to some of the EDIIIs; specifically, Z004mature, Z006mature, and Z034mature with DENV2 EDIII, Z004mature with DENV4 EDIII, and all mature IgGs with WNV EDIII (KDs >100 µM) (Table 1 and SI Appendix, Figs. S8–S12). Although the binding of Abs to DENV2, DENV4, and WNV is weak (SI Appendix, Figs. S8A, S10A, and S11A), it is clearly detectable compared to negative controls that show no binding (SI Appendix, Figs. S8B, S9, S10B, S11B, and S12). However, reporter viral particle (RVP)-based neutralization assays with a subset of VH3-23/VK1-5 Abs showed mature Abs neutralize ZIKV and DENV1, but not WNV (SI Appendix, Fig. S13). Additionally, RVP-based ADE assays showed no ability of mature Abs to induce ADE of DENV2 or WNV (SI Appendix, Fig. S14).
Fig. 2.
SPR binding assays with ZIKV EDIII. IgGs were captured on a protein A biosensor chip, and the indicated concentrations of ZIKV EDIII were injected. Sensorgrams are indicated in colors representing different injected concentrations. (A) Mature IgGs binding to ZIKV EDIII. Fits to a 1:1 binding model are in black; since the models very closely fit the data, the models are only slightly visible. The legend shown in the Bottom Left applies to all sensorgrams. Residual plots for the 1:1 binding model fitting are shown in SI Appendix, Fig. S7A. Two independent experiments were performed; the other set of sensorgrams is shown in SI Appendix, Fig. S5. (B) iGL IgGs binding to ZIKV EDIII. Fitting curves for equilibrium binding responses are shown in Fig. 3. The y axes show RU. The legend shown (Right) applies to both sensorgrams.
Table 1.
KDs (nM) of mature and iGL Ab binding to EDIIIs determined by SPR
| IgG | EDIII | ||||||
| ZIKV | DENV1 | DENV2 | DENV3 | DENV4 | WNV | YFV | |
| Z004mature | 0.28 | 0.47 | >100,000 | n.b. | >100,000 | >100,000 | n.b. |
| Z006mature | 0.50 | 1.7 | >100,000 | n.b. | n.b. | >100,000 | n.b. |
| Z031mature | 3.0 | 0.33 | n.b. | n.b. | n.b. | >100,000 | n.b. |
| Z032mature | 0.30 | 1.1 | n.b. | n.b. | n.b. | >100,000 | n.b. |
| Z034mature | 0.059 | 0.80 | >100,000 | n.b. | n.b. | >100,000 | n.b. |
| Z035mature | 0.78 | 5.2 | n.b. | n.b. | n.b. | >100,000 | n.b. |
| Z036mature | 0.29 | 0.53 | n.b. | n.b. | n.b. | >100,000 | n.b. |
| Z004iGL | 1,200 | >100,000 | n.b. | n.b. | n.b. | n.b. | n.b. |
| Z03XiGL | 92 | 2000 | >100,000 | n.b. | >100,000 | n.b. | n.b. |
n.b: No detectable binding at concentrations ≤150 µM.
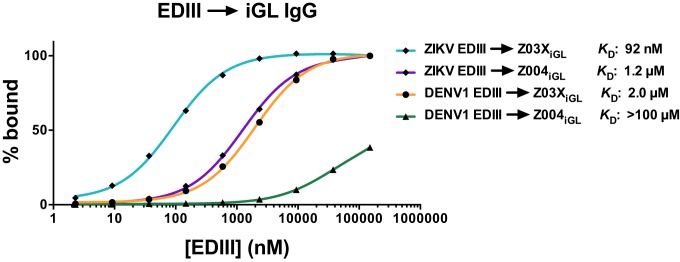
As expected, the iGL IgGs bound EDIII with lower affinities (e.g., low micromolar to high nanomolar KDs for the interactions of Z004iGL and Z03XiGL with ZIKV and DENV1 EDIII, respectively) than the mature IgGs (Fig. 2B, Table 1, and SI Appendix, Fig. S4B). The iGL IgGs bound ZIKV EDIII with ∼22 to 83× higher affinity than DENV1 EDIII, similar to the trend shown by mature IgGs (Fig. 3). The only other EDIIIs that showed detectable interactions with iGL IgGs were Z03XiGL–DENV2 EDIII and Z03XiGL–DENV4 EDIII (Table 1 and SI Appendix, Figs. S8 and S10). While the Z004iGL and Z03XiGL IgGs neutralized ZIKV RVPs (NT50s [concentration at 50% neutralization] of 8.8 ng/mL and 0.82 ng/mL, respectively) and DENV1 RVPs (NT50s: 1,400 ng/mL and 40 ng/mL), these iGL IgGs also showed ability to induce some ADE of ZIKV and DENV1 (but not DENV2 or WNV) (SI Appendix, Figs. S13 and S14).
Fig. 3.
Comparison of qualitative ZIKV and DENV EDIII binding to iGL IgGs. Normalized Req from the sensorgrams in Fig. 2B and SI Appendix, Fig. S4B are plotted versus the log of the concentration of the indicated injected proteins with best fit binding curves to the experimental data points shown as continuous lines. The SEs of the fit and 95% confidence intervals (CI) of the KDs are as follows: ZIKV EDIII → Z03XiGL IgG (KD: 92 nM; SE: 4.9 nM; CI: 79 to 100 nM), ZIKV EDIII → Z004iGL IgG (KD: 1.2 µM; SE: 61 nM; CI: 1,100 to 1,400 nM), and DENV1 EDIII → Z03XiGL IgG (KD: 2.0; SE: 100 nM; CI: 1,700 to 2,200 nM). Since DENV1 EDIII → Z004iGL binding reaction did not reach equilibrium, the KD is approximated as greater than the highest concentration of analyte injected.
To characterize affinity maturation of anti-ZIKV Abs and the structural correlates of Ab cross-reactivity, we set up crystallization screens for all 7 Fabs and for the 31 Fab–EDIII complexes that exhibited detectable binding interactions (Table 1). Crystals were obtained and X-ray structures determined for two complexes: Z004iGL Fab with ZIKV EDIII (3.1 Å resolution) and Z032mature Fab with WNV EDIII (2.9 Å resolution) (SI Appendix, Table S1).
Comparing iGL and Mature Fab Interactions with Flavivirus EDIIIs.
