Significance
IDH1 mutation is a common genetic abnormality in human malignancies, whereas selective therapeutics for IDH1-mutated malignancies remain unavailable. Our present study reveals that IDH1-mutated cancer cells exhibit addiction to Nrf2-governed antioxidative pathways. Blockade of Nrf2 transcriptional activity achieved synergistic lethality with a neomorphic IDH1 mutation. We also showed that triptolide serves as a potent Nrf2 inhibitor, which inhibited Nrf2 transcriptional activity and led to apoptotic changes in IDH1-mutated cells via redox catastrophe. We believe that our findings highlight the broad scope and significance of Nrf2 blockade as a therapeutic strategy and provide an actionable strategy for IDH1-mutated malignancies.
Keywords: triptolide, IDH1 mutation, Nrf2, glutathione, reactive oxygen species
Abstract
Isocitrate dehydrogenase (IDH) mutation is a common genetic abnormality in human malignancies characterized by remarkable metabolic reprogramming. Our present study demonstrated that IDH1-mutated cells showed elevated levels of reactive oxygen species and higher demands on Nrf2-guided glutathione de novo synthesis. Our findings showed that triptolide, a diterpenoid epoxide from Tripterygium wilfordii, served as a potent Nrf2 inhibitor, which exhibited selective cytotoxicity to patient-derived IDH1-mutated glioma cells in vitro and in vivo. Mechanistically, triptolide compromised the expression of GCLC, GCLM, and SLC7A11, which disrupted glutathione metabolism and established synthetic lethality with reactive oxygen species derived from IDH1 mutant neomorphic activity. Our findings highlight triptolide as a valuable therapeutic approach for IDH1-mutated malignancies by targeting the Nrf2-driven glutathione synthesis pathway.
Mutations in isocitrate dehydrogenase (IDH1/2) are common genetic abnormalities in human malignancies such as glioma, acute myeloid leukemia, cholangiocarcinoma, and chondrosarcoma (1–6). Amino acid substitutions in arginine residue (R132) of the catalytic center establish neomorphic activity for the enzyme. In contrast to the wild-type enzymes, which convert isocitrate into α-ketoglutarate (α-KG) through an NADP+ dependent way, the IDH1 mutant enzymes consume α-KG for 2-hydroxyglutarate (2-HG) production, which also transforms NADPH into NADP+ (7, 8). Alterations in enzyme catalytic function result in reprogramming of the Krebs cycle, redox homeostasis, and energy metabolism (9–13). Moreover, growing evidence shows that IDH1 mutations lead to a distinctive pattern in cancer metabolism, cell biology, and resistance signatures, suggesting that selective therapeutics may be possible for this type of malignancy (14–18).
The development of an IDH1 mutant enzyme inhibitor showed promising therapeutic effect and durable remission in IDH1-mutated hematopoietic malignancies (18). However, the inhibitor showed less efficacy and response rate in solid tumors such as glioma and chondrosarcoma. Moreover, suppression of the mutant enzyme may relieve metabolic stress in cancer cells, which supports cancer resistance to genotoxic therapies (19, 20). On the other hand, targeting distinctive molecular patterns in IDH1-mutated malignancies is effective. For example, we and our colleagues showed that acquisition of IDH1 mutation disrupts NAD+ metabolism and homologous DNA repair, which establishes vulnerability to NAD+/PARP inhibitors (15, 21). Inhibition of glutaminase appears to slow the proliferation of IDH1-mutated glioma cells (22). The application of 5-azacytidine rectifies the epigenetic reprogramming and induces tumor regression in patient-derived IDH1-mutated xenografts (23).
Nuclear factor (erythroid-derived 2)-like 2 (NFE2L2, Nrf2) is a transcriptional factor that governs the antioxidant pathway in eukaryotic cells (24, 25). Our previous study showed that Nrf2 plays a critical protective role in IDH1-mutated glioma and supports oncogenesis (14, 26). However, an ideal chemical inhibitor for Nrf2 remains unavailable. In the present study, we demonstrated that triptolide, a diterpenoid epoxide from Tripterygium wilfordii, exhibited robust and selective inhibitory effect in patient-derived IDH1-mutated cells and a preclinical xenograft model. Mechanistically, triptolide reduces the affinity of Nrf2 to the promoter regions of essential regulatory genes for glutathione metabolism, such as GCLC, GCLM, and SLC7A11. Further, triptolide disrupts glutathione de novo synthesis, which establishes synthetic lethality with intrinsic oxidative stress in IDH1-mutated cells, which leads to oxidation of macromolecules, such as DNA oxidative damage and lipid peroxidation, and subsequently, cytotoxicity and tumor suppression.
Results
Elevated Oxidative Stress in IDH1-Mutated Cells.
