Significance
Autism is a complex neurodevelopmental disorder whose causative mechanisms are unclear. Taking advantage of a unique cohort with recessively inherited autism, we identified six families with biallelic mutation of the neuronal-specific subunit of the BAF complex, ACTL6B (also known as BAF53b). Relative to all other genes, ACTL6B was the most statistically significant mutated gene in the recessive autism cohort. We describe autism-relevant phenotypes in human brain organoids and in mouse and fly models. We foresee the outcomes from this study will be the following: 1) a link between neuronal activity-dependent transcriptional repression and autism; 2) a characterization of mouse and fly models to study ACTL6B mutant autism; and 3) an understanding the role of ACTL6B and nBAF complexes in neuronal transcriptional regulation.
Keywords: autism, mouse model, recessive, BAF, activity dependent
Abstract
Synaptic activity in neurons leads to the rapid activation of genes involved in mammalian behavior. ATP-dependent chromatin remodelers such as the BAF complex contribute to these responses and are generally thought to activate transcription. However, the mechanisms keeping such “early activation” genes silent have been a mystery. In the course of investigating Mendelian recessive autism, we identified six families with segregating loss-of-function mutations in the neuronal BAF (nBAF) subunit ACTL6B (originally named BAF53b). Accordingly, ACTL6B was the most significantly mutated gene in the Simons Recessive Autism Cohort. At least 14 subunits of the nBAF complex are mutated in autism, collectively making it a major contributor to autism spectrum disorder (ASD). Patient mutations destabilized ACTL6B protein in neurons and rerouted dendrites to the wrong glomerulus in the fly olfactory system. Humans and mice lacking ACTL6B showed corpus callosum hypoplasia, indicating a conserved role for ACTL6B in facilitating neural connectivity. Actl6b knockout mice on two genetic backgrounds exhibited ASD-related behaviors, including social and memory impairments, repetitive behaviors, and hyperactivity. Surprisingly, mutation of Actl6b relieved repression of early response genes including AP1 transcription factors (Fos, Fosl2, Fosb, and Junb), increased chromatin accessibility at AP1 binding sites, and transcriptional changes in late response genes associated with early response transcription factor activity. ACTL6B loss is thus an important cause of recessive ASD, with impaired neuron-specific chromatin repression indicated as a potential mechanism.
Autism spectrum disorder (ASD) represents a heterogeneous group of neurodevelopmental disorders that are characterized by social deficits and restricted or repetitive behaviors, and affect ∼1% of children worldwide (1, 2). The heritability of ASD is estimated to be ∼80% (3), implicating genetic mutation as the prominent cause of autism. Indeed, exome-sequencing studies have identified hundreds of genetic mutations that substantially increase ASD risk (4). Among the most frequently mutated genes in ASD are subunits of the mammalian SWI/SNF (BAF) ATP-dependent chromatin remodeling complex (4). BAF complexes facilitate dynamic changes in gene expression by controlling DNA accessibility to the transcriptional machinery (5). To accomplish this, BAF complexes mobilize nucleosomes, evict polycomb repressive complexes, and recruit type II topoisomerases that decatenate DNA (6–9).
Although present in all cells, BAF complexes orchestrate cell type-specific functions through combinatorial assembly of ∼15 subunits from the products of 29 genes (5). During neural development, exit from the cell cycle is accompanied by BAF subunit exchange: neural progenitor (np) subunits ACTL6A (BAF53a), DPF2/PHF10 (BAF45a/d), and SS18 are exchanged for neuron-specific (n) subunits ACTL6B (BAF53b), DPF1/3 (BAF45b/c), and SS18L1 (CREST), respectively (10–13). Subunit exchange is critical for neuronal function, as genetic deletion of either ACTL6B or SS18L1 impairs activity-dependent dendritic arborization (14, 15). Furthermore, expression of the two microRNAs, miR9* and miR124, which control BAF subunit switching, is sufficient to convert fibroblasts into neurons (13).
Recent findings link mutations in nearly every constitutive member of the BAF complex to ASD or intellectual disability (ID), including syndromic forms such as Coffin–Siris and Nicolaides–Baraitser syndromes (16–18). Implicated subunits include the following (protein/gene): BAF250b/ARID1B, BAF250a/ARID1A, BAF200/ARID2, BCL11A/BCL11A, BRG1/SMARCA4, BRM/SMARCA2, BAF155/SMARCC1, BAF170/SMARCC2, BAF45a/PHF10, BAF45d/DPF2 BAF47/SMARCB1, BAF57/SMARCE1, BAF53a/ACTL6A, BAF53b/ACTL6B, BAF60a/SMARCD1, and β-actin/ACTB (17, 19–29). BAF mutant forms of ASD share overlapping clinical features such as corpus callosal hypoplasia, epilepsy, ID, lack of speech, craniofacial abnormalities, developmental delays, and fifth-digit shortening (17). The mechanisms through which BAF subunit mutations give rise to ASD are unclear.
Results
Biallelic Inherited Mutations in ACTL6B Cause Recessive Autism.
Phenotypic and genotypic heterogeneity in ASD make this a challenging disorder to study at the molecular level. Distinct molecular mechanisms may underlie social deficits and repetitive behaviors, as well as ID, epilepsy, sleep and mood disorders, hyperactivity, and systemic issues that are frequently comorbid with autism (30). Because autism mutations are predominantly de novo and can occur in genes that function in a variety of tissues during development (4, 16, 21, 31), it can also be difficult to define the relevant developmental and cellular contexts in which to study ASD mechanisms. Finally, many ASD mutations increase susceptibility but do not consistently cause autism phenotypes in humans or in animal models (32, 33).
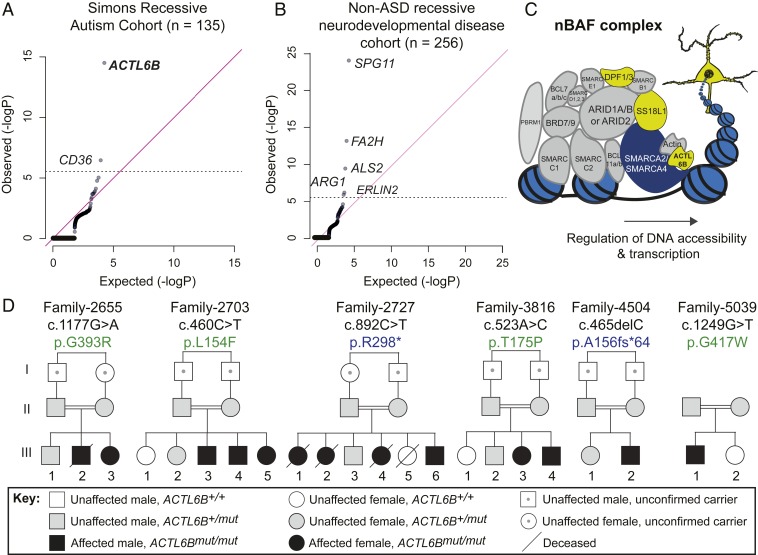
One strategy to uncover causative mechanisms in ASD is to study families with recessively inherited autism, since it is rare for two copies of a mutant gene to segregate perfectly with recessive autism by chance. Thus, we studied 135 ASD probands from consanguineous marriages recruited for the Simons Recessive Autism Cohort (SRAC) (34–37). Genomic DNA underwent whole-exome sequencing and were compared with a cohort of 256 controls with recessive neurodevelopmental disease (NDD) without ASD (SI Appendix, Supplementary Text). To identify significantly mutated genes, we generated quartile–quartile (Q-Q) plots comparing the observed to expected number of damaging recessive genotypes (deletion, frameshift/stop, or damaging missense) in the SRAC vs. non-ASD recessive NDD cohorts. When considering all coding genes, only ACTL6B, a subunit of the nBAF complex, and CD36, a fatty acid translocase and scavenger receptor (38), showed genome-wide significance for mutations in the SRAC (Fig. 1 A and C). Limiting to just loss of function (LoF)-intolerant genes (i.e., those with pLI ≥ 0.9) left only ACTL6B with genome-wide significance (P < 10−14) (SI Appendix, Fig. S1A). A similar statistical analysis was conducted on the non-ASD NDD cohort of 256 individuals of similar genetic background, matched for consanguinity. The non-ASD NDD cohort showed enrichment for genes previously implicated in non-ASD NDD (39) but not ACTL6B (Fig. 1B and SI Appendix, Fig. S1B). To test whether ACTL6B mutations were enriched in a genetically distinct recessive autism cohort, we rank ordered 409 genes with recessive missense variants in the Autism Sequencing Consortium by their RAFT P value and found that ACTL6B was the sixth most significantly mutated gene (SI Appendix, Fig. S1C) (25). Individuals with recessive ASD-like phenotypes linked to biallelic ACTL6B mutations have also been identified in recent case studies (29, 40, 41). Cumulatively, these findings implicate mutations in ACTL6B as an important cause of recessive autism.
Fig. 1.