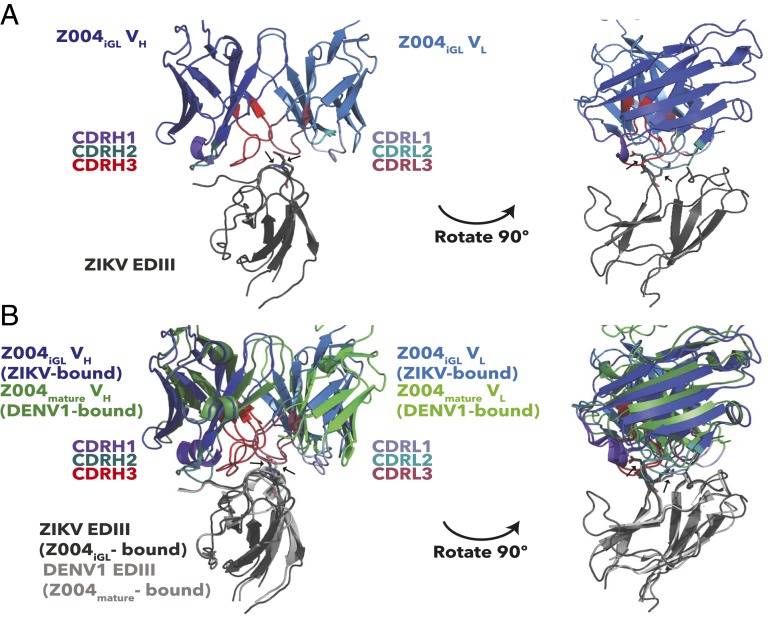
Crystals of the Z004iGL Fab–ZIKV EDIII complex contained two Fab–EDIII complexes in the asymmetric unit (root mean square deviation [rmsd] of 0.37 Å for 279 Cα atoms in the VH-VL and EDIII domains) (SI Appendix, Table S1). The structure revealed that Z004iGL binds the same epitope on the EDIII lateral ridge as Z004mature. Alignment of the EDIII portions of both complexes revealed similar binding interactions, including recognition of the EDIII EK amino acid motif (E393-K394) that is central to the binding epitope in the Z004mature Fab–DENV1 EDIII (PDB ID code 5VIC) and Z006mature–ZIKV EDIII (PDB ID code 5VIG) crystal structures (Fig. 4 and SI Appendix, Fig. S15A and Table S2) (28).
Fig. 4.
Recognition of ZIKV EDIII by Z004 iGL and mature Fabs. Fab–EDIII structures are shown as cartoon representations with Fab VH-VL domains only and the EK motifs of ZIKV and DENV1 EDIIIs highlighted as sticks (indicated by arrows). (A) Z004iGL Fab–ZIKV EDIII structure. (B) Superimposition of the Z004iGL Fab–ZIKV EDIII and Z004mature Fab–DENV1 EDIII (PDB 5VIC) structures (28). Structures were superimposed on the EDIII.
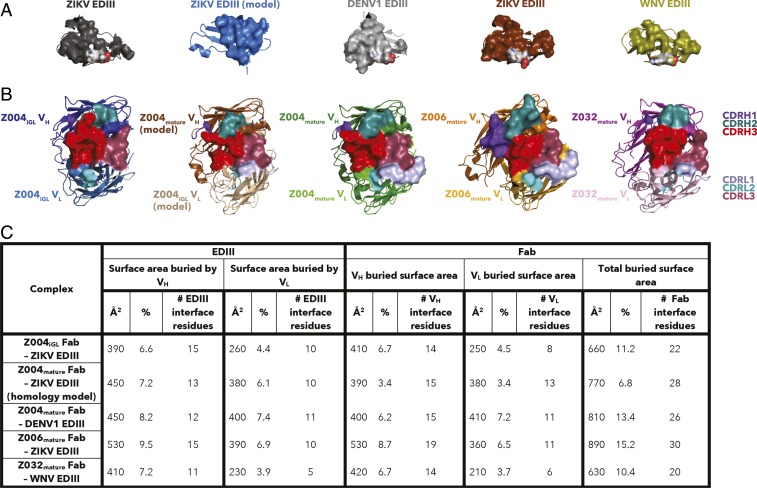
To compare the binding interfaces of the iGL and mature Abs with EDIII, we calculated buried surface areas (BSAs) on the Fab and EDIII in each complex (Fig. 5). Increased BSA generally correlates with a larger number of interface residues and a higher binding affinity (55). As expected from the low affinity of the Z004iGL Fab interaction with EDIII, less surface area was buried by EDIII on Z004iGL Fab (∼660 Å2) than on Z004mature (∼810 Å2) or Z006mature (∼890 Å2) (Fig. 5). The difference in Fab BSA between Z004iGL and Z004mature was largely accounted for by interactions with VL rather than VH. Specifically, the VH BSA was similar for Z004iGL (∼410 Å2) and Z004mature (∼400 Å2), whereas the VL BSA was greater for Z004mature (∼410 Å2) than Z004iGL (∼250 Å2). There was more EDIII surface area buried by VH than by VL for all complexes (Fig. 5). Since the Z004iGL–ZIKV EDIII and Z004mature–DENV1 EDIII structures do not directly compare iGL and mature Z004 binding to the same EDIII, we also made a homology model of Z004mature–ZIKV EDIII binding by threading the sequence of ZIKV EDIII onto the structure DENV1 EDIII in the Z004mature–DENV1 EDIII structure. The BSA on the Fab (770 Å2) for the Z004mature–ZIKV EDIII model showed the same trend as Z004mature–DENV1 EDIII: The VH BSA of Z004mature (∼390 Å2) was similar to Z004iGL (∼410 Å2), whereas the VL BSA was greater for Z004mature (∼380 Å2) than Z004iGL (∼250 Å2) (Fig. 5).
Fig. 5.
Comparison of Fab–EDIII binding interfaces. Surface representations of the (A) EDIII epitopes contacted by Fabs and (B) Fab binding epitopes contacted by EDIII in the Z004iGL Fab–ZIKV EDIII (PDB ID code 6UTA), Z004mature Fab–ZIKV EDIII homology model, Z004mature Fab–DENV1 EDIII (PDB ID code 5VIC), Z006mature Fab–ZIKV EDIII (PDB ID code 5VIG), and Z032mature Fab–WNV EDIII (PDB ID code 6UTE) structures (left to right) (28). The Z004mature Fab–ZIKV EDIII homology model was made by threading the sequence of ZIKV EDIII onto the structure DENV1 EDIII in the Z004mature–DENV1 EDIII structure. Binding epitopes are shown as surfaces over cartoon representations. CDRs are colored as indicated (Right). (C) Quantification of BSA (Å2 and percentage of total surface area) and number of interface residues buried on the entire EDIII (Left) and VH and VL of the Fab (Right) based on the interfaces mapped in A and B. The column labeled total buried surface area for Fabs includes the sum of BSA for the VH and VL.
Z004iGL showed only 5 residues that interact with the ZIKV EDIII antigen compared with 10 interacting residues for Z004mature (with DENV1 EDIII in the crystal structure and with ZIKV EDIII in the homology model) and 13 residues in Z006mature Fabs that interact with ZIKV EDIII (SI Appendix, Fig. S16 A–D and Table S2). The finding of fewer interacting residues is consistent with the weaker binding of Z004iGL, demonstrated by SPR (Figs. 2 and 3 and Table 1). Similar to the trends with differences in BSA, differences in the number of Fab residues predicted to interact with EDIII was pronounced for VL: one by Z004iGL VL, four by Z004mature VL (with ZIKV in the model), five by Z004mature VL (with DENV1), six by Z006mature VL (with ZIKV) (SI Appendix, Fig. S16 A–D and Table S2). Among Fab residues that interact with EDIII by either Z004iGL or Z004mature, we observe that the only residues that differ in sequence between the iGL and mature version are in the VL CDRL3: F91VL, Y92VL and V94VL in Z004mature, compared with Y91VL, N92VL, and Y94VL in Z004iGL (SI Appendix, Figs. S16 and S17A and Table S2). Two of these residues, F91VL and Y92VL, interact with the ZIKV EDIII EK motif. In contrast, the only residues that interact with EDIII by both Z004mature and Z004iGL are in the VH and share the same sequence: S56VH (CDRH2), Y58VH (framework region [FWR]H3), and E100CVH (CDRH3). Z006mature Fab also shared one of the same VH interacting residues: Y58VH (FWRH3) (SI Appendix, Fig. S16 and Table S2).