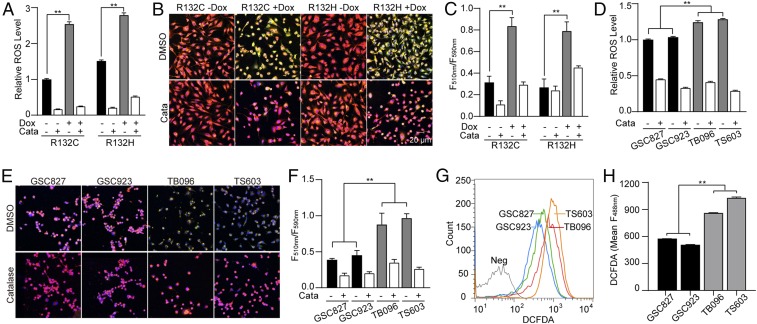
To investigate the redox homeostasis in IDH1-mutated cells, doxycycline-induced IDH1-mutated cells were established based on the U251 MG cell line. Remarkable accumulation of cellular reactive oxygen species (ROS) was recorded at 24 h after IDH1 mutant enzymes (IDH1R132C or IDH1R132H) were induced (Fig. 1A, **P < 0.01). Moreover, we investigated accumulated oxidized lipids through live-cell imaging. Significantly elevated lipid oxidation was recorded after the acquisition of IDH1 mutants (Fig. 1 B and C, **P < 0.01). We further investigated the oxidative stress in patient-derived brain tumor-initiating cells (BTIC). Consistent with previous findings, elevated ROS accumulation was observed in BTIC with somatic IDH1 mutations, TB096 and TS603, compared with IDH1 wild-type BTIC, GSC827, and GSC923 (Fig. 1D, **P < 0.01). Remarkable lipid peroxidation was also observed in IDH1-mutated BTIC (Fig. 1 E and F, **P < 0.01). Finally, H2DCFDA flow cytometry analysis confirmed the accumulation of cellular ROS in IDH1-mutated BTIC (Fig. 1 G and H, **P < 0.01). The elevated oxidative stress in IDH1-mutated cells was found reversible with the presence of exogenous hydrogen peroxide scavenger catalase.
Fig. 1.
IDH1 mutation increases ROS burden. (A) ROS level was quantified using the ROS-Glo H2O2 kit in doxycycline (Dox)-induced IDH1-mutated U251 cells. Catalase (0.1 mg/mL) was used as an ROS scavenger. n = 3. **P < 0.01. (B) Lipid peroxidation assay of Dox-induced IDH1-mutated U251 cells. Before lipid peroxidation, the emission fluorescence showed at 590 nm (red). After lipid peroxidation, the emission fluorescence showed at 510 nm (green). (C) Lipid peroxidation was quantified using the ratio of F510 nm to F590 nm. n = 5. **P < 0.01. (D) ROS level was quantified using ROS-Glo H2O2 assay in BTIC lines treated with DMSO or 0.1 mg/mL catalase. n = 3. **P < 0.01. (E) Lipid peroxidation assay of BTIC lines. (F) Lipid peroxidation was quantified using the ratio of F590 nm to F510 nm. n = 5. Data are presented as mean ± SEM. **P < 0.01. (G) Flow cytometry analysis of ROS level using CM-H2DCFDA staining in BTIC lines. (H) Quantification of DCFDA using fluorescence intensity at 488 nm. n = 3. **P < 0.01.
Nrf2-Governed Glutathione Synthesis Pathway Is Up-Regulated in IDH1-Mutated Cells.
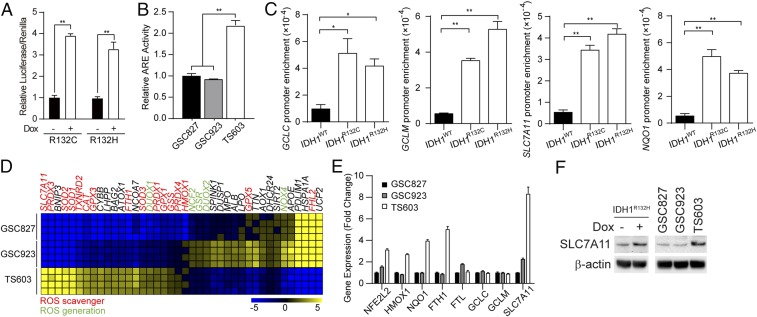
Cellular oxidative stress is strictly regulated by the Nrf2-governed gene transcription (27, 28). In accordance with the elevation of cellular oxidative stress, we recorded enhanced Nrf2 transcriptional activity by an ARE luciferase reporter assay in both IDH1-induced U251 cells and patient-derived BTIC. Cells harboring IDH1 mutations exhibited 2- to threefold higher levels of Nrf2 transcriptional activity (Fig. 2 A and B, **P < 0.01). This finding is further confirmed by chromatin immunoprecipitation assay (ChIP), showing that Nrf2 exhibited higher affinity to the promoter regions of antioxidant genes, such as GCLC, GCLM, SLC7A11, and NQO1, in IDH1-mutated cells (Fig. 2C, *P < 0.05, **P < 0.01). Furthermore, gene expression analysis showed that in IDH1-mutated BTIC TS603, ROS scavenger genes were up-regulated, suggesting the Nrf2-governed antioxidant pathway is activated. Notably, the cystine-glutamate antiporter xCT (SLC7A11) showed the most remarkable up-regulation in IDH1-mutated BTIC, which increased sevenfold compared with IDH1 wild-type BTIC (Fig. 2 D and E). Finally, immunoblotting confirmed that the protein expression of SLC7A11 was significantly up-regulated in induced IDH1-mutated U251 cells as well as IDH1-mutated BTIC TS603 cells (Fig. 2F).
Fig. 2.
Enhanced Nrf2-driven pathway activity in IDH1-mutated cells. (A) Relative luciferase activity of Dox-induced IDH1-mutated U251 cells transfected with the pGL4.37 ARE-luciferase reporter vector. The luciferase activity was normalized by Renilla luciferase. n = 3. **P < 0.01. (B) Nrf2 activity was determined using a Cignal ARE-luciferase reporter assay in BTIC lines. n = 3. **P < 0.01. (C) Promoter affinity of Nrf2 was measured by ChIP-PCR in U251 WT, R132C, and R132H cells. n = 3. **P < 0.01, *P < 0.05. (D) Gene expression assay quantified ROS generating and scavenging genes in BTIC lines. (E) qRT-PCR analysis of Nrf2-associated expression in BTIC lines. Each gene was normalized to its control. n = 3. (F) Immunoblotting of SLC7A11 expression in BTIC and Dox-induced IDH1-mutated U251 cells. β-actin was used as an internal control.
Glutathione de Novo Synthesis Supports Cellular Homeostasis in IDH1-Mutated Cells.
