Significance
Poorly irrigated tumors exhaust nutrients from their environment. On the other hand, innate immune cells, such as neutrophils and macrophages, have been frequently shown to infiltrate tumors and promote an immunosuppressive environment. Our results show that, when nutrients are low, tumor cells secrete factors related to wound healing that stimulate endothelial cells and attract innate immune cells. This suggests that targeting signals elicited by metabolic stress may help the immune system target malignant cells and that metabolites regulate the communication between immune and tumor cells also through modulation of peptidic signals.
Keywords: cancer metabolism, cytokines, cancer immunity, glucose
Abstract
Cellular starvation is typically a consequence of tissue injury that disrupts the local blood supply but can also occur where cell populations outgrow the local vasculature, as observed in solid tumors. Cells react to nutrient deprivation by adapting their metabolism, or, if starvation is prolonged, it can result in cell death. Cell starvation also triggers adaptive responses, like angiogenesis, that promote tissue reorganization and repair, but other adaptive responses and their mediators are still poorly characterized. To explore this issue, we analyzed secretomes from glucose-deprived cells, which revealed up-regulation of multiple cytokines and chemokines, including IL-6 and IL-8, in response to starvation stress. Starvation-induced cytokines were cell type-dependent, and they were also released from primary epithelial cells. Most cytokines were up-regulated in a manner dependent on NF-κB and the transcription factor of the integrated stress response ATF4, which bound directly to the IL-8 promoter. Furthermore, glutamine deprivation, as well as the antimetabolic drugs 2-deoxyglucose and metformin, also promoted the release of IL-6 and IL-8. Finally, some of the factors released from starved cells induced chemotaxis of B cells, macrophages, and neutrophils, suggesting that nutrient deprivation in the tumor environment can serve as an initiator of tumor inflammation.
Numerous pathological conditions involve nutrient starvation. Upon mild starvation, proliferating cells undergo growth arrest and down-regulate their anabolic metabolism. Nonproliferating tissues are also sensitive to nutrient restriction, and, if starvation is prolonged, cells undergo cell death, which damages the tissue in ischemic diseases, such as stroke and myocardial infarction. Tissue injury due to blunt force trauma or cuts also disturbs nutrient flux, promoting hypoxia and starvation. This type of injury, as well as ischemic damage, engages the wound-healing response to promote tissue repair.
Solid tumor development is also accompanied by a microenvironment low in nutrients (1). When blood vessels are not delivering enough oxygen or nutrients, an increase in vascular endothelial growth factor (VEGF) and other proangiogenic molecules ensures the growth of blood vessels. This response is also coopted by tumors to cope with low oxygen and nutrients in a process known as neoangiogenesis that is associated with tumor invasion and metastasis (2). While neoangiogenesis has been widely studied as a response to hypoxia, several reports indicate that nutrient restriction evokes the same angiogenic responses through the release of proangiogenic factors (3, 4). Another well-characterized response to ischemia in noncancerous tissues is inflammation, which is associated with rejection of transplanted ischemic organs and contributes to injury, but also to tissue repair, in stroke and other ischemic diseases (5, 6). Inflammation, in this context, is generally thought to be driven by the necrotic cell death occurring in ischemic tissues due to the lack of oxygen or nutrients. However, some reports suggest that nutrient restriction, even without cell death, can be sufficient to promote the synthesis and/or secretion of select proinflammatory cytokines (7–9). This suggests that cells in solid tissues possess an evolutionarily conserved program to react to starvation that involves engagement of surrounding cells, including the stroma and resident immune system, to counteract the damage.
We hypothesized that starving cells, including tumor cells, promote adaptive responses to nutrient deprivation that go beyond angiogenesis and resemble a wound-healing response. Starving cells could promote tissue reorganization to restore nutrients and dispose of “waste” products like lactate. To identify possible cell–cell responses, we analyzed the secretome of starved cells and found that the release of inflammatory cytokines and chemokines is a primary response to starvation in multiple cancer cell types, which has implications in cancer immunity and for treatment with antimetabolic drugs.
Results
Cells Subjected to Glucose Deprivation Release Immune Cytokines and Chemokines.
We hypothesized that a reduction in the availability of nutrients induces cell–cell signals, which would include protein mediators (cytokines). To analyze this, we subjected two different cell lines, HeLa (cervical cancer) and Rh4 (alveolar rhabdomyosarcoma), to glucose deprivation and to treatment with 2-deoxyglucose (2-DG), a glycolytic inhibitor (10). Supernatants were then subjected to biotin label-based antibody arrays, which detect a total of 1,000 proteins involved in a variety of functions (SI Appendix, Materials Table). Because dead cells release their content and serum interferes with arrays of secreted proteins, we lowered serum, while selecting times with low amounts of cell death (maximum of 10% cell death) (SI Appendix, Fig. S1A). Results of the arrays are shown in Datasets S1 and S2.
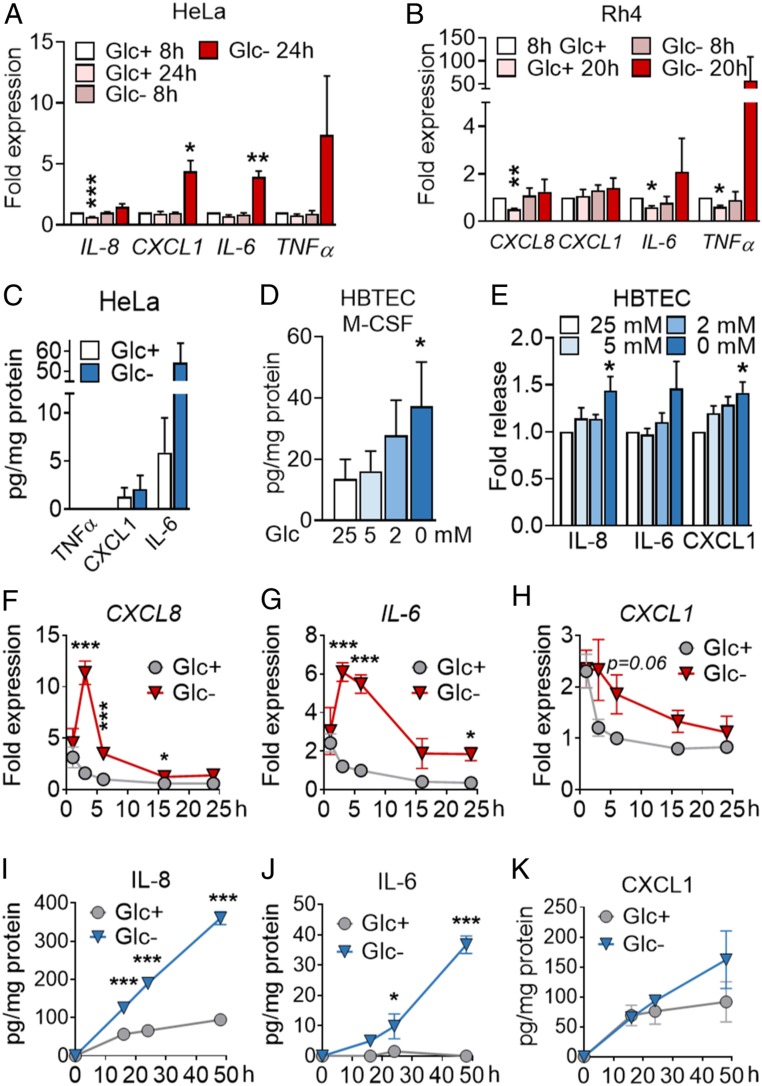
To identify pathways, rather than individual proteins subjected to regulation upon starvation, we performed gene set enrichment analysis (GSEA). This analysis revealed a number of biological processes and molecular functions significantly regulated by glucose deprivation or 2-DG (SI Appendix, Fig. S1 B and C and Dataset S3). Selected proteins from these pathways, such as interleukin-8 (IL-8) (gene name CXCL8), IL-6, CXCL1, and TNFα, were confirmed to be regulated at the messenger RNA (mRNA) level in HeLa (Fig. 1A and SI Appendix, Fig. S1D), and glucose deprivation induced the mRNA of IL-6 and TNFα in Rh4 cells (Fig. 1B). IL-6 and IL-8 were indeed secreted from starved HeLa (Fig. 1C and SI Appendix, Fig. S1D). Importantly, cytokine secretion was not exclusive of tumor cells, and we could also observe the release of M-CSF, CXCL1, IL-6, and IL-8 from primary pulmonary epithelial cells (HBTECs) upon glucose deprivation (Fig. 1 D and E).