To further investigate the effects of affinity maturation in the VH versus the VL domain for high-affinity EDIII binding, we prepared two Z004 chimeric IgGs for SPR and neutralization assay analysis: One with mature VH and iGL VL and the other with iGL VH and mature VL. The SPR sensorgrams for ZIKV EDIII binding to the Z004 chimeras were fit to a 1:1 binding model (SI Appendix, Fig. S18) and showed 10-fold higher affinity binding to the ViGL HC-Vmature LC chimera (KD: 2.5 nM) than to the Vmature HC-ViGL LC chimera (KD: 29 nM) (SI Appendix, Table S3). The ViGL HC-Vmature LC sensorgrams appeared similar to mature VH-VL sensorgrams (slow off-rate), whereas the Vmature HC-ViGL LC sensorgrams were more similar to the iGL VH-VL sensorgrams (fast off-rate), consistent with the importance of light-chain (LC) maturation in the development of high-affinity recognition of Zika EDIII. Neutralization assays comparing the two chimeric Abs showed that while both Z004 chimeras can neutralize ZIKV and DENV1, the ViGL HC-Vmature LC chimera neutralizes just as potently (NT50: 0.34) as fully mature Z004 IgG (NT50: 0.55), while Vmature HC-ViGL LC chimera neutralizes with slightly lower potency (NT50: 1.00) (SI Appendix, Fig. S13).
To assess which residues are important for high-affinity binding, we first prepared ZIKV EDIII with the EK motif (central to the binding epitope and involved in several interactions with Z004), mutated to alanines (E393A-K394A) for binding studies with SPR. Binding of Z004mature IgG to ZIKV EDIIIAA mutant was nearly abolished with a KD of >>100 µM (SI Appendix, Fig. S17B and Table S3). To verify that the Z004 VL residues that interact with EDIII and differ between Z004iGL and Z004mature (F91VL, Y92VL, and V94VL) are important for high-affinity binding to ZIKV EDIII, we prepared Z004mature IgG variants with two or all three residues mutated to alanines. When the two EK-interacting residues were mutated (Z004mature IgG: VL F91A-Y92A), we observed 100-fold reduced binding affinity to ZIKV EDIII (KD: 35 nM). When all three residues were mutated (Z004mature IgG: VL F91A-Y92A-V94A), we observed 1,000-fold reduced binding affinity (KD: 230 nM) (SI Appendix, Fig. S17 C and D and Table S3).
Structural Correlates of Weak Ab Cross-Reactivity.
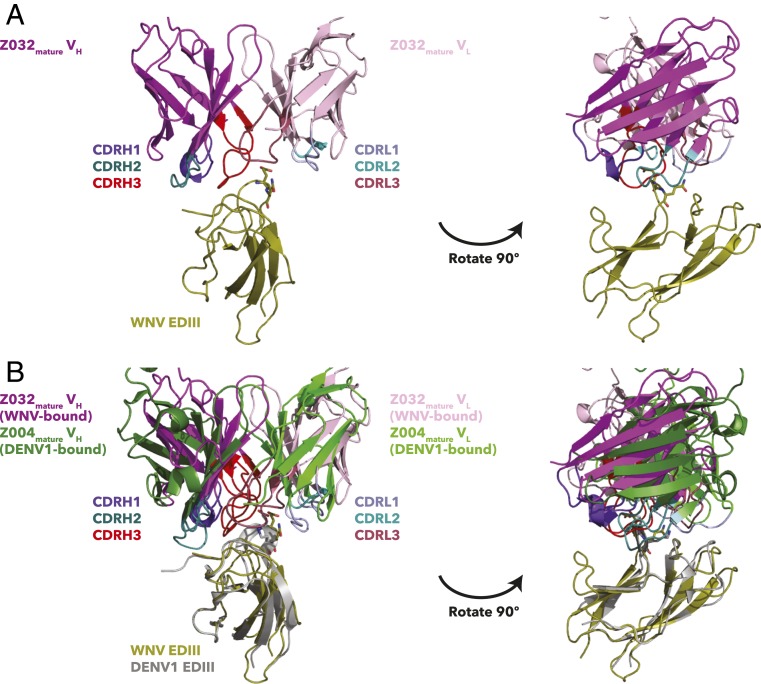
We were able to crystallize a complex of Z032mature Fab bound to WNV EDIII despite the low affinity of this interaction (KD ≥ 100 µM) (Table 1). Perhaps correlating with the low affinity of the complex, the crystallographic asymmetric unit contained one Fab–WNV EDIII complex and four unbound Fabs (Fig. 6A and SI Appendix, Table S1). The four unbound Fabs were similar to each other (rmsds ranging from 0.22 Å to 0.47 Å for pairwise superimpositions of 203 to 214 Cα residues in the VH-VL domains) (SI Appendix, Table S4). WNV EDIII-bound and -unbound Fabs were also similar (rmsds ranging from 0.35 Å to 0.45 Å for pairwise superimpositions of 210 to 217 Cα residues in the VH-VL domains), indicating no major structural changes upon EDIII binding (SI Appendix, Fig. S19 and Table S4).
Fig. 6.
Cross-reactive recognition of WNV EDIII by Z032mature Fab. Fab–EDIII complex structures are shown as cartoon representations with Fab VH-VL domains only and the EQ or EK motifs of WNV and DENV1 EDIIIs highlighted as sticks. (A) Z032mature Fab–WNV EDIII structure. (B) Superimposition of the Z032mature Fab–WNV EDIII and Z004mature Fab–DENV1 EDIII (PDB 5VIC) structures (28). Structures were superimposed on the EDIII domains.
The Z032mature Fab–WNV EDIII structure showed that Z032mature Fab interacts with WNV EDIII at the lateral ridge epitope recognized by Z004mature, Z006mature, and Z004iGL, although with a low Fab BSA (∼630 Å2) (Figs. 5 and 6 and SI Appendix, Fig. S14). Similar to Z004iGL, the VH (420 Å2) contributes more to the total Z032mature Fab BSA than VL (210 Å2). The low Fab BSA correlates with fewer interacting residues at the binding interface: Only eight Z032 Fab residues were found to interact with six WNV EDIII residues (SI Appendix, Fig. S16 and Table S2). Some Z032mature Fab interacting residues were also involved in interactions in other Fab–EDIII structures: S56VH (in Z004iGL and Z004mature), Y58VH (FWR3) (in Z004iGL, Z004mature, and Z006mature), R96VH (CDRH3) (in Z004mature and Z006mature), E100CVH (CDRH3) (in Z004iGL and Z004mature), Y91VL (in Z006mature), and Y94VL (in Z004iGL) (SI Appendix, Fig. S16 and Table S2). This suggests that these residues contribute to enhanced cross-reactivity of anti-ZIKV VH3-23/VK-15 Abs.