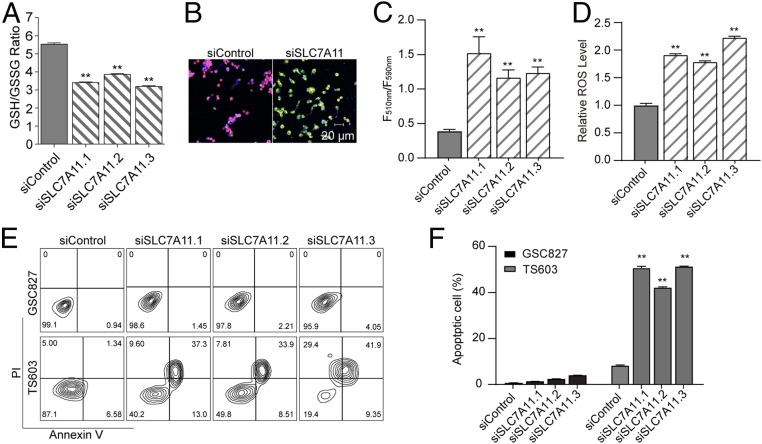
The xCT cystine/glutamate transporter (SLC7A11) is a member of heterodimeric sodium-dependent amino acid transporter that imports cystine and exports glutamate. Several hallmark investigations showed that the SLC7A11 fuels glutathione de novo synthesis through internalizing cysteine from the microenvironment (29). To understand the role of SLC7A11 in IDH1-mutated cells, we investigated the alteration of glutathione pool and redox status in IDH1-mutated cells. With small interfering RNA suppressing SLC7A11 expression, a significant decrease of both cellular GSH/GSSG ratio and the total pool of glutathione was observed (Fig. 3A, **P < 0.01; SI Appendix, Fig. S1 A and B). Furthermore, genetic silencing of SLC7A11 led to enhanced lipid peroxidation in IDH1-mutated cells (Fig. 3 B and C, **P < 0.01). The ROS quantification assay confirmed that the loss of SLC7A11 expression resulted in substantial ROS accumulation in IDH1-mutated cells (Fig. 3D, **P < 0.01; SI Appendix, Fig. S1C). Additionally, genetic silencing of SLC7A11 resulted in apoptotic changes in IDH1-mutated cells, whereas this phenomenon is not seen in cells with wild-type IDH1 (Fig. 3 E and F, **P < 0.01; SI Appendix, Fig. S1 D and E). These findings suggest that SLC7A11 played vital roles in supporting glutathione de novo synthesis, which protects IDH1-mutated cells from apoptotic cell death.
Fig. 3.
SLC7A11 plays a key role in the redox balance and survival of IDH1-mutated cells. (A) GSH/GSSG ratio was measured using GSH/GSSG-Glo assay in TS603 cells with siRNA targeting SLC7A11. n = 3. **P < 0.01. (B) Lipid peroxidation assay of TS603 cells with siRNA targeting SLC7A11. Before lipid peroxidation, the emission fluorescence showed at 590 nm (red). After lipid peroxidation, the emission fluorescence showed at 510 nm (green). (C) Lipid peroxidation was quantified using the ratio of F510 nm to F590 nm. n = 5. **P < 0.01. (D) ROS-Glo H2O2 assay of TS603 cells with siRNA targeting SLC7A11. (E) Annexin V/PI apoptosis assay of BTIC lines (IDH1WT GSC827 and IDH1R132H TS603 cells) with siRNA targeting SLC7A11. (F) Quantification of apoptosis assay. n = 3. **P < 0.01.
Triptolide Is a Potent Nrf2 Inhibitor that Compromises Glutathione Synthesis.
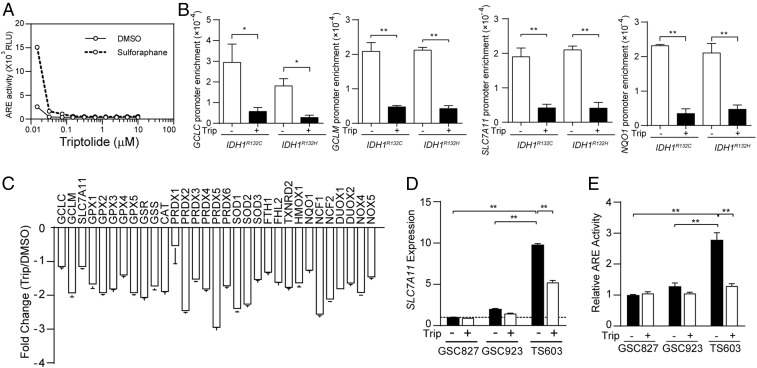
Considering the critical role of glutathione de novo synthesis in IDH1-mutated glioma cells, targeting the Nrf2/SLC7A11 axis may be effective against this type of malignancy. Several chemical inhibitors for Nrf2 or SLC7A11 have been reported in previous studies. For example, brusatol, a plant-derived quassinoid, has been reported for its potent inhibitory effect on Nrf2 (30), whereas the substantial molecule weight and toxicity profile prevents further clinical applications of this compound. In the present study, we discovered that triptolide, a diterpenoid epoxide from Tripterygium wilfordii, exhibited robust inhibition to ARE-driven transcriptional activity at nanomolar levels (Fig. 4A). A ChIP assay confirmed that the affinity of Nrf2 to the promoter regions of GCLC, GCLM, SLC7A11, and NQO1 was strikingly compromised with the presence of triptolide (Fig. 4B, *P < 0.05, **P < 0.01). Gene expression analysis in TS603 showed that most of Nrf2 downstream genes were affected by triptolide, suggesting that triptolide is a broad-spectrum inhibitor for Nrf2-governed gene transcription (Fig. 4C). Consistently, the messenger ribonucleic acid (mRNA) level of SLC7A11 decreased in IDH1-mutated BTIC TS603 and TB096 after triptolide treatment, but not the IDH1 wild-type BTICs (Fig. 4D, **P < 0.01; SI Appendix, Fig. S2A). Similarly, triptolide exhibited a potent inhibitory effect on Nrf2 transcriptional activity in IDH1-mutated BTIC TS603 and TB096 (Fig. 4E, **P < 0.01; SI Appendix, Fig. S2B).