Biallelic mutations in ACTL6B cause recessive autism. (A) Q-Q plot showing the observed/expected number of mutations for all coding genes in the SRAC. ACTL6B and CD36 were significantly mutated. (B) Q-Q plot showing the observed/expected number of mutations for all coding genes in a genetically matched non-ASD recessive neurodevelopmental cohort, where ACTL6B was not found to be enriched. (C) ACTL6B encodes a tissue-restricted subunit of the neuronal (nBAF) BAF complex. Representation of the multisubunit nBAF complex containing ubiquitously expressed subunits (gray), a core ATPase subunit (dark blue), and neuronal-specific subunits (yellow) including ACTL6B in bold. The balls on a string represent nucleosomes. (D) Recessive ASD inheritance with ACTL6B mutations in six independent consanguineous families. Double lines, first cousin status; squares, males; circles, females; slash-through, mortality; black fill, ASD. Missense variants (green) and truncating variants (blue). Obligate carriers depicted with dot at center of symbol.
Focusing on ACTL6B, we more closely examined the patient phenotype and sought to understand how mutations in this gene give rise to autism. Six families representing 4.4% of the SRAC cohort demonstrated homozygous variants in ACTL6B with fully penetrant recessive inheritance (Fig. 1D and SI Appendix, Table S1). Variants were not present in ExAC or gnomAD databases, nor in ethnically matched controls from the Greater Middle East Variome or the cohort of 256 controls (42, 43), indicating that they are rare variants. Subjects exhibited nonverbal autism with stereotypies, as well as ID, developmental delay, hyperactivity, and mild spasticity (SI Appendix, Table S2 and Movie S1). There were no syndromic features that would have identified these patients as unique a priori from other SRAC subjects (SI Appendix, Table S2 and Supplementary Text). Epilepsy was seen in all subjects, presenting at 2 wk to 5 y of age, and generally responded to anticonvulsant medication. Males and females were similarly affected, consistent with fully penetrant inheritance. While the SRAC patients were of Middle Eastern descent, similar phenotypes have been reported for ACTL6B mutant individuals of French-Canadian, Sicilian, and Finnish descent (29, 40, 41, 44, 45). The high penetrance and perfect segregation of mutant alleles with ASD indicate a causal relationship between ACTL6B mutation and autism. This distinguishes ACTL6B from most other autism-associated genes, which substantially increase risk when mutated, but alone may not be sufficient to cause disease.
Patient Mutations Destabilize ACTL6B and Reduce Its Incorporation into the BAF Complex.
Consistent with the recessive mode of inheritance, patient variants were protein truncating, frame shifting, or missense for highly conserved residues (SI Appendix, Fig. S2 A–C). Variants occurred throughout the open reading frame, arguing against dominant-negative effects. Gibbs free energy calculations revealed that the missense variants likely disrupt protein folding, in line with PolyPhen-2 “damaging” predictions (SI Appendix, Fig. S2D). To visualize how the patient mutations are oriented in relation to the BAF complex, we generated a protein model of human ACTL6B binding to the conserved HSA domain of SMARCA4. This revealed two patient missense variants (L154F and G393R) located in residues that are critical for stabilizing the hydrophobic binding interface between ACTL6B and SMARCA4 (SI Appendix, Fig. S2E), consistent with a LoF disease mechanism.
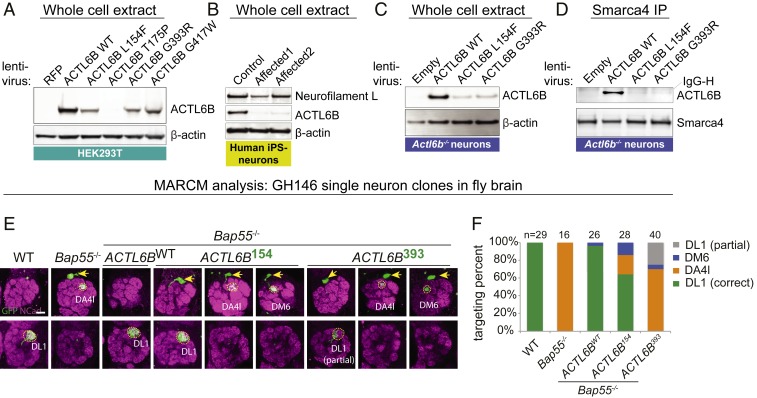
To test these predictions in vitro, we assessed ACTL6B missense mutant protein expression and incorporation into BAF complexes in the HEK293T cell line, human embryonic stem cells, and Actl6b−/− primary mouse neurons. To rule out the effects of species or cellular context, we also derived induced pluripotent stem cells (iPSCs) from control or affected humans in family 2703 (ACTL6BL154F/L154F) and conducted neural differentiation over 8 wk. RNA sequencing (RNA-seq) of (n = 3) control individuals confirmed that the switch of progenitor BAF subunits for nBAF subunits (11) was conserved in humans, as indicated from down-regulation of nonneuronal ACTL6A and up-regulation of ACTL6B (SI Appendix, Fig. S3 A and B). In each cellular context, ACTL6B mutant protein expression was dramatically reduced relative to wild type, with little to no mutant protein incorporated into neuronal or nonneuronal BAF (Fig. 2 A–D and SI Appendix, Fig. S4 A–E). These data suggest that the patient mutations destabilize ACTL6B protein and result in the formation of nBAF complexes that lack ACTL6B.
Fig. 2.
Patient mutations destabilize ACTL6B protein and cause a loss-of-function “perfect dendritic retargeting” phenotype in the fly olfactory system. (A) Patient missense mutant proteins were less stable than wild-type ACTL6B when expressed in HEK293T cells; n = 2 for T175P and G417W; n = 4 for wild type, L154F, and G393R. (B) Endogenous ACTL6B protein was decreased in human neurons derived from iPSCs of two affected individuals (ACTL6BL154F/L154F) in family 2703 relative to the wild-type control; n = 3 replicates. Neurofilament light chain, a mature neuronal marker that confirms differentiation status. (C) Missense mutant proteins were less stable than wild-type ACTL6B when expressed for 2 wk in cultured striatal neurons from Actl6b−/− E18.5 mice; n = 2. (D) Missense mutant proteins showed decreased recovery following coimmunoprecipitation with SMARCA4 from striatal neurons shown in C; n = 1 (see similar results in cortical neurons, human ESCs, and HEK293Ts shown in SI Appendix, Fig. S4 B–E). (E) MARCM (mosaic analysis with a repressible cell marker) was used to fluorescently label and replace fly Bap55 with human ACTL6B alleles in olfactory projection neurons (PNs). PNs always project a dendrite to the dorsolateral glomerulus (DL1) in the posterior antennal lobe. Clones were induced in GH146-GAL4 flies at a time when DL1 PNs were born. Replacement of the fly ortholog with human wild-type ACTL6B but not patient variants (L154F or G393R) rescued the perfect dendritic retargeting phenotypes in single-neuron clones. Arrows, cell bodies; Ncad, neuropil marker. (Scale bar, 20 µm.) (F) Quantification of E where single PNs were correctly targeted to DL1 or mistargeted to DA4l or DM6; n = animals per condition.
Patient Mutations Produce LoF “Perfect Dendritic Retargeting” Phenotype in Fly Brain.
Social communication is a key defect in ASD (30). In insects, social communication is dependent upon olfactory cues (46) and specifically olfactory projection neurons, which transmit signals to specific glomeruli in a stereotyped manner. Previously, human ACTL6B was shown to genetically complement the fly ortholog Bap55 and rescue a LoF “perfect dendritic retargeting” phenotype in the fly olfactory system, where dendritic trees project cell autonomously to the wrong glomerulus with 100% expressivity (47). This results in a switch in synaptic specificity that may affect how the fly interprets chemical cues. Using the mosaic analysis with a repressible cell marker (MARCM) method (48), we simultaneously replaced the fly ortholog with human wild-type or ASD mutant ACTL6B and expressed GFP in single neurons or neuroblast clones (SI Appendix, Fig. S5A). We found that wild type but not patient ACTL6B missense alleles quantitatively rescued targeting (Fig. 2 E and F and SI Appendix, S5 B and C), confirming that patient mutations show LoF in vivo and suggesting that altered synaptic specificity may contribute to the ACTL6B autism phenotype.
Defects in Callosal Anatomy in Humans and Mice Lacking ACTL6B.