Discussion
Understanding the structural basis of Ab recognition of ZIKV and other flavivirus antigens informs considerations and precautions for vaccine design to elicit EDIII-specific Abs. Here we conducted binding and structural studies comparing interactions of mature and iGL VH3-23/VK1-5 Abs isolated from ZIKV-exposed donors with a panel of flavivirus EDIII domains to provide insight into the affinity maturation process of this class of potently neutralizing ZIKV Abs. In addition to revealing interactions critical for potent binding to ZIKV, we also identified weaker interactions that may contribute to cross-reactivity and potentially ADE.
By comparing mature and iGL VH3-23/VK1-5 Ab binding to EDIIIs, we identified the mutations introduced through somatic hypermutation that facilitate tight binding of the Z004 Ab to ZIKV. As expected, a crystal structure of a Z004iGL–EDIII complex showed fewer Fab residues that interact with EDIII than observed for EDIII complexes including mature Fabs, such as Z004mature, Z006mature, and Z032mature (28). In a direct comparison of the Z004mature–DENV1 EDIII structure and Z004mature–ZIKV EDIII homology model with the Z004iGL–ZIKV EDIII structure, the only three Fab residues involved in interactions with EDIII that differed in sequence between Z004mature and Z004iGL are in VL. In contrast, the three interacting residues that are shared by both Z004mature and Z004iGL are in VH. This suggests that affinity maturation of VL CDRL3 may be particularly important for higher affinity binding to EDIII. This is further supported by the finding that the increased BSA on Z004mature compared to Z004iGL was largely accounted for by an increase in the VL BSA. Comparison of the binding affinities and neutralization potencies of Z004 mature/iGL chimeras also suggests the importance of VL maturation for EDIII recognition.
Through investigation of the effects of site-directed mutations in Z004mature IgG and ZIKV EDIII on binding, we showed that interactions with the EDIII EK motif are critical for high-affinity binding. While both VH and VL residues interact with the EK motif, the only EDIII-interacting residues that differ between Z004mature and Z004iGL are in VL (F91, Y92, and V94). The reduced binding affinity when these residues are mutated to alanines supports that affinity maturation of the VL is important for high-affinity binding.
The importance of VL somatic hypermutation was also observed in a previous longitudinal analysis of a ZIKV-infected patient to trace the lineage of ZK2B10, a protective VH1-8/VL1-47 Ab against ZIKV that binds a different part of the EDIII lateral ridge (56). Two residues in the VL1-47 germline-coded λ VL, N31 (CDRL1) and S91 (CDRL3), were shown to be necessary and sufficient for functional maturation of the VH1-8/VL1-47 Ab lineage to achieve potent ZIKV neutralization. They observed low somatic hypermutation in germline-like somatic variants of VH, concluding that restricted VH gene segment usage, rather than somatic hypermutation in the VH domain, was important to achieve high affinity and potency. While ZK2B10 is derived from different germline genes and binds a different part of the EDIII lateral ridge than the VH3-23/VK1-5 Abs studied here, we observed a similar trend for the VH3-23/VK1-5 Ab Z004: Affinity maturation of VL was important for strong binding and neutralization, while VH interactions were restricted to residues already present in the germline gene.
Through assessment of VH3-23/VK1-5 Ab binding to a panel of flavivirus EDIIIs using SPR, the Abs tightly bound ZIKV and DENV1 and weakly bound DENV2, DENV4, and WNV EDIII, indicating a potential for cross-reactivity. Although this class of mature Abs was shown to neutralize DENV1 in addition to ZIKV (28), the ability of iGL Abs to bind DENV1 EDIII as well as the apparent weak binding of both mature and iGL Abs to other flaviviruses suggests the possibility of ADE upon subsequent infection with a different flavivirus in humans. The EK motif, which is only present in ZIKV and DENV1 EDIII, likely contributes to initial recognition by germline Abs that leads to the tighter binding and neutralization of these two flaviviruses by VH3-23/VK1-5 Abs. The importance of E393 in the EK motif for neutralization of ZIKV strains was also previously described for the VH3-23/VK1-5 Ab ZIKV-116 (57). In contrast to ZIKV and DENV1, the lateral ridges of DENV2, DENV4, and WNV EDIIIs all contain motifs other than EK, yet still showed weak binding to at least one mature or iGL Ab by SPR. This suggests the interactions that contribute to cross-reactive binding of Abs to these flaviviruses are different from, or only partially overlap with, the interactions that contribute to high affinity and neutralizing binding to ZIKV and DENV1 EDIIIs. Apart from DENV1, WNV EDIII was the only flavivirus for which all seven mature VH3-23/VK1-5 Abs showed cross-reactivity (Table 1). Given that binding to DENV2, DENV4, and WNV EDIIIs is weak (KD ≥ 100 µM), it is unclear whether this cross-reactivity could facilitate ADE of these flaviviruses upon infection. However, our RVP-based assays showing no ADE for DENV2 or WNV infection suggests this is of low concern. The potential for cross-reactivity of other Abs with ZIKV and WNV EDIII was shown in studies demonstrating that previous exposure to WNV enhances subsequent ZIKV infection in mice, although immunodominant DII-specific fusion loop Abs might explain this in vivo enhancement (41).
The crystal structure of Z032mature Fab complexed with WNV EDIII provides an example of the structural basis of cross-reactive recognition. This structure shows that the Z03X Abs bind the same lateral ridge epitope as other VH3-23/VK1-5 Abs (28). A few Z032mature Fab-interacting residues (S56VH [CDRH2], Y58VH [FWRH3], R96VH [CDRH3], E100CVH [CDRH3], Y91VL [CDRL3], and Y94VL [CDRL3]) are also involved in at least one other interaction in the Z004iGL, Z004mature, and Z006mature complexes with EDIII domains. This suggests these residues, including the one in a conserved FWR (Y58VH), may contribute to cross-reactivity of VH3-23/VK-15 Abs, potentially even precursor germline versions, with different flaviviruses. These residues are present in iGL sequences, suggesting that residues that contribute to cross-reactivity may already be present prior to affinity maturation. The weak, cross-reactive binding of Z004iGL to DENV1 EDIII and of Z03XiGL to DENV1, DENV2, and DENV4 further supports this suggestion. Five of the eight Z032mature Fab residues predicted to bind WNV EDIII are in the VH, suggesting the VH may contribute more to weak, cross-reactive binding, whereas somatic hypermutation in the VL may contribute more to tight binding of mature VH3-23/VK1-5 Abs to ZIKV.
Comparison of the structures of bound and unbound Z032mature Fabs indicated the VH3-23/VK1-5 class of Abs does not require major conformational changes for binding. Superimposition of the bound and unbound Z032mature VH-VL revealed a low calculated rmsd and no major differences in the backbone structure, suggesting that conformations were preformed prior to binding. This suggests that VH3-23/VK1-5 mature Abs use a lock-and-key mode of binding, involving minimal conformational changes between the bound and unbound states of antigen and Ab (58–60).
