Significance
Increasing legume use in agriculture is a key requirement for the sustainable intensification of global farming systems. Maximizing the yield of legumes requires matching of the plant to elite rhizobia that are both competitive for nodulation and capable of high rates of nitrogen fixation. We have developed a high-throughput method that allows identification of elite strains at the single nodule level with the potential to revolutionize the search for elite indigenous rhizobia.
Keywords: nodulation, legume, rhizobium, competition, nitrogen-fixing
Abstract
Legumes tend to be nodulated by competitive rhizobia that do not maximize nitrogen (N2) fixation, resulting in suboptimal yields. Rhizobial nodulation competitiveness and effectiveness at N2 fixation are independent traits, making their measurement extremely time-consuming with low experimental throughput. To transform the experimental assessment of rhizobial competitiveness and effectiveness, we have used synthetic biology to develop reporter plasmids that allow simultaneous high-throughput measurement of N2 fixation in individual nodules using green fluorescent protein (GFP) and barcode strain identification (Plasmid ID) through next generation sequencing (NGS). In a proof-of-concept experiment using this technology in an agricultural soil, we simultaneously monitored 84 different Rhizobium leguminosarum strains, identifying a supercompetitive and highly effective rhizobial symbiont for peas. We also observed a remarkable frequency of nodule coinfection by rhizobia, with mixed occupancy identified in ∼20% of nodules, containing up to six different strains. Critically, this process can be adapted to multiple Rhizobium-legume symbioses, soil types, and environmental conditions to permit easy identification of optimal rhizobial inoculants for field testing to maximize agricultural yield.
Legumes have evolved the remarkable ability to host N2 fixing bacteria, known as rhizobia, in specialized organs called root nodules. In a symbiotic partnership within the root nodule, legumes supply nutrients to rhizobia that fix N2 gas from the atmosphere into reduced forms that are supplied to the legume (1). These symbiotic associations have been estimated to fix ∼80% of the biologically fixed N2 in agricultural areas (2). Rhizobia have been studied for more than a 100 years because of their ability to increase yields of legumes crops. The first industrial production of rhizobial inoculants began by the end of the 19th century (3). Since then, farmers have applied rhizobial inoculants as an alternative to economically expensive chemical fertilizers and in organic crop systems. The bioavailable nitrogen that is generated in nodules of legumes benefits nonlegume crops grown in rotation or at the same time (3). In some cases, rhizobial inoculants that can significantly improve legume crop growth under controlled conditions fail to compete against native strains with inferior N2 fixing abilities, known as the “rhizobial competition problem” (4). In other cases, native rhizobia perform better at N2 fixation than commercial inoculants (5) or even N-fertilized controls (6), showing the importance of developing inoculants for local crops that take advantage of strains preadapted to thrive in local environments (tailor-made inoculants). A potential breakthrough in the use of inoculants is the recent development of a seed coating based on silk fibronin and trehalose that stabilizes rhizobia in saline and, possibly, arid environments (7). While this is an exciting development, to improve inoculants, the rhizobia coated onto seeds must be both competitive for nodule occupancy and effective in N2 fixation.
Since N2 fixation happens inside nodules, several indirect measurements of rhizobial effectiveness have been developed such as shoot dry weight (DW), acetylene reduction assays (ARA), total nitrogen content, 15N2 fixation, and many others (8). These techniques are not amenable for the analysis of individual nodules and, as a result, when screening for effective rhizobial strains, only one strain per plant is usually tested. In the case of nodule occupancy, different strategies have been used to assess the competitiveness of rhizobial strains. These include chromosomally encoded marker genes (9), naturally occurring or induced antibiotic-resistance genes (10), the use of PCR fingerprints which target specific genes (11), or metagenomic sequencing to monitor strain-level changes (12). Many of these methods are highly time-consuming, since they involve a complex procedure for chromosomal integration of genes. Currently available metagenomic sequencing approaches require very complex computational analyses and are dependent on the availability of reference genomes for strains of interest. Marker systems using plasmids are easier to introduce into rhizobia than chromosomal integrations and are not dependent on preexisting genome data. However, concerns about plasmid stability in the absence of antibiotic selection during plant assays have limited the use of plasmids to study competitiveness (13). Plasmids that show stability in the absence of antibiotic selection have now been developed, such as the stable promoter-probe and low-copy vector pJP2 (14), and recently adapted to Golden Gate cloning to enable high-throughput plasmid assembly (15).
Effectiveness at N2 fixation and competitiveness for nodulation are independent rhizobial traits (12, 16) and, due to the complexity of competition assays, there are few studies evaluating these two traits simultaneously. Therefore, strains that are both highly effective and competitive (elite strains) can take years to identify (17, 18). To improve the identification of elite rhizobial inoculants, it is crucial to increase the number of rhizobial strains that can be evaluated simultaneously in the presence of native rhizobia and concomitantly assess their N2 fixing ability (19, 20). To overcome these limitations, we present a plasmid-based library allowing simultaneous assessment of competitiveness and effectiveness for rhizobial strains. To achieve this goal, plasmids were developed with two key modules: 1) a module for evaluating rhizobial effectiveness that included a consensus nifH promoter (PsnifH) driving nodule-specific expression of green fluorescent protein (GFP) as reporter gene, and 2) a module for measuring bacterial competitiveness, consisting of a unique synthetic nucleotide sequence (Plasmid ID), allowing the identification of multiple rhizobial strains in a single experiment using next generation sequencing (NGS). Plasmids were assembled by high-throughput Golden Gate cloning (21) in the broad host-range vector pOGG026, which has excellent stability in rhizobia, even in the absence of antibiotics (15).
Results
A Synthetic nifH Promoter (PsnifH) Adapted for Use in Diverse Rhizobia.
When considering a molecular tool to measure effectiveness, we hypothesized that the expression of superfolder GFP (sfGFP) from a nifH promoter (which drives the expression of the genes encoding the nitrogenase complex, nifHDK) (22) would correlate with N2 fixation in individual nodules. The fluorescence signal from the nodules containing such a bioreporter should reflect the expression of nifH, as well as the size of the bacteroid population, which is related to nodule size that is associated with effectiveness of N2 fixation (23, 24). A promoter that is active only following nodule formation will also avoid fitness penalties prior to the invasion of the plant that might alter strain competitiveness. The well-studied region upstream of the genes encoding nitrogenase (nifHDK), which constitutes the nifH promoter (PnifH), has the characteristics of a type -24/-12 promoter recognized by the RNA polymerase sigma factor RpoN (σ54) (25). This promoter has been used for many years in studying Rhizobium competition and also as a symbiosis-induced promoter (15, 26, 27). To make a nifH promoter as broadly useful as possible, a 258-bp consensus promoter (PsnifH) was derived from alignment of 48 strains of the genus Rhizobium (SI Appendix, Fig. S1). It includes an upstream activator sequence (UAS) (TGT-N10-ACA), a binding sequence for RpoN σ54 (sigma factor for expression of all nif genes in nodules) (TGGCACG-N4-TTG), and a ribosome-binding site (RBS) (GAAGGAA) (SI Appendix, Fig. S2A) (25, 28, 29). The synthetic nifH promoter was synthesized as a “PU” module with BsaI restriction enzyme sites at either ends (SI Appendix, Fig. S2B), allowing Golden Gate assembly into the stable plasmid pOGG026 (15).
