Significance
Identification of triple-negative breast cancer (TNBC)-specific genetic events that could guide the treatment decisions represents an unmet clinical need, whereas recent genomic sequencing studies have revealed a paucity of TNBC-specific mutations. In this study, analysis of whole-genome sequencing data for 215 breast tumors catalogued 99 recurrent gene fusions. Among different types of rearrangements, we found that a special class of cryptic adjacent gene rearrangements (AGRs) may occur more frequently than realized in breast cancer. The most frequent AGRs are preferentially found in more aggressive forms of breast cancers, among which BCL2L14–ETV6 is exclusively detected in TNBC. BCL2L14–ETV6 is enriched in high-grade, necrotic, mesenchymal TNBC tumors, and endows increased invasiveness and paclitaxel resistance via inducing partial epithelial mesenchymal transition.
Keywords: BCL2L14–ETV6, gene fusion, triple-negative breast cancer, recurrent adjacent gene rearrangements, chemoresistance
Abstract
Triple-negative breast cancer (TNBC) accounts for 10 to 20% of breast cancer, with chemotherapy as its mainstay of treatment due to lack of well-defined targets, and recent genomic sequencing studies have revealed a paucity of TNBC-specific mutations. Recurrent gene fusions comprise a class of viable genetic targets in solid tumors; however, their role in breast cancer remains underappreciated due to the complexity of genomic rearrangements in this cancer. Our interrogation of the whole-genome sequencing data for 215 breast tumors catalogued 99 recurrent gene fusions, 57% of which are cryptic adjacent gene rearrangements (AGRs). The most frequent AGRs, BCL2L14–ETV6, TTC6–MIPOL1, ESR1–CCDC170, and AKAP8–BRD4, were preferentially found in the more aggressive forms of breast cancers that lack well-defined genetic targets. Among these, BCL2L14–ETV6 was exclusively detected in TNBC, and interrogation of four independent patient cohorts detected BCL2L14–ETV6 in 4.4 to 12.2% of TNBC tumors. Interestingly, these fusion-positive tumors exhibit more aggressive histopathological features, such as gross necrosis and high tumor grade. Amid TNBC subtypes, BCL2L14–ETV6 is most frequently detected in the mesenchymal entity, accounting for ∼19% of these tumors. Ectopic expression of BCL2L14–ETV6 fusions induce distinct expression changes from wild-type ETV6 and enhance cell motility and invasiveness of TNBC and benign breast epithelial cells. Furthermore, BCL2L14–ETV6 fusions prime partial epithelial–mesenchymal transition and endow resistance to paclitaxel treatment. Together, these data reveal AGRs as a class of underexplored genetic aberrations that could be pathological in breast cancer, and identify BCL2L14–ETV6 as a recurrent gene fusion in more aggressive form of TNBC tumors.
Recurrent gene fusions that result from chromosome translocations comprise a critical class of genetic cancer-causing aberrations, which have fueled modern cancer therapeutics. In the past decade, the discovery of novel gene fusions in epithelial tumors have generated great therapeutic impact in recent years. This is represented by the discovery of an EML4–ALK fusion in ∼4% of lung cancer and the FGFR–TACC fusion in ∼3% of glioblastomas that have culminated in effective targeted therapies in these tumors (1, 2). Most recently, larotrectinib targeting the NTRK gene fusions accounting for up to ∼1% of solid tumors have received Food and Drug Administration approval for pan-cancer use, which is considered as the first targeted therapy with tissue-agnostic indication (3). Although low in percentages, these neoplastic gene fusions will likely move toward genetic subtyping of solid tumors that may be possibly curable by fusion-targeted therapies.
In our previous study, analysis of a Cancer Genome Atlas (TCGA) RNA-sequencing (RNA-seq) dataset identified a recurrent gene fusion between the 5′ region of ESR1 and the coding region of the adjacent CCDC170 gene, which was subsequently verified by several other studies (4–8). This fusion represents a cryptic class of genomic rearrangements between adjacent genes (genes within 500-kb distance), which we termed as adjacent gene rearrangements (AGRs). ESR1–CCDC170 is detected in 6 to 8% of luminal B breast tumors and promotes increased aggressiveness (9), which suggests that AGRs can meaningfully contribute to breast cancer development, pathogenesis, and resistance to cancer therapies. Nonetheless, AGRs have been frequently overlooked by fusion detection tools based on RNA-seq data due to the overwhelming number of adjacent chimeras resulting from intergenic splicing events. In addition, such cryptic genomic changes cannot be detected by conventional cytogenetic assays, such as spectral karyotyping or fluorescence in situ hybridization due to the proximity of the rearranged DNAs and the limited resolutions of these assays. For these reasons, AGRs remain an underexplored area of breast cancer genetics.
Here, we performed a landscape study of adjacent gene rearrangements in breast cancer catalogued by whole-genome sequencing (WGS) data, and identified a recurrent fusion, BCL2L14–ETV6, that is preferentially present in triple-negative breast cancer (TNBC). The fusion partners, an ETS family transcription factor gene (ETV6) and an apoptosis facilitator Bcl-2–like protein 14 gene (BCL2L14) are neighboring genes of ∼154 kb apart on the same strand of chromosome 12, with BCL2L14 positioned at the 3′ of ETV6. BCL2L14 encodes a protein member of the Bcl-2 family and was previously described as a novel proapoptotic factor (10). ETV6 encodes a ubiquitously expressed transcriptional repressor that is generally considered as a tumor suppressor unless it forms oncogenic fusions (11) [i.e., ETV6–NTRK3 fusion in secretory breast carcinoma (12)]. In this study, we further investigated the pathological role of BCL2L14–ETV6 in TNBC.
Results
AGRs Comprise the Most Frequent Form of Intergenic Rearrangements in Breast Cancer.
To provide a systematic picture of AGR events in breast cancer, we first analyzed the full spectrum of experimentally confirmed somatic translocations in 9 breast cancer cell lines and 15 breast tumors catalogued from WGS data in a previous study (13). Among 9,408 authentic somatic rearrangements, about half are intrachromosomal rearrangements between adjacent genes located within 500 kb of each other on the chromosome (Fig. 1A). As shown in Fig. 1A, a majority of the intrachromosomal translocations are within 500 kb apart, with a median distance of ∼100 kb. This suggests that AGRs may be a more frequent genetic event than realized. Although most of these rearrangements are likely the consequence of genomic instability, it is plausible that a subset of them could be recurrent genetic events that are pathological in breast cancer.
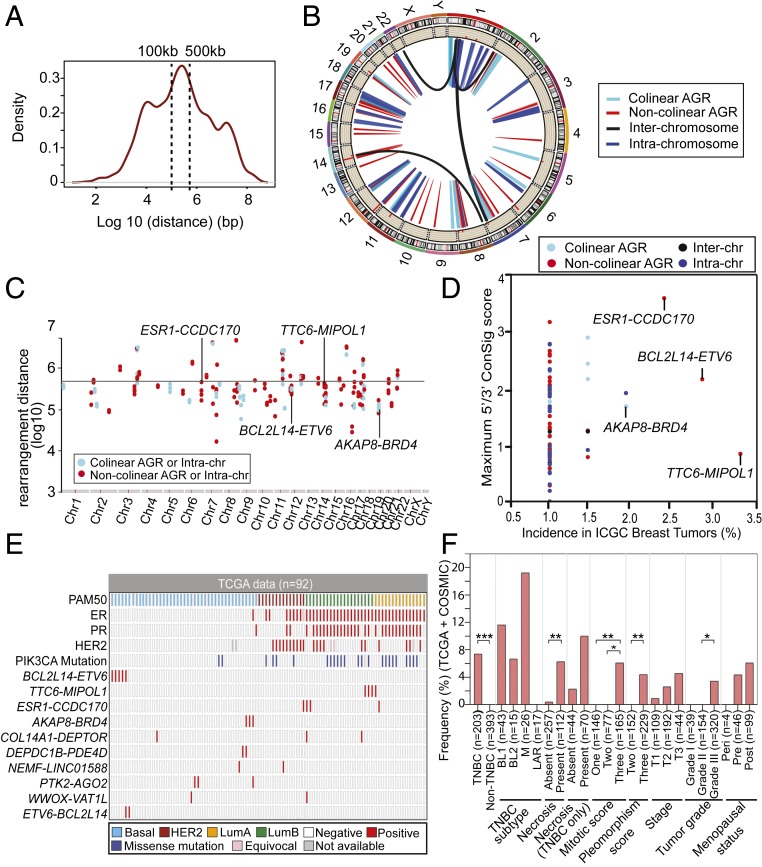
Fig. 1.