Germline versions of VH3-23/VK1-5 ZIKV-neutralizing Abs showed detectable binding to ZIKV and DENV1 EDIII in the nanomolar and low micromolar range. This was also previously observed for the germline version of the VH3-23/VK1-5 Ab ZIKV-116, which bound (KDs of 48.9 nm-10 µM) and neutralized ZIKV and DENV1 strains (57). This ability of germline versions of VH3-23/VK1-5 Abs to bind ZIKV contrasts with germline-reverted forms of most broadly neutralizing Abs against HIV-1, which generally do not bind HIV-1 envelope (61). The ability of germline versions of neutralizing Abs to bind antigens is of particular interest for vaccine design, as this suggests immunogens may effectively elicit precursors of the desired Ab class, which could then mature into neutralizing Abs. Initial studies of the potential of ZIKV EDIII to serve as a safe and effective immunogen are underway, and indicate potential to elicit a specific and potent neutralizing Ab response to ZIKV in mice (62–64).
Increased understanding of the differences in the interactions that contribute to neutralization versus cross-reactivity leading to ADE may enable strategic immunogen design. The in vitro ability of germline and mature (65) VH3-23/VK1-5 Abs to induce some ADE for ZIKV and DENV1, but not for DENV2 or WNV, suggests there may be minimal risk of ADE due to weak cross-reactivity for this class of Abs. While this is not indicative of in vivo ability to enhance infection, there may be concern that Ab titers falling below neutralizing levels may be a risk for ADE. These findings indicate the importance of examining the cross-reactivity and ADE-potential of other anti-ZIKV classes of Abs under consideration for vaccine design or passive delivery. Understanding which residues contribute to cross-reactivity versus potent neutralization may also inform the necessity of modifying passively delivered Abs to reduce cross-reactivity and prevent ADE by introducing Fc of mutations that prevent FcγR binding.
Materials and Methods
Design of iGL Versions of Abs against ZIKV.
Sequences of iGL versions of anti-ZIKV Abs are based on V, D, and joining J gene segment assignments from IgBlast (28, 54). All mature IGHV3-23/IGKV1-5 Ab sequences from donors MEX 18 and BRA 112 were considered for design of the Z004 and Z03X iGLs, respectively. CDR1 and CDR2 of the iGLs were based on the V gene segment assignment of the mature sequences, specifically IGHV3-23 for the heavy chain (HC) and IGKV1-5 for the LC. The CDRH3 of the iGL was based on a consensus of the V, D, and J gene segment assignments for the mature HC sequences, and CDRL3 of the iGL was based on a consensus of V and J gene segment assignments for the mature LC sequences (SI Appendix, Figs. S1–S3).
Protein Expression.
Abs were produced as previously described (28, 66). Briefly, Z006mature, Z031mature, Z032mature, Z034mature, Z035mature, Z036mature, and Z03XiGL IgGs were expressed by transient transfection of HEK293-6E cells with equal amounts of Ig HC and LC expression vectors. After 7 d, IgGs were purified from supernatants using Protein G Sepharose 4 Fast Flow (GE Healthcare). Z004mature, Z004iGL, and N6 IgGs were expressed by transient transfection and purified from supernatants using a HiTrap MabSelect column (GE Healthcare) and size-exclusion chromatography (SEC) using a Superdex 200 column (GE Healthcare) in 20 mM Tris pH 8.0, 150 mM NaCl.
Fabs with C-terminal His-tags were produced by transient transfection of Expi293F cells with equal amounts of HC and LC expression vectors. The Fabs were purified from supernatants with Ni-NTA affinity chromatography and SEC with a Superdex 200 column in 20 mM Tris, 150 mM NaCl.
Flavivirus EDIIIs were expressed in Escherichia coli and purified from inclusion bodies as previously described (28, 36, 67). Briefly, EDIII genes from ZIKV (H/PF/2013 strain, GenBank KJ776791), DENV1 (45AZ5 strain, National Center for Biotechnology Information [NCBI] reference NC_001477), DENV2 (NCBI reference NC_001474), DENV3 (NCBI reference NC_001475.2), DENV4 (NCBI reference NC_002640.1), YFV (Asibi strain, GenBank KF769016), and WNV (GenBank KX547539.1) in pET21 expression plasmids were transformed into BL21 (DE3) competent cells and cultures were grown in LB with carbenicillin at 37 °C. Expression was induced with isopropyl-β-d-thiogalactopyranoside at an OD of ∼0.6, and the culture was harvested after 4 h and stored overnight at −20 °C. The pellet was resuspended in 20 mM Tris pH 8.0, 150 mM NaCl, cells were lysed and centrifuged at 21,000 × g for 30 min, and the pellet was resuspended in 6 M guanidine hydrochloride, 100 mM Tris⋅HCl pH 8.0. This suspension was centrifuged again at 21,000 × g for 30 min, and then 20 mM β-mercaptoethanol was added to the supernatant. EDIII in the supernatant was refolded by dropwise, rapid dilution into 400 mM l-arginine, 100 mM Tris-base pH 8.0, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, and 10% glycerol at 4 °C. The protein was then concentrated and purified by SEC with a Superdex 75 column (GE Healthcare) in 20 mM Tris pH 8.0, 150 mM NaCl, 0.02% NaN3.
SPR Binding Assays.
SPR experiments were performed using a Biacore T200 instrument (GE Healthcare). Binding assays were done by flowing EDIII analytes over IgG ligands bound to a protein A-coupled biosensor chip in 0.2-µm–filtered HBS-EP+ running buffer. The protein A-coupled chip was prepared from a CM5 chip (GE Healthcare) by coupling 1 µM His-tagged Protein A at pH 4.5 to each flow cell to a final density of ∼3,000 response units (RUs). IgGs were then injected onto each flow cell at 50 nM. An irrelevant HIV-1 Ab, N6 (68), was used as a control on a reference flow cell. To remove IgGs from the chip between runs, a solution of 10 mM glycine 50% (vol/vol) pH 1.5/pH 2.5, 1 M guanidine hydrochloride was applied.
For interactions with measurable on- and off-rates, kinetic constants were derived from sensorgram data using global fitting of the association and dissociation phases of binding curves in the working set using Biacore T200 Evaluation Software (GE Healthcare). The sensorgrams were fit to a binding model that assumed a single class of noninteracting binding sites in a 1:1 binding interaction. KD values were derived as KD = kd/ka, the ratio of the association (ka) and dissociation (kd) constants. For interactions with fast on- and off-rates, KD values were derived by nonlinear regression analysis of plots of Req (the equilibrium binding response) versus the log of the injected protein concentration. Data were fit to a 1:1 binding model, and each KD was determined as the concentration at which half-maximal binding was observed. If a saturated binding response was not achieved at the highest injected concentration of analyte, we approximated the saturated response as the highest Req achieved.
For SPR runs used to determine KD values, the EDIIIs for ZIKV, DENV1-4, WNV, and YFV were dialyzed into HBS-EP+ running buffer using a Slide-A-Lyzer MINI Dialysis Device with 3,500 Da molecular mass cutoff (ThermoFisher) to minimize refractive index changes between the association and dissociation phases. We used concentrations of ZIKV and DENV1 EDIII ranging from 0.04 to 10 nM in a threefold dilution series flowed over immobilized mature IgG, and concentrations of ZIKV, DENV1, and ZIKVAA mutant EDIIIs ranging from 2.3 nM to 150 µM in a fourfold dilution series flowed over immobilized iGL IgGs or Z004mature IgG and the control IgG. Concentrations of DENV2, DENV3, DENV4, WNV, and YFV EDIIIs from 2.3 nM to 150 µM in a fourfold dilution series were flowed over immobilized mature and iGL IgGs. Concentrations of ZIKV EDIII ranging from 0.036 nM to 150 µM in a fourfold dilution series were flowed over immobilized Z004 iGL/mature chimeric IgGs in two independent experiments. Concentrations of ZIKV EDIII ranging from 0.036 nM to 37.5 µM in a fourfold dilution series were flowed over immobilized Z004 IgG VL site-directed mutants. The injection flow rate was 10 µL/min over 3 min and the dissociation time was 2 min. For repeated SPR runs for ZIKV and DENV1 EDIII flowed over mature IgGs, concentrations of EDIII from 1.4 to 1,000 nM in a threefold dilution series were injected at a flow rate of 30 µL/min over 1 min and a dissociation time of 5 min (SI Appendix, Figs. S5 and S6). KDs for mature IgG binding to ZIKV EDIII and DENV1 EDIIIs were calculated as the average of two independent experiments.