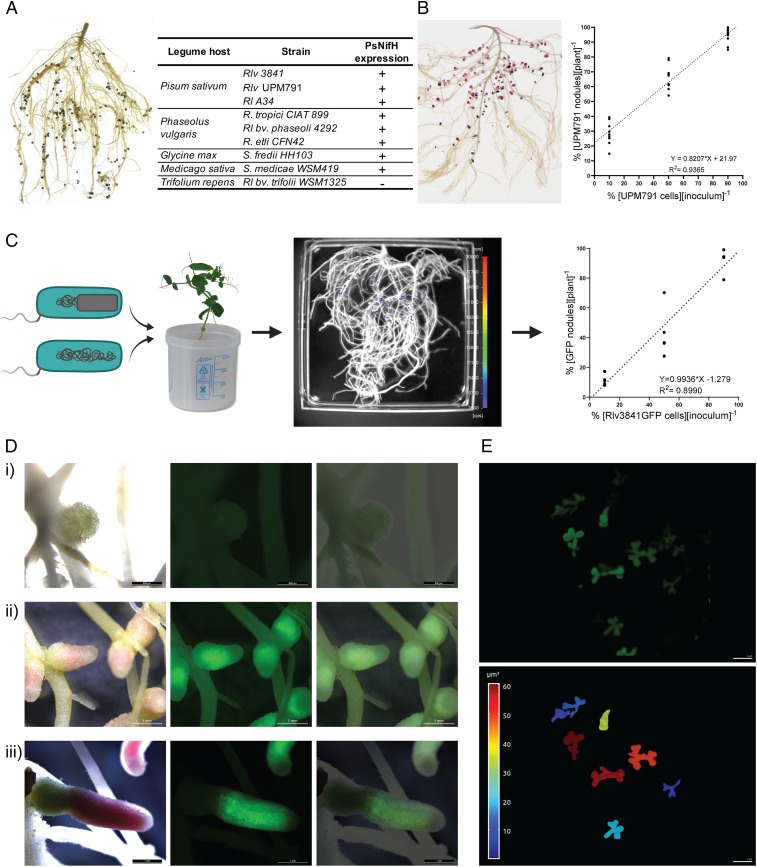
To evaluate the function of PsnifH in various rhizobial strains, the chromogenic markers gusA (β-glucuronidase) and celB (thermostable β-galactosidase) (26) were assembled downstream of the nifH promoter generating pOPS0262 and pOPS0263, respectively. They enable tracking of gene expression by staining GusA with Magenta-GlcA (pink) or CelB with X-Gal (blue). These plasmids were conjugated into 11 rhizobial strains and inoculated onto their respective host (Fig. 1A). Rhizobium leguminosarum bv. trifolii WSM1325 was used as a negative control since it has a different nifH promoter to the other R. leguminosarum strains (24). All mature nodules expected to show nifH promoter activity stained correctly after enzymatic reaction with Magenta-GlcA (for GusA) or X-Gal (for CelB) (Fig. 1A) in several Rhizobium and even members of the Sinorhizobium genus. Since GusA and CelB are useful markers for competition assays that are not dependent on expensive laboratory equipment, the efficacy of the plasmids in competition assays were compared to gusA or celB integrated in the chromosome (9). Pea plants were coinoculated at ratios: 10, 50, and 90% of RlvUPM791[pOPS0263] versus Rlv3841[pOPS0262]. At 21 days postinoculation (dpi), pea roots were sequentially double stained with Magenta-GlcA and X-Gal after thermal treatment (Fig. 1B and SI Appendix, Table S5). As found by ref. 9, RlvUPM791 was significantly more competitive than Rlv3841 (y intercept = 21.97; P < 0.0001 different from 0) (Fig. 1B), supporting the use of stable plasmids based on pOGG0026 to mark R. leguminosarum strains.
Fig. 1.
Design and validation of a PsnifH consensus promoter. (A) Common bean root system inoculated with CIAT899[pOPS0262] (celB under the control of PsnifH) stained with X-Gal after thermal treatment and results obtained from the full qualitative assessment of PsnifH performance assessing celB and gusA expression in nodules. (B) Competition assays for nodule formation, pea root system inoculated with UPM791[pOPS0263], and Rlv3841[pOPS0262] after sequential double-staining (n = 10). (C) Competition assays for nodule formation between Rlv3841[pOPS0381] (marked) and Rlv3841-WT (unmarked) strains on pea roots. GFP detection (scale, 3,000–30,000 cps). Analysis of competitiveness was performed by comparing percent of marked strain cells in the original mix inoculum with percent of the obtained marked nodules in each plant. Results (n = 5) showed that marked nodules were not significantly different from unmarked nodules. (D) Microscope images of pea nodules formed by Rlv3841sfGFP harboring a plasmid containing sfGFP under control of PsnifH: immature (i), mature (ii), and senescent nodule (iii). Bright-field (Left), fluorescence (GFP filter) (Center), and merged images (Right). (E) Maximum intensity projection of a confocal stack of GFP-expressing bacteroids formed by Rlv3841[pOPS0381] from crushed pea root nodules (Upper), and 3D-segmented bacteroids from the same confocal stack color-coded for volume (scale from 0 to 60 μm3) (Lower). Video of 3D structures in SI Appendix.
Next, the fluorescent proteins, sfGFP (30) and mCherry (31) were assembled with PsnifH, generating pOPS0381 and pOPS0382, respectively. The native Rlv3841 PnifH promoter driving sfGFP (pOPS0379) and mCherry (pOPS0380) were used as positive controls (15). These plasmids were conjugated into Rlv3841, inoculated onto pea plants and their roots analyzed using a NightOWL charge coupled device (CCD) camera to quantify green and red fluorescence expression. Nodule-specific expression of both sfGFP and mCherry occurred in the positive control and with PsnifH (SI Appendix, Fig. S3). Additionally, the Indigo software permitted automatic counting of nodules expressing either GFP or mCherry. However, as fluorescence of sfGFP was almost 10 times greater than mCherry, only sfGFP was used for subsequent experiments. Rlv3841 marked with sfGFP under control of PsnifH (Rlv3841[pOPS0381]), and unmarked Rlv3841 (WT) (Fig. 1C) were inoculated together on pea roots (Fig. 1C). The percentage of 21 dpi nodules containing marked strains was not significantly different from their percentage in the inoculum (Fig. 1C). The slope of the regression line was 0.9936 ± 0.096 (95% CI [0.7841, 1.203]), not significantly different from 1 (P = 0.9476), and the y intercept = −1.279 ± 5.475 (95% CI [13.21, 10.65]) is not significantly different from 0 (P = 0.8192), demonstrating pOPS0381 does not alter the competitiveness of Rlv3841.
PsnifH Promoting sfGFP Expression Is a Biosensor for Nitrogenase Activity.