Landscape of recurrent adjacent gene rearrangements in breast cancer revealed by WGS data. (A) Frequency chart of experimentally validated interchromosome, intrachromosome distant, and intrachromosome adjacent translocations in nine breast cancer cell lines and 15 breast tumors revealed by WGS data (13). Among 9,408 confirmed somatic translocations, about half are intrachromosomal translocations within 500-kb distance. (B) CIRCOS plot showing the landscape of 99 recurrent gene rearrangements detected in 215 breast tumors based on WGS data from the ICGC. The histogram inside the CIRCOS plot represents the recurrence of the gene rearrangements in the chromosome position, indicating the number of patients that harbor the gene fusions. (C) Genomic hotspots of colinear and noncolinear AGR or intrachromosomal gene rearrangements. Adjacent and intrachromosomal rearrangements in the genomes are displayed in rainfall plot, with each dot representing a respective positive sample. The x axis shows 24 chromosomes in the human genome, and y axis shows the distance between the rearrangement points (base pairs at log10 scale). The horizontal line indicates the cutoff for adjacent gene rearrangements (500 kb in distance). (D) Scatter plot showing the incidences of 99 recurrent gene rearrangements and their concept signature scores, which were detected from 215 ICGC breast tumors profiled by WGS. The x axis indicates the incidence of gene rearrangements in the cohort. The y axis indicates the maximum ConSig scores of 5′ or 3′ partner genes. (E) Tile plot showing the top recurrent AGRs and the known breast cancer oncogenes, including ER, PR, HER2, and PI3KCA mutations in TCGA 92 breast tumors. The AGRs detected in at least two TCGA tumors and >1% of all ICGC tumors are shown in the figure. Group-wise mutual exclusivity test using discrete independence statistics called “Discover” that take into account the distribution of all somatic gene rearrangements, suggests that there are significant number of tumors that harbor only one of these AGRs (P < 0.001). (F) Bar graph showing the association between BCL2L14–ETV6 fusion and different clinicopathological features of 608 breast tumors in the TCGA (92 tumors) and COSMIC (516 tumors) cohort. The y axis shows the incidence of BCL2L14–ETV6 fusion in different clinicopathological groups. *P < 0.05; **P < 0.01; ***P < 0.001. Significance was determined using Fisher’s exact test (two-tailed).
To discover AGRs in breast cancer systematically, we further analyzed the somatic structural mutations catalogued by the International Cancer Genome Consortium (ICGC) based on WGS data for 215 breast tumors. The somatic structural mutations were first mapped with the human exome to reveal genes and exons affected by the rearrangements. The fusion partners were determined based on the strands and genomic regions retained in the rearrangements. To explore if the intergenic rearrangements are enriched in specific breast cancer subtypes, we isolated the 92 ICGC breast tumors contributed by TCGA that have detailed histopathological data from a recent report (14) (SI Appendix, Fig. S1). Overall Her2 and Basal subtypes show significantly higher total number of rearrangements compared to luminal A tumors. Luminal B tumors also exhibit a trend of increased total rearrangements than luminal A tumors. In addition, the breast tumors with high nuclear pleomorphism show significantly higher number of rearrangements. Next, we identified the recurrent gene fusions and classified them into AGRs (local rearrangements involving genes less than 500 kb apart), distant intrachromosomal rearrangements (involving partner genes more than 500 kb apart), or interchromosomal rearrangements (Fig. 1B). In total, we identified 99 recurrent gene fusions that occur in at least two breast tumors, including 57 adjacent gene fusions (57.6%), 35 intrachromosome fusions (35.4%), and 7 interchromosome fusions (7.1%) (Dataset S1). The AGR events spread throughout the genome, with some genomic regions harboring higher incidence of recurrent gene rearrangements (Fig. 1C). Among the 57 recurrent AGRs, 20 are between colinear genes with 5′ located upstream of the 3′ partner (35.1%) and 35 are between noncolinear genes with the 5′ partner located downstream of the 3′ partner (61.4%), which are likely the results of intergenic deletions or tandem duplications, respectively.
Systematic Discovery of Recurrent AGRs in Breast Cancer.
We ranked the recurrent gene rearrangements based on their incidence in the ICGC breast tumor patient cohort, and their concept signature (ConSig) scores (Fig. 1D and Dataset S1). The ConSig scores were developed in our previous study to compute the functional relevance of fusion genes underlying cancer based on their associations with molecular concepts associated with known cancer causal genes (15). The top four most frequent gene fusions identified by our analysis include BCL2L14–ETV6, TTC6–MIPOL1, ESR1–CCDC170, and AKAP8–BRD4, all of which are AGRs (Fig. 1D). To test if the top recurrent AGRs could be a function of genomic instability, we isolated the 92 TCGA breast tumors that have available DNA damage repair (DDR) deficiency scores (16), and sorted these tumors by their genomic instability index (GII) (SI Appendix, Fig. S2). Overall these fusions showed modest enrichment in the tumors with high GII scores, suggesting that DDR deficiency may facilitate formation of a subset of the rearrangements generating these fusions. Further clinicopathological association analysis of these lead recurrent AGRs revealed their preferential presence in the more aggressive forms of breast cancers, including basal-like and luminal B breast cancers (Fig. 1E and Dataset S2). Among the top AGRs, BCL2L14–ETV6 and AKAP8–BRD4 are exclusively found in basal-like breast cancers, while TTC6–MIPOL1 and ESR1–CCDC170 (9) are preferentially present in luminal B tumors. While basal-like and luminal B tumors tend to have a higher number of rearrangements, the specific enrichment of these fusions in either of these subtypes but not in all genomically unstable entities implies their potential function in these tumors. To test if the lead recurrent AGRs display alteration patterns in which most tumors only have one of these fusions, we performed mutual exclusivity tests using discrete independence statistics, called “Discover,” that account for the heterogeneous rearrangement rates across tumors (17). A group-wise mutual exclusivity test for the top recurrent AGRs suggests that there are significant number of tumors that harbor only one of these rearrangements (P < 0.001) (Fig. 1E). This suggests that these recurrent AGRs tend not to cooccur in the same tumor, as opposed to typical DDR-driven rearrangements coexisting in DDR-deficient tumors. Next, we surveyed the incidences of rearrangements based on fusion partner genes and stratified these incidences based on TCGA clinicopathological features (SI Appendix, Fig. S3). The result revealed that most of the lead fusion genes are preferentially present in high-grade tumors, except for TENM4, SHANK2, and TPM3P9. Among these lead fusion genes, we detected several kinase fusion genes, such as DLG2, BRD4, and TNIK. Taken together, the preferential presence of these recurrent AGRs in specific aggressive forms of breast tumors and their tendency not to coexist in genomically unstable tumors imply their potential pathological roles in breast cancer.
Characterization of the Lead Recurrent AGRs in Breast Cancer Samples.
To explore if the most frequent gene rearrangements are significantly associated with specific histopathological features, we analyzed the detailed histopathological data of TCGA breast tumors available from a recent report (14). Interestingly, our analysis revealed that BCL2L14–ETV6 and AKAP8–BRD4 tend to occur in breast tumors with gross necrosis (particularly, extensive necrosis), higher tubule formation score, and higher nuclear pleomorphism (SI Appendix, Fig. S4 and Dataset S3). Tumor necrosis is defined as the morphological changes following cell death (18). The presence of necrosis in breast cancer indicates more aggressive tumors that are associated with early recurrence, poor prognosis (19), and ∼35% of TNBC tumors present necrosis features (20). To further verify the above histopathological associations in a larger cohort of TNBC tumors, we analyzed the somatic rearrangements detected by WGS data in 516 breast tumors, which are provided by the Catalogue of Somatic Mutations in Cancer (COSMIC) (21, 22). From a total of 162 TNBCs in this cohort, we detected 10 BCL2L14–ETV6+ cases, but there is no AKAP8–BRD4+ case (Table 1 and Dataset S4). In both TCGA and COSMIC cohorts of TNBC tumors, the BCL2L14–ETV6+ tumors tend to have a higher level of ETV6 expression than fusion-negative cases, but not all ETV6-overexpressing tumors harbor the BCL2L14–ETV6 fusion (SI Appendix, Fig. S5). The BCL2L14–ETV6 fusions are exclusively detected in TNBC, and correlate with more aggressive features, including presence of necrosis, high mitotic and nuclear pleomorphism scores, advanced tumor stage, and high pathology grade, consistent with the above findings (Fig. 1F). In addition, among TNBC subtypes, BCL2L14–ETV6 fusions most frequently present in the mesenchymal subtype characterized by enriched cell motility and epithelial-to-mesenchymal transition (EMT) pathways, accounting for ∼19.2% of these tumors in the TCGA+COSMIC cohort (Fig. 1F). In addition, BCL2L14–ETV6 is also detected in 11.6% of the basal-like 1 (BL1) tumors characterized by enriched cell cycle and cell-division pathways (23).