Fig. 4.
Triptolide suppresses glutathione metabolism via the Nrf2 blockade. (A) ARE luciferase reporter assay in TS603 with triptolide treatment for 24 h. Sulforaphane was used to induce Nrf2 activation. (B) The affinity of Nrf2 to the promoter of GCLC, GCLM, SLC7A11 was determined by ChIP-PCR assay in Dox-induced IDH1-mutated U251 cells treated with DMSO or 30 nM triptolide. NQO1 promoter was used as a positive control. *P < 0.05, **P < 0.01. (C) Gene expression assay quantified ROS generating and scavenging genes in TS603 cells treated with 30 nM triptolide for 24 h. n = 4. Data are presented as mean ± SEM. (D) SLC7A11 expression was quantified by qRT-PCR in BTIC lines treated with DMSO or 30 nM triptolide. n = 3. **P < 0.01. (E) Nrf2 transcriptional activity was measured by ARE-luciferase reporter assay in BTIC lines (IDH1WT GSC827, IDH1WT GSC923, and IDH1R132H TS603 cells). Cells were treated with DMSO or 30 nM triptolide for 24 h. n = 3. **P < 0.01.
Triptolide Induces Oxidative Damage in IDH1-Mutated Cells.
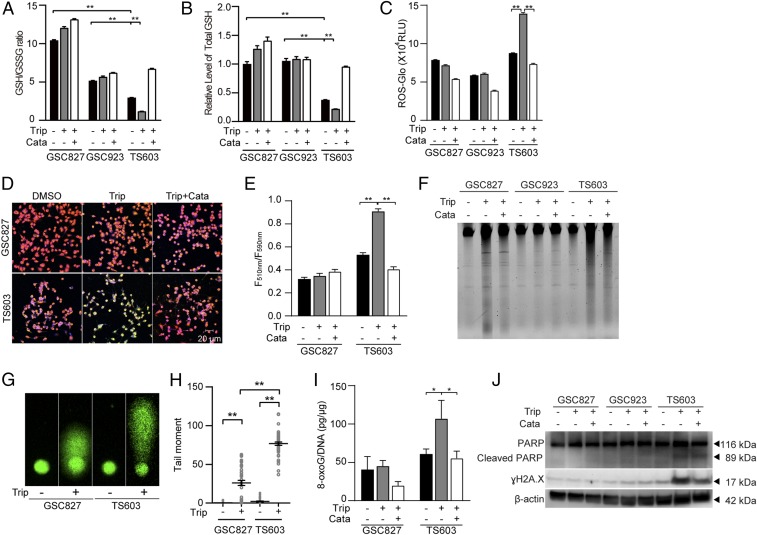
To further analyze the impact of triptolide in IDH1-mutated cells, we measured glutathione levels in BTICs under triptolide treatment. The ratio of GSH/GSSG, as well as total amount of glutathione, were depleted by triptolide treatment in IDH1-mutated BTIC TS603 cells, but the effect was minimal in IDH1 wild-type counterparts GS827 and GSC923 cells (Fig. 5 A and B, **P < 0.01). The depletion of the glutathione pool was in accordance with substantially elevated ROS, as well as remarkable lipid peroxidation (Fig. 5 C–E and SI Appendix, Fig. S2C). Furthermore, the accumulation of ROS translated to oxidative damage in IDH1-mutated cells. DNA electrophoresis and comet assay revealed triptolide-induced DNA damage and DNA fragmentation in IDH1-mutated cells, whereas IDH1 wild-type cells exhibited a minimal response to the same treatment (Fig. 5 F–H, **P < 0.01). We also confirmed DNA oxidation and DNA damage in IDH1-mutated cells, evidenced by elevated 8-oxoG, PARP cleavage, and γH2A.X staining (Fig. 5 I and J, *P < 0.05; SI Appendix, Fig. S2D).
Fig. 5.
Triptolide disrupts ROS homeostasis and leads to oxidative DNA damage in IDH1-mutated cells. (A) GSH/GSSG ratio and (B) Total GSH quantification were measured using GSH/GSSG-Glo assay in BTIC lines (IDH1WT GSC827, IDH1WT GSC923, and IDH1R132H TS603 cells). n = 3. **P < 0.01. (C) ROS-Glo H2O2 assay of BTIC lines. n = 3. **P < 0.01. (D) Lipid peroxidation assay of BTIC lines. Before lipid peroxidation, the emission fluorescence showed at 590 nm (red). After lipid peroxidation, the emission fluorescence showed at 510 nm (green). (E) Lipid peroxidation was quantified using the ratio of F510 nm to F590 nm. n = 5. **P < 0.01. (F) Total genomic DNA electrophoresis analysis of BTIC lines after triptolide treatment. (G) The DNA fragmentation was measured by comet assay in BTIC lines after triptolide treatment. (H) Quantification of the tail moments in G. n = 50. **P < 0.01. (I) The 8-oxoG level was measured by oxidative DNA damage ELISA in BTIC lines. n = 3. *P < 0.05. (J) DNA damage was measured by immunoblotting of γH2A.X and cleaved PARP in BTIC lines. In this part, cells were treated with DMSO, 30 nM triptolide, or triptolide combined with 0.1 mg/mL catalase.
Triptolide Exhibits Selective Cytotoxicity for IDH1-Mutated Cells.