Fig. 1.
Inhibition of glycolysis promotes cytokine synthesis and secretion. (A) HeLa cells were treated for 8 or 24 h with media containing 25 mM (Glc+) or 2 mM (Glc−) glucose. qPCR for indicated genes is shown. Fold expression was calculated by normalizing to control sample Glc+ at 8 h. Data are represented as mean ± SEM (n = 3). Asterisks denote significant differences between treated cells and the 8-h Glc+ sample for each mRNA. (B) Rh4 cells were treated and analyzed for 8 or 20 h as in A. Data represent mean ± SEM (n = 3). Asterisks denote significant differences to the control sample at 8 h in Glc+ of each siRNA. (C) HeLa cells were treated for 24 h with 25 mM or 0 mM Glc, and supernatants were analyzed by ELISA of TNFα, CXCL1, and IL-6. Data represent mean ± SEM (n = 3). (D and E) HBTECs were treated for 24 h with media containing indicated glucose concentrations. Supernatants were analyzed by ELISA for M-CSF (D) and IL-8, IL-6, and CXCL1 (E). Data represent mean ± SEM (n = 3). Asterisks represent significant differences between the 25-mM control sample and each treatment analyzed by one-tailed paired t test. (F–H) A549 cells were treated for 1, 3, 6, 16, and 24 h with media containing 25 (Glc+) or 0 mM (Glc−) glucose. qPCR of CXCL8 (F), IL-6 (G), CXCL1 (H) is shown. Fold expression was calculated by normalizing to cells treated for 6 h with Glc+. Data are represented as mean ± SEM (n = 3–9). Asterisks denote significant differences versus the samples in Glc+ for each time point analyzed by two-way ANOVA. (I–K) A549 cells were treated as in F for indicated time points. ELISA of IL-8 (I), IL-6 (J), and CXCL1 (K) is shown. Data are represented as mean ± SEM (n = 3–4). Asterisks denote significance between the Glc+ and Glc− sample for each time point analyzed by two-way ANOVA. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
To detect more inflammatory cytokines that may have been overlooked in the first array, we performed specific arrays for immune cytokines and chemokines. To this end, we employed A549 non-small cell lung adenocarcinoma (LUAC) cells, which were less sensitive to glucose deprivation than HeLa or Rh4, thus allowing the minimization of cell death in supernatants (SI Appendix, Fig. S1F). Chemokine and cytokine arrays revealed that a number of immune cytokines and chemokines were also up-regulated in A549 cells upon glucose deprivation (SI Appendix, Figs. S1 J and K and S2 A–G and Dataset S4). Among them, we found induction of chemokines like CXCL8 (IL-8), CCL5 (RANTES), CCL20 (MIP-3β), and CCL19, as well as immune cytokines, including IL-6, IL-2, IL-11, M-CSF, and CD14. Cytokines with other functions, like VEGF, CTGF, or adiponectin, were also induced while some chemokines like CCL2 were down-regulated. The mRNA coding for some of the proteins analyzed peaked at 3 h and returned to nearly normal levels after 24 h (Fig. 1 F–H and SI Appendix, Fig. S2 A–G), thereby showing a different pattern than observed in HeLa cells (Fig. 1A). We verified using enzyme-linked immunosorbent assay (ELISA) the inducible synthesis and release of IL-6 and IL-8 whereas CXCL1 and M-CSF levels did not change in the supernatant (Fig. 1 I–K and SI Appendix, Fig. S2H).
Given the functional relevance of IL-8 and IL-6 in cancer and that these cytokines were consistently up-regulated, we focused on the regulation of these two proteins to understand the signaling pathways that promote their synthesis and secretion. These cytokines are likely regulated differently, based on different kinetics of mRNA synthesis shown in Fig. 1. IL-8 showed a much higher release than IL-6 from A549 (Fig. 1 I and J) (Fig. 1 I and J), which allowed us to study its secretion at short times within the ELISA detection range. IL-8 is involved in a variety of physiological functions and pathologies, such as cancer. Notably, it is a proangiogenic factor, and it confers survival, chemoresistance, and immunosuppressive function (11). We verified that, even though its messenger peaked at 3 h (Fig. 1F), the protein was continuously being synthesized and released at longer time points, as previously observed in HeLa cells. Analysis of protein content in the cell lysate indicated that IL-8 was being secreted as it was synthesized (SI Appendix, Fig. S2 I–K). Importantly, its secretion correlated inversely with the amount of glucose present in the medium (SI Appendix, Fig. S2L). We extended the analysis of IL-8 and IL-6 secretion to other LUAC cell lines, H460 and H1299, and to two squamous lung carcinoma cell lines, H520 and SW900. These cell lines displayed heterogeneous sensitivity to glucose deprivation (SI Appendix, Fig. S1) and heterogeneous patterns of cytokine release (SI Appendix, Fig. S2 M–T), with H460 and SW900 showing a high constitutive secretion of IL-6 and IL-8 that was further increased upon glucose deprivation. H1299 and H520 did not secrete either cytokine under basal or starved conditions, indicating that not all cell lines exhibit the same pattern. These data indicate that starvation promotes the induction of protumorigenic cytokines, which prompted the question whether targeting cancer metabolism might trigger the same response.
2-DG, Metformin, and Glutamine Depletion, but Not Complete Starvation, Induce IL-6 and IL-8 Secretion.
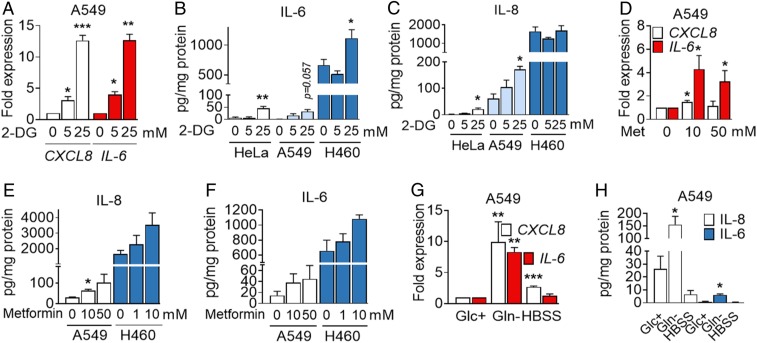
Drugs targeting cancer metabolism are currently being used in clinical trials. 2-DG is an inhibitor of glycolysis and glycosylation frequently used to mimic glucose deprivation that has been tested in patients (10, 12). Metformin targets mitochondrial complex I and is being proposed as an anticancer agent due to epidemiological observations of reduction of cancer incidence, as well as due to associations between diabetes and cancer prognosis (13, 14). We observed that, in general, 2-DG was not toxic to HeLa or A549 cells in contrast to other cell lines that undergo cell death with 2-DG added in the millimolar range (15) (SI Appendix, Figs. S1A and S3A). Exposure to 2-DG in complete medium promoted the induction of CXCL8 and IL-6 in A549 (Fig. 2A), release of IL-6 from HeLa, A549, and H460 cells (Fig. 2B), and release of IL-8 from HeLa and A549 (Fig. 2C), without promoting cell death. Other cell lines that showed little or no cytokine induction upon glucose deprivation did not secrete IL-8 upon treatment with 2-DG (SI Appendix, Fig. S3 B and C), and primary HBTECs did not respond to 2-DG in the same way than to glucose deprivation (SI Appendix, Fig. S3D). Intriguingly, 2-DG prevented cell death of glucose-deprived A549 cells, but it could not substitute for glucose in terms of IL-8 release (SI Appendix, Fig. S3 E and F). This suggests that 2-DG is partially metabolized and can contribute to some essential metabolic or signaling pathway upon glucose depletion. This phenomenon has been previously observed in other cancer cell lines (16, 17).