Table 1.
Incidence of BCL2L14–ETV6 gene fusion detected in four different patient cohorts of 942 breast tumors
| Cohort | Method | Total | Fusion-positive frequency by TNBC (%) | Frequency by tumor grade in TNBC (%) | Frequency by TNBC subtypes (%)* | ||||||
| Non-TNBC | TNBC | Necrotic TNBC | Low | High | BL1 | BL2 | M | LAR | |||
| TCGA | WGS | 92 | 0/48 (0) | 5/41 (12.2) | 3/23 (13.0) | 0/5 (0) | 4/29 (13.8) | 3/16 (18.8) | 0/5 (0) | 2/11 (18.2) | 0/7 (0) |
| COSMIC | WGS | 516 | 0/345 (0) | 10/162 (6.2) | 4/48 (8.3) | 0/14 (0) | 7/133 (5.3) | 2/27 (7.4) | 1/10 (10) | 3/15 (20.0) | 0/10 (0) |
| PITT | RT-PCR | 89 | – | 4/89 (4.5) | 4/4† | 0/10 (0) | 4/79 (5.1) | – | |||
| BCM | RT-PCR | 245 | 0/200 (0) | 2/45 (4.4) | – | 0/12 (0) | 2/26 (7.7) | – | |||
| Total | 942 | 0/593 (0) | 21/337 (6.2) | 7/71 (9.9) | 0/41 (0) | 17/267 (6.4) | 5/43 (11.6) | 1/15 (6.7) | 5/26 (19.2) | 0/17 (0) | |
BL1 and BL2, basal-like 1 and 2; M, mesenchymal; LAR, luminal androgen receptor.
Only four BCL2L14–ETV6+ cases from the Pitt cohort are analyzed for pathological features, which are not counted in the overall frequencies in necrotic TNBC.
We then proceeded to validate the lead recurrent AGR fusions, including BCL2L14–ETV6, TTC6–MIPOL1, and AKAP8–BRD4, in a panel of breast cancer cell lines and human breast cancer tissues by RT-PCR. The validation of the most frequent AGR, BCL2L14–ETV6, will be detailed below. Since TTC6–MIPOL1 is preferentially expressed in luminal breast tumors, we first screened this fusion in 141 estrogen receptor (ER)+ breast tumors from the University of Pittsburgh (Pitt) cohort using primers located on the first exon of TTC6 and the last exon of MIPOL1, which identified one positive case in this cohort (SI Appendix, Fig. S6A). In addition, we also detected this fusion in one luminal cell line (MDA-MB-361) (SI Appendix, Fig. S6B) and verified the presence of the AKAP8–BRD4 fusion in one patient-derived xenograft (PDX) tumor through screening a panel of 34 TNBC PDX tumors (24, 25) (SI Appendix, Fig. S7A).
BCL2L14–ETV6 Is Exclusively Detected in TNBC.
Next, we focused our study on the BCL2L14–ETV6 rearrangements, which were identified in 12.2% and 6.2% of TNBC cases in the TCGA and COSMIC cohorts, respectively (Fig. 1F and Table 1). We first detected BCL2L14–ETV6 fusion transcripts by RT-PCR in 134 TNBC tumors from two available patient cohorts. To detect most fusion variations, we designed a pair of primers located on exon 2 of BCL2L14 and the last exon of ETV6, respectively. This primer set detected BCL2L14–ETV6 fusion in 4 of the 89 TNBC tumors from the Pitt cohort (Fig. 2A), and 2 of the 45 TNBC tumors from the Baylor College of Medicine (BCM) cohort (Fig. 2B). The clinicopathology features for all of the 134 TNBC patients from Pitt and BCM cohorts are provided in Dataset S5. The fusion-positive cases were subsequently verified by capillary sequencing. Next, we tested the expression of BCL2L14–ETV6 in a panel of 44 breast cancer cell lines and 34 TNBC PDX tumors. We detected one PDX tumor that expresses BCL2L14–ETV6 but not in the cell lines tested (SI Appendix, Fig. S8). The most common fusion variant detected is the fusion between exon 4 of BCL2L14 and exon 2 of ETV6 (referred to as E4E2) that present in two patient cases and one PDX tumor. We also tested BCL2L14–ETV6 by RT-PCR in 200 ER+ breast tumor tissues from the BCM cohort but did not detect any fusion-positive ER+ tumors, which supports its TNBC specificity (SI Appendix, Fig. S9).
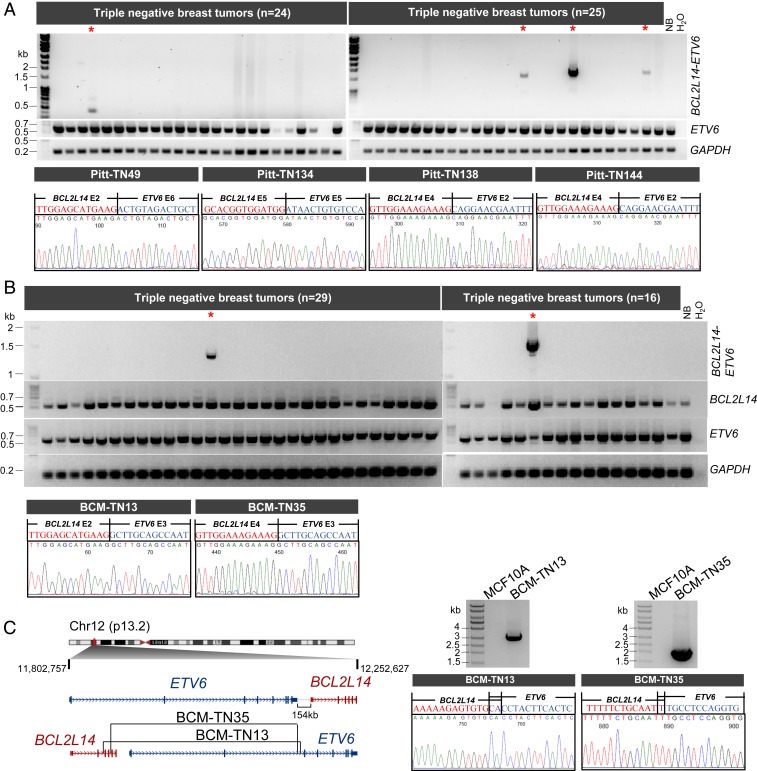
Fig. 2.
Characterization of the BCL2L14–ETV6 fusions in 134 triple-negative breast tumors from two different patient cohorts. (A) RT-PCR analyses of BCL2L14–ETV6 fusion and wtETV6 in triple-negative tumors from the Pitt cohort (n = 89). A five-donor normal breast pool (NB) was used as a negative control. Representative gel images are shown. Fusion-positive cases are marked with red asterisks. The chromatograms (Lower) show the junction sequences of BCL2L14–ETV6 fusion variants detected in Pitt-TN49, Pitt-TN134, Pitt-TN138, and Pitt-TN144 tumors. (B) RT-PCR analyses of BCL2L14–ETV6 fusion, wild-type BCL2L14, wild-type ETV6, and GAPDH in 45 triple-negative breast tumors from the BCM cohort. A five-donor normal breast pool (NB) was used as a negative control. Fusion-positive cases are labeled with red asterisks. Chromatograms (Lower) show the junction sequences of BCL2L14–ETV6 fusion variants detected in BCM-TN13 and BCM-TN35 tumors. (C) Genomic PCR analysis of the BCL2L14–ETV6+ TNBC tumor samples from the BCM cohort (BCM-TN13 and BCM-TN35) identified the precise genomic fusion points. (Left) Schematic of the genomic breakpoints identified in BCM-TN13 and BCM-TN35 tumors. (Right) Gel images and chromatograms of BCL2L14–ETV6 genomic PCR products. Genomic DNA from MCF10A cells was used as a negative control.
