Significance
C9ORF72, together with SMCR8 and WDR41, can form a stable complex that regulates membrane trafficking. We report the cryo-EM structure of the C9ORF72–SMCR8–WDR41 complex at atomic resolution. Notably, the stoichiometry of the three subunits in the C9ORF72–SMCR8–WDR41 complex is 2:2:2. Interestingly, the C termini of C9ORF72 and the DENN domain of SMCR8 mediate the dimerization of the two C9ORF72–SMCR8–WDR41 protomers in the complex. Moreover, WDR41 binds to the DENN domain of SMCR8 by the C-terminal helix without direct contact with C9ORF72. Most importantly, the C9ORF72–SMCR8 complex works as a GAP for Rab8a and Rab11a in vitro, and the Arg147 of SMCR8 is the arginine finger.
Keywords: C9ORF72, SMCR8, WDR41, GAP activity, cryo-EM
Abstract
A massive intronic hexanucleotide repeat (GGGGCC) expansion in C9ORF72 is a genetic origin of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Recently, C9ORF72, together with SMCR8 and WDR41, has been shown to regulate autophagy and function as Rab GEF. However, the precise function of C9ORF72 remains unclear. Here, we report the cryogenic electron microscopy (cryo-EM) structure of the human C9ORF72–SMCR8–WDR41 complex at a resolution of 3.2 Å. The structure reveals the dimeric assembly of a heterotrimer of C9ORF72–SMCR8–WDR41. Notably, the C-terminal tail of C9ORF72 and the DENN domain of SMCR8 play critical roles in the dimerization of the two protomers of the C9ORF72–SMCR8–WDR41 complex. In the protomer, C9ORF72 and WDR41 are joined by SMCR8 without direct interaction. WDR41 binds to the DENN domain of SMCR8 by the C-terminal helix. Interestingly, the prominent structural feature of C9ORF72–SMCR8 resembles that of the FLNC–FNIP2 complex, the GTPase activating protein (GAP) of RagC/D. Structural comparison and sequence alignment revealed that Arg147 of SMCR8 is conserved and corresponds to the arginine finger of FLCN, and biochemical analysis indicated that the Arg147 of SMCR8 is critical to the stimulatory effect of the C9ORF72–SMCR8 complex on Rab8a and Rab11a. Our study not only illustrates the basis of C9ORF72–SMCR8–WDR41 complex assembly but also reveals the GAP activity of the C9ORF72–SMCR8 complex.
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are the most common neurodegenerative diseases and show overlapping pathology, genetic abnormalities, and patient symptoms (1–3). Disrupted RNA and protein homeostasis have been indicated to be the general causes of ALS-FTD (4). In 2011, the expanded intronic hexanucleotide repeat (GGGGCC) in the 5′ noncoding region of the gene C9ORF72 was found to accounted for most cases of familial ALS and FTD as well as some sporadic cases of both diseases, representing a historic discovery (5–7). The two potential mechanisms of disease onset related to repeat expansion identified to date are gain of function and loss of function (3, 8). Studies suggest that neurotoxic materials, including dipeptide repeat proteins (DRPs) and RNA G-quadruplexes, are generated based on the expanded hexanucleotide repeat, which is described as “gain of function” (9–11). In addition, the expanded hexanucleotide repeat can also inhibit the transcription of C9ORF72 and, thereby, decrease the production of C9ORF72 protein (5, 6, 12, 13). However, the precise function of C9ORF72 is unclear.
Recently, C9ORF72, together with SMCR8 (Smith–Magenis syndrome chromosomal region candidate gene 8) and WDR41 (WD40 repeat-containing protein 41), has been shown to form a stable complex that participates in regulating macroautophagy (hereafter referred to as autophagy) by directly interacting with the ULK1 complex (14–16). C9ORF72 knockout causes a defect in starvation-induced autophagy, indicating that C9ORF72 regulates autophagy positvely (14, 16–18). SMCR8 was so named because its gene location on the chromosome is close to the gene related to Smith–Magenis syndrome; however, it has no relationship with this syndrome (19). Interestingly, SMCR8 knockout results in an increase in ULK1 gene expression, which suggests that SMCR8 is a negative regulator of autophagy (20, 21). WDR41, the function of which is unknown, is a WD40 repeat-containing protein located on the endoplasmic reticulum (ER) (Fig. 1A) (22, 23). The subunits of the ULK1 complex, to which the C9ORF72–SMCR8–WDR41 complex (hereafter referred to as CSW) binds, are still controversial. Although studies have suggested that the CSW complex interacts with the ULK1 complex via ATG101 or FIP200 (14, 16), the details of the interaction between these complexes have yet to elucidated.
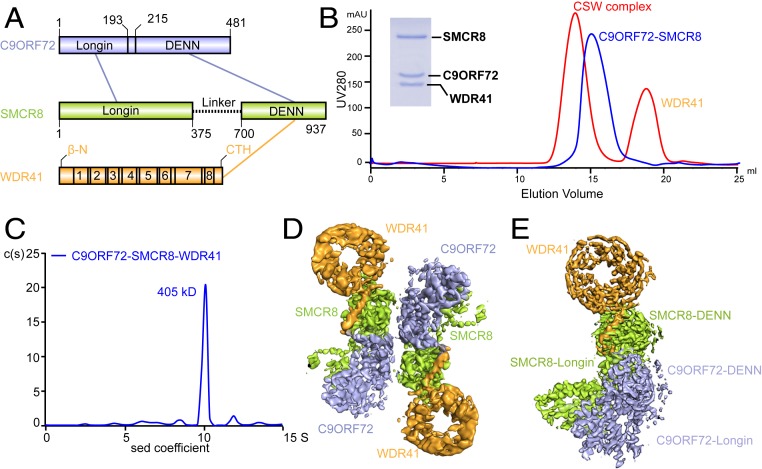
Fig. 1.