Fig. 2.
2-DG, metformin (Met), and glutamine deprivation induces IL-6 and IL-8. (A) A549 cells were treated with 2-DG for 6 h. qPCR for CXCL8 and IL-6 is shown. Values were normalized to control sample at 0 mM 2-DG. Data are represented as mean ± SEM (n = 3–4). Asterisks denote significant differences with the 0-mM sample for each cytokine. (B and C) A549, HeLa, and H460 cells were treated with 2-DG for 24 h at indicated concentrations. ELISA values for IL-6 (B) and IL-8 (C) are shown. Data represent mean ± SEM (n = 3–4). Asterisks denote significant differences versus the 0-mM control sample for each cell line. (D) A549 cells were treated for 6 h with metformin at indicated concentrations. qPCR for CXCL8 and IL-6 is shown. Values are normalized to cells treated without the drug. Data are represented as mean ± SEM (n = 4). Asterisks denote significant differences versus the 0-mM control sample. (E and F) A549 and H460 cells were treated for 24 h as in D. ELISA is shown for IL-8 (E) and IL-6 (F). Data are represented as mean ± SEM (n = 3). Asterisks denote significant differences vs. the control for each cell line. (G) A549 cells were incubated in 25 mM (Glc+) glucose, 0 mM glutamine (Gln−) medium or HBSS for 6 h. qPCR is shown for CXCL8 and IL-6. Values were normalized to Glc+. Data represent mean ± SEM (n = 3). Asterisks denote significant differences vs. Glc+. (H) A549 cells were incubated with Glc+, Gln− medium, or HBSS for 24 h. ELISA is shown for IL-8 and IL-6. Data indicate mean ± SEM (n = 3–4). Asterisks denote significant differences versus Glc+. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Mannose, a glucose isomer that can substitute for glucose in some cell lines or inhibit glucose metabolism in others (18, 19), prevented both cell death and IL-8 release in these cells (SI Appendix, Fig. S3 E and F). Metformin in the millimolar range induced mRNA expression and cytokine release of IL-8 and IL-6 in both A549 and H460 cells (Fig. 2 D–F). In addition, glutamine depletion also promoted IL-6 and IL-8 mRNA induction and protein release, as previously described in other cell lines (7, 8). Complete starvation through incubation in a saline solution, Hanks’ balanced salt solution (HBSS), led to induction of CXCL8 mRNA, but it did not lead to secretion of IL-6 or IL-8 (Fig. 2 G and H), indicating that different starvation stimuli lead to different outcomes.
Glucose Deprivation Induces the Unfolded Protein Response (UPR) and Promotes mTOR Inactivation.
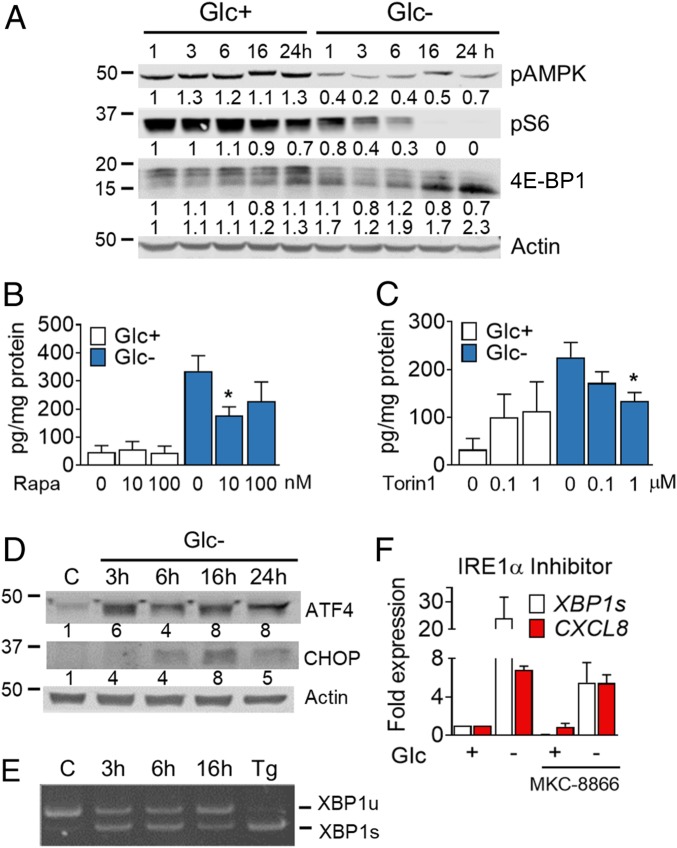
Glucose deprivation inactivates mTORC1, in part through activation of AMPK (20). mTOR inactivation down-regulates protein synthesis and other anabolic pathways. Many tumors and cell lines, including A549, are deficient for the kinase LKB1 that activates AMPK upon glucose withdrawal. Fig. 3A shows mTORC1 inactivation upon glucose deprivation in A549, possibly due to secondary loss of nonessential amino acids. Since mTORC1 inactivation is a common feature of most forms of starvation, we next evaluated whether the use of mTOR inhibitors would be sufficient to promote cytokine release. Rapamycin, an inhibitor of mTORC1, did not promote IL-8 release at doses that inactivate mTORC1 (Fig. 3B and SI Appendix, Fig. S4A). Torin-1, which also inhibits mTORC2, induced IL-8 (Fig. 3C and SI Appendix, Fig. S4B). However, when combined with glucose deprivation, both inhibitors reduced cytokine release, indicating that mTOR was required for IL-8 release (Fig. 3 B and C), as described previously in experiments of glutamine deprivation (7).
Fig. 3.
Glucose deprivation induces the UPR and promotes mTORC1 inhibition. (A) A549 cells were treated for 1, 3, 6, 16 and 24 h in media containing 25 mM (Glc+) or 0 mM (Glc−) glucose. A representative Western blot for pAMPK, pS6, and 4E-BP1 is shown. Protein bands were quantified and normalized to actin; numbers underneath each image represent normalized band intensity. (B and C) Cells were treated for 24 h with rapamycin (Rapa) (B) or Torin1 (C) in media containing 25 mM (Glc+) or 0 mM (Glc−) glucose. ELISA of IL-8 was performed. Data are represented as mean ± SEM (n = 3–4). Asterisks denote significant differences of rapamycin- or torin-treated cells versus the drug-free sample for each culture medium. (D) Cells were treated for indicated times with 25 mM glucose (C) or 0 mM (Glc−). A representative Western blot (n = 3) for ATF4 and CHOP is shown. Protein bands were quantified and normalized to actin. (E) Cells were treated as described in D or for 24 h with 4 µM thapsigargin (Tg) and lysed for mRNA extraction. Retrotranscription was performed followed by RT-PCR for XBP1. Representative PCR is shown out of 3. (F) Cells were pretreated with 10 μM of the IRE-1 inhibitor MKC-8866 for 16 h and for another 6 h in combination with media containing 25 mM (Glc+) or 0 mM (Glc−) glucose. XBP1s and CXCL8 mRNA was analyzed by qPCR. Values are normalized to the Glc+ control sample of each treatment. Data show mean ± SEM (n = 3). Error bars represent the SEM. The significance was indicated as follows: *P < 0.05.