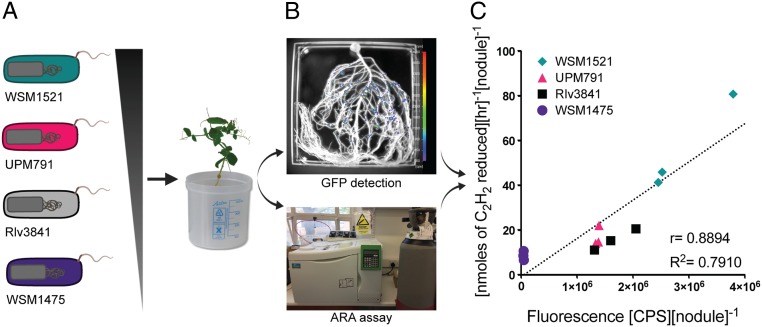
Pea nodules formed by Rlv3841 marked with sfGFP under control of PsnifH (Fig. 1D) showed high GFP in the N2 fixing zone of mature nodules, but no expression in immature nodules. Furthermore, individual bacteroids of Rlv3841 expressing GFP from crushed nodules were visualized using confocal microscopy (Fig. 1E) and their three-dimensional (3D) shapes reconstructed. Individual bacteroids are coralloid in shape with volumes of approximately 10–60 μm3. These qualitative results confirmed the correlation of sfGFP expression from PsnifH with N2 fixation. To determine if nitrogenase activity correlates quantitatively with GFP expression from PsnifH, four R. leguminosarum strains with different levels of effectiveness at fixing nitrogen were used. WSM1475 and WSM1521 were previously identified with a low and high effectiveness through the analysis of biomass (32). Additionally, Rlv3841 and UPM791 were added as inoculants as they are well-studied strains and their behavior is broadly characterized. The plasmids pOPS0491, pOPS0503, pOPS0504, and pOPS0515 (differing only by Plasmid ID to pOPS0381) were conjugated into WSM1475, WSM1521, Rlv3841, and UPM791, and transconjugants inoculated on pea (Fig. 2A). Effectiveness for nitrogenase activity of the strains was determined by ARA at 28 dpi and 35 dpi (Fig. 2B) and shoot dry biomass (DW) at 35 dpi and 42 dpi (SI Appendix, Table S6). The high effectiveness of strain WSM1521 had been confirmed before (33) through glasshouse evaluation of dry matter yields. Therefore, WSM1521 was used as a positive control for nitrogenase activity, with the prediction that GFP fluorescence from WSM1521 nodules should be higher than WSM1475, Rlv3841, and UPM791. GFP detection, ARA, and shoot DW biomass were normalized by the number of nodules per plant (SI Appendix, Fig. S4). A two-tailed Pearson test shows a positive correlation for GFP fluorescence and ARA at 28 dpi of r = 0.8894 (95% CI [0.6444–0.9688]), P = 0.0001 and a R2 = 0.7910. At 35 dpi, the positive correlation was r = 0.8160 (95% CI [0.4234–0.9506]), P = 0.0022 and a R2 = 0.6659. Results from 28 dpi were grouped by strain in Fig. 2C. At both 28 dpi and 35 dpi, GFP fluorescence and nitrogenase activity correlated, with a peak at 28 dpi (Fig. 2C), in agreement with maximum ARA occurring at flowering in peas (34). Dry biomass measurements at 35 dpi and 42 dpi (SI Appendix, Table S6) showed that strain WSM1521 is highly effective and confirm that sfGFP fluorescence is an easy method for quantification of nitrogenase activity in individual nodules.
Fig. 2.
PsnifH promoting sfGFP expression is a biosensor for nitrogenase activity. (A) Four strains with different effectiveness class were used to inoculate pea plants. (B) At 28 and 35 dpi, root systems were simultaneously analyzed for their GFP detection from nodules and for their nitrogenase activity by ARA. (C) Correlation of GFP (fluorescence, CPS) and ARA ([nmoles of C2H2 reduced][h−1]) at 28 dpi (n = 3).
Plasmid IDs Allow High-Throughput Identification of Rhizobial Strains.
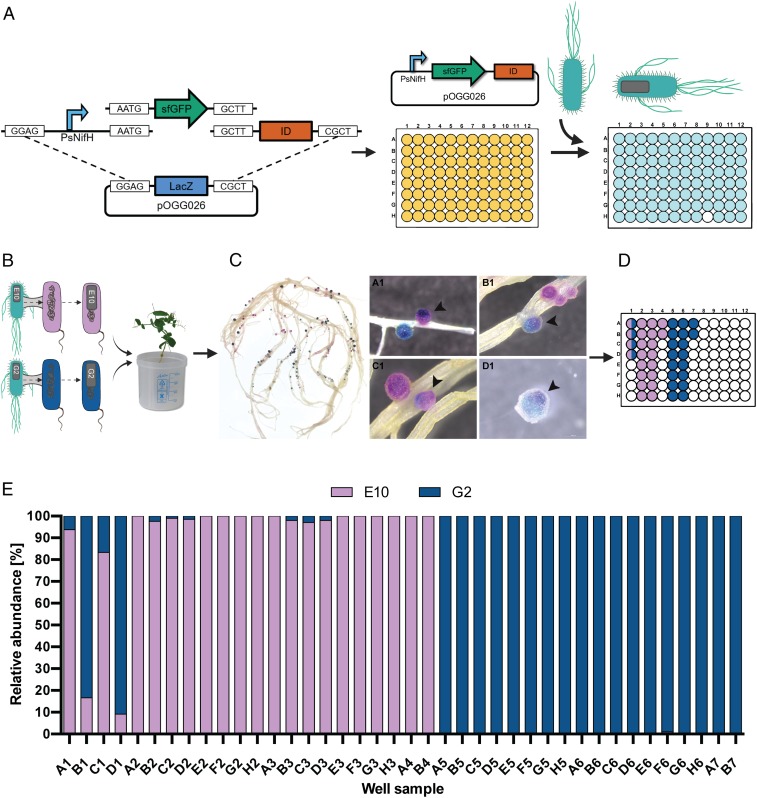
Due to limitations in the number of strains able to be tested for competitiveness in traditional marker systems, we developed a system utilizing NGS technology to monitor the competitiveness of many strains simultaneously. Plasmids were therefore tagged with 96 unique 12-nt Golay barcodes (35), incorporated into a 57-bp Level 0 Golden Gate “T” module (SI Appendix, Table S3 and Fig. S5). The overall features are terminal BsaI enzyme recognition sites, Plasmid ID (Golay barcode) at nucleotides 12–23, and a universal primer binding site at nucleotides 24–46. Ninety-five individual Golden Gate Level 1 cloning one-tube one-step reactions were set up (BsaI digestion and ligation) (21) in a 96-well plate, where the only difference was the T module (Plasmid ID). (Fig. 3A and SI Appendix, Figs. S6 and S7). To test that Plasmid IDs allow multiple rhizobial strains to be marked and identified, strains UPM791gusA and Rlv3841celB, with marker genes integrated into their genomes, respectively, had Plasmid ID E10 (pOPS0548) and Plasmid ID G2 (pOPS0564) introduced by conjugation (Fig. 3B). UPM791gusA-E10 and Rlv3841celB-G2 were set to compete 1:1 for pea nodulation and the strain occupying each nodule identified by sequential double-staining with Magenta-GlcA and X-Gal after thermal treatment (Fig. 3C). Forty nodules were analyzed by NGS, 36 singly infected nodules and four mixed nodules as determined by Magenta-GlcA and X-Gal staining (Fig. 3D). The Plasmid IDs of all 40 samples matched the Magenta-GlcA and X-Gal staining. For the four mixed nodules, the percentages of IDs matched qualitatively with the relative colors of each nodule (Fig. 3E).