The CSW complex is a dimer of C9ORF72–SMCR8–WDR41. (A) Schematic diagram of the domain arrangement of C9ORF72 (light blue), SMCR8 (light green), and WDR41 (orange). The names and boundaries of domains are labeled. CTR, C-terminal helix of WDR41. The numbers in WDR41 represent WD40 domains: WD1 (41–81), WD2 (88–131), WD3 (137–168), WD4 (177–281), WD5 (226–276), WD6 (281–314), WD7 (326–401), and WD8 (411–432, and β-N). The interactions between different domains are shown in lines: light-blue line, C9ORF72–SMCR8 interaction; orange line, WDR41–SMCR8 interaction. (B) Gel filtration (superpose 6 10/300 GL) profile of reconstituted C9ORF72–SMCR8–WDR41 and C9ORF72–SMCR8 complex. The horizontal axis is elution volume, and the vertical axis is ultraviolet (UV) absorption. The UV absorbance is shown in red (C9ORF72–SMCR8–WDR41) and blue (C9ORF72–SMCR8) lines. The peaks of proteins are labeled. The Coomassie blue-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) gel shows the peak fraction of the CSW from gel filtration. (C) Analysis of peak fraction from B by sedimentation velocity AUC. C(S) functions calculated from sedimentation velocity data are shown in blue curve. The calculated molecular mass is denoted. Horizontal axis, sedimentation coefficient; vertical axis, continuous sedimentation coefficient distribution. (D and E) Cryo-EM density map of the CSW complex. (D) The overall map for the dimer. (E) The final map of one protomer of the CSW complex. The components of the CSW complex are indicated in different colors. Light blue, C9ORF72; light green, SMCR8; orange, WDR41.
Both C9ORF72 and SMCR8 are predicted to be members of the DENN (differentially expressed in normal and neoplastic cells) family, although they share low sequence similarity (Fig. 1A) (24–26). DENN domain-containing proteins are well known for functioning as guanine nucleotide exchange factors (GEFs) for many Rab GTPases (27–30). Hence, C9ORF72 and SMCR8 may participate in regulating membrane trafficking by mediating the activity of Rab GTPases (28). Consistent with this hypothesis, studies showed that the CSW complex but not C9ORF72 alone has a significant ability to stimulate the exchange of GDP for GTP of Rab8a and Rab39b in vitro (14, 17). Besides, C9ORF72 has also been suggested to interact weakly with Rab11a (31). However, the physiological targets of the CSW complex need further analysis. Additionally, C9ORF72 and SMCR8 have been predicted to be similar to FLCN and FNIP. Before being shown to function as a GTPase activating protein (GAP) of RAG on lysosomes, the FLCN–FNIP complex was indicated to be a Rab35 GEF in vitro (32–34). Therefore, understanding the structure and biochemical properties of the CSW complex may shed light on the functions of this neurodegenerative disease-related complex.
In this work, we determined the cryogenic electron microscopy (cryo-EM) structure of the CSW complex, which revealed that the complex is composed of two copies of the three proteins, consistent with the biophysical analysis results. The C-terminal tail of C9ORF72 mediates the dimerization of two protomers of the CSW complex by binding to the DENN domain of SMCR8 in the other protomer, this observation was corroborated by biochemical analysis. In the protomer, C9ORF72 and WDR41 are held together by SMCR8 without direct contact with each other. WDR41 binds to the DENN domain of SMCR8 by the C-terminal helix. The overall structure of C9ORF72–SMCR8 resembles that of the FLNC–FNIP2 complex. Biochemical analysis revealed that C9ORF72–SMCR8 has GAP activity for Rab8a and Rab11a and that Arg147 of SMCR8 is critical to the GAP function. Together, our results reveal the assembly basis of the CSW complex and sheds light on the mechanism of its Rab GAP activity.
Results
The CSW Complex Is a Dimer of Trimer.
The CSW complex was previously reported to form a stable complex at a stoichiometry of 1:1:1 (17). After testing different expression systems, the well-behaved CSW complex was expressed and purified using Sf9 cells (Fig. 1B and SI Appendix, Fig. S1). Curiously, the elution volume of the CSW complex in gel filtration corresponded to a molecular mass of ∼400 kDa, a mass larger than that of the CSW complex with a 1:1:1 stoichiometry of the three subunits (Fig. 1B and SI Appendix, Fig. S1). To determine the accurate molecular mass of the CSW complex, an analytical ultracentrifugation (AUC) experiment was carried out. Interestingly, the measured sedimentation coefficient of the CSW complex was ∼10 S, which corresponded to a molecular mass of ∼405 kDa (Fig. 1C). The theoretical molecular masses of C9ORF72, SMCR8, and WDR41 are ∼54 kDa, ∼105 kDa, and ∼48.5 kDa, respectively. The intensity of Coomassie blue suggested that the molar ratio of the three subunits was 1:1:1 (Fig. 1B and SI Appendix, Fig. S1). Collectively, these observations suggested that the CSW complex might contain two copies of each of the three subunits to yield a total molecular mass of 415 kDa.
We carried out single-particle cryo-EM analysis of the CSW complex (SI Appendix, Figs. S2 and S3 and Table S1) and solved the cryo-EM density map at a resolution of 3.2 Å (Fig. 1 D and E). The resolution of the core domain ranges from 3.4 Å to 3.2 Å, which clearly shows the side chains at the core region (SI Appendix, Fig. S3), and provides us with the details of the CSW complex at atomic resolution. The map revealed that the CSW complex is a dimer of C9ORF72–SMCR8–WDR41, which is consistent with the data obtained from analytical gel filtration and AUC analysis (Fig. 1 B–D). The model of the CSW complex was built by combining homology modeling and de novo model building, which allowed us to visualize the structure of the CSW complex in details.
Both the C-Terminal Region of C9OFR72 and the DENN Domain of SMCR8 Are Required for the Dimerization of the CSW Protomers.
The CSW complex is a homodimer of C9ORF72–SWCR8–WDR41 with a width of ∼130 Å and a height of 150 Å (Fig. 2A and SI Appendix, Fig. S4). The two protomers present an imperfect twofold symmetry and basically agree with each other after 180° rotation (Fig. 2A). The observed buried area between the two protomers is ∼1,544 Å2.
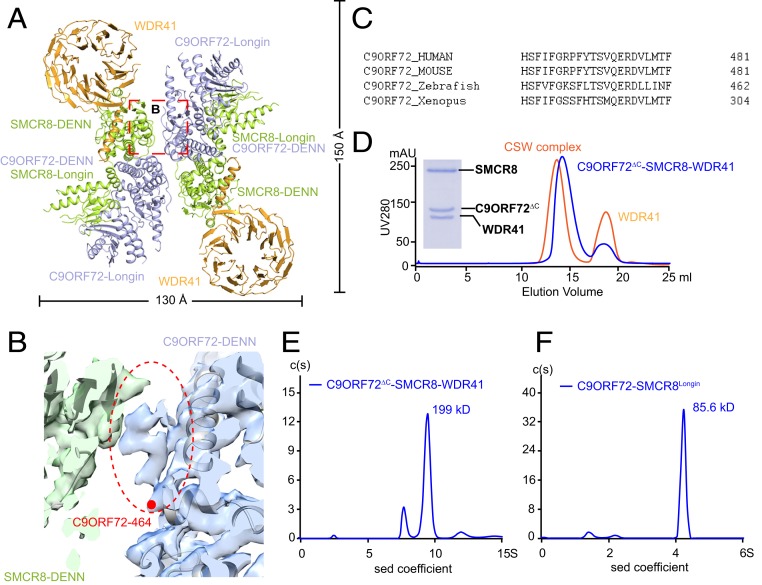
Fig. 2.