Another related outcome of starvation, which promotes mTOR inactivation in a context-dependent manner, is the activation of the UPR and/or the integrated stress response (ISR), which attenuates 5′ cap-dependent protein translation. These responses converge toward the phosphorylation of the translation initiation factor eIF2α and are engaged upon endoplasmic reticulum (ER) stress as part of the UPR, or upon amino acid/heme depletion and viral double-stranded RNA (dsRNA) detection as part of the ISR. Glucose deprivation engages both the UPR and the ISR in multiple cell lines, depending on activation of the kinases PERK or GCN2, respectively, which phosphorylate eIF2α (21–23). Phosphorylation of this translation initiation factor is mainly involved in translational attenuation which in turn may result in selective translation of certain mRNA, including that encoding the transcription factor ATF4. This transcription factor targets the expression of metabolic genes whose products restore cell homeostasis. But ATF4 also promotes cell death upon glucose deprivation or 2-DG (15, 18). GCN2 was shown to be required for the induction of ATF4 upon glucose deprivation in A549 cells (23). Analysis of ATF4 expression in these cells revealed the fast (3 h) accumulation of this protein upon glucose starvation (Fig. 3D and SI Appendix, Fig. S4C), a time in which we could also detect a strong induction of one of its target genes, CHOP (SI Appendix, Fig. S4D). Besides activation of GCN2, possibly by uncharged transfer RNA, it is likely that glucose-deprived cells activate the UPR because they undergo ER stress in the absence of sugar derivatives required for N-glycosylation. In this sense, XBP1 mRNA splicing was also detected at short times (Fig. 3E and SI Appendix, Fig. S4E). Since the endoribonuclease responsible for XBP1 mRNA splicing (IRE1) and XBP1 have previously been involved in cytokine synthesis in immune cells and animals subjected to restrictive diets (24, 25) or in cytokine release by cancer cells (26, 27), we analyzed their role in cytokine synthesis. The use of a chemical inhibitor of IRE1α RNase activity, MKC-8866, prevented XBP1 splicing, but not CXCL8 induction (Fig. 3F). Silencing of XBP1 with two different small interfering RNA (siRNA) sequences reduced the level of XBP1s and its downstream target Erdj4. However, IL-8 was not consistently down-regulated (SI Appendix, Fig. S4 G–I). In summary, the IRE1/XBP1 pathway does not play a relevant role in IL-8 induction in our experimental settings.
ATF4 and NF-κB Mediate Cytokine Induction and Release.
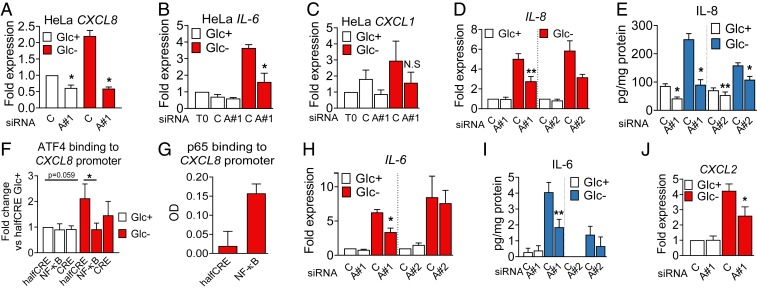
Glucose deprivation, 2-DG, and metformin induced ATF4 (Fig. 3D and SI Appendix, Fig. S5 A–C). A role for ATF4 in cytokine induction and secretion upon glucose deprivation was investigated by using siRNA. HeLa cells transfected with siRNA against ATF4 showed less accumulation of the mRNA of three cytokines tested: CXCL1, CXCL8 (IL-8), and IL-6 (Fig. 4 A–C and SI Appendix, Fig. S5D). In A549 cells, the use of two different siRNAs against ATF4 showed that CXCL8 expression and IL-8 secretion were ATF4-dependent (Fig. 4 D and E and SI Appendix, Fig. S5E). Moreover, quantitative DNA-protein interaction ELISA (qDPI-ELISA) and chromatin immunoprecipitation analysis unveiled that CXCL8 is a direct target of ATF4 (Fig. 4 F and G and SI Appendix, Fig. S5G; see promoter scheme in SI Appendix, Fig. S5F). Similar results were obtained when analyzing IL-6, which is an ATF4 target in macrophages (28), and CXCL2 (Fig. 4 H–J). The use of a chemokine array allowed us to detect other chemokines regulated by ATF4, like CCL2 and CXCL5 (SI Appendix, Fig. S5 H and I). CCL2 was down-regulated by glucose deprivation, and its levels were recovered by silencing ATF4 (SI Appendix, Fig. S5J). CXCL5 was secreted in high quantities by A549 cells, and its mRNA was regulated by ATF4, in the presence or absence of glucose, although the reduction of the protein in cell supernatants was not significant (SI Appendix, Fig. S5 K and L). Other cytokines and chemokines like CXCL3 were only mildly or not regulated by ATF4 (SI Appendix, Fig. S5 N–Q).
Fig. 4.
ATF4 regulates cytokine mRNA and secretion in HeLa and A549 cells. (A–C) HeLa cells were transfected for 24 h with nontargeting siRNA (labeled as ‘’C” for Control siRNA) or siRNA against ATF4 (labeled “A#1” for ATF4 sequence 1). After 24 h, cells were washed and incubated in 25 (Glc+) or 2 mM glucose (Glc−) media. qPCR for CXCL8 (A), IL-6 (B), and CXCL1 (C) is shown. Data are represented as mean ± SEM (n = 3). Fold expression was calculated by normalizing to the control siRNA in Glc+ conditions or time = 0 (T0) (before treatment). Asterisks denote significant differences of ATF4-transfected cells versus the control siRNA in each culture medium. N.S. means not significant. (D, E, and H–J). A549 cells were transfected for 40 h as described in A as well as with siRNA against ATF4 (labeled “A#2” for ATF4 sequence 2) and treated with 25 mM (Glc+) or 0 mM (Glc−) glucose for 6 h (D, H, and J) or 24 h (E and I). qPCR data for CXCL8 (D), IL-6 (H), and CXCL2 (J) were normalized to the control siRNA of Glc+. ELISA is shown for IL-8 (E) and IL-6 (I). Data are represented as mean ± SEM (n = 3–5). Asterisks denote significant differences of ATF4-transfected cells versus the control siRNA in each culture medium. (F and G) A549 cells were treated for 6 h with 25 mM (Glc+) or 0 mM (Glc−) glucose (F), or with 25 mM glucose and 10 ng/mL TNFα (G). Cells were harvested and processed for qDPI-ELISA as described in SI Appendix for ATF4 (F) and p65 (G). Values represent mean ± SEM after normalization to the value of halfCRE-containing oligo Glc+ (F, n = 4) or average of optic density values (OD) (G, n = 2). Asterisks indicate significant differences by two-way ANOVA. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01.