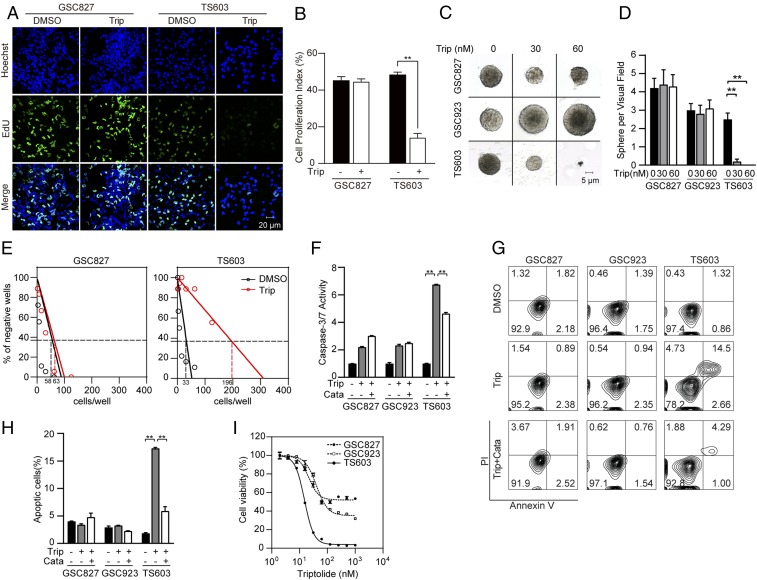
In accordance with the elevated oxidative damage, our finding revealed that triptolide effectively suppresses patient-derived IDH1-mutated glioma cells. An EdU incorporation assay confirmed that triptolide reduced cellular proliferation by 75% in IDH1-mutated cells, whereas minimal reduction was observed in IDH1 wild-type cells (Fig. 6 A and B, **P < 0.01). Moreover, sphere-formation assays showed that triptolide treatment significantly reduced both sphere size and number in IDH1-mutated, but not IDH1 wild-type cells (Fig. 6 C and D, **P < 0.01). A limited dilution assay showed decreased colony formation capability in IDH1-mutated BTIC TS603 cells (Fig. 6E; 0.37 intercept, dimethyl sulfoxide, DMSO = 33 cells per well, triptolide = 196 cells per well), but not in IDH1 wild-type BTIC GSC827 cells (0.37 intercept, DMSO = 58 cells per well, triptolide = 63 cells per well). A caspase 3/7 activity assay showed that triptolide treatment induced remarkable apoptosis in IDH1-mutated cells, which can be rescued by ROS scavenger catalase (Fig. 6F, **P < 0.01). Similarly, an annexin V/PI flow cytometry assay recorded more apoptotic cells in IDH1-mutated cells compared with IDH1 wild-type cells after triptolide treatment. Moreover, cytotoxicity could be reduced by combined treatment with catalase (Fig. 6 G and H, **P < 0.01; SI Appendix, Fig. S2 E and F). These findings indicate that the disruption of redox homeostasis is one of the principal causes of reduced cellular proliferation and apoptotic cell death. Furthermore, a cell viability assay showed that IDH1-mutated cells were more vulnerable to triptolide treatment, with an IC50 of 15 nM, as opposed to an IC50 of 60 nM for IDH1 wild-type cells, indicating that triptolide exhibited stronger cytotoxicity for IDH1-mutated cells (Fig. 6I and SI Appendix, Fig. S2G).
Fig. 6.
Triptolide selectively suppresses IDH1-mutated cells. (A) EdU (green) incorporation assay of BTIC lines (IDH1WT GSC827 and IDH1R132H TS603 cells) treated with DMSO or 30 nM triptolide for 24 h. Cell nuclei were labeled with Hoechst 33342 (blue). Bar = 20 μm. (B) Quantification of EdU positive cells in A. n = 10. **P < 0.01. (C) Sphere formation assay of BTIC lines treated with DMSO or triptolide. Bar = 5 μm. (D) The number of spheres was recorded. n = 10. **P < 0.01. (E) Limited dilution assay of BTIC lines treated with DMSO or 30 nM triptolide for 3 wk. The 0.37 intercept was labeled in dashed lines. (F) Caspase 3/7-Glo assay of BTIC lines with DMSO, 30 nM triptolide, or triptolide combined with 0.1 mg/mL catalase for 24 h. Luminescence was normalized to protein quantification. n = 3. Data are presented as mean ± SEM. **P < 0.01. (G) Annexin V/PI apoptosis analysis of BTIC lines with DMSO, 30 nM triptolide, or combined with 0.1 mg/mL catalase for 96 h. (H) Quantification of apoptotic cells from G. n = 3. **P < 0.01. (I) Cell viability assay shows the dose–response curve of triptolide in BTIC lines.
Triptolide as a Therapeutic Approach for IDH1-Mutated Malignancy via Nrf2-Driven Glutathione Metabolism.