To assess if the BCL2L14–ETV6+ tumors present the histopathological features discussed above, we performed histopathological evaluations for the four index tumors from the Pitt cohort for which the tissue sections are available. All four tumors are reported as grade 3 tumors with a high nuclear pleomorphism score and high mitotic count score (Dataset S6). In addition, two of four fusion-positive tumors present extensive necrosis and the remaining two fusion-positive tumors present focal necrosis (SI Appendix, Fig. S10), consistent with the above findings.
Characterization of BCL2L14–ETV6 Genomic Rearrangements and Protein Products.
To verify the genomic origin of BCL2L14–ETV6 in the positive cases, we performed genomic PCR using tiling primers designed specifically for BCL2L14 or ETV6 intron regions predicted to harbor the rearrangement based on the fusion variants detected in the index cases from the BCM cohort. This assay successfully amplified the genomic fusion points in both of the BCL2L14–ETV6+ tumors in the the BCM cohort (Fig. 2C). The breakpoint junctions in the genomic DNA were further verified by capillary sequencing. Next, we explored the association of BCL2L14–ETV6 with copy number aberrations in the TCGA cohort. Copy number data revealed frequent somatic tandem duplications in the ETV6/BCL2L14 loci, which are present in four of the five positive TCGA tumors detected by WGS data (SI Appendix, Fig. S11A). In addition, copy number data also revealed tandem duplications delineating the ETV6/BCL2L14 loci in the TCGA tumors that were not profiled by WGS, suggesting these as potential positive cases. These data suggest that BCL2L14–ETV6 fusions may be the result of either tandem duplications or reciprocal rearrangements that generate both BCL2L14–ETV6 and ETV6–BCL2L14 fusions (Fig. 1E), as with the ESR1–CCDC170 fusion we identified previously (9).
Next, we investigated the structure of BCL2L14–ETV6 proteins. Among five variants detected, three variants (E2E3, E2E6, and E4E2) encode chimeric proteins containing the amino terminus (N terminus) of BCL2L14 and the carboxyl-terminus (C terminus) of ETV6 (Fig. 3A). The ETV6 protein contains an N-terminal pointed (PNT) domain responsible for protein partner binding, and a C-terminal DNA-binding (ETS) domain critical for DNA binding-dependent transcriptional repressor function. Both the most common variant, E4E2, and the E2E3 variants retain the PNT domain and ETS domain, whereas the E2E6 protein only retains the ETS domain. E4E3 and E5E5, on the other hand, do not translate the protein sequence of ETV6 due to a frameshift after the fusion junction, resulting in expression of C-terminus–truncated BCL2L14 proteins.
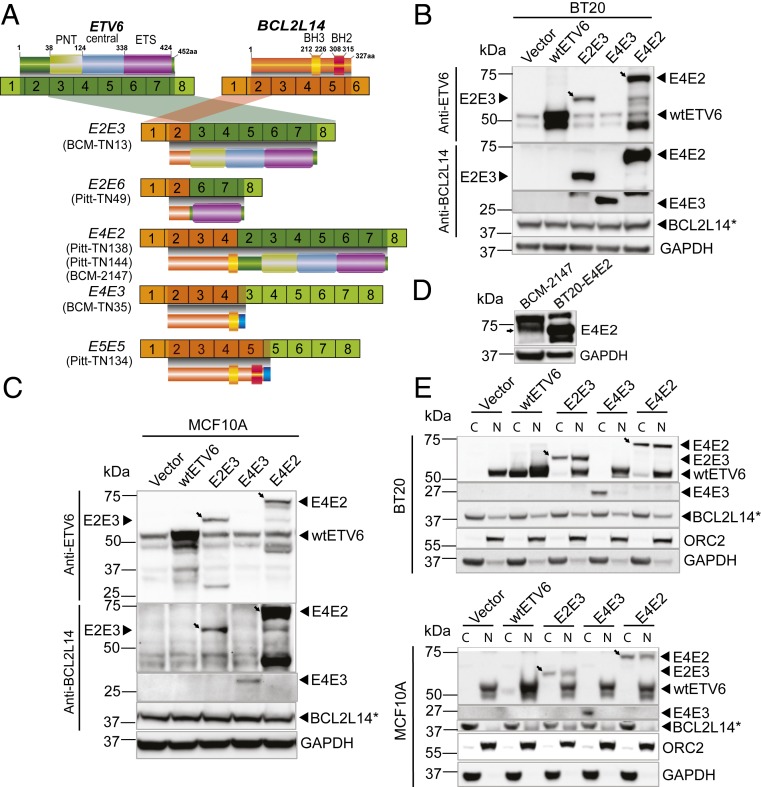
Fig. 3.
Characterization of the protein products encoded by BCL2L14–ETV6 fusion variants. (A) Schematic of BCL2L14–ETV6 fusion variants and encoded proteins identified in the positive cases of the BCM and Pitt cohorts (BCM-TN13, BCN-TN35, Pitt-TN49, Pitt-TN134, Pitt-TN138, Pitt-TN144, and BCM-2147). ORFs of BCL2L14 and ETV6 are depicted in dark shades. Amino acid numbers of BCL2L14 and ETV6 are derived from reference sequence NP_620048 and NP_001978, respectively. Functional protein domains are annotated on top of each gene. (B and C) Western blots detecting BCL2L14–ETV6 fusions (E2E3, E4E3, and E4E2), wtETV6 (ectopic or endogenous), and endogenous BCL2L14 in the engineered BT20 TNBC cells (B) and engineered MCF10A benign mammary epithelial cells (C). Oblique arrows denote the band for E4E2 or E2E3 fusion protein. The fusion variants E4E2 and E2E3 were detected by both polyclonal antibodies of BCL2L14 and ETV6 (Sigma), while the E4E3 variant that does not have an ETV6-encoded sequence was detected only by the BCL2L14 polyclonal antibody (Sigma). The E4E3 fusion variant encodes a much smaller protein (27 kDa) than the E4E2 (74 kDa) and E2E3 (62 kDa) proteins, which is hard to detect on the same blot, and was thus detected separately. An asterisk (also in D and E) denotes the wild-type BCL2L14 protein that was detected by the BCL2L14 monoclonal antibody (Abcam), which identifies a unique band. (D) Western blot using anti-ETV6 polyclonal antibody (Sigma) detected the endogenous protein (pointed by the arrow) encoded by BCL2L14-ETV6 E4E2 variant in the BCM-2147 triple-negative PDX sample. (E) Subcellular localization of wtETV6, BCL2L14, and BCL2L14–ETV6 fusion proteins, in engineered BT20 and MCF10A cells. Oblique arrows point out the fusion protein (E4E2, E2E3). The nuclear protein ORC2 and cytoplasmic protein GAPDH are used as positive controls for fractionation. C, cytoplasm; N, nucleus.
Next, we ectopically expressed the open-reading frames (ORFs) of the fusion variants E2E3, E4E3, and E4E2 in the fusion-negative MCF10A breast epithelial cell line and the BT20 basal-like breast cancer cell line, both of which are triple-negative in (ER, progesterone receptor [PR], and HER2) receptor expression (26). Cells transduced with the vector containing the lacZ gene or the vector containing the wild-type (wt) ETV6 ORF were used as controls. Western blot using polyclonal antibodies against the C terminus of ETV6 or the N terminus of BCL2L14 detected strong expression of the E2E3 (62 kDa) and E4E2 (74 kDa) proteins in the transduced BT20 and MCF10A cells (Fig. 3 B and C). The 27-kDa E4E3 fusion protein was detected by the BCL2L14 antibody, but not by the ETV6 antibody, suggesting that this variant encodes a truncated BCL2L14 protein, which does not contain the ETV6 protein sequence. Next, we sought to detect the endogenous BCL2L14–ETV6 fusion protein in the PDX tumor expressing the E4E2 variant (BCM-2147). Western blot using the ETV6 antibody detected the same-sized band of E4E2 protein as in the engineered BT20 cells (Fig. 3D).