Fig. 3.
Plasmid IDs allow high-throughput identification of rhizobial strains. (A) Plasmid ID Library built by Golden Gate cloning. Promoter, marker gene, and backbone vector were identical, with the T module (Plasmid ID) as the unique identifier in each case. (B) Experimental design: Plasmid ID-E10 (pOPS0548) and Plasmid ID-G2 (pOPS0564) were conjugated into UPM791gusA and Rlv3841celB, respectively and used to coinoculate pea plants. (C) Pea roots that were sequentially double-stained with Magenta-GlcA and X-Gal after thermal treatment, resulting in pink nodules formed by UPM791gusA-E10[pOPS0548] (gusA constitutively expressed) and blue nodules formed by Rlv3841celB-G2[pOPS0564] (celB constitutively expressed). Four mixed nodules were identified (indicated by black arrows). (D) Forty nodules were selected and prepared for analysis by NGS with Ion Torrent, including the four mixed nodules on wells A1, B1, C1, and D1. (E) Sequencing results showing ID relative abundance per well sample.
Simultaneous High-Throughput Assessment of Nodulation Competitiveness and N2 Fixation Effectiveness.
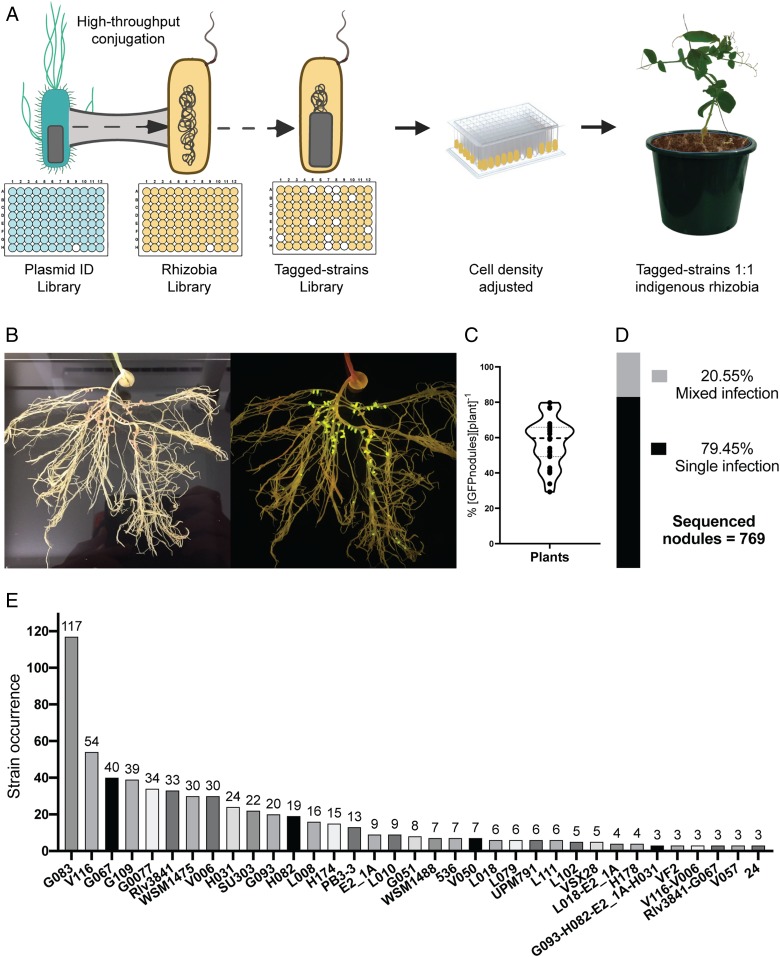
Measuring effectiveness and competitiveness of Rhizobium strains in sterile conditions, deprived of N, limits the finding’s relevance to agricultural conditions. Therefore, a competition assay was performed by adding tagged-rhizobial strains to nonsterile nutrient-rich field soil containing indigenous rhizobia. Soil was from Yatesbury House Farm (Yatesbury, Calne, Wiltshire, United Kingdom). Soil was analyzed for chemical composition (SI Appendix, Tables S8 and S9) and by most probable number (MPN) (8) for rhizobial population size. Yatesbury soil contained ∼100 cells of indigenous rhizobia per gram, so that competition assays contained 6.20 × 104 rhizobial cells in 620.4 ± 1.96 g of Yatesbury soil. Ninety-five well-characterized R. leguminosarum bv. viciae (Rlv) strains were selected from different geographical locations (SI Appendix, Fig. S11) and tested for effective symbiosis with pea plants (SI Appendix, Fig. S12 A and B). The plasmid library (SI Appendix, Fig. S7) was grown in liquid culture in a 96-deepwell plate and then conjugated into the strains (SI Appendix, Fig. S11) (Fig. 4A), with 84 strains reaching the desired growth. The final Tagged-strains library was grown in liquid culture in a 96-deepwell plate, total cell density adjusted to ∼6.20 × 104 cells, and the combined Tagged-strains library inoculated onto pea seedlings in 0.5 L of Yatesbury soil (Fig. 4A). At 21 dpi (flowering time), pea plants were harvested (SI Appendix, Fig. S14A) and roots were analyzed (SI Appendix, Fig. S14 B and C). Plants (n = 30) formed an average of 54.3 nodules, with 57.4% (±2.6 SEM) expressing PsnifH GFP and 42.6% occupied by indigenous rhizobia (Fig. 4C). The IDs from GFP-expressing nodules were PCR-amplified using the conserved primer binding sites, sequenced by Ion Torrent and Plasmid IDs cross-referenced to strains (SI Appendix, Fig. S8). For a nodule to be scored as a mixed infection, a threshold for the second strain was set at 10%. After high-quality filtering (SI Appendix, Fig. S8), 769 sequenced nodules were identified, of which 611 nodules resulted from a single clonal infection and 158 contained more than one strain (i.e., mixed nodules). Even with such a conservative strain identification method (10% threshold), nodules with six strains were identified (SI Appendix, Table S10). Mixed nodules represent 20.6% of the total population (Fig. 4D), which is much higher than the ∼1% that we observed in competition assays when only two strains were inoculated in sterile vermiculite/sand (SI Appendix, Table S5). Furthermore, 20.6% is a lower estimate since indigenous rhizobia occupied 42.6% of nodules and mixtures of GFP-expressing tagged strains and indigenous rhizobia would not be identified. Remarkably, 15.2% of all sequenced nodules were occupied by strain G083, followed by strains V116 (7.02%), G067 (5.20%), G109 (5.07%) G0077 (4.42%), and Rlv3831 (4.29%).
Fig. 4.