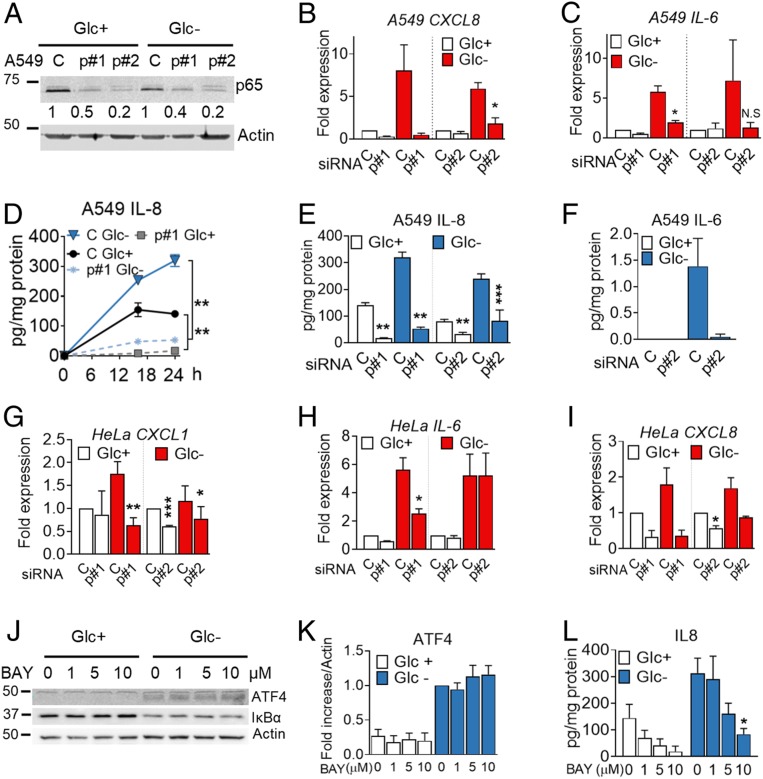
Inflammatory cytokines and chemokines are generally regulated by the transcription factor NF-κB. This transcription factor is activated downstream of the UPR/ISR via multiple mechanisms which include IRE1-mediated TRAF/JNK activation and degradation of IκB when translation is reduced (29). Down-regulation of p65, one of the canonical NF-κB subunits, using siRNA, indicated that IL-8 was regulated at the mRNA and protein level upon glucose deprivation and also in basal conditions in A549 cells (Fig. 5 A, B, D, and E). Similarly, silencing p65 reduced IL-6 induction and secretion (Fig. 5 C and F). p65 NF-κB participated in regulation of the mRNA of M-CSF, CXCL1, CXCL2, and CCL20 (SI Appendix, Fig. S6 C–F) while the effect on CXCL3 and CXCL5 was minor (SI Appendix, Fig. S6 G and H). NF-κB was also required for induction of CXCL8, CXCL1, and IL-6 in HeLa cells (Fig. 5 G–I and SI Appendix, Fig. S6 A and B).
Fig. 5.
The p65 NF-κB subunit regulates cytokine synthesis and secretion. (A–F) A549 cells were transfected for 40 h with control nontargeting siRNA (labeled as “C”) or siRNA for p65 (labeled as “p#1” for sequence 1 and “p#2” for sequence 2). Western blot of p65 is shown in A. Cells were subsequently incubated with media containing 25 mM (Glc+) or 0 mM (Glc−) glucose for 6 h (B and C), for 16 and 24 h (D), or for 24 h (E and F). qPCR for CXCL8 (B) or IL-6 (C) was performed. Values were normalized versus control siRNA in Glc+. IL-8 (D and E) and IL-6 (F) were analyzed by ELISA. Results show mean ± SEM (n = 3–4). Asterisks denote significant differences of p65-transfected cells versus the control siRNA in each culture medium. N.S. means not significant. (G–I) HeLa cells were transfected for 24 h with siRNA (as in A) and treated for 24 h with 25 mM (Glc+) or 2 mM (Glc-) glucose. qPCR for CXCL1 (G), IL-6 (H) and CXCL8 (I) is shown. Values are normalized versus the control siRNA in Glc+. Results show mean ± SEM (n = 4). Asterisks denote significant differences of p65-transfected cells versus the control siRNA in each culture medium. (J–L) A549 cells were treated with the NF-κB inhibitor (BAY11-7082 [BAY]) at concentrations shown with media containing 25 mM (Glc+) or 0 mM (Glc−) of glucose for 16 h. Western blot of ATF4 and IκBα is shown in J. ATF4 (K) (n = 4) was quantified and normalized to actin. Fold induction was calculated vs. Glc− control sample. IL8 (L) was analyzed by ELISA after treating as in J. Results show average ± SEM (n = 4). Asterisks denote significant differences of treated cells vs. the 0-mM control of each of the culture media. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
We investigated a possible cross-talk between ATF4 and NF- κB in the regulation of IL-8 and found that inhibition of NF-κB using BAY 11-7082 or silencing p65 did not alter ATF4 induction (Fig. 5 J–L and SI Appendix, Fig. S6 I–K) and silencing ATF4 had only a minor effect on IκBα degradation (SI Appendix, Fig. S6 L–N). In summary, a subset of immune cytokines and chemokines is induced transcriptionally and translationally by NF-κB p65 and/or ATF4 during glucose deprivation.
Conditioned Medium from Starved Cells Promotes Paracrine Effects on Cancer Cells.
The pattern of cytokine induction was conserved among multiple cancer cell types and even nontransformed cells. This suggests that this is a conserved response that may play wound healing or alarm roles in the tissue and which could influence the cross-talk between cancer cells and the stroma. Additionally, our results suggested the possibility that proteins secreted from cancer cells subjected to starvation protect neighboring cancer cells or stimulate their own migration. To test which roles could the cytokine mixture play, we collected conditioned medium from A549 cells subjected to glucose deprivation or grown in control medium. A fraction of the conditioned medium was filtered and washed with fresh medium to concentrate the cytokines and eliminate metabolites secreted from starved cells (see scheme in SI Appendix, Fig. S7A). The conditioned medium was applied to the lower part of a Boyden chamber in which A549 cells were plated on top, and migration was measured. We observed that A549 cells migrate toward conditioned media. However, there was no major difference in the migration between the conditioned media from cells grown in the presence or absence of glucose (SI Appendix, Fig. S7 B and C). We also incubated A549 cells in conditioned media and measured cell death induced by glucose deprivation. Filtered conditioned media without glucose, containing secreted cytokines but not metabolites, induced more cell death than control media without glucose (SI Appendix, Fig. S7 D and E), and it did not promote epithelial–mesenchymal transition (EMT) or matrigel invasion (SI Appendix, Fig. S7 F–I). However, the factors released by starved cells changed A549 adhesion in collagen and stimulated their proliferation (Fig. 6 A and B). A predicted effect of the conditioned medium, according to the cytokines secreted, was angiogenesis. We tested the effects of conditioned medium on human umbilical vein endothelial cells (HUVECs) and observed that the fraction larger than 3 kDa from starved A549 cells stimulated their proliferation and migration compared to medium from nonstarved cells (Fig. 6 C and D)
Fig. 6.