The dimer interface of the two protomers in the CSW complex. (A) The overall structure of the CSW complex is shown in cartoon. The CSW complex shows a twofold symmetry. The domains are labeled. The width and height are denoted. C9ORF72, SMCR8, and WDR41 are colored in light blue, light green, and orange, respectively. (B) The density map of the interface between C9ORF72CTR and SMCR8DENN. The unoccupied ambiguous map is highlighted with an oval circle. The last residue presented in the model was denoted with a red dot and labeled. (C) The sequence alignment of C9ORF72CTR in different species. Human, Homo sapiens; mouse, Mus musculus; Xenopus, Xenopus tropicalis; zebrafish, Danio rerio. (D) Gel filtration (superpose 6 10/300 GL) profile of reconstituted C9ORF72∆C–SMCR8–WDR41 complex. C9ORF72∆C represents the construct of C9ORF72 with 461–481 deleted. The UV280 absorption curve of C9ORF72∆C–SMCR8–WDR41 complex is shown in blue, whereas WT C9ORF72–SMCR8–WDR41 complex is shown in orange as comparison. The peak fraction of C9ORF72∆C–SMCR8–WDR41 complex is examined by Coomassie blue-stained SDS/PAGE. (E and F) Analysis of by sedimentation velocity AUC of C9ORF72∆C–SMCR8–WDR41 (E) and C9ORF72–SMCR8Longin (F). C(S) functions calculated from sedimentation velocity data are shown in a blue curve. The calculated molecular mass is denoted. Horizontal axis, sedimentation coefficient; vertical axis, continuous sedimentation coefficient distribution.
The structure model reveals that the dimerization of the two CSW protomers is mainly mediated by the C-terminal region of C9ORF72 and the DENN domain of SMCR8 (Fig. 2A and SI Appendix, Fig. S4A). The density of the C9ORF72 C-terminal region (461–481, hereafter C9ORF72CTR) is ambiguous in both protomers (Fig. 2B). Moreover, C9ORF72CTR is highly conserved cross species (Fig. 2C). To assess the contribution of C9ORF72CTR to dimerization, a mutant with C9ORF72CTR deleted (C9ORF72∆C) was constructed. As expected, the C9ORF72∆C–SMCR8 complex is capable of binding to WDR41 as tightly as the C9ORF72–SMCR8 complex (Fig. 2D and SI Appendix, Fig. S1). However, the elution volume of the C9ORF72∆C–SMCR8–WDR41 complex in gel filtration was delayed by ∼0.9 mL relative to that of the CSW complex, which corresponded to a molecular mass of ∼200 kDa (Fig. 2D and SI Appendix, Fig. S1). Consistent with this observation, the AUC experiment showed that the sedimentation coefficient of the C9ORF72∆C–SMCR8–WDR41 complex is 9.87 S, corresponding to a molecular mass of ∼199 kDa (Fig. 2E). Together, these observations indicate that the C9ORF72∆C–SMCR8–WDR41 complex is monomeric in solution and that C9ORF72CTR is critical to the dimerization of two CSW protomers.
The structure of CSW does not show where the C9ORF72CTR packs on the DENN domain of SMCR8 (700–937, SMCR8DENN); therefore, the assembly status of the complex of C9ORF72 and SMCR8 Longin domain (1–349, SMCR8Longin) was evaluated. AUC analysis showed that the sedimentation coefficient of C9ORF72–SMCR8Longin was ∼4.35 S (Fig. 2F), which corresponds to a molecular mass of ∼85.6 kDa and suggests C9ORF72–SMCR8Longin is monomeric in solution. Gel filtration analysis indicated that C9ORF72–SMCR8Longin is a monomer is solution as well (SI Appendix, Fig. S5).
Collectively, the data support that both the C-terminal region of C9OFR72 and the DENN domain of SMCR8 together mediate the dimerization of the CSW protomers together.
Organization of the CSW Protomer.
The C9ORF72–SMCR8–WDR41 complex in one protomer adopts an elongated rod shape, in which the C9ORF72–SMCR8 complex resembles the FLCN–FNIP2 complex (Fig. 3A) (35, 36). Structural comparison shows that C9ORF72 corresponds to FNIP2, whereas SMCR8 resembles FLCN (Fig. 3B and SI Appendix, Fig. S6A), which is consistent with the bioinformatic analysis (24, 37).
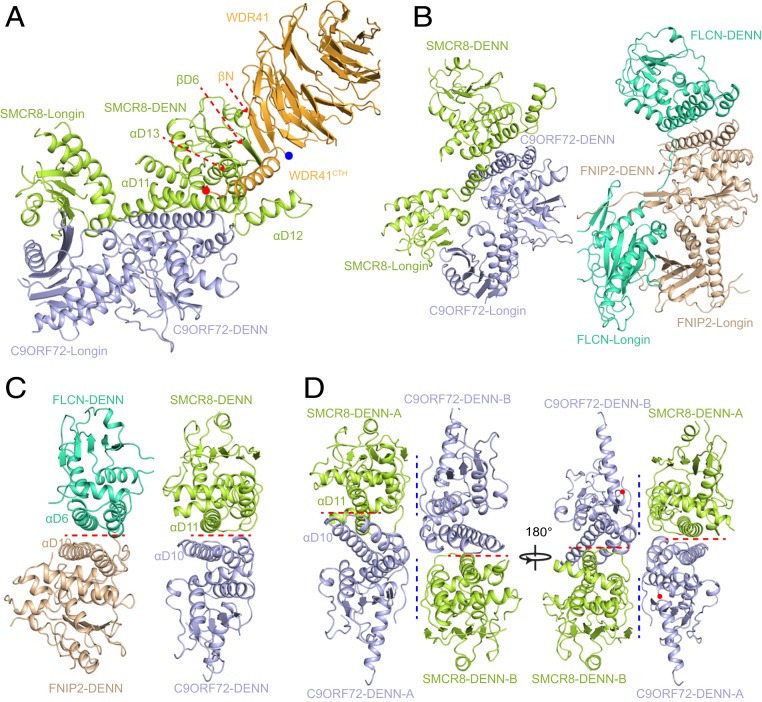
Fig. 3.