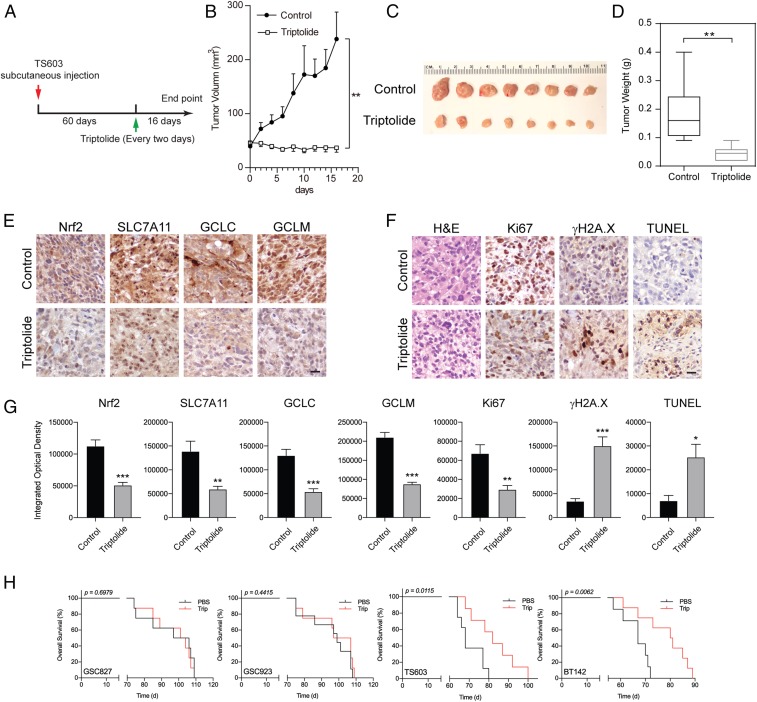
Our in vitro findings suggest that triptolide resulted in effective cytotoxicity in IDH1-mutated glioma cells through a compromised Nrf2-driven glutathione synthetic pathway. To further evaluate whether triptolide could be used for cancer therapeutics, we established a preclinical model with IDH1-mutated glioma xenograft. NSG mice were s.c. injected with TS603 cells and randomly allocated into two groups. Mice have received 0.5 mg/kg triptolide through i.p. every 2 d for a total of 16 d after tumor formation (Fig. 7A). We found that triptolide significantly suppressed the growth of TS603 xenograft compared with the solvent control (Fig. 7 B–D, **P < 0.01). Moreover, immunohistochemistry staining showed that triptolide inhibited the expression of Nrf2 and glutathione metabolism-related genes, such as SLC7A11, GCLC, and GCLM (Fig. 7E). We also recorded decreased proliferation marker Ki67 in triptolide-treated xenograft tissues, while DNA damage marker γH2A.X was found elevated. Increased TUNEL signal suggested the elevation of cellular apoptosis by triptolide treatment (Fig. 7 F and G). Finally, through orthopotic xenograft models, we showed that triptolide established strong suppressive effects in patient-derived, IDH1-mutated intracranial tumors (Fig. 7H. Overall survival: TS603, phosphate-buffered saline, PBS = 68 d, Trip = 82 d, *p = 0.0115; BT142, PBS = 67 d, Trip = 80.5 d, **p = 0.0062). In contrast, triptolide showed minimal benefit to the disease outcomes in both IDH1 wild-type intracranial xenografts (Overall survival: GSC827, PBS = 101.5 d, Trip = 102.5 d, P = 0.6979; GSC923, PBS = 99 d, Trip = 102 d, P = 0.4415). Overall, the data acquired from the orthotopic preclinical models strongly suggest the therapeutic effect of triptolide in IDH1-mutated malignancies.
Fig. 7.
Triptolide suppresses IDH1-mutated xenograft expansion in vivo. (A) Schematic illustration for the xenograft modeling and triptolide treatment. (B) The tumor growth curve shows that triptolide suppresses tumor growth by measuring tumor volume. n = 8. **P < 0.01. (C) Tumor imaging shows that triptolide treatment suppressed IDH1-mutated xenograft in vivo. (D) Quantification of tumor weight shown in C. n = 8. **P < 0.01. (E) Immunohistochemistry staining of the expression of Nrf2, SLC7A11m GCLC, and GCLM in TS603 xenograft tissues after mice received triptolide treatment. (F) immunohistochemistry staining of H&E, Ki67, γH2A.X, and TUNEL assay in TS603 xenograft tissues after mice received triptolide. Bar = 50 μm. (G) Quantification of integrated optical density of immunohistochemistry staining in E and F. n = 6. ***P < 0.001, **P < 0.01, *P < 0.05. (H) Kaplan-Meier analysis of intracranial xenografts in mice that received triptolide treatment.
Discussion
In the current study, we identified Nrf2 blockade using triptolide as a selective therapeutic for IDH1-mutated malignancies. We showed that the IDH1 mutant neomorphic activity prompted the accumulation of ROS. Cancer cells develop an addiction to Nrf2-governed pathways, such as the glutathione de novo synthesis. Moreover, we found that triptolide potently suppressed Nrf2 protein stability and transcriptional activity, which established synthetic lethality with intrinsic oxidative stress in IDH1-mutated cells. Our results showed that triptolide exhibited selective cytotoxicity to patient-derived IDH1-mutated cells in vitro and in vivo, with significant suppression of antioxidant pathways and tumor expansion.
The neomorphic activity of the IDH1 mutant causes numerous impacts in cellular biology, including metabolism reprogramming, disrupted redox homeostasis, and genome-wide hypermethylation. Our previous study demonstrated that IDH1-mutated glioma cells exhibited high levels of ROS stress compared with IDH1 wild-type cells. Accordingly, Nrf2-associated ROS scavenging pathways, like glutathione de novo synthesis, are highly activated to maintain redox homeostasis and cell survival in IDH1-mutated cells (Fig. 6) (14, 26, 31). Under the basal condition, Nrf2 is regulated by Kelch-like ECH-associated protein 1 (Keap1), which binds to Nrf2 as a negative regulator to form CuI3-Rbx1-E3 complex, leading to Nrf2 ubiquitination and proteasomal degradation. With the presence of oxidative stress, such as ROS and reactive nitrogen species, the conformational change of Keap1 blocks the ubiquitination of Nrf2, which led to the activation of Nrf2 (25). We believed that the activation of Nrf2 is a consequence of ROS overload, as Nrf2 activity was abolished by ROS scavengers in IDH1-mutated cells (Fig. 5). However, the correlation between 2-HG–induced hypermethylation in IDH1-mutated cells and the activation of Nrf2 pathways needs further investigations.