High-throughput competition assay among tagged-rhizobial strains in a nonsterile nutrient-rich field soil containing indigenous rhizobia. (A) Strategy for the competition assay among Tagged-strains in the presence of indigenous rhizobia. (B) Example of roots exposed to a blue-light transilluminator to check nodules expressing GFP belonged to Tagged-strains. (C) Average of nodules expressing GFP per plant (n = 30). (D) Percentage of single and mixed infection from total sequenced nodules. (E) Total occurrence of Tagged-strains identified from a total of 769 sequenced nodules.
Identification of a Supercompetitive and Highly Effective Rhizobial Strain.
We calculated a competitiveness index (CI) of each strain by comparing their own occurrence (the number of nodules in which the strains were identified from the 769 sequenced nodules) to the number of GFP-tagged nodules per plant:
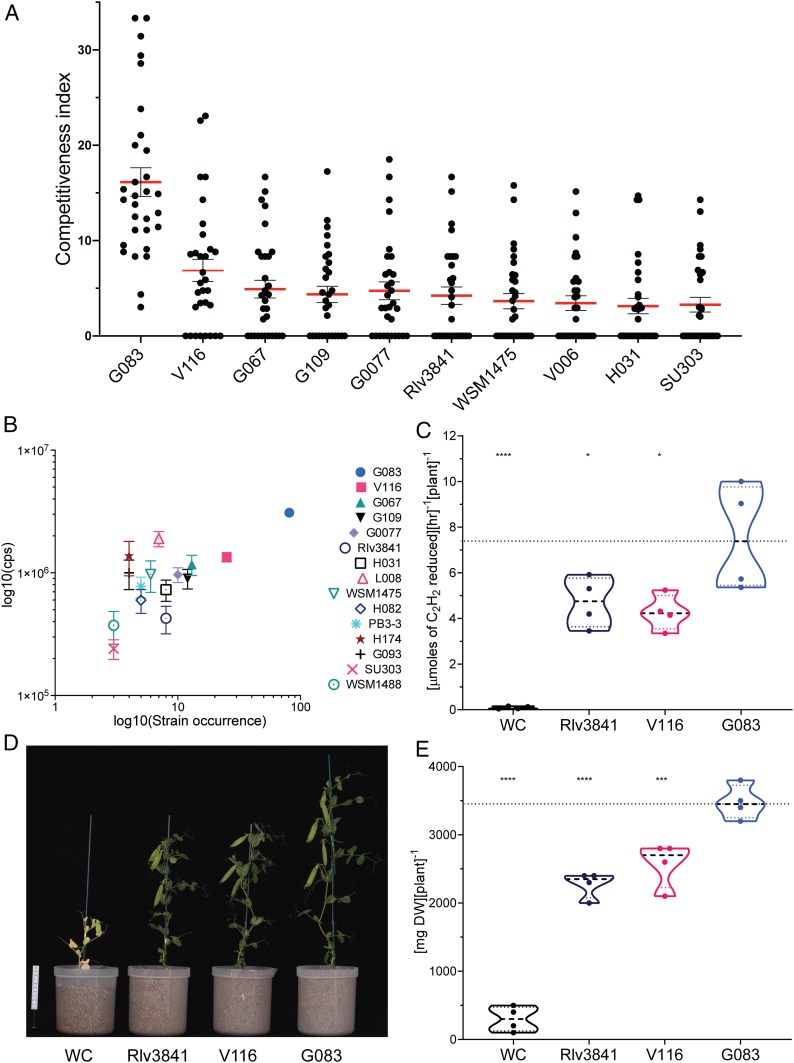
Statistical analyses of the 10 strains with the highest CI (Fig. 5A) was performed and after ordinary one-way ANOVA, a Tukey’s multiple comparisons test showed that the CI of G083 (CI = 16.14) was higher (P < 0.0001) than any of the other strains (SI Appendix, Table S11). Additionally, G083 was the only strain that appeared in each of the 30 plants tested (Fig. 5A).
Fig. 5.
Identification of a supercompetitive and highly effective rhizobial strain. (A) Competitiveness index (CI) of the top 10 strains. Dots represent CI of each strain per plant, and bars represent the SEM. (B) Comparison of the effectiveness of nodules occupied by different strains measured by GFP detection (cps) and their occurrence (number of nodules in which the strains were identified from the 240 most effective nodules); error bar represent SEM. (C) Effectiveness assessment of selected strains by ARA (n = 4). (D) Examples of 6-wk-old pea plants inoculated with selected strains. (E) Assessment of effectiveness by measurement of DW. WC; water control (n = 4).
All individual nodules were quantified for fluorescence while still on the plant (SI Appendix, Fig. S15A). The eight brightest nodules (i.e., the most effective) from each of the 30 plants analyzed (n = 240) were picked and gridded in microtiter plates, and their fluorescence was quantified (SI Appendix, Fig. S15B). DNA was then extracted, and Plasmid IDs determined by sequencing (Materials and Methods).
After ordinary one-way ANOVA and Tukey’s multiple comparisons, among the most effective nodules, G083 had the highest occurrence (n = 81) and showed significantly higher GFP fluorescence than nodules containing other strains (Fig. 5B), with the exception of L008 (n = 7), where the difference was marginally nonsignificant (P = 0.056) (SI Appendix, Table S12). These data suggested that in addition of being the most competitive strain, G083 is also a highly effective strain. While our high-throughput methodology represents a significant advance in simultaneous assessment of competitiveness and N2 fixing ability in Rhizobium-legume symbioses, can this be extrapolated to plant performance and growth yield? To answer this, the effectiveness of G083 was assessed by ARA at 21 dpi and by DW at 42 dpi. Along with G083, strains V116 and Rlv3841 were selected as controls—V116 as it had the second highest occurrence of the most effective nodules (n = 25) and Rlv3841 (n = 8) as the most well-studied strain of R. leguminosarum bv viciae. In both assays, ARA (Fig. 5C and SI Appendix, Table S13) and DW (Fig. 5 D and E and SI Appendix, Table S15) values for G083 were significantly higher than V116 and Rlv3841, while the latter strains did not show a significant difference from each other (ordinary one-way ANOVA and Dunnett’s multiple comparisons test) (SI Appendix, Tables S14 and S16). Additionally, G083 produced fewer but larger nodules and more pea pods per plant at 42 dpi (SI Appendix, Table S15). Both ARA and DW measurements show that the results obtained by high-throughput GFP detection in individual nodules correlates with plant performance and growth. Overall, the molecular toolkit developed here allows the identification of elite strains, such as G083 in pea, which is both supercompetitive and highly effective at N2 fixation.
Discussion
PsnifH as a Consensus Promoter for Rhizobium Strains.