Organization of the CSW protomer. (A) The overall structure of the CSW protomer. The key secondary structures are labeled. The colors are consistent with the previous figures. (B) The structure of the C9ORF72–SMCR8 complex adopts a similar organization to the FLCN–FNIP2 complex. (B, Left) C9ORF72–SMCR8 complex. (B, Right) FLCN–FNIP2 complex. C9ORF72 and SMCR8 are colored as in A. FLCN and FNIP2 are colored in green-cyan and wheat, respectively. The details of the comparison are presented in SI Appendix, Fig. S6. (C and D) Two interfaces between C9ORF72 DENN and SMCR8DENN are shown. (C) Comparison of DENN pair of C9ORF72–SMCR8 and FLCN-FNIP. The key helixes are labeled. The red dash line indicates the position of the interface. (D) The two interfaces between C9ORF72 DENN and SMCR8DENN are shown. The red dash line indicates the positions of the intraprotomer interface, while the blue dash line indicates the positions of the interprotomer interface.
Superimposition of the C9ORF72–SMCR8 complex and FLCN–FLIP2 dimer revealed that the position of the Longin domain shifts by ∼40 Å when the DENN domains are superimposed well (SI Appendix, Fig. S6A). Although the C9ORF72–SMCR8 complex and FLCN–FNIP2 dimer show a slight difference in overall conformation, the Longin and DENN dimer of the C9ORF72–SMCR8 complex can be superimposed well on their counterparts in FLCN–FNIP2 (Fig. 3B and SI Appendix, Fig. S6 B and C). Intriguingly, SMCR8Longin and SMCR8DENN are connected by αL5, αL6, and the following loop, whereas the Longin domain and DENN domain of FLCN are connected by a flexible loop (SI Appendix, Fig. S4B). This observation might explain the structural discrepancy between the two complexes.
The interaction of C9ORF72 and SMCR8 in one protomer is mediated by the Longin domains and DENN domains, respectively, which is consistent with the observations in the FLCN–FNIP2 structure (Fig. 3 B and C and SI Appendix, Fig. S6 A–C) (35, 36). Importantly, C9ORF72CTR and SMCR8DENN also mediate the dimerization of the CSW protomers (Fig. 2 A and B). Thus, there are two interfaces between the DENN domains of C9ORF72 and SMCR8: the interface consisting of αD10 of C9ORF72 and αD11 of SMCR8, which mediates the intraprotomer interaction, and the interface between C9ORF72CTR and SMCR8DENN mediates the dimerization of the CSW protomer (Fig. 3D). This feature distinguished the DENN domains of the C9ORF72–SMCR8 protomer from other members of the DENN domain-containing proteins.
The structure of WDR41 reveals an eight-blade β-propeller with the N-terminal first strand and C-terminal last three strands coming together to form the first propeller, generating a “Velcro” closure that stabilizes the architecture of WDR41 (Fig. 3A and SI Appendix, Fig. S4D) (22). WDR41 binds to the SMCR8 DENN domain via its N-terminal β-strand (βN) and C-terminal helix (CTH) without direct physical contact with C9ORF72 (Fig. 3A and SI Appendix, Fig. S4 A and D).
The Interface of SMCR8–WDR41.
As mentioned above, WDR41 binds to SMCR8 mainly via its C-terminal helix (WDR41CTH) and the very BN (Fig. 3A). WDR41CTH packs against the groove formed by αD11, αD12, and αD13 of SMCR8, whereas the BN of WDR41 forms an antiparallel β-sheet with βD6 of SMCR8 (Figs. 3A and 4A).
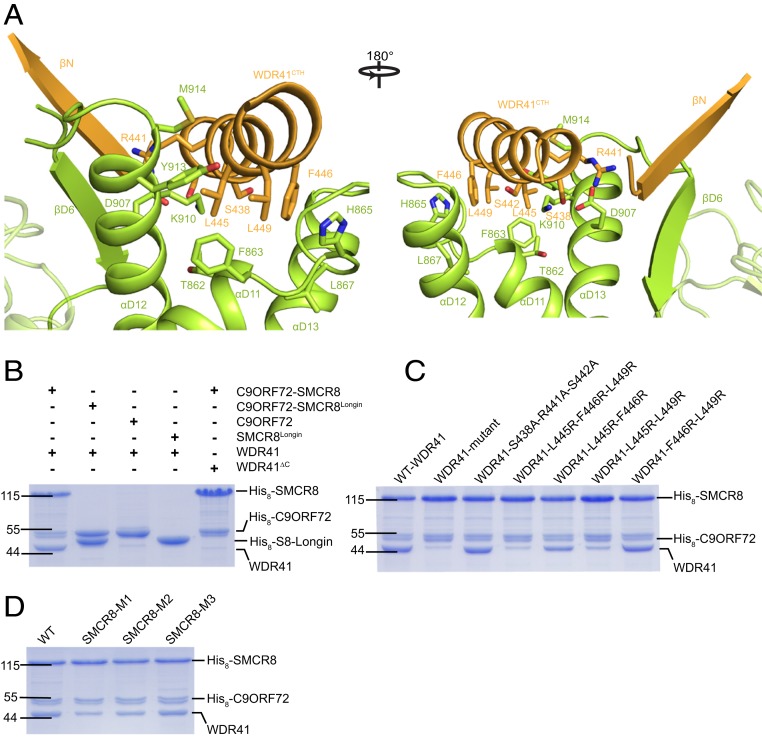
Fig. 4.
The interface of SMCR8 and WDR41. (A) The details of the interface of SMCR8 and WDR41. The key residues on the interface are shown in stick model. Secondary structures are shown as cartoon. (A, Left) Front view. (A, Right) Back view. (B) C9ORF72, SMCR8Longin, C9ORF72–SMCR8Longin complex, and C9ORF72–SMCR8 complex were used to pulldown WDR41 and WDR41∆C. (C and D) Mutational analysis of key residues on WDR41CTH (C) and SMCR8 (D) using pulldown assay. The results are visualized by Coomassie blue-stained SDS/PAGE. Protein markers are labeled at left (unit: kilodaltons). (C) WT C9ORF72–SMCR8 complex with N-terminal His-tag was used to pull down untagged WT or indicated mutants of WDR41. WDR41-mutant: WDR41with mutations of S438A-R441A-S442A-L445R-F446R-L449R. Other mutants are shown as the label. (D) Indicated mutants of SMCR8 with N-terminal His-tag were used to pull down untagged WT WDR41. SMCR8M1, C9ORF72–SMCR8M1 (SMCR8 with mutations of T862A, F863A, H865A, L867A, E907A, K910A, Y913A, M914A); SMCR8M2, C9ORF72–SMCR8M2 (SMCR8 with mutations of T862A, F863A, H865A, L867A); SMCR8M3, C9ORF72–SMCR8M3 (SMCR8 with mutations of E907A, K910A, Y913A, M914A); WT, WT C9ORF72–SMCR8.