Since gene fusions tend to translocate to abnormal cellular compartments (9), we investigated the cellular localization of the fusion proteins compared to wtETV6 protein in the transduced BT20 and MCF10A cells. Due to the lack of specific antibody against BCL2L14–ETV6 that can be used for immunofluorescence, we performed fractionation of the fusion overexpressing cells and detected the fusion protein localizations by Western blots. Interestingly, the E2E3 and E4E2 fusion proteins tend to be enriched in the cytoplasm fraction, while wtETV6 mainly presents in the nucleus, in line with its role as a transcription factor. The E4E3 fusion that expresses the truncated BCL2L14 protein was found to be enriched in the cytoplasm as well (Fig. 3E). Differential localization of the fusion proteins from wtETV6 suggests that BCL2L14–ETV6 fusion proteins may function in a distinct cellular mechanism compared to wtETV6. It is possible that the BCL2L14 portion of the fusion variants may promote cytoplasm localization of the fusion proteins.
BCL2L14–ETV6 Endows Enhanced Invasiveness and Paclitaxel Resistance.
We sought to examine the function of the BCL2L14–ETV6 fusion in the engineered BT20 and MCF10A cell lines. Among TNBC cell lines, BT20 is a nonmetastatic, chemo-sensitive line (27, 28) overexpressing E-cadherin (29). We thus selected this line for studying the more aggressive and chemo-resistant phenotypes driven by this fusion. MCF10A is an immortal but untransformed human mammary epithelial cell (HMEC) line. Both MCF10A and BT20 cell lines express endogenous ETV6 and BCL2L14 proteins (Fig. 3 B and C). Transwell migration and invasion assays revealed that ectopic expression of the E2E3, E4E3, or E4E2 fusion variants but not wtETV6 significantly enhanced cell motility and invasion in BT20 cells, when compared to vector control (Fig. 4A). Similarly, enhanced cell motility and invasion (Fig. 4B) were also observed in the engineered MCF10A cells expressing these fusion variants. On the other hand, ectopic expression of the fusion variants in BT20 cells did not result in significant changes in cell viability or cell cycle progression, whereas the wtETV6-expressing BT20 cells showed decreased viability and increased G0/G1 phase (SI Appendix, Fig. S12).
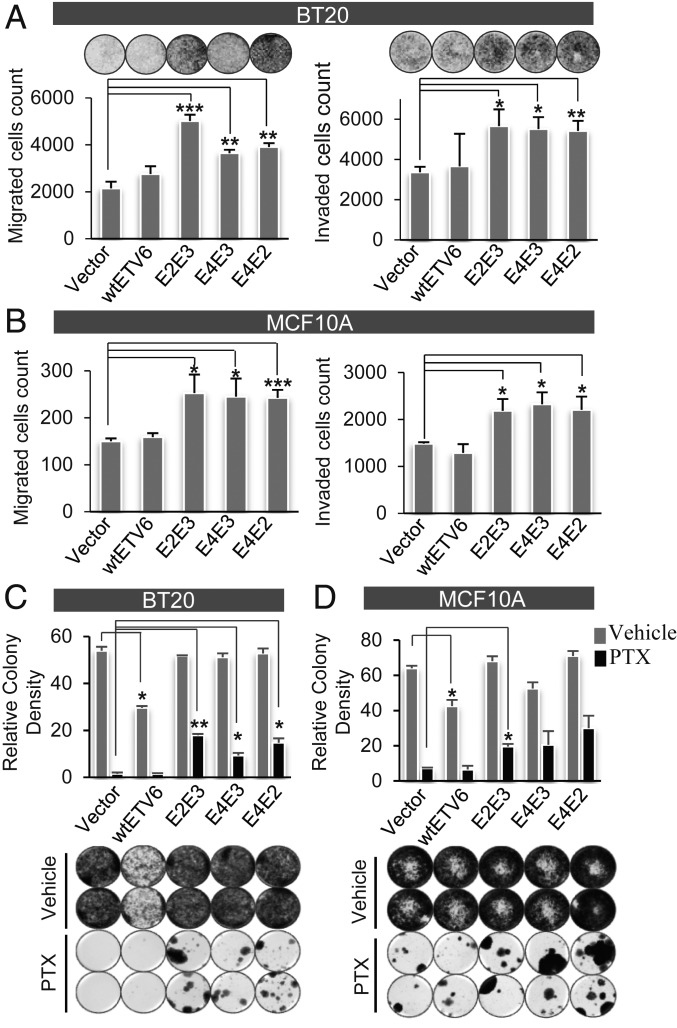
Fig. 4.
Ectopic expression of BCL2L14–ETV6 endows increased cell migration, invasion, and paclitaxel resistance. (A and B) Ectopic expression of BCL2L14–ETV6 fusion variants in BT20 TNBC cells (A) and MCF10A benign mammary epithelial cells (B) significantly enhanced cell migration as revealed by Boyden chamber assay (Left), and increased cell invasion as revealed by transwell Matrigel assay (Right), relative to the vector control. Results were summarized from experimental triplicates. (C) BCL2L14–ETV6 fusions endows clonal resistance in BT20 cells following prolonged paclitaxel treatment for one month as shown by clonogenic assay. Here a low dosage of 5 nM paclitaxel is used for treatment to observe long-term treatment effect. (D) BCL2L14–ETV6 fusions endows clonal resistance in MCF10A cells following prolonged paclitaxel treatment for 1 mo as shown by clonogenic assay. Here 15 nM paclitaxel is used for treatment since MCF10A is less sensitive to paclitaxel. The quantitative results in the Upper panels of C and D are based on two replicates of each condition. The vehicle-treated cells were harvested in 14 d for the BT20 model, and 7 d for the MCF10A model, while the PTX-treated cells were harvested in 1 mo due to their different growth rates. The comparing cell models (i.e., vector, wtETV6, fusion variants) were harvested at the same time point. PTX: paclitaxel; Vehicle: 0.1% dimethyl sulfoxide. *P < 0.05; **P < 0.01; ***P < 0.001, significance was determined using Student’s t test (two-tailed) and error bars reflect mean ± SD.
Taxane-based chemotherapy remains the cornerstone for the treatment of TNBC patients; however, the effectiveness is severely limited by intrinsic and acquired resistance. Since BCL2L14–ETV6 most frequently present in the mesenchymal TNBC tumors that are relatively resistant to chemotherapy (30), we explored the role of BCL2L14–ETV6 in chemoresistance. We first treated the engineered BT20 cells with various doses of paclitaxel, a widely used taxane drug for TNBC patients. BCL2L14–ETV6 fusion-expressing BT20 cells displayed modest reduced sensitivity to paclitaxel following short-term (72 h) treatment, compared to the vector- or wtETV6-expressing cells (SI Appendix, Fig. S13A). We then tested the effect of low-dose prolonged paclitaxel treatment to observe acquired resistance. Following paclitaxel treatment for 1 mo, BT20 cells expressing wtETV6 or vector control were almost eradicated, whereas all fusion-expressing BT20 cells showed evident clonal resistance (Fig. 4C). Similarly, the engineered MCF10A cells expressing BCL2L14–ETV6 fusions also showed increased clonal resistance to paclitaxel, compared to vector- or wtETV6-expressing MCF10A cells (Fig. 4D). These results suggest the role of BCL2L14–ETV6 fusions in endowing paclitaxel resistance in TNBC. Since BCL2L14 is an apoptosis facilitator, we wonder if the fusion may act through impairing the apoptotic pathway. We thus examined the changes in apoptosis biomarkers following paclitaxel treatment. The BT20 cells overexpressing BCL2L14–ETV6 fusions did not show evident reduced apoptosis compared to wtETV6-expressing cells (SI Appendix, Fig. S13B). This suggests that the paclitaxel resistance driven by this fusion may not be attributed to the apoptotic pathway.
BCL2L14–ETV6 Fusions Induce Distinctive Expression Changes from wtETV6.