Conditioned (cond) medium promotes migration of immune cells and HUVECs. (A) A549 cells were pretreated for 16 h with conditioned (yellow) or washed conditioned (orange) media derived from other A549 cells which were cultured for 24 h with 25 (+) or 0 (−) mM glucose. Fresh Glc was added when indicated. Cells were harvested and plated in a collagen-coated plate, and attached cells were counted by crystal violet. Results were normalized to each Glc+ control and show mean ± SEM (n = 4). Asterisks denote significant differences between each pair. (B and C) A549 (B) and HUVECs (C) were cultured for 24 h with media containing 25 mM glucose or with washed conditioned (orange) media of other A549 cells which were cultured as described in A; 10% FBS was added in all conditions. Fresh Glc was added when indicated. Proliferation was measured by crystal violet staining. Values were normalized to cells cultured in fresh glucose containing media (white bar). Results show mean ± SEM of A549 (n = 6) and HUVEC (n = 7) experiments. Asterisks denote significant differences using one-tailed (A549) or two-tailed (HUVEC) t test as indicated. (D) HUVECs were allowed to migrate for 16 h toward washed conditioned (orange) media derived from A549 cells treated as in A. Fresh Glc was added when indicated. Migrated cells were stained by crystal violet and counted by ImageJ. Results show mean ± SEM (n = 4). (E and F) Human PBMCs were allowed to migrate for 20 h toward fresh glucose-containing (25 mM) or glucose-free medium (white bars), or conditioned media (yellow bars) from A549 cells grown in glucose-containing or glucose-free medium for 24 h. Conditioned media which were refreshed are designated in orange, and elution in blue (SI Appendix, Fig. S7). Migrated cells were stained for indicated markers and analyzed by fluorescence-activated cell sorter (FACS). Data are represented as mean ± SEM. CD3− CD56+ (n = 5–7) and CD19+ (n = 6–9). The dashed line indicates the level of basal migration in fresh Glc+ medium. Asterisks denote significant differences between cells migrated toward the Glc+ control sample (white bar) or as indicated. (G) THP-1 cells were differentiated, allowed to migrate for 20 h toward media as described in E, and counted. Data are represented as mean ± SEM (n = 3–4). Asterisks denote significant differences of cells migrated toward fresh Glc+ and conditioned media or as indicated. (H) HL60 cells were differentiated and cells migrated for 2 h toward fresh (white bars) or conditioned media of H460 cells (yellow bars) which were grown for 24 h in media containing 25 (+) or 0 (−) mM glucose and 2% FBS. “−/+” indicates conditioned, glucose-free medium to which 25 mM glucose was added before the experiment. Data represent mean ± SEM (n = 3). Asterisks denote significant differences of cells migrated toward fresh Glc− media using one-tailed Student t test. (I) Neutrophils were extracted from human blood and allowed to migrate for 1 h toward conditioned media as described in H. Data are represented as mean ± SEM (n = 6). Asterisks denote significant differences versus Glc+. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01.
Conditioned Medium from Starved Cells Promotes Migration of Immune Cells.
Inflammation occurs during wound healing, and it involves the recruitment of immune cells to injured or infected sites. Our results suggested that glucose deprivation could promote a similar response. We first tested whether conditioned medium from starved A549 cells would stimulate migration of lymphoid cells from healthy donors. Cells incubated in Boyden chambers showed a tendency to migrate more toward conditioned medium from starved cells than to medium from control cells. This tendency was conserved when the conditioned medium was filtered and washed. The trend was observed for T lymphocytes (CD3+), natural killer T cells (CD3+ CD56+), and natural killer cells (CD3− CD56+), but not for monocytes (Fig. 6E and SI Appendix, Fig. S7 J–L), and was more obvious and significant for B cells (CD19+) (Fig. 6F).
The known roles of some of the chemokines studied include chemoattraction of myeloid cells. We performed migration experiments using THP-1 cells differentiated to macrophage-like cells using phorbol 12-myristate 13-acetate (PMA). We observed a higher migration toward conditioned media of starved cells regardless of glucose availability (Fig. 6G). Moreover, migration was not due to secreted metabolites (Fig. 6G, blue) but due to the fraction larger than 3 kDa, which also induced migration of THP-1 when replenished with fresh complete media.
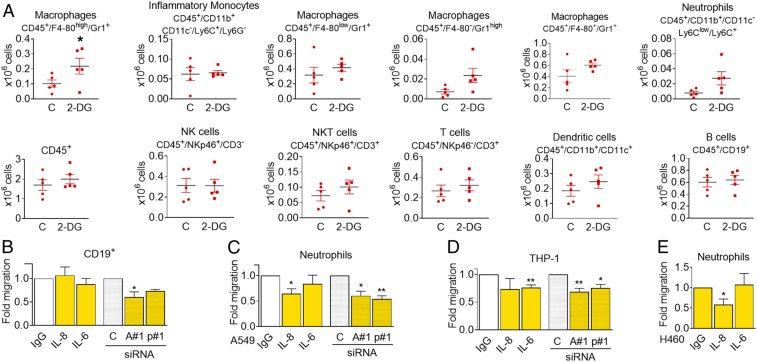
The first responders to an injury site are neutrophils. We differentiated HL60 cells to neutrophil-like cells and observed that, although they do not migrate toward conditioned media of A549 (SI Appendix, Fig. S7M), conditioned media from starved H460 cells, which release higher amounts of cytokines (SI Appendix, Fig. S2), attracted HL60 cells (Fig. 6H). We also purified neutrophils from human blood and let them migrate toward conditioned media of A549 or H460 cells. Primary neutrophils migrated preferentially toward conditioned media from starved cells (Fig. 6I and SI Appendix, Fig. S7N). These media did not promote differential effects on cell death or activation of neutrophils (SI Appendix, Fig. S7 O and P). Additionally, we injected conditioned medium from murine CT26 cells in the peritoneal cavity of syngeneic immunocompetent mice. The conditioned medium was collected from cells that had been incubated in the presence or absence of 2-DG. Afterward, the conditioned media were washed with fresh media to remove 2-DG. Forty-eight hours after the injection, the number of macrophages and neutrophils retrieved was higher in mice injected with the 2-DG conditioned medium (Fig. 7A) although the effect was lost at 72 h (SI Appendix, Fig. S8).
Fig. 7.
Conditioned medium from starved cells alters immune cell composition in the peritoneal cavity. (A) Conditioned medium from CT26 cells that had been incubated for 48 h with or without 25 mM 2-DG was cleared by centrifugation and further concentrated 20 times. Then 350 µL of this medium were injected intraperitoneally every 24 h. Forty-eight hours after the first injection, peritoneal cells were isolated and subjected to FACS analysis using the indicated markers. Asterisks denote significance using Student t test (P < 0.05). (B) PBMCs were plated in the top of Boyden chambers and allowed to migrate to conditioned media from glucose-deprived A549 cells (0 mM glucose, 24 h) previously transfected for 40 h with nontargeting siRNA (control, “C”), ATF4#1 (“A#1”), or p65#1 (“p#1”), or toward conditioned medium from glucose-deprived A549 cells containing control unspecific immunoglobulin G (IgG) or neutralizing antibodies against ("α") IL-6 or IL-8. CD19+ cells were measured by FACS analysis 20 h later. Results show mean ± SEM of four to six experiments. Asterisks denote significant differences between each treatment and its siRNA or antibody control. (C and D) Neutrophils or differentiated THP-1 were allowed to migrate to media described in B for 2 or 20 h, respectively, and counted. Data show mean ± SEM of five to six experiments. Asterisks denote significant differences between each treatment and its siRNA or antibody control. (E) Conditioned media of H460 cells starved for 24 h (0 mM Glc, 2% FBS) were incubated with unspecific IgG or neutralizing antibodies against IL-6 or IL-8 and placed in the bottom of a Boyden chamber. Neutrophils extracted from human blood were allowed to migrate for 1 h and counted. Results show mean ± SEM (n = 4–6) experiments. Asterisks denote significant differences between IgG and each treatment. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01.
Finally, we tested whether ATF4 and p65 played a role in the paracrine effects of conditioned media and whether some of the migratory effects were mediated by IL-8 and/or IL-6. We verified that neutralization of IL-6 or IL-8 affected migration of THP-1 cells or neutrophils while silencing p65 or ATF4 reduced migration of CD19+, neutrophils, and macrophage-like THP-1 cells (Fig. 7 B–E).
Discussion
Nutrient limitation occurs during physiological tissue growth or as a consequence of various pathological events, such as wounds or vascular occlusions that lead to transient ischemia. To recover, ischemic tissues employ mechanisms that promote wound healing and the restoration of the nutrient flux. These mechanisms include angiogenesis and inflammation, which have been classically attributed to hypoxia or to necrotic inflammatory signals, respectively. We show here that nutrient deprivation could be, by itself, a powerful signal to induce inflammation as part of a normal wound-healing response as pathogens and sterile injuries trigger similar events which include the recruitment of the innate immune system. It is possible that nutrient deprivation followed by the ISR is one of the many signals that the body uses to detect infections as pathogens also deprive nutrients from the site of infection. In fact, the kinase PKR, that detects viral dsRNA, is one of the kinases that activate the ISR (30).