To verify this observation, a mutant of WDR41 with a C-terminal helix deleted (436–459, WDR41∆C) and SMCR8Longin were purified successfully and subjected to the pulldown assay. In the pulldown assay, C9ORF72–SMCR8 was able to interact with WDR41 but not WDR41∆C (Fig. 4B and SI Appendix, Figs. S7A and S8). In contrast, C9ORF72 was not capable of binding to WDR41 or WDR41∆C (Fig. 4B and SI Appendix, Fig. S7A). Moreover, SMCR8Longin was not able to bind to either WDR41 or WDR41∆C (Fig. 4B and SI Appendix, Fig. S7A). Collectively, these observations support that WDR41 binds to the DENN domain of SMCR8 via its C-terminal helix.
The density map of the interface was clear enough to carry out a detailed alanine mutagenesis analysis. The residues on WDR41CTH facing SMCR8, including S438, R441, S442, L445, F446, and L449, were individually mutated to either alanine or arginine (SI Appendix, Fig. S7B). However, none of the mutated residues disrupted the interaction between WDR41 and the C9ORF72–SMCR8 complex (SI Appendix, Fig. S7B). Therefore, combinations of these mutations were examined. The mutant (WDR41-mutant) with all six key residues mutated abolished the interaction of WDR41 and SMCR8 (Fig. 4C and SI Appendix, Fig. S7C). Interestingly, WDR41L445R-F446R-L449R blocked binding to the C9ORF72–SMCR8 complex, whereas WDR41S438A-R441A-S442A had little effect on binding (Fig. 4C and SI Appendix, Fig. S7C). The double mutant WDR41L445R-L449R had obvious effects on binding, whereas WDR41L445R-F446R and WDR41F446R-L449R had weaker phenotypic effects than WDR41L445R-F446R-L449R (Fig. 4C and SI Appendix, Fig. S7C).
On the SMCR8 side, all residues (T862-F863-H865-L867-E907A-K910-Y913-M914) involved in binding to L445, F446, and L449 of WDR41 were mutated to alanine, and the resulting construct was termed SMCR8M1. The C9ORF72–SMCR8M1 complex was purified and subjected to a pulldown assay. Compared with WT C9ORF72–SMCR8, C9ORF72–SMCR8M1 reduced the binding ability to WDR41 by ∼50% (Fig. 4D and SI Appendix, Fig. S7D). Two other mutants, SMCR8M2 (T862A-F863A-H865A-L867A) and SMCR8M3 (E907A-K910A-Y913A-M914A), were also tested and showed little effect on the binding to WDR41 (Fig. 4D and SI Appendix, Fig. S7D). The alanine mutational analysis was probably unable to completely destroy the hydrophobic cavity. Hence, we attempted to mutate these residues to arginine, however, a single mutation of any residues to arginine caused C9ORF72–SMCR8 to precipitate. We also tested the effect of βD6 of SMCR8 on the binding to WDR41 by deleting the C-terminal region of SMCR8 (923-937, SMCR8∆C). The pulldown assay showed that SMCR8∆C had little effect on the interaction of C9ORF72–SMCR8 and WDR41 (SI Appendix, Fig. S7E), indicating that the antiparallel β-sheet formed between βD6 of SMCR8 and βN of WDR41 is not essential to the binding of SMCR8 and WDR41.
Collectively, these observations demonstrate that the hydrophobic interaction of the C-terminal helix of WDR41 and the DENN domain of SMCR8 is critical to the binding of WDR41 to SMCR8.
The C9ORF72–SMCR8 Complex Is a GAP for Rab8a and Rab11a In Vitro.
The CSW complex has been reported to function as a GEF of Rab7a, Rab8a, Rab11a, Rab39a, and Rab39b (14, 17, 31). Hence, a bioluminescence-based GTPase activity assay was carried out to assess whether the CSW complex stimulates these Rabs (38). In this type of assay, unhydrolyzed GTP can be transformed into a bioluminescence signal. A higher bioluminescence signal indicates less GTP hydrolyzation by Rabs. Stimulated by either the CSW complex or the C9ORF72–SMCR8 complex but not C9ORF72 alone, the amount of GTP consumed by Rab8a and Rab11a increased by ∼100% and 50%, respectively (Fig. 5A). However, C9ORF72, the CSW complex, and the C9ORF72–SMCR8 complex had little effect on Rab7a, Rab39a, and Rab39b (Fig. 5A). Interestingly, the CSW complex and C9ORF72–SMCR8 complex showed similar effects on Rab8a and Rab11a, indicating that WDR41 is not essential for the stimulation of target Rabs (Fig. 5A). Moreover, Rab8a and Rab11a play critical roles in coordinating primary ciliogenesis and axon growth (39, 40). Therefore, we focused on the relationships between Rab8a/11a and the CSW complex and the C9ORF72–SMCR8 complexes in vitro.
Fig. 5.