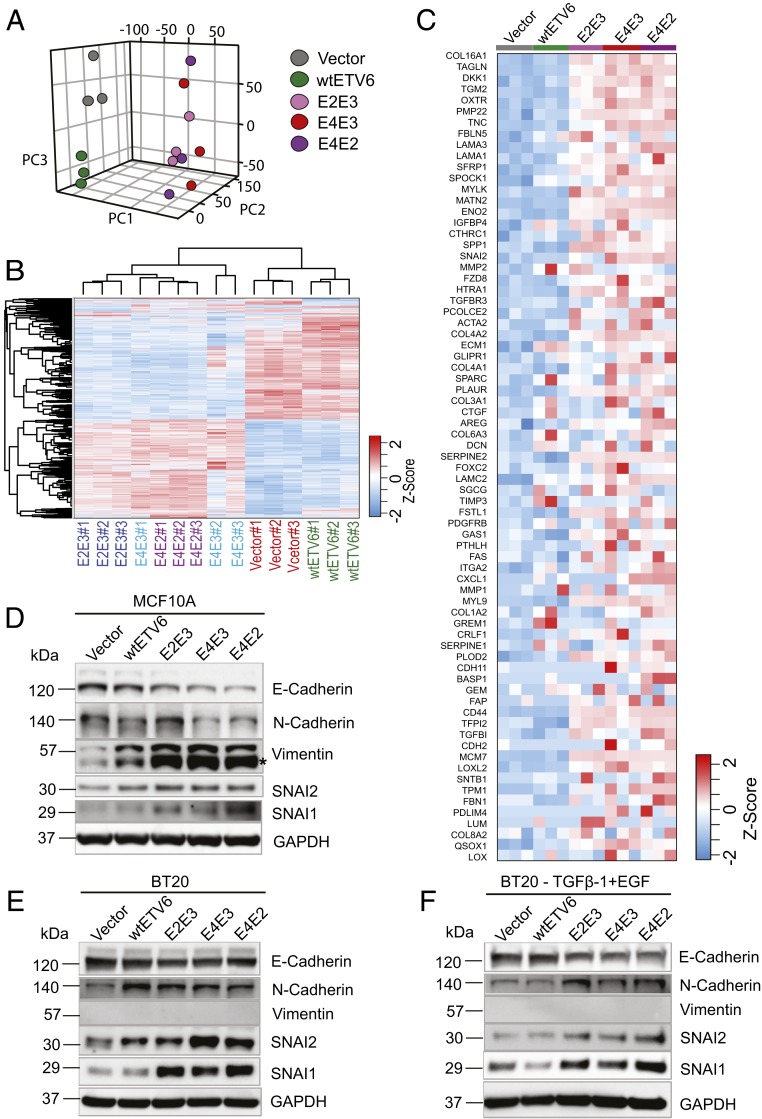
To systematically profile the expression changes induced by BCL2L14–ETV6, we performed transcriptome sequencing of BT20 cells stably expressing the vector, wtETV6, or BCL2L14–ETV6 variants. Principal component analysis (PCA) revealed that the vector- and wtETV6-expressing cells form distinctive and independent clusters, whereas the BT20 cells expressing the different fusion variants are clustered together far from both the vector- and wtETV6-expressing cells (Fig. 5A). Furthermore, hierarchical clustering analysis revealed that the engineered BT20 cells were clustered into two main clusters, with the vector control or wtETV6-expressing cells as one major cluster and fusion-expressing cells as the other major cluster (Fig. 5B). These data suggest that BCL2L14–ETV6 fusions induced distinct gene-expression changes from wtETV6 and vector control in BT20 cells. It is interesting to note that while the E4E3 fusion variant encodes the C-terminus–truncated BCL2L14 protein, this variant induced a similar pattern of expression changes as the E2E3 and E4E2 variants that encode chimeric BCL2L14–ETV6 protein, suggesting that these distinct fusion variants may play a coherent functional role. To identify the pathways characteristic of BCL2L14–ETV6-expressing BT20 cells, we performed gene set enrichment analysis (GSEA) comparing the three fusion variants with the vector control in pairwise. Interestingly, the EMT pathway, known to promote paclitaxel resistance and invasiveness, is among the top up-regulated pathways in BT20 cells expressing BCL2L14–ETV6 (SI Appendix, Fig. S14 and Datasets S7 and S8). Among the core enrichment genes, 73 EMT pathway genes were up-regulated in the fusion-expressing BT20 cells (Fig. 5C and Dataset S9). These results indicate that BCL2L14–ETV6 fusions may induce up-regulation of EMT gene signature. To investigate the transcriptional regulatory mechanisms that regulate the EMT gene signature driven by BCL2L14–ETV6, we constructed breast cancer cell line BT20-specific transcriptional regulatory network using the ARACNe algorithm (31), and performed master regulator analysis (MRA). Among the 13 predicted master regulator candidates, SNAI2 is an established EMT-inducing transcription factor (SI Appendix, Fig. S15). The snail family genes SNAI1 (also denoted as SNAIL) and SNAI2 (also denoted as SLUG) are known to activate EMT and repress epithelial genes in tumors, including in breast cancer (29, 32).
Fig. 5.
BCL2L14–ETV6 fusions induce coherent gene-expression changes distinctive from wtETV6, and prime partial EMT. (A) Unsupervised PCA separated the BT20 cells expressing BCL2L14–ETV6 variants and the BT20 cells expressing the vector or wtETV6 into distinct clusters. We used the first three principal components to present the samples in the 3-dimensional PCA plot. (B) Hierarchical clustering showing the global gene-expression differences between the engineered BT20 cells expressing vector, wtETV6, or BCL2L14–ETV6 fusion variants. (C) Gene-expression heatmap of the 73 core enrichment genes of the EMT signature in BCL2L14–ETV6 fusion variant expressing BT20 cells compared to vector- and wtETV6-expressing BT20 cells. The genes are sorted by their ranks from GSEA analysis. (D–F) Western blots detecting the EMT markers including E-cadherin, N-cadherin, vimentin, and EMT transcription factors including SNAI1 and SNAI2 in the engineered stable cell lines of (D) MCF10A cells, (E) BT20 cells, and (F) TGFβ-1– and EGF-treated BT20 cells. Engineered BT20 cells were treated with 10 ng/mL of TGFβ-1 and 20 ng/mL of EGF for 72 h before being harvested. GAPDH was used as the loading control. An asterisk indicates a nonspecific band.
BCL2L14–ETV6 Fusions Prime EMT.
Next, we explored the expression of EMT biomarkers in the engineered MCF10A and BT20 cells by Western blots, including E-cadherin, N-cadherin, and vimentin. Loss of E-cadherin represents the first step of EMT transition (33). Both MCF10A and BT20 expressing vector control strongly express E-cadherin, suggesting their epithelial states (Fig. 5 D–F). In fusion-expressing MCF10A cells, the expression level of E-cadherin is repressed, whereas the expression level of vimentin, an end-stage marker in EMT (34), but not N-cadherin, was increased (Fig. 5D). In addition, consistent with MRA results, we observed increased protein levels of SNAI2 and its family member SNAI1 in fusion-expressing MCF10A cells. In the engineered BT20 cells, E-cadherin is repressed in all fusion-expressing models, but not in the wtETV6 model. Up-regulation of N-cadherin and SNAI1/SNAI2 were also observed in fusion-expressing BT20 cells; however, there is no induction of vimentin following fusion overexpression (Fig. 5E). The fusion-specific induction of SNAI1/2 transcriptional factors and EMT markers became more obvious when the BT20 cells were treated with TGFβ-1 and EGF, known to induce EMT (35) (Fig. 5F). Loss of the epithelial marker E-cadherin and gain of one of the mesenchymal markers, N-cadherin or vimentin, in MCF10A or BT20 cells suggest that the cells are likely having partial instead of full activation of EMT.
Since EMT is often associated with stemness properties (36) known to promote clonal chemoresistance (37), we examined the expression of the known stemness biomarkers for breast cancer, CD44 and ALDH1A3 (38), in the BT20 models. Our RNA-seq data revealed increased expression of CD44 and ALDH1A3 in fusion-expressing BT20 cells compared to vector or wtETV6-expressing BT20 cells (SI Appendix, Fig. S16A). Consistently, flowcytometry analysis revealed higher number of CD44+/ALDH1high cell populations in fusion-expressing BT20 cells, compared to vector or wtETV6 controls (SI Appendix, Fig. S16 B and C). Together, these results support the role of BCL2L14–ETV6 in inducing partial EMT in TNBC cells.