In cancer, starvation can occur in three major scenarios. During tumor growth, the rapid expansion of tumor cells results in poor vascularization. Second, some oncogenes promote the massive use of nutrients, which create a state of “pseudo-starvation” that has been proposed to contribute to metastasis (31). Third, antimetabolic drugs being used or tested for cancer therapy cause starvation-like signals in cancer cells. Given the role of several of the cytokines described here in tumor progression, our results suggest a note of caution when treating patients with these drugs. On the other hand, tumorigenic progression is often associated to chronic inflammation. Our data suggest that one of the major promoters of tissue inflammation in tumors may be the lack of nutrients, which induces the secretion of chemokines and cytokines that in turn may promote tumor survival through suppressive immune infiltration.
We performed RNA and protein arrays in several cell lines and confirmed that glucose deprivation leads to the induction and release of a broad range of proteins, such as VEGF and IL-8, that are associated with angiogenesis, or IL-6 and IL-11, that promote tumor growth and survival. Paracrine signaling could transmit noncell autonomous “adaptation to stress” signals to neighboring cells to promote escape or adapt to the nutrient shortage and overcome the starvation. Interestingly, some cytokines, like IGFBP-2, or chemokines, like CXCL5 and CCL2, were down-regulated upon glucose deprivation even if their mRNA was induced, which was the case of TNF. This could be due to impaired N-glycosylation in the absence of glucose, and it should be tested under other starvation conditions that do not impair this protein modification.
Focusing on chemokines and cytokines involved in inflammation, we found IL-8 and IL-6 as proteins consistently secreted from different cell lines (HeLa, A549, H460, and SW900), indicating that the response is conserved among distinct cancer cell types. Some cells analyzed here showed high basal levels of IL-6 and IL-8 (H460, SW900), which has been attributed to their mutations in KRAS (32). IL-8 promotes angiogenesis, metastasis, and proliferation in lung cancer, and it is also a chemoattractant for neutrophils. Higher levels of circulating IL-8 are associated with poor prognosis and overall survival of non-small cell lung cancer patients (32). IL-6 is well-described as an activator of tumor-associated signaling cascades promoting tumor growth, immune cell infiltration, and cachexia (33). This suggests that induction of IL-6 and IL-8 by starvation signals present in poorly irrigated tumors may worsen patient outcome.
Glucose deprivation induces ER stress and the UPR, frequently due to the lack of flux through the glucosamine pathway, which impairs N-glycosylation. In some cell lines, the UPR and cell death can thus be prevented by readdition of N-acetylglucosamine and substrates for glycosylation, like mannose. In A549 cells, mannose prevented cell death and IL-8 induction, but N-acetylglucosamine did not, which suggests, in accordance to a previous report, that another pathway than the PERK-driven UPR might activate ATF4 in A549 cells (23). In parallel, however, we have observed activation of the ER stress sensor IRE1, which mediates immune infiltration in cancer. In our experimental conditions, we could not detect the involvement of the IRE1–XBP1 pathway in IL-8 induction, but its effects on the overall response to starvation should be tested more extensively by analyzing other cytokines. ATF4 had been previously found to mediate IL-6 induction upon metabolic stress in macrophages and human tumor cells (3, 28), as well as VEGF induction (3, 4, 34). In A549, we observed that ATF4 was induced upon treatment with all starvation-promoting stimuli, and it was linked to cytokine production. IL-6 and IL-8 contain CRE/ATF4 binding sites in their promoters, and we describe here that ATF4 binds the IL-8 (CXCL8) promoter, so it is possible that ATF4 acts directly as a transcription factor of several of the cytokines. However, it could also be working in combination with NF-κB or CHOP, which have been shown to participate together in the induction of IL-8 transcription upon infection (35), or indirectly on some of the cytokines. Moreover, nutrients regulate epigenetic modifiers that could be contributing to rewiring transcriptional pathways under nutrient scarcity (36). On the other hand, glucose deprivation inactivates mTORC1 in our setting. While in some cells the mTORC1 inhibitor rapamycin has been shown to induce IL-6, suggesting that this would be the cause for cytokine induction (9), our experiments and those from others indicate that some mTORC1 activity is required for cytokine secretion (7). Enhanced translation of specific genes during the UPR/ISR may also be contributing to promote synthesis of the cytokines (8). The transcriptional and translational links between mTOR, the UPR/ISR, and starvation are, however, far from simple, and more experiments should be performed to establish the precise role for each signaling pathway in the regulation of each cytokine upon different forms of starvation.
Several recent reports indicate that the metabolic rewiring of tumors interferes with antitumor immunity in many ways (reviewed by ref. 37). On one hand, tumor-derived metabolites, such as lactate or adenosine, influence the immune response. On the other hand, in the tumor microenvironment low in glucose, immunity is impaired due to a “metabolic competition” between the tumor and the T cells (38, 39). However, not only T cells are affected: The innate immune system could also play a role in immune escape. In this sense, tumor hypoxia rewires tumor-associated macrophages (40), and our data suggest that low nutrients could be promoting macrophage and neutrophil attraction. It was also shown that inhibition of glycolysis in the tumor by using 2-DG in vitro or silencing lactate dehydrogenase A in vivo prevents the secretion of a cytokine required for the development of myeloid-derived suppressor cells, which have yet to be analyzed in the starvation settings used in our study (41). Since tumor-intrinsic factors, including chemokines, are often responsible for the differences of a T cell inflamed versus an inflammatory suppressor environment (42), we propose that cell–cell effects of cancer starvation are one of the suppressive factors impairing immune activity. In summary, our data indicate that nutrient shortage promotes a response that resembles wound healing. This response could be required to coordinate tissue repair after an episode of ischemia, but, in the case of cancer, it could contribute to maintenance of the tumor.
Methods
Cell Lines and Human-Derived Primary Cultures.
Human blood samples (buffy coats) were obtained from the Banc de Sang i Teixits (BST) (Blood and Tissue Bank). Experiments were approved by the Clinical Research Ethical Committee from the Hospital of Bellvitge (Comitè d’Ètica d’Investigació Clínica [CEIC]). Isolation of peripheral blood mononuclear cells (PBMCs) and neutrophils was performed immediately after arrival of buffy coats, and primary cultures did not exceed 24 h, including experiments. Primary immune cells were cultured in pyruvate-free Dulbecco’s modified Eagle’s medium (DMEM) containing 25 mM glucose, decomplemented 10% fetal bovine serum (FBS) (heated at 57 °C for 30 min), and 2 mM fresh l-glutamine at 37 °C and 5% CO2 atmosphere. Primary endothelial cells (HUVEC/TERT) were cultured in EGMTM-2 Endothelial Cell Growth Medium-2 BulletKit containing additional 10% FBS. Primary bronchial/tracheal epithelial cells (HBTECs) were cultured in Airway Epithelial Cell Basal Medium supplemented with the Bronchial Epithelial Cell Growth Kit at 37 °C and 5% CO2 atmosphere.
Different cancer cell lines (lung: A549, H1299, H460, H520, SW900; cervix: HeLa; rhabdomyosarcoma: Rh4; and leukemia: HL60) were cultured in DMEM supplemented as for immune cells. Cells were maintained at 37 °C and 5% CO2 atmosphere, and adherent cells were split twice a week when they reached 90% confluence using a 0.05% trypsin ethylenediaminetetraacetic acid solution. THP-1 (leukemia) cells were grown in RPMI supplemented with 10% FBS and 1% l-glutamine and diluted once a week with fresh RPMI at a ratio of 1:20. Cells were grown in antibiotic-free medium.
Cell Treatments.