We designed a synthetic consensus promoter (PsnifH) compatible with Golden Gate cloning to drive nodule-specific expression of CelB, GusA, mCherry, and sfGFP, which confirmed that PsnifH can be used in a broad range of R. leguminosarum strains and potentially other species of rhizobia (e.g., R. tropici, S. fredii) (Fig. 1A). Fluorescent proteins do not need cofactors or substrates (36), and additionally, sfGFP has a greater resistance to chemical denaturants than GFP (30). This could have been problematic if sfGFP fluorescence accumulated in the nodules over time. However, senescent areas of nodules did not show fluorescence, which indicates that the signal was restricted to the N2 fixing zone of nodules (Fig. 1 D, iii). Although we were able to show a strong positive correlation between fluorescence detection and ARA (r = 0.8894, Fig. 2C), this correlation is not strictly linear since the fluorescence detection from WSM1475 was lower compared to its results from ARA. These results do not invalidate the use of florescence detection as an easy and quantitative proxy for nitrogenase activity in individual nodules, but rather define the limit of the assay. There are more accurate measurements of total nitrogenase activity than GFP florescence, such as flow-through ARA systems and 15N2 assays, which will improve the linearity of correlation, but they are very laborious, time-consuming, and require expensive special equipment that is not accessible for low-income laboratories. Therefore, assessing a handful of strains is a major undertaking; moreover, assessing tens or hundreds of strains is almost impossible using these standard techniques.
For a rhizobial strain to be selected as a suitable inoculant, it must first be evaluated for its tolerance to environmental stresses, its N2 fixation effectiveness, its soil persistence, and most importantly, its ability to compete against indigenous strains for nodule formation in a directly relevant soil (37). The addition of the Plasmid IDs to the PsnifH sfGFP reporter plasmids provides the opportunity to assess the competitiveness of up to 95 strains simultaneously, although the number can be increased by building more Plasmid IDs since all parts are compatible with our recently developed bacterial expression vector archive (BEVA) bacterial cloning system (15).
Identification of a Supercompetitive and Highly Effective Rhizobial Strain.
Soil pH has been shown to affect the competitiveness of R. leguminosarum strains (38), which may explain the high competitiveness of G083 in our test soil. G083 was isolated from a soil with a pH 7.6 (39), which is very similar to that of Yatesbury soil (pH 7.75) (SI Appendix, Table S9). A key breakthrough of such high-throughput screening is that multiple soils, environmental conditions, and plant cultivars can be tested all at the same time (20). While G083 is undoubtedly an elite strain in our test soil, other strains may perform better under different conditions; e.g., of soil type, pH, moisture content, or nutrient status. High-throughput screening will enable matching of inoculant to field conditions.
One possible explanation for the competitive advantage of strain G083 is that it may migrate to root hairs faster and initiate infection much earlier than any other tested strains. This ability could be related to communication between the first strain that enters in the infection threads (ITs) and the plant itself, triggering an immune response that obstructs entry of further strains (40). For instance, necrosis of cortical cells of ITs in alfalfa is observed after S. meliloti infection (41). Alternatively, G083 may have active mechanisms of preventing other strains from infecting or of disabling them in a competitive race. Examples of such mechanisms include type VI secretion systems (T6SS), which secrete effector proteins (42) that may have antibacterial activity (43) as well a positive role in Rhizobium–legume symbioses (44). G083 was only found in nine mixed nodules (5.7% of mixed nodules), a very low occurrence compared to its presence in single-infection nodules (19.1% of single infection nodules). This suggests a supercompetitive strain may trigger autoregulation of nodulation (AON) much earlier. AON involves long-distance root-shoot signaling that inhibits further nodulation events (45–47). We therefore presume that from the moment G083 began to form nodules, and fix N2 at high rates, it restricted less competitive strains from successful infection (45).
A remarkable observation from comparing nodulation of multiple strains in soil is that mixed nodules represent 20.6% of the total sequenced nodules. This percentage is much higher than the ∼1% observed in previous competition assays where only two strains were inoculated at a time in sterile vermiculite/sand (SI Appendix, Table S5). Furthermore, some of the nodules containing one tagged strain are likely to contain indigenous rhizobia that would not be detected. Tagged strain identification was also conservative with no mismatches allowed in either sample or Plasmid ID and a 10% threshold of total reads used for the presence of a strain. Although there are existing reports of high levels of mixed nodules, such as 36% reported in lentils (48), this tends to be the exception. Our results showing that inoculation of low numbers (i.e., same number as indigenous rhizobia in the soil) of multiple strains into an agricultural soil increases the mixed nodule occupancy from a few percent to over 20% has significant implications for the evolution and stability of Rhizobium-legume symbioses. Bacteroids of R. leguminosarum are terminally differentiated and do not regrow from nodules of peas due to the carefully orchestrated production of antimicrobial peptides by the plant (49–51). Plants cannot select effective N2 fixing strains from the soil (40), instead they must sanction ineffective nodules, presumably by reducing resource allocation to a nodule and shutting it down. The result is that fewer undifferentiated bacteria are released from ineffective nodules compared to effective nodules (52). In legumes that produce antimicrobial peptides, this biased release of effective strains is thought to be a primary driver of the evolution of symbiosis. However, one way in which ineffective rhizobia can cheat on the symbiosis is to form mixed nodules with more effective strains. Therefore, the demonstration that the presence of a rich microbiota in soil promotes a large increase in mixed nodule occupancy suggests that less effective strains may benefit by “hitch hiking” with effective strains. Many strains also only appear in mixed nodules, suggesting there may be cooperative partnerships between strains. While the work here has only scratched the surface of such ecological investigations, it is clear that tagged rhizobial strain sets represent a powerful tool to address the mechanisms underlying competitive cooperation and exclusion.
Although we quantified GFP using the sensitive, but expensive, NightOWL CCD camera, it is possible to use low-cost equipment that is widely available. For example, by using a smart phone with a blue-light transilluminator and an orange filter, it is possible to obtain good quality photos (Fig. 4B) suitable for fluorescence image analysis with Fiji software (53). The only specialized equipment required is a PCR thermocycler to amplify the Plasmid IDs, and the samples can then be posted to one of the many low-cost sequencing centers around the world.
Until now, the search for elite strains has been a time-consuming process involving numerous studies in the laboratory before the strains could be assessed in greenhouse and field trials. The existing tagged PsnifH sfGFP plasmid library can be transferred to many other Rhizobium species for strain selection for improvement of plant performance or use in ecological studies of strain competition. By using our BEVA construction system, it is also easy to exchange the nifH promoter, IDs, or resistance markers, to make it more suitable for other rhizobia. While we have developed a plasmid system to facilitate its transfer, it could be adapted to Tn7 (54) for chromosomal integration in species of rhizobia where plasmid maintenance might be a problem.
In this work, we demonstrate a molecular approach to address the challenge of improving elite rhizobial inoculants for sustainable agriculture. It is further apparent that the tools developed here also have powerful applications to microbial ecology. We believe that the next crucial step in translating this research to a real-world benefit would be to integrate these findings into coordinated multidisciplinary studies with agronomists and breeders.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains and plasmids used in this study are listed in See SI Appendix, Table S1. Escherichia coli strains were grown in liquid or solid Luria–Bertani medium at 37 °C supplemented with appropriate antibiotics (µg·mL−1): ampicillin, 100; tetracycline, 10; and kanamycin, 20. For E. coli ST18 strains, 5‐aminolevulinic acid (ALA) was added at 50 µg·mL−1. All plasmids are available from Addgene (https://www.addgene.org) with the Addgene ID given in SI Appendix, Table S1. Rhizobial strains, listed in SI Appendix, Table S1, were grown at 28 °C in Tryptone-Yeast (TY) extract or Universal Minimal Salts (UMS) (55) with appropriate carbon and nitrogen sources at 10 mM unless otherwise stated. Antibiotics were used at the following concentrations (µg·mL−1) unless otherwise stated: gentamicin, 20; kanamycin, 20; neomycin, 40; nitrofurantoin, 10; rifampicin, 50; spectinomycin, 50; streptomycin 500; tetracycline (2 in UMS, 5 in TY).