Discussion
TNBC comprises 10 to 20% of all breast cancers. Due to lack of well-defined molecular targets, treatment of TNBC tumors relies on taxane and platinum-based chemotherapies. Despite the distinctive receptor status, recent genomic sequencing studies have revealed a paucity of TNBC-specific mutations, apart from a distinctive mutational enrichment pattern from other breast cancers, such as more frequent TP53 mutations and less frequent PIK3CA mutations (39). While recent transcriptomic and genomic sequencing studies have revealed oncogenic gene fusions in TNBC patients, some of these may be nonrecurrent and might be considered individual fusions, such as MAGI3–AKT3 and FGFR3–TACC3 (40–42), whereas others tend to fuse with promiscuous partners, such as Notch and MAST fusions, which may be considered as gene family fusions (43). To date, canonical gene fusions of the same fusion partners that recur in a significant subset of TNBC patients have not been reported. Identification of TNBC-specific genetic events that could guide the treatment decisions in this aggressive subtype of breast cancer represents an unmet clinical need.
Despite the complexity and heterogeneity of structural rearrangements in breast cancer (7, 13), our systematic analyses of somatic structural rearrangements based on WGS data catalogued 99 recurrent gene fusions in breast cancer. Among the different types of rearrangements, we found that AGR represents a special type of cryptic rearrangement that may occur more frequently than realized in breast cancer. Such cryptic genomic changes are hardly detectable by conventional cytogenetic assays or by transcriptome sequencing. For these reasons, AGRs can only be confidently detected from WGS datasets. Further studies revealed that the top recurrent AGRs are more frequently enriched in specific more aggressive forms of breast cancer that lack well-defined drivers, such as basal or luminal B breast cancer. These AGRs tend not to aggregate in the genomically unstable tumors, suggesting that they are potential pathological events instead of merely the consequence of genomic instability. Among the top four confirmed recurrent gene rearrangements (BCL2L14–ETV6, AKAP8–BRD4, TTC6–MIPOL1, and ESR1–CCDC170) BCL2L14–ETV6 is frequently and specifically detected in TNBC, and with which we chose to perform further functional studies. For the TTC6–MIPOL1 rearrangement, while the tandem duplication delineating this fusion encompasses the immediately proximal FOXA1 gene, it is unlikely that one copy number gain can significantly enhance FOXA1 expression. In addition, two of four TTC6–MIPOL1+ TCGA tumors do not exhibit copy number changes in the FOXA1 locus (SI Appendix, Fig. S11B). Future studies will be required to further evaluate the function of this fusion in luminal breast cancer.
Next, we performed in-depth functional studies on the BCL2L14–ETV6 fusion. We first experimentally validated this fusion in two independent TNBC patient cohorts, which identified 6 BCL2L14–ETV6+ cases of a total of 134 TNBC cases. Taking together WGS data and RT-PCR validation results, this fusion was detected in 4.4 to 12.2% of TNBC tumors (with an average of 6.2%) from four independent patient cohorts (Table 1). Further investigation of histopathological associations in the TCGA and COSMIC cohorts revealed that this fusion is preferentially present in the TNBC tumors with gross necrosis and more aggressive histopathological features, such as marked nuclear pleomorphism, numerous mitoses, and high tumor grade (Fig. 1F). Such association is further verified by evaluating pathological slides for the fusion-positive cases from the Pitt cohort. All of these cases are grade III TNBCs with extensive or focal necrosis. It is interesting to note that RT-PCR of wtETV6 also revealed ETV6 exon duplications in TNBC cell lines or PDX tumors. These include exon 2 duplication of ETV6 in two PDX tumors and in HCC1187, and exon 4 duplication of ETV6 in one PDX tumor (SI Appendix, Fig. S8). This suggests that ETV6 genetic aberrations could involve both intergenic and intragenic rearrangements. Future studies will be required to explore the function of ETV6 exon duplications in TNBC.
While it remains to be addressed whether DNA repair deficiency may promote the formation of this fusion, our biological studies suggest that BCL2L14–ETV6 fusions appear to enhance cell mobility and invasiveness, and promote paclitaxel resistance when ectopically expressed in basal-like HMEC cell line and nonmetastatic, chemo-sensitive TNBC cell line models. In addition, transcriptome sequencing revealed that despite encoding distinct protein products, the three fusion variants induced a coherent transcriptional program that is distinctive from wtETV6. Of note, while TCGA copy number data suggest genomic amplifications of the ETV6 genomic loci in a subset of breast tumors harboring BCL2L14–ETV6 tandem duplications (SI Appendix, Fig. S11A), ectopic overexpression of wtETV6 did not elicit increased cell migration, invasion, or paclitaxel resistance in TNBC cells (Fig. 4). It is possible that the observed genomic amplifications could be secondary events following formation of this fusion to enhance its function. In our previous study we leveraged the secondary amplifications of oncogenic fusions to identify pathological recurrent fusions (44).
Furthermore, our data suggest that the breast cancer cells overexpressing BCL2L14–ETV6 show a characteristic enrichment of EMT signature. EMT is known to confer stemness features and thus induce invasiveness and chemoresistance in TNBC (32, 45). Interestingly, our data suggest that BCL2L14–ETV6 fusion proteins may prime for partial EMT instead of full activation of EMT. Tumor cells in partial EMT state are in a state of plasticity that favors metastasis and chemoresistance (46), and are frequently observed in TNBC (47). Consistently, BCL2L14–ETV6 fusions are most frequently detected in the mesenchymal subtype of TNBC tumors that is closely associated with EMT (23, 30). In this study, we compared the function of BCL2L14–ETV6 with wtETV6 as the major fusion variant E4–E2 and E2–E3 retain most of the ETV6 domains, whereas the C-terminal–truncated BCL2L14 portion lacks intact BCL2-like domain. Furthermore, the paclitaxel resistance driven by this fusion does not seem to be attributable to the changes in apoptosis signaling (SI Appendix, Fig. S13B). Nonetheless, future studies will be required to elucidate the function of the BCL2L14 portion in the fusion.
While it will be interesting to study the endogenously expressed fusion protein in the BCM-2147 PDX model, technical difficulties exist for genetic inhibition studies in many PDX tumors, including BCM-2147. First, the knockdown studies will require rescue experiments to verify the specificity of the siRNAs, which need to be performed on stable cell lines. There are no fewer than six laboratories attempting to generate cell lines from our BCM PDX models, including laboratories that have generated stable cell lines from primary tissue previously. Thus far, it has not been possible to generate cell lines from any PDX model tested. Although we have established methods for lentiviral transduction for shRNA-mediated knockdown in PDX, the transduction rate is about 30 to 50%, unlike established cell lines where the infection rate typically exceeds 95%. Given this low transduction rate, shRNA-mediated knockdown and genome editing with CRISPR is very inefficient. Furthermore, whereas a majority PDX models will retransplant after dissociation to single cells, which is required for lentiviral transduction, BCM-2147 does not retransplant under all of the dissociation conditions tested. Future studies will be required to overcome these technical challenges and elucidate the function of the endogenous BCL2L14–ETV6 in vivo.
In summary, this study revealed adjacent gene rearrangements as a class of cryptic genetic events that is more frequent than realized in breast cancer. Future investigation of molecular mechanisms of the pathological AGRs, such as BCL2L14–ETV6, may pave the way to new precision medicine against these genetic targets and improve the clinical outcome. Furthermore, immune checkpoint inhibition was most recently found to significantly prolong the survival of metastatic TNBC patients (48). The potential benefit of immunotherapy in the BCL2L14–ETV6+ patient cohort merits future investigations.