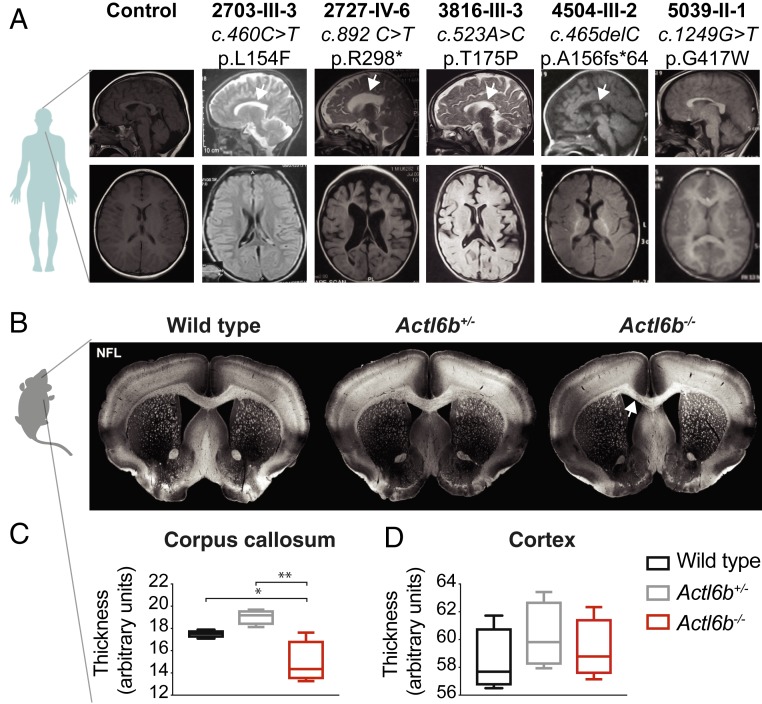
Perfect dendritic retargeting in ACTL6B mutant flies may be mechanistically related to a distinctive clinical feature of BAF mutant ASD: reduced or absent corpus callosum (17, 49). This defect involves a failure of axons to cross to the opposite cerebral hemisphere and is often associated with mutations in guidance molecules such as semaphorins, which also direct dendritic and axonal targeting in flies (50–52). Hypogenesis of the corpus callosum may further reflect altered functional connectivity, as has been detected by resting-state functional MRI in some ASD patients (53, 54). We compared available clinical brain MRIs (axial and midline sagittal) to identify evidence of altered brain anatomy in autistic patients with ACTL6B mutations. Compared with a healthy control, affected individuals showed reduced cerebral white matter volume and most showed corpus callosal hypoplasia (Fig. 3A).
Fig. 3.
Loss of ACTL6B causes corpus callosum hypoplasia in humans and mice. (A) Human brain MRI scans of control compared with ACTL6B mutated subjects. (Top row) Midline sagittal. White arrows: corpus callosum hypoplasia. (Bottom row) Axial images at the level of the foramen of Monroe showing paucity of white matter compared with control. (B) Adult mouse coronal brain slices from wild type, Actl6b+/−, or Actl6b−/− were stained with antibodies to neurofilament light chain (NFL) to visualize callosal axon tracks. Representative images of male wild-type, Actl6b+/−, and Actl6b−/− brain slices. Actl6b−/− corpus callosum was visibly thinner, indicated with a white arrow. (C) Quantification of relative callosal thickness showing significantly thinner callosum in knockouts. Blinded measurements from n = 2 slices were averaged for each animal and compared for n = 4 mice per genotype. (D) Quantification of relative thickness of the somatosensory cortex in slices used for C showing no significant difference. Significance was assessed by ordinary one-way ANOVA with Tukey’s correction for multiple comparisons; F(2,9) = 10.73 for C; F(2,9) = 0.62 for D. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Actl6b knockout mice have thinner myelin sheaths (14), consistent with the reduction in white matter in ACTL6B patients. This prompted us to examine Actl6b knockout mice for evidence of reduced or absent corpus callosum. We immunostained coronal sections of adult mouse brain for neurofilament and measured relative corpus callosum thickness using ImageJ. Actl6b−/− mice showed a ∼20% reduction in corpus callosum thickness compared to wild type (Fig. 3 B and C). Heterozygous mice showed no difference in corpus callosum thickness, indicating that this is a recessive phenotype in mice. No significant differences in cortical thickness were observed (Fig. 3D). Reduced corpus callosum volume has also been reported in mice lacking one copy of the ASD-related BAF subunit Arid1b (55). Thus, like other BAF subunits, ACTL6B is required for corpus callosum formation in humans and mice.
Actl6b−/− Mice Exhibit Autism-Related Behaviors in Two Genetic Backgrounds.
Because the patient mutations were LoF and Actl6b−/− mice showed corpus callosum hypoplasia like ACTL6B patients, we asked whether Actl6b−/− mice exhibit ASD-related behaviors. In 2007, we generated Actl6b−/− mice with mixed 129/Sv; C57BL/6 background and found that few survived past weaning (14). Subsequent backcrossing over many generations to C57BL/6 produced litters with Actl6b−/− at the expected 25% ratio and with most mice living to adulthood when provided with recovery gel after weaning. In parallel, a second cohort of Actl6b+/− mice were crossed for many generations to either 129S6/SvEv or C57BL/6 strains at Fujita Health University, and the F1 cross of these congenic strains also produced Actl6b−/− mice that survived to adulthood. We conducted our behavioral characterization on both cohorts of Actl6b−/− mice and their littermates separately, and then compared results after data were acquired.
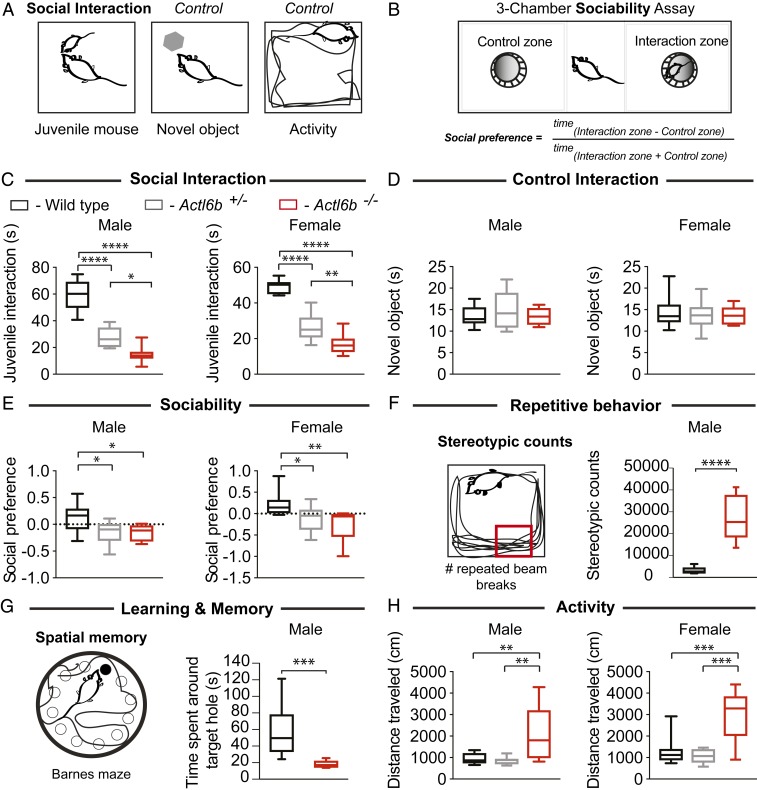
We assessed social behavior, a hallmark of autism, using a juvenile interaction test and three-chamber sociability assay (Fig. 4 A and B). The juvenile interaction test measured the amount of time 7-wk-old test mice spent interacting with a 4-wk-old juvenile wild-type mouse of the same sex. This revealed significant and gene dosage-dependent decreases in social interaction in both male and female C57BL/6 mice (P < 0.0001 for male or female wild type vs. Actl6b+/− or Actl6b−/−, P = 0.011 for Actl6b+/− vs. Actl6b−/− males, P = 0.0047 for Actl6b+/− vs. Actl6b−/− females by one-way ANOVA; Fig. 4C). These results were specific for social defects, because replacing the juvenile mouse with a novel toy mouse revealed no differences in interaction across genotypes (Fig. 4D). Similarly, male knockout mice on C57BL/6 × 129S6/SvEv background showed significantly reduced social interactions with an adult mouse of the same sex and genotype (P = 0.0208, Student’s t test; SI Appendix, Fig. S6 A and B), suggesting a role for Actl6b in murine social interaction that is independent of genetic background.
Fig. 4.