Designing of a PsnifH Consensus Promoter and Plasmid ID.
To determine a consensus sequence for a nodule-specific promoter expressed in a wide variety of biovars and strains of R. leguminosarum and designed as a PU module for Golden Gate cloning, we followed the process described in SI Appendix, Fig. S1 and detailed in SI Appendix.
For the Plasmid ID, a unique 12-nt, error-correcting Golay barcode (35) was selected as suitable for strain identification (the 96 selected Golay barcodes are given in SI Appendix, Table S1) and the full design is in SI Appendix.
Plasmid ID Library.
A total of 95 individual and unique reporter plasmids were built by Golden Gate Level 1 cloning detailed in SI Appendix.
Primer Design for Multiplex Ion Torrent Sequencing Strategy.
We designed primers (SI Appendix, Table S4) for a two-step PCR. Full design is in SI Appendix.
Thermostable β-Galactosidase Activity from Cloned celB.
Roots were submerged upside down in a 50-mL Falcon tube containing 40 mL of phosphate buffer and kept at 70 °C for 1 h in order to destroy endogenous β-galactosidases. After cooling, 500 μL of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) solution (250 μg/mL) was added. Tubes were covered with aluminum foil and incubated overnight at 37 °C.
Sequential Double Staining to Detect both β-Glucuronidase and Thermostable β-Galactosidase Activity.
The gusA activity assay was run first with Magenta-GlcA (5-bromo-6-chloro-3-indolyl-β-d-glucuronide acid) solution (200 μg/mL) as substrate and, after overnight incubation at 28 °C, the thermostable β-galactosidase activity for celB was done overnight at 37 °C with X-Gal as indicated above (26).
Competition Assay.
Coinoculation with two strains.
Protocol is modified from ref. 9 and fully described in SI Appendix.
Coinoculation with multiple rhizobial strains.
Rhizobium cultures to be used as inoculum were grown to early stationary phase (OD600 < 0.6–0.8). The OD600 of a 200-μL sample from each strain was measured in 96-well plate using a FLUOStar Omega Microplate Reader (BMG). Cultures were adjusted to the desired OD and mixed in a flask. Inoculum was prepared by adding 1 mL of the Tagged-strain mix in 74 mL of 2.5× concentrated nitrogen-free rooting solution (55) and then distributed homogenously into 500-mL pots. Five-day-old similar-sized pea (Avolar) seedlings were transferred to preinoculated pots. Pea plants were grown without inoculum as negative controls (water control [WC]).
Analysis of Agricultural Soil for Competition Assays.
Agricultural soil was collected in mid-August 2017 from Yatesbury House Farm (Yatesbury, Calne, Wiltshire, UK). This is a 600-ha farm maintained under European Union organic regulations and certified by The Soil Association Certification Limited (SA Certification). The crops grown in this farm are wheat, barley, oats, field beans, and lentils are cultivated with legume-based mixtures of clover. Full details of the analysis are in SI Appendix. MPN of indigenous rhizobia from Yatesbury soil was carried out following the methodology described in ref. 8 and fully described in SI Appendix.
Image Acquisition.
For green and red fluorescence expression, fluorescence CCD images were acquired with a ring-light epi illumination accessory for NightOWL LB-983 camera and for full root system, a blue-light transilluminator (VWR) together with a Kodak Wratten Gelatin Filter no. 49 were used (specific parameters in SI Appendix). Dissecting microscope images were taken using a microscope Leica M165 FC and the digital camera Leica DFC310 FX (software LAS v4.5). Visible light filter, green fluorescent, and red fluorescent filters were used.
Confocal Microscopy for Bacteroids Analysis.
Confocal microscope images were acquired using an HCX PL APO CS 20x/0.7 IMM ultraviolet lens on a Leica SP 5. sfGFP was excited at 488 nm and detected at 500–600 nm. Three-dimensional segmentation was performed in MorphographX (56) (specific parameters in SI Appendix).
Bioinformatic Analysis and Availability of Materials.
Paired-end merging of amplicon reads was done with USEARCH 10.0.240 (57). Quality filtering of amplicon reads was carried out with Trimmomatic-0.36 (58) (steps fully described in SI Appendix), and the Plasmid ID was determined using a custom script available in: https://github.com/marcelamendoza/Plasmid-ID.
Data Manipulation and Statistical Analysis.
Data manipulation of PivotTables and conditional tables were done in Microsoft Excel, and data were processed in either the statistics program Prism8 (GraphPad Software) or The R project for Statistical computing (59).
Data Availability.
The data supporting the findings of the study are available in this article and its SI Appendix. Raw reads for this study have been deposited in the ENA at EMBL-EBI under accession no. PRJEB37539
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council Grants BB/L011484/1, BB/N003608/1, and BB/N013387/1 (to P.S.P.). M.A.M.-S was recipient of the Mexican Government Grant Consejo Nacional de Ciencia y Tecnología 266954/399852. We thank John Howieson, Euan James, and Juan Imperial for rhizobial strains; Vanessa De Sousa Vieira for technical assistance funded by the University of Oxford Disability Advisory Service; Richard Ganlett for farm soil; and Alison East for critical revision of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw reads for this study have been deposited in the European Nucleotide Archive (ENA) at European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI) under accession no. PRJEB37539.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921225117/-/DCSupplemental.