Materials and Methods
To systematically characterize recurrent AGRs in breast cancer, we analyzed the somatic structural mutation data catalogued by the ICGC based on WGS data for 215 breast tumors. To detect BCL2L14–ETV6 fusion transcripts, we designed a pair of primers located on exon 2 of BCL2L14 and the last exon of ETV6, respectively, and performed RT-PCR on 134 triple-negative breast tumors, including 45 tumors procured from the Tumor Bank at Baylor College of Medicine and 89 tumors procured from the Health Sciences Tissue Bank of University of Pittsburgh. The primer sequences and PCR conditions are provided in Dataset S10. The full-length cDNAs of BCL2L14–ETV6 fusion variants (E2E3, E4E3, and E4E2) were amplified from fusion-positive tumors, and engineered into a lentiviral pLenti7.3 vector (Invitrogen). BCL2L14–ETV6 protein products were detected by Western blots and the antibodies are provided in Dataset S11. Transwell migration and Matrigel invasion assays were performed to assess cell invasiveness, and clonogenic assays were performed to assess cell viability following paclitaxel treatment. Transcriptome sequencing of the engineered BT20 cells was performed on the Novaseq 6000 system. The RNA-seq data are made available through Gene Expression Omnibus (GSE120919). The protocols, codes, and materials used in this study are available upon request to the corresponding author. More detailed methods are provided in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Fangping Mu for the kind assistance of the research computing. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas project established by the National Cancer Institute and National Human Genome Research Institute (dbGaP accession: phs000178.v6.p6). This study was supported by NIH Grants 1R01CA183976 (to X.-S.W.) and 1R01CA181368 (to X.-S.W.), the Commonwealth of Pennsylvania Tobacco Phase 15 Formula Fund (X.-S.W.), the Shear Family Foundation, and the Hillman Foundation. Breast tumor tissues were provided by the Health Science Tissue Bank at the University of Pittsburgh and the Tumor Bank of Lester and Sue Smith Breast Center at Baylor College of Medicine (supported by SPORE Grant P50 CA186784 and Grant P30CA125123 to the Dan L. Duncan Cancer Center). M.T.L. is supported by a Cancer Prevention and Research Institute of Texas Core Facilities Support Grant RP170691, and by Patient-Derived Xeograft Development and Trial Center Grant U54CA224076. This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided. This work also used the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation Grant OCI-1053575. Specifically, it used the Bridges system, which is supported by National Science Foundation Award ACI-1445606, at the Pittsburgh Supercomputing Center. This project used the Hillman Tissue and Research Pathology Services/Health Sciences Tissue Bank and Cytometry Facility that are supported in part by award P30CA047904.
Footnotes
Competing interest statement: M.T.L. is a founder and limited partner in StemMed Ltd., and a manager in StemMed Holdings, its general partner. He is a founder and equity stake holder in Tvardi Therapeutics Inc.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE120919 for the RNA-seq data on BT20 models and GSE123917 for combined BT20 expression profile data).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921333117/-/DCSupplemental.
References
- 1.Koivunen J. P., et al. , EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin. Cancer Res. 14, 4275–4283 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh D., et al. , Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337, 1231–1235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocco E., Scaltriti M., Drilon A., NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15, 731–747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matissek K. J., et al. , Expressed gene fusions as frequent drivers of poor outcomes in hormone receptor-positive breast cancer. Cancer Discov. 8, 336–353 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Hartmaier R. J., et al. , Recurrent hyperactive ESR1 fusion proteins in endocrine therapy-resistant breast cancer. Ann. Oncol. 29, 872–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giltnane J. M., et al. , Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci. Transl. Med. 9, eaai7993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fimereli D., et al. , Genomic hotspots but few recurrent fusion genes in breast cancer. Genes Chromosomes Cancer 57, 331–338 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Lei J. T., et al. , Functional annotation of ESR1 gene fusions in estrogen receptor-positive breast cancer. Cell Rep. 24, 1434–1444.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veeraraghavan J., et al. , Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. 5, 4577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo B., Godzik A., Reed J. C., Bcl-G, a novel pro-apoptotic member of the Bcl-2 family. J. Biol. Chem. 276, 2780–2785 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Rasighaemi P., Ward A. C., ETV6 and ETV7: Siblings in hematopoiesis and its disruption in disease. Crit. Rev. Oncol. Hematol. 116, 106–115 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Tognon C., et al. , Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell 2, 367–376 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Stephens P. J., et al. , Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462, 1005–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heng Y. J., et al. , The molecular basis of breast cancer pathological phenotypes. J. Pathol. 241, 375–391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X. S., et al. , An integrative approach to reveal driver gene fusions from paired-end sequencing data in cancer. Nat. Biotechnol. 27, 1005–1011 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquard A. M., et al. , Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark. Res. 3, 9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canisius S., Martens J. W., Wessels L. F., A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol. 17, 261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Cruchten S., Van Den Broeck W., Morphological and biochemical aspects of apoptosis, oncosis and necrosis. Anat. Histol. Embryol. 31, 214–223 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Leek R. D., Landers R. J., Harris A. L., Lewis C. E., Necrosis correlates with high vascular density and focal macrophage infiltration in invasive carcinoma of the breast. Br. J. Cancer 79, 991–995 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urru S. A. M., et al. , Clinical and pathological factors influencing survival in a large cohort of triple-negative breast cancer patients. BMC Cancer 18, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nik-Zainal S., et al. , Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes S. A., et al. , COSMIC: High-resolution cancer genetics using the catalogue of somatic mutations in cancer. Curr. Protoc. Hum. Genet. 91, 10.11.1–10.11.37 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Lehmann B. D., et al. , Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., et al. , A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 73, 4885–4897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neelakantan D., et al. , EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat. Commun. 8, 15773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez K. J., Garimella S. V., Lipkowitz S., Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 32, 35–48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottewell P. D., O’Donnell L., Holen I., Molecular alterations that drive breast cancer metastasis to bone. Bonekey Rep. 4, 643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucantoni F., Lindner A. U., O’Donovan N., Düssmann H., Prehn J. H. M., Systems modeling accurately predicts responses to genotoxic agents and their synergism with BCL-2 inhibitors in triple negative breast cancer cells. Cell Death Dis. 9, 42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajra K. M., Chen D. Y., Fearon E. R., The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 (2002). [PubMed] [Google Scholar]
- 30.Park J. H., Ahn J. H., Kim S. B., How shall we treat early triple-negative breast cancer (TNBC): From the current standard to upcoming immuno-molecular strategies. ESMO Open 3 (suppl. 1), e000357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin A. A., et al. , ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinf. 7 (suppl. 1), S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani S. A., et al. , The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsubakihara Y., Moustakas A., Epithelial-mesenchymal transition and metastasis under the control of transforming growth factor β. Int. J. Mol. Sci. 19, E3672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brabletz T., Kalluri R., Nieto M. A., Weinberg R. A., EMT in cancer. Nat. Rev. Cancer 18, 128–134 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Buonato J. M., Lan I. S., Lazzara M. J., EGF augments TGFβ-induced epithelial-mesenchymal transition by promoting SHP2 binding to GAB1. J. Cell Sci. 128, 3898–3909 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Brabletz T., et al. , Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. U.S.A. 98, 10356–10361 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Ejeh F., et al. , Breast cancer stem cells: Treatment resistance and therapeutic opportunities. Carcinogenesis 32, 650–658 (2011). [DOI] [PubMed] [Google Scholar]
- 38.de Beça F. F., et al. , Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J. Clin. Pathol. 66, 187–191 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Shi Y., Jin J., Ji W., Guan X., Therapeutic landscape in mutational triple negative breast cancer. Mol. Cancer 17, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaver T. M., et al. , Diverse, biologically relevant, and targetable gene rearrangements in triple-negative breast cancer and other malignancies. Cancer Res. 76, 4850–4860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosquera J. M., et al. , MAGI3-AKT3 fusion in breast cancer amended. Nature 520, E11–E12 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Banerji S., et al. , Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson D. R., et al. , Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 17, 1646–1651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X. S., et al. , Characterization of KRAS rearrangements in metastatic prostate cancer. Cancer Discov. 1, 35–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedele M., Cerchia L., Chiappetta G., The epithelial-to-mesenchymal transition in breast cancer: Focus on basal-like carcinomas. Cancers (Basel) 9, E134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karaosmanoğlu O., Banerjee S., Sivas H., Identification of biomarkers associated with partial epithelial to mesenchymal transition in the secretome of slug over-expressing hepatocellular carcinoma cells. Cell Oncol. (Dordr.) 41, 439–453 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Sarrió D., et al. , Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 68, 989–997 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Schmid P., et al. ; IMpassion130 Trial Investigators , Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.