Actl6b−/− mice exhibit autism-related behaviors. (A) Social interaction and control tests: relative interaction time between the adult test mouse and a juvenile mouse (3–5 wk) or novel object. Open-field test measured activity over 20 min as total distance traveled. (B) Schematic of three-chamber sociability assay: Test mice may enter the zone with a novel object or the zone with a novel juvenile mouse. Social preference scores were calculated from time spent in each zone using the formula shown. (C) Box plots showing male and female littermates of Actl6b+/− × Actl6b+/− crosses with gene dosage-dependent impairment in social interaction with a juvenile mouse but not with (D) a control novel object. (E) Actl6b+/− and Actl6b−/− mice of both sexes showed defects in sociability, which were most severe in female knockouts. (F) Single, repeated photobeam breaks or “stereotypic counts” represented by a small red box in an open field. Male Actl6b−/− mice showed repetitive movements indicated from increased stereotypic counts over 120 min. (G) Diagram of Barnes maze test of spatial memory: an elevated, white, circular open field containing 12 holes, with one “target” escape hole leading to a comfortable cage. Visual cues provided the mouse with a frame of reference for the location of the target hole. Male Actl6b−/− mice spent less time around the target hole, indicating impaired memory as to the location of the target hole. (H) Actl6b−/− mice of both sexes showed increased activity, not observed in Actl6b+/−. Tests shown in C, E, and H were conducted at Stanford University on a cohort of mice highly backcrossed to the C57BL/6 background; mice (+/+, +/−, −/−): n = 9, 12, 7 males, n = 11, 12, 10 females for C; n = 10, 13, 6 males, n = 11, 12, 9 females for D; n = 10, 12, 7 males, n = 11, 12, 9 females for E; n = 10, 13, 6 males, n = 11, 11, 5 females for H. Values for each test in this cohort were as follows: F(2,30)Females = 79.87 and F(2,25)Males = 59.49 for C; F(2,29)Females = 0.29 and F(2,26)Males = 0.74 for D; F(2,29)Females = 6.82 and F(2,26)Males = 4.83 for E; and F(2,24)Females = 13.44 and F(2,26)Males = 8.60 for H. Stereotypic counts in F and spatial memory in G were assessed at Fujita Health University on adult male mice that were the F1 offspring of a cross between Actl6b+/− 129S6/SvEv × Actl6b+/− C57BL/6. Mice (+/+, −/−): n = 22, 21 in F, and n = 22, 14 in G. Significance was calculated for B, E, and H using a one-way analysis of variance (ANOVA) with Tukey’s multiple-comparison post hoc. Significance for F and G was calculated using Student’s t test: t41 = 8.68 in F and t34 = 3.99 in G. Whiskers indicate 10th and 90th percentiles. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In the three-chamber assay for sociability, animals were given the choice of inhabiting a chamber containing a novel juvenile mouse in a cage or a novel, empty cage (Fig. 4B). Social preference scores, calculated from the ratio of time spent in each chamber, indicated that Actl6b+/− and Actl6b−/− mice were less sociable compared to wild types (for males, P = 0.025 for wild type vs. Actl6b+/− and P = 0.048 for wild type vs. Actl6b−/−; for females, P = 0.038 for wild type vs. Actl6b+/− and P = 0.0037 for wild type vs. Actl6b−/− by one-way ANOVA; Fig. 4E). Female mice exhibited a trend toward increasing social avoidance with decreasing Actl6b gene dosage. These results suggest a direct role for Actl6b in social interaction and sociability.
Patients with autism also display restricted or stereotypic (repeated) behaviors. We thus assessed stereotypy in the open-field test in male littermates (Actl6b+/− 129S6/SvEv × Actl6b+/− C57BL/6 F1 cross) using repeated breaks of the same photobeam (stereotypic counts). Actl6b−/− mice showed significantly elevated stereotypic counts, consistent with increased repetitive behaviors (P < 0.0001, Student’s t test; Fig. 4F).
ID affects 45% of all autism patients (30), is a consistent feature of BAF mutant ASD (17), and was present in affected individuals with ACTL6B mutations. We therefore conducted the Barnes maze test for spatial memory (Fig. 4G) and the T-maze forced alternation test for working memory (SI Appendix, Fig. S6C). These forms of memory depend upon neural circuits involving the hippocampus and prefrontal cortex, respectively (56, 57). Both tests revealed significant memory impairment in male Actl6b−/− mice (129S6/SvEv × C57BL/6 F1 cross) (P = 0.0003 for Barnes maze and P < 0.0001 for T-maze forced alternation, Student’s t test; Fig. 4G and SI Appendix, Fig. S6 D and E), consistent with previous studies showing impaired hippocampal memory consolidation, striatum-dependent cocaine-conditioned place preference, and amygdala-based fear learning in heterozygous Actl6b+/− mice (58–60). We conclude that Actl6b is required for memory formation in multiple neuronal systems.
Hyperactivity and anxiety are common comorbidities in ASD that show bias toward males and females, respectively (30). However, all affected individuals with ACTL6B mutations showed hyperactivity. We recorded the location and distance mice traveled in an open field to assess anxiety and activity levels (Fig. 4A and SI Appendix, Fig. S6F). Male Actl6b−/− mice showed no clear trend in anxiety as indicated from relative time spent in the open field center; however, female Actl6b−/− were more anxious than their heterozygous or wild-type sisters (P = 0.0017 for Actl6b−/− vs. Actl6b+/− and P = 0.064 for Actl6b−/− vs. wild type, one-way ANOVA; SI Appendix, Fig. S6 G and H). Total distance traveled in the open field indicated that Actl6b−/− mice of both cohorts and sexes traveled more than three times greater distance than their littermates, suggestive of a recessive hyperactive phenotype (P < 0.001 in all comparisons to Actl6b−/− for each sex, one-way ANOVA; Fig. 4H, SI Appendix, Fig. S6 I–L, and Movies S2 and S3). We conclude that Actl6b−/− mice robustly model general autism-related behaviors and behaviors that are characteristic of ACTL6B mutant ASD.
Actl6b−/− nBAF Complexes Retain the Nonneuronal Variant Actl6a and Have Reduced Affinity for Neuronal Chromatin.
Having established conserved roles for ACTL6B in mammalian social behavior and neural circuit formation, we sought to uncover the molecular function of ACTL6B and explore disease-relevant mechanisms. In mouse and human, ACTL6B expression is restricted to neurons, with lower expression in testes (10, 61). Previous studies have indicated that the majority of ACTL6B protein is likely associated with the nBAF complex (11, 12, 14). Because ACTL6B mutant proteins were not incorporated into nBAF complexes, we considered that the assembly of nBAF might be affected by the loss of Actl6b. We immunoprecipitated Actl6b−/− or wild-type nBAF complexes with the J1 antibody, which recognizes both Smarca2 and Smarca4 (62), and then digested and labeled peptides with “heavy” ∆4 (13C15N) or “light” ∆0 (12C14N) mTRAQ reagent, respectively. Peptides from each complex were pooled 1:1 and analyzed by mass spectrometry, where +4-Da shifted peaks indicated Actl6b−/− peptides (SI Appendix, Fig. S7A). Normalized automated statistical analysis of protein (ASAP) ratios of heavy knockout to light wild-type peptides were generated (Protein Prophet Probability cutoff > 0.9) such that values <1 indicated reduced interactions and >1 indicated stabilized interactions in Actl6b−/− nBAF complexes (SI Appendix, Fig. S7 B–D). The total number of peptides from the pool of wild-type and knockout complexes was also recorded. Because BAF complexes are highly abundant in the nucleus (300,000 per cell), its interactions with low abundance proteins (e.g., transcription factors) of <10,000 per cell are difficult to detect. Thus, we included proteins recovered as only a single peptide (62) as a resource, but with cautious interpretation.
Proteomic analysis of Actl6b−/− nBAF complexes indicated complete assembly of nBAF subunits, with peptides specific to the nonneuronal homolog Actl6a recovered in place of Actl6b. The Actl6b ASAP ratio ∼2 and nearly equivalent peptide counts for Actl6a (in knockout) and Actl6b (in wild type) suggested that Actl6b−/− nBAF complexes fully retain Actl6a (SI Appendix, Fig. S7B). Mutant nBAF complexes showed reduced interaction with the abundant chromatin protein histone H1 and with autism-related proteins including Kat6a/Myst3 and Adnp, both confirmed BAF interacting proteins (63, 64) (SI Appendix, Fig. S7C). Several proteins exhibited stronger interactions with mutant than wild-type complexes (SI Appendix, Fig. S7D), including Wiz, a promoter binding protein previously linked to abnormal social behavior in mouse (65). The proteomic data indicate that Actl6b loss results in the formation of an abnormal nBAF complex containing Actl6a with reduced interactions with established binding partners and gained interactions with novel partners.
Actl6a and Actl6b proteins share 95% sequence similarity except in the 43-residue subdomain 2, which shows only 53% sequence similarity (66). We previously showed that Actl6a cannot rescue neuronal function of nBAF in Actl6b−/− cells, and neither can a chimeric Actl6b containing the subdomain 2 of Actl6a (14). To confirm misincorporation of Actl6a into nBAF in the absence of Actl6b, we assessed Actl6a expression and incorporation into nBAF complexes in postnatal day 0 (P0) brain tissues from wild-type, Actl6b+/−, or Actl6b−/− mouse littermates. We found elevated Actl6a protein in Actl6b−/− cerebellar whole-cell extracts and in nuclear extracts from hippocampal tissue at P0 (SI Appendix, Fig. S7 E and G). BAF complexes immunoprecipitated with nBAF-specific Ss18l1 antibody from P0 cerebellar, cortical, or hippocampal nuclear extracts showed Actl6b gene dosage-dependent increases in Actl6a incorporation (SI Appendix, Fig. S7 F and G). Given that microRNAs switch off Actl6a expression during neural differentiation (13), we tested whether the persistence of Actl6a in nBAF was transcriptionally mediated. In cultured primary cortical neurons, we found significantly increased (∼1.2-fold) Actl6a transcripts by RT-qPCR from Actl6b−/− compared with wild type (SI Appendix, Fig. S7H). This observation suggested that Actl6a was retained in nBAF in part due to a failure to repress Actl6a transcription in postmitotic neurons.
To learn whether ACTL6A was transcriptionally up-regulated in affected human neurons, we measured expression in brain organoids, an improved culture method for modeling human brain development (67). We first confirmed that human brain organoids express ACTL6A and ACTL6B during maturation in controls at day 69 (SI Appendix, Fig. S8A). Brain organoids from unaffected (ACTL6B+/L154F) or two affected (ACTL6BL154F/L154F) individuals of family 2703 were cultured to day 28 and harvested for RNA or immunostained for developmental markers of neural progenitors (Sox2) and immature neurons (Tuj1). No gross differences in development were observed between unaffected and affected brain organoids (SI Appendix, Fig. S8B). However, brain organoids from two affected individuals displayed about a threefold increase in ACTL6A transcript compared to the unaffected control (SI Appendix, Fig. S7I). These data support a conserved mechanism in which ACTL6A expression is increased transcriptionally in the absence of functional ACTL6B, leading to the formation of an abnormal nBAF complex.