References
- 1.Sprent J. I., James E. K., Legume evolution: Where do nodules and mycorrhizas fit in? Plant Physiol. 144, 575–581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Hara G. W., The role of nitrogen fixation in crop production. J. Crop Prod. 1, 115–138 (1998). [Google Scholar]
- 3.Parnell J. J., et al. , From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front. Plant Sci. 7, 1110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates R. J., Howieson J. G., Reeve W. G., O’Hara G. W., A re-appraisal of the biology and terminology describing rhizobial strain success in nodule occupancy of legumes in agriculture. Plant Soil 348, 255–267 (2011). [Google Scholar]
- 5.Chibeba A. M., Kyei-Boahen S., Guimarães M. F., Nogueira M. A., Hungria M., Isolation, characterization and selection of indigenous Bradyrhizobium strains with outstanding symbiotic performance to increase soybean yields in Mozambique. Agric. Ecosyst. Environ. 246, 291–305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulas D., et al. , Distribution and efficiency of Rhizobium leguminosarum strains nodulating Phaseolus vulgaris in Northern Spanish soils: Selection of native strains that replace conventional N fertilization. Soil Biol. Biochem. 43, 2283–2293 (2011). [Google Scholar]
- 7.Zvinavashe A. T., Lim E., Sun H., Marelli B., A bioinspired approach to engineer seed microenvironment to boost germination and mitigate soil salinity. Proc. Natl. Acad. Sci. U.S.A. 116, 25555–25561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howieson J. G., Dilworth M. J., Working with Rhizobia, Howieson J. G., Dilworth M. J., Eds. (ACIAR, 2016), pp. 97–105. [Google Scholar]
- 9.Sánchez-Cañizares C., Palacios J., Construction of a marker system for the evaluation of competitiveness for legume nodulation in Rhizobium strains. J. Microbiol. Methods 92, 246–249 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Geddes B. A., Oresnik I. J., Inability to catabolize galactose leads to increased ability to compete for nodule occupancy in Sinorhizobium meliloti. J. Bacteriol. 194, 5044–5053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duodu S., Brophy C., Connolly J., Svenning M. M., Competitiveness of a native Rhizobium leguminosarum biovar trifolii strain for nodule occupancy is manifested during infection. Plant Soil 318, 117–126 (2009). [Google Scholar]
- 12.Burghardt L. T., et al. , Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc. Natl. Acad. Sci. U.S.A. 115, 2425–2430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corich V., et al. , Aspects of marker/reporter stability and selectivity in soil microbiology. Microb. Ecol. 41, 333–340 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Pini F., et al. , Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol. 174, 1289–1306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geddes B. A., Mendoza-Suárez M. A., Poole P. S., A bacterial expression vector archive (BEVA) for flexible modular assembly of golden gate-compatible vectors. Front. Microbiol. 9, 3345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso A. A., Andraus M. P., Borba T. C., Martin-Didonet C. C., Ferreira E. P., Characterization of rhizobia isolates obtained from nodules of wild genotypes of common bean. Braz. J. Microbiol. 48, 43–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourion V., et al. , Co-inoculation of a pea core-collection with diverse rhizobial strains shows competitiveness for nodulation and efficiency of nitrogen fixation are distinct traits in the interaction. Front. Plant Sci. 8, 2249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irisarri P., et al. , Selection of competitive and efficient rhizobia strains for white clover. Front. Microbiol. 10, 768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Checcucci A., DiCenzo G. C., Bazzicalupo M., Mengoni A., Trade, diplomacy, and warfare: The quest for elite rhizobia inoculant strains. Front. Microbiol. 8, 2207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenlon A., et al. , Global-level population genomics reveals differential effects of geography and phylogeny on horizontal gene transfer in soil bacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 15200–15209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler C., Kandzia R., Marillonnet S., A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubio L. M., Ludden P. W., Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Regus J. U., et al. , Cell autonomous sanctions in legumes target ineffective rhizobia in nodules with mixed infections. Am. J. Bot. 104, 1299–1312 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Melino V. J., et al. , Identifying abnormalities in symbiotic development between Trifolium spp. and Rhizobium leguminosarum bv. trifolii leading to sub-optimal and ineffective nodule phenotypes. Ann. Bot. 110, 1559–1572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thöny B., Hennecke H., The -24/-12 promoter comes of age. FEMS Microbiol. Rev. 5, 341–357 (1989). [DOI] [PubMed] [Google Scholar]
- 26.Sessitsch A., Wilson K. J., Akkermans A. D., de Vos W. M., Simultaneous detection of different Rhizobium strains marked with either the Escherichia coli gusA gene or the Pyrococcus furiosus celB gene. Appl. Environ. Microbiol. 62, 4191–4194 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denton M. D., Reeve W. G., Howieson J. G., Coventry D. R., Competitive abilities of common field isolates and a commercial strain of Rhizobium leguminosarum bv. trifolii for clover nodule occupancy. Soil Biol. Biochem. 35, 1039–1048 (2003). [Google Scholar]
- 28.Sullivan J. T., Brown S. D., Ronson C. W., The NifA-RpoN regulon of Mesorhizobium loti strain R7A and its symbiotic activation by a novel LacI/GalR-family regulator. PLoS One 8, e53762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brito B., et al. , Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein nifA. Proc. Natl. Acad. Sci. U.S.A. 94, 6019–6024 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Shaner N. C., et al. , Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Howieson J., Loi A., Carr S., Biserrula pelecinus L.-a legume pasture species with potential for acid, duplex soils which is nodulated by unique root-nodule bacteria. Aust. J. Agric. Res. 46, 997 (1995). [Google Scholar]
- 33.Slattery J., Pearce D., Development of elite inoculant Rhizobium strains in Southeastern Australia. ACIAR Proc. 109e, 86–94 (2002). [Google Scholar]
- 34.Mulley G., et al. , Pyruvate is synthesized by two pathways in pea bacteroids with different efficiencies for nitrogen fixation. J. Bacteriol. 192, 4944–4953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso J. G., et al. , Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C., Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Dilworth M. J., James E. K., Sprent J. I., Newton W. E., Nitrogen-fixing Leguminous Symbioses, Dilworth M. J., James E. K., Sprent J. I., Newton W. E., Eds. (Springer Netherlands, 2008), p. 88. [Google Scholar]
- 38.Frey S. D., Blum L. K., Effect of pH on competition for nodule occupancy by type I and type II strains of Rhizobium leguminosarum bv. phaseoli. Plant Soil 163, 157–164 (1994). [Google Scholar]
- 39.Jorrin Rubio B., “Genomics of Specificity in the Symbiotic Interaction between Rhizobium leguminosarum and Legumes,” PhD thesis, Universidad Politécnica de Madrid (2016). 10.20868/UPM.thesis.43380. Accessed 25 September 2018. [DOI] [Google Scholar]
- 40.Westhoek A., et al. , Policing the legume-Rhizobium symbiosis: A critical test of partner choice. Sci. Rep. 7, 1419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasse J., Billy F., Truchet G., Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 4, 555–566 (1993). [Google Scholar]
- 42.Gallique M., Bouteiller M., Merieau A., The type VI secretion system: A dynamic system for bacterial communication? Front. Microbiol. 8, 1454 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell A. B., et al. , Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salinero-Lanzarote A., et al. , The Type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol. Ecol. 95, fiz054 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Mortier V., Holsters M., Goormachtig S., Never too many? How legumes control nodule numbers. Plant Cell Environ. 35, 245–258 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Ryu H., Cho H., Choi D., Hwang I., Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells 34, 117–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferguson B. J., et al. , Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76 (2010). [DOI] [PubMed] [Google Scholar]
- 48.May S. N., Bohlool B. B., Competition among Rhizobium leguminosarum strains for nodulation of Lentils (Lens esculenta). Appl. Environ. Microbiol. 45, 960–965 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mergaert P., et al. , A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van De Velde W., et al. , Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Montiel J., et al. , Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. U.S.A. 114, 5041–5046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daubech B., et al. , Spatio-temporal control of mutualism in legumes helps spread symbiotic nitrogen fixation. eLife 6, e28683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romero-Jiménez L., Rodríguez-Carbonell D., Gallegos M. T., Sanjuán J., Pérez-Mendoza D., Mini-Tn7 vectors for stable expression of diguanylate cyclase PleD* in Gram-negative bacteria. BMC Microbiol. 15, 190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poole P. S., Schofield N. A., Reid C. J., Drew E. M., Walshaw D. L., Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140, 2797–2809 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Barbier de Reuille P., et al. , MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 4, 05864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R Core Team , R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2014). http://www.R-project.org/. Accessed 14 April 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of the study are available in this article and its SI Appendix. Raw reads for this study have been deposited in the ENA at EMBL-EBI under accession no. PRJEB37539