Crystallization Trials.
For the Z004iGL Fab–ZIKV EDIII complex, Fabs and EDIII were incubated at a 1:1.8 molar ratio for 3 d and then purified by SEC on Superdex 200 10/300 Increase column (GE Healthcare). For the Z032mature Fab–WNV EDIII complex, Fabs and EDIII were incubated at a 1:1 molar ratio for 3 d. Crystallization trials were set up at ∼5 mg/mL (Z004iGL Fab–ZIKV EDIII) and ∼10 mg/mL (Z032mature Fab–WNV EDIII) in sitting-drop plates with a Mosquito microcrystallization robot and stored at room temperature. Crystals were cryoprotected with 25% glycerol. Crystallization conditions corresponding to determined structures include 1% (wt/vol) tryptone, 0.001 M sodium azide, 0.05 M Hepes sodium pH 7.0, 20% (wt/vol) polyethylene glycol 3350 (Z004iGL Fab–ZIKV EDIII) and 0.2 M sodium bromide, 20% (wt/vol) polyethylene glycol 3350 (Z032mature Fab–WNV EDIII).
X-Ray Structure Determinations.
X-ray data for the Z004iGL Fab–ZIKV EDIII structure were collected at the Stanford Synchrotron Radiation Lightsource. Data for the Z032mature Fab–WNV EDIII complex were collected at the Advanced Photon Source, Argonne, IL, using the GM/CA 23-ID-D beamline. Z004iGL Fab–ZIKV EDIII diffraction data were processed using the XDS package (69), and Z032mature Fab–WNV EDIII complex data were processed using iMosflm (70). Data were scaled using Pointless and Aimless (71, 72).
Structures were solved by molecular replacement (MR) using Phaser-MR (73). For the Z004iGL Fab–ZIKV EDIII structure, the structure was first determined at 3.3 Å using Z004mature Fab–DENV1 EDIII (PDB ID code 5VIC) as the initial search model (28). This gave a partial solution with two Z004mature Fab–DENV1 EDIII molecules in the asymmetric unit, with one of the Fabs containing an incorrectly placed constant (CHCL) domain. This CHCL domain was removed and MR was repeated using the initial partial solution and one Z004mature CHCL as search models. The structure was then determined at 3.1 Å using a similar method, except the Fab with the incorrectly placed CHCL domain was completely removed and MR was repeated using the initial partial solution and the corresponding Z004mature Fab–DENV1 EDIII from the 3.3 Å structure as a search model. For the Z032mature Fab–WNV EDIII complex, Z006mature VHVL and Z006mature CHCL (PDB ID code 5VIG) domains were used as the initial search models (28), which produced a partial solution with four VHVL and four CHCL. MR was repeated using this partial solution and WNV EDIII (PDB ID code 1ZTX) as search models (33). This generated a partial solution with one WNV EDIII placed correctly and three placed incorrectly, so all molecules were removed except for one Fab bound to WNV EDIII. For this Fab–EDIII partial solution, Fab residues were mutated to match the Z032mature sequence, and then to generate Z032mature Fab, Z032mature VHVL, and Z032mature CHCL search models. MR was repeated using the Z032mature Fab–WNV EDIII partial solution and Z032mature VHVL and CHCL as search models. This gave a partial solution with two unbound Fabs and one EDIII-bound Fab correctly placed; again, incorrectly placed molecules were removed. MR was repeated with this partial solution and Z032mature Fab as a search model. This gave a partial solution with four correctly placed Fabs, one EDIII-bound and three unbound. MR was repeated with this partial solution and an additional Z032mature Fab as search models, resulting in the final structure with four unbound and one EDIII-bound Fab. Protein models were refined with phenix.refine using torsion angle refinement, group B factors, and noncrystallography symmetry restraints (74). PDB ID codes and X-ray data collection and refinement statistics are in SI Appendix, Table S1.
Figures were prepared and rmsds of superimposed C⍺ atoms were calculated using Pymol (75). A Z004 Fab–Zika EDIII homology model was created using SWISS-MODEL (76). A control for the homology model was prepared by threading ZIKV EDIII onto DENV1 EDIII in the Z021 Fab–DENV1 EDIII structure (PDB ID code 6DFJ); comparison of VHVL–EDIIIs from the Z021 Fab–ZIKV EDIII homology model and the known Z021 Fab–ZIKV EDIII structure (PDB ID code 6DFI) resulted in an rmsd of 0.28 for 239 C⍺ atoms (29). Fab–EDIII binding interfaces were mapped as residues within 4 Å in Pymol. BSAs (calculated using a 1.4 Å probe) and the contact residues at Fab–EDIII interfaces (calculated using a distance of <3.89 Å and an A-D-H angle >90° for H-bonds and a distance <4 Å for salt bridges) were determined with PDBePISA (77).
Plasmid Construction.
pWNV/TX02/CprME was generated by assembly PCR. Using pZIKV/HPF/CprME [obtained from Ted Pierson, Viral Pathogenesis Section, Laboratory of Viral Diseases, National Institutes of Health, Bethesda, MD (28)] as template and oligos RU-O-24611 (5′- CTTGACCGACAATTGCATGAAG) and RU-O-24620 (5′- CCTCCTGGTTTCTTAGACATAGCCTGCTTTTTTGTACAAAC), the CMV promoter region linked to the beginning of the WNV capsid protein was amplified. The WNV CprME region with upstream overlap with the CMV promoter was amplified using Oligos RU-O-24619 (5′- GTTTGTACAAAAAAGCAGGCTATGTCTAAGAAACCAGGAGG) and RU-O-24621 (5′- TTCGAACCGCGGCTGGGTCCTATTAAGCGTGCACGTTCACGGAGAG) and a full-length WNV strain TX02 infectious clone [obtained from Ilya Frolov, Department of Microbiology, University of Alabama at Birmingham, Birmingham, AL (78)] as a template. The two fragments were assembled by PCR using oligos RU-O-24611 and RU-O-24621, and the product was digested with SnaBI and SacII and cloned into similarly digested pZIKV/HPF/CprME. All PCR-derived DNA regions were verified by sequencing.
RVP Production.