Constitutive activation of Nrf2 was observed in a variety of malignancies, including breast, liver, lung, bladder, pancreatic, ovarian, and endometrial cancers (32–38). Nrf2 serves numerous supportive roles for cancer cells, including enhanced cancer progression and therapy resistance (33, 35, 39, 40). Several seminal studies proposed that the Nrf2 blockade has synergistic toxicity with chemotherapy, resulting in attenuated cancer progression (41–43). We and several other investigations indicated that triptolide shows the potent suppressive effect on Nrf2 transcriptional activity (Fig. 4) (44–46). Our results showed that triptolide induced potent cytotoxicity in IDH1-mutated cells in vitro and in vivo (Figs. 6 and 7), which likely resulted from the synthetic lethality effect derived from IDH1 neomorphic activity and Nrf2 blockade. We found that the nanomolar level of triptolide mono treatment was sufficient to induce severe DNA fragmentation and oxidative damage, which led to cell apoptosis in IDH1-mutated cells (Figs. 5 and 6). Importantly, the cytotoxicity was found much less often in non-IDH1-mutated cells, suggesting that triptolide may serve as a selective therapy for IDH1-mutated cancers, with less cytotoxicity to other somatic cells with intact metabolic pathways. Several previous reports showed that triptolide might suppress several oncogenes such as NF-κB and c-Myc (47–49), suggesting that multiple mechanisms may be involved in triptolide-induced cytotoxicity. Besides these seminal findings, our study demonstrates triptolide acts on the governing transcriptional factor Nrf2 and synergizes with an intrinsic deficiency in cancer cells, such as IDH1 mutant-associated metabolic depletion.
Overall, the present study provides a proof-of-concept preclinical study suggesting the potential value of triptolide as a selective therapy for IDH1 mutated malignancies. However, limitations such as the unclear mechanism of triptolide-induced Nrf2 blockade, as well as the uncertain specificity of triptolide, still need to be further investigated. The side effects and blood–brain barrier penetration should also be validated to translate our current study into clinical applications.
Methods
Cell Culture and Reagents.
The U251 MG cell line was obtained from Sigma-Aldrich. Cells were cultured in DMEM/F12 (Life Technologies) supplemented with 10% fetal bovine serum (FBS, Cellgro) and 1% penicillin/streptomycin (Invitrogen).
The BTIC lines GSC827 and GSC923 were reported previously (50). TB096 was kindly provided by Hai Yan, Duke School of Medicine, Durham, NC (51). TS603 cells was kindly provided by Timothy Chan, Memorial Sloan Kettering Cancer Center, New York, NY (17). GSC827, GSC923, BT142, and TS603 cells were cultured in the NBE medium, as previously described (52). The TB096 cell line was cultured in media containing a 1:1 combination of (complete NeuroCult media supplemented with 20 ng/mL EGF, 10 ng/mL FGF, and 2 μg/mL heparin) : (DMEM + 10% Invitrogen FBS). Triptolide (MedChemExpress), brusatol (Sigma), and AGI-5198 (Selleckchem) were dissolved in DMSO. ROS scavenger catalase was purchased from Sigma.
Lentivirus Production and Stable Cell Line Generation.
The doxycycline-inducible pLVX-TetOne-Puro vector (Clontech) carrying IDH1 R132C or R132H variants was transduced into U251 MG by lentivirus transduction. U251 cells with stably expressed wild-type IDH1, R132C, or R132H variants were generated using lentivirus transduction, as previously described (14).
Luciferase Reporter Assay.
Nrf2-driven transcriptional activity was determined using a reporter plasmid pGL4.37 [luc2P/ARE/Hygro] or Cignal Lenti ARE luciferase reporter assay (QIAGEN) with Dual-Luciferase Reporter Assay System (Promega) as previously described (53, 54). Nine hundred nanograms of the reporter plasmid and 100 nanograms of pRL-TK plasmid were transfected to 100,000 cells by Lipofectamine 2000 (Invitrogen). The luminescence signal was recorded using a microplate reader and normalized to the protein or Renilla luciferase activity.
Animal Modeling and Treatment.
All animal experiments were approved by the National Cancer Institute (NCI) Animal Use and Care Committee. TS603 cells (5 × 106) were s.c. injected into 6-wk-old nonobese diabetic scid gamma mouse (NSG) mice. When the tumor sizes were above 50 mm3, mice were randomly divided into two groups and treated intraperitoneally with DMSO or triptolide (0.5 mg/kg) every other day. Each group contained eight mice. Tumor size was measured using Vernier calipers. The mice were sacrificed at day 16 after treatment, and tumors were harvested for analysis. For intracranial xenograft model, 5 × 105 cells were injected into the cerebral cortex of NSG mice. The mice were randomized into two groups (n = 7 or 8 for each group) 30 d after injection. The mice were treated intraperitoneally with triptolide (0.5 mg/kg) every other day for 16 d. The mice were sacrificed when they reach the experimental end point. Kaplan-Meier analysis was used to analyze the mice survival over time.
Statistical Analysis.
Statistical analysis was performed with GraphPad Prism software (Version 6.01). A one-way ANOVA or Student t test was applied for statistical comparisons. All statistical tests were two-tailed. Results are shown as mean ± SEM, and P < 0.05 was considered statistically significant.
Data Availability Statement.
All data discussed in the paper can be found within the main text and SI Appendix.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NCI, and the foundations (81472374) from the National Natural Science Foundation of China. We appreciate Dr. Mark R. Gilbert, Dr. Wei Zhang, Dr. Hua Song, and Ms. Dionne L. Davis for providing essential laboratory resources and techniques.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913633117/-/DCSupplemental.