The incorporation of Actl6a into Actl6b−/− nBAF was associated with reduced interactions between mutant complexes and histone H1 (SI Appendix, Fig. S7C), suggesting that mutant nBAF complexes may have abnormal interactions with chromatin. To test this, we fractionated chromatin from embryonic day 18.5 (E18.5)/day in vitro 7 (DIV7) wild-type vs. Actl6b−/− cortical neurons using increasing concentrations of salt (300–700 mM NaCl) and blotted for nBAF in the resulting soluble nuclear or chromatin fractions. We found that Actl6b−/− nBAF complexes were not as stably associated with chromatin as wild type across a wide range of salt concentrations (68) (SI Appendix, Fig. S7J), suggesting impaired BAF–chromatin interactions as a result of altered nBAF constituency.
ACTL6B Suppresses the Activity-Responsive Transcriptional Program in Resting Neurons.
Actl6b is required in neurons for activity-dependent dendritic outgrowth and long-term potentiation (14, 58), processes that require transcription and facilitate learning and memory (69). Targeted chromatin immunoprecipitation experiments in neurons have suggested that nBAF complexes are bound to the promoter of the immediate early gene Fos and may regulate its expression (70). However, genome-wide expression studies in postmitotic neurons have yielded inconsistent insights into the molecular function of Actl6b and the neuronal BAF complex (58, 71).
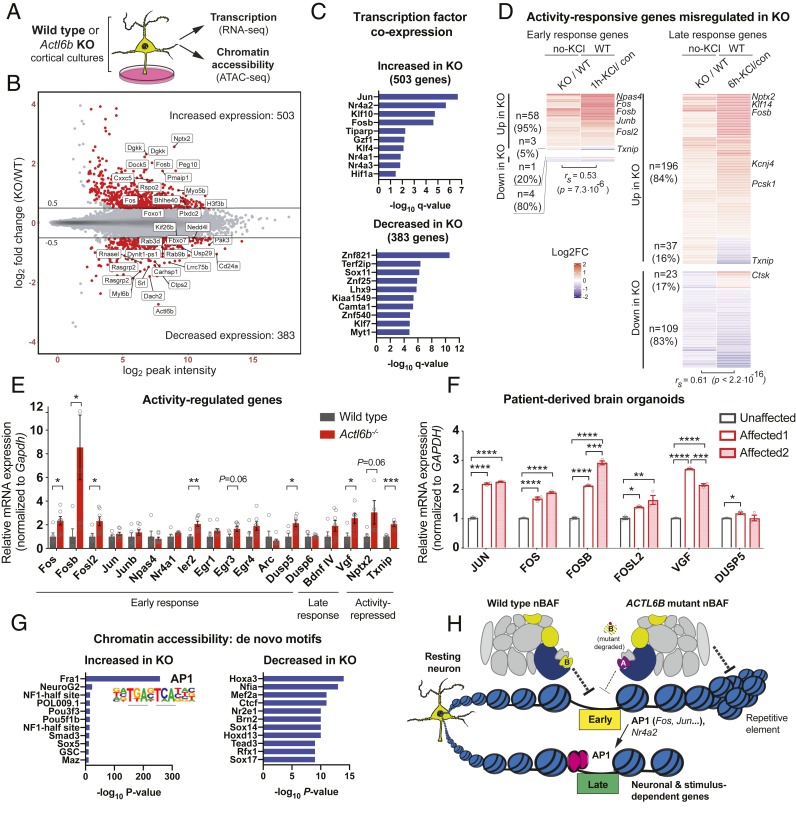
To clarify the role of ACTL6B in transcriptional regulation, we conducted RNA-seq and ATAC sequencing (ATAC-seq) on primary cortical neurons from wild-type or Actl6b−/− E16.5 embryos cultured for 7 DIV (72). Neurons from n = 5 wild-type and n = 7 knockout biological replicates (littermates) were collected after 2-h treatment with TTX/APV, which silenced network activity in order to reflect the resting state (73). RNA libraries were prepared from a fraction of the neurons in each biological sample, while another fraction was transposed with Tn5 to cut and tag accessible chromatin for sequencing (ATAC-seq) (74) (Fig. 5A and Datasets S1 and S2). The quality of collected data were confirmed by principal-component analysis, which demonstrated separation of biological samples by genotype in both RNA-seq and ATAC-seq datasets (SI Appendix, Fig. S9 A and B). We also confirmed Actl6b deletion in Actl6b−/− neurons (SI Appendix, Fig. S9C).
Fig. 5.
ACTL6B suppresses the activity-responsive transcriptional program in resting neurons. (A) Experimental design: Primary E16.5/DIV7 cortical cultures from (n = 5) wild-type or (n = 7) Actl6b−/− littermates were treated for 2 h with TTX/APV to silence action potentials and represent the “resting” neuronal state. RNA was collected to measure transcription by RNA-seq and DNA was transposed with Tn5 to measure chromatin accessibility by ATAC-seq. (B) MA plot of transcriptional changes in resting Actl6b−/− (KO) neurons relative to wild type (WT). Differentially expressed genes showing FDR < 5% and absolute log2 fold change >0.5 were defined as “significant” and highlighted in red. Gene names are shown for the top 30 most significant genes. (C) Transcription factor coexpression analysis was performed on the significantly up- or down-regulated genes in resting Actl6b−/− (KO) neurons. Up-regulated genes commonly coexpress with early response transcription factors such as Jun, Nr4a2, and Fosb, indicating possible activity-dependent transcription factor activity. (D) Heat maps showing log2 fold changes for genes that were both significantly differentially expressed in resting Actl6b−/− (KO) neurons and in wild-type (WT) neurons that were stimulated for 1 or 6 h with 55 mM KCl. Early response genes showed altered expression after 1-h KCl stimulation in wild type (SI Appendix, Fig. S10A) and late response genes showed altered expression after 6-h KCl stimulation in wild type (SI Appendix, Fig. S10B). Representative genes from each group are labeled. Transcriptional changes in resting Actl6b−/− neurons significantly correlated with activity-induced responses in wild type. (E) mRNA expression of activity-responsive genes in the biological samples used for RNA-seq, measured by RT-qPCR. AP1 transcription factors Fos, Fosb, Fosl2, and their late response target gene Vgf were significantly increased in resting Actl6b−/− neurons. (F) mRNA expression of activity-responsive genes in 28-d-old cerebral brain organoids cultured from induced pluripotent cells of an unaffected father (ACTL6BL154F/+) and his two affected children (ACTL6BL154F/L154F) in family 2703, measured by RT-qPCR; n = 3 technical replicates per individual. AP1 transcription factors FOS, FOSB, FOSL2, JUN, and their late response target gene VGF were significantly increased in affected human brain organoids. (G) Chromatin accessibility was assayed by ATAC-seq as described in A, and HOMER de novo motif analysis was performed on the significantly increased or decreased sites in Actl6b−/− (KO) neurons. Sites with increased chromatin accessibility were selectively enriched for the AP1 transcription factor binding motif, indicated by FRA1. (H) Summary model: Autism mutant ACTL6B “B” proteins are unstable and rapidly degraded, leading to retention of the nonneuronal homolog ACTL6A “A” in the nBAF complex. The loss of ACTL6B relieves transcriptional repression on early response transcription factors but increases repression on repetitive elements in resting neurons. mRNAs encoding early response transcription factors, particularly those in the AP1 family, are translated into proteins that regulate the expression of late response neuronal genes. Multisubunit nBAF complexes containing ubiquitously expressed subunits (gray), a core ATPase subunit (dark blue), neuronal-specific subunits (yellow), and neural-progenitor subunit ACTL6A (purple). AP1 transcription factor proteins are shown in pink. Balls on a string indicate nucleosomes. The arrow represents transcriptional activation; T represents transcriptional repression. Dashed lines in H indicate that the mechanism of repression may be direct or indirect. Significance was calculated by Spearman rank correlation in D, individual Student’s t tests in E, and two-way analysis of variance (ANOVA) in F. Error bars indicate SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Comparing mRNA expression in wild-type vs. Actl6b−/− neurons, we identified 503 genes with increased expression and 383 genes with reduced expression in the absence of Actl6b (false-discovery rate [FDR] < 5%; absolute log2 fold change > 0.5) (Fig. 5B). Gene coexpression analysis revealed that down-regulated genes frequently coexpress with ZNF821 (Fig. 5C), a transcription factor that was previously linked to methamphetamine-associated psychosis (75). Up-regulated genes in Actl6b−/− frequently coexpress with early response transcription factors Nr4a2 and members of the AP1 family including Fosb and Jun. This was surprising because early response transcription factors are normally expressed at very low levels in resting neurons. During neural activity, they are rapidly induced and subsequently regulate the expression of “late response” genes, which encode proteins that support neural plasticity (76).