RVPs were generated as previously described (28) by cotransfection of two plasmids: A luciferase-expressing WNV replicon plasmid (pWNVII-Rep-REN-IB, obtained from Ted Pierson) and a C-prM-E expression plasmid encoding structural proteins of ZIKV strain HPF with the PRVABC59 E protein (pZIKV/HPF/CprM*PRVABC59E*) (79), DENV1 strain WP (pDENV1/WP/CprME, obtained from Ted Pierson), DENV2 strain 16681 (pDENV2/16681/CprME, obtained from Ted Pierson), or WNV strain TX02 (pWNV/TX02/CprME). Lenti-X 293T cells were seeded at 5 × 105 cells per well in collagen-coated six-well plates 1 d before DNA transfection. One microgram of pWNVII-Rep-REN-IB (WNV replicon expression construct) and 3 μg of the flavivirus CprME expression construct were cotransfected with Lipofectamine 2000 (Invitrogen). After incubation at 37 °C for 4 to 5 h, media (containing lipid–DNA complexes) was removed and replaced with Dulbecco’s modified Eagle’s medium (DMEM) containing 20 mM Hepes, 3% fetal bovine serum (FBS). Cells were then incubated at 34 °C for 48 to 72 h before supernatant (containing RVPs) was harvested, filtered through a 0.22-µm filter, and frozen at −80 °C.
RVP-Based Neutralization Assays.
Antibodies were diluted in triplicate in Medium-199 with 17% BSA, 1% P/S (BA-1 diluent) to 40 μg/mL and then serially diluted in BA-1 diluent using fivefold dilutions. RVPs were diluted in OPTI-MEM to a concentration that results in ∼1 × 106 relative light units (RLU) per 25 μL (determined based on a titration of the harvested RVPs on Huh-7.5 cells). Serially diluted antibodies were mixed with equal volumes of RVPs and incubated for 1 h at 37 °C. Fifty microliters of RVP–Ab complex was added to Huh-7.5 cells seeded in 96-well half-area plates at 7.5 × 103 cells per well in 50 μL the day prior. After incubation at 37 °C for 24 h, media was removed, cells were lysed in 35 μL 1× Lysis Buffer, and 20 μL was used for Renilla luciferase measurement on a FLUOstar Omega luminometer (BMG LabTech) using the Renilla Luciferase Assay System (Promega). Luciferase activity, measured as RLUs, was normalized as the percentage of luciferase activity relative to activity from RVPs incubated without Ab. Experiments were repeated twice and plotted on the same graph. The N6 Ab (negative control) was assessed at 10 μg/mL final concentration in the well. IC50 values (the Ab concentration that resulted in 50% inhibition) were determined by nonlinear regression fitting of the curve in GraphPad Prism.
ADE Assays.
Antibodies were diluted in triplicate in BA-1 diluent to 40 μg/mL and then serially diluted by threefold dilutions. RVPs were diluted in OPTI-MEM with a goal of achieving ∼1 × 106 Huh-7.5 cell RLU per 12.5 μL (determined based on a titration of the harvested RVPs on Huh-7.5 cells in the absence of antibody). For some RVPs this goal was not attainable and lower RLU were used. Serially diluted antibodies were mixed with equal volumes of RVPs, and incubated for 1 h at 37 °C. Twenty-five microliters of RVP-Ab complex was added to K562 cells seeded the prior day in poly-l-lysine–coated 96-well half-area plates at 5.0 × 103 cells per well in 25 μL of DMEM containing 10% FBS and 1× nonessential amino acids solution (NEAA). Every plate included a no antibody control, an N6 negative control, and Z004 wild-type antibody (10 ng/mL final concentration) with ZIKV RVPs as a positive control. Each plate also included Huh-7.5 cells seeded the day prior at 7.5 × 103 cells per well in 50 μL to serve as a positive control for RVP activity. After incubation at 37 °C for 24 h, media was removed, cells were lysed in 35 μL 1× Lysis Buffer, and 20 μL was used for Renilla luciferase measurement on a FLUOstar Omega luminometer (BMG LabTech) using the Renilla Luciferase Assay System (Promega). Luciferase activity, measured as RLUs, was normalized to respective RVP luciferase activity determined on fully permissive Huh-7.5 cells (positive control). Experiments were repeated twice and plotted on the same graph. The HIV-1 Ab N6 (negative control) and cross-reactive Ab Z015mature (WNV positive control) were assessed at the highest concentration (10 µg/mL). Z004mature Ab (positive control) (65) was assessed at 0.01 µg/mL, a concentration known to show ADE for ZIKV.
Data Availability.
Crystallographic coordinates for structures Z004iGL Fab–ZIKV EDIII and Z032mature Fab–WNV EDIII are available from the Protein Data Bank under ID codes 6UTA and 6UTE.
Supplementary Material
Acknowledgments
We thank Dr. Jost Vielmetter at the Caltech Protein Expression Center funded by the Beckman Institute at the California Institute of Technology for protein expression and surface plasmon resonance instrument usage; Dr. Jens Kaiser at the California Institute of Technology Molecular Observatory for assistance with X-ray data collection; Dr. Anthony West for discussions regarding germline gene assignments; Alex Cohen for suggestions on EDIII production; Dr. Christopher Barnes for coordinating Advanced Photon Source (APS) beamline collection and beamline scientists at Stanford Synchrotron Radiation Lightsource and APS GM/CA 23-ID-D; and Dr. Ted Pierson for providing pDENV1/WP/CprME and pDENV2/16681/CprME plasmids. The California Institute of Technology Molecular Observatory is supported by the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program. Operations at Stanford Synchrotron Radiation Lightsource are supported by the US Department of Energy and NIH. Operations at APS are supported by US Department of Energy. This work was supported by NIH Grant P01AI138938 (to D.F.R., C.M.R., M.C.N., P.J.B.); NIH Training Grant 5-T32-GM007616-40 (to S.R.E.); NIH National Research Service Award Fellowship F30AI147579 (to S.R.E.); and NIH National Institute of General Medical Sciences Training Grant T32-GM008042 (to S.R.E.) through the University of California, Los Angeles–California Institute of Technology Medical Scientist Training Program.
Footnotes
Competing interest statement: D.F.R., M.C.N., and The Rockefeller University have filed a patent application for antibody Z004.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6UTA and 6UTE).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919269117/-/DCSupplemental.