References
- 1.Yan H., et al. , IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki H., et al. , Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 47, 458–468 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Amary M. F., et al. , IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 224, 334–343 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Borger D. R., et al. , Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17, 72–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farshidfar F., et al. ; Cancer Genome Atlas Network , Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 19, 2878–2880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel J. P., et al. , Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366, 1079–1089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alp P. R., Newsholme E. A., Zammit V. A., Activities of citrate synthase and NAD+-linked and NADP+-linked isocitrate dehydrogenase in muscle from vertebrates and invertebrates. Biochem. J. 154, 689–700 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L., et al. , Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W., et al. , Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koivunen P., et al. , Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turcan S., et al. , IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latini A., et al. , D-2-hydroxyglutaric acid induces oxidative stress in cerebral cortex of young rats. Eur. J. Neurosci. 17, 2017–2022 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Shi J., et al. , Decreasing GSH and increasing ROS in chemosensitivity gliomas with IDH1 mutation. Tumour Biol. 36, 655–662 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , Targeting IDH1-mutated malignancies with NRF2 blockade. J. Natl. Cancer Inst. 111, 1033–1041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y., et al. , Chemosensitivity of IDH1-mutated gliomas due to an impairment in PARP1-mediated DNA repair. Cancer Res. 77, 1709–1718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kernytsky A., et al. , IDH2 mutation-induced histone and DNA hypermethylation is progressively reversed by small-molecule inhibition. Blood 125, 296–303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohle D., et al. , An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., et al. , Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 340, 622–626 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Sulkowski P. L., et al. , 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 9, eaal2463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molenaar R. J., et al. , Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res. 75, 4790–4802 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Tateishi K., et al. , Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell 28, 773–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seltzer M. J., et al. , Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 70, 8981–8987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borodovsky A., et al. , 5-azacytidine reduces methylation, promotes differentiation and induces tumor regression in a patient-derived IDH1 mutant glioma xenograft. Oncotarget 4, 1737–1747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motohashi H., Yamamoto M., Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549–557 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi M., Yamamoto M., Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 7, 385–394 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Cai S. J., Liu Y., Han S., Yang C., Brusatol, an NRF2 inhibitor for future cancer therapeutic. Cell Biosci. 9, 45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh K., et al. , Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaramillo M. C., Zhang D. D., The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih A. Y., et al. , Cystine/glutamate exchange modulates glutathione supply for neuroprotection from oxidative stress and cell proliferation. J. Neurosci. 26, 10514–10523 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren D., et al. , Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. U.S.A. 108, 1433–1438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang X., et al. , Blockade of glutathione metabolism in IDH1-mutated glioma. Mol. Cancer Ther. 19, 221–230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh A., et al. , Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3, e420 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata T., et al. , Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135, 1358–1368, 1368.e1-4 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Wang R., et al. , Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 373, 151–154 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Jiang T., et al. , High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 70, 5486–5496 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y. R., et al. , Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 220, 446–451 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Solis L. M., et al. , Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 16, 3743–3753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A., et al. , RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 68, 7975–7984 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang P., et al. , Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 9, 336–346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X. J., et al. , Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J., et al. , An overview of chemical inhibitors of the Nrf2-ARE signaling pathway and their potential applications in cancer therapy. Free Radic. Biol. Med. 99, 544–556 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Panieri E., Saso L., Potential applications of NRF2 inhibitors in cancer therapy. Oxid. Med. Cell. Longev. 2019, 8592348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telkoparan-Akillilar P., Suzen S., Saso L., Pharmacological applications of Nrf2 inhibitors as potential antineoplastic drugs. Int. J. Mol. Sci. 20, E2025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan J., et al. , Triptolide induces oxidative damage in NRK-52E cells through facilitating Nrf2 degradation by ubiquitination via the GSK-3β/Fyn pathway. Toxicol. In Vitro 58, 187–194 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Chen F., et al. , Triptolide, a Chinese herbal extract, enhances drug sensitivity of resistant myeloid leukemia cell lines through downregulation of HIF-1α and Nrf2. Pharmacogenomics 14, 1305–1317 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Zhu J., et al. , Triptolide enhances chemotherapeutic efficacy of antitumor drugs in non-small-cell lung cancer cells by inhibiting Nrf2-ARE activity. Toxicol. Appl. Pharmacol. 358, 1–9 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Lee K. Y., Park J. S., Jee Y. K., Rosen G. D., Triptolide sensitizes lung cancer cells to TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by inhibition of NF-kappaB activation. Exp. Mol. Med. 34, 462–468 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Yang S., et al. , Triptolide inhibits the growth and metastasis of solid tumors. Mol. Cancer Ther. 2, 65–72 (2003). [PubMed] [Google Scholar]
- 49.Ding X., Zhou X., Jiang B., Zhao Q., Zhou G., Triptolide suppresses proliferation, hypoxia-inducible factor-1α and c-Myc expression in pancreatic cancer cells. Mol. Med. Rep. 12, 4508–4513 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Ene C. I., et al. , Histone demethylase Jumonji D3 (JMJD3) as a tumor suppressor by regulating p53 protein nuclear stabilization. PLoS One 7, e51407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moure C. J., et al. , CRISPR editing of mutant IDH1 R132H induces a CpG methylation-low state in patient-derived glioma models of G-CIMP. Mol. Cancer Res. 17, 2042–2050 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang C., et al. , β-Catenin signaling initiates the activation of astrocytes and its dysregulation contributes to the pathogenesis of astrocytomas. Proc. Natl. Acad. Sci. U.S.A. 109, 6963–6968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y., et al. , Therapeutic targeting of SDHB-mutated pheochromocytoma/paraganglioma with pharmacologic ascorbic acid. Clin. Cancer Res. clincanres.2335.2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y., et al. , Targeting NRF2-governed glutathione synthesis for SDHB-mutated pheochromocytoma and paraganglioma. Cancers (Basel) 12, 280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper can be found within the main text and SI Appendix.