The results from coexpression analysis prompted us to explore a connection between transcriptional changes in Actl6b−/− and transcriptional changes due to neural activity. To define activity-responsive genes, we conducted RNA-seq on wild-type E16.5/DIV7 cortical cultures that were silenced with TTX/APV for 1 h and then stimulated for 1 or 6 h with 55 mM KCl to model neural activity (n = 5 biological replicates each for 1-h KCl and 1-h control, n = 3 for 6-h KCl, and n = 4 for 6 h control). In wild-type mice, we identified 534 up-regulated and 237 down-regulated “early response” genes at 1-h KCl; and 2,603 up-regulated and 2,287 down-regulated late response genes at 6-h KCl (FDR < 5%, absolute log2 fold change > 0.5) (SI Appendix, Fig. S10 A and B). Over 40% of differentially expressed genes in Actl6b−/− neurons could be classified as “activity responsive” (Fig. 5D). Of the 66 early response genes that were differentially expressed in resting Actl6b−/− neurons, 94% followed a pattern of expression expected during neural activity (Spearman correlation coefficient rs = 0.53, P = 7.3 × 10−6; Fig. 5D and SI Appendix, Fig. S10C). Up-regulated genes in resting Actl6b−/− neurons included immediate early transcription factors Fos, Fosb, Fosl2, Junb, Nr4a2, Npas4, Arc, Egr1, Egr2, Egr4, and Klf10 (Fig. 5D and Dataset S1).
A predicted consequence of immediate early transcription factor activation is that late response genes will show an expression pattern mimicking activity. Of 365 late response genes that were differentially expressed in Actl6b−/− neurons, 84% were altered as if by activity (Spearman correlation coefficient = 0.61; P < 2.2 × 10−16, Fig. 5D and SI Appendix, Fig. S10D). Expression changes in early and late response genes were validated by RT-qPCR in E16.5/DIV7 murine cortical cultures (Fig. 5E) and in 65-d-old brain organoids derived from affected individuals in family 2703 (ACTL6BL154F/L154F) (Fig. 5F). These findings indicate a conserved role for ACTL6B in suppressing the activity-responsive transcriptional program in resting neurons.
A functional readout for chromatin remodeling activity and transcription factor binding is chromatin accessibility, which we measured by ATAC-seq. Accessible sites in resting neurons were predominantly found in intergenic and intronic regions, with ∼20% in promoters (SI Appendix, Fig. S11A). Chromatin was more accessible in Actl6b−/− than in wild-type neurons, with 843 sites showing increased accessibility and 174 sites showing reduced accessibility (SI Appendix, Fig. S11B). To learn which DNA binding proteins might occupy differentially accessible sites, we conducted de novo motif analysis using HOMER. Sites with increased accessibility in Actl6b−/− neurons were significantly and selectively enriched for the AP1 transcription factor motif (Fig. 5G), consistent with increased expression of AP1 subunits Fos, Fosb, Fosl2, and Junb. These sites were disproportionally located at distal intergenic regions, suggestive of enhancer activation (SI Appendix, Fig. S11C). Decreased sites were modestly enriched for DNA binding proteins Hoxa3, Nfia, Mef2a, and Ctcf (Fig. 5G), and were inordinately found in introns (SI Appendix, Fig. S11D). The profound enrichment of AP1 motifs at gained accessible sites suggested increased AP1 transcription factor activity in resting Actl6b−/− neurons.
To learn how elevated early response gene expression in resting Actl6b−/− neurons affected transcriptional responses to neural activity, we measured mRNA levels for activity-responsive genes in wild-type or Actl6b−/− E16.5/DIV7 cortical cultures stimulated with or without KCl, as above. KCl stimulation induced the expression of immediate early genes and reduced the expression of Txnip in wild-type neurons, as expected (SI Appendix, Fig. S12A). However, the expression of activity-regulated genes in Actl6b−/− neurons was significantly higher than in wild type (SI Appendix, Fig. S12B). Thus, early response gene expression was abnormally elevated in resting and active Actl6b−/− neurons.
Repetitive element transcription has been used as a measure of global transcriptional noise in resting neurons (77). However, neural activity can also induce repetitive element transcription (78). To test whether increased early response gene expression in Actl6b−/− neurons was associated with global transcriptional noise, we measured repetitive element transcripts in wild-type or Actl6b−/− neurons shown in Fig. 5E, whose network activity had been blocked for 2 h with TTX/APV. Intriguingly, repetitive element transcript levels were reduced in Actl6b−/− neurons compared to wild type (SI Appendix, Fig. S12C). Following 1-h KCl stimulation, repetitive element transcripts were strongly induced in wild-type but not in Actl6b−/− neurons, resulting in much lower expression of repetitive elements in Actl6b−/− neurons in the context of neural activity (SI Appendix, Fig. S12D). These results indicate that early response gene activation in Actl6b−/− neurons was not a consequence of globally increased transcriptional noise. Rather, early response genes were specifically de-repressed while repetitive elements were more silenced in the absence of Actl6b.
Since activity-dependent transcription is required for long-term memory formation (69, 76), the inappropriate activation of activity responsive genes in resting Actl6b−/− neurons is a plausible explanation for impaired memory in Actl6b mutant mice (Fig. 4G and SI Appendix, Fig. S6 C–E) (58–60, 79) and in affected humans (SI Appendix, Table S1) (29, 40, 41, 44, 45, 80–82). However, it is unclear whether this mechanism can account for other aspects of the ACTL6B patient phenotype, such as social deficits or corpus callosum hypogenesis. To gain deeper insight into the molecular changes in Actl6b−/− neurons and how these relate to aspects of the ACTL6B mutant phenotype, we used the Enrichr (83, 84) tool to perform a set of enrichment analyses on up-regulated and down-regulated genes in mutant neurons. The most enriched term for molecular functions of up-regulated genes in Actl6b−/− neurons was transcription factor activity, consistent with the DNA binding activity of many early response genes (SI Appendix, Fig. S13A). Enrichment analysis for down-regulated genes highlighted terms related to the nucleosome remodeling and histone deacetylase (NuRD) complex, semaphorin signaling, and genes involved in ion balance. These terms are consistent with altered chromatin repression, impaired axon guidance, and seizure activity, respectively.
Biological pathway analysis revealed that up-regulated genes in Actl6b−/− neurons were involved in neuroactive ligand–receptor interactions and in serotonin and anxiety-related events (SI Appendix, Fig. S13 B and C). Genes associated with these terms included stress-related corticotrophin releasing hormone (Crh, also known as CRF) and its receptor Crhr1, as well as serotonin receptors 1b (Htr1b), 2c (Htr2c), and 3a (Htr3a). CRF administration can induce seizures in rats (85), and both CRF and serotonin have been implicated in memory and social behaviors (86, 87). Pathway analysis of down-regulated genes identified roles in axon guidance and fatty acid biosynthesis, in line with callosal defects and loss of white matter observed in ACTL6B mutant humans and mice (Fig. 3 A–C). Enrichment analysis thus revealed potential mechanisms underlying the corpus callosum defects, seizures, memory, and social impairments in ACTL6B mutant humans and mice.
Discussion
We identified ACTL6B as the most significantly mutated gene in the SRAC, indicating that mutations in this gene may be a relatively common cause of recessive autism. In line with this finding, ACTL6B was the sixth most significantly mutated gene out of 409 genes with recessive missense variants in the Autism Sequencing Consortium (25), and recent case studies have identified individuals with biallelic ACTL6B variants showing similar recessive neurodevelopmental phenotypes to the patients we describe. Severe epilepsy, ID, lack of speech, developmental delays, feeding difficulties, microcephaly, hypomyelination, and corpus callosum hypogenesis were consistent clinical features associated with ACTL6B LoF (29, 40, 41, 44, 45, 82). One patient was diagnosed with Rett syndrome (82). Several patients passed away before 10 y of age (29, 41). Patients were not uniformly diagnosed with autism, possibly due to the severity of the phenotype or to differences in diagnostic criteria. ACTL6B mutations segregated perfectly with recessive phenotypes in a variety of genetic backgrounds (Middle Eastern, French Canadian, Finnish, Sicilian) and in children of both consanguineous and nonconsanguinous parents, affirming high penetrance.
Interestingly, two dominant ACTL6B alleles (G343R or D77G) were recently identified in patients with autism who did not experience seizures but otherwise exhibited characteristics of the recessive ACTL6B patients (29). In a separate study of genetic causes of ID, two affected individuals were found to have inherited the G343R mutation from their intellectually disabled father (80). The dominant G343R mutation was also identified as a de novo mutation in large screens for genetic causes of developmental delays, ID, and seizures (81, 88, 89). These dominant alleles may act on the wild-type copy protein or may poison the function of the nBAF complex, although further work is needed to explore these possibilities. In addition, one patient diagnosed with ASD was found to have a postzygotic mosaic, synonymous mutation in ACTL6B (90). Including the present study, there are at least 38 recessively affected and 19 dominantly affected individuals documented with ASD-related phenotypes caused by mutations in ACTL6B (25, 29, 40, 41, 44, 45, 80–82, 90, 91).