Cell treatments were performed by plating 300,000 (6-well plate) or 150,000 (12-well plate) cells 24 h prior to treatment. At a confluence of 80%, cells were washed twice with pyruvate-free DMEM, 0 mM glucose, and 0% FBS, and glucose-deprived cells were treated with pyruvate-free DMEM containing 0 mM glucose, 10% dialyzed FBS, and 1% l-glutamine while the control cells were supplemented with glucose at concentrations indicated in the figures. For FBS dialysis, the dialyzing membrane was washed according to the manufacturer’s recommendations and filled with 100 mL of heat-inactivated FBS. The pipe was washed twice for 1 h and once overnight in 1 L of PBS at 4 °C while stirring. Dialyzed FBS was sterilized by using a 22-µm syringe filter.
For other treatments, cells were incubated in pyruvate-free DMEM with 25 mM glucose, 10% FBS, and 1% l-glutamine, unless indicated. For HBSS treatments, cells were washed twice and incubated in HBSS with calcium, magnesium and glucose 5.5 mM. For glutamine free treatments, cells were washed twice with glutamine-free, pyruvate-free DMEM containing 10% FBS and incubated with this medium.
Statistical Analysis.
The significance of the data was calculated using the two-tailed, paired Student’s t test unless indicated differently. Two-way ANOVA followed by Sidak’s multiple comparison test was performed using Prism GraphPad. Error bars represent the SEM. The significance was indicated as follows: P < 0.05, one asterisk (*); P < 0.01, two asterisks (**) ; P < 0.001, three asterisks (***). N.S. means not significant.
For detailed materials and methods, see SI Appendix.
Data Availability Statement.
All data are included in the manuscript and supporting information.
Supplementary Material
Acknowledgments
We are grateful to M. Sánchez Céspedes, E. Ballestar, M. Molina, I. Fabregat, M. Graupera, V. Soto, and R. Pérez-Tomás (Bellvitge Biomedical Research Institute) for providing cell lines; J. Moreno and M. Hernández for assistance in performing experiments; M. A. Pujana, S. Logue, I. Zuazo-Gaztelu, T. Avril, J. Obacz, and E. Hernández for help with methods; and S. J. Martin, F. Viñals, and J. Aramburu for conceptual suggestions. We thank the CERCA Program/Generalitat de Catalunya for institutional support. This study has been funded by Ministerio de Ciencia, Innovación y Universidades, part of Agencia Estatal de Investigación, through Grant BFU2016-78154-R (cofunded by the European Regional Development Fund, “A way to build Europe,” ISCIII PI13-00139); and the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreements 675448 (TRAINERS) and 766214 (META-CAN).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913707117/-/DCSupplemental.
References
- 1.Sullivan L. B., et al. , Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiménez-Valerio G., Casanovas O., Angiogenesis and metabolism: Entwined for therapy resistance. Trends Cancer 3, 10–18 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., et al. , The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res. 72, 5396–5406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longchamp A., et al. , Amino acid restriction triggers angiogenesis via GCN2/ATF4 regulation of VEGF and H2S production. Cell 173, 117–129.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhu S. D., Frangogiannis N. G., The biological basis for cardiac repair after myocardial infarction: From inflammation to fibrosis. Circ. Res. 119, 91–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenski M., Scherer M. A., Analysis of synovial inflammatory markers to differ infectious from gouty arthritis. Clin. Biochem. 47, 49–55 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Shanware N. P., et al. , Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat. Commun. 5, 4900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gameiro P. A., Struhl K., Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep. 24, 1415–1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon S., et al. , NF-κB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene 31, 3467–3481 (2012). [DOI] [PubMed] [Google Scholar]
- 10.El Mjiyad N., Caro-Maldonado A., Ramírez-Peinado S., Muñoz-Pinedo C., Sugar-free approaches to cancer cell killing. Oncogene 30, 253–264 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Alfaro C., et al. , Interleukin-8 in cancer pathogenesis, treatment and follow-up. Cancer Treat. Rev. 60, 24–31 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Xi H., Kurtoglu M., Lampidis T. J., The wonders of 2-deoxy-D-glucose. IUBMB Life 66, 110–121 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Levy A., Doyen J., Metformin for non-small cell lung cancer patients: Opportunities and pitfalls. Crit. Rev. Oncol. Hematol. 125, 41–47 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Bergamino M., et al. , Fasting plasma glucose is an independent predictor of survival in patients with locally advanced non-small cell lung cancer treated with concurrent chemoradiotherapy. BMC Cancer 19, 165 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramírez-Peinado S., et al. , 2-deoxyglucose induces Noxa-dependent apoptosis in alveolar rhabdomyosarcoma. Cancer Res. 71, 6796–6806 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Lowman X. H., et al. , The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol. Cell 40, 823–833 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Lee H. Y., et al. , Ca2+-dependent demethylation of phosphatase PP2Ac promotes glucose deprivation–induced cell death independently of inhibiting glycolysis. Sci. Signal 11, eaam7893 (2018). [DOI] [PubMed] [Google Scholar]
- 18.León-Annicchiarico C. L., et al. , ATF4 mediates necrosis induced by glucose deprivation and apoptosis induced by 2-deoxyglucose in the same cells. FEBS J. 282, 3647–3658 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez P. S., et al. , Mannose impairs tumour growth and enhances chemotherapy. Nature 563, 719–723 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Dibble C. C., Manning B. D., Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15, 555–564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muaddi H., et al. , Phosphorylation of eIF2α at serine 51 is an important determinant of cell survival and adaptation to glucose deficiency. Mol. Biol. Cell 21, 3220–3231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S., et al. , ERK2 mediates metabolic stress response to regulate cell fate. Mol. Cell 59, 382–398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaveroux C., et al. , Nutrient shortage triggers the hexosamine biosynthetic pathway via the GCN2-ATF4 signalling pathway. Sci. Rep. 6, 27278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obacz J., et al. , Regulation of tumor-stroma interactions by the unfolded protein response. FEBS J. 286, 279–296 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Rubio-Patiño C., et al. , Low-protein diet induces IRE1α-dependent anticancer immunosurveillance. Cell Metab. 27, 828–842.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Obacz J., et al. , IRE1-UBE2D3 signaling controls the recruitment of myeloid cells to glioblastoma. bioRxiv:10.1101/533018 (31 January 2019).
- 27.Logue S. E., et al. , Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat. Commun. 9, 3267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki Y., et al. , Activating transcription factor 4 links metabolic stress to interleukin-6 expression in macrophages. Diabetes 63, 152–161 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Zhang K., Kaufman R. J., From endoplasmic-reticulum stress to the inflammatory response. Nature 454, 455–462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong K. L., et al. , Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 11, 1553–1562 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Jiménez C., Goding C. R., Starvation and pseudo-starvation as drivers of cancer metastasis through translation reprogramming. Cell Metab. 29, 254–267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunaga N., et al. , Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer. Int. J. Cancer 130, 1733–1744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson D. E., O’Keefe R. A., Grandis J. R., Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15, 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malabanan K. P., Kanellakis P., Bobik A., Khachigian L. M., Activation transcription factor-4 induced by fibroblast growth factor-2 regulates vascular endothelial growth factor-A transcription in vascular smooth muscle cells and mediates intimal thickening in rat arteries following balloon injury. Circ. Res. 103, 378–387 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Bezzerri V., et al. , Mapping the transcriptional machinery of the IL-8 gene in human bronchial epithelial cells. J. Immunol. 187, 6069–6081 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Reid M. A., Dai Z., Locasale J. W., The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19, 1298–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ecker C., Riley J. L., Translating in vitro T cell metabolic findings to in vivo tumor models of nutrient competition. Cell Metab. 28, 190–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C. H., et al. , Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho P.-C., et al. , Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henze A.-T., Mazzone M., The impact of hypoxia on tumor-associated macrophages. J. Clin. Invest. 126, 3672–3679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W., et al. , Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 28, 87–103.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., et al. , Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity 49, 178–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and supporting information.