References
- 1.Zanluca C., et al. , First report of autochthonous transmission of Zika virus in Brazil. Mem. Inst. Oswaldo Cruz 110, 569–572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy M. R., et al. , Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 360, 2536–2543 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Cao-Lormeau V.-M., et al. , Zika virus, French Polynesia, South Pacific, 2013. Emerg. Infect. Dis. 20, 1085–1086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oehler E., et al. , Zika virus infection complicated by Guillain-Barre syndrome—Case report, French Polynesia, December 2013. Euro Surveill. 19, 20720 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Musso D., Zika virus transmission from French Polynesia to Brazil. Emerg. Infect. Dis. 21, 1887 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick G. W. A., Zika virus. II. Pathogenicity and physical properties. Trans. R. Soc. Trop. Med. Hyg. 46, 521–534 (1952). [DOI] [PubMed] [Google Scholar]
- 7.Dick G. W. A., Kitchen S. F., Haddow A. J., Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 46, 509–520 (1952). [DOI] [PubMed] [Google Scholar]
- 8.Simpson D. I. H., Zika virus infection in man. Trans. R. Soc. Trop. Med. Hyg. 58, 335–338 (1964). [PubMed] [Google Scholar]
- 9.Foy B. D., et al. , Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg. Infect. Dis. 17, 880–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suy A., et al. , Prolonged Zika virus viremia during pregnancy. N. Engl. J. Med. 375, 2611–2613 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Barzon L., et al. , Infection dynamics in a traveller with persistent shedding of Zika virus RNA in semen for six months after returning from Haiti to Italy, January 2016. Euro Surveill. 21, 30316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray K. O., et al. , Prolonged detection of Zika virus in vaginal secretions and whole blood. Emerg. Infect. Dis. 23, 99–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckham J. D., Pastula D. M., Massey A., Tyler K. L., Zika virus as an emerging global pathogen: Neurological complications of Zika virus. JAMA Neurol. 73, 875–879 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasil P., et al. , Guillain-Barré syndrome associated with Zika virus infection. Lancet 387, 1482 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Miner J. J., Diamond M. S., Zika virus pathogenesis and tissue tropism. Cell Host Microbe 21, 134–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C., et al. , Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19, 120–126 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Brasil P., et al. , Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 375, 2321–2334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miner J. J., et al. , Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165, 1081–1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyne C. B., Lazear H. M., Zika virus—Rreigniting the TORCH. Nat. Rev. Microbiol. 14, 707–715 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Chang H.-H., et al. , Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bull. World Health Organ. 95, 517–525I (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Q., et al. , Genomic characterization and phylogenetic analysis of Zika virus circulating in the Americas. Infect. Genet. Evol. 43, 43–49 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Heinz F. X., Stiasny K., The antigenic structure of Zika virus and its relation to other flaviviruses: Implications for infection and immunoprophylaxis. Microbiol. Mol. Biol. Rev. 81, e00055-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierson T. C., Fremont D. H., Kuhn R. J., Diamond M. S., Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: Implications for vaccine development. Cell Host Microbe 4, 229–238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barba-Spaeth G., et al. , Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Dai L., et al. , Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Oliphant T., et al. , Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11, 522–530 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., et al. , Structural basis for neutralization and protection by a Zika virus-specific human antibody. Cell Rep. 26, 3360–3368.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Robbiani D. F., et al. , Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell 169, 597–609.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeffe J. R., et al. , A combination of two human monoclonal antibodies prevents Zika virus escape mutations in non-human primates. Cell Rep. 25, 1385–1394.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beasley D. W., Barrett A. D., Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J. Virol. 76, 13097–13100 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crill W. D., Roehrig J. T., Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75, 7769–7773 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H., et al. , Structural basis of Zika virus-specific antibody protection. Cell 166, 1016–1027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nybakken G. E., et al. , Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437, 764–769 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L., et al. , Delineating antibody recognition against Zika virus during natural infection. JCI Insight 2, 93042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stettler K., et al. , Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353, 823–826 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Sapparapu G., et al. , Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540, 443–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahala W. M. P. B., Kraus A. A., Haymore L. B., Accavitti-Loper M. A., de Silva A. M., Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology 392, 103–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukupolvi-Petty S., et al. , Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J. Virol. 81, 12816–12826 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Victora G. D., Nussenzweig M. C., Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Vaughn D. W., et al. , Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181, 2–9 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Bardina S. V., et al. , Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356, 175–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison S. C., Immunogenic cross-talk between dengue and Zika viruses. Nat. Immunol. 17, 1010–1012 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Priyamvada L., et al. , Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. U.S.A. 113, 7852–7857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers T. F., et al. , Zika virus activates de novo and cross-reactive memory B cell responses in dengue-experienced donors. Sci. Immunol. 2, eaan6809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halstead S. B., Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60, 421–467 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Dejnirattisai W., et al. , Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 17, 1102–1108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahala W. M., Silva A. M., The human antibody response to dengue virus infection. Viruses 3, 2374–2395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morens D. M., Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin. Infect. Dis. 19, 500–512 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Scharf L., et al. , Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. U.S.A. 110, 6049–6054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scharf L., et al. , Structural basis for germline antibody recognition of HIV-1 immunogens. eLife 5, e13783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West A. P. Jr, Diskin R., Nussenzweig M. C., Bjorkman P. J., Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. U.S.A. 109, E2083–E2090 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu X., et al. ; NISC Comparative Sequencing Program , Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simonich C. A., et al. , Kappa chain maturation helps drive rapid development of an infant HIV-1 broadly neutralizing antibody lineage. Nat. Commun. 10, 2190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye J., Ma N., Madden T. L., Ostell J. M., IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J., Sawyer N., Regan L., Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 22, 510–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao F., et al. , Development of a potent and protective germline-like antibody lineage against Zika virus in a convalescent human. Front. Immunol. 10, 2424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao H., et al. , Mechanism of differential Zika and dengue virus neutralization by a public antibody lineage targeting the DIII lateral ridge. J. Exp. Med. 217, e20191792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foote J., Milstein C., Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. U.S.A. 91, 10370–10374 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorpe I. F., Brooks C. L. 3rd, Molecular evolution of affinity and flexibility in the immune system. Proc. Natl. Acad. Sci. U.S.A. 104, 8821–8826 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wedemayer G. J., Patten P. A., Wang L. H., Schultz P. G., Stevens R. C., Structural insights into the evolution of an antibody combining site. Science 276, 1665–1669 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Escolano A., Dosenovic P., Nussenzweig M. C., Progress toward active or passive HIV-1 vaccination. J. Exp. Med. 214, 3–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tai W., et al. , Critical neutralizing fragment of Zika virus EDIII elicits cross-neutralization and protection against divergent Zika viruses. Emerg. Microbes Infect. 7, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang M., Dent M., Lai H., Sun H., Chen Q., Immunization of Zika virus envelope protein domain III induces specific and neutralizing immune responses against Zika virus. Vaccine 35, 4287–4294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang M., Lai H., Sun H., Chen Q., Virus-like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci. Rep. 7, 7679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robbiani D. F., et al. , Risk of Zika microcephaly correlates with features of maternal antibodies. J. Exp. Med. 216, 2302–2315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein F., et al. , Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J. Exp. Med. 211, 2361–2372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nelson C. A., Lee C. A., Fremont D. H., “Oxidative refolding from inclusion bodies.” in Structural Genomics and Drug Discovery: Methods and Protocols, Anderson W. F., Ed. (Springer, New York, 2014), pp. 145–157. [DOI] [PubMed] [Google Scholar]
- 68.Huang J., et al. , Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity 45, 1108–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Battye T. G. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. W., iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Evans P. R., An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schrödinger, LLC, The PyMOL Molecular Graphics System (Version 1.8, Schrödinger, LLC, 2015).
- 76.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 78.McGee C. E., et al. , Infection, dissemination, and transmission of a West Nile virus green fluorescent protein infectious clone by Culex pipiens quinquefasciatus mosquitoes. Vector Borne Zoonotic Dis. 10, 267–274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Rompay K. K. A., et al. , A combination of two human monoclonal antibodies limits fetal damage by Zika virus in macaques. bioRxiv:2020.01.31.926899 (posted 2 Feburary 2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystallographic coordinates for structures Z004iGL Fab–ZIKV EDIII and Z032mature Fab–WNV EDIII are available from the Protein Data Bank under ID codes 6UTA and 6UTE.