Deletion of the fly ortholog of ACTL6B in olfactory projection neurons resulted in cell-autonomous rewiring of the olfactory system in a manner that might cause the fly to misinterpret chemical social cues. Altered semaphorin signaling may underlie this defect, as deletion of Sema1a in fly olfactory neurons results in similar but less specific dendritic retargeting to ACTL6B mutants (47, 50) and we found that semaphorin genes (Sema4c, Sema4g, Sema6c) and their receptors (Plxna3, Plxnb1, Plxnc1) showed reduced expression in Actl6b−/− neurons. Polymorphisms in PLXNA3 have been identified in individuals with abnormal postnatal corpus callosum development (92) and Plxnb1/2 double-knockout mice exhibit corpus callosum agenesis (93), suggesting a connection with the callosum hypogenesis we observed in humans and mice lacking ACTL6B.
We also found that Actl6b knockout mice exhibit similar behaviors to ACTL6B patients, including social behavior deficits, increased repetitive movements, memory impairments, and hyperactivity. Autism mouse models 16p11.2, Shank3, Cntnap2, Pten, Tsc1, En2, and Gabrb3, show similar deficits to Actl6b knockout mice in the juvenile interaction and three-chamber assays (32, 87, 94), but unlike some of these models, behaviors in Actl6b−/− mice were robust on two genetic backgrounds. Hyperactivity and callosal defects showed recessive inheritance, while the severity of social behavior deficits increased with decreasing Actl6b gene dosage. Previous studies showed memory defects in heterozygous mice (58–60), suggesting dosage sensitivity; however, the heterozygous relatives of patients here showed no overt symptoms.
Approximately 40% of gene expression changes in electrically silenced Actl6b−/− neurons reflected transcriptional responses to neural activity, indicating that the de-repression of early response transcription factors like AP1 may be a primary defect in ACTL6B mutant neurons (Fig. 5H). Similarly, heterozygous mutations in the BAF ATPase SMARCA2 that cause autism-related Nicolaides–Baraitser syndrome were found to de-repress FOSL2 and lead to AP1 pioneer factor activity and BAF recruitment to enhancers in human neural progenitor cells (95). That mutation of either ACTL6B or SMARCA2 results in elevated AP1 expression suggests that the mechanism for de-repression involves a functional breakdown of the BAF complex. This is consistent with the fact that BAF mutations cause similar forms of ASD (23, 29, 96). We speculate that abnormal AP1 activation may be a common mechanism in BAF mutant ASD.
In summary, we show that LoF mutations in ACTL6B cause recessive autism in humans, patient-related phenotypes in mice and flies, and abnormal expression of early response genes, repetitive elements, semaphorins, CRF, and serotonin receptors in mouse neurons. The mechanism may involve the formation of nBAF complexes containing the nonneuronal homolog Actl6a, which have reduced affinity for neuronal chromatin. The genetic background and gender dependence of many genes implicated in autism have made biochemical analysis difficult because one would expect that any biochemical event underlying behavior would be subject to undetermined variables. In contrast, the gender and genetic background independence of ACTL6B mutations in both humans and mice should provide a powerful model for analysis of the role of chromatin regulation in autism.
Materials and Methods
This study was approved by the institutional review boards of University of California, San Diego, for human subjects, and by the animal review board of Stanford University and Fujita Health University. Blood, skin punch, and clinical data were obtained from research subjects after obtaining informed consent. Detailed description of the patient clinical history, generation of iPSCs, methods for study of Bap55 fly model using MARCM, behavioral assessment of mouse Actl6b mutants, primary neuronal cultures, transcriptional and proteome profiling, and assessment of chromatin are provided in SI Appendix, Supplementary Text.
Data Availability.
Exome-sequencing data from individuals in this study have been deposited to the Database of Genotypes and Phenotypes (dbGaP) under accession number phs000288.v2.p2. BAF53b and ACTL6B are used interchangeably. The GenBank accession ID is NR_134539.1. Raw RNA- and ATAC-seq data have been deposited in the Gene Expression Omnibus under GSE147056. The processed RNA-seq dataset is included as Dataset S1 and ATAC-seq as Dataset S2. Bap55 mutant flies expressing autism mutants for MARCM, patient-derived iPS cells, ACTL6B mutant plasmids, α-ACTL6A/B antibodies, and other resources developed or used in the paper are available upon request.
Supplementary Material
Acknowledgments
We thank Lei Chen, Alex Valdefiera, and Claire M. Ellis for their help with mouse breeding. We thank Kyle Loh, Lay Teng Ang, Alon Goren, and Ian Maze for thoughtful discussion. This work was supported by Simons Foundation for Autism Research Grant 514863 (to J.G.G. and G.R.C.); National Institutes of Health Grant NS046789 (to G.R.C.); Grant R01NS048453 and Qatar National Research Fund National Priorities Research Program Grant 6-1463-3-351 (to J.G.G. and T.B.-O.); Grants 1F31MH116588-01 and 2T32GM007790-38 (to W.W.); Grant 5T32GM008666 (to S.G.); and Grants U54HG003067 (to the Broad Institute) and U54HG006504 (to the Yale Center for Mendelian Disorders). Behavior studies were supported by grants from the Wu Tsai Neurosciences Institute and NIH P50 DA042012 (to R.C.M. and J.J.W.) and by the Joint Usage/Research Center for Genes, Brain and Behavior (Institute for Comprehensive Medical Science, Fujita Health University) accredited by the Ministry of Education, Culture, Sports, Science and Technology of Japan (to H.S. and T.M.). J.G.G., G.R.C., and L.L. are investigators with the Howard Hughes Medical Institute. J.G.G. received support from Rady Children’s Institute for Genomic Medicine.
Footnotes
Competing interest statement: G.R.C. is a founder of Foghorn Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: The exome-sequencing data from individuals in this study have been deposited in the Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/ (accession no. phs000288.v2.p2) and are available upon reasonable request. The raw RNA- and ATAC-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE147056). The processed RNA-sequencing dataset is included as Dataset S1 and ATAC-sequencing dataset as Dataset S2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908238117/-/DCSupplemental.
References
- 1.Baio J., et al. , Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2014. MMWR Surveill. Summ. 67, 1–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elsabbagh M., et al. , Global prevalence of autism and other pervasive developmental disorders. Autism Res. 5, 160–179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai D., et al. , Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry, 10.1001/jamapsychiatry.2019.1411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams B. S., et al. , SFARI gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges C., Kirkland J. G., Crabtree G. R., The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb. Perspect. Med. 6, a026930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller E. L., et al. , TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 24, 344–352 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadoch C., Crabtree G. R., Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 153, 71–85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadoch C., et al. , Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 49, 213–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanton B. Z., et al. , Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 49, 282–288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olave I., Wang W., Xue Y., Kuo A., Crabtree G. R., Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16, 2509–2517 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard J., et al. , An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 55, 201–215 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staahl B. T., et al. , Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J. Neurosci. 33, 10348–10361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo A. S., Staahl B. T., Chen L., Crabtree G. R., MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J. I., et al. , Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron 56, 94–108 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Aizawa H., et al. , Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303, 197–202 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Son E. Y., Crabtree G. R., The role of BAF (mSWI/SNF) complexes in mammalian neural development. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 333–349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosho T., et al. , Clinical correlations of mutations affecting six components of the SWI/SNF complex: Detailed description of 21 patients and a review of the literature. Am. J. Med. Genet. A. 161A, 1221–1237 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Miyakawa T., et al. , Neurogranin null mutant mice display performance deficits on spatial learning tasks with anxiety related components. Hippocampus 11, 763–775 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Bramswig N. C., et al. , Heterozygosity for ARID2 loss-of-function mutations in individuals with a Coffin-Siris syndrome-like phenotype. Hum. Genet. 136, 297–305 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Dias C., et al. ; DDD Study , BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am. J. Hum. Genet. 99, 253–274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim E. T., et al. ; Autism Sequencing Consortium , Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 20, 1217–1224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neale B. M., et al. , Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasileiou G., et al. ; Deciphering Developmental Disorders Study , Mutations in the BAF-complex subunit DPF2 are associated with coffin-Siris syndrome. Am. J. Hum. Genet. 102, 468–479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon K. C. J., et al. ; DDD Study , A syndromic neurodevelopmental disorder caused by mutations in SMARCD1, a core SWI/SNF subunit needed for context-dependent neuronal gene regulation in flies. Am. J. Hum. Genet. 104, 596–610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doan R. N., et al. ; Autism Sequencing Consortium , Recessive gene disruptions in autism spectrum disorder. Nat. Genet. 51, 1092–1098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieczorek D., et al. , A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum. Mol. Genet. 22, 5121–5135 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Marom R., et al. , Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum. Mutat. 38, 1365–1371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuvertino S., et al. ; DDD Study , ACTB loss-of-function mutations result in a pleiotropic developmental disorder. Am. J. Hum. Genet. 101, 1021–1033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell S., et al. , Mutations in ACTL6B cause neurodevelopmental deficits and epilepsy and lead to loss of dendrites in human neurons. Am. J. Hum. Genet. 104, 815–834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeste S. S., Geschwind D. H., Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat. Rev. Neurol. 10, 74–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López A. J., Wood M. A., Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Front. Behav. Neurosci. 9, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellegood J., Crawley J. N., Behavioral and neuroanatomical phenotypes in mouse models of autism. Neurotherapeutics 12, 521–533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rylaarsdam L., Guemez-Gamboa A., Genetic causes and modifiers of autism spectrum disorder. Front. Cell. Neurosci. 13, 385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarlungeanu D. C., et al. , Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell 167, 1481–1494.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novarino G., et al. , Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 338, 394–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen A., et al. , Mutations in MBOAT7, encoding lysophosphatidylinositol acyltransferase I, lead to intellectual disability accompanied by epilepsy and autistic features. Am. J. Hum. Genet. 99, 912–916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., et al. , Whole exome and whole genome sequencing of human families with recessive autism. Database of Genotypes and Phenotypes (dbGaP). https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000288.v2.p2. Deposited 22 June 2010.