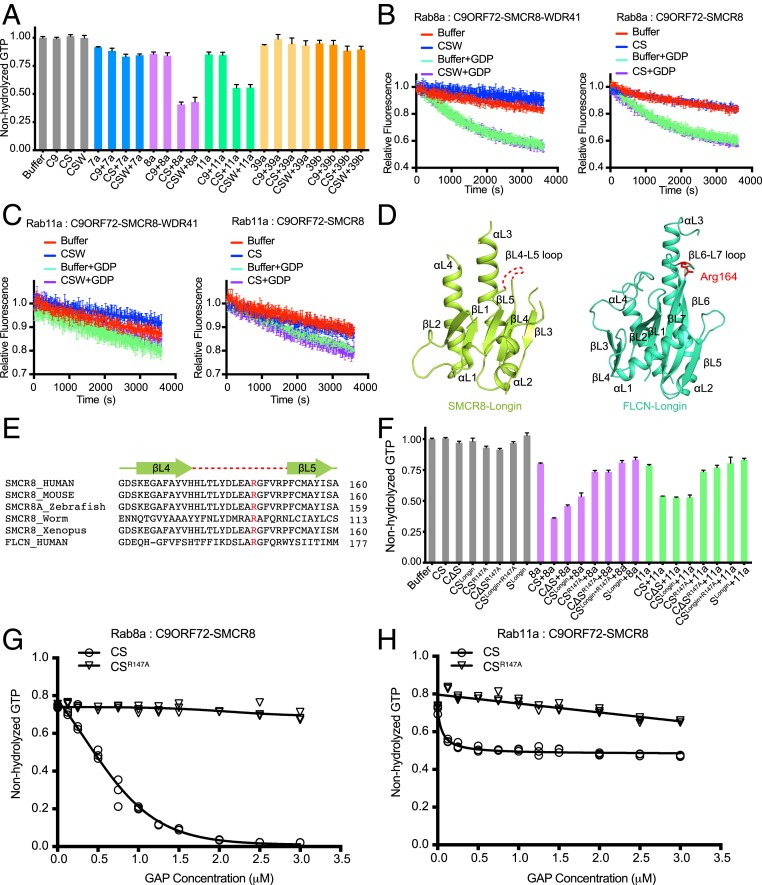
CSW and C9ORF72–SMCR8 complex are GAPs of Rab8a and Rab11a in vitro. (A) The screen of Rabs that can be stimulated by CSW or C9ORF72–SMCR8 complex using bioluminescence-based GTPase activity assay. In this assay, the final concentration of Rabs and C9/CS/CSW was 1.5 µM and 0.75 µM, respectively. The GTP in the reaction system containing no protein was normalized to “1.0.” The proteins or protein mixtures added in the reaction system are indicated below. Buffer, buffer control; C9, C9ORF72; CS, C9ORF72–SMCR8; CSW, WT C9ORF72–SMCR8–WDR41; 7a, Rab7a; 8a, Rab8a; 11a, Rab11a; 39a, Rab39a; 39b, Rab39b. The error bars represent mean ± SD (n = 3). (B and C) MANT-GDP–based nucleotide exchange assay for Rab8a (B) and Rab11a (C). The abbreviations of protein names are the same as A. One hundred micomolar GDP was used to initiate the exchange reaction. Reactions without GDP were monitored as a control. The error bars represent mean ± SD (n = 3). The intrinsic nucleotide exchange rate of Rab11a is negligible. (D) Comparison of the Longin domains of SMCR8 and FLCN. The secondary structures are labeled. The Arg164 of FLCN-Longin domain is highlighted in red. The missing loop of SMCR8Longin is indicated as a red dash line. (E) Sequence alignment of the βL4-βL5 loop of SMCR8. Arg147 of human SMCR8 is conserved across species and corresponds to Arg164 of FLCN. Human, Homo sapiens; mouse, Mus musculus; worm, Caenorhabditis elegans; Xenopus, Xenopus tropicalis zebrafish, Danio rerio. The secondary structures are shown on top of the sequence in light-green, and the βL4-βL5 is shown in red dash line. (F) Test the effect of Arg147 of SMCR8 on the stimulation of Rab8a and Rab11a. The experiment was carried out as in A, and the labels are the same as in A. C∆S, C9ORF72∆C-SMCR8; CSLongin, C9ORF72–SMCR8Longin; CSR147A, C9ORF72–SMCR8R147A; C∆SR147A, C9ORF72∆C-SMCR8R147A; CSLongin+R147A, C9ORF72–SMCR8Longin+R147A; SLongin, SMCR8Longin. The error bars represent mean ± SD (n = 3). (G and H) Measurement of GAP activity to Rab8a (G) and Rab11a (H) using different concentrations of C9ORF72–SMCR8 or C9ORF72–SMCR8Arg147. The final concentration of Rabs in this assay was 2 µM. The concentrations of C9ORF72–SMCR8 or C9ORF72–SMCR8Arg147 are indicated in the horizontal axis. The relative amount of nonhydrolyzed GTP in the system is shown in the vertical axis. Each concentration of GAP was measured in triplicate. The value of each measurement is shown in as “○” (C9ORF72–SMCR8) and “▽” (C9ORF72–SMCR8Arg147) since the error bars of several measurements are too small to be shown clearly in the figure. The curve was fitted to using the Stimulation Model in Graphpad.
Since the CSW complex was previously identified as Rab GEF, we investigated whether the CSW complex and C9ORF72–SMCR8 complex promote the nucleotide exchange rate of Rab8a and Rab11a. Hence, a fluorescence-based GEF activity assay was performed using N-methylanthraniloyl (MANT)-GDP (41, 42). Compared with buffer, the CSW complex and the C9ORF72–SMCR8 complex were unable to accelerate MANT-GDP release from Rab8a and Rab11a (Fig. 5 B and C), suggesting that neither complex functions as a GEF for Rab8a and Rab11a.
As previously mentioned, the C9ORF72–SMCR8 complex shares a similar overall structure with the FLCN–FNIP2 heterodimer, the GAP of RagC/D(33-36). Arg164 on the loop of β6-β7 of FLCN is critical to the GAP activity of the FLCN–FNIP2 complex (35, 36). Although the density of the β6-β7 loop in the C9ORF72–SMCR8 complex is not clear, sequence alignment shows that Arg147 of SMRC8 is conserved across species and corresponds to Arg164 of FLCN (Fig. 5 D and E). Hence, we investigated whether the C9ORF72–SMCR8 complex has GAP activity for Rab8a and Rab11a. The effect of Arg147 of SMCR8 was probed by the mutation R147A using a bioluminescence assay.
Excitingly, compared with WT C9ORF72–SMCR8, C9ORF72–SMCR8R147A showed no obvious stimulatory effect on Rab8a and Rab11a (Fig. 5F). Intriguingly, C9ORF72∆C–SMCR8 showed a stimulatory effect on Rab8a and Rab11a similar to that of WT C9ORF72–SMCR8, suggesting that the dimerization of CSW protomers is not essential for the stimulatory effect on Rab8a and Rab11a (Fig. 5F). Moreover, C9ORF72∆C–SMCR8R147A eliminated the stimulatory effect on Rab8a and Rab11a (Fig. 5F).
C9ORF72 showed no stimulatory activity against Rab8a and Rab11a. Hence, we attempted to test whether SMCR8 alone had GAP activity against Rab8a and Rab11a. However, SMCR8 cannot be purified alone. Fortunately, we were able to successfully purify SMCR8Longin and C9ORF72–SMCR8Longin. Interestingly, SMCR8Longin showed no stimulatory effects on Rab8a and Rab11a (Fig. 5F), whereas C9ORF72–SMCR8Longin showed an effect on Rab8a and Rab11a similar to that of C9ORF72–SMCR8 (Fig. 5F). As expected, C9ORF72–SMCR8Longin-R147A showed no GAP stimulatory effects on Rab8a and Rab11a (Fig. 5F).
Together, the data indicate that CSW and the C9ORF72–SMCR8 complex but not C9ORF72 or SMCR8 alone can exhibit GAP activity for Rab8a and Rab11a in vitro. Additionally, the data support that the dimerization of C9ORF72–SMCR8 is not required for the complex’s GAP activity.
To further assess the responses of Rab8a and Rab11a to stimulation by the C9ORF72–SMCR8 complex, a titration experiment was performed at a fixed concentration of Rab. With increasing concentration of the WT C9ORF72–SMCR8 complex, Rab8a hydrolyzed more GTP and almost exhausted the GTP when the molar ratio of Rab8a and C9ORF72–SMCR8 complex reached 1:1. In contrast, the C9ORF72–SMCR8R147A complex showed no obvious stimulatory effect on Rab8a (Fig. 5G).
Similarly, Rab11a consumed more GTP with increasing concentration of the C9ORF72–SMCR8 complex but not that of the C9ORF72–SMCR8R147A complex (Fig. 5H). However, the limit of GTP hydrolyzed by stimulated Rab11a was ∼50% of the total amount of GTP (Fig. 5H). This phenomenon might have been caused by the slow intrinsic nucleotide exchange rate of Rab11a (Fig. 5C), which might have prevented Rab11a from efficiently accessing GTP when the ratio between GTP:GDP in the system was 1:1 (43).
In a word, C9ORF72–SMCR8 showed GAP activity against Rab8a and Rab11a in vitro, which was contrary to our expectation. Furthermore, Arg147 of SMCR8 is critical to the GAP activity of C9ORF72–SMCR8.
Discussion
The CSW complex is critical to a variety of cellular processes and is strongly associated with familial ALS and FTD. In addition, the CSW complex is highly conserved in animals (SI Appendix, Figs. S9–S11). Nevertheless, the exact functions of the CSW complex remain unclear. Here, we investigated this important complex using recombinant proteins, biochemical assays, and single-particle cryo-EM analysis.
First, we determined that the stoichiometry of C9ORF72, SMCR8, and WDR41 in the CSW complex is 2:2:2. Both biochemical analysis and the structure of the CSW complex revealed that the CSW complex is a dimer of C9ORF72–SMCR8–WDR41. Although the density map of the dimerization interface is unclear, mutagenesis analysis, AUC analysis, and analytical gel filtration indicated that C9ORF72CTR and SMCR8DENN are necessary for the dimerization of C9ORF72–SMCR8–WDR41. Notably, dimerization of C9ORF72–SMCR8–WDR41 or C9ORF72–SMCR8 is not essential to the complex’s GAP activity (Fig. 5F). To obtain detailed information on the dimer interface of C9ORF72–SMCR8–WDR41, a higher-resolution structure is needed, and determining the biological significance of dimerization requires in vivo genetic and functional analyses.
Additionally, we clarified the relationship among C9ORF72, SMCR8, and WDR41 based on the structure of CSW. WDR41 binds to a groove on SMCR8DENN without direct contact with C9ORF72 (Fig. 4). WDR41 and C9ORF72 are joined together by SMCR8. A triad of L445, F446, and L449 from WDR41CTH is critical to the binding of WDR41 to SMCR8. Mutants containing L445R, F446R, and L449R do not have the ability to bind to SMCR8. Eight residues of SMCR8DENN involved in the binding to WDR41CTH were assessed by alanine mutagenesis analysis since arginine mutagenesis crashed SMCR8. All eight residues have to be mutated into alanine simultaneously to cripple the binding of SMCR8 to WDR41. Interestingly, WDR41 is not essential to the GAP activity of the CSW complex. Previous studies showed that WDR41 locates on ER and participates in the autophagosome-lysosome pathway (15, 16, 23). Thus, the role of WDR41 in the CSW complex might be localizing the complex on appropriate positions.
We found that the DENN domain of C9ORF72 can interact with two DENN domains of SMCR8 simultaneously and vice versa (Fig. 3D). Not only does such a binding mode enable the DENN dimer of C9ORF72–SMCR8 to mediate intraprotomer interaction as well as the interprotomer interaction, it distinguishes these DENN domains from others, including the DENN domains of FLCN and FNIP2. These observations enhance our understanding of the functional evolution of the DENN domain.
Most importantly, the GAP activity of C9ORF72–SMCR8 was identified based on the structure of C9ORF72–SMCR8 and a GTPase activity assay. The structure of C9ORF72–SMCR8 resembles that of FLCN–FNIP2, which is a GAP of RagC/D. The Longin dimers in both structures adopt similar organization patterns (SI Appendix, Fig. S4C). Intriguingly, Lst7, the homolog of FLCN in yeast, contains only the Longin domain (44, 45). Moreover, the catalytic arginine of FLCN–FNIP2 is located in the Longin domain of FLCN (35, 36). Together, these data indicate that the Longin dimer might be functionally conserved during evolution. Based on the structure and sequence alignment, Arg147 of SMCR8 was found to correspond to Arg164. However, previous studies indicate that the CSW complex might function as a GEF of many Rabs. Hence, we screened Rabs that can be stimulated by CSW or the C9ORF72–SMCR8 complex in vitro using reconstituted proteins. Serendipitously, CSW and C9ORF72–SMCR8 were found able to stimulate Rab8a and Rab11a at similar levels, which indicated that WDR41 is not essential for the stimulating effect of CSW on Rab8a and Rab11a. Curiously, neither CSW nor C9ORF72–SMCR8 accelerated the nucleotide exchange rate of Rab8a and Rab11a in vitro, indicating that CSW and C9ORF72–SMCR8 are not GEFs for Rab8a and Rab11a. Mutagenesis analysis combined with GAP-stimulated GTPase activity assay confirmed that Arg147 of SMCR8 is essential to its GAP activity for Rab8a and Rab11a in vitro. In addition, both C9ORF72 and SMCR8 are required for GAP activity, as evidenced by the finding that neither C9ORF72 nor SMCR8Longin shows GAP activity toward Rab8a and Rab11a. It is of interest to dissect how C9ORF72 and SMCR8 recognize and collaboratively activate Rab8a/11a. Unfortunately, we have not yet been able to obtain a stable complex of C9ORF72–SMCR8–Rab8a/11a, indicating that the interaction of C9ORF72–SMCR8 and Rab8a/11a might be weak or ephemeral.
Our results demonstrate that CSW is a dimer of C9ORF72–SMCR8–WDR41 and is able to stimulate the GTPase activity of Rab8a and Rab11a as a GAP. Rab8a and Rab11a function coordinately in many fundamental cellular processes, including receptor recycling, ciliogenesis, and axon growth (39, 40, 46). It is tempting to propose the following model: CSW takes two endosomes containing Rab8a/11a via WDR41 simultaneously and then activates the Rabs located on the surface to prompt the fusion of the two endosomes.
Although the GAP activity of the C9ORF72–SMCR8 complex has been tested in vitro, the specific biological processes in which CSW and the C9ORF72–SMCR8 complex participate as a GAP require further exploration. The findings and the biochemical data presented in our work will definitely facilitate future functional studies of CSW and the C9ORF72–SMCR8 complex.
Materials and Methods
Protein Purification.
Protein were expressed in Escherichia coli BL21 (DE3) cells or Sf9 cells and purified using fast protein liquid chromatography (FPLC).
Analytical Ultracentrifugation.
Analytical ultracentrifugation sedimentation experiments were carried out at 20 °C in an XL-I analytical ultracentrifuge (Beckman Coulter) equipped with Rayleigh Interference detection (655 nm).
Structure Determination.
The structure of CSW complex were determined using single-particle cryo-EM.
GTPase Activity Assay.
GTPase activity assays were carried out using GTPase-Glo assay kit (Promega, V7681).
Nucleotide Exchange Assay.
Releasing of MANT-GDP was recorded by monitoring the decrease in fluorescence emission at 448 nm (excited at 360 nm) in intervals of 15 s at 25 °C for 3,600 s. Data were collected using 384-well plate (Corning, 3701) in BioTek Synergy2.
Data Availability.
Coordinates and cryo-EM map of the structure reported here have been deposited into the Protein Data Bank (PDB) with PDB ID code 6LT0, and the Electron Microscopy Data Bank with accession no. EMD-0966. Details of methods are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the cryo-EM facility support; the computational facility support on the cluster of Bio-Computing Platform (Tsinghua University Branch of China National Center for Protein Sciences Beijing); Jianhua He, Bo Sun, Wenming Qin, Huan Zhou, and Feng Yu from the beamline stations of BL17U, BL18U, and BL19U at National Facility for Protein Science in Shanghai, Zhangjiang Laboratory, China for providing technical support and assistance in crystal testing; and staff from Protein Preparation and Identification Facility at Technology Center for Protein Science, Tsinghua University, for the assistance with AUC data collection. This work was supported by National Key R&D Program of China Grants 2017YFA0506300 (to S.Q.) and 2018YFC1004601 (to S.Q.) and National Natural Science Foundation of China Grants 81671388 (to S.Q.) and 31770820 (to K.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6LT0). The cryo-EM map has been deposited in the Electron Microscopy Data Bank (accession no. EMD-0966).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002110117/-/DCSupplemental.
References
- 1.Dion P. A., Daoud H., Rouleau G. A., Genetics of motor neuron disorders: New insights into pathogenic mechanisms. Nat. Rev. Genet. 10, 769–782 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Rowland L. P., Shneider N. A., Amyotrophic lateral sclerosis. N. Engl. J. Med. 344, 1688–1700 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Balendra R., Isaacs A. M., C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling S. C., Polymenidou M., Cleveland D. W., Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M., et al. , Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renton A. E., et al. ; ITALSGEN Consortium , A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majounie E., et al. ; Chromosome 9-ALS/FTD Consortium ; French research network on FTLD/FTLD/ALS ; ITALSGEN Consortium , Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: A cross-sectional study. Lancet Neurol. 11, 323–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amick J., Ferguson S. M., C9orf72: At the intersection of lysosome cell biology and neurodegenerative disease. Traffic 18, 267–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen X., et al. , Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon I., et al. , Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conlon E. G., et al. , The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife 5, e17820 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haeusler A. R., et al. , C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., et al. , Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 24, 313–325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M., et al. , A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci. Adv. 2, e1601167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amick J., Roczniak-Ferguson A., Ferguson S. M., C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol. Biol. Cell 27, 3040–3051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan P. M., et al. , The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol. Commun. 4, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sellier C., et al. , Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 35, 1276–1297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ugolino J., et al. , Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 12, e1006443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slager R. E., Newton T. L., Vlangos C. N., Finucane B., Elsea S. H., Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet. 33, 466–468 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., et al. , The C9orf72-interacting protein Smcr8 is a negative regulator of autoimmunity and lysosomal exocytosis. Genes Dev. 32, 929–943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung J., et al. , Multiplex image-based autophagy RNAi screening identifies SMCR8 as ULK1 kinase activity and gene expression regulator. eLife 6, e23063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C., Min J., Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ge L., Melville D., Zhang M., Schekman R., The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2, e00947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D., Iyer L. M., He F., Aravind L., Discovery of novel DENN proteins: Implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3, 283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine T. P., Daniels R. D., Gatta A. T., Wong L. H., Hayes M. J., The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29, 499–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levivier E., et al. , uDENN, DENN, and dDENN: Indissociable domains in Rab and MAP kinases signaling pathways. Biochem. Biophys. Res. Commun. 287, 688–695 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Chaineau M., Ioannou M. S., McPherson P. S., Rab35: GEFs, GAPs and effectors. Traffic 14, 1109–1117 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Barr F., Lambright D. G., Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X., et al. , Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc. Natl. Acad. Sci. U.S.A. 108, 18672–18677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marat A. L., Dokainish H., McPherson P. S., DENN domain proteins: Regulators of Rab GTPases. J. Biol. Chem. 286, 13791–13800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farg M. A., et al. , C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 23, 3579–3595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nookala R. K., et al. , Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2, 120071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petit C. S., Roczniak-Ferguson A., Ferguson S. M., Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202, 1107–1122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsun Z. Y., et al. , The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52, 495–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen K., et al. , Cryo-EM structure of the human FLCN-FNIP2-Rag-ragulator complex. Cell 179, 1319–1329 e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrence R. E., et al. , Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex. Science 366, 971–977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine T. P., et al. , Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4, 62–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mondal S., Hsiao K., Goueli S. A., A homogenous bioluminescent system for measuring GTPase, GTPase activating protein, and guanine nucleotide exchange factor activities. Assay Drug Dev. Technol. 13, 444–455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knödler A., et al. , Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 6346–6351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furusawa K., et al. , Cdk5 regulation of the GRAB-mediated Rab8-Rab11 cascade in axon outgrowth. J. Neurosci. 37, 790–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiontke S., et al. , Architecture and mechanism of the late endosomal Rab7-like Ypt7 guanine nucleotide exchange factor complex Mon1-Ccz1. Nat. Commun. 8, 14034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Lu A., Chen X., Wei L., Ding J., RABEX-5 is upregulated and plays an oncogenic role in gastric cancer development by activating the VEGF signaling pathway. PLoS One 9, e113891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bos J. L., Rehmann H., Wittinghofer A., GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129, 865–877 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Pacitto A., et al. , Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 5, 150174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Péli-Gulli M. P., Sardu A., Panchaud N., Raucci S., De Virgilio C., Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Rep. 13, 1–7 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Vogel G. F., et al. , Abnormal Rab11-Rab8-vesicles cluster in enterocytes of patients with microvillus inclusion disease. Traffic 18, 453–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and cryo-EM map of the structure reported here have been deposited into the Protein Data Bank (PDB) with PDB ID code 6LT0, and the Electron Microscopy Data Bank with accession no. EMD-0966. Details of methods are included in SI Appendix.