- 38.Silverstein R. L., Febbraio M., CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2, re3 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber I., et al. , SPG11 mutations cause widespread white matter and basal ganglia abnormalities, but restricted cortical damage. Neuroimage Clin. 19, 848–857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yüksel Z., Yazol M., Gümüş E., Pathogenic homozygous variations in ACTL6B cause DECAM syndrome: Developmental delay, epileptic encephalopathy, cerebral Atrophy, and abnormal myelination. Am. J. Med. Genet. A. 179, 1603–1608 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Fichera M., et al. , Mutations in ACTL6B, coding for a subunit of the neuron-specific chromatin remodeling complex nBAF, cause early onset severe developmental and epileptic encephalopathy with brain hypomyelination and cerebellar atrophy. Hum. Genet. 138, 187–198 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Lek M., et al. ; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott E. M., et al. ; Greater Middle East Variome Consortium , Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 48, 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddirevula S., et al. , Autozygome and high throughput confirmation of disease genes candidacy. Genet. Med. 21, 736–742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karaca E., et al. , Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron 88, 499–513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L., et al. , Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat. Neurosci. 14, 757–762 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tea J. S., Luo L., The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 6, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J. S., Luo L., A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat. Protoc. 1, 2583–2589 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Sokpor G., Xie Y., Rosenbusch J., Tuoc T., Chromatin remodeling BAF (SWI/SNF) complexes in neural development and disorders. Front. Mol. Neurosci. 10, 243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komiyama T., Sweeney L. B., Schuldiner O., Garcia K. C., Luo L., Graded expression of semaphorin-1a cell-autonomously directs dendritic targeting of olfactory projection neurons. Cell 128, 399–410 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Van Battum E. Y., Brignani S., Pasterkamp R. J., Axon guidance proteins in neurological disorders. Lancet Neurol. 14, 532–546 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Yu H. H., Araj H. H., Ralls S. A., Kolodkin A. L., The transmembrane Semaphorin Sema I is required in Drosophila for embryonic motor and CNS axon guidance. Neuron 20, 207–220 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Hull J. V., et al. , Resting-state functional connectivity in autism spectrum disorders: A review. Front. Psychiatry 7, 205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liska A., et al. , Homozygous loss of autism-risk gene CNTNAP2 results in reduced local and long-range prefrontal functional connectivity. Cereb. Cortex 28, 1141–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Celen C., et al. , Arid1b haploinsufficient mice reveal neuropsychiatric phenotypes and reversible causes of growth impairment. eLife 6, e25730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Leary T. P., Brown R. E., Optimization of apparatus design and behavioral measures for the assessment of visuo-spatial learning and memory of mice on the Barnes maze. Learn. Mem. 20, 85–96 (2013). [DOI] [PubMed] [Google Scholar]
- 57.McNab F., Klingberg T., Prefrontal cortex and basal ganglia control access to working memory. Nat. Neurosci. 11, 103–107 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Vogel-Ciernia A., et al. , The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat. Neurosci. 16, 552–561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White A. O., et al. , BDNF rescues BAF53b-dependent synaptic plasticity and cocaine-associated memory in the nucleus accumbens. Nat. Commun. 7, 11725 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoo M., et al. , BAF53b, a neuron-specific nucleosome remodeling factor, is induced after learning and facilitates long-term memory consolidation. J. Neurosci. 37, 3686–3697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhan X., et al. , Generation of BAF53b-Cre transgenic mice with pan-neuronal Cre activities. Genesis 53, 440–448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho L., et al. , An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arboleda V. A., et al. ; UCLA Clinical Genomics Center , De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am. J. Hum. Genet. 96, 498–506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vandeweyer G., et al. , The transcriptional regulator ADNP links the BAF (SWI/SNF) complexes with autism. Am. J. Med. Genet. C. Semin. Med. Genet. 166C, 315–326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isbel L., et al. , Wiz binds active promoters and CTCF-binding sites and is required for normal behaviour in the mouse. eLife 5, e15082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muller J., et al. , Sequence and comparative genomic analysis of actin-related proteins. Mol. Biol. Cell 16, 5736–5748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arlotta P., Organoids required! A new path to understanding human brain development and disease. Nat. Methods 15, 27–29 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Nakayama R. T., et al. , SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet. 49, 1613–1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alberini C. M., Kandel E. R., The regulation of transcription in memory consolidation. Cold Spring Harb. Perspect. Biol. 7, a021741 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu Z., Ghosh A., A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron 60, 775–787 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z., et al. , Autism-associated chromatin regulator Brg1/SmarcA4 is required for synapse development and myocyte enhancer factor 2-mediated synapse remodeling. Mol. Cell Biol. 36, 70–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wenderski W., Krokhotin A., and Crabtree G. R., Loss of the neural-specific BAF subunit ACTL6B relieves repression of early response genes and causes recessive autism. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147056. Deposited 17 May 2020. [DOI] [PMC free article] [PubMed]
- 73.Maze I., et al. , Critical role of histone turnover in neuronal transcription and plasticity. Neuron 87, 77–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breen M. S., et al. , Candidate gene networks and blood biomarkers of methamphetamine-associated psychosis: An integrative RNA-sequencing report. Transl. Psychiatry 6, e802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yap E. L., Greenberg M. E., Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skene P. J., et al. , Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noh K. M., et al. , ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc. Natl. Acad. Sci. U.S.A. 112, 6820–6827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Τ., et al. , Histone methyltransferase Ash1L mediates activity-dependent repression of neurexin-1α. Sci. Rep. 6, 26597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lecoquierre F., et al. , Variant recurrence in neurodevelopmental disorders: The use of publicly available genomic data identifies clinically relevant pathogenic missense variants. Genet. Med. 21, 2504–2511 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Lelieveld S. H., et al. , Spatial clustering of de novo missense mutations identifies candidate neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 101, 478–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sajan S. A., et al. , Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet. Med. 19, 13–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen E. Y., et al. , Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuleshov M. V., et al. , Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weiss S. R., et al. , CRF-induced seizures and behavior: Interaction with amygdala kindling. Brain Res. 372, 345–351 (1986). [DOI] [PubMed] [Google Scholar]
- 86.Hupalo S., Berridge C. W., Working memory impairing actions of corticotropin-releasing factor (CRF) neurotransmission in the prefrontal cortex. Neuropsychopharmacology 41, 2733–2740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walsh J. J., et al. , 5-HT release in nucleus accumbens rescues social deficits in mouse autism model. Nature 560, 589–594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heyne H. O., et al. ; EuroEPINOMICS RES Consortium , De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 50, 1048–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Deciphering Developmental Disorders Study , Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krupp D. R., et al. , Exonic mosaic mutations contribute risk for autism spectrum disorder. Am. J. Hum. Genet. 101, 369–390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deciphering Developmental Disorders Study , Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belyk M., Kraft S. J., Brown S.; Pediatric Imaging, Neurocognition and Genetics Study , PlexinA polymorphisms mediate the developmental trajectory of human corpus callosum microstructure. J. Hum. Genet. 60, 147–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Daviaud N., Chen K., Huang Y., Friedel R. H., Zou H., Impaired cortical neurogenesis in plexin-B1 and -B2 double deletion mutant. Dev. Neurobiol. 76, 882–899 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Bey A. L., Jiang Y. H., Overview of mouse models of autism spectrum disorders. Curr. Protoc. Pharmacol. 66, 5.66.1-5.66.26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao F., et al. , Heterozygous mutations in SMARCA2 reprogram the enhancer landscape by global retargeting of SMARCA4. Mol. Cell 75, 891–904.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsurusaki Y., et al. , Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376–378 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Exome-sequencing data from individuals in this study have been deposited to the Database of Genotypes and Phenotypes (dbGaP) under accession number phs000288.v2.p2. BAF53b and ACTL6B are used interchangeably. The GenBank accession ID is NR_134539.1. Raw RNA- and ATAC-seq data have been deposited in the Gene Expression Omnibus under GSE147056. The processed RNA-seq dataset is included as Dataset S1 and ATAC-seq as Dataset S2. Bap55 mutant flies expressing autism mutants for MARCM, patient-derived iPS cells, ACTL6B mutant plasmids, α-ACTL6A/B antibodies, and other resources developed or used in the paper